Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A palindromic triplex architecture for DNA-templated synthesis designed for the core of a synthetic ribosome
Rana Abdul Razzakab,
Jonathan Bath ab,
Rachel K. O'Reilly
ab,
Rachel K. O'Reilly c and
Andrew J. Turberfield
c and
Andrew J. Turberfield *ab
*ab
aDepartment of Physics, University of Oxford, Parks Road, Oxford OX1 3PU, UK. E-mail: Andrew.Turberfield@physics.ox.ac.uk
bKavli Institute for Nanoscience Discovery, University of Oxford, South Parks Road, Oxford OX1 3QU, UK
cSchool of Chemistry, University of Birmingham, Edgbaston, Birmingham, B15 2TT, UK
First published on 26th January 2026
Abstract
We demonstrate a triplex-based architecture for DNA-templated synthesis. This study is motivated by progress towards the development of a synthetic ribosome – autonomous, genetically programmable, molecular machinery for synthesis. Such schemes for the creation and evolution of chemically diverse DNA-tagged chemical libraries rely on hybridization reactions of oligonucleotide adapters to control sequential, DNA-templated reactions of covalently attached building blocks. To enable parallel one-pot library synthesis it is desirable that any building block can be incorporated at any position in a product oligomer: this is incompatible with geometries commonly used for DNA-templated synthesis which require alternate reactants to be attached to 3′ and 5′ termini of their adapters. Our triplex-based architecture overcomes this problem by templating reactions between building blocks attached to adapters with identical structures. It is intended to form the core of programmable molecular machinery for multistep synthesis. Here, we use single-step coupling reactions to characterize the triplex reaction template.
Introduction
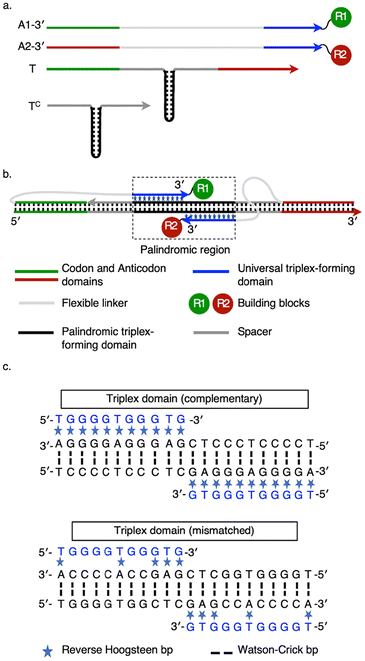
Combinatorial synthesis has transformed lead compound development.1 Split-and-pool techniques can generate libraries with millions of compounds but identifying active members of a complex library presents a challenge.2 DNA-encoded chemical libraries (DECLs) address this issue by associating each product with a DNA tag which records synthetic steps:3,4 tags can be amplified and sequenced to identify selected library members. Autonomous DECLs streamline multistep synthesis by employing the DNA tag to direct or encode the sequential addition of DNA-labelled building blocks without external intervention.5,6 Building blocks attached to identifying DNA adapters (analogous to tRNAs) remain effectively unreactive, due to their low concentrations4 or protective sequestration,7 until held in close proximity through hybridization reactions of their adapters. Colocalization increases the effective concentrations of the selected building blocks and thus promotes their reaction, enabling control of reaction sequence.5,6 If the identifying DNA tag acts as a program to direct synthesis, then multiple rounds of in vitro selection could be achieved by amplifying and reusing selected tags. Mutation and recombination of DNA-encoded programs would allow evolution of products not present in the original library.8,9An ideal architecture for programmed DNA-templated oligomer synthesis would allow any building block to be incorporated at any position in the product. It would also provide an identical environment for each reaction10,11 so the efficiency of chain extension would be independent of chain length. DNA-templated reactions often occur across the end of a double helix, formed by hybridization of two adapters to each other, or where adapters are brought together by hybridization to contiguous domains of a common template, i.e., across a nick in the backbone of one strand of a duplex.10–14 In either case, if an oligomer is to be created by a defined sequence of transfer reactions (as in the ribosome) then alternate building blocks must be attached to the 5′ and 3′ ends of their respective DNA adapters (Fig. S1). This is inelegant and has the practical disadvantage that a given building block can only be incorporated at odd or even positions in the product. To avoid this restriction, parallel duplexes, triplexes or quadruplexes15–18 could be used to juxtapose like ends of oligonucleotide adapters. Like ends of two adapters have been juxtaposed to template reactions at a triplex–duplex junction;18 however, this architecture is intrinsically asymmetric – one adapter forms a triplex, one a duplex – and is ill-adapted to template sequential reactions. The principle that like ends of two triplex-forming oligonucleotides (TFOs) can be colocalized was demonstrated in experiments in which alkylating agents tethered to the 5′-ends of TFOs bound to adjacent inverted purine tracts were used to cleave dsDNA at the junction between the triplexes.19,20 Here, we introduce and test a similar triplex architecture that juxtaposes like ends (5′ or 3′) of adapters for DNA-templated synthesis (Fig. 1). This architecture enables specific recruitment of any two DNA-conjugated reactants from a diverse pool and places them in an identical reaction environment. Adapters comprise two functional domains: an anticodon that identifies the attached building block and a universal triplex-forming sequence used to colocalize reactants.
Results
Fig. 1 shows an implementation of the templating architecture that juxtaposes building blocks attached to 3′ termini (for 5′-functionalized adapters see Fig. S2). The templating gene (T) is designed to hybridize to its partial complement (Tc) to create a duplex with, at its centre, two identical, inverted (palindromic) polypurine/polypyrimidine duplexes flanked by 10 bp spacer duplexes. Codons at either end of the gene hybridize to the anticodons of two DNA adapters (A1 and A2) which carry the corresponding reactive building blocks at their 3′ termini. Triplex-forming domains at the functionalized ends of the adapters bind, by reverse Hoogsteen base pairing, to the polypurine domains at the centre of the template duplex to form identical, inverted triplexes. Triplex formation juxtaposes the 3′ termini of the adapters in the major groove of the templating duplex, holding the reactive building blocks in close proximity and thus promoting their reaction. A key feature of the design is that the reaction-templating triplexes should not be sufficiently stable to recruit adapters without the cooperative binding of the codon domains. Both adapters have the same structure comprising anticodon, spacer, universal triplex-forming domain and reactive building block. This means that any one of a pool of adapters could be recruited by a matching codon to either position (red or green) on the templating complex. The reaction-templating triplex could thus form the core of a synthetic molecular machine that brings together adapters, recruited from a pool of reactants by specific binding of their anticodon domains, in a programmed sequence. In such a machine, after the nth synthesis step the growing chain (concatenated building blocks 1 … n + 1) would be attached to the n + 1th adapter bound at one codon, with the spent nth adapter bound to the other. The machinery would then replace the nth codon with the n + 2th, recruiting adapter n + 2 and promoting transfer of the growing chain to building block n + 2. The cycle would be repeated, with each transfer reaction taking place in the same environment at the end of the growing oligomer.The base sequence of the triplex-forming domain is purine-rich and binds in an antiparallel orientation to the polypurine tract of the dsDNA template in a pH-independent manner.20–23 The antiparallel triplex motif is stabilized by specific reverse Hoogsteen hydrogen bonds: thymine recognises adenine–thymine base pairs and guanine recognises guanine–cytosine.22,23 The specific sequence chosen was adapted from those studied by the Dervan group;20–23 weaker A·AT bonds could also be incorporated.21,22 The two adapters bind to opposite strands of the templating duplex on either side of the major groove with their 3′ (or 5′) ends juxtaposed, in the required configuration for DNA-templated synthesis. We chose not to investigate the alternative parallel triplex motif,24 which incorporates protonated cytosines, because the requirement for low pH would constrain the chemistries used for DNA-templated synthesis.
To test the ability of the triplex-forming sequence to form juxtaposed, inverted triplexes in this duplex–triplex hybrid architecture,25 we used a restriction endonuclease protection assay (Fig. S3).26 The palindromic domain in the dsDNA template is designed to overlap a 6 bp sequence recognised and cut by a restriction endonuclease. Triplex-forming oligonucleotides inhibit the activity of DNA-binding proteins, including endonucleases.26–29 Adapters (whether juxtaposing 3′ or 5′-ends) thus protect the template from digestion; adapters with mismatched triplex-forming domains provide no protection. These measurements demonstrate binding of the triplex-forming domains of the adapters, as designed.
We also evaluated the stability of the architecture using Förster resonant energy transfer (FRET) between Cy3 and Cy5 fluorophores in place of the building blocks (Fig. S4 & S5). A single, well-defined melting transition of the four-strand complex at approximately 70 °C is observed. In contrast, when the triplex-forming domains of the template were mutated to prevent triplex formation a broader transition with a lower melting temperature was observed. These observations are consistent with cooperative binding of the anticodon and triplex domains of the adapters.
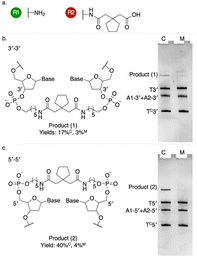
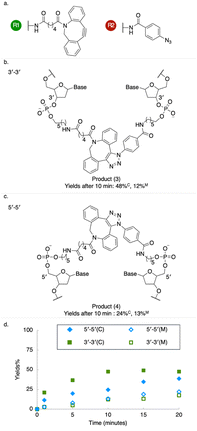
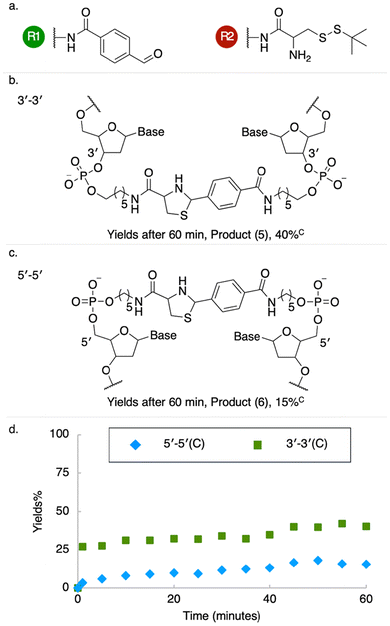
We tested the effectiveness of the palindromic triplex architecture at templating synthesis, with reactants tethered to juxtaposed 3′–3′ or 5′–5′ termini, using three established DNA-templated coupling reactions: amide bond formation, copper-free azide–alkyne cycloaddition (copper-free click) and thiazolidine synthesis4,14,30 (Fig. 2–4). For 3′ modifications we employed amino-C6 linkers to the oligonucleotide adapters, while amino-C5 linkers were used for 5′ modifications (Fig. S6) to compensate for the additional 4′C–5′C bond between the functionalized phosphate group and the ribose sugar at the 5′ terminus; the linkers therefore had closely similar lengths. Results obtained using a longer amino-C12 linker are presented in SI (Fig. S6, S9–S14). Reactions were analysed using denaturing polyacrylamide gel electrophoresis (PAGE) (SI Methods). Templated amide bond formation between 5′-linked reactants resulted in higher yields than for 3′ modifications (Fig. 2, S9–S14); copper-free click (Fig. 3 and S7) and thiazolidine synthesis (Fig. 4 and S8) showed the opposite bias.
Energy-minimized structural models of the adjacent inverted triplexes show that the distance between juxtaposed 5′ termini is significantly shorter than that between 3′ termini (Fig. S15), as observed in studies of alternate-strand triplexes formed by TFOs with polarity-inverting backbone linkages.31,32 This is consistent with our measurement of greater amide coupling yields with 5′-linked reactants (Fig. 2). The opposite bias observed with copper-free click and thiazolidine synthesis may reflect additional localizing interactions between building blocks (or their linkers) and nearby nucleobases.19,33–35
Exploration of other factors affecting the yield of the templated amide coupling reaction is presented in SI. Higher yields, up to 62% for 5′–5′ and 28% for 3′–3′ reactions, were obtained using a longer linker to the amine group (Fig. S9–S14). The efficiency of amide bond formation between 5′-linked reactants (Fig. S9) is comparable to the across-the-nick architecture, widely used to template synthesis, in which adapters functionalized at 3′ and 5′ termini are brought together by hybridization to a common template (Fig. S1).12–14 Non-templated controls (NTC) in which the template duplex was absent or mismatched in the triplex-forming region produced significantly lower yields. Triplex-templated yields are insensitive to temperature up to 55 °C; yields with mismatched triplex domains decrease strongly with increasing temperature (Fig. S9). The relationship between ionic concentrations and templated amide bond formation is complex (Fig. S10–S13). At 55 °C yield is increased by increasing magnesium ion concentration but decreased by increasing sodium ion concentration. At 25 °C, increasing sodium ion concentration increases yield for magnesium ion concentrations less than approximately 15 mM but decreases yield at higher magnesium concentrations. Optimum combinations of sodium and magnesium are different for 5′–5′ and 3′–3′ coupling. Screening, most effectively by dications, is necessary to stabilize both double and triple helices; the complex dependence of reaction yield on salt conditions observed36 is likely to reflect the stabilities of the designed structure and of competing patterns of base pairing.
When intervening base pairs were introduced between the two triplex domains the yield of triplex-templated amide bond formation was reduced to below the yield with mismatched triplex-forming domains (Fig. S14). This provides supporting evidence that the triplex-forming domains of the adapters are bound to the corresponding regions of the templating duplex and for the hypothesis that the yield of the templated reaction depends strongly on the separation of the tethered reactants.
Conclusions
The proposed triplex-based architecture is capable of templating diverse reactions between reactive building blocks coupled to identically structured DNA adapters. Reaction yields are comparable to more conventional architectures for DNA-templated synthesis but without the requirement for alternating 5′- and 3′-conjugated adapters, allowing any building block to be incorporated at any position in multistep oligomer synthesis. The triplex architecture fulfils a key requirement of the functional core of a synthetic ribosome, that it be capable of controlling multiple reactions between DNA-labelled building blocks in an identical environment in an arbitrarily programmable sequence.Author contributions
J. B., R. K. O'R. and A. J. T. initiated and developed the research goals of the project. A. J. T. and R. K. O'R. were responsible for funding acquisition. A. J. T. conceived the triplex design. R. A.-R., with support from J. B., carried out all experimental work and analysis. R. A.-R. and A. J. T. wrote the manuscript with helpful comments and critical input from J. B. and R. K. O'R.Conflicts of interest
There are no conflicts to declare.Data availability
Data supporting this article have been uploaded as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5py01159k.Acknowledgements
We thank Dr Seham Helmi for helpful discussions. This research was funded by the UKRI [EP/T000562/1, EP/T000481/1].References
- R. A. Houghten, C. Pinilla, S. E. Blondelle, J. R. Appel, C. T. Dooley and J. H. Cuervo, Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery, Nature, 1991, 354, 84–86 CrossRef CAS PubMed.
- E. Erb, K. D. Janda and S. Brenner, Recursive Deconvolution of Combinatorial Chemical Libraries, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 11422–11426 CrossRef CAS PubMed.
- D. R. Halpin and P. B. Harbury, DNA display II. Genetic manipulation of combinatorial chemistry libraries for small-molecule evolution, PLoS Biol., 2004, 2, E174–E174 CrossRef PubMed.
- Z. J. Gartner and D. R. Liu, The Generality of DNA-Templated Synthesis as a Basis for Evolving Non-Natural Small Molecules, J. Am. Chem. Soc., 2001, 123, 6961–6963 CrossRef CAS PubMed.
- W. Meng, R. A. Muscat, M. L. McKee, P. J. Milnes, A. H. El-Sagheer, J. Bath, B. G. Davis, T. Brown, R. K. O'Reilly and A. J. Turberfield, An autonomous molecular assembler for programmable chemical synthesis, Nat. Chem., 2016, 8, 542–548 CrossRef CAS PubMed.
- Y. He and D. R. Liu, Autonomous Multistep Organic Synthesis in a Single Isothermal Solution Mediated by a DNA Walker, Nat. Nanotechnol., 2010, 5, 778–782 CrossRef CAS PubMed.
- J. Frommer, R. Oppenheimer, B. M. Allott, S. Núñez-Pertíñez, T. R. Wilks, L. R. Cox, J. Bath, R. K. O'Reilly and A. J. Turberfield, A New Architecture for DNA–Templated Synthesis in Which Abasic Sites Protect Reactants from Degradation, Angew. Chem., Int. Ed., 2024, 63, e202317482 Search PubMed.
- R. K. O'Reilly, A. J. Turberfield and T. R. Wilks, The Evolution of DNA-Templated Synthesis as a Tool for Materials Discovery, Acc. Chem. Res., 2017, 50, 2496–2509 Search PubMed.
- B. R. Vummidi, L. Farrera-Soler, J.-P. Daguer, M. Dockerill, S. Barluenga and N. Winssinger, A mating mechanism to generate diversity for the Darwinian selection of DNA-encoded synthetic molecules, Nat. Chem., 2022, 14, 141–152 CrossRef CAS PubMed.
- M. L. McKee, P. J. Milnes, J. Bath, E. Stulz, A. J. Turberfield and R. K. O'Reilly, Multistep DNA-Templated Reactions for the Synthesis of Functional Sequence Controlled Oligomers, Angew. Chem., Int. Ed., 2010, 49, 7948–7951 CrossRef CAS.
- P. J. Milnes, M. L. McKee, J. Bath, L. Song, E. Stulz, A. J. Turberfield and R. K. O'Reilly, Sequence-specific synthesis of macromolecules using DNA-templated chemistry, Chem. Commun., 2012, 48, 5614–5616 RSC.
- M. L. McKee, P. J. Milnes, J. Bath, E. Stulz, R. K. O'Reilly and A. J. Turberfield, Programmable One-Pot Multistep Organic Synthesis Using DNA Junctions, J. Am. Chem. Soc., 2012, 134, 1446–1449 CrossRef CAS PubMed.
- M. L. McKee, A. C. Evans, S. R. Gerrard, R. K. O'Reilly, A. J. Turberfield and E. Stulz, Peptidomimetic bond formation by DNA-templated acyl transfer, Org. Biomol. Chem., 2011, 9, 1661–1666 RSC.
- M. Shelbourne, X. Chen, T. Brown and A. H. El-Sagheer, Fast Copper-Free Click DNA Ligation by the Ring-Strain Promoted Alkyne-Azide Cycloaddition Reaction, Chem. Commun., 2011, 47, 6257–6259 Search PubMed.
- V. R. Parvathy, S. R. Bhaumik, K. V. R. Chary, G. Govil, K. Liu, F. B. Howard and H. T. Miles, NMR structure of a parallel-stranded DNA duplex at atomic resolution, Nucleic Acids Res., 2002, 30, 1500–1511 Search PubMed.
- Y. Wang and D. J. Patel, Solution Structure of a Parallel-stranded G-Quadruplex DNA, J. Mol. Biol., 1993, 234, 1171–1183 CrossRef CAS PubMed.
- K. Gehring, J.-L. Leroy and M. Guéron, A tetrameric DNA structure with protonated cytosine-cytosine base pairs, Nature, 1993, 363, 561–565 Search PubMed.
- Y. Chen and C. Mao, Reprogramming DNA-Directed Reactions on the Basis of a DNA Conformational Change, J. Am. Chem. Soc., 2004, 126, 13240–13241 CrossRef CAS PubMed.
- T. J. Povsic, S. A. Strobel and P. B. Dervan, Sequence-Specific Double-Strand Alkylation and Cleavage of DNA Mediated by Triple-Helix Formation, J. Am. Chem. Soc., 1992, 114, 5934–5941 CrossRef CAS.
- K. B. Grant and P. B. Dervan, Sequence-Specific Alkylation and Cleavage of DNA Mediated by Purine Motif Triple Helix Formation, Biochemistry, 1996, 35, 12313–12319 CrossRef CAS PubMed.
- P. A. Beal and P. B. Dervan, Second Structural Motif for Recognition of DNA by Oligonucleotide-Directed Triple-Helix Formation, Science, 1991, 251, 1360–1363 CrossRef CAS PubMed.
- W. A. Greenberg and P. B. Dervan, Energetics of Formation of Sixteen Triple Helical Complexes Which Vary at a Single Position Within a Purine Motif, J. Am. Chem. Soc., 1995, 117, 5016–5022 CrossRef CAS.
- P. A. Beal and P. B. Dervan, The Influence of Single Base Triplet Changes on the Stability of a Pur·Pur·Pyr Triple Helix Determined by Affinity Cleaving, Nucleic Acids Res., 1992, 20, 2773–2776 CrossRef CAS PubMed.
- H. E. Moser and P. B. Dervan, Sequence-Specific Cleavage of Double Helical DNA by Triple Helix Formation, Science, 1987, 238, 645–650 CrossRef CAS.
- S. Rhee, Z.-j. Han, K. Liu, H. T. Miles and D. R. Davies, Structure of a Triple Helical DNA with a Triplex–Duplex Junction, Biochemistry, 1999, 38, 16810–16815 CrossRef CAS PubMed.
- S. A. Strobel, L. A. Doucette-Stamm, L. Riba, D. E. Housman and P. B. Dervan, Site-Specific Cleavage of Human Chromosome 4 Mediated by Triple-Helix Formation, Science, 1991, 254, 1639–1642 CrossRef CAS PubMed.
- L. J. Maher, B. Wold and P. B. Dervan, Inhibition of DNA Binding Proteins by Oligonucleotide-Directed Triple Helix Formation, Science, 1989, 245, 725–730 CrossRef CAS PubMed.
- S. A. Strobel and P. B. Dervan, Single-site enzymatic cleavage of yeast genomic DNA mediated by triple helix formation, Nature, 1991, 350, 172–174 CrossRef CAS PubMed.
- L. J. Maher, P. B. Dervan and B. Wold, Analysis of Promoter-Specific Repression by Triple-Helical DNA Complexes in a Eukaryotic Cell-free Transcription System, Biochemistry, 1992, 31, 70–81 CrossRef CAS PubMed.
- C. Cao, P. Zhao, Z. Li, Z. Chen, Y. Huang, Y. Bai and X. Li, A DNA-Templated Synthesis of Encoded Small Molecules by DNA Self-Assembly, Chem. Commun., 2014, 50, 10997–10999 RSC.
- A. Ono, C. N. Chen and L. S. Kan, DNA triplex formation of oligonucleotide analogs consisting of linker groups and octamer segments that have opposite sugar-phosphate backbone polarities, Biochemistry, 1991, 30, 9914–9921 CrossRef CAS PubMed.
- B.-W. Zhou, C. Marchand, U. Asseline, N. T. Thuong, J.-S. Sun, T. Garestier and C. Helene, Recognition of alternating oligopurine/oligopyrimidine tracts of DNA by oligonucleotides with base-to-base linkages, Bioconjugate Chem., 1995, 6, 516–523 CrossRef CAS PubMed.
- J. Nakao, Y. Mikame, H. Eshima, T. Yamamoto, C. Dohno, T. Wada and A. Yamayoshi, Unique Crosslinking Properties of Psoralen–Conjugated Oligonucleotides Developed by Novel Psoralen N–Hydroxysuccinimide Esters, ChemBioChem, 2023, 24, e202200789 CrossRef CAS.
- J. Hennessy, B. McGorman, Z. Molphy, N. P. Farrell, D. Singleton, T. Brown and A. Kellett, A Click Chemistry Approach to Targeted DNA Crosslinking with cis–Platinum(II)–Modified Triplex–Forming Oligonucleotides, Angew. Chem., Int. Ed., 2022, 61, e202110455 CrossRef CAS PubMed.
- S. Sasaki, S. Nakashima, F. Nagatsugi, Y. Tanaka, M. Hisatome and M. Maeda, Design of a novel artificial nucleobase for the selective formation of a triple-complex with a cytosine-guanine base pair, Tetrahedron Lett., 1995, 36, 9521–9524 CrossRef CAS.
- L. J. Maher, P. B. Dervan and B. J. Wold, Kinetic Analysis of Oligodeoxyribonucleotide-Directed Triple-Helix Formation on DNA, Biochemistry, 1990, 29, 8820–8826 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |