Cleavable energy transfer labeled oligonucleotide probe for enhanced isothermal amplification detection and nano digital chip-based readout†
Li
Liu
a,
Stephen J.
Dollery
b,
Gregory J.
Tobin
b,
Guoyu
Lu
c and
Ke
Du
 *a
*a
aDepartment of Chemical and Environmental Engineering, University of California, Riverside, CA, USA. E-mail: kdu@ucr.edu
bBiological Mimetics, Inc., 124 Byte Drive, Frederick, MD, USA
cIntelligent Vision and Sensing Lab, University of Georgia, Athens, GA, USA
First published on 26th November 2024
Abstract
Quantitative analysis of human papillomavirus (HPV)-infected cervical cancer is essential for early diagnosis and timely treatment of cervical cancer. Here, we introduce a novel energy transfer-labeled oligonucleotide probe to enhance the loop-mediated isothermal amplification (LAMP) assay for highly sensitive and specific detection of HPV 16. Conducted as a single-step assay within a digital nanofluidic chip featuring numerous reaction reservoirs, our method facilitates target amplification under isothermal conditions. Targeting an HPV 16 gene, our chip demonstrates the capability to detect HPV DNA at concentrations as low as 1 fM, spanning a dynamic range of five orders of magnitude. Importantly, our nano digital chip enables for highly quantitative detection at low concentrations, with the correlation between target concentration and the number of microwells exhibiting fluorescence signals. Furthermore, we have developed a computer vision method for automated and 100% accurate quantification of target concentrations. This research holds promising applications in clinical diagnosis and is poised for seamless integration into both hospital and point-of-care settings.
Introduction
Cervical cancer is the fourth most common cancer in women globally, and in almost all instances this begins with human papillomavirus (HPV) infection.1 The two most common high-risk genotypes, HPV16 and 18, are responsible for over 70% of cervical cancer cases.2 Due to their elevated sensitivity and specificity, HPV DNA tests have become the gold standard for cervical cancer diagnosis.3 However, these PCR-based DNA tests rely on bulky and expensive instruments such as thermal cyclers, making them impractical for point-of-care (POC) settings.4 In recent years, nucleic acid isothermal amplification detection has emerged as a promising alternative to traditional PCR due to its simplicity, speed, cost-effectiveness, and high sensitivity.5 Various nucleic acid isothermal amplification methods have been developed, including loop-mediated isothermal amplification (LAMP),6–8 recombinase polymerase amplification (RPA),9–11 rolling circle amplification (RCA),12–14 and nucleic acid sequence-based amplification (NASBA).15,16 Even though highly promising, most isothermal amplification detection methods have limitations such as a lack of quantitative detection capabilities, occasional unexpected non-specific amplification signals, and relatively high background signals.17Microfluidic platforms have revolutionized the field of molecule detection by offering a highly miniaturized, cost-effective, and efficient approach for handling fluids at the microscale and even the nanoscale. These platforms—including droplet-based microfluidics, paper-based microfluidics, electrokinetic microfluidics, surface plasmon resonance (SPR) microfluidics, and optofluidics—each offer unique advantages that make them suitable for a wide range of applications. For example, droplet-based microfluidic was high throughput, low reagent consumption and have excellent isolation of reactions.18 Paper-based microfluidic device are particularly attractive as low-cost, portable and user-friendly platforms, ideal for POC testing.19 Electrokinetic microfluidics, on the other hand, excels in sensitivity and offers precise control over sample movement without relying on moving mechanical parts.20 SPR microfluidic platforms leverage the interaction between light and a metallic surface, allowing real-time, label-free monitoring of biomolecular interactions with high sensitivity.21 Similarly, optofluidic platforms combine optical and fluidic technologies, enabling particle manipulation via optical tweezers, thus facilitating high-sensitivity detection and control over individual particles.22 These diverse microfluidic platforms not only enhance sensitivity and processing speed but also require minimal sample volumes, making them promising tools in the continuous evolution of molecular detection technologies.
LAMP is one of the most widely applied isothermal amplification technologies in pathogen diagnostics.23 The advantages of the LAMP reaction include high sensitivity and the use of multiple primers, which have exceptionally high specificity because a set of four primers with six binding sites must hybridize correctly to their target sequence before DNA biosynthesis occurs.24 The reaction temperature is around 63 °C, which avoids undesirable pre-amplification at room temperature.25 In addition, LAMP exhibits a strong tolerance to inhibitors. Traditional detection methods relying on intercalating dyes like EvaGreen or SYBR Green can directly detect amplified targets but have drawbacks such as low selectivity and the potential inhibition of the amplification reaction,26 thus requiring post-reaction analysis and causing false positives.27 Recent progress in integrating LAMP with microfluidic platform has propelled POC diagnostics by enabling compact, portable devices with enhanced efficiency, suitable for applications like for pathogen detection. For instance, Bai et al. developed a portable all-in-one device (PAD) for colorimetric detection of nucleic acids, capable of rapidly identifying various type of viral.28 Cao et al. developed a LAMP-based microfluidic chip for both colorimetric and fluorescent readout for the simultaneous detection of multiple foodborne bacteria.29 However, these methods lacked quantitative detection of nucleic acids. To address these issues, energy transfer-labeled oligonucleotide probes for sequence-specific fluorescence detection have been developed to improve the amplification process. Here, we introduce an innovative ribonuclease-dependent cleavable FQ LB (fluorophore–quencher Loop B) primer designed to augment the sensitivity and specificity of LAMP detection. This primer undergoes cleavage exclusively upon recognizing a specific nucleic acid sequence, thereby markedly enhancing detection specificity and minimizing background signals.
Digital micro-and nanofluidic detection chips have been developed in recent years with advantages over traditional methods, such as greater tolerance to inhibitory substances, higher sensitivity, and more accurate detection.30 The combination of digital chips and nucleic acid amplification methods enables absolute quantification analysis of nucleic acid targets by distributing target molecules into small wells or droplets. When performing limited dilution into aliquots, those aliquots contain no target molecule or only one molecule. The target molecule concentration can then be derived by counting the number of positive aliquots. The isolation of aliquots eliminates the competition of primers and probes, which is especially important for detecting minute DNA targets.31 Currently, there are two main strategies for generating isothermal reaction units, one of which involves using chamber microfluidic chips to generate reaction units. However, due to manufacturing process limitations, the number of chambers in microfluidic chips is limited as the chamber volume is relatively large, leading to a restricted dynamic range.32 The other strategy involves using droplet microfluidic chips to generate monodisperse droplets. However, the process of droplet formation is relatively complex, and the stability of the droplets poses issues, making real-time reaction monitoring a significant challenge.33
In this work, we show a novel energy transfer-labeled oligonucleotide probe with improved fluorescence properties to create a highly sensitive and specific isothermal amplification of nucleic acids for sensitive and quantitative detection of HPV 16 sequences in plasmids that mimic dsDNA HPV genomes. The digital warm start assay is established through a LAMP-based reaction in sub-microliter aliquots within a digital nanofluidic chip. This reaction is a one-pot format FQ labeled primer isothermal amplification-based detection, preventing premature target amplifications at room temperature and enabling accurate digital quantification of nucleic acids. Further, separation of aliquots eliminates competition between primers and probes and largely reduces false positive signals. We apply machine learning algorithm to facilitate the straightforward derivation of quantitative relationships for target concentrations. Combining these advantages enables our system to be adapted in hospitals or POC settings for the quantification of sexually transmitted infections.
Results
The schematic of the digital nanofluidic chip based on the one-step LAMP assay is shown in Fig. 1a. The primers were designed to amplify an HPV 16 L1 fragment gene sequence (GenBank accession MT316211.1). The FQ-LAMP reaction mixture was first prepared in an Eppendorf tube. The prepared reaction mixture was then distributed into a QuantStudio 3D digital chip. The 3D digital chip is a 10 mm2 high-density reaction plate that has a single array of 20000 reaction microwells. Each microwell has a diameter of 60 μm and a depth of 500 μm (Fig. 1b). The chip was pretreated with a hydrophobic coating on the chip surface to enable the loading and isolation of the LAMP reactions within the microwells. After the addition of the reaction reagent, an oil layer was applied to cover the chip. This step not only facilitates the straightforward isolation of each reaction microwell but also serves as a preventive measure against contamination and reagent evaporation. Following a 45 min incubation at 63 °C, microwells containing the target DNA exhibit a green fluorescence signal attributed to successful target amplification, while microwells lacking the target remain devoid of such fluorescence.
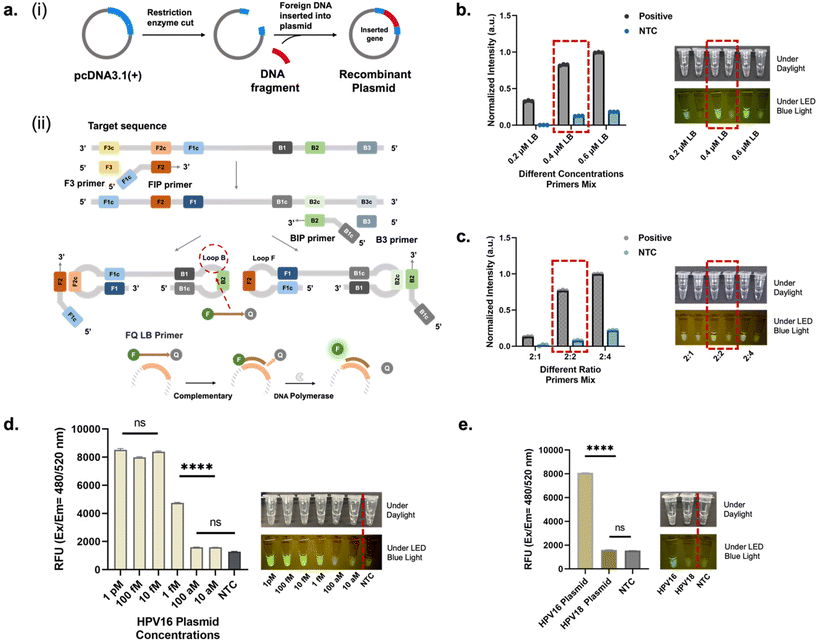
Fig. 2a shows the schematic of the FQ probe-based LAMP reaction. This assay was developed with the designation of two sets of primers to specifically detect the HPV 16 DNA target. Five LAMP primers (FIP, BIP, F3, B3, and LB) were designed according to the distinct regions of the HPV 16. In the present study, two different types of LBs were constructed: the LB probe is the traditional probe with unlabeled ends. The other probe, named the FQ LB probe, has the same sequence as the LB probe but was tagged with FAM fluorophore at the 5′-end and Iowa Black® RQ quencher at the 3′-end. The quencher functions to inhibit the fluorophore from emitting signals when they are close to each other. In its unbound state, this probe is quenched and does not emit fluorescence. However, when it binds to a specific complementary target sequence during amplification, DNA polymerase degrades the probe, separating the fluorophore from the quencher. This separation enables the fluorophore to emit a signal, indicating the presence of target. The fluorophore and quencher were placed further from each other to allow them to anneal the stem-loop region of the dumbbell-like LAMP amplicons specifically. As the probe is longer than 20 base pairs, it was designed with an additional internal quencher, that is, an internal quencher/ZEN/positioned in the middle of the strand. This design was intended to reduce the assay's crosstalk signals, increase the amplification signal, and produce a lower background noise. The concentrations and ratio of the LB and FQ LB primer reactions were optimized, as illustrated in Fig. 2b and c, the optimal concentration for both LB and FQ LB primer was determined to be 0.4 μM, ensuring efficient amplification and detection. Furthermore, the ideal ratio of LB primers to FQ LB primers was established as 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which optimizes the interaction between the primers and the target sequence. This balance ratio enhances both the sensitivity and specificity of the reaction, leading to more accurate resulting during the amplification process. The fluorescence intensity was measured for the target DNA concentration ranging from 10 aM to 1 pM (Fig. 2d), and the positive reaction was clearly observed in the 1 fM sample with both the naked eye and UV illumination. Fig. 2e shows the comparison of the fluorescence intensity for positive and negative targets (1 pM), and a clear difference between positive (HPV16) and negative groups (HPV 18 and NTC) is observed.
1, which optimizes the interaction between the primers and the target sequence. This balance ratio enhances both the sensitivity and specificity of the reaction, leading to more accurate resulting during the amplification process. The fluorescence intensity was measured for the target DNA concentration ranging from 10 aM to 1 pM (Fig. 2d), and the positive reaction was clearly observed in the 1 fM sample with both the naked eye and UV illumination. Fig. 2e shows the comparison of the fluorescence intensity for positive and negative targets (1 pM), and a clear difference between positive (HPV16) and negative groups (HPV 18 and NTC) is observed.
After optimizing the reaction conditions off-chip, the analytical performance of the microwell chip was investigated. Fig. 3a illustrates the typical workflow of the FQ-LAMP assay, encompassing the synthesis of DNA Plasmid which mimics the HPV circular dsDNA genome, preparation of a one-pot LAMP reaction mixture, distribution of the reaction mixture into the chip, and on-chip incubation at 63 °C. Fig. 3b shows the images of on-chip reactions under a microscope with various incubation times (e.g., 0, 15, 30, 45, 60, and 90 min) using 1 pM HPV plasmid as a target. As shown in Fig. 3c, a 45 min incubation is enough for the FQ probe LAMP assay to reach the maximum percentage of the positive spots. The uniformity of the microwell volume was also characterized by ImageJ. As shown in Fig. 3d, the average well area was 2371 ± 28.01 μm2 at a target concentration of 100 pM, which conforms to a Gaussian distribution (R2 = 0.9702). These results indicate that our chip has high uniformity across all wells and is ideal for digital fluorescence sensing.
By testing various concentrations of HPV 16 DNA plasmid target, the on-chip detection sensitivity was also investigated. As shown in Fig. 4a, the digital FQ LAMP assay was performed with a target concentration ranging from 100 aM to 100 pM, and signals can directly be observed by counting the positive spots shown under the microscope. We are able to detect the HPV 16 target with a concentration as low as 1 fM, and the signal is saturated with a concentration greater than 10 pM. The specificity of the digital LAMP assay was also carried out by using similar strains. As shown in Fig. 4c, positive spots are observed in the chip loaded with the HPV 16 positive control, whereas not for those negative control samples, such as the HPV 18 control and the no template control, which is consistent with our off-chip results. Additionally, the digital nanofluidic chip provides highly quantitative readouts across target concentrations from 1 fM to 10 pM (Fig. 4b), demonstrating an excellent linear relationship (R2 = 0.9764) between target concentrations (X) and the number of positive spots (Y) (Fig. S1†), with a dynamic range spanning 5 orders of magnitude. Moreover, notable distinctions exist in the specificity of HPV target detection when compared to other strains (Fig. 4d).
The entire pipeline for processing images and classifying the testing samples is shown in Fig. 5. To estimate target concentrations from sample images, we first inputted a dataset of training images showcasing results. These images are crucial for creating a baseline for the system's understanding of target appearances at various concentrations. Next, template matching algorithms were applied to detect samples within these images due to the spatial hexagon shape property of each individual sample. We slide this hexagon template across the entire sample image to detect individual samples. The detected samples were sorted based on image intensity levels, ranging from high intensity to low intensity. This sorting is essential to standardizing the analysis across different samples, as the sample image intensity is highly correlated to the target concentration. Subsequently, the intensity values of all samples were normalized to account for variations in target concentration. This normalization ensures consistent and reliable data fed into the neural network. Finally, a Convolutional Neural Network (CNN) was trained using these normalized images. The CNN learns the patterns and correlations between the visual characteristics of the target samples and their known concentrations. This learning enables the network to accurately estimate target concentrations in new, unseen images. Eventually, we composed the target concentration result for each individual sample to the entire training sample image with thousands of individual samples and visualized the target concentration distribution. With this learning algorithm, our system can correctly identify all the positive and negative samples, and the network can accurately estimate the target concentration in unseen images.
Discussion
In this study, we developed a novel energy transfer-labeled oligonucleotide probe to create a highly sensitive and specific isothermal amplification assay for nucleic acid detection. We applied it in a digital nanofluidic chip to enable rapid quantitative detection of HPV targets. FQ LB is a single fluorophore and quencher-labeled oligonucleotide probe that can be cleaved by ribonuclease, simultaneously initiating rapid nucleic acid amplification and generating sequence-specific fluorescence. The designed detection method is a process conducted in a closed one-pot system, integrating multi-step reactions within the same enclosed reservoir, thereby mitigating the risk of carryover or cross-contamination arising from post-amplification steps. The quencher restricts the signals by the fluorophore when they are in close proximity. The probes employed in our approach are quenched in the unbound state and emit fluorescence exclusively upon annealing to a specific complementary region during the amplification process, ensuring high sequence specificity. For LB probes shorter than 20 nucleotides, labelling is limited to the ends, while those exceeding 20 nucleotides incorporate an internal quencher at the midpoint, which would otherwise not be quenched due to the length of the DNA strand. This design reduces crosstalk, enhances amplification, and diminishes background noise.34 In addition, the use of FQ probes does not require a complex pre-processing procedure. Due to the complexity of distinguishing non-specific LAMP amplicons from target amplicons, FQ probes are more specific than conventional LAMP assays using DNA intercalating dyes and can significantly reduce background signals. Therefore, this study reveals the benefits of using FQ LB probes to improve amplicon detection in LAMP assays.The novel FQ LB probe design not only simplifies the reaction steps but also improves the specificity of the assay. Compared to the commonly used calcein/manganese chloride or SYBR green dyes, these traditional choices carry the potential risk of interfering with the reaction components or the detection system and are extremely sensitive to factors such as pH and temperature.41 Another common detection method is to combine CRISPR with LAMP, however this introduces additional steps and components that increase the complexity of the reaction. This complexity can lead to more errors, thus requiring more extensive optimization and inevitably increasing off-target effects.42 Our new assay avoids competition between by different substrates minimizing reagent species and can be used for the development of multiplexing LAMP reacttions.43
We developed a robust digital amplification system for easy identification of detection targets. This method is a simple patterning method that is unique in its ability to concentrate reagents in a small size. Because the target molecules are assigned to numerous small, separated reaction reservoirs, the positive and negative reservoirs can easily be distinguished from each other without interference. The system is more tolerant to reaction inhibitors, as the potential reaction inhibitors are separated from the reaction mixture, reducing the amplification reaction inhibition in the digital assay and making it well-suited for the detection of low-level targets. We found that due to the multiprobing of the conventional LAMP assays, reactions in Eppendorf tubes are more susceptible to non-specific amplifications due to the cross-linking of amplicons, and aerosols generated during the reaction may cause contamination, leading to false positives.44 In contrast, no false-positive signals were found in the reaction system of our digital nanofluidic chip. Therefore, digital detection can directly detect nucleic acid in many samples without complicated sample pre-treatment and nucleic acid purification processes.
Our microliter digital microwells enable the quantitative detection of nucleic acids without the need for calibration curves, thereby improving the accuracy of detecting low-copy nucleic acid templates. Conventional tube-based reactions, such as colorimetric,45 fluorescence detection,46 and turbidity-based assays,35 pose challenges for accurate quantitative detection. Therefore, they are more appropriate for qualitative assessments, primarily serving to differentiate between positive and negative results without offering precise concentration measurements. In contrast, the nano digital chip enables straightforward and highly accurate quantitative detection, as it facilitates individual target molecule counting, which translates into precise measurements of target concentrations. This feature makes digital chip technology highly effective for applications requiring detailed quantification over a broad dynamic range. Table 1 presents a comparison of representative LAMP-based HPV POC testing methods. The ability to quantitatively detect nucleic acids without relying on calibration curves is a key feature that enhances accuracy, particularly in low-copy nucleic acid template detection. In addition, compared to droplet microfluidic chips that produce monodisperse droplets, fixed-structure chambers are more stable and suitable for real-time monitoring.47 The compartmentalization of nucleic acid in digital detection enables individual amplification and detection, enhancing sensitivity to the single-molecule level. This surpasses the sensitivity achievable with traditional isothermal amplification reactions, which often require intricate primer screening and probe optimization.
| Reaction Principle | Sensing strategy | Labelled material or chemical | Sample-to-result time | Reaction volume | Quantitative or not | Limit of detection per reaction | Ref. |
|---|---|---|---|---|---|---|---|
| LAMP | Turbidity | Magnesium pyrophosphate | 70 min | 25 μL | Not | 104 copies | 35 |
| LAMP | Fluorescence | EvaGreen dye | 30 min | 20 μL | Not | 5.5 × 104 copies | 36 |
| LAMP | Colorimetry | Gold-nanoparticle | 30 min | 12.5 μL | Not | 104 copies | 37 |
| LAMP | Electrochemical | Methylene blue | 2 h | 30 μL | Not | 1.2 × 104 copies | 38 |
| LAMP | Electrochemical | AntiDIG-HRP | 2.5 h | 75 μL | Not | 0.1 ng | 39 |
| LAMP | Digital fluorescence | Calcein | 110 min | 10 μL | Yes | 600 copies per μL | 40 |
| LAMP | Digital fluorescence | Fluorescent dye | <45 min | 15 μL | Yes | 1 fM | This work |
We have developed a computer vision-based data analysis system. Through this learning process, our system correctly identifies both positive and negative samples, and the network can also accurately estimate the concentration of targets in unseen images. In the end, the classification accuracy of separating detected positive samples from negative samples is 100%. Our method integrates both deep learning and classical computer vision techniques to achieve optimal outcomes. Classical computer vision methods are computationally efficient and well-suited for sample detection. After sorting the samples based on intensity values, they can be evaluated in a standardized range in later deep learning stage. This approach makes our method a robust and powerful tool for analysing target concentrations, with important applications in areas such as molecular detection and clinical diagnosis. The application of computer vision in biosensors has great prospects, as it enables large-scale, automated, high-throughput, and multi-target detection compared with the traditional analysis methods.48 The prospect of enhancing sensitivity, specificity, and efficiency in biosensing through computer vision signifies a significant leap forward in the realm of diagnostic and analytical methodologies. It stands as a vital tool for the future development of traditional biosensors toward intelligent biosensors.49
HPVs are associated with a variety of malignancies, of which cervical cancer is the most important and prevalent. There are many HPV types; HPV 16/18 are the most carcinogenic. The HPV tests that are currently most widely applied are based on two principles. The first, as used in the HCII assay, the analytical sensitivity of the HCII assay is at the picogram level of HPV DNA. Another method is PCR assays which can reach the sub-picogram level.50 A quantitative assay might be more effective in identifying women at risk of progression, thus improving both negative and positive predictive values.51 Our innovative detection approach represents a major improvement. Not only does it surpass the previously established femtomolar detection limit, but it also offers the added advantage of enabling precise quantification, particularly crucial when dealing with samples containing lower concentrations of HPV DNA. In clinical settings, viral load is typically measured in copies per 103 cells. We also observed that when the clinically relevant HPV16 viral load exceeds 22000 copies per 103 cells, women with prevalent high-grade lesions can be identified.52 A Pap smear slide should have at least 5000 squamous epithelial cells to be considered adequate.53 In our study, we demonstrated that the detection limit for a 2 μL input target was 1 fM (600 copies per μL), offering a dynamic range across five orders of magnitude, making it applicable for future cancer staging diagnostics. This heightened sensitivity and quantification capability promise to revolutionize HPV screening protocols, ultimately leading to more effective early detection and prevention of HPV-related malignancies, particularly cervical cancer. Recent research demonstrates that cervical cells can be simply prepared by suspending them in normal saline oral rinses and then boiling at 100 °C for 5 min, followed by LAMP-based detection for HPV target.54 In the future, we plan to apply our nano digital chip to assess patient samples across different stages of cervical lesions.
Experimental
Materials and reagents
Deoxynucleotide (dNTP) solution mix (10 mM of each), Bst 2.0 WarmStart DNA polymerase (8 U μL−1), MgSO4 (100 mM), 10× Isothermal Amplification Buffer (200 mM Tris–HCl, 500 mM KCl, 100 mM (NH4)2SO4, and 20 mM MgSO4, 1.0% Tween 20 and pH 8.8 at 25 °C) were purchased from New England BioLabs (Ipswich, MA). Primers, FQ probe, and the plasmid pCDNA3.1(+) containing 450 bp HPV 16 and HPV 18 gene sequences were purchased from either Integrated DNA Technologies (Coralville, IA) or synthesized by Genscript. All the sequence information of the primers and target sequences is listed in Table S1.† The primer sequences were designed according to the LAMP Primer Design Tool from the New England BioLabs website. The QuantStudio 3D digital 20K chip kit (Version 2) was purchased from Thermo Fisher Scientific (Waltham, MA).Preparation of the target plasmids
The 450-base pair (bp) target sequences of the HPV-L1 gene from HPV16 and HPV18 were synthesized by GenScript (GenScript USA, Inc., Piscataway, NJ, USA) with HindIII and NotI restriction enzyme sites flanking the sequence. These restriction sites were incorporated to facilitate subsequent cloning steps. The synthesized gene was designed to yield a 463 bp product that was then inserted into the cDNA3.1(+) expression vector to yield a 5820 bp plasmid. The insert sequence was verified post-synthesis through sequencing analysis (QuintaraBio, Frederick, MD, USA).Off-chip isothermal amplification assay
We performed all the HPV 16 target detection with FQ LB (fluorescence quenched) probe-based LAMP assay. Briefly, 25 μl of the reaction for each experiment contained 2.5 μl of 10× Isothermal Amplification Buffer, 1.5 μL of MgSO4 (100 mM), 3.5 μL dNTP mix (10 mM), 2.5 μL of 10× target-specific primer mix (FIP/BIP 16 μM, F3/B3 2 μM, LoopB 4 μM, FQ LB probe 4 μM), 1 μL of Bst 2.0 WarmStart DNA Polymerase (8 U μL−1), and 2 μL of HPV target with different concentrations. The rest of the reaction contained 12 μL of nuclease-free water. The LAMP conditions were optimized for the reaction with a mixture of LB/FQ LB primers at concentrations of 0.2, 0.4, and 0.6 μM, respectively. We also optimized the ratio of LB/FQ LB primers with 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, and 2
2, and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, respectively. The reaction was incubated at 63 °C for 45 min. After incubation, the tubes were placed under a Maestrogen UltraSlim LED blue light illuminator (Pittsburgh, PA) to take images (Canon EOS 6D, Canon, Japan). The endpoint fluorescence was measured by a plate reader (BioTek Cytation 5, Agilent, CA, USA).
4, respectively. The reaction was incubated at 63 °C for 45 min. After incubation, the tubes were placed under a Maestrogen UltraSlim LED blue light illuminator (Pittsburgh, PA) to take images (Canon EOS 6D, Canon, Japan). The endpoint fluorescence was measured by a plate reader (BioTek Cytation 5, Agilent, CA, USA).
On-chip isothermal digital amplification assay
The digital nanofluidic chip is a 10 mm2 high-density reaction plate that has a single array of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 reaction microwells with 15 μL volume for each well. The 15 μL reaction mixture contained 1.5 μL of 10× Isothermal Amplification Buffer, 0.9 μL of MgSO4 (100 mM), 2.1 μL dNTP mix (10 mM), 1.5 μL of 10× target-specific primer mix (FIP/BIP 16 μM, F3/B3 2 μM, LoopB 4 μM, FQ LB probe 4 μM), 0.6 μL of Bst 2.0 WarmStart DNA Polymerase (8000 U mL−1), 1.2 μL of target DNA, and 7.2 μL of nuclease-free water. After assay mixing, the 15 μL reaction mixture was loaded into the chip, and mineral oil was used to seal the loading port. The device was placed on a heat block and incubated at 63 °C for 45 min.
000 reaction microwells with 15 μL volume for each well. The 15 μL reaction mixture contained 1.5 μL of 10× Isothermal Amplification Buffer, 0.9 μL of MgSO4 (100 mM), 2.1 μL dNTP mix (10 mM), 1.5 μL of 10× target-specific primer mix (FIP/BIP 16 μM, F3/B3 2 μM, LoopB 4 μM, FQ LB probe 4 μM), 0.6 μL of Bst 2.0 WarmStart DNA Polymerase (8000 U mL−1), 1.2 μL of target DNA, and 7.2 μL of nuclease-free water. After assay mixing, the 15 μL reaction mixture was loaded into the chip, and mineral oil was used to seal the loading port. The device was placed on a heat block and incubated at 63 °C for 45 min.
Data acquisition and analysis
After incubation, the chip was quantified using a BioTek Cytation 5 Cell Imaging Multimode Reader (Agilent, CA, USA) with a 4× magnification objective. Every image was captured in ∼30 s of laser irradiation, and the irradiation was turned off until the next imaging. Five distinct regions (3.5 × 3.5 mm) without overlapping areas were randomly captured by microscopy to cover approximately 2750 microwells. The number of positive spots and corresponding fluorescence intensity were measured by using the ImageJ software (bit depth: 8 bits; 1992 × 1992 pixels). GraphPad Software Prism 9.5.1 was used to plot real-time fluorescence curves, analyse linear regression, and verify statistical significance between different assay groups.Conclusions
In summary, our study presents a comprehensive approach to nucleic acid detection, utilizing an innovative energy transfer-labeled oligonucleotide probe in tandem with a digital nanofluidic chip and advanced data analysis technique. The incorporation of FQ LB probes offers advantages like enhanced specificity, reduced background noise, and simplified processing, while the closed one-pot procedure minimizes the risk of contamination. Our robust digital amplification system enables rapid and quantitative detection of nucleic acid targets, even at low concentrations, facilitated by microliter digital microwells that enhance sensitivity and enable easy quantitative analysis without calibration curves.Additionally, integration of a computer vision-based data analysis system improves target identification and concentration estimation, promising large-scale, automated, and high-throughput molecular detection applications. Together, these innovations signify significant advancements in sensitivity, specificity, and efficiency, holding promise for the development of intelligent biosensors and transforming molecular detection in clinical diagnosis and beyond.
Data availability
Data for this article are available at UC-Riverside Google Driver system.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by NIH R35GM 142763, NIH 75N93023C00056-0-9999-1, and USDA NIFA 2022-67021-41478.References
- Cervical cancer,https://www.who.int/health-topics/cervical-cancer, (accessed January 28, 2024).
- Human Papillomavirus (HPV), https://www.who.int/teams/health-product-policy-and-standards/standards-and-specifications/vaccine-standardization/human-papillomavirus, (accessed January 24, 2024).
- M. Arbyn, M. Simon, E. Peeters, L. Xu, C. J. L. M. Meijer, J. Berkhof, K. Cuschieri, J. Bonde, A. Ostrbenk Vanlencak, F.-H. Zhao, R. Rezhake, M. Gultekin, J. Dillner, S. de Sanjosé, K. Canfell, P. Hillemanns, M. Almonte, N. Wentzensen and M. Poljak, Clin. Microbiol. Infect., 2021, 27, 1083–1095 CrossRef CAS PubMed.
- J. Wang, K. M. Elfström, C. Lagheden, C. Eklund, K. Sundström, P. Sparén and J. Dillner, PLoS Med., 2023, 20, e1004304 CrossRef PubMed.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed.
- C. Yan, J. Cui, L. Huang, B. Du, L. Chen, G. Xue, S. Li, W. Zhang, L. Zhao, Y. Sun, H. Yao, N. Li, H. Zhao, Y. Feng, S. Liu, Q. Zhang, D. Liu and J. Yuan, Clin. Microbiol. Infect., 2020, 26, 773–779 Search PubMed.
- T. Notomi, Y. Mori, N. Tomita and H. Kanda, J. Microbiol., 2015, 53, 1–5 Search PubMed.
- L. Becherer, N. Borst, M. Bakheit, S. Frischmann, R. Zengerle and F. von Stetten, Anal. Methods, 2020, 12, 717–746 RSC.
- I. M. Lobato and C. K. O'Sullivan, TrAC, Trends Anal. Chem., 2018, 98, 19–35 Search PubMed.
- R. Peng, X. Chen, F. Xu, R. Hailstone, Y. Men and K. Du, Nanoscale Horiz., 2023, 8, 1677–1685 Search PubMed.
- M. Bao, S. Zhang, C. ten Pas, S. J. Dollery, R. V. Bushnell, F. N. U. Yuqing, R. Liu, G. Lu, G. J. Tobin and K. Du, Lab Chip, 2022, 22, 4849–4859 Search PubMed.
- L. Gu, W. Yan, L. Liu, S. Wang, X. Zhang and M. Lyu, Pharmaceuticals, 2018, 11, 35 CrossRef PubMed.
- M. M. Ali, F. Li, Z. Zhang, K. Zhang, D.-K. Kang, J. A. Ankrum, X. C. Le and W. Zhao, Chem. Soc. Rev., 2014, 43, 3324–3341 RSC.
- R. Wang, X. Zhao, X. Chen, X. Qiu, G. Qing, H. Zhang, L. Zhang, X. Hu, Z. He, D. Zhong, Y. Wang and Y. Luo, Anal. Chem., 2020, 92, 2176–2185 Search PubMed.
- B. Deiman, P. van Aarle and P. Sillekens, Mol. Biotechnol., 2002, 20, 163–179 CrossRef CAS PubMed.
- N. Cook, J. Microbiol. Methods, 2003, 53, 165–174 Search PubMed.
- R. Asadi and H. Mollasalehi, Anal. Biochem., 2021, 631, 114260 CrossRef CAS PubMed.
- W. Liu and Y. Zhu, Anal. Chim. Acta, 2020, 1113, 66–84 CrossRef CAS PubMed.
- D. M. Cate, J. A. Adkins, J. Mettakoonpitak and C. S. Henry, Anal. Chem., 2015, 87, 19–41 CrossRef CAS PubMed.
- N. Van Thanh Nguyen, M. Taverna, C. Smadja and T. D. Mai, Chem. Rec., 2021, 21, 149–161 CrossRef CAS PubMed.
- M. Song, L. Feng, P. Huo, M. Liu, C. Huang, F. Yan, Y. Lu and T. Xu, Nat. Nanotechnol., 2023, 18, 71–78 CrossRef CAS PubMed.
- A. Stollmann, J. Garcia-Guirado, J.-S. Hong, P. Rüedi, H. Im, H. Lee, J. Ortega Arroyo and R. Quidant, Nat. Commun., 2024, 15, 4109 CrossRef CAS PubMed.
- M. Parida, S. Sannarangaiah, P. K. Dash, P. V. L. Rao and K. Morita, Rev. Med. Virol., 2008, 18, 407–421 Search PubMed.
- L. Niessen, J. Luo, C. Denschlag and R. F. Vogel, Food Microbiol., 2013, 36, 191–206 CrossRef CAS PubMed.
- M. Soroka, B. Wasowicz and A. Rymaszewska, Cells, 2021, 10, 1931 CrossRef CAS PubMed.
- T. L. Quyen, T. A. Ngo, D. D. Bang, M. Madsen and A. Wolff, Front. Microbiol., 2019, 10, 2234 Search PubMed.
- K. Nath, J. W. Sarosy, J. Hahn and C. J. Di Como, J. Biochem. Biophys. Methods, 2000, 42, 15–29 Search PubMed.
- H. Bai, Y. Liu, L. Gao, T. Wang, X. Zhang, J. Hu, L. Ding, Y. Zhang, Q. Wang, L. Wang, J. Li, Z. Zhang, Y. Wang, C. Shen, B. Ying, X. Niu and W. Hu, Biosens. Bioelectron., 2024, 248, 115968 CrossRef CAS PubMed.
- Y. Cao, C. Ye, C. Zhang, G. Zhang, H. Hu, Z. Zhang, H. Fang, J. Zheng and H. Liu, Food Control, 2022, 134, 108694 Search PubMed.
- W. Yin, J. Zhuang, J. Li, L. Xia, K. Hu, J. Yin and Y. Mu, Small, 2023, 19, 2303398 CrossRef CAS PubMed.
- B. Coelho, B. Veigas, E. Fortunato, R. Martins, H. Águas, R. Igreja and P. V. Baptista, Sensors, 2017, 17, 1495 CrossRef PubMed.
- Z. Yu, L. Xu, W. Lyu and F. Shen, Lab Chip, 2022, 22, 2954–2961 RSC.
- F. X. Liu, J. Q. Cui, H. Park, K. W. Chan, T. Leung, B. Z. Tang and S. Yao, Anal. Chem., 2022, 94, 5883–5892 Search PubMed.
- J. M. Wong Tzeling, E. A. R. Engku Nur Syafirah, A. A. Irekeola, W. Yusof, N. N. Aminuddin Baki, A. Zueter, A. Harun and Y. Y. Chan, Anal. Chim. Acta, 2021, 1171, 338682 CrossRef CAS PubMed.
- C. Saetiew, T. Limpaiboon, P. Jearanaikoon, S. Daduang, C. Pientong, A. Kerdsin and J. Daduang, J. Virol. Methods, 2011, 178, 22–30 CrossRef CAS PubMed.
- F. Sun, A. Ganguli, J. Nguyen, R. Brisbin, K. Shanmugam, D. L. Hirschberg, M. B. Wheeler, R. Bashir, D. M. Nash and B. T. Cunningham, Lab Chip, 2020, 20, 1621–1627 RSC.
- N. M. Rodriguez, W. S. Wong, L. Liu, R. Dewar and C. M. Klapperich, Lab Chip, 2016, 16, 753–763 RSC.
- M. Zamani, J. M. Robson, A. Fan, M. S. Bono Jr., A. L. Furst and C. M. Klapperich, ACS Cent. Sci., 2021, 7, 963–972 CrossRef CAS PubMed.
- M. Bartosik, L. Jirakova, M. Anton, B. Vojtesek and R. Hrstka, Anal. Chim. Acta, 2018, 1042, 37–43 CrossRef CAS PubMed.
- T. D. Rane, L. Chen, H. C. Zec and T.-H. Wang, Lab Chip, 2015, 15, 776–782 RSC.
- D. Das, C.-W. Lin and H.-S. Chuang, Biosensors, 2022, 12, 1068 CrossRef CAS PubMed.
- X. Ding, K. Yin, Z. Li, M. M. Sfeir and C. Liu, Biosens. Bioelectron., 2021, 184, 113218 CrossRef CAS PubMed.
- A. Bektaş, Food Anal. Methods, 2018, 11, 686–692 CrossRef.
- K. Karthik, R. Rathore, P. Thomas, T. R. Arun, K. N. Viswas, K. Dhama and R. K. Agarwal, MethodsX, 2014, 1, 137–143 CrossRef CAS PubMed.
- W. E. Huang, B. Lim, C.-C. Hsu, D. Xiong, W. Wu, Y. Yu, H. Jia, Y. Wang, Y. Zeng, M. Ji, H. Chang, X. Zhang, H. Wang and Z. Cui, Microb. Biotechnol., 2020, 13, 950–961 CrossRef CAS PubMed.
- Z. Fan, X. Feng, W. Zhang, N. Li, X. Zhang and J.-M. Lin, Talanta, 2020, 217, 121015 CrossRef CAS PubMed.
- S.-Y. Teh, R. Lin, L.-H. Hung and A. P. Lee, Lab Chip, 2008, 8, 198–220 RSC.
- R. Antiochia, Biosens. Bioelectron., 2021, 173, 112777 CrossRef CAS PubMed.
- L. Liu and K. Du, Biomicrofluidics, 2024, 18, 011301 CrossRef CAS PubMed.
- P. J. Snijders, A. J. van den Brule and C. J. Meijer, J. Pathol., 2003, 201, 1–6 Search PubMed.
- A. Molijn, B. Kleter, W. Quint and L.-J. van Doorn, J. Clin. Virol., 2005, 32, 43–51 Search PubMed.
- M. Saunier, S. Monnier-Benoit, F. Mauny, V. Dalstein, J. Briolat, D. Riethmuller, B. Kantelip, E. Schwarz, C. Mougin and J.-L. Prétet, J. Clin. Microbiol., 2008, 46, 3678–3685 CrossRef PubMed.
- H. C. Kitchener, M. Gittins, M. Desai, J. H. Smith, G. Cook, C. Roberts, L. Turnbull, H. C. Kitchener, M. Gittins, M. Desai, J. H. Smith, G. Cook, C. Roberts and L. Turnbull, A study of cellular counting to determine minimum thresholds for adequacy for liquid-based cervical cytology using a survey and counting protocol, NIHR Journals Library, 2015.
- D. T. Vo and M. D. Story, Mol. Cell. Probes, 2021, 59, 101760 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr03142c |
| This journal is © The Royal Society of Chemistry 2025 |