Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Supramolecular delivery of dinuclear ruthenium and osmium MCU inhibitors†
Nicholas P. Bigham a,
Robyn J. Novorolskybc,
Keana R. Davis
a,
Robyn J. Novorolskybc,
Keana R. Davis e,
Haipei Zou
e,
Haipei Zou af,
Samantha N. MacMillan
af,
Samantha N. MacMillan a,
Michael J. Stevenson
a,
Michael J. Stevenson e,
George S. Robertson
e,
George S. Robertson bcd and
Justin J. Wilson
bcd and
Justin J. Wilson *af
*af
aDepartment of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853, USA. E-mail: jjw275@cornell.edu
bDepartment of Pharmacology, Faculty of Medicine, Dalhousie University, 6th Floor Sir Charles Tupper Medical Building, Halifax, B3H 4R2, Canada
cBrain Repair Centre, Faculty of Medicine, Dalhousie University, Life Sciences Research Institute, Halifax, NS B3H 4R2, Canada
dDepartment of Psychiatry, Faculty of Medicine, Dalhousie University, Halifax, NS B3H 2E2, Canada
eDepartment of Chemistry, University of San Francisco, San Francisco, CA 94117, USA
fDepartment of Chemistry & Biochemistry, University of California Santa Barbara, Santa Barbara, CA 93106, USA
First published on 11th July 2024
Abstract
The transmembrane protein known as the mitochondrial calcium uniporter (MCU) mediates the influx of calcium ions (Ca2+) into the mitochondrial matrix. An overload of mitochondrial Ca2+ (mCa2+) is directly linked to damaging effects in pathological conditions. Therefore, inhibitors of the MCU are important chemical biology tools and therapeutic agents. Here, two new analogues of previously reported Ru- and Os-based MCU inhibitors Ru265 and Os245, of the general formula [(C10H15CO2)M(NH3)4(μ-N)M(NH3)4(O2CC10H15)](CF3SO3)3, where M = Ru (1) or Os (2), are reported. These analogues bear adamantane functional groups, which were installed to act as guests for the host molecule cucurbit-[7]-uril (CB[7]). These complexes were characterized and analyzed for their efficiency as guests for CB[7]. As shown through a variety of spectroscopic techniques, each adamantane ligand is encapsulated into one CB[7], affording a supramolecular complex of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry. The biological effects of these compounds in the presence and absence of two equiv. CB[7] were assessed. Both complexes 1 and 2 exhibit enhanced cellular uptake compared to the parent compounds Ru265 and Os245, and their uptake is increased further in the presence of CB[7]. Compared to Ru265 and Os245, 1 and 2 are less potent as mCa2+ uptake inhibitors in permeabilized cell models. However, in intact cell systems, 1 and 2 inhibit the MCU at concentrations as low as 1 μM, marking an advantage over Ru265 and Os245 which require an order of magnitude higher doses for similar biological effects. The presence of CB[7] did not affect the inhibitory properties of 1 and 2. Experiments in primary cortical neurons showed that 1 and 2 can elicit protective effects against oxygen-glucose deprivation at lower doses than those required for Ru265 or Os245. At low concentrations, the protective effects of 1 were modulated by CB[7], suggesting that supramolecular complex formation can play a role in these biological conditions. The in vivo biocompatibility of 1 was investigated in mice. The intraperitoneal administration of these compounds and their CB[7] complexes led to time-dependent induction of seizures with no protective effects elicited by CB[7]. This work demonstrates the potential for supramolecular interactions in the development of MCU inhibitors.
2 stoichiometry. The biological effects of these compounds in the presence and absence of two equiv. CB[7] were assessed. Both complexes 1 and 2 exhibit enhanced cellular uptake compared to the parent compounds Ru265 and Os245, and their uptake is increased further in the presence of CB[7]. Compared to Ru265 and Os245, 1 and 2 are less potent as mCa2+ uptake inhibitors in permeabilized cell models. However, in intact cell systems, 1 and 2 inhibit the MCU at concentrations as low as 1 μM, marking an advantage over Ru265 and Os245 which require an order of magnitude higher doses for similar biological effects. The presence of CB[7] did not affect the inhibitory properties of 1 and 2. Experiments in primary cortical neurons showed that 1 and 2 can elicit protective effects against oxygen-glucose deprivation at lower doses than those required for Ru265 or Os245. At low concentrations, the protective effects of 1 were modulated by CB[7], suggesting that supramolecular complex formation can play a role in these biological conditions. The in vivo biocompatibility of 1 was investigated in mice. The intraperitoneal administration of these compounds and their CB[7] complexes led to time-dependent induction of seizures with no protective effects elicited by CB[7]. This work demonstrates the potential for supramolecular interactions in the development of MCU inhibitors.
10th anniversary statementPrior to joining Inorganic Chemistry Frontiers as an associate editor in 2023, my literature searches within bioinorganic medicinal chemistry would often lead me to this new journal. I was impressed by the high-quality of the manuscripts within this field, as well as their increasing number and visibility. This new journal was arising to a top-tier level. I was a consistent reviewer for this journal, and I saw many interesting and high-impact studies. Now, as an associate editor, I have been pleased to see the submission of such manuscripts first-hand. In all of my roles at Inorganic Chemistry Frontiers as a reader, reviewer, author, and editor, I have been impressed and excited about the science within this journal. In particular, I think it has arisen as a fantastic forum to disseminate studies in bioinorganic chemistry to the broader community. |
Introduction
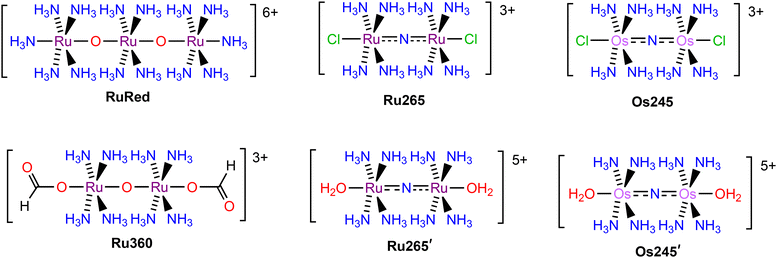
The biochemistry of calcium ions (Ca2+) has been widely studied due to its importance in cellular function and signaling.1–3 Notably, the mitochondria are critical for regulating intracellular Ca2+ levels, and mitochondrial Ca2+ (mCa2+) uptake is an important process for maintaining cellular homeostasis.4–6 The dysregulation of mCa2+ levels, however, can lead to damaging effects and pathological conditions. Most prominently, excessive levels of Ca2+ in the mitochondria lead to mCa2+ overload, which triggers cell death.7,8 The phenomenon of mCa2+ overload and its damaging effects have been implicated in a number of pathological conditions such as heart disease,9–11 cancer,12,13 cystic fibrosis,14,15 neurodegenerative disorders,16,17 and ischemia-reperfusion injury.18–20 Given the physiological importance of mCa2+ levels, substantial research has been committed to understand how the mitochondria regulate Ca2+ uptake. These efforts have revealed the mitochondrial calcium uniporter (MCU) complex to be responsible for mediating Ca2+ uptake into this organelle.6,21–24 This tetrameric, transmembrane protein consists of the pore-forming MCU subunit25–28 and three regulatory subunits – EMRE,29 MICU1,30 and MICU2.31 Both MICU1 and MICU2 contain EF-hand domains to sense extramitochondrial Ca2+ levels and interact with the MCU subunit at a highly conserved solvent-exposed DXXE motif near the pore opening.25,26 Upon exposure to high cytosolic Ca2+ concentrations, these EF-hand domains dissociate from the MCU subunit,31–34 allowing for Ca2+ to traverse the pore and enter the mitochondrial matrix.35,36 Therefore, targeting the DXXE motif with chemical inhibitors could be a viable approach to prevent mCa2+ overload as a potential therapeutic strategy.A number of organic37–42 and inorganic43–50 inhibitors of the MCU have been reported to date. The most well-known and widely used MCU inhibitors are the polynuclear Ru complexes, ruthenium red (RuRed) and Ru360 (Chart 1). Subsequent work on this compound class led to the discovery of the dinuclear nitrido-bridged Ru compound named Ru265 (Chart 1).47 In contrast to the earlier RuRed and Ru360, this complex, which exhibits nanomolar MCU-inhibitory activity, is cell permeable and stable towards biological reduction,51,52 properties that make it a useful tool for intact cell systems. Given the significant promise of Ru265 as an MCU inhibitor, our group has developed analogues in order to understand structure–activity relationships for this compound class and to identify those with improved properties for biological applications. A key observation is that the axial chloride ligands of Ru265 dissociate within a timescale of minutes under physiological conditions to give the aquated complex Ru265′ (Chart 1), which itself is a potent MCU inhibitor.51 Notably, replacing these chlorides with carboxylates substantially slows the aquation process. Careful modification of the carboxylates has conferred these compounds with prodrug capabilities,53 higher cell uptake,54 and fluorogenic properties.55 In addition, we have reported and studied the MCU-inhibitory properties of the Os-analogue of Ru265, named Os245 (Chart 1).50 The aquation of the axial chloride ligands of this complex to form Os245′ (Chart 1) occurs at a rate nearly 100× slower than that of Ru265. The slower axial chloride substitution rate of Os245 is consistent with the known greater kinetic inertness of 5d transition metals compared to 4d transition metals.56,57 In conjunction with this slow substitution rate, the use of alternative axial ligands, like carboxylates, on the Os245 scaffold could result in new analogues with very long biological activation times.
Although we have successfully shown in earlier studies that modification of the axial carboxylates of Ru265 analogues could sufficiently alter their properties, we sought alternative approaches to finetune their behavior in biological solutions. In this regard, the use of supramolecular complexes and interactions has recently been demonstrated to have promising biological applications. For example, biologically active guest molecules can exhibit substantially different solubilities, cell permeabilities, and pharmacokinetic properties once encapsulated in a supramolecular host.58–60 The most widely used host molecules for these applications are cyclodextrins and cucurbiturils. In particular, cucurbit-[n]-uril (CB[n]) has been shown to be a highly effective host for a number of different molecular architectures and has had substantial impacts on their biological properties.58–63 A significant example demonstrated that the encapsulation of pentylenetetrazol, a GABA-A receptor antagonist, within CB[7] was able to attenuate its dose-limiting side of effect of seizure induction in vivo.64 Given that the in vivo applications of both Ru265 and Os245 are also dose-limited by seizure induction,50,52 we hypothesized that the encapsulation of these MCU inhibitors within CB[7] would be a promising approach to modify and potentially improve their biological properties. To make these compounds amenable for CB[7] encapsulation, we appended them with axial carboxylate adamantane ligands. Adamantane, a well-known lipophilic moiety in drug development,65 is a high affinity guest molecule for CB[7], making the resulting host–guest complex valuable for biological applications.66–78 In prior studies, the covalent attachment of this group to a cisplatin derivative enabled its encapsulation in CB[7], validating this approach for metal-based compounds.79 Thus, we sought to perform a similar functionalization of Ru265 and Os245 for their encapsulation into CB[7]. In this work, we report the synthesis and characterization of adamantane-functionalized derivatives of Ru265 and Os245 (1 and 2, Chart 2), an examination of their supramolecular complex formation with CB[7], and in vitro and in vivo investigations of the biological ramifications of this complex formation. Collectively, these studies reveal that the addition of CB[7] to these complexes increase their cellular accumulation without sacrificing their MCU-inhibitory activities.
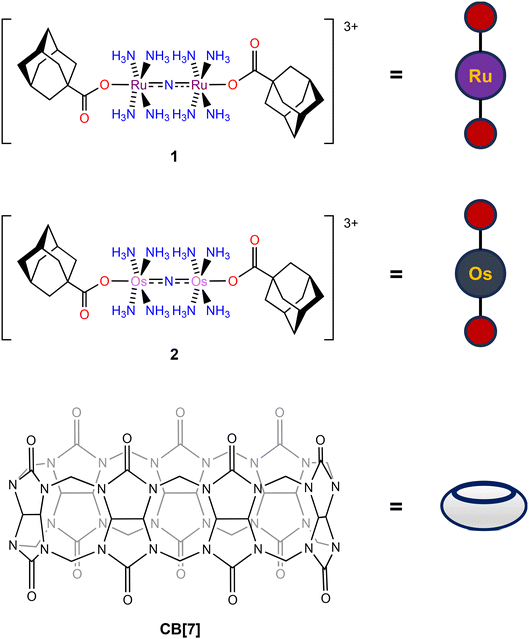 | ||
| Chart 2 Structures of 1, 2, and CB[7] with their pictorial representations used within this manuscript. | ||
Results and discussion
Synthesis and characterization
Compounds 1 and 2 were designed to be guest molecules for CB[7] by incorporation of 1-adamantanecarboxylate (OAd−) ligands on the axial positions. The adamantane group provides a high-affinity guest for CB[7], and the carboxylate has been established by us to be a suitable axial ligand for this compound class. The syntheses of these complexes commenced from Ru265′![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 51,53 and Os245′.50 The ligand 1-adamantanecarboxylic acid was converted to the sodium 1-adamantanecarboxylate (NaOAd) salt via the reaction with one equiv. sodium hydroxide in ethanol. To access 1, a procedure that we had previously developed for related carboxylate-functionalized analogues was applied.53–55 Specifically, Ru265′ was treated with two equiv. of NaOAd in MeOH, while heating the mixture at 60 °C for 16 h. Subsequent crystallization of the crude product from this reaction via vapor diffusion of hexane into THF afforded analytically pure 1. Compound 2 was synthesized under similar conditions by treating Os245′ with two equiv. NaOAd in MeOH at 60 °C. Given the greater inertness of Os, however, the 48 h reaction time required for formation of 2 was substantially longer than that needed for 1. Upon purification of the crude product by low-temperature evaporation of THF, analytically pure 2 was obtained as a yellow crystalline solid. Importantly, 2 is the first example of an Os245 analogue bearing axial carboxylate ligands, and this synthetic procedure should therefore be broadly applicable for making new derivatives of this MCU inhibitor. CB[7] used within these studies was synthesized according to previous literature procedures.80,81
51,53 and Os245′.50 The ligand 1-adamantanecarboxylic acid was converted to the sodium 1-adamantanecarboxylate (NaOAd) salt via the reaction with one equiv. sodium hydroxide in ethanol. To access 1, a procedure that we had previously developed for related carboxylate-functionalized analogues was applied.53–55 Specifically, Ru265′ was treated with two equiv. of NaOAd in MeOH, while heating the mixture at 60 °C for 16 h. Subsequent crystallization of the crude product from this reaction via vapor diffusion of hexane into THF afforded analytically pure 1. Compound 2 was synthesized under similar conditions by treating Os245′ with two equiv. NaOAd in MeOH at 60 °C. Given the greater inertness of Os, however, the 48 h reaction time required for formation of 2 was substantially longer than that needed for 1. Upon purification of the crude product by low-temperature evaporation of THF, analytically pure 2 was obtained as a yellow crystalline solid. Importantly, 2 is the first example of an Os245 analogue bearing axial carboxylate ligands, and this synthetic procedure should therefore be broadly applicable for making new derivatives of this MCU inhibitor. CB[7] used within these studies was synthesized according to previous literature procedures.80,81
Both compounds, as well as NaOAd and CB[7], were characterized via IR and NMR (Fig. S1–S14†) spectroscopies, elemental analysis, and X-ray crystallography (Fig. 1 and Tables S1–S2†). The 1H NMR spectra of 1 and 2 in DMSO-d6 showed diagnostic resonances arising from the OAd− ligand. In particular, the hydrogen atoms of this ligand appear at 1.92, 1.71, and 1.63 ppm for both 1 and 2. Consistent with coordination to the metal centers, these chemical shifts are slightly downfield of the resonances for free NaOAd salt, which appear at 1.87, 1.70, and 1.60 ppm. In addition, the 1H NMR spectra also reveal broad singlets at 3.90 and 4.74 ppm for 1 and 2, which can be assigned to the protons of the coordinated NH3 ligands. For 1, the resonances are slightly shifted upfield by 0.2 ppm compared to the parent compound Ru265, but are comparable to other carboxylate-capped analogues whose resonances span the range of 3.90–3.95 ppm.53–55 For 2, the resonance of these NH3 hydrogen protons is similarly shifted 0.2 ppm upfield, comparable to that of Os245′. Because 2 is the first example of carboxylate-capped Os analogue of this compound class, this resonance can provide a diagnostic means of characterization. Within the IR spectra of 1 and 2, the asymmetric M–N–M stretching modes are tentatively assigned to be 1049 and 1116 cm−1, respectively. These energies are comparable to the stretching modes found in the parent Ru265 and Os245 complexes at 1050 and 1108 cm−1, indicating that axial ligand substitution has little effect on the metal-nitrido bond. The higher energy of the M–N–M stretching mode of 2 compared to 1 suggests stronger π-bonding in the Os–N–Os moiety than that of the Ru–N–Ru.
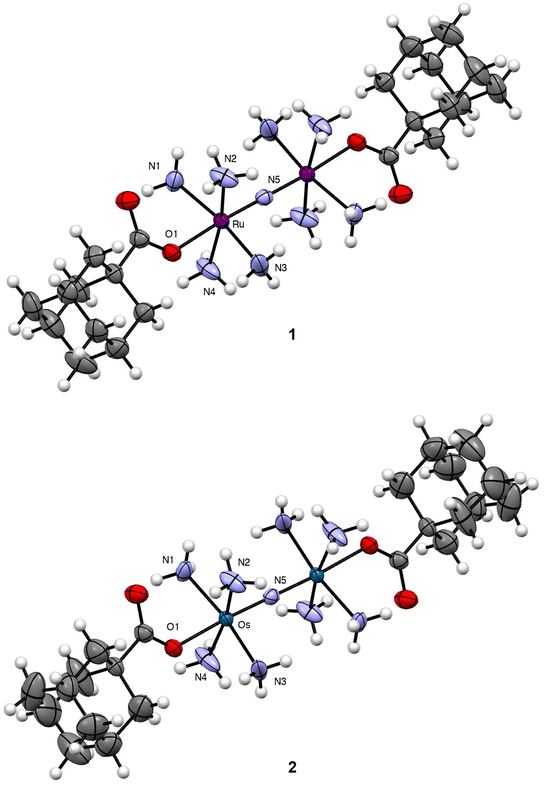 | ||
| Fig. 1 Crystal structures of 1 and 2. Outer-sphere counterions and solvent molecules are omitted for clarity. Thermal ellipsoids are shown at the 50% probability level. Crystallographic information for both complexes is given in Table S1.† Key interatomic distances and angles are given in Table 1. All interatomic distances and angles are given in Table S2.† | ||
Single crystals of both complexes were analyzed by X-ray diffraction. The crystal structures of their Ru- and Os-containing cations are shown in Fig. 1 with relevant interatomic distances and angles shown in Table 1 and Table S2.† Both complexes form isomorphous crystals, attaining the same space group and nearly identical unit cell parameters. Consequently, their structural features are largely similar. The bridging N3− of both complexes resides on a crystallographic inversion center. As such, the M–N–M angle is perfectly linear for 1 and 2, and the four NH3 ligands on each metal center are arranged in an eclipsed conformation. The OAd− ligands occupy the position trans to the N3− and are supported by additional intramolecular hydrogen bonds with the equatorial NH3 ligands. Within 1, the Ru–N and Ru–O interatomic distances are similar to those for previously reported carboxylate-functionalized derivatives of Ru265.53–55 As noted previously, the strong trans influence of the N3− bridging ligand leads to elongation of the axial Ru–O distances compared to mononuclear analogues. The interatomic distances found within 2 are nearly the same as those of 1, a property that reflects the similar ionic radii of these metals.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a
a
Encapsulation into CB[7]
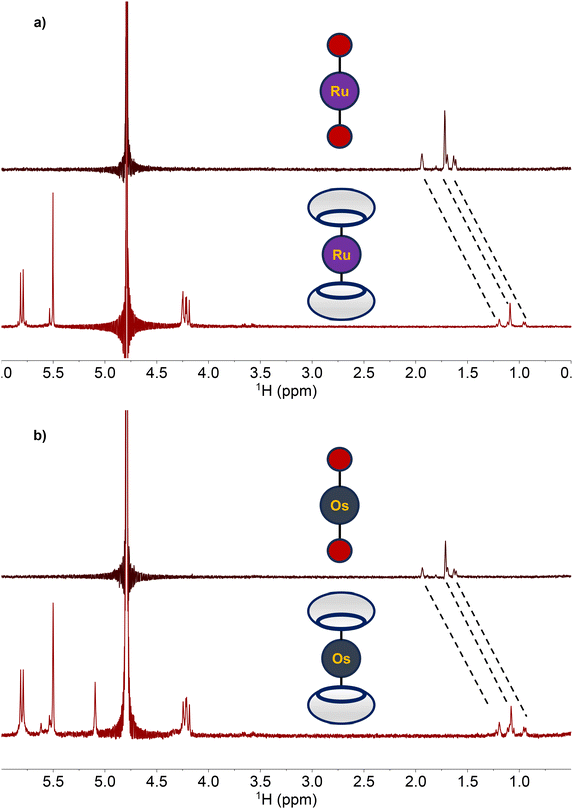
With the adamantane-functionalized 1 and 2 fully characterized, their capacity to act as guests for the supramolecular host molecule CB[7] was investigated. Upon the addition of two equiv. CB[7] to either 1 and 2 in D2O, substantial changes in the 1H NMR resonances of these complexes were observed. Specifically, resonances of the adamantane functional group in both 1 and 2 shifted upfield by nearly 0.6 ppm (Fig. 2). This upfield shift is a consequence of the encapsulation of adamantane within these hosts, as observed previously.79,82,83Encapsulation of 1 and 2 within CB[7] was further confirmed through 2D diffusion-ordered NMR experiments (Fig. S15 and S16†). Using this method, we measured the diffusion constants (D) of the free hosts and guests, as well as the putative host–guest complex, by implementing a pulse-gradient spin echo experiment (PGSE) in conjunction with the mathematical relationship outlined in eqn (1) and (2),84
| ln(I/I0) = −DG2 | (1) |
| G2 = γ2δ2g2(Δ − (δ/3)) | (2) |
| Compound | D (× 10−6 cm2 s−1) |
|---|---|
| a Determined at 25 °C in D2O at 1 mM for 1 and 2 and 2 mM for CB[7]. | |
| CB[7] | 3.14 ± 0.09 |
| 1 | 3.40 ± 0.14 |
| 1 + CB[7] | 2.10 ± 0.04 |
| 2 | 3.11 ± 0.18 |
| 2 + CB[7] | 2.18 ± 0.05 |
Binding constant determination
To quantify the strengths of the interactions of 1 and 2 with CB[7], isothermal titration calorimetry (ITC) was used to measure the binding constants (Ka) and thermodynamic properties of each complex with CB[7] (Fig. S18–S20†). ITC has been widely used in biophysical chemistry to study the interactions between biomolecules, but is also broadly applicable to other systems like supramolecular host–guest complexes.86 The binding analysis of 1 and 2 with CB[7] by this method was carried out using 50 mM MOPS as a buffer, which has an affinity for CB[7].87 Accordingly, the data analysis and fitting to acquire thermodynamic parameters were adjusted to account for this competitive equilibrium.88 The ITC thermograms (Fig. S18–S20†) reveal steep binding isotherms that are characteristic of large Ka values. Fitting of these thermograms, accounting for competitive MOPS binding, was used to obtain the Ka values and binding stoichiometries (n), shown in Table 3. The free ligand NaOAd interacts with CB[7] via the formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex and a binding constant of 8 × 106 M−1. Consistent with the presence of two adamantyl guest moieties on both 1 and 2, their binding stoichiometries were determined to be 2
1 complex and a binding constant of 8 × 106 M−1. Consistent with the presence of two adamantyl guest moieties on both 1 and 2, their binding stoichiometries were determined to be 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (n ≈ 0.5), a property that was also confirmed by NMR spectroscopy (Fig. S21–S23†). Additionally, the encapsulation of these complexes were not affected by the presence of biologically relevant metal ions Mg2+, Ca2+, and Zn2+, as evidenced by NMR spectroscopy (Fig. S24†). We also note that some biologically relevant organic molecules, such as cholesterol, are guests of CB[7]. The large Ka values of our complexes, conferred by the adamantane functional group, should limit their displacement under biological conditions. Notably, the Ka values for the interaction of 1 and 2 with CB[7] were similar and roughly two orders of magnitude larger than that of free NaOAd. In addition, only a single Ka value was measured for both complexes, indicating that the addition of the second CB[7] is most likely not cooperative and occurs concurrently with the first. The Ka values for CB[7] interacting with 1 and 2 are two orders of magnitude smaller compared to the cisplatin-aminoadamantane complex mentioned previously, which has a Ka with CB[7] of 7.26 × 1010 M−1.79 Aminoadamantane itself has a substantially higher affinity for CB[7] (Ka = 1.7 × 1014 M−1)89 than NaOAd, most likely due to the additional hydrogen-bonding NH2 groups that are available on this compound to stabilize the host–guest complex. In any case, the 108 M−1 of these Ka values are still sufficiently large for biological applications.
1 (n ≈ 0.5), a property that was also confirmed by NMR spectroscopy (Fig. S21–S23†). Additionally, the encapsulation of these complexes were not affected by the presence of biologically relevant metal ions Mg2+, Ca2+, and Zn2+, as evidenced by NMR spectroscopy (Fig. S24†). We also note that some biologically relevant organic molecules, such as cholesterol, are guests of CB[7]. The large Ka values of our complexes, conferred by the adamantane functional group, should limit their displacement under biological conditions. Notably, the Ka values for the interaction of 1 and 2 with CB[7] were similar and roughly two orders of magnitude larger than that of free NaOAd. In addition, only a single Ka value was measured for both complexes, indicating that the addition of the second CB[7] is most likely not cooperative and occurs concurrently with the first. The Ka values for CB[7] interacting with 1 and 2 are two orders of magnitude smaller compared to the cisplatin-aminoadamantane complex mentioned previously, which has a Ka with CB[7] of 7.26 × 1010 M−1.79 Aminoadamantane itself has a substantially higher affinity for CB[7] (Ka = 1.7 × 1014 M−1)89 than NaOAd, most likely due to the additional hydrogen-bonding NH2 groups that are available on this compound to stabilize the host–guest complex. In any case, the 108 M−1 of these Ka values are still sufficiently large for biological applications.
| Compound | ΔHITC (kJ mol−1) | n | Ka (M−1) |
|---|---|---|---|
| a Values are obtained from data fits (Fig. S18–S20†) and a post hoc analysis for Ka accounts for the competitive binding of the MOPS buffer. | |||
| NaOAd | −6.3 ± 0.4 | 1.01 ± 0.01 | (8.7 ± 0.8) × 106 |
| 1 | −31 ± 2 | 0.6 ± 0.1 | (4 ± 2) × 108 |
| 2 | −32 ± 1 | 0.7 ± 0.1 | (6 ± 2) × 108 |
Aquation kinetics
Carboxylate-functionalized analogues of Ru265 undergo aquation with half-lives ranging from 2–10 h. Accordingly, the aquation kinetics of 1 and 2 were determined by UV-vis spectroscopy in pH 7.4 MOPS buffer at 37 °C (Fig. S25 and S26†). The aquation half-lives of 1 and 2 were determined by monitoring the decrease in the peaks at 258 nm and 244 nm, respectively, over time. As noted previously, UV-vis spectroscopy cannot suitably resolve the individual aquation steps of both the first and second axial ligands for this compounds class.54,55,90 However, an exponential fit of these data, corresponding to a simplified first-order process, is able to accurately capture the pseudo-first order rate constants (kobs) for the aquation of the first carboxylate, which we have previously found to be substantially smaller than that of the second.53 An exponential fit of the UV-vis spectra of 1 and 2 revealed aquation half-lives (t1/2) of 7.5 and 29.9 h (Table 4). The 7.5 h half-life of 1 is within the range previously observed for other carboxylate-capped Ru compounds of this class.53–55 The slower aquation kinetics for 2 are expected based on the greater inertness of 5d transition metals. The extended half-life of 2 suggests that it may stay intact longer in biological settings compared to 1. To assess the impact of encapsulation within CB[7], the aquation kinetics of 1 and 2 were measured in the presence of two equiv. of this host molecule. As shown in Table 4, encapsulation of 1 and 2 had no significant effect on the rates of aquation of these compounds. This result is consistent with a comparatively rapid on–off preequilibrium of encapsulation, followed by the slow rate-determining aquation of the first carboxylate ligands.| Compound | kobs (× 10−5 s−1) | t1/2 (h) |
|---|---|---|
| a Data were collected using UV-vis spectroscopy in 100 mM MOPS-buffered solution (pH 7.4) at 37 °C. All data are presented as the average of three independent trials ± SD. | ||
| 1 | 2.77 ± 0.33 | 7.96 ± 0.85 |
| 1 + CB[7] | 2.48 ± 0.31 | 7.85 ± 0.98 |
| 2 | 0.645 ± 0.33 | 29.91 ± 1.56 |
| 2 + CB[7] | 0.676 ± 0.41 | 28.48 ± 1.69 |
Cytotoxicity and cellular uptake
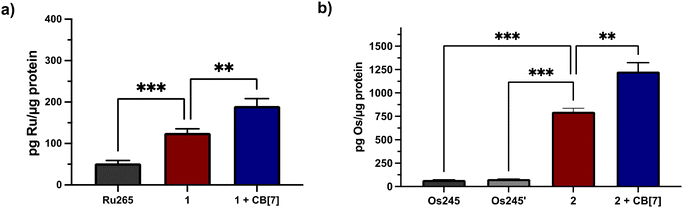
Having characterized 1 and 2 and studied their supramolecular encapsulation properties, their in vitro biological activities were probed. To effectively use these compounds as tools or therapeutic agents related to mCa2+ overload, they need to ideally be non-cytotoxic in human cells. Thus, their cytotoxicity was assessed in HeLa cells using the colorimetric thiazolyl blue tetrazolium bromide (MTT) assay (Fig. S27†). After a 72 h incubation period, 2 was effectively non-toxic even at its highest administered concentration of 100 μM. By contrast, 1 was moderately cytotoxic under these conditions with a 50% growth inhibitory concentration (IC50) of 21.9 ± 2.0 μM. The addition of two equiv. of CB[7] did not change the lack of cytotoxicity of 2; however, two equiv. of CB[7] did attenuate the cytotoxicity of 1. As a further control experiment, the cytotoxicity of the ligand, NaOAd in the presence and absence of CB[7] was determined, revealing it to be non-toxic under any of these conditions. The moderate cytotoxicity of 1 is therefore enigmatic, given that both Ru265′ and NaOAd are non-toxic, but is unlikely to have a significant impact for downstream applications of this compound as a nM inhibitor of mCa2+ uptake. In addition, the JC-1 assay was employed to verify that compounds 1 and 2 do not negatively affect the mitochondrial membrane potential of HeLa cells treated with 50 μM for 24 h. The presence of two equiv. of CB[7] with these complexes also did not negatively affect mitochondrial function (Fig. S28 and S29†).Given the need for cell-permeable MCU inhibitors, the cell uptake in HeLa cells of complexes 1 and 2 in the presence and absence of two equiv. CB[7] was investigated. The HeLa cells were treated with 50 μM of either 1 and 2 with or without 100 μM CB[7] for 3 h. The cells were then washed, collected, and lysed, and the protein content was measured via the bicinchoninic acid (BCA) assay. For 1, the Ru content was determined via graphite furnace atomic absorption spectroscopy (GFAAS), whereas for 2, the Os content was determined via inductively-coupled plasma mass spectrometry (ICP-MS). The cell uptake of these complexes is reported as the total metal concentrations measured normalized to the protein content of the cells, and these data are shown in Fig. 3. Both 1 and 2 are taken up at higher levels under these conditions compared to Ru265 and Os245, consistent with the presence of the lipophilic adamantane group. In the presence of two equiv. of CB[7], the uptakes of 1 and 2 increase further, suggesting that the CB[7] facilitates their transport through the cell membrane. Additionally, in both the presence and absence of CB[7], 1 is significantly accumulated within the mitochondria of HeLa cells dosed with the complex (Fig. S30†). As the cellular uptake properties of 1 and 2 are similar, we did not measure the subcellular localization of 2, but expect it to also accumulate within the mitochondria. These similar uptake properties bring into question the mechanism of cellular uptake of these complexes with and without CB[7]. We have previously reported that Ru265′ and Os245′ are readily taken up into cells through the organic cation transporter 3 (OCT3).50,51 Our studies on other carboxylate derivatives have suggested an alternative mechanism.53 To probe mechanistic interactions for these complexes, we co-incubated 1 ± two equiv. CB[7] with a variety of uptake mechanism inhibitors – specifically an inhibitor of OCT3, an inhibitor of an energy-dependent uptake pathway, and CuSO4 to saturate the CTR1 transporter (Fig. S31†). In all scenarios, the cellular accumulation was not significantly reduced, indicating that further mechanistic studies need to be probed to determine the cellular uptake pathway of these complexes. Given the favorable properties associated with the CB[7] supramolecular complexes, this host molecule was assessed in more detail in subsequent investigations described below.
mCa2+ uptake inhibition
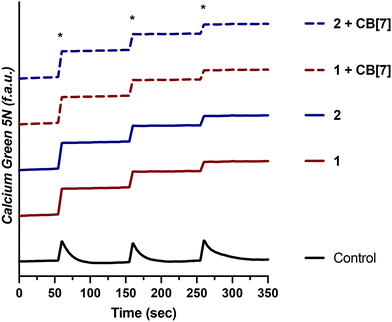
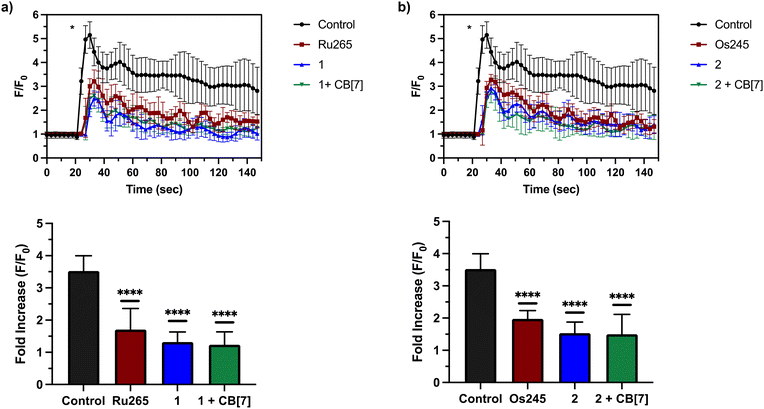
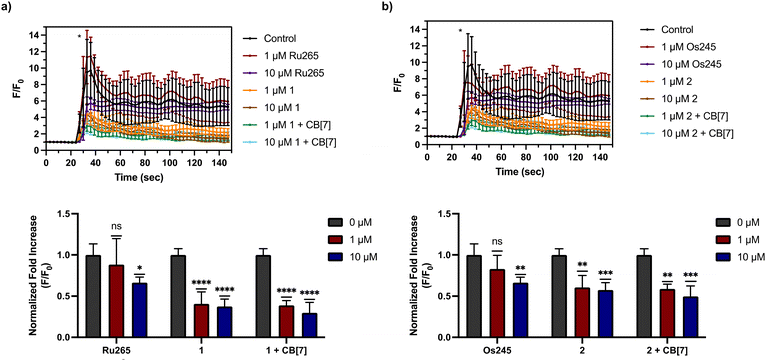
Having demonstrated that 1 and 2, as well as their supramolecular complexes with CB[7], exhibit good cell uptake and minimal cytotoxicity, we next sought to examine the MCU-inhibitory activity of these complexes in both permeabilized and intact cells. In digitonin-permeabilized HEK293T cells, the parent complexes Ru265, Os245, and Os245′ all show potent, nanomolar mCa2+ uptake inhibition, and within intact cells they can abrogate mCa2+ uptake completely when administered at a concentration of 50 μM.47,50 To assess the mCa2+ uptake of the new analogues 1 and 2, as well as their supramolecular complexes with CB[7], they were first tested at a single-dose concentration of 25 μM in the presence or absence of 50 μM CB[7] in permeabilized HEK293T cells. As shown in Fig. 4, both 1 and 2 completely inhibit mCa2+ uptake with and without CB[7] at this concentration. Notably, the free ligand (NaOAd) showed no inhibitory activity of mCa2+ uptake up to 50 μM (Fig. S32†). Building upon this initial result, a dose–response analysis of inhibitory activity was carried out to determine the relative potencies of 1, 2, and their supramolecular CB[7] complexes, in comparison to Ru265 and Os245′ (Fig. S33†). As shown in Table 5, both 1 and 2, when administered immediately, inhibit mCa2+ with nanomolar efficacies, but are significantly less potent than Ru265 and Os245′. In addition, the presence of CB[7] does not significantly affect the potency of these complexes in this permeabilized cell model, suggesting that the complexes can readily dissociate from this host and interact with the putative MCU target. The release of supramolecular materials from metal-based complexes has previously been established,91 confirming that this property is common in these constructs. The IC50 value of 1 is comparable to those of other carboxylate-derivatives of Ru265 reported, at least when comparing their activities prior to aquation.54,55 Complex 2 is the first carboxylate-capped analogue of Os245, and therefore its activity cannot be compared to related derivatives. In comparison to the aquated Os245′, however, compound 2 is approximately 40× less potent. By contrast, the difference in potencies between the Ru analogues is less substantial; compound 1 is only 5× less effective than Ru265. The larger inhibitory activity differential for the Os complexes is attributed to the slower aquation kinetics of 2 in comparison to 1. As the aquation data discussed above show, the aquation of Os245 occurs with a half-life of 12 h,50 whereas the half-life for 2 is 30 h, 2.5 times longer. In comparing the IC50 values, it is also apparent that that of 2 is 2.5 times larger than Os245. Thus, the inhibition of mCa2+ uptake primarily mediated by the aquated form Os245′.After assessing their permeabilized cell inhibitory activities, 1, 2, and their CB[7] complexes were evaluated in intact cell systems. In these studies, HeLa cells were incubated with the Ca2+-responsive mitochondria-localizing fluorescent sensor Rhod2-AM, stimulated with histamine to trigger mCa2+ uptake, and then imaged by confocal fluorescence microscopy to measure mCa2+ levels. When these HeLa cells were treated with 50 μM of 1 or 2, a significant decrease of mCa2+ uptake was detected after stimulation with histamine (Fig. 5). Furthermore, the presence of two equiv. of CB[7] did not negatively impact the inhibitory properties of these compounds under these conditions. After demonstrating 50 μM to be an effective concentration, we investigated the abilities of these complexes to operate at lower concentrations. As shown in Fig. 6, both 1 and 2, in the presence or absence of CB[7], were able to inhibit mCa2+ uptake, whereas the parent complexes Ru265 and Os245 were not effective at these lower concentrations. This result is consistent with the cellular uptake studies that show 1 and 2 to be taken up by cells at much higher concentrations than Ru265 and Os245.
Protection against oxygen glucose deprivation
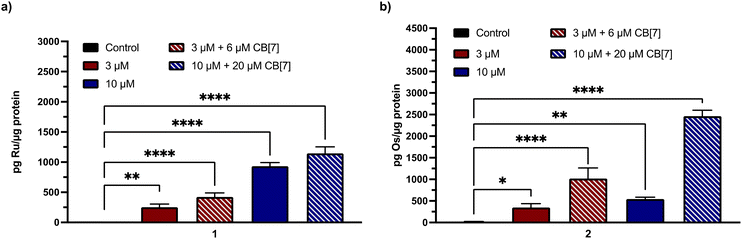
To determine the functional significance of the mCa2+ uptake inhibitory properties of 1 and 2, we investigated them in an in vitro model of ischemic stroke, oxygen glucose deprivation (OGD).50,52 Because mCa2+ overload is a key factor that leads to neuronal cell death in ischemic stroke,92 we sought to assess the cytoprotective effects of 1, 2, and their CB[7] complexes. Previously, we have demonstrated that Ru265 and Os245 protect cortical neurons against an OGD model, a consequence of their ability to prevent mCa2+ overload. As shown in Fig. 6, lower concentrations of 1 and 2 in the presence or absence of CB[7] are more effective mCa2+ uptake inhibitors in intact cells than Ru265 or Os245, suggesting that they might also exhibit better cytoprotective effects against this in vitro OGD model.To validate that the cell uptake properties of these complexes remain consistent in primary cortical neurons, we performed GFAAS and ICP-MS studies on cells treated with 3 or 10 μM 1 or 2 in the presence or absence of CB[7] for 24 h. After normalization to the protein content by the BCA assay, the data in Fig. 7 show that both 1 and 2 are taken up effectively by these cortical neurons. Significantly, the addition of two equiv. of CB[7] further increased the neuronal uptake of these compounds, consistent with the cellular uptake data measured within HeLa cells (Fig. 3).
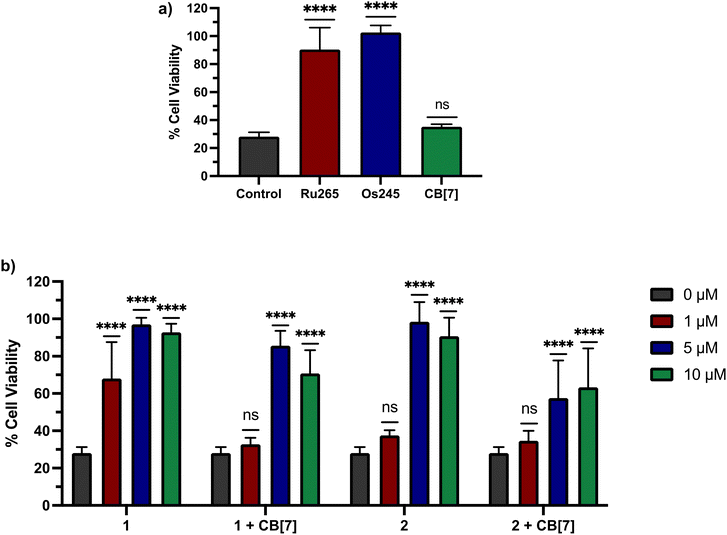
Based on these cell uptake data, we proceeded to analyze the protective effects of these compounds against OGD in primary cortical neurons. Consistent with previous results, Ru265 (10 μM) and Os245 (50 μM) preserve the cell viability of cortical neurons subjected to OGD (Fig. 8a). By contrast, CB[7] (20 μM) showed no protective effects (Fig. 8a). We next investigated 1 and 2 in the presence or absence of two equiv. CB[7] within this model. Both compounds 1 and 2 show protective effects against OGD, even at concentrations as low as 1 and 5 μM, respectively (Fig. 8b). The protective effects of 1 at this concentration are consistent with its ability to inhibit mCa2+ uptake in intact cells at this concentration as well (Fig. 6). By contrast, although 2 is also able to inhibit mCa2+ uptake in intact cells at 1 μM (Fig. 6), it fails to elicit any protection under these conditions. The poorer cytoprotection conferred by 2 is consistent with the similarly poorer protective effects of Os245 compared to Ru265.50 Thus, this trend appears to be a recurring phenomenon in comparing Ru and Os analogues of these inhibitors, and it may be a consequence of their different ligand substitution kinetics. The addition of two equiv. CB[7] to 1 and 2 altered their protective effects. At 1 μM, the protective effects observed by 1 are completely eliminated in the presence of two equiv. CB[7], indicating that supramolecular encapsulation can have an impact on its biological properties. However, at higher concentrations, the addition of CB[7] did not affect the cytoprotective properties of 1 and 2. This result is somewhat unexpected because we demonstrated that CB[7] can increase the cell uptake of these complexes. A possible explanation is that the intracellular localization of the CB[7]-encapsulated compounds is unfavorable for mCa2+ uptake inhibition.
In vivo biological properties
Finally, we sought to investigate in vivo properties of these complexes in the presence and absence of CB[7]. As noted above, when administered to mice at doses greater than 10 mg kg−1, Ru265 and Os245 cause seizures.50,52 This side effect significantly attenuates the therapeutic windows of these compounds. The mechanism by which these compounds induce convulsions is currently unknown, but this phenomenon has also been observed with RuRed.50,52,93–95 Furthermore, a recent study has suggested that the off-target inhibition P/Q-type96 Ca2+ and KCNQ97 K+ channels by Ru265 may be an important contributing factor to this seizure activity.98 In this context, a related study, mentioned previously, showed that CB[7] can attenuate the seizure-induction of pentylenetetrazol in mice, thus prompting us to investigate the ability of this host molecule to modulate the in vivo properties of 1. Male C57/BI6 mice were intraperitoneally (i.p.) injected with 1 at a dose of 10 mg kg−1 in the absence and presence of 20 mg kg−1 CB[7], which coincides with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar stoichiometry of 1 to CB[7]. These mice were then monitored for seizure-like behaviors, including whisker trembling, motionless staring, facial jerking, and clonic convulsions. As shown in Fig. 9, the duration of the convulsions increased with time after the injection of 1, showing the enhanced severity of the side effects. Notably, the presence of CB[7] failed to elicit any change in this time course of events, showing that it failed to protect mice against the seizure induction of 1. Thus, although we demonstrated that in vitro CB[7] can modulate the biological properties, these effects do not translate in vivo. This result may be a consequence of faster ligand substitution of the guest adamantyl carboxylate ligands of 1 in vivo compared to in vitro. Based on these results, further studies involving 2 were not undertaken in order to minimize animal usage and distress. Because of its structural similarity to 1, as well as the seizure-inducing side effects of both Os245 and Ru265,50,52 we do not expect CB[7] to improve the in vivo tolerability of 2. Installing a guest ligand on the non-leaving ammine ligands of 1 may be a better strategy for enabling the benefits of this host–guest chemistry in vivo, as it is likely that the parent Ru265′ or Os245′ compounds are directly responsible for the seizure-inducing behavior in vivo.
2 molar stoichiometry of 1 to CB[7]. These mice were then monitored for seizure-like behaviors, including whisker trembling, motionless staring, facial jerking, and clonic convulsions. As shown in Fig. 9, the duration of the convulsions increased with time after the injection of 1, showing the enhanced severity of the side effects. Notably, the presence of CB[7] failed to elicit any change in this time course of events, showing that it failed to protect mice against the seizure induction of 1. Thus, although we demonstrated that in vitro CB[7] can modulate the biological properties, these effects do not translate in vivo. This result may be a consequence of faster ligand substitution of the guest adamantyl carboxylate ligands of 1 in vivo compared to in vitro. Based on these results, further studies involving 2 were not undertaken in order to minimize animal usage and distress. Because of its structural similarity to 1, as well as the seizure-inducing side effects of both Os245 and Ru265,50,52 we do not expect CB[7] to improve the in vivo tolerability of 2. Installing a guest ligand on the non-leaving ammine ligands of 1 may be a better strategy for enabling the benefits of this host–guest chemistry in vivo, as it is likely that the parent Ru265′ or Os245′ compounds are directly responsible for the seizure-inducing behavior in vivo.
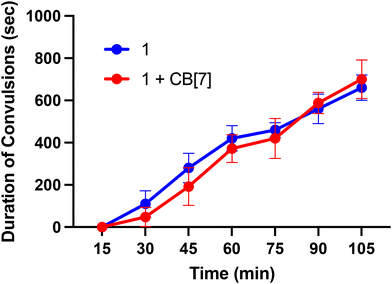 | ||
| Fig. 9 Seizure duration in male C57/BI6 mice injected i.p. with 10 mg kg−1 of 1 or 1 + two equiv. CB[7]. | ||
Conclusions
In this study, we reported the synthesis and characterization of two new derivatives of Ru265 and Os245 with adamantane carboxylate groups appended to the axial site. Compound 1 expands upon the list of previously reported carboxylate-functionalized analogues of Ru265,53–55 and compound 2 is the first reported carboxylate-functionalized analogue of Os245. Both 1 and 2 interact strongly with CB[7], forming 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes. When investigated in biological models, the role of the CB[7] complexation is somewhat enigmatic. CB[7] enhances the cellular uptake of both compounds but does not appear to influence their permeabilized or intact cell mCa2+ uptake-inhibitory properties. Within the in vitro OGD model, only the protective effects of 1 at low concentrations are affected by CB[7]. Lastly, CB[7] fails to attenuate in vivo seizure activity of 1 in mice. The poor success of this CB[7] encapsulation approach therefore suggests that alternatives are needed. It is well-documented in other systems that CB[7] can improve cell uptake, decrease toxic side effects, and enhance biological activities of promising drug candidates that are strongly encapsulated by it.58–63 A possible reason for the poor performance of the CB[7] complexes of 1 and 2 in the biological models may be a consequence of the fact that the guest adamantyl moiety is attached to the carboxylate leaving group ligands of these complexes. As such, the effects of the CB[7] encapsulation may be negated via departure of these axial ligands. This hypothesis suggests that an alternative approach involving the attachment of adamantyl groups to the amine ligands that remain bound to the metal centers may be more fruitful. In any case, a remarkable discovery within this study is that the adamantyl group of 1 and 2 provide substantial advantages over the parent complexes Ru265 and Os245. Most notably, 1 and 2 are effective mCa2+ uptake inhibitors and cytoprotective agents at concentrations lower than those needed for Ru265 and Os245. These results highlight that optimization of the pharmacokinetic properties of these compounds via axial ligand modification provides a promising strategy for accessing more effective and biologically useful inhibitors.
1 complexes. When investigated in biological models, the role of the CB[7] complexation is somewhat enigmatic. CB[7] enhances the cellular uptake of both compounds but does not appear to influence their permeabilized or intact cell mCa2+ uptake-inhibitory properties. Within the in vitro OGD model, only the protective effects of 1 at low concentrations are affected by CB[7]. Lastly, CB[7] fails to attenuate in vivo seizure activity of 1 in mice. The poor success of this CB[7] encapsulation approach therefore suggests that alternatives are needed. It is well-documented in other systems that CB[7] can improve cell uptake, decrease toxic side effects, and enhance biological activities of promising drug candidates that are strongly encapsulated by it.58–63 A possible reason for the poor performance of the CB[7] complexes of 1 and 2 in the biological models may be a consequence of the fact that the guest adamantyl moiety is attached to the carboxylate leaving group ligands of these complexes. As such, the effects of the CB[7] encapsulation may be negated via departure of these axial ligands. This hypothesis suggests that an alternative approach involving the attachment of adamantyl groups to the amine ligands that remain bound to the metal centers may be more fruitful. In any case, a remarkable discovery within this study is that the adamantyl group of 1 and 2 provide substantial advantages over the parent complexes Ru265 and Os245. Most notably, 1 and 2 are effective mCa2+ uptake inhibitors and cytoprotective agents at concentrations lower than those needed for Ru265 and Os245. These results highlight that optimization of the pharmacokinetic properties of these compounds via axial ligand modification provides a promising strategy for accessing more effective and biologically useful inhibitors.
Data availability
The data supporting this article have been included as part of the ESI.† Crystallographic data for 1 and 2 have been deposited at the CCDC under 2352958 and 2352959.†Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the College of Arts & Sciences at Cornell University. Dr Ivan Keresztes is thanked for his assistance with all NMR experiments. This work made use of the Cornell NMR facility, which is supported in part by the NSF (CHE-1531632), and the Cornell Institute of Biotechnology Imaging Facility, which is supported in part by the National Institute of Health (NIH) under award number S10RR025502. Work in the lab of M. J. Stevenson is funded in part by the Faculty Development Fund and Startup Funds at the University of San Francisco. K. Davis is funded in part through the Whitehead Summer Research Fellowship. Work in the lab of G. S. Robertson is supported by the Heart and Stroke Foundation of Canada (G-22-0031944; GSR) and the Dalhousie University Brain Repair Centre (Knowledge and Translation Grant; GSR).References
- T. Pozzan, R. Rizzuto, P. Volpe and J. Meldolesi, Molecular and Cellular Physiology of Intracellular Calcium Stores, Physiol. Rev., 1994, 74, 595–636 CrossRef CAS.
- M. J. Berridge, M. D. Bootman and H. L. Roderick, Calcium Signalling: Dynamics, Homeostasis and Remodelling, Nat. Rev. Mol. Cell Biol., 2003, 4, 517–529 CrossRef CAS PubMed.
- D. E. Clapham, Calcium Signaling, Cell, 2007, 131, 1047–1058 CrossRef CAS.
- E. Carafoli, Historical Review: Mitochondria and Calcium: Ups and Downs of an Unusual Relationship, Trends Biochem. Sci., 2003, 28, 175–181 CrossRef CAS.
- G. S. B. Williams, L. Boyman, A. C. Chikando, R. J. Khairallah and W. J. Lederer, Mitochondrial Calcium Uptake, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 10479–10486 CrossRef CAS.
- K. J. Kamer and V. K. Mootha, The Molecular Era of the Mitochondrial Calcium Uniporter, Nat. Rev. Mol. Cell Biol., 2015, 16, 545–553 CrossRef CAS.
- E. Carafoli and M. Brini, Calcium Signalling and Disease: Molecular Pathology of Calcium, in Subcell. Biochem., Springer, New York, NY, 2007, pp. 481–506 Search PubMed.
- S. Orrenius, V. Gogvadze and B. Zhivotovsky, Calcium and Mitochondria in the Regulation of Cell Death, Biochem. Biophys. Res. Commun., 2015, 460, 72–81 CrossRef CAS.
- Å. B. Gustafsson and R. A. Gottlieb, Heart Mitochondria: Gates of Life and Death, Cardiovasc. Res., 2008, 77, 334–343 CrossRef PubMed.
- G. Santulli, W. Xie, S. R. Reiken and A. R. Marks, Mitochondrial Calcium Overload Is a Key Determinant in Heart Failure, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11389–11394 CrossRef CAS.
- B. Zhou and R. Tian, Mitochondrial Dysfunction in Pathophysiology of Heart Failure, J. Clin. Invest., 2018, 128, 3716–3726 CrossRef PubMed.
- A. Vultur, C. S. Gibhardt, H. Stanisz and I. Bogeski, The Role of the Mitochondrial Calcium Uniporter (MCU) Complex in Cancer, Pflug. Arch. Eur. J. Physiol., 2018, 470, 1149–1163 CrossRef CAS.
- C. Delierneux, S. Kouba, S. Shanmughapriya, M. Potier-Cartereau, M. Trebak and N. Hempel, Mitochondrial Calcium Regulation of Redox Signaling in Cancer, Cells, 2020, 9, 432 CrossRef CAS PubMed.
- A. Rimessi, C. Pozzato, L. Carparelli, A. Rossi, S. Ranucci, I. de Fino, C. Cigana, A. Talarico, M. R. Wieckowski, C. M. P. Ribeiro, C. Trapella, G. Rossi, G. Cabrini, A. Bragonzi and P. Pinton, Pharmacological Modulation of Mitochondrial Calcium Uniporter Controls Lung Inflammation in Cystic Fibrosis, Sci. Adv., 2020, 6, eaax9093 CrossRef CAS PubMed.
- A. Rimessi, V. A. M. Vitto, S. Patergnani and P. Pinton, Update on Calcium Signaling in Cystic Fibrosis Lung Disease, Front. Pharmacol., 2021, 12, 581645 CrossRef CAS.
- F. Celsi, P. Pizzo, M. Brini, S. Leo, C. Fotino, P. Pinton and R. Rizzuto, Mitochondria, Calcium and Cell Death: A Deadly Triad in Neurodegeneration, Biochim. Biophys. Acta, Bioenerg., 2009, 1787, 335–344 CrossRef CAS.
- R. Abeti and A. Y. Abramov, Mitochondrial Ca2+ in Neurodegenerative Disorders, Pharmacol. Res., 2015, 99, 377–381 CrossRef CAS.
- A. P. Halestrap, Calcium, Mitochondria and Reperfusion Injury: A Pore Way to Die, Biochem. Soc. Trans., 2006, 34, 232–237 CrossRef CAS.
- K. Shintani-Ishida, M. Inui and K. Yoshida, Ischemia-Reperfusion Induces Myocardial Infarction through Mitochondrial Ca2+ Overload, J. Mol. Cell. Cardiol., 2012, 53, 233–239 CrossRef CAS.
- T. H. Sanderson, C. A. Reynolds, R. Kumar, K. Przyklenk and M. Huttemann, Molecular Mechanisms of Ischemia-Reperfusion Injury in Brain: Pivotal Role of the Mitochondrial Membrane Potential in Reactive Oxygen Species Generation, Mol. Neurobiol., 2013, 47, 9–23 CrossRef CAS PubMed.
- Y. Kirichok, G. Krapivinsky and D. E. Clapham, The Mitochondrial Calcium Uniporter Is a Highly Selective Ion Channel, Nature, 2004, 427, 360–364 CrossRef CAS.
- J. M. Baughman, F. Perocchi, H. S. Girgis, M. Plovanich, C. A. Belcher-Timme, Y. Sancak, X. R. Bao, L. Strittmatter, O. Goldberger, R. L. Bogorad, V. Koteliansky and V. K. Mootha, Integrative Genomics Identifies MCU as an Essential Component of the Mitochondrial Calcium Uniporter, Nature, 2011, 476, 341–345 CrossRef CAS PubMed.
- D. De Stefani, A. Raffaello, E. Teardo, I. Szabó and R. Rizzuto, A Forty-Kilodalton Protein of the Inner Membrane Is the Mitochondrial Calcium Uniporter, Nature, 2011, 476, 336–340 CrossRef CAS.
- N. Nemani, S. Shanmughapriya and M. Madesh, Molecular Regulation of MCU: Implications in Physiology and Disease, Cell Calcium, 2018, 74, 86–93 CrossRef CAS.
- R. Baradaran, C. Wang, A. F. Siliciano and S. B. Long, Cryo-EM Structures of Fungal and Metazoan Mitochondrial Calcium Uniporters, Nature, 2018, 559, 580–584 CrossRef CAS PubMed.
- C. Fan, M. Fan, B. J. Orlando, N. M. Fastman, J. Zhang, Y. Xu, M. G. Chambers, X. Xu, K. Perry, M. Liao and L. Feng, X-Ray and, Cryo-EM Structures of the Mitochondrial Calcium Uniporter, Nature, 2018, 559, 575–579 CrossRef CAS.
- N. X. Nguyen, J.-P. Armache, C. Lee, Y. Yang, W. Zeng, V. K. Mootha, Y. Cheng, X.-C. Bai and Y. Jiang, Cryo-EM Structure of a Fungal Mitochondrial Calcium Uniporter, Nature, 2018, 559, 570–574 CrossRef CAS.
- J. Yoo, M. Wu, Y. Yin, M. A. Herzik, G. C. Lander and S.-Y. Lee, Cryo-EM Structure of a Mitochondrial Calcium Uniporter, Science, 2018, 361, 506–511 CrossRef CAS.
- Y. Wang, N. X. Nguyen, J. She, W. Zeng, Y. Yang, X.-C. Bai and Y. Jiang, Structural Mechanism of EMRE-Dependent Gating of the Human Mitochondrial Calcium Uniporter, Cell, 2019, 177, 1252–1261 CrossRef CAS.
- K. J. Kamer, Y. Sancak, Y. Fomina, J. D. Meisel, D. Chaudhuri, Z. Grabarek and V. K. Mootha, MICU1 Imparts the Mitochondrial Uniporter with the Ability to Discriminate between Ca2+ and Mn2+, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E7960–E7969 CrossRef CAS.
- K. J. Kamer, W. Jiang, V. K. Kaushik, V. K. Mootha and Z. Grabarek, Crystal Structure of MICU2 and Comparison with MICU1 Reveal Insights into the Uniporter Gating Mechanism, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 3546–3555 CrossRef CAS.
- M. Paillard, G. Csordás, K. T. Huang, P. Várnai, S. K. Joseph and G. Hajnóczky, MICU1 Interacts with the D-Ring of the MCU Pore to Control Its Ca2+ Flux and Sensitivity to Ru360, Mol. Cell, 2018, 72, 778–785 CrossRef CAS.
- Y. Xing, M. Wang, J. Wang, Z. Nie, G. Wu, X. Yang and Y. Shen, Dimerization of MICU Proteins Controls Ca2+ Influx through the Mitochondrial Ca2+ Uniporter, Cell Rep., 2019, 26, 1203–1212 CrossRef CAS.
- C. B. Phillips, C.-W. Tsai and M.-F. Tsai, The Conserved Aspartate Ring of MCU Mediates MICU1 Binding and Regulation in the Mitochondrial Calcium Uniporter Complex, eLife, 2019, 8, e41112 CrossRef.
- K. Oxenoid, Y. Dong, C. Cao, T. Cui, Y. Sancak, A. L. Markhard, Z. Grabarek, L. Kong, Z. Liu, B. Ouyang, Y. Cong, V. K. Mootha and J. J. Chou, Architecture of the Mitochondrial Calcium Uniporter, Nature, 2016, 533, 269–273 CrossRef CAS PubMed.
- C. Cao, S. Wang, T. Cui, X.-C. Su and J. J. Chou, Ion and Inhibitor Binding of the Double-Ring Ion Selectivity Filter of the Mitochondrial Calcium Uniporter, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E2846–E2851 CAS.
- N. Kon, M. Murakoshi, A. Isobe, K. Kagechika, N. Miyoshi and T. Nagayama, DS16570511 Is a Small-Molecule Inhibitor of the Mitochondrial Calcium Uniporter, Cell Death Discovery, 2017, 3, 17045 CrossRef CAS PubMed.
- D. M. Arduino, J. Wettmarshausen, H. Vais, P. Navas-Navarro, Y. Cheng, A. Leimpek, Z. Ma, A. Delrio-Lorenzo, A. Giordano, C. Garcia-Perez, G. Médard, B. Kuster, J. García-Sancho, D. Mokranjac, J. K. Foskett, M. T. Alonso and F. Perocchi, Systematic Identification of MCU Modulators by Orthogonal Interspecies Chemical Screening, Mol. Cell, 2017, 67, 711–723 CrossRef CAS PubMed.
- J. Schwartz, E. Holmuhamedov, X. Zhang, G. L. Lovelace, C. D. Smith and J. J. Lemasters, Minocycline and Doxycycline, but Not Other Tetracycline-Derived Compounds, Protect Liver Cells from Chemical Hypoxia and Ischemia/Reperfusion Injury by Inhibition of the Mitochondrial Calcium Uniporter, Toxicol. Appl. Pharmacol., 2013, 273, 172–179 CrossRef CAS PubMed.
- G. Di Marco, F. Vallese, B. Jourde, C. Bergsdorf, M. Sturlese, A. De Mario, V. Techer-Etienne, D. Haasen, B. Oberhauser, S. Schleeger, G. Minetti, S. Moro, R. Rizzuto, D. De Stefani, M. Fornaro and C. Mammucari, A High-Throughput Screening Identifies MICU1 Targeting Compounds, Cell Rep., 2020, 30, 2321–2331 CrossRef CAS PubMed.
- V. T. Thu, H.-K. Kim, L. T. Long, S.-R. Lee, T. M. Hanh, T. H. Ko, H.-J. Heo, N. Kim, S. H. Kim, K. S. Ko, B. D. Rhee and J. Han, NecroX-5 Prevents Hypoxia/Reoxygenation Injury by Inhibiting the Mitochondrial Calcium Uniporter, Cardiovasc. Res., 2012, 94, 342–350 CrossRef CAS PubMed.
- J. Santo-Domingo, L. Vay, E. Hernández-SanMiguel, C. D. Lobatón, A. Moreno, M. Montero and J. Alvarez, The Plasma Membrane Na+/Ca2+ Exchange Inhibitor KB-R7943 Is Also a Potent Inhibitor of the Mitochondrial Ca2+ Uniporter, Br. J. Pharmacol., 2007, 151, 647–654 CrossRef CAS.
- J. Emerson, M. J. Clarke, W.-L. Ying and D. R. Sanadi, The Component of “Ruthenium Red” Responsible for Inhibition of Mitochondrial Calcium Ion Transport. Spectra, Electrochemistry, and Aquation Kinetics. Crystal Structure of μ-O-[(HCO2)(NH3)4Ru]2Cl3, J. Am. Chem. Soc., 1993, 115, 11799–11805 CrossRef CAS.
- M. A. Matlib, Z. Zhou, S. Knight, S. Ahmed, K. M. Choi, J. Krause-Bauer, R. Phillips, R. Altschuld, Y. Katsube, N. Sperelakis and D. M. Bers, Oxygen-Bridged Dinuclear Ruthenium Amine Complex Specifically Inhibits Ca2+ Uptake into Mitochondria in Vitro and in Situ in Single Cardiac Myocytes, J. Biol. Chem., 1998, 273, 10223–10231 CrossRef CAS PubMed.
- J. F. Unitt, K. L. Boden, A. V. Wallace, A. H. Ingall, M. E. Coombs and F. Ince, Novel Cobalt Complex Inhibitors of Mitochondrial Calcium Uptake, Bioorg. Med. Chem., 1999, 7, 1891–1896 CrossRef CAS.
- S. R. Nathan, N. W. Pino, D. M. Arduino, F. Perocchi, S. N. MacMillan and J. J. Wilson, Synthetic Methods for the Preparation of a Functional Analogue of Ru360, a Potent Inhibitor of Mitochondrial Calcium Uptake, Inorg. Chem., 2017, 56, 3123–3126 CrossRef CAS.
- J. J. Woods, N. Nemani, S. Shanmughapriya, A. Kumar, M. Zhang, S. R. Nathan, M. Thomas, E. Carvalho, K. Ramachandran, S. Srikantan, P. B. Stathopulos, J. J. Wilson and M. Madesh, A Selective and Cell-Permeable Mitochondrial Calcium Uniporter (MCU) Inhibitor Preserves Mitochondrial Bioenergetics after Hypoxia/Reoxygenation Injury, ACS Cent. Sci., 2019, 5, 153–166 CrossRef CAS.
- J. J. Woods, M. X. Rodriguez, C.-W. Tsai, M.-F. Tsai and J. J. Wilson, Cobalt Amine Complexes and Ru265 Interact with the DIME Region of the Mitochondrial Calcium Uniporter, Chem. Commun., 2021, 57, 6161–6164 RSC.
- J. Cervinka, A. Gobbo, L. Biancalana, L. Markova, V. Novohradsky, M. Guelfi, S. Zacchini, J. Kasparkova, V. Brabec and F. Marchetti, Ruthenium(II)−Tris-Pyrazolylmethane Complexes Inhibit Cancer Cell Growth by Disrupting Mitochondrial Calcium Homeostasis, J. Med. Chem., 2022, 65, 10567–10587 CrossRef CAS PubMed.
- J. J. Woods, R. J. Novorolsky, N. P. Bigham, G. S. Robertson and J. J. Wilson, Dinuclear Nitrido-Bridged Osmium Complexes Inhibit the Mitochondrial Calcium Uniporter and Protect Cortical Neurons against Lethal Oxygen–Glucose Deprivation, RSC Chem. Biol., 2023, 4, 84–93 RSC.
- J. J. Woods, J. Lovett, B. Lai, H. H. Harris and J. J. Wilson, Redox Stability Controls the Cellular Uptake and Activity of Ruthenium-Based Inhibitors of the Mitochondrial Calcium Uniporter (MCU), Angew. Chem., Int. Ed., 2020, 59, 6482–6491 CrossRef CAS PubMed.
- R. J. Novorolsky, M. Nichols, J. S. Kim, E. V. Pavlov, J. J. Woods, J. J. Wilson and G. S. Robertson, The Cell-Permeable Mitochondrial Calcium Uniporter Inhibitor Ru265 Preserves Cortical Neuron Respiration after Lethal Oxygen Glucose Deprivation and Reduces Hypoxic/Ischemic Brain Injury, J. Cereb. Blood Flow Metab., 2020, 40, 1172–1181 CrossRef CAS PubMed.
- N. P. Bigham, Z. Huang, J. A. Spivey, J. J. Woods, S. N. MacMillan and J. J. Wilson, Carboxylate-Capped Analogues of Ru265 Are MCU Inhibitor Prodrugs, Inorg. Chem., 2022, 61, 17299–17312 CrossRef CAS.
- Z. Huang, J. A. Spivey, S. N. Macmillan and J. J. Wilson, A Ferrocene-Containing Analogue of the MCU Inhibitor Ru265 with Increased Cell Permeability, Inorg. Chem. Front., 2023, 10, 591–599 RSC.
- Z. Huang, S. N. Macmillan and J. J. Wilson, A Fluorogenic Inhibitor of the Mitochondrial Calcium Uniporter, Angew. Chem., Int. Ed., 2023, 62, e202214920 CrossRef CAS PubMed.
- D. T. Richens, Ligand Substitution Reactions at Inorganic Centers, Chem. Rev., 2005, 105, 1961–2002 CrossRef CAS PubMed.
- F. P. Rotzinger, Treatment of Substitution and Rearrangement Mechanisms of Transition Metal Complexes with Quantum Chemical Methods, Chem. Rev., 2005, 105, 2003–2038 CrossRef CAS PubMed.
- X. Ma and Y. Zhao, Biomedical Applications of Supramolecular Systems Based on Host–Guest Interactions, Chem. Rev., 2015, 115, 7794–7839 CrossRef CAS.
- M. J. Webber and R. Langer, Drug Delivery by Supramolecular Design, Chem. Soc. Rev., 2017, 46, 6600–6620 RSC.
- W.-C. Geng, J. L. Sessler and D.-S. Guo, Supramolecular Prodrugs Based on Host–Guest Interactions, Chem. Soc. Rev., 2020, 49, 2303–2315 RSC.
- J. Murray, K. Kim, T. Ogoshi, W. Yao and B. C. Gibb, The Aqueous Supramolecular Chemistry of Cucurbiturils, Pillar[n]Arenes and Deep-Cavity Cavitands, Chem. Soc. Rev., 2017, 46, 2479–2496 RSC.
- D. Das, K. I. Assaf and W. M. Nau, Applications of Cucurbiturils in Medicinal Chemistry and Chemical Biology, Front. Chem., 2019, 7, 619 CrossRef CAS.
- A. Gu and N. J. Wheate, Macrocycles as Drug-Enhancing Excipients in Pharmaceutical Formulations, J. Inclusion Phenom. Macrocyclic Chem., 2021, 100, 55–69 CrossRef CAS.
- Q. Huang, K. I. Kuok, X. Zhang, L. Yue, S. M. Y. Lee, J. Zhang and R. Wang, Inhibition of Drug-Induced Seizure Development in Both Zebrafish and Mouse Models by a Synthetic Nanoreceptor, Nanoscale, 2018, 10, 10333–10336 RSC.
- L. Wanka, K. Iqbal and P. R. Schreiner, The Lipophilic Bullet Hits the Targets: Medicinal Chemistry of Adamantane Derivatives, Chem. Rev., 2013, 113, 3516–3604 CrossRef CAS PubMed.
- B. Gong, B.-K. Choi, J.-Y. Kim, D. Shetty, Y. H. Ko, N. Selvapalam, N. K. Lee and K. Kim, High Affinity Host–Guest FRET Pair for Single-Vesicle Content-Mixing Assay: Observation of Flickering Fusion Events, J. Am. Chem. Soc., 2015, 137, 8908–8911 CrossRef CAS PubMed.
- S.-R. Wang, Y.-Y. Song, L. Wei, C.-X. Liu, B.-S. Fu, J.-Q. Wang, X.-R. Yang, Y.-N. Liu, S.-M. Liu, T. Tian and X. Zhou, Cucurbit[7]Uril-Driven Host–Guest Chemistry for Reversible Intervention of 5-Formylcytosine-Targeted Biochemical Reactions, J. Am. Chem. Soc., 2017, 139, 16903–16912 CrossRef CAS.
- X. Zhou, X. Su, P. Pathak, R. Vik, B. Vinciguerra, L. Isaacs and J. Jayawickramarajah, Host–Guest Tethered DNA Transducer: ATP Fueled Release of a Protein Inhibitor from Cucurbit[7]Uril, J. Am. Chem. Soc., 2017, 139, 13916–13921 CrossRef CAS.
- K. L. Kim, G. Sung, J. Sim, J. Murray, M. Li, A. Lee, A. Shrinidhi, K. M. Park and K. Kim, Supramolecular Latching System Based on Ultrastable Synthetic Binding Pairs as Versatile Tools for Protein Imaging, Nat. Commun., 2018, 9, 1712 CrossRef.
- M. Li, A. Lee, K. L. Kim, J. Murray, A. Shrinidhi, G. Sung, K. M. Park and K. Kim, Autophagy Caught in the Act: A Supramolecular FRET Pair Based on an Ultrastable Synthetic Host–Guest Complex Visualizes Autophagosome–Lysosome Fusion, Angew. Chem., Int. Ed., 2018, 57, 2120–2125 CrossRef CAS.
- L. Zou, A. S. Braegelman and M. J. Webber, Spatially Defined Drug Targeting by in Situ Host–Guest Chemistry in a Living Animal, ACS Cent. Sci., 2019, 5, 1035–1043 CrossRef CAS.
- C. Sun, Z. Wang, L. Yue, Q. Huang, Q. Cheng and R. Wang, Supramolecular Induction of Mitochondrial Aggregation and Fusion, J. Am. Chem. Soc., 2020, 142, 16523–16527 CrossRef CAS.
- Z. Yu, M. Ai, S. K. Samanta, F. Hashiya, J. Taniguchi, S. Asamitsu, S. Ikeda, K. Hashiya, T. Bando, G. N. Pandian, L. Isaacs and H. Sugiyama, A Synthetic Transcription Factor Pair Mimic for Precise Recruitment of an Epigenetic Modifier to the Targeted DNA Locus, Chem. Commun., 2020, 56, 2296–2299 RSC.
- A. Lee, M. Li, Y. H. Ko, S. Park, J. Seo, K. M. Park and K. Kim, Visualization of Lipophagy Using a Supramolecular FRET Pair, Chem. Commun., 2021, 57, 12179–12182 RSC.
- Z. Zhang, X. Wang, X. Wei, S. W. Zheng, B. J. Lenhart, P. Xu, J. Li, J. Pan, H. Albrecht and C. Liu, Multiplex Quantitative Detection of SARS-CoV-2 Specific IgG and IgM Antibodies Based on DNA-Assisted Nanopore Sensing, Biosens. Bioelectron., 2021, 181, 113134 CrossRef CAS.
- D. V. D. W. Kankanamalage, J. H. T. Tran, N. Beltrami, K. Meng, X. Zhou, P. Pathak, L. Isaacs, A. L. Burin, M. F. Ali and J. Jayawickramarajah, DNA Strand Displacement Driven by Host–Guest Interactions, J. Am. Chem. Soc., 2022, 144, 16502–16511 CrossRef CAS.
- V. I. J. Jallinoja, C. H. Abbriano, K. Bhatt, A. Kaur, D. J. Schlyer, P. J. Yazaki, B. D. Carney and J. L. Houghton, Pretargeting with Cucurbituril–Adamantane Host–Guest Pair in Xenograft Models, J. Nucl. Med., 2023, 64, 1203–1209 CrossRef CAS PubMed.
- X. Yang, K. Varini, M. Godard, F. Gassiot, R. Sonnette, G. Ferracci, B. Pecqueux, V. Monnier, L. Charles, S. Maria, M. Hardy, O. Ouari, M. Khrestchatisky, P. Lécorché, G. Jacquot and D. Bardelang, Preparation and In Vitro Validation of a Cucurbit[7]Uril-Peptide Conjugate Targeting the LDL Receptor, J. Med. Chem., 2023, 66, 8844–8857 CrossRef CAS.
- M. Sojka, J. Chyba, S. S. Paul, K. Wawrocka, K. Hönigova, B. J. R. Cuyacot, A. C. Castro, T. Vaculovič, J. Marek, M. Repisky, M. Masařik, J. Novotny and R. Marek, Supramolecular Coronation of Platinum(II) Complexes by Macrocycles: Structure, Relativistic DFT Calculations, and Biological Effects, Inorg. Chem., 2021, 60, 17911–17925 CrossRef CAS.
- A. C. Gomes, C. I. R. Magalhães, T. S. M. Oliveira, A. D. Lopes, I. S. Gonçalves and M. Pillinger, Solid-State Study of the Structure and Host–Guest Chemistry of Cucurbituril-Ferrocene Inclusion Complexes, Dalton Trans., 2016, 45, 17042–17052 RSC.
- A. C. Garcia, L. N. Zakharov and M. D. Pluth, Supramolecular Activation of S8 by Cucurbiturils in Water and Mechanism of Reduction to H2S by Thiols: Insights into Biological Sulfane Sulfur Trafficking, J. Am. Chem. Soc., 2022, 144, 15324–15332 CrossRef CAS.
- J. Lagona, P. Mukhopadhyay, S. Chakrabarti and L. Isaacs, The Cucurbit[n]Uril Family, Angew. Chem., Int. Ed., 2005, 44, 4844–4870 CrossRef CAS.
- N. J. Wheate, P. G. A. Kumar, A. M. Torres, J. R. Aldrich-Wright and W. S. Price, Examination of Cucurbit[7]Uril and Its Host-Guest Complexes by Diffusion Nuclear Magnetic Resonance, J. Phys. Chem. B, 2008, 112, 2311–2314 CrossRef CAS PubMed.
- E. O. Stejskal, Use of Spin Echoes in a Pulsed Magnetic-Field Gradient to Study Anisotropic, Restricted Diffusion and Flow, J. Chem. Phys., 1965, 43, 3597–3603 CrossRef.
- M. P. Grant, N. J. Wheate and J. R. Aldrich-Wright, Diffusion Coefficient of Cucurbit[n]Urils (n = 6 or 7) at Various Concentrations, Temperatures, and pH, J. Chem. Eng. Data, 2009, 54, 323–326 CrossRef CAS.
- R. O'Brien, N. Markova and G. A. Holdgate, Thermodynamics in Drug Discovery, in Appl. Biophys. Drug Discovery, John Wiley & Sons, Ltd, 2017, pp. 7–28 Search PubMed.
- A. J. Selinger and D. H. Macartney, Cucurbit[7]Uril Complexations of Good's Buffers, RSC Adv., 2017, 7, 42513–42518 RSC.
- N. E. Grossoehme, A. M. Spuches and D. E. Wilcox, Application of Isothermal Titration Calorimetry in Bioinorganic Chemistry, J. Biol. Inorg. Chem., 2010, 15, 1183–1191 CrossRef CAS PubMed.
- S. Moghaddam, C. Yang, M. Rekharsky, Y. H. Ko, K. Kim, Y. Inoue and M. K. Gilson, New Ultrahigh Affinity Host-Guest Complexes of Cucurbit[7]Uril with Bicyclo[2.2.2]Octane and Adamantane Guests: Thermodynamic Analysis and Evaluation of M2 Affinity Calculations, J. Am. Chem. Soc., 2011, 133, 3570–3581 CrossRef CAS.
- J. J. Woods, J. A. Spivey and J. J. Wilson, A [1H,15N] Heteronuclear Single Quantum Coherence NMR Study of the Solution Reactivity of the Ruthenium-Based Mitochondrial Calcium Uniporter Inhibitor Ru265, Eur. J. Inorg. Chem., 2022, e202100995 CrossRef CAS.
- F. Schibilla, A. Holthenrich, B. Song, A. L. L. Matos, D. Grill, D. R. Martir, V. Gerke, E. Zysman-Colman and B. J. Ravoo, Phosphorescent Cationic Iridium(III) Complexes Dynamically Bound to Cyclodextrin Vesicles: Applications in Live Cell Imaging, Chem. Sci., 2018, 9, 7822–7828 RSC.
- E. L. Que, D. W. Domaille and C. J. Chang, Metals in Neurobiology: Probing Their Chemistry and Biology with Molecular Imaging, Chem. Rev., 2008, 108, 1517–1549 CrossRef CAS.
- R. Tapia, G. Meza-Ruíz, L. Durán and R. Drucker-Colín, Convulsions or Flaccid Paralysis Induced by Ruthenium Red Depending on Route of Administration, Brain Res., 1976, 116, 101–109 CrossRef CAS.
- G. García-Ugalde and R. Tapia, Convulsions and Wet-Dog Shakes Produced by Systemic or Intrahippocampal Administration of Ruthenium Red in the Rat, Exp. Brain Res., 1991, 86, 633–640 CrossRef.
- C. Bas-Orth, J. Schneider, A. Lewen, J. McQueen, K. Hasenpusch-Theil, T. Theil, G. E. Hardingham, H. Bading and O. Kann, The Mitochondrial Calcium Uniporter Is Crucial for the Generation of Fast Cortical Network Rhythms, J. Cereb. Blood Flow Metab., 2020, 40, 2225–2239 CrossRef PubMed.
- V. Nimmrich and G. Gross, P/Q–type Calcium Channel Modulators, Br. J. Pharmacol., 2012, 167, 741–759 CrossRef CAS PubMed.
- T. J. Jentsch, Neuronal KCNQ Potassium Channels: Physiology and Role in Disease, Nat. Rev. Neurosci., 2000, 1, 21–30 CrossRef CAS.
- P. Xu, S. Swain, R. J. Novorolsky, E. Garcia, Z. Huang, T. P. Snutch, J. J. Wilson, G. S. Robertson and R. B. Renden, The Mitochondrial Calcium Uniporter Inhibitor Ru265 Increases Neuronal Excitability and Reduces Neurotransmission via Off-Target Effects, Br. J. Pharmacol., 2024 DOI:10.1111/bph.16425.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures and supporting data. CCDC 2352958 and 2352959. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4qi01102c |
| This journal is © the Partner Organisations 2024 |