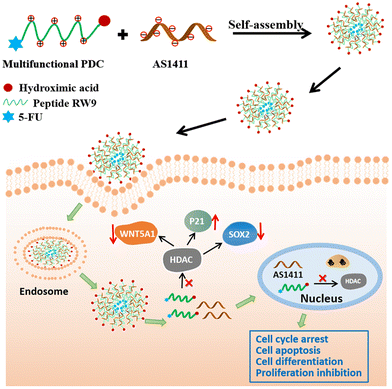
An assembly-inducing PDC enabling the efficient nuclear delivery of nucleic acid for cancer stem-like cell suppression†
Dongyuan
Wang
*ab,
Yuan
Tian
 e,
Yu
Zhang
ab,
Xiaona
Sun
fg,
Yuxuan
Wu
fg,
Ruping
Liu
g,
Fang
Zeng
ab,
Jingjing
Du
*d and
Kuan
Hu
e,
Yu
Zhang
ab,
Xiaona
Sun
fg,
Yuxuan
Wu
fg,
Ruping
Liu
g,
Fang
Zeng
ab,
Jingjing
Du
*d and
Kuan
Hu
 *cf
*cf
aDepartment of Pharmacy, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China. E-mail: wangdy2019@hust.edu.cn
bHubei Province Clinical Research Center for Precision Medicine for Critical Illness, Wuhan, China
cDepartment of Advanced Nuclear Medicine Sciences, Institute of Quantum Medical Science, National Institutes for Quantum Science and Technology, Chiba, Japan. E-mail: kuan.hu@qst.go.jp
dHubei Key Laboratory of Kidney Disease Pathogenesis and Intervention, College of Medicine, Hubei Polytechnic University, Huangshi, Hubei 435003, China
eSchool of Life Science and Engineering, Southwest Jiaotong University, Chengdu, Sichuan, China
fInstitute of Materia Medica, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100050, P. R. China
gBeijing Institute of Graphic Communication, Beijing 102600, China
First published on 6th October 2022
Abstract
Nucleic acid therapy is attracting great attention in diverse clinical translations because of its therapeutic advantages. As a renowned oligonucleotide therapeutical candidate in the clinical stage, AS1411 has shown outstanding tumor suppressing effects; however, its efficient delivery to the cell nucleus is critical for its anticancer effect. Herein, we identified a multifunctional peptide drug conjugate (PDC) as a safe and efficient carrier to achieve the nuclear delivery of AS1411. This PDC consists of the cell penetration peptide RW9, an HDAC inhibitor warhead (peptide C-terminus), and 5-FU (peptide N-terminus), which can coassemble with AS1411 to form nanospheres. The PDC efficiently delivered AS1411 to the nucleus of several types of cancer cells. Moreover, it reversed the stemness of a cancer stem-like cell line. Significantly, due to the assembly-induced accumulation enhancement and retention, a safe single agent concentration of PDC showed unexpected synergy with AS1411 to augment the cancer cell suppression efficiency, exemplified by the downregulation of the stemness-related proteins and the upregulation of apoptosis-related proteins. Therefore, our work presents a powerful strategy for the nuclear delivery of nucleic acid drugs by leveraging cancer-suppressing PDC as assembly inducers, which provides a powerful combination regimen in treating cancer stem-like cells.
Introduction
Nucleic acid-derived therapeutics have attracted global attention in recent decades as they offer the opportunity to address a wide range of diseases that were previously considered undruggable or lacked treatment options.1–3 To this end, numerous strategies have been developed for the encapsulation or stabilization of various nucleic acid structures to achieve successful intracellular delivery.4–7 The anticancer aptamer AS1411 is a 26-mer guanine-rich oligonucleotide DNA that has been in phase II clinical trials for relapsed or refractory acute myeloid leukemia and renal cell carcinoma.8–10 Although AS1411 is widely attached to nanoparticles as a cancer-targeting ligand by interacting with nucleolin on the cancer cell membrane, AS1411 kills cancer cells through cellular mechanisms.11–13 Thus, there is an urgent need for the development of delivery systems that can effectively transport AS1411 into the cell nucleus.In recent decades, numerous strategies have been developed for nucleic acid delivery, such as liposomes, silica nanoparticles, metal-based nanoparticles, and inorganic nanoparticles.14–16 Many of them have shown great promise in clinic due to their delivery efficiency.17,18 Nevertheless, concerns regarding the long-term toxicity of these nanomaterials have not been dispelled. In contrast, peptides are considered “ideal” nucleic acid delivery vehicles due to their remarkable biocompatibility, biodegradability, and flexibility of chemical modification for various clinical demands.4,19–21 Compared to peptides alone, peptide drug conjugates (PDCs) provide additional functionality, such as improving the drug solubility and target selectivity and circumventing multidrug resistance (MDR).22–24 Thus, PDCs are now attracting rapidly growing interest in the drug delivery community. Many studies have revealed that the electrostatic interactions between short nucleic acids (such as siRNA or DNA aptamers) and cell penetrating peptides or amphiphilic peptides can promote the formation of a stable complex and cellular uptake.25–30 Thus, PDCs can also take advantage of positively charged peptides to achieve nucleic acid drug delivery through DNA-peptide self-assembly by electrostatic interactions. As a result, remarkable synergistic effects of nucleic acid drugs and PDC on cancer proliferation inhibition are observed. This characteristic of PDC could make it a good candidate for multifunctional combination cancer therapy.
Combination therapy with two or more drugs has attracted increasing attention in cancer therapy due to its improved therapeutic efficiency and reduced toxicity.31–33 HDACs are popular anticancer targets that can modulate the gene expression and cellular functions, thus regulating different altered pathways in cancer, such as apoptosis, cell cycle, and DNA repair.34,35 5-Fluorouracil (5-FU) is a first-line anticancer drug for many cancer patients, such as colon cancer, and has been widely used for combination therapy with other drugs or nanodrugs.36,37 Piro et al. revealed that the approved HDACi vorinostat could induce synergistic antitumor and proapoptotic effects in combination with 5-FU in colorectal cancer, breast cancer, or squamous cancer cell models via multiple signaling pathways.38 Crea et al. reported that the HDACi trichostatin A can reverse 5-FU resistance in human cancer cells by downregulating thymidylate synthase (TS).39 In addition, the AS1411–FUdR conjugate can cause enhanced anticancer potency due to the synergistic effect induced by the simultaneous activation of p53 by AS1411 and the inhibition of thymidylate synthase by FUdR, indicating their potential combination therapeutic effect.40 Thus, in this study, we hypothesize that the combination of HDACI, 5-FU, and AS144 could be a new and promising option for cancer therapy via multiple anticancer signaling pathways.
Herein, we designed a multifunctional peptide drug conjugate (PDC) using a cell penetrating peptide in combination with HDACI and 5-FU. The PDC could coassemble with AS1411 through electrostatic interactions to form nanospheres with a diameter of approximately 100 nm (Scheme 1). The PDC/DNA nanospheres showed excellent anticancer effects toward various cancer cells, especially PA-1 stem-like cancer cells, with negligible toxicity toward normal cells. We also found that these multifunctional nanoparticles inhibited cancer cell proliferation by inducing cell apoptosis, cell cycle arrest, downregulation of the expression of SOX2 and WNT5A1, and upregulation of the cell cycle-related protein P21. We believe that this multifunctional PDC-based nanodelivery system could provide a safe and efficient therapeutic modality for malignant cancer treatment.
Experimental methods
Materials
All solvents and reagents used for solid phase peptide synthesis were purchased from GL Biochem (Shanghai) Ltd or Energy Chemical Co., and were used without further purification, unless otherwise stated. 2-Chlorotityl-N-Fmoc-hydroxylamine resin (loading capacity: 0.53 mmol g−1) was purchased from HECHENG S&T Ltd. Antibodies against Ac-H3, GAPDH, H3, Sox2, p21, and WNT5A1 were obtained from Proteintech (Wuhan). The reagents used for biological assays were purchased from Sigma Aldrich, Thermo Fisher, or other companies approved by the Huazhong University of Science and Technology. Cells were purchased through ATCC and cultured according to the ATCC guidelines. DMEM, MEM, fetal bovine serum (FBS), 0.25% (w/v) trypsin solution, and penicillin–streptomycin solution were purchased from Gibco BRL (USA). AS1411 and scrambled nucleotides were purchased from ShangHai Genepharma company.Peptide synthesis
Peptides were synthesized on 2-chlorotityl-N-Fmoc-hydroxylamine resin (loading capacity: 0.53 mmol g−1) via standard Fmoc-based solid-phase peptide synthesis. First, the resin was swollen in DMF for 30 min, filtered, and then treated with 50% (vol/vol) morpholine in NMP for 30 min × 2. The resin was washed sequentially with DCM and NMP (5×). Fmoc-protected amino acids (5.0 equiv.) and HATU (4.9 equiv.) were dissolved in DMF, followed by DIPEA (10.0 equiv.). The mixture was preactivated for 1 min and added to the resin for 2 h with N2 bubbling. The resin was washed sequentially with DCM and NMP (5×). The final resin was treated with TFA/DCM/TIS/water (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5) at room temperature for 3 h. The peptide was precipitated and washed three times in cold diethyl ether and isolated by centrifugation. The dried precipitate was then dissolved in 50% (vol/vol) water/acetonitrile, and the solution was purified by reversed-phase HPLC immediately (Waters 600, Agilent Zorbax SB-Aq: 4.6 × 250 mm, 220 nm & 254 nm) and analyzed by LC-MS (AB SCIEX Elite QSTAR or Shimadzu LC-MS 2020).
2.5) at room temperature for 3 h. The peptide was precipitated and washed three times in cold diethyl ether and isolated by centrifugation. The dried precipitate was then dissolved in 50% (vol/vol) water/acetonitrile, and the solution was purified by reversed-phase HPLC immediately (Waters 600, Agilent Zorbax SB-Aq: 4.6 × 250 mm, 220 nm & 254 nm) and analyzed by LC-MS (AB SCIEX Elite QSTAR or Shimadzu LC-MS 2020).
Tumor cell lines and culture
The human liver cancer cell line Huh7, human colon cancer cell line HT29, and human kidney cell line 293T were cultured in DMEM with 10% (v/v) fetal bovine serum FBS and penicillin/streptomycin (final concentration: 50–100 IU mL−1 penicillin and 50–100 μg mL−1 streptomycin). The human ovarian teratocarcinoma cell line PA-1 was cultured in MEM with 10% (v/v) fetal bovine serum (FBS) and penicillin/streptomycin (final concentration: 50–100 IU mL−1 penicillin and 50–100 μg mL−1 streptomycin). All these cells were maintained in a humidified incubator containing 5% CO2 at 37 °C.In vitro deacetylation assays
The HeLa nuclear extract was obtained according to the instructions of the Nuclear Extraction Kit (Solarbio). The assays were carried out in the 384-well format using the fluorometric histone deacetylase kit according to the manufacturer's protocol (BioVision and Caymanchem). HDAC activity was detected at an excitation wavelength of 350 nm and an emission wavelength of 450 nm using Envision (PerkinElmer). The analytical software GraphPad Prism 8.0 (GraphPad Software, Inc., USA) was used to generate IC50 values via nonlinear regression analysis.Cell viability assay
Cell viability for different peptides in different cell lines was measured by CCK8 assay. The cells were incubated on a 96-well plate for 24 h in the growth medium prior to peptide treatment (four peptide concentrations: 10, 20, 40, 80, and 160 μM). DMSO at 0.1% was used as the negative control. Then, the media was removed, and peptides were added to the medium with 5% FBS (fetal bovine serum, v/v) for 48 h of incubation. CCK8 (10 μL) was added, and the cells were incubated for 1 h at 37 °C with 5% CO2. The absorbance was measured with a microplate reader (Bio-Rad) at a wavelength of 450 nm.The cell viability for different nanoparticles in different cell lines was also measured by CCK8 assays. The cells were incubated on a 96-well plate for 24 h in the growth medium prior to treatment with different nanoparticles. For nanoparticle preparation, peptides (20 μM in PA-1 cells or 80 μM in other cells) were incubated with AS1411 (0.134 μg) or scrambled DNA (0.134 μg) named NC for 30 min before being added to 96-well plates with the medium containing 0% FBS. The N/P ratio is approximately 25 for PA-1 cells and 100 for other cell lines. The medium was removed, followed by the addition of peptide-based nanoparticles to the medium with 0% FBS (v/v) for 4 h of incubation and then adding FBS to a final concentration of 5% FBS medium for another 40 h of incubation. CCK8 (10 μL) was added, and the cells were incubated for 1 h at 37 °C with 5% CO2. The absorbance was measured with a microplate reader (Bio-Rad) at a wavelength of 450 nm.
Gel retardation
Peptide FU-NHOH (24 μg) was dissolved properly in ddH2O and diluted in a gradient concentration. AS1411 (0.3 μg) was incubated with diluted peptides at room temperature for 30 min. Then, the samples were added to a 1.5% agarose gel and subjected to 110 V electrophoresis for 15 min in TAE buffer. The blocking effect of the peptide–AS1411 complex was analyzed by a gel imaging analyzer, and then the position of the peptide–AS1411 complex and free AS1411 was compared.The characteristics of FU-NHOH–AS1411 nanoparticles
The average size and zeta potential of the nanoparticles were measured by a Malvern Zetasizer (Nano-ZS90, England). FU-NHOH (160 μg) was incubated with AS1411 (2 μg) for 30 min to form nanoparticles, and then the samples were dispersed in deionized water with a proper volume for detection. Each sample was measured in triplicate at 25 °C. The morphology of the nanoparticles was observed by TEM (transmission electron microscopy) (JEOL-2100F, JEOL Ltd, Japan).The stability of FU-NHOH–AS1411 nanoparticles in protein environment
To test the integration and stability of PDC-induced nanoparticles, the nanoparticles were incubated in PBS (pH 7.4), 2% FBS in PBS and 10% FBS in PBS for 4 h at 37 °C, respectively, and then this reaction system was tested using DLS (dynamic light scattering) or TEM.Transfection efficiency of the nanoparticles by flow cytometry analysis
Cells (30% density) were grown in a medium with 10% FBS (v/v) and cultured in a 37 °C, 5% CO2 incubator in 24-well plates. The medium was replaced with Opti-MEM medium for 2 h. For the PA-1 cells, 0.3 μg FAM-labeled AS1411 and peptide FU-NHOH (10 μM (N/P 35), 20 μM (N/P 70), and 40 μM (N/P 140)) were mixed for 30 min and then added to 24-well plates containing 300 μL MEM medium with 0% FBS (v/v) for 2 h incubation before being harvested and analyzed by FACS. For Huh7 cells or HT-29 cells, 0.3 μg FAM-labeled AS1411 and peptide FU-NHOH (20 μM, 40 μM, and 80 μM (N/P 280)) were mixed for 30 min and then added to the 24-well plates containing 300 μL DMEM with 0% FBS (v/v) for 2 h incubation before being harvested and analyzed by FACS. The medium fluorescence intensity of the cells was analyzed by the FlowJo 7.6.1 software.Cell distribution of the nanoparticles by the confocal microscopy image
Cells (20–30% density) were grown in a medium with 10% FBS (v/v) and cultured in a 37 °C, 5% CO2 incubator in 24-well plates on glass. Then, the cells were washed and treated with peptide-based nanoparticles (0.3 μg FAM-labeled AS1411 and FU-NHOH (10 μM, 20 μM, 40 μM in PA-1 cells or 20 μM, 40 μM, 80 μM in HT-29 or Huh7 cells) for 30 min in 300 μL medium with 0% FBS for 2 h incubation. The transfection agent Oligofectamine™ was used as the positive control. The cells were washed with PBS three times and fixed with 4% formaldehyde for 15 min at room temperature. After fixation, the cells were washed three times with PBS, and DAPI (1 μg mL−1 4′,6-diamidino-2-phenylindole)-containing reagent (Invitrogen, CA) was used to seal the slides. Finally, the cells were analyzed using confocal microscopy.Cell apoptosis detection
The apoptosis assay was performed according to the manufacturer's instructions using an Annexin V: FITC Apoptosis Detection Kit I (BD Pharmingen™). Briefly, PA-1 cells were seeded in a 12-well plate and allowed to grow for 24 h in the medium with 10% FBS. Then, 0.6 μg AS1411 or scrambled DNA was mixed with 20 μM (N/P 70) FU-NHOH for 30 min and added to 600 μL MEM medium with 0% FBS for 2 h, followed by another 20 h of incubation in 5% FBS medium. The transfection agent Oligofectamine™ was used as the positive control. The cells were harvested by trypsinization, washed twice with PBS, and suspended in 1× binding buffer. The suspended cells were stained with propidium iodide (PI) and Annexin V and then analyzed by flow cytometry to determine the apoptotic cells. The cells with positive fluorescence intensity signals for both FITC and PI were used for the apoptotic cell count.Cell cycle detection
For the cell cycle arrest experiments, PA-1 cells were seeded in a 12-well plate and allowed to grow for 24 h in the medium with 10% FBS. The cells were treated with peptide-based nanoparticles (0.6 μg AS1411 or scrambled DNA mixed with 20 μM FU-NHOH for 30 min) or 20 μM FU-NHOH alone in the medium with 0% FBS for 2 h, followed by another 20 h of incubation in 5% FBS medium. Then, the cells were washed twice with PBS and harvested by trypsinization. The cells were fixed with cold 70% ethanol for 4 h at 4 °C and isolated by centrifugation at 2000 rpm for 5 min. Then, the precipitant cells were suspended in PBS with 1% Triton-100, 5 mg mL−1 PI, and 1 mg mL−1 RNase and stained at 37 °C for 30 min. The samples were detected by flow cytometry, and the percentages of cells in the G0/G1, S, and G2/M phases were analyzed by the FlowJo software.Western blot
For western blot analysis, PA-1 cells were seeded in 12-well plates and incubated for 24 h. Then, the cells were treated with 40 μM FU-NHOH alone or 20 μM (N/P 70) FU-NHOH-based nanoparticles for 2 h of incubation, followed by another 20 h of incubation in 5% FBS medium. The cells were harvested and lysed in the lysis buffer (50 mM Tris–Cl, pH = 6.8, 2% SDS, 6% glycerol, 1% β-mercaptoethanol, 0.004% bromophenol blue). The extracted protein concentrations were measured by a spectrophotometer (Nano-Drop ND-2000). Equal amounts of protein were loaded onto an SDS-page gel and resolved by electrophoresis. Protein bands were then transferred to the nitrocellulose blotting membranes, followed by incubation with the appropriate primary antibody overnight at 4 °C. The antibody dilutions were used as follows: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 for rabbit monoclonal anti-Ac-H3 (Proteintech), rabbit monoclonal anti-H3 (Proteintech), mouse monoclonal anti-GAPDH (Proteintech), 1
1000 for rabbit monoclonal anti-Ac-H3 (Proteintech), rabbit monoclonal anti-H3 (Proteintech), mouse monoclonal anti-GAPDH (Proteintech), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 for rabbit monoclonal anti-Sox2 (Proteintech), rabbit monoclonal anti-WNT5A1 and anti-P21. Then, the membranes were washed with a buffer (1× TBS, 0.05% Tween 20). Goat anti-rabbit or anti-mouse secondary antibodies were used for 1 h of incubation at room temperature. The proteins were then visualized with chemiluminescent substrates.
500 for rabbit monoclonal anti-Sox2 (Proteintech), rabbit monoclonal anti-WNT5A1 and anti-P21. Then, the membranes were washed with a buffer (1× TBS, 0.05% Tween 20). Goat anti-rabbit or anti-mouse secondary antibodies were used for 1 h of incubation at room temperature. The proteins were then visualized with chemiluminescent substrates.
Results and discussion
Design and screening of multifunctional peptide–drug conjugates
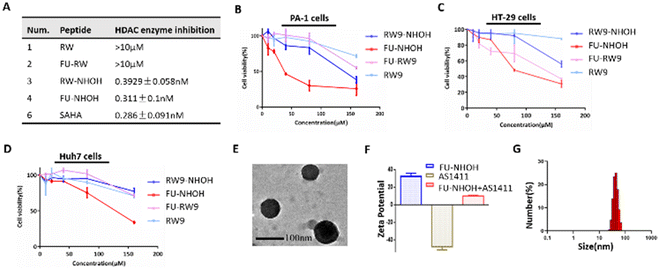
We chose the short cell penetrating peptide RW9 (sequence: RRWWRRWRR) as this peptide was previously reported as an efficient carrier to deliver peptides or nucleic acids to the cell nucleus.41–43 We attached a hydroxamic acid group at the C-terminus for HDAC inhibition and 5-FU at the N-terminus to increase its antitumor effect, named FU-NHOH, as shown in Scheme 1 and Fig. S1.† The peptides RW, FU-RW, and RW-NHOH were designed as negative controls. All the designed peptides were synthesized based on solid-phase peptide synthesis reported previously, as shown in Fig. S2.†44,45 Peptides RW-NHOH and FU-NHOH, both of which have a hydroxamic acid functional group, showed HDAC enzyme inhibition activity at nanomolar concentrations (Fig. 1A and Fig. S3†), suggesting their potential function as HDACi. We then tested the anticancer effect of the four different peptides using the CCK-8 reagent. As expected, FU-NHOH had the most toxic effect among these four designed peptides, as shown in Fig. 1B–D, on PA-1, Huh7, and HT29 cells. Interestingly, FU-NHOH showed a better anticancer effect on PA-1 cells than on the other two cancer cell lines, partly because this cancer stem-like cell line is reported to be more sensitive to HDAC inhibition. The selective toxicity toward different cancer cells suggests the therapeutic potential in various cancer types. Thus, we chose the most toxic PDC FU-NHOH as a carrier for AS1411 delivery and anticancer therapy.First, we tested the self-assembly of FU-NHOH with AS1411 by the gel retardation assay shown in Fig. S4.† AS1411 (approximately 0.3 μg) could be fully loaded with 24 μg FU-NHOH (the N/P ratio was approximately 72). The characterization of the peptide–AS1411 complex was further conducted using TEM. As shown in Fig. 1E, the peptide–AS1411 complex formed nanospheres approximately 100 nm in diameter. Furthermore, the peptide–AS1411 nanoparticles were positively charged according to the zeta potential, as shown in Fig. 2F. The hydrodynamic size of the peptide/AS1411 complexes was determined and displayed a diameter of approximately 100 nm, which is consistent with the TEM observation. Meanwhile, the nanoparticles were stable in PBS, 2% FBS in PBS, and 10% FBS in PBS, as shown as Fig. S5.† All these results demonstrated that our designed PDC could readily co-assemble with AS1411 to form uniform nanoparticles.
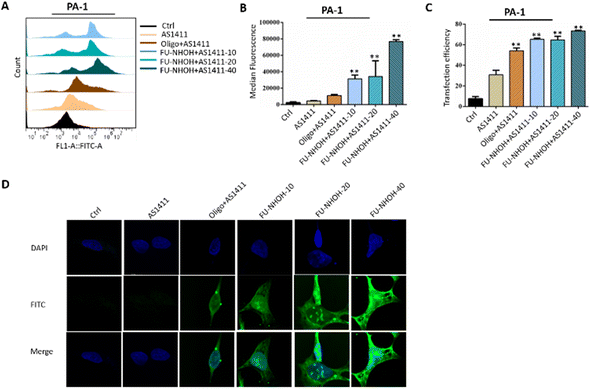 | ||
| Fig. 2 Cellular uptake of different nanoparticles in PA-1 cells. (A) Statistical cell counts by flow cytometry, where the y-axis represents cell counts and the x-axis the FAM fluorescence intensities. (B) Median fluorescence intensity of these nanoparticles according to (A). (C) Transfection efficiency of these nanoparticles according to Fig. 3A. (D) Fluorescence images of PA-1 cells after incubation with different samples. AS1411 means 0.3 μg of FAM-labeled AS1411 alone. Oligo + AS1411 means 0.3 μg of FAM-labeled AS1411 assembled with oligofectamin™. Peptide-10, -20, and -40 indicate 0.3 μg of FAM-labeled AS1411 assembled with FU-NHOH at 10 μM (N/P 36), 20 μM (N/P 72), and 40 μM (N/P 144), respectively. **p < 0.01 vs. Ctrl. | ||
Efficient delivery of AS1411 by FU-NHOH into the PA-1 cells
The cellular uptake efficiency of AS1411 was first monitored using a flow cytometry assay. PA-1 cells were treated with FAM-labeled AS1411 nanoparticles for 2 h before flow cytometric analysis, and Oligofectamine™ was used as the positive control. Fig. 2A shows the fluorescence plots of PA-1 cells treated with different AS1411 nanoparticles. Compared to PBS-treated cells, AS1411 showed a weak fluorescence signal, and cells treated with Oligofectamine™/AS1411 nanoparticles or FU-NHOH/AS1411 nanoparticles had a much stronger fluorescence signal than cells treated with AS1411 alone. FU-NHOH/AS1411 nanoparticles showed a concentration-dependent transfection efficiency when AS1411 was incubated with different concentrations of FU-NHOH (10 μM for N/P 36, 20 μM for N/P 72, 40 μM for N/P 144), as shown in Fig. 2B and C. We also tested the transfection efficiency of these nanoparticles in Huh7 and HT-29 cells, as shown in Fig. S6 and 7.† We found that the transfection efficiency of these nanoparticles was weaker in both HT-29 and Huh7 cells than in the PA-1 cells, partly because of the different cell membrane penetration properties in different cell lines.The intracellular distribution of FAM-labeled AS1411 in different cell lines was monitored using fluorescence microscopy, as shown in Fig. 2D and Fig. S8, 9.† Similar to the FACS assay, the cancer cells were treated with Oligofectamine™/AS1411 nanoparticles or FU-NHOH/AS1411 nanoparticles for 2 h before being tested by LSCM. FAM-labeled AS1411 could be successfully transported to PA-1 cells and distributed in the cell nucleus by the FU-NHOH-based delivery system. In addition, the fluorescence intensity was stronger with increasing concentrations of FU-NHOH. In comparison, AS1411 alone or Oligofectamine™/AS1411 nanoparticles had a much weaker fluorescence signal both in the cytoplasm and cell nucleus in PA-1 cells. The fluorescence intensity of FU-NHOH-based nanoparticles was much weaker in Huh7 cells and HT-29 cells than in PA-1 cells, which was consistent with the results of the FACS assay. All these data demonstrated that our designed multifunctional PDC is an efficient vehicle for AS1411 nucleus delivery.
FU-NHOH–AS1411 nanoparticles suppress cell proliferation in different cancer cells
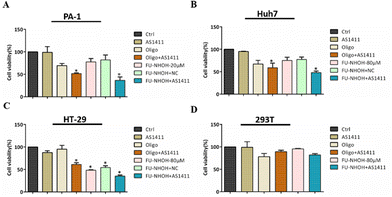
Having demonstrated that FU-NHOH successfully delivered AS1411 to the cancer cell nucleus, we then tested their anticancer effect on different cells. We chose 20 μM (N/P 72) FU-NHOH for AS1411 delivery in PA-1 cells as this ratio was confirmed to be successfully assembled with AS1411 and had good transfection efficiency, according to Fig. 2. As shown in Fig. 3A, AS1411, 20 μM FU-NHOH alone or assembled with scrambled DNA (FU-NHOH/NC) had a weak anticancer effect on the PA-1 cells, while when 20 μM FU-NHOH was assembled with AS1411, the anticancer effect was significantly increased. In addition, we found that the IC50 of FU-NHOH–AS1411 particles was approximately 10 μM in the PA-1 cells (shown in Fig. S10†), while the IC50 of FU-NHOH was approximately 40 μM (shown in Fig. 2B), indicating the synergistic effect of the combination therapy. The positive control Oligofectamine™/AS1411 showed better cell toxicity than Oligofectamine™ alone, indicating the cellular toxicity of AS1411. We also tested the anticancer effect of FU-NHOH/AS1411 nanoparticles on Huh 7 cells or HT-29 cells, as shown in Fig. 3B, C and Fig. S10B, C.† We chose 80 μM (N/P 288) FU-NHOH for AS1411 delivery considering its transfection efficiency and peptide toxicity in these two cell lines. Similar to PA-1 cells, FU-NHOH/AS1411 nanoparticles showed a synergistic effect of the combination therapy as the IC50 of FU-NHOH was approximately 120 μM and 90 μM in Huh-7 cells and HT-29 cells, whereas, the IC50 of FU-NHOH-AS1411 nanoparticles in those cells were approximately 80 μM and 50 μM, respectively. The anticancer effect in PA-1 cells was much stronger than that in the other two cancer cell lines partly because the cancer stem-like PA-1 cells were more sensitive to this combination therapy. The negligible toxicity toward normal 293T cells suggested that our delivery system had low toxicity, which may have good potential for clinical application.PDC–AS1411 assembly caused cell apoptosis and cell cycle arrest in cancer stem-like PA-1 cells
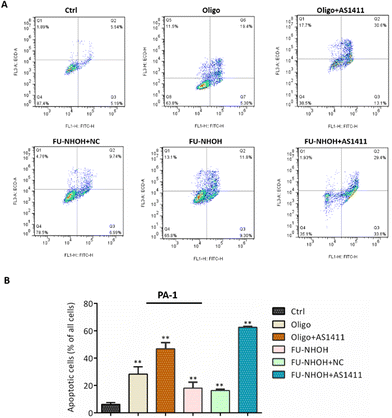
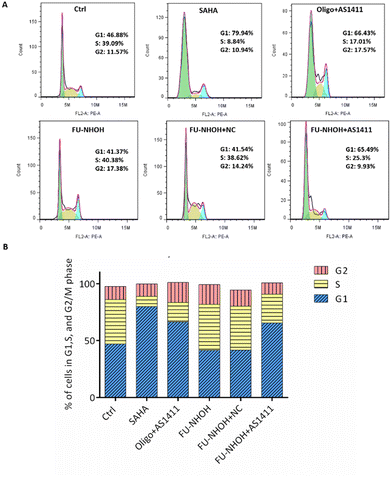
After confirming the potent anticancer effect of FU-NHOH/AS1411 on PA-1 cells, we chose PA-1 cells as a model to study the cellular function influenced by the nanoparticle system. We tested whether the multifunctional nanoparticles could induce cancer cell apoptosis using an Annexin V/PI assay. As shown in Fig. 4, 20 μM FU-NHOH alone or FU-NHOH/NC could induce less than 30% cell apoptosis in PA-1 cells, while FU-NHOH/AS1411 could induce more than 50% apoptosis, which is consistent with the viability results, demonstrating that the multifunctional nanoparticle had a more potent ability to induce cancer cell apoptosis.We further studied whether these peptide/AS1411 nanoparticles induced cell cycle arrest in PA-1 cells. As shown in Fig. 5, PDC alone or PDC/NC nanoparticles had little effect on the cell cycle arrest of PA-1 cells. Oligofectamine™/AS1411 nanoparticles could induce cell cycle arrest at the G1 phase. Similarly, peptide/AS1411 nanoparticles could induce cell cycle arrest at the G1 phase, which was also consistent with the approved drug SAHA (HDACi, approved in 2006). All these data demonstrated that our designed peptide/AS1411 system could have an obvious influence on the cellular function of cancer stem-like cells.
Peptide–AS1411 assembly could inactivate the cellular HDAC signaling pathway in cancer stem-like cells
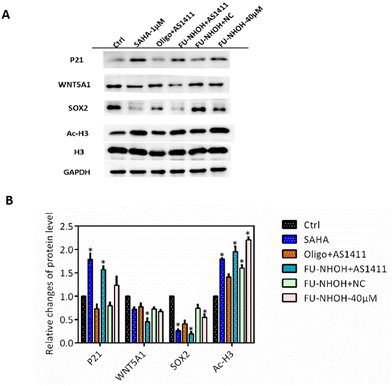
Many studies have revealed that HDAC inhibition can induce cell differentiation and the expression of genes associated with the cell cycle, such as P21, in cancer stem-like cells.44,46 In addition, HDAC inhibition could cause significant downregulation of the stemness gene Sox2, which is convincing evidence of CSC differentiation.44,46 In this study, we first tested the cellular HDAC inhibition of these nanoparticles. As shown in Fig. 6, the acetylation level of H3 was significantly increased when PA-1 cells were treated with 40 μM FU-NHOH, 20 μM FU-NHOH/AS1411, or 20 μM FU-NHOH/NC nanoparticles, similar to the approved HDAC drug SAHA (1 μM). In addition, FU-NHOH/AS1411 nanoparticles could upregulate the expression of P21, explaining the effect of FU-NHOH/AS1411 on G1 phase arrest. Many reports have revealed that HDAC inhibition could influence the WNT signaling pathway, such as WNT5A1, which is highly associated with malignant cancer.44,47,48 We found that FU-NHOH/AS1411 nanoparticles could significantly decrease the level of WNT5A1 and were more potent than SAHA or 40 μM FU-NHOH alone. The stemness protein Sox2 was significantly decreased when treated with FU-NHOH/AS1411 nanoparticles or the positive control SAHA (1 μM), suggesting its potent effect on inducing cell differentiation. All these data demonstrated that FU-NHOH/AS1411 nanoparticles had a more obvious effect on cancer-related signaling pathways than FU-NHOH, even with higher concentrations or FU-NHOH/NC nanoparticles, on cancer stem-like cells, indicating the potent synergistic effect of this combination therapy.Conclusion
Combination therapy is the key trend for malignant cancer treatment. In this study, we designed a multifunctional PDC FU-NHOH that combined HDACi and 5-FU and exhibited potent anticancer effects on cancer stem-like PA-1 cells. Meanwhile, this PDC could be self-assembled with AS1411 to successfully deliver AS1411 to the cell nucleus and enhance their synergistic anticancer effect. We found that the multifunctional nanoparticles could not only have selective toxicity to cancer stem-like cells with negligible toxicity to normal 293T cells but also inhibit cancer proliferation via multiple pathways, such as inducing cell apoptosis, cell cycle arrest, downregulation of stemness protein Sox2, and cancer-related protein WNT5A1. Our work demonstrated that this multifunctional delivery system had promising potential for malignant cancer therapy, especially for chemoresistant cancer types via different pathways, and represents a potentially alternative treatment option for advanced cancer patients.Author contributions
Dongyuan Wang: Conceptualization, investigation, writing – original draft, funding acquisition, and writing – original draft. Kuan Hu: Conceptualization, validation, supervision and writing – review & editing. Jingjing Du: Investigation, data curation, formal analysis, resources and writing – review and editing. Yuan Tian: Data curation, methodology, software and validation. Yu Zhang: Data curation, project administration, resources and funding acquisition, supervision. Fang Zeng: Investigation, methodology, software and validation. Xiaona Sun, Yuxuan Wu, and Ruping Liu helped collect TEM and DLS data, and performed the mass analysis of the hydrolyzed peptide.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (22007033), the Key R&D Program of Hubei Province of China (2020BCA060), the Talent Introduction Project of Hubei Polytechnic University (21xjz32R) and CAMS Innovation Fund for Medical Science of China (No. 02120101, 2021-I2M-1-026-1 and 2022-I2M-1-026-1), and the State Key Laboratory of Bioactive Substance and Function of Natural Medicines, Institute of Materia Medica, the Chinese Academy of Medical Sciences and Peking Union Medical College (No. 0504002).References
- S. M. Hammond, A. Aartsma-Rus, S. Alves, S. E. Borgos, R. A. M. Buijsen, R. W. J. Collin, G. Covello, M. A. Denti, L. R. Desviat, L. Echevarria, C. Foged, G. Gaina, A. Garanto, A. T. Goyenvalle, M. Guzowska, I. Holodnuka, D. R. Jones, S. Krause, T. Lehto, M. Montolio, W. Van Roon-Mom and V. Arechavala-Gomeza, EMBO Mol. Med., 2021, 13, e13243 CrossRef CAS PubMed.
- J. A. Kulkarni, D. Witzigmann, S. B. Thomson, S. Chen, B. R. Leavitt, P. R. Cullis and R. van der Meel, Nat. Nanotechnol., 2021, 16, 630–643 CrossRef CAS PubMed.
- M. Herkt and T. Thum, Mol. Ther., 2021, 29, 521–539 CrossRef CAS PubMed.
- Y. Liu and L. Yin, Adv. Drug Delivery Rev., 2021, 171, 139–163 CrossRef CAS PubMed.
- J. Karlsson, K. M. Luly, S. Y. Tzeng and J. J. Green, Adv. Drug Delivery Rev., 2021, 179, 113999 CrossRef CAS PubMed.
- A. S. Piotrowski-Daspit, A. C. Kauffman, L. G. Bracaglia and W. M. Saltzman, Adv. Drug Delivery Rev., 2020, 156, 119–132 CrossRef CAS PubMed.
- H. J. Vaughan, J. J. Green and S. Y. Tzeng, Adv. Mater., 2020, 32, e1901081 CrossRef PubMed.
- R. Yazdian-Robati, P. Bayat, F. Oroojalian, M. Zargari, M. Ramezani, S. M. Taghdisi and K. Abnous, Int. J. Biol. Macromol., 2020, 155, 1420–1431 CrossRef CAS PubMed.
- J. Carvalho, J. L. Mergny, G. F. Salgado, J. A. Queiroz and C. Cruz, Trends Mol. Med., 2020, 26, 848–861 CrossRef CAS PubMed.
- S. Shigdar, B. Schrand, P. H. Giangrande and V. de Franciscis, Mol. Ther., 2021, 29, 2396–2411 CrossRef CAS PubMed.
- X. Tong, L. Ga, J. Ai and Y. Wang, J. Nanobiotechnol., 2022, 20, 57 CrossRef CAS PubMed.
- D. Xiao, Y. Li, T. Tian, T. Zhang, S. Shi, B. Lu, Y. Gao, X. Qin, M. Zhang, W. Wei and Y. Lin, ACS Appl. Mater. Interfaces, 2021, 13, 6109–6118 CrossRef CAS PubMed.
- L. Dai, D. Wei, J. Zhang, T. Shen, Y. Zhao, J. Liang, W. Ma, L. Zhang, Q. Liu and Y. Zheng, Cell Proliferation, 2021, 54, e13130 CrossRef CAS PubMed.
- Y. Eygeris, M. Gupta, J. Kim and G. Sahay, Acc. Chem. Res., 2022, 55, 2–12 CrossRef CAS PubMed.
- B. Kim, J. H. Park and M. J. Sailor, Adv. Mater., 2019, 31, e1903637 CrossRef PubMed.
- R. A. Revia, Z. R. Stephen and M. Zhang, Acc. Chem. Res., 2019, 52, 1496–1506 CrossRef CAS PubMed.
- N. Chaudhary, D. Weissman and K. A. Whitehead, Nat. Rev. Drug Discovery, 2021, 20, 817–838 CrossRef CAS PubMed.
- J. A. Kulkarni, D. Witzigmann, S. Chen, P. R. Cullis and R. van der Meel, Acc. Chem. Res., 2019, 52, 2435–2444 CrossRef CAS PubMed.
- J. Geng, X. Xia, L. Teng, L. Wang, L. Chen, X. Guo, B. Belingon, J. Li, X. Feng, X. Li, W. Shang, Y. Wan and H. Wang, J. Controlled Release, 2022, 341, 166–183 CrossRef CAS PubMed.
- H. Wang, Z. Feng and B. Xu, Theranostics, 2019, 9, 3213–3222 CrossRef CAS PubMed.
- J. Wang, G. Chen, N. Liu, X. Han, F. Zhao, L. Zhang and P. Chen, Adv. Colloid Interface Sci., 2022, 302, 102638 CrossRef CAS PubMed.
- B. M. Cooper, J. Iegre, D. H. O' Donovan, M. Ö. Halvarsson and D. R. Spring, Chem. Soc. Rev., 2021, 50, 1480–1494 RSC.
- Y. Fang and H. Wang, Pharmaceutics, 2022, 14, 212 CrossRef CAS PubMed.
- M. Alas, A. Saghaeidehkordi and K. Kaur, J. Med. Chem., 2021, 64, 216–232 CrossRef CAS PubMed.
- K. Le Vay, E. Y. Song, B. Ghosh, T. D. Tang and H. Mutschler, Angew. Chem., Int. Ed., 2021, 60, 26096–26104 CrossRef CAS PubMed.
- J. Kim, A. Narayana, S. Patel and G. Sahay, Theranostics, 2019, 9, 3191–3212 CrossRef CAS PubMed.
- A. Varanko, S. Saha and A. Chilkoti, Adv. Drug Delivery Rev., 2020, 156, 133–187 CrossRef CAS PubMed.
- D. Mumcuoglu, M. Sardan, T. Tekinay, M. O. Guler and A. B. Tekinay, Mol. Pharm., 2015, 12, 1584–1591 CrossRef CAS PubMed.
- M. Mazza, M. Hadjidemetriou, I. de Lazaro, C. Bussy and K. Kostarelos, ACS Nano, 2015, 9, 1137–1149 CrossRef CAS PubMed.
- X. Zhao, K. Zhang, Y. Wang, W. Jiang, H. Cheng, Q. Wang, T. Xiang, Z. Zhang, J. Liu and J. Shi, Adv. Funct. Mater., 2022, 32, 2108883 CrossRef CAS.
- L. Huang, S. Zhao, F. Fang, T. Xu, M. Lan and J. Zhang, Biomaterials, 2021, 268, 120557 CrossRef CAS PubMed.
- J. Galon and D. Bruni, Nat. Rev. Drug Discovery, 2019, 18, 197–218 CrossRef CAS PubMed.
- M. Zhao, D. van Straten, M. L. D. Broekman, V. Preat and R. M. Schiffelers, Theranostics, 2020, 10, 1355–1372 CrossRef CAS PubMed.
- M. Zhou, M. Yuan, M. Zhang, C. Lei, O. Aras, X. Zhang and F. An, Eur. J. Med. Chem., 2021, 226, 113825 CrossRef CAS PubMed.
- M. K. Ediriweera, K. H. Tennekoon and S. R. Samarakoon, Drug Discovery Today, 2019, 24, 685–702 CrossRef CAS PubMed.
- J. Guo, Z. Yu, M. Das and L. Huang, ACS Nano, 2020, 14, 5075–5089 CrossRef CAS PubMed.
- S. Vodenkova, T. Buchler, K. Cervena, V. Veskrnova, P. Vodicka and V. Vymetalkova, Pharmacol. Ther., 2020, 206, 107447 CrossRef CAS PubMed.
- G. Piro, M. S. Roca, F. Bruzzese, C. Carbone, F. Iannelli, A. Leone, M. G. Volpe, A. Budillon and E. Di Gennaro, Mol. Cancer Ther., 2019, 18, 1405–1417 CrossRef CAS PubMed.
- F. Crea, S. Nobili, E. Paolicchi, G. Perrone, C. Napoli, I. Landini, R. Danesi and E. Mini, Drug Resistance Updates, 2011, 14, 280–296 CrossRef CAS PubMed.
- B. T. Tran, J. Kim and D. R. Ahn, Nanoscale, 2020, 12, 22945–22951 RSC.
- N. A. Alhakamy, P. Dhar and C. J. Berkland, Mol. Pharm., 2016, 13, 1047–1057 CrossRef CAS PubMed.
- E. Mathieu, A. S. Bernard, H. Y. V. Ching, A. Somogyi, K. Medjoubi, J. R. Fores, H. C. Bertrand, A. Vincent, S. Trepout, J. L. Guerquin-Kern, A. Scheitler, I. Ivanovic-Burmazovic, P. Seksik, N. Delsuc and C. Policar, Dalton Trans., 2020, 49, 2323–2330 RSC.
- C. Cordier, F. Boutimah, M. Bourdeloux, F. Dupuy, E. Met, P. Alberti, F. Loll, G. Chassaing, F. Burlina and T. E. Saison-Behmoaras, PLoS One, 2014, 9, e104999 CrossRef PubMed.
- D. Wang, W. Li, R. Zhao, L. Chen, N. Liu, Y. Tian, H. Zhao, M. Xie, F. Lu, Q. Fang, W. Liang, F. Yin and Z. Li, Cancer Res., 2019, 79, 1769–1783 CrossRef CAS PubMed.
- D. Wang, M. Yu, N. Liu, C. Lian, Z. Hou, R. Wang, R. Zhao, W. Li, Y. Jiang, X. Shi, S. Li, F. Yin and Z. Li, Chem. Sci., 2019, 10, 4966–4972 RSC.
- M. S. Roca, E. Di Gennaro and A. Budillon, J. Clin. Med., 2019, 8, 1769 CrossRef PubMed.
- B. G. Debeb, L. Lacerda, W. Xu, R. Larson, T. Solley, R. Atkinson, E. P. Sulman, N. T. Ueno, S. Krishnamurthy, J. M. Reuben, T. A. Buchholz and W. A. Woodward, Stem Cells, 2012, 30, 2366–2377 CrossRef CAS PubMed.
- W. Yang, Y. Li, R. Gao, Z. Xiu and T. Sun, Oncogene, 2020, 39, 1098–1111 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nr02118h |
| This journal is © The Royal Society of Chemistry 2022 |