Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Blending recombinant amyloid silk proteins generates composite fibers with tunable mechanical properties†
Shri Venkatesh
Subramani
a,
Jingyao
Li
a,
Kok Zhi
Lee
a,
Natalie
Fisher
b and
Fuzhong
Zhang
 *acd
*acd
aDepartment of Energy, Environmental and Chemical Engineering, Washington University in St. Louis, Saint Louis, MO 63130, USA. E-mail: fzhang@seas.wustl.edu
bDepartment of Materials Science and Mechanical Engineering, Washington University in St. Louis, Saint Louis, MO 63130, USA
cInstitute of Materials Science & Engineering, Washington University in St. Louis, Saint Louis, MO 63130, USA
dDivision of Biological & Biomedical Sciences, Washington University in St. Louis, Saint Louis, MO 63130, USA
First published on 21st February 2024
Abstract
Recent efforts in microbial production of recombinant silk and silk-inspired proteins have yielded fibers with excellent mechanical performances on par with or even superior to their natural counterparts. However, most previous recombinant fibers comprise a single protein. Studies exploring composite fibers of recombinant proteins with varied sequences and properties have been limited. Here, we explored the impact of blending different amyloid-silk proteins on the mechanical properties of the resulting composite fibers. Amyloid-silk hybrid proteins contain repetitive sequences of an amyloid zipper-forming peptide flanked by flexible glycine-rich peptides from a spider silk protein. We pursued three blending strategies, by mixing amyloid-silk proteins of different molecular weights, opposite protein charges, and distinct mechanical behaviors. Our findings revealed that the ultimate strength of composite fibers consistently fell within the range of the pure protein fibers across all three blending strategies. However, composite fibers comprising oppositely charged proteins displayed toughness higher than both pure protein fibers. Additionally, mixing amyloid-silk proteins of different molecular weights or mechanical properties allowed us to fine-tune the mechanical properties of composite fibers by controlling the protein ratios. The findings highlight the potential of composite protein fibers as a versatile platform for achieving diverse yet precisely tunable mechanical properties. The trends in fiber properties and blending strategies observed in this study open up exciting prospects for future engineering of protein materials tailored to specific characteristics.
Introduction
Biology has long served as a source of inspiration for materials design.1 A wide array of natural protein-based materials (PBMs) are found to exhibit remarkable performance characteristics, finely tuned through millions of years of evolution.2 Examples include insect silks,3 muscle titin,4 mussel byssal threads,5 and insect resilin6 which display unique and attractive mechanical properties. The exceptional properties of these natural materials have motivated decades of research into their large-scale production from rapidly growing micro-organisms.7,8 Despite the challenges posed by their repetitive sequences and biased amino acid compositions, recent advances in synthetic biology have overcome many obstacles in the heterologous synthesis of mechanically robust PBMs.9–11 Several microbially produced PBMs have displayed mechanical performances comparable to or even higher than their natural counterparts.12–15While most recombinant PBMs typically comprise a single purified protein—an approach crucial for studying material sequence-structure–property relationships—natural PBMs commonly contain a mixture of different proteins with unique attributes in their amino acid compositions, secondary structures, and molecular weights (MWs).16–24 The overall integrity and properties of such composite materials stem from the intricate interplay among these distinct component proteins. For instance, the silk cocoon filaments spun by the Bombyx mori silkworm comprise sericin and fibroin proteins.21 Fibroin forms β-sheets and is responsible for the strength of silk filaments, while sericin cements the fibroin proteins together and toughens the fiber structure. This synergistic effect, brought about by the molecular interactions between serine residues of sericin and glycine residues within fibroin, contributes to the observed mechanical behavior of silkworm silk.21 Likewise, all known dragline spider silk fibers contain multiple proteins,19 suggesting that spiders tune fiber mechanical properties using different protein compositions.25
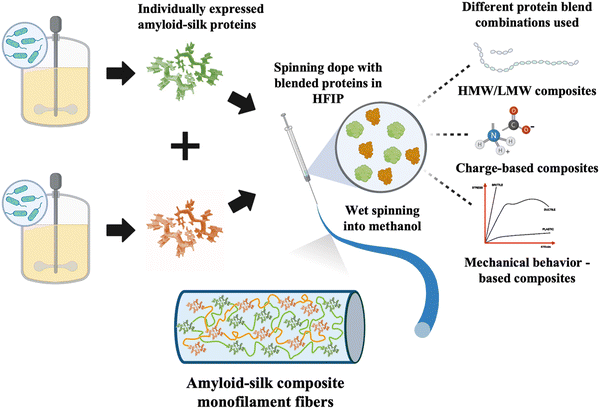
Similar to natural composite materials, blending recombinant proteins may offer strategies for controlling PBM mechanical properties. While some previous studies have explored composite fibers made of different recombinant proteins, they have often focused on mixing proteins with similar properties.26–29 Composite fibers consisting of proteins with different mechanical properties have been rarely explored. In this study, we investigated the influence of different proteins and their compositions on the mechanical properties of composite fibers. We created composite fibers using recombinant amyloid-silk proteins of high/low molecular weights (HMW/LMW), proteins with opposite charges, and those exhibiting different mechanical behaviors (Fig. 1), at varying protein ratios. Our findings revealed that all three composite fibers, with varying MWs, opposing protein charges and contrasting mechanical behavior, displayed a linear trend in ultimate fiber strength, determined by the mixture ratio of each component (Fig. S1, ESI†). Moreover, we demonstrated that the mechanical properties of such composite fibers can be precisely tuned by varying the composition of each protein, presenting an exciting opportunity to control fiber mechanical characteristics.
Results
Blending amyloid-silk proteins of different MW yields tailored composite fibers
To explore the mechanical traits of composite PBM fibers, we chose to blend different amyloid-silk hybrid proteins due to their advantageous mechanical properties. Amyloid-silk hybrid proteins contain repetitive sequences of an amyloid zipper-forming peptide flanked by a flexible glycine-rich peptide from spider silk proteins.30 The strong β-sheet forming propensity of amyloid peptides contributed to increased crystallinity in the resulting protein fibers.31 This inherent characteristic offered an advantage in tailoring the mechanical performance of the composite fibers. Moreover, these hybrid proteins boast relatively higher yield compared to recombinant silk and other proteins, due to reduced repetitiveness of their protein sequences (Table S1, ESI†).32 Additionally, the large library of diverse amyloid zipper-forming peptides offers extensive design possibilities. Each amyloid-silk protein was individually expressed in Escherichia coli shake flask cultures and purified (Fig. S2, ESI†). Subsequently, composite fibers as well as pure protein fibers were prepared and analyzed under identical experimental conditions. Optical and scanning electron microscopy revealed cylindrical fibers with consistent diameters (Fig. 2–4b and Fig. S3, S4, ESI†).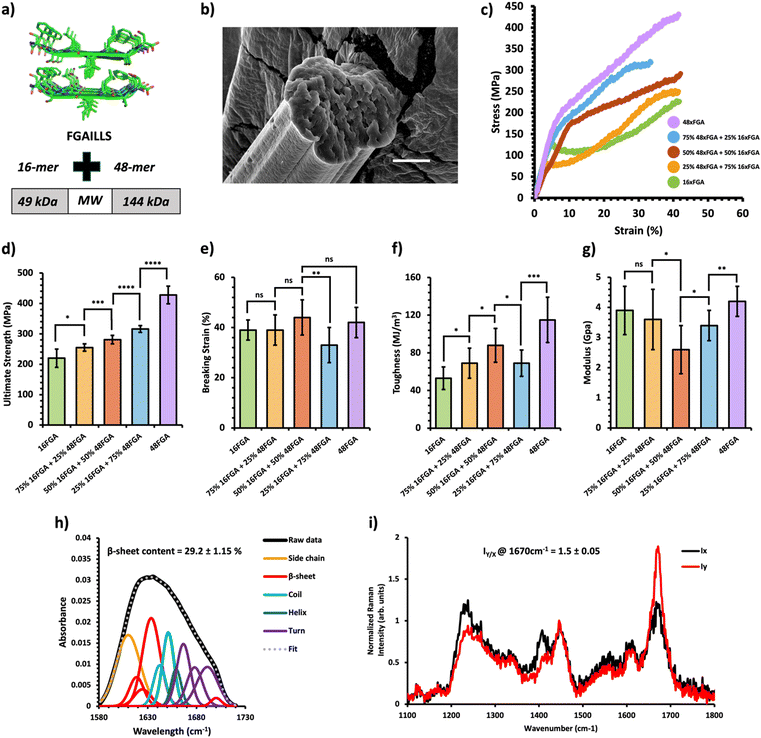 | ||
Fig. 2 Mechanical and structural properties of HMW/LMW protein composite fibers. (a) Crystal structure of the FGAILSS (PDB: 5E61) amyloid peptide. Polymeric amyloid-silk proteins containing 16 (MW = 49 kDa) and 48 (MW = 144 kDa) repeats of the FGAILSS peptide were mixed to generate HMW/LMW protein composite fibers. (b) Representative S.E.M. image of the 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 HMW/LMW protein composite fibers. Stretched fiber pieces were imaged after tensile testing. Scale bars represent 10 μm. (c) Representative stress–strain curves from tensile testing of the HMW/LMW protein composite fibers at different mixture ratios. (d)–(g) Ultimate tensile stress (d), breaking strain (e), toughness (f), and Young's modulus (g) of the HMW/LMW protein composite fibers. Error bars represent standard deviation. nsP > 0.05, *0.01 < P < 0.05, **0.001 < P < 0.01, ***0.0001 < P < 0.001, ****P < 0.0001, two-tailed unpaired t test. (h) FTIR spectrum of stretched 50 50 HMW/LMW protein composite fibers. Stretched fiber pieces were imaged after tensile testing. Scale bars represent 10 μm. (c) Representative stress–strain curves from tensile testing of the HMW/LMW protein composite fibers at different mixture ratios. (d)–(g) Ultimate tensile stress (d), breaking strain (e), toughness (f), and Young's modulus (g) of the HMW/LMW protein composite fibers. Error bars represent standard deviation. nsP > 0.05, *0.01 < P < 0.05, **0.001 < P < 0.01, ***0.0001 < P < 0.001, ****P < 0.0001, two-tailed unpaired t test. (h) FTIR spectrum of stretched 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 HMW/LMW protein composite fibers. The amide 1 band was deconvoluted into 11 Gaussian peaks and the average β-sheet content was determined using steps outlined in the ‘Methods’ section. (i) Raman spectra of stretched 50 50 HMW/LMW protein composite fibers. The amide 1 band was deconvoluted into 11 Gaussian peaks and the average β-sheet content was determined using steps outlined in the ‘Methods’ section. (i) Raman spectra of stretched 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 HMW/LMW protein composite fibers oriented perpendicular (IY; red line) or parallel (IX; blue line) to the direction of the incident laser polarization. Spectra shown were acquired by averaging spectra from three separate fibers. Orientation sensitivity was determined by calculating the average peak intensity ratio at 1670 cm−1 (IY/X). 50 HMW/LMW protein composite fibers oriented perpendicular (IY; red line) or parallel (IX; blue line) to the direction of the incident laser polarization. Spectra shown were acquired by averaging spectra from three separate fibers. Orientation sensitivity was determined by calculating the average peak intensity ratio at 1670 cm−1 (IY/X). | ||
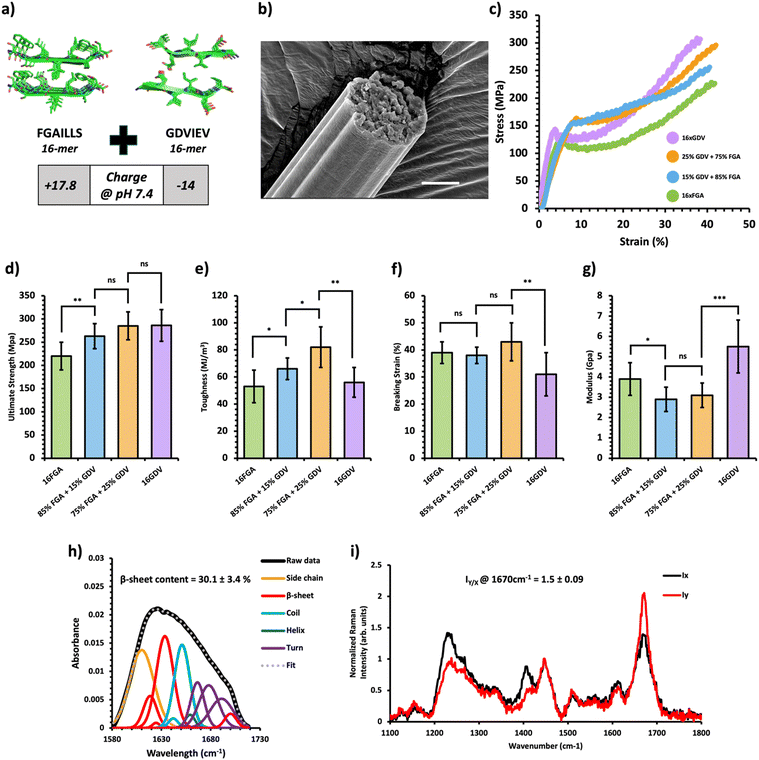 | ||
Fig. 3 Mechanical and structural properties of protein charge-based composite fibers. (a) Crystal structure of the FGAILSS (PDB: 5E61) and GDVIEV (PDB: 3SGS) amyloid peptides. Polymeric amyloid-silk proteins containing 16 repeats of the FGAILSS peptide (+17.8 charged at pH 7.4) or 16 repeats of the GDVIEV peptide (−14 charged at pH 7.4) were mixed to generate protein charge-based composite fibers. (b) Representative S.E.M. image of the 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 16xFGA/16xGDV protein composite fibers. Stretched fiber pieces were imaged after tensile testing. Scale bars represent 10 μm. (c) Representative stress–strain curves from tensile testing of the protein charge-based composite fibers at different mixture ratios. (d)–(g) Ultimate tensile stress (d), toughness (e), breaking strain (f), and Young's modulus (g) of the protein charge-based composite fibers. Error bars represent standard deviation. nsP > 0.05, *0.01 < P < 0.05, **0.001 < P < 0.01, ***0.0001 < P < 0.001, ****P < 0.0001, two-tailed unpaired t test. (h) FTIR analysis of post-spin drawn 75 25 16xFGA/16xGDV protein composite fibers. Stretched fiber pieces were imaged after tensile testing. Scale bars represent 10 μm. (c) Representative stress–strain curves from tensile testing of the protein charge-based composite fibers at different mixture ratios. (d)–(g) Ultimate tensile stress (d), toughness (e), breaking strain (f), and Young's modulus (g) of the protein charge-based composite fibers. Error bars represent standard deviation. nsP > 0.05, *0.01 < P < 0.05, **0.001 < P < 0.01, ***0.0001 < P < 0.001, ****P < 0.0001, two-tailed unpaired t test. (h) FTIR analysis of post-spin drawn 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 16xFGA/16xGDV protein composite fibers. The amide 1 band was deconvoluted into 11 Gaussian peaks and the average β-sheet content was determined using steps outlined in the ‘Methods’ section. (i) Raman spectra of stretched 75 25 16xFGA/16xGDV protein composite fibers. The amide 1 band was deconvoluted into 11 Gaussian peaks and the average β-sheet content was determined using steps outlined in the ‘Methods’ section. (i) Raman spectra of stretched 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 FGA/GDV protein composite fibers oriented perpendicular (IY; red line) or parallel (IX; blue line) to the direction of the incident laser polarization. Spectra shown were acquired by averaging spectra from three separate fibers. Orientation sensitivity was determined by calculating the average peak intensity ratio at 1670 cm−1 (IY/X). 25 FGA/GDV protein composite fibers oriented perpendicular (IY; red line) or parallel (IX; blue line) to the direction of the incident laser polarization. Spectra shown were acquired by averaging spectra from three separate fibers. Orientation sensitivity was determined by calculating the average peak intensity ratio at 1670 cm−1 (IY/X). | ||
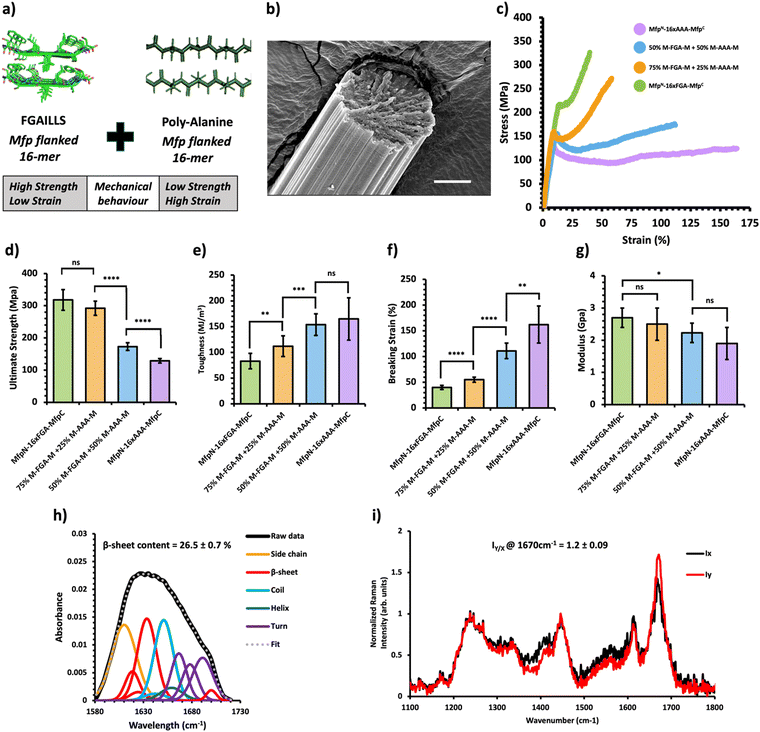 | ||
Fig. 4 Mechanical and structural properties of protein mechanical behavior-based composite fibers. (a) Crystal structure of the FGAILSS (PDB: 5E61) amyloid peptide and representative poly-alanine peptide. Polymeric silk proteins, flanked on their ends by the split mussel foot protein (Mfp5), containing 16 repeats of the FGAILSS peptide and 16 repeats of the penta-alanine peptide were mixed to generate protein mechanical behavior-based composite fibers. (b) Representative S.E.M. image of the 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 NM-16xFGA-CM/NM-16xAAA-CM protein composite fibers. Stretched fiber pieces were imaged after tensile testing. Scale bars represent 10 μm. (c) Representative stress–strain curves from tensile testing of the protein mechanical behavior-based composite fibers at different mixture ratios. (d)–(g) Ultimate tensile stress (d), toughness (e), breaking strain (f), and Young's modulus (g) of the protein charge-based composite fibers. Error bars represent standard deviation. nsP > 0.05, *0.01 < P < 0.05, **0.001 < P < 0.01, ***0.0001 < P < 0.001, ****P < 0.0001, two-tailed unpaired t test. (h) FTIR analysis of post-spin drawn 50 50 NM-16xFGA-CM/NM-16xAAA-CM protein composite fibers. Stretched fiber pieces were imaged after tensile testing. Scale bars represent 10 μm. (c) Representative stress–strain curves from tensile testing of the protein mechanical behavior-based composite fibers at different mixture ratios. (d)–(g) Ultimate tensile stress (d), toughness (e), breaking strain (f), and Young's modulus (g) of the protein charge-based composite fibers. Error bars represent standard deviation. nsP > 0.05, *0.01 < P < 0.05, **0.001 < P < 0.01, ***0.0001 < P < 0.001, ****P < 0.0001, two-tailed unpaired t test. (h) FTIR analysis of post-spin drawn 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 NM-16xFGA-CM/NM-16xAAA-CM protein composite fibers. The amide 1 band was deconvoluted into 11 Gaussian peaks and the average β-sheet content was determined using steps outlined in the ‘Methods’ section. (i) Raman spectra of stretched 50 50 NM-16xFGA-CM/NM-16xAAA-CM protein composite fibers. The amide 1 band was deconvoluted into 11 Gaussian peaks and the average β-sheet content was determined using steps outlined in the ‘Methods’ section. (i) Raman spectra of stretched 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 NM-16xFGA-CM/NM-16xAAA-CM protein composite fibers oriented perpendicular (IY; red line) or parallel (IX; blue line) to the direction of the incident laser polarization. Spectra shown were acquired by averaging spectra from three separate fibers. Orientation sensitivity was determined by calculating the average peak intensity ratio at 1670 cm−1 (IY/X). 50 NM-16xFGA-CM/NM-16xAAA-CM protein composite fibers oriented perpendicular (IY; red line) or parallel (IX; blue line) to the direction of the incident laser polarization. Spectra shown were acquired by averaging spectra from three separate fibers. Orientation sensitivity was determined by calculating the average peak intensity ratio at 1670 cm−1 (IY/X). | ||
A previous study on amyloid-silk hybrid protein fibers showed clear MW dependent mechanical properties.31 Higher MW amyloid-silk proteins (up to 378 kDa) formed fibers with superior ultimate tensile strength.31 We first prepared HMW/LMW composite fibers by blending two amyloid-silk hybrid proteins of differing MWs: one with 16 repeats of the FGAILLS-silk peptide (16xFGA) (MW = 49 kDa) and the other with 48 repeats of the same peptide (48xFGA) (MW = 144 kDa) (Fig. 2a). The HMW protein fibers (48xFGA) displayed greater ultimate strength and toughness compared to the LMW fibers (16xFGA), while exhibiting similar extensibility and elastic moduli (Fig. 2c–g). We then varied the mixture ratios to 75/25, 50/50, and 25/75 of the constituent proteins, aimed at elucidating key trends in their mechanical behavior (Fig. 2c, Table 1 and Fig. S5, Tables S2–S6, ESI†).
| Blend type | Sample name | Ultimate strength (MPa) | Toughness (MJ m−3) | Breaking strain (%) | Modulus (GPa) | Diameter (μm) |
|---|---|---|---|---|---|---|
| Molecular weight based | 100 LMW | 221 ± 32 | 53 ± 12 | 38 ± 4 | 3.8 ± 0.9 | 18 ± 1.5 |
75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 LMW/HMW 25 LMW/HMW |
256 ± 12 | 69 ± 16 | 39 ± 6 | 3.6 ± 1.0 | 18 ± 0.9 | |
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 LMW/HMW 50 LMW/HMW |
281 ± 14 | 88 ± 18 | 44 ± 7 | 2.6 ± 0.8 | 19 ± 0.9 | |
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75 LMW/HMW 75 LMW/HMW |
316 ± 11 | 69 ± 14 | 33 ± 7 | 3.4 ± 0.5 | 17 ± 0.7 | |
| 100 HMW | 428 ± 29 | 115 ± 24 | 42 ± 6 | 4.2 ± 0.5 | 14 ± 0.3 | |
| Protein charge based | FGA | 221 ± 32 | 53 ± 12 | 38 ± 4 | 3.8 ± 0.9 | 18 ± 1.5 |
85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 FGA/GDV 15 FGA/GDV |
264 ± 27 | 66 ± 08 | 38 ± 3 | 2.9 ± 0.6 | 27 ± 2.2 | |
75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 FGA/GDV 25 FGA/GDV |
286 ± 30 | 82 ± 15 | 43 ± 7 | 3.1 ± 0.6 | 27 ± 1.1 | |
| GDV | 286 ± 34 | 56 ± 11 | 31 ± 9 | 5.5 ± 1.3 | 11 ± 0.7 | |
| Mechanical behavior based | M-FGA-M | 318 ± 32 | 83 ± 15 | 40 ± 4 | 2.7 ± 0.3 | 31 ± 1.1 |
75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 M-FGA-M/M-AAA-M 25 M-FGA-M/M-AAA-M |
292 ± 22 | 112 ± 20 | 55 ± 5 | 2.5 ± 0.5 | 31 ± 1.3 | |
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 M-FGA-M/M-AAA-M 50 M-FGA-M/M-AAA-M |
173 ± 12 | 154 ± 21 | 111 ± 15 | 2.2 ± 0.3 | 30 ± 2.2 | |
| M-AAA-M | 129 ± 07 | 165 ± 41 | 163 ± 37 | 1.9 ± 0.5 | 39 ± 1.5 |
We found that as the HMW 48xFGA composition increases, the fibers’ ultimate tensile strength and toughness showed almost linear growth. Fibers containing a minimum of 25% HMW protein exhibited a slightly elevated ultimate tensile strength (256 ± 12 MPa) compared to those comprising only the LMW protein (221 ± 32 MPa) (Fig. 2d). Further addition of HMW proteins led to significant enhancements in ultimate strength, reaching up to 281 ± 14 MPa at a 50% HMW ratio, and 316 ± 11 MPa at a 75% HMW ratio (Fig. 2d). Composite fibers of HMW/LMW amyloid-silk proteins displayed similar ultimate strains (∼38–44%), with the exception of the 75% HMW fibers, which exhibited a slightly reduced breaking strain of ∼33 ± 7% (Fig. 2e). This minor fluctuation in the breaking strain could be attributed to microscale defects within fibers as well as strain localization during mechanical loading which are beyond manual control. Fiber toughness followed a similar trend to fiber strength, exhibiting remarkable enhancement with increased compositions of HMW proteins. For instance, fibers from a 50% HMW blend (88 ± 18 MJ m−3) showed a 69% increase in toughness compared to LMW 16xFGA fibers (52 ± 10 MJ m−3) (Fig. 2f). Additionally, we observed that the initial moduli of the composite fibers were noticeably lower than those of pure protein fibers. Notably, the modulus of the 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 HMW/LMW fibers (2.6 ± 0.8 GPa) suffered a substantial decline of about 33% and 38% compared to that of 100% LMW (3.9 ± 0.8 GPa) and 100% HMW (4.2 ± 0.5 GPa) fibers, respectively (Fig. 2g). This decline in modulus in composite fibers has been previously observed and might be attributed to disrupted individual crystallization of each protein compared to pure protein fibers.26 Meanwhile, co-crystallization of the two proteins may lead to the formation of mixed crystals with less dense packing and increased molecular irregularities,33 as suggested by the increased diameters of our composite fibers. In general, composite amyloid-silk fibers of HMW/LMW exhibit increasing ultimate strength and toughness as the composition of HMW proteins increases, while the ultimate strains of the composite fibers are not affected by protein composition.
50 HMW/LMW fibers (2.6 ± 0.8 GPa) suffered a substantial decline of about 33% and 38% compared to that of 100% LMW (3.9 ± 0.8 GPa) and 100% HMW (4.2 ± 0.5 GPa) fibers, respectively (Fig. 2g). This decline in modulus in composite fibers has been previously observed and might be attributed to disrupted individual crystallization of each protein compared to pure protein fibers.26 Meanwhile, co-crystallization of the two proteins may lead to the formation of mixed crystals with less dense packing and increased molecular irregularities,33 as suggested by the increased diameters of our composite fibers. In general, composite amyloid-silk fibers of HMW/LMW exhibit increasing ultimate strength and toughness as the composition of HMW proteins increases, while the ultimate strains of the composite fibers are not affected by protein composition.
Blending amyloid-silk proteins of opposite charges enhanced fiber toughness
Next, we created composite fibers by blending amyloid-silk proteins with opposite charges to investigate the influence of electrostatic interactions on composite fibers. We selected the 16xFGA protein as the positively charged component (+17.8 charge at pH 7.4) and the 16xGDV protein, featuring the GDVIEV amyloid sequence, as the negatively charged component (−14 charged at pH 7.4) (Note S1, ESI†). Pure protein fibers from 16xFGA and 16xGDV displayed similar toughness while the 16xGDV fibers exhibited a 30% increase in ultimate strength over the 16xFGA fibers (Fig. 3d and e). Mixing these two proteins with over 50% 16xGDV did not lead to fiber formation, likely due to pronounced electrostatic effects causing protein aggregation within the spinning dope.Consequently, we explored composite fibers comprising only 15% and 25% 16xGDV (Fig. 3c, Table 1 and Fig. S6, Tables S7–S9, ESI†). These composite fibers displayed significantly higher toughness (66–82 MJ m−3) compared to both pure 16xFGA (53 ± 12 MJ m−3) and 16xGDV (56 ± 11 MJ m−3) fibers (Fig. 3e). Notably, fibers containing 25% 16xGDV were 47–55% tougher than both 16xFGA and 16xGDV fibers. Additionally, the composite fibers displayed significantly higher ultimate tensile strength compared to pure 16xFGA fibers (Fig. 3e). Incorporating 15% 16xGDV protein increased the fiber strength by 20% (263 ± 27 MPa) compared to pure 16xFGA fibers (221 ± 32 MPa) (Fig. 3d). Further addition of 16xGDV up to 25% resulted in fibers (285 ± 34 MPa) closely resembling the ultimate strength of pure 16xGDV fibers (286 ± 34 MPa) (Fig. 3d). This enhancement in ultimate strength, even with a marginal addition of the 16xGDV protein, likely stems from electrostatic attractions between the oppositely charged proteins. Moreover, the breaking strains of the composite fibers were comparable (38–43%) to pure 16xFGA fibers and slightly higher than those of pure 16xGDV fibers (31%) (Fig. 3f). The initial moduli of charge-based composites (2.9–3.1 GPa) were significantly lower than those of both pure 16xFGA (3.9 ± 0.8 GPa) and 16xGDV (5.5 ± 1.3 GPa) fibers (Fig. 3g). This trend mirrors observations in HMW/LMW composite fibers. In summary, all charge-based composites demonstrated substantial improvements in fiber toughness, with ultimate strengths and breaking strains falling between those of pure protein fibers.
Blending proteins with distinct mechanical characteristics leads to composite fibers with customized mechanical properties
Previous research has shown that bi-terminal fusion of cohesive mussel foot protein (bt-Mfp) fragments enhances fiber strength.32 Intriguingly, these bt-Mfp fibers exhibit different stress–strain curves. The bt-Mfp fused spider silk protein containing 16 repeats of the polyalanine peptide (16xAAA), denoted as NM-16xAAA-CM, exhibited high ultimate strain but low ultimate strength, whereas the bt-Mfp fused 16xFGA (NM-16xFGA-CM) displayed higher ultimate strength but lower ultimate strain, demonstrating distinct mechanical behaviors. Notably, the strain-hardening behavior observed in NM-16xFGA-CM fibers post the yielding point is absent in NM-16xAAA-CM fibers. Consequently, blending NM-16xAAA-CM and NM-16xFGA-CM proteins enabled an investigation into the effects of combining proteins with contrasting mechanical behaviors (Fig. 4a).Upon mixing these proteins, the resultant composite fibers displayed intermediate performance, with their stress–strain curves located in between the two pure protein fibers (Fig. 4c, Table 1 and Fig. S7, Tables S10–S13, ESI†). As the proportion of the NM-16xFGA-CM protein increased in the composite fibers, they exhibited higher ultimate strength but lower ultimate strain compared to pure NM-16AAA-CM fibers (Fig. 4d and f). Moreover, the toughness of composite fibers also decreased with increasing proportion of the NM-16xFGA-CM protein in the fibers. Notably, fibers containing 50% NM-16xFGA-CM displayed a weak strain-hardening behavior with an ultimate tensile strength of 173 ± 12 MPa, 34% higher than that of the pure NM-16AAA-CM protein fibers (129 ± 7 MPa) and a toughness of 154 ± 21 MJ m−3, 86% higher than that of the pure NM-16xFGA-CM fibers (83 ± 15 MJ m−3, Fig. 4d). These data show that each composite fiber displayed a unique mechanical behavior heavily modulated by the ratio of the component proteins. Overall, we observed that ultimate strength and modulus were dictated by NM-16xFGA-CM while extensibility and toughness were controlled by NM-16xAAA-CM proteins. Thus, mixing the NM-16xFGA-CM and the NM-16xAAA-CM proteins provides an opportunity to tailor fibers with specific mechanical properties and stress–strain curves, merely by adjusting the protein ratio.
Composite fibers display a similar molecular structure to pure fibers
The tunable mechanical behavior of amyloid-silk protein composite fibers motivated us to investigate into the underlying structural features that contribute to their unique characteristics. We employed Fourier-Transform Infrared (FT-IR) Spectroscopy and polarized Raman spectromicroscopy to examine the underlying mechanisms of these composite fibers, revealing information on the relative β-sheet content and their alignment along the fiber axis, respectively. FT-IR analysis unveiled a significant presence of β-sheet secondary structures within the composite fibers, estimating a β-sheet content ranging from 26.5–30% across all composite fibers through deconvolution of the amide I peak (Fig. 2–4h). Notably, there was no significant disparity in β-sheet content between the composite fibers and pure protein fibers (Fig. S8 and S9, ESI†), indicating that blending these proteins prior to the spinning process does not appreciably affect β-sheet formation within the constituent proteins. This consistent formation of β-sheets suggests that other underlying mechanisms contribute to the variations in mechanical properties among these composite fibers. Using polarized Raman spectromicroscopy, we further examined the amide I β-sheet peak (1670 cm−1) and compared the Raman spectra when fibers were oriented either parallel (x-direction) or perpendicular (y-direction) to the direction of laser polarization. Interestingly, all three types of composite fibers displayed a strong alignment of β-sheets along the fiber axes, with the amide I β-sheet peak exhibiting significantly higher intensity at the perpendicular position (Iy) compared to the parallel position (Ix) (Fig. 2–4i). Specifically, the amide I β-sheet peak of both HMW/LMW composite fibers and charge-based composite fibers exhibited a substantial difference between the perpendicular and parallel orientations, as indicated by a peak intensity ratio (IY/X) of 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ±
± ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.09 (Fig. 2i and 3i). Remarkably, the peak intensity ratios of all composite fibers were similar to those of pure protein fibers reinforcing that blending processes do not disrupt the alignment of β-sheets within the fiber (Fig. S10 and S11, ESI†). In essence, these findings revealed that composite fibers maintain similar molecular structures to pure protein fibers, emphasizing the pivotal role of protein composition in fine-tuning the mechanical properties of blended fibers.
0.09 (Fig. 2i and 3i). Remarkably, the peak intensity ratios of all composite fibers were similar to those of pure protein fibers reinforcing that blending processes do not disrupt the alignment of β-sheets within the fiber (Fig. S10 and S11, ESI†). In essence, these findings revealed that composite fibers maintain similar molecular structures to pure protein fibers, emphasizing the pivotal role of protein composition in fine-tuning the mechanical properties of blended fibers.
Discussion
Our findings demonstrated that blending different proteins impacts fiber properties differently, particularly in terms of ultimate tensile strength, toughness, and initial modulus. The observed differences in fiber strength arise from the protein blend composition and their mixing ratio rather than the β-sheet content and orientation. In the case of molecular weight-based composites, an increase in the HMW protein ratio increases the probability of inter-chain interactions and physical entanglements while decreasing chain-end defects, all of which contribute to an improvement in strength. In the case of charge-based composites, the addition of an oppositely charged protein promotes the formation of inter-chain interactions due to electrostatic attraction, thus improving fiber strength. In mechanical behavior-based composites, the addition of the NM-16xFGA-CM protein enhances fiber strength due to its increased stiffness compared to the NM-16xAAA-CM protein.When blending oppositely charged 16xFGA and 16xGDV proteins, the behavior of protein mixtures was found to be influenced by the ratio of the two proteins. At smaller ratios (<25% of 16xGDV), the blended proteins successfully formed composite fibers with increased toughness due to the additional energy required to break the electrostatic interactions between the oppositely charged protein chains. However, at higher ratios (>25% of 16xGDV), the intense interactions between the two proteins result in protein aggregation in the dope, rendering it impossible to spin these aggregates into fibers. Furthermore, blending proteins with different MWs or mechanical behaviors offered a straightforward strategy to fine tune fiber mechanical properties. By simply adjusting protein proportions, we precisely controlled properties such as fiber ultimate strength, strain, toughness, modulus, and strain-hardening behavior to meet specific requirements, enabling diverse applications.
Taken together, our results validated the concept of blending amyloid-silk proteins to create composite protein fibers with tailored properties, showcasing exceptional tunability in fiber mechanical properties without the need for synthesizing complex hybrid proteins. This protein blending strategy can be potentially extended to other fiber proteins. The observed trends in mechanical properties offer valuable guidance for future engineering of composite fibers, leading to tunable material properties. Furthermore, multi-functional materials that combine the attractive properties of multiple proteins have broad applications. Current methods in creating multi-functional materials focus on engineering hybrid proteins containing peptide segments of different origins and properties.30,34–36 While powerful, this strategy often encounters challenges in both protein design and synthesis.9 Despite recent advancements in synthetic biology addressing issues related to material protein expression9,11 and facilitating microbial production of numerous complex products,37–41 engineering multifunctional hybrid proteins sometime involve trade-offs between different material properties.42 Here we show that blending proteins with different properties offer a simple alternative, bypassing the limitations in designing and synthesizing complex hybrid proteins. These precisely controlled mechanical properties of composite protein fibers hold promise across structural materials and biomedical applications, signaling potential advancements in various domains.
Methods
Chemicals and reagents
All chemicals and reagents were purchased from MilliporeSigma (Burlington, MA), unless explicitly stated otherwise. All proteins were purified using Ni-NTA columns purchased from GE Healthcare (Chicago, IL).Strains, plasmids, and growth conditions
Escherichia coli strain NEB 10-beta (NEB10β) transformed with a suitable plasmid was used for protein production. Proteins 16xFGA, 48xFGA, 16xGDV, NM-16xFGA-CM and NM-16xAAA-CM were expressed from plasmids pJL464, pJL48, pJL523, pJL85 and pJL83 reported in previous studies.31,32 During protein expression, E. coli strains were grown in Terrific Broth (TB) containing 24 g L−1 of yeast extract, 20 g L−1 of tryptone, 0.4% v/v glycerol, 17 mM KH2PO4, and 72 mM K2HPO4, with incubation at 37 °C and constant shaking. Appropriate antibiotics (50 μg mL−1 ampicillin) were added as needed.Protein expression in shake flasks
A single colony of a suitable strain was inoculated into TB medium and cultivated at 37 °C on an orbital shaker. Subsequently, this culture was used to inoculate a fresh TB medium, which was allowed to grow until it reached an OD600 of 0.6. To induce protein expression, 0.04% arabinose was added, and the culture was continued at 37 °C for 24 hours. Afterward, cells were harvested by centrifugation at 3500 rpm, and the resulting cell pellets were preserved at −80 °C for later use.Protein purification
Cell pellets were subjected to lysis in buffer A (6 M guanidine hydrochloride, 300 mM NaCl, 50 mM K2HPO4 at pH 8.0) for a duration of 12 hours at 4 °C with continuous stirring. The lysate was then centrifuged, and the resulting supernatant was loaded onto a Ni-NTA column. This column was subsequently washed sequentially with buffer B (8 M urea, 300 mL of NaCl, 50 mM K2HPO4 at pH 8.0) using imidazole concentrations of 0, 20, and 50 mM. The polymeric amyloid-silk proteins were eluted using buffer B containing 300 mM imidazole. All the purified proteins were dialyzed in 1% acetic acid, lyophilized, and stored at −80 °C for later use.SDS-PAGE analysis
All SDS-PAGE gels contain a 5% stacking gel at the top and a separation gel with indicated percentages at the bottom. Samples were prepared in Laemmli sample buffer (2% SDS, 10% glycerol, 60 mM Tris at pH 6.8, 0.01% bromophenol blue, and 100 μM DTT). The gels were electrophoresed using Mini-PROTEAN Tetra Cells from Bio-Rad in 1× Tris-glycine S.D.S. buffer (25 mM Tris, 250 mM glycine, and 0.1% w/v SDS) until the dye front exits the gel.Protein charge calculation
The protein charges reported in this study were determined using an online calculator tool (https://www.protpi.ch). The protein charge was calculated for 16-mer amyloid-silk proteins including the amyloid peptides, linker regions and His-tag.Fiber spinning
Lyophilized protein powders were dissolved in HFIP to create spinning dopes at a concentration of 15% w/w. For protein blends, the two proteins were mixed in desired ratios before dissolving in HFIP. These dopes were loaded into a 100 μL syringe from Hamilton Robotics and extruded slowly into a bath of 95% v/v methanol. The extrusion was controlled by a Harvard Apparatus Pump 11 Elite Syringe Pump from Harvard Apparatus, which delivered the dopes at a rate of 10 μL min−1. The extruded fibers were then placed in a 75% v/v methanol bath and gently elongated up to 4–6 times their original length just before reaching the point of fracture. Specifically, MW-based composite fibers and charge-based composite fibers were stretched to about 4.5 times their original length while mechanical behavior-based composite fibers were stretched to about 5.5 times their original length. After extension, the fibers were removed from the methanol bath and allowed to air-dry.Light microscopy
Fiber diameters were assessed utilizing a Zeiss Axio Observer ZI inverted microscope, which was equipped with a 20× phase-contrast objective lens. The measurements were quantified using Axiovision LE software from Zeiss.SEM (scanning electron microscopy)
After tensile tests, fibers were affixed to a sample holder using conductive tapes. This holder was then coated with a 10 nm layer of gold using a Leica EM ACE600 high-vacuum sputter coater (Leica Microsystems). Subsequently, the fibers were imaged with a Nova NanoSEM 230 field emission scanning electron microscope from the Field Electron and Ion Company (FEI). These images were acquired at an accelerating voltage of 10 kV.Tensile testing
Sections of post-drawn fibers, each measuring 20 mm in length, were deliberately placed in a precisely vertical orientation across a rectangular opening cut into a 20 mm by 20 mm piece of cardstock. This opening had dimensions of 5 mm in height (vertical) and 15 mm in width (horizontal). The fibers were secured in place using adhesive tape at both ends of the opening. Tensile tests were carried out using an MTS Criterion Model 41 Universal Test Frame equipped with a 1 N load cell from MTS Systems Corporation. The tests were conducted at a room temperature of 25 °C and a relative humidity of 20%, with a constant pulling speed of 10 mm min−1. The ultimate tensile strength was calculated as the maximum load applied to the initial cross-sectional area of the fiber, assuming that the cross-section was circular. The modulus was determined as the slope of the initial elastic region on the stress–strain curve using linear least-squares fitting. The ultimate breaking strain was calculated as the percentage of fiber elongation relative to the initial gauge length (5 mm) before the fiber broke. The toughness was computed as the area under the entire stress–strain curve. Stress–strain curves were recorded by the MTS TW Elite Test Suite at a sampling rate of 50 Hz. All statistical analyses were performed by determining the p-values using two-tailed unpaired t-test between each pair of fibers.Fourier transform infrared (FT-IR) spectroscopy
FT-IR spectra were obtained using a Thermo Nicolet 470 FT-IR spectrometer equipped with a Smart Performer ATR accessory containing a Ge crystal. Spectra were recorded in the range of 1415–1780 cm−1 with a resolution of 2 cm−1. Each single fiber sample was subjected to 254 scans. All spectra were processed using Fityk 0.9.865, where baseline corrections were applied using Fityk's convex hull algorithm.43 The amide I band (1600–1700 cm−1) was deconvoluted into eleven Gaussian peaks centered at specific wavenumbers (1610, 1618.5, 1624.5, 1632.5, 1642, 1651, 1659, 1666.5, 1678, 1690.5, and 1700 cm−1) corresponding to various secondary structures such as α-sheet, random coil, β-helix, or β-turn structures.30,44 Peak areas were integrated, and the component percentages were calculated by dividing the area of each component peak by the sum of all peak areas. These percentages were averaged based on measurements of three fibers for each condition.Polarized Raman spectromicroscopy
The method used for polarized Raman spectromicroscopy was adapted from previous studies on molecular alignment in spider silk fibers.45,46 Single composite fiber pieces were securely affixed to glass microscope slides marked with microscale indicators to ensure that spectra were acquired from the same location before and after stage rotation. Raman spectra were collected using a Renishaw RM1000 InVia Confocal Raman Spectrometer connected to a Leica DM LM microscope featuring a rotating stage. Initially, fibers were oriented along the x-axis, parallel to the laser polarization. A fixed point on the fibers was irradiated using the 514 nm argon laser line with the laser polarization oriented along the x-axis. This was done through a 50× objective (NA = 0.75), and spectra were recorded in the range of 1100–1800 cm−1 with an 1800 lines per mm grating. Each acquisition involved accumulating a total of 10 spectra, each recorded for 10 seconds. Subsequently, the stage was rotated to align the fibers along the y-axis, while the laser polarization remained the same, and spectra were recorded again at the same fixed point. No signs of thermal degradation were observed, either visually or within the recorded spectra. Fityk 0.9.865 was used for baseline corrections, employing its automatic convex hull algorithm.43 For intensity ratio calculations, all spectra were normalized to the intensity of the 1450 cm−1 peak, stemming from CH2 bending and insensitive to protein conformation.45 For each fiber, the normalized intensity of the peak at 1670 cm−1 when oriented along the Y-axis was divided by the normalized intensity of the same peak when oriented along the X-axis, resulting in the intensity ratio I(Y/X). This procedure was conducted on three separate fibers for each condition, and the calculated intensity ratios were averaged.Author contributions
FZ and SVS conceived the project. SVS performed cell culture, protein expression, protein purification, SDS-PAGE, fiber spinning, light microscopy, and fiber mechanical testing. SVS and JL performed FTIR and Raman spectroscopy. KZL performed Scanning Electron Microscopy. NF performed cell culture, protein expression, and protein purification. SVS and FZ prepared the manuscript with comments from all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank Dr Srikanth Singamaneni and Yixuan Wang for their assistance with the Raman spectrometry. SEM images were acquired within the facilities of IMSE at Washington University. This work is funded by the United States Department of Agriculture (grant number 20196702129943 to FZ) and the National Science Foundation (award numbers DMR-2105150, DMR-2207879, and OIA-2219142 to FZ).References
- U. G. K. Wegst, H. Bai, E. Saiz, A. P. Tomsia and R. O. Ritchie, Bioinspired structural materials, Nat. Mater., 2015, 14(1), 23–36, DOI:10.1038/nmat4089
.
- K. Numata, How to define and study structural proteins as biopolymer materials, Polym. J., 2020, 52(9), 1043–1056, DOI:10.1038/s41428-020-0362-5
.
- F. G. Omenetto and D. L. Kaplan, New Opportunities for an Ancient Material, Science, 2010, 329(5991), 528–531, DOI:10.1126/science.1188936
.
- M. S. Z. Kellermayer, S. B. Smith, H. L. Granzier and C. Bustamante, Folding-Unfolding Transitions in Single Titin Molecules Characterized with Laser Tweezers, Science, 1997, 276(5315), 1112–1116, DOI:10.1126/science.276.5315.1112
.
- T. Priemel, E. Degtyar, M. N. Dean and M. J. Harrington, Rapid self-assembly of complex biomolecular architectures during mussel byssus biofabrication, Nat. Commun., 2017, 8(1), 14539, DOI:10.1038/ncomms14539
.
- G. Qin, X. Hu, P. Cebe and D. L. Kaplan, Mechanism of resilin elasticity, Nat. Commun., 2012, 3(1), 1003, DOI:10.1038/ncomms2004
.
- Y. J. Yang, A. L. Holmberg and B. D. Olsen, Artificially Engineered Protein Polymers, Annu. Rev. Chem. Biomol. Eng., 2017, 8, 549–575 CrossRef CAS PubMed
.
- J. C. M. Van Hest and A. Tirrell, Protein-based materials, toward a new level of structural control, Chem. Commun., 2001, 1897–1904, 10.1039/B105185G
.
- J. Jeon, S. V. Subramani, K. Z. Lee, B. Jiang and F. Zhang, Microbial Synthesis of High-Molecular-Weight, Highly Repetitive Protein Polymers, Int. J. Mol. Sci., 2023, 24(7), 6416, DOI:10.3390/ijms24076416
.
- K. Z. Lee, J. Jeon, B. Jiang, S. V. Subramani, J. Li and F. Zhang, Protein-Based Hydrogels and Their Biomedical Applications, Molecules, 2023, 28(13), 4988, DOI:10.3390/molecules28134988
.
- C. H. Bowen, T. J. Reed, C. J. Sargent, B. Mpamo, J. M. Galazka and F. Zhang, Seeded Chain-Growth Polymerization of Proteins in Living Bacterial Cells, ACS Synth. Biol., 2019, 8(12), 2651–2658, DOI:10.1021/acssynbio.9b00362
.
- C. H. Bowen, B. Dai and C. J. Sargent,
et al., Recombinant Spidroins Fully Replicate Primary Mechanical Properties of Natural Spider Silk, Biomacromolecules, 2018, 19(9), 3853–3860, DOI:10.1021/acs.biomac.8b00980
.
- E. Kim, B. Dai, J. B. Qiao, W. Li, J. D. Fortner and F. Zhang, Microbially Synthesized Repeats of Mussel Foot Protein Display Enhanced Underwater Adhesion, ACS Appl. Mater. Interfaces, 2018, 10(49), 43003–43012, DOI:10.1021/acsami.8b14890
.
- W. Bai, C. J. Sargent, J. M. Choi, R. V. Pappu and F. Zhang, Covalently-assembled single-chain protein nanostructures with ultra-high stability, Nat. Commun., 2019, 10(1), 3317, DOI:10.1038/s41467-019-11285-8
.
- C. H. Bowen, C. J. Sargent and A. Wang,
et al., Microbial production of megadalton titin yields fibers with advantageous mechanical properties, Nat. Commun., 2021, 12(1), 5182, DOI:10.1038/s41467-021-25360-6
.
- E. Novaes, M. Kirst, V. Chiang, H. Winter-Sederoff and R. Sederoff, Lignin and Biomass: A Negative Correlation for Wood Formation and Lignin Content in Trees, Plant Physiol., 2010, 154(2), 555–561, DOI:10.1104/pp.110.161281
.
- X. Feng, Chemical and Biochemical Basis of Cell-Bone Matrix Interaction in Health and Disease, Curr. Chem. Biol., 2009, 3(2), 189–196, DOI:10.2174/187231309788166398
.
- A. Hagenau, H. A. Scheidt, L. Serpell, D. Huster and T. Scheibel, Structural Analysis of Proteinaceous Components in
Byssal Threads of the Mussel Mytilus galloprovincialis, Macromol. Biosci., 2009, 9(2), 162–168, DOI:10.1002/mabi.200800271
.
- A. Sponner, W. Vater, S. Monajembashi, E. Unger, F. Grosse and K. Weisshart, Composition and Hierarchical Organisation of a Spider Silk, PLoS One, 2007, 2(10), e998, DOI:10.1371/journal.pone.0000998
.
- D. L. Kaplan, Mollusc shell structures: novel design strategies for synthetic materials, Curr. Opin. Solid State Mater. Sci., 1998, 3(3), 232–236, DOI:10.1016/S1359-0286(98)80096-X
.
- S. Du, J. Zhang and W. T. Zhou,
et al., Interactions between fibroin and sericin proteins from Antheraea pernyi and Bombyx mori silk fibers, J. Colloid Interface Sci., 2016, 478, 316–323, DOI:10.1016/j.jcis.2016.06.030
.
-
L. E. Murr, in Examples of Natural Composites and Composite Structures, Handbook of Materials Structures, Properties, Processing and Performance, ed. L. E. Murr, Springer International Publishing, 2015, pp. 425–449 Search PubMed
.
-
S. Mitra, P. Bhattacharya, S. Sen and T. N. Patel, Protein-based composites and their applications, Green Sustainable Process for Chemical and Environmental Engineering and Science, 2023, pp. 205–250 DOI:10.1016/B978-0-323-95183-8.00006-8
.
- X. Hu, P. Cebe, A. S. Weiss, F. Omenetto and D. L. Kaplan, Protein-based composite materials, Mater. Today, 2012, 15(5), 208–215, DOI:10.1016/S1369-7021(12)70091-3
.
- C. Y. Hayashi, N. H. Shipley and R. V. Lewis, Hypotheses that correlate the sequence, structure, and mechanical properties of spider silk proteins, Int. J. Biol. Macromol., 1999, 24(2–3), 271–275, DOI:10.1016/S0141-8130(98)00089-0
.
- Y. Yao, B. J. Allardyce and R. Rajkhowa,
et al., Spinning Regenerated Silk Fibers with Improved Toughness by Plasticizing with Low Molecular Weight Silk, Biomacromolecules, 2021, 22(2), 788–799, DOI:10.1021/acs.biomac.0c01545
.
- B. Zhu, W. Li, R. V. Lewis, C. U. Segre and R. Wang, E-Spun Composite Fibers of Collagen and Dragline Silk Protein: Fiber Mechanics, Biocompatibility, and Application in Stem Cell Differentiation, Biomacromolecules, 2015, 16(1), 202–213, DOI:10.1021/bm501403f
.
- J. M. Caves, V. A. Kumar and A. W. Martinez,
et al., The use of microfiber composites of elastin-like protein matrix reinforced with synthetic collagen in the design of vascular grafts, Biomaterials, 2010, 31(27), 7175–7182, DOI:10.1016/j.biomaterials.2010.05.014
.
- L. Zhao, J. Li and L. Zhang,
et al., Biosynthetic protein and nanocellulose composite fibers with extraordinary mechanical performance, Nano Today, 2022, 44, 101485, DOI:10.1016/j.nantod.2022.101485
.
- B. Dai, C. J. Sargent, X. Gui, C. Liu and F. Zhang, Fibril Self-Assembly of Amyloid–Spider Silk Block Polypeptides, Biomacromolecules, 2019, 20(5), 2015–2023, DOI:10.1021/acs.biomac.9b00218
.
- J. Li, Y. Zhu, H. Yu, B. Dai, Y. S. Jun and F. Zhang, Microbially Synthesized Polymeric Amyloid Fiber Promotes β-Nanocrystal Formation and Displays Gigapascal Tensile Strength, ACS Nano, 2021, 15(7), 11843–11853, DOI:10.1021/acsnano.1c02944
.
- J. Li, B. Jiang, X. Chang, H. Yu, Y. Han and F. Zhang, Bi-terminal fusion of intrinsically-disordered mussel foot protein fragments boosts mechanical strength for protein fibers, Nat. Commun., 2023, 14(1), 2127, DOI:10.1038/s41467-023-37563-0
.
-
M. Pracella, in Crystallization of Polymer Blends, Handbook of Polymer Crystallization, ed. E. Piorkowska, G. C. Rutledge, Wiley, 1st edn, 2013, pp. 287–326 Search PubMed
.
- K. Koh, J. K. Wang and J. X. Y. Chen,
et al., Squid Suckerin-Spider Silk Fusion Protein Hydrogel for Delivery of Mesenchymal Stem Cell Secretome to Chronic Wounds, Adv. Healthcare Mater., 2023, 12(1), 2201900 CrossRef CAS PubMed
.
- E. Kim, J. Jeon and Y. Zhu,
et al., A Biosynthetic Hybrid Spidroin-Amyloid-Mussel Foot Protein for Underwater Adhesion on Diverse Surfaces, ACS Appl. Mater. Interfaces, 2021, 13(41), 48457–48468, DOI:10.1021/acsami.1c14182
.
- M. Saric and T. Scheibel, Two-in-One Spider Silk Protein with Combined Mechanical Features in All-Aqueous Spun Fibers, Biomacromolecules, 2023, 24(4), 1744–1750, DOI:10.1021/acs.biomac.2c01500
.
- W. Bai, W. Geng, S. Wang and F. Zhang, Biosynthesis, regulation, and engineering of microbially produced branched biofuels, Biotechnol. Biofuels, 2019, 12(1), 84, DOI:10.1186/s13068-019-1424-9
.
- W. Jiang, P. Gu and F. Zhang, Steps towards ‘drop-in’ biofuels: focusing on metabolic pathways, Curr. Opin. Biotechnol., 2018, 53, 26–32, DOI:10.1016/j.copbio.2017.10.010
.
- W. Jiang, J. B. Qiao, G. J. Bentley, D. Liu and F. Zhang, Modular pathway engineering for the microbial production of branched-chain fatty alcohols, Biotechnol. Biofuels, 2017, 10(1), 244, DOI:10.1186/s13068-017-0936-4
.
- A. Cravens, J. Payne and C. D. Smolke, Synthetic biology strategies for microbial biosynthesis of plant natural products, Nat. Commun., 2019, 10(1), 2142, DOI:10.1038/s41467-019-09848-w
.
- J. V. Pham, M. A. Yilma and A. Feliz,
et al., A Review of the Microbial Production of Bioactive Natural Products and Biologics, Front Microbiol., 2019, 10, 1404, DOI:10.3389/fmicb.2019.01404
.
- J. Jeon, K. Z. Lee and X. Zhang,
et al., Genetically Engineered Protein-Based Bioadhesives with Programmable Material Properties, ACS Appl Mater Interfaces, 2023, 15(49), 56786–56795, DOI:10.1021/acsami.3c12919
.
- M. Wojdyr, Fityk: a general-purpose peak fitting program, J. Appl. Cryst., 2010, 43(5), 1126–1128, DOI:10.1107/S0021889810030499
.
- B. D. Lawrence, F. Omenetto, K. Chui and D. L. Kaplan, Processing methods to control silk fibroin film biomaterial features, J. Mater. Sci., 2008, 43(21), 6967–6985, DOI:10.1007/s10853-008-2961-y
.
- T. Lefèvre, F. Paquet-Mercier, J. F. Rioux-Dubé and M. Pézolet, Review structure of silk by Raman spectromicroscopy: from the spinning glands to the fibers, Biopolymers, 2012, 97(6), 322–336, DOI:10.1002/bip.21712
.
- Z. Shao, F. Vollrath, J. Sirichaisit and R. J. Young, Analysis of spider silk in native and supercontracted states using Raman spectroscopy, Polymer, 1999, 40(10), 2493–2500, DOI:10.1016/S0032-3861(98)00475-3
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ma01056b |
| This journal is © The Royal Society of Chemistry 2024 |