Open Access Article
Open Access ArticleMultidisciplinary approaches for enzyme biocatalysis in pharmaceuticals: protein engineering, computational biology, and nanoarchitectonics
Suhyeon
Kim
 abc,
Seongmin
Ga
abc,
Seongmin
Ga
 bc,
Hayeon
Bae
bc,
Hayeon
Bae
 bc,
Ronald
Sluyter
bc,
Ronald
Sluyter
 d,
Konstantin
Konstantinov
d,
Konstantin
Konstantinov
 a,
Lok Kumar
Shrestha
a,
Lok Kumar
Shrestha
 ef,
Yong Ho
Kim
ef,
Yong Ho
Kim
 *bcg,
Jung Ho
Kim
*bcg,
Jung Ho
Kim
 *a and
Katsuhiko
Ariga
*a and
Katsuhiko
Ariga
 *eh
*eh
aInstitute for Superconducting and Electronic Materials, Australian Institute for Innovative Materials, University of Wollongong, Squires Way, North Wollongong, NSW 2500, Australia. E-mail: jhk@uow.edu.au
bSKKU Advanced Institute of Nanotechnology (SAINT), Sungkyunkwan University (SKKU), 2066 Seobu-ro, Jangan-gu, Suwon, Gyeonggi-do 16419, Republic of Korea. E-mail: yhkim94@skku.edu
cDepartment of Nano Science and Technology, Sungkyunkwan University (SKKU), Suwon, Republic of Korea
dMolecular Horizons and School of Chemistry and Molecular Bioscience, University of Wollongong, Northfields Avenue, Wollongong, NSW 2522, Australia
eResearch Center for Materials Nanoarchitectonics (MANA), National Institute for Materials Science (NIMS), Tsukuba 305-0044, Japan. E-mail: ARIGA.Katsuhiko@nims.go.jp
fDepartment of Materials Science, Faculty of Pure and Applied Sciences, University of Tsukuba, 1-1, Tennodai, 305-8573 Tsukuba, Ibaraki, Japan
gDepartment of Nano Engineering, Sungkyunkwan University (SKKU), Suwon, Republic of Korea
hGraduate School of Frontier Sciences, The University of Tokyo, Kashiwa 277-8561, Japan
First published on 28th October 2023
Abstract
Enzyme biocatalysis is reshaping pharmaceutical synthesis, offering sustainable and efficient pathways for drug discovery and production. This paradigm shift towards eco-friendly methodologies addresses concerns inherent in traditional chemical synthesis. Enzymes, celebrated for their precision and adaptability to mild conditions, are poised as ideal candidates for pharmaceutical applications. Their versatility facilitates the synthesis of diverse pharmaceutical compounds, ensuring precise drug design and minimizing environmental impact. The integration of multidisciplinary approaches, including protein engineering, computational biology, and nanoarchitectonics, holds the potential to propel enzyme biocatalysis even further. Protein engineering utilizes directed evolution and rational design to customize enzymes, enhancing their stability and efficacy. Computational biology aids in deciphering enzymatic mechanisms, while nanoarchitectonics introduces innovative enzyme integration strategies into continuous flow systems. This comprehensive review explores how these multidisciplinary approaches can revolutionize pharmaceutical research and production. The synergy among these disciplines promises to expedite pharmaceutical processes, promote sustainability, optimize efficiency, and elevate precision—aligning perfectly with the evolving requirements of the pharmaceutical industry.
Broader contextEnzyme biocatalysis is propelling a significant revolution in pharmaceutical synthesis, offering eco-friendly and highly efficient pathways for drug discovery and production. This departure from traditional chemical methods not only aligns with sustainability goals but also reflects a broader global trend towards greener practices. Enzymes, renowned for their precision and adaptability under mild conditions, are poised to play a pivotal role in shaping the pharmaceutical industry's future. Their inherent versatility allows for the precise synthesis of a wide spectrum of pharmaceutical compounds, simultaneously reducing the environmental impact—a critical consideration in contemporary research and development. In this evolving landscape, multidisciplinary approaches, reminiscent of recent catalyst development strategies, hold the key to unlocking the full potential of enzyme biocatalysis. These approaches encompass protein engineering, computational biology, and nanoarchitectonics, which collectively contribute to enhancing enzyme stability, deciphering complex enzymatic processes, and introducing innovative integration methods within continuous flow systems. This comprehensive review not only underscores the remarkable impact of multidisciplinary approaches but also highlights their potential to streamline pharmaceutical processes, promote sustainability, optimize efficiency, and elevate precision—a paradigm shift aligned with the evolving demands of the pharmaceutical industry and broader environmental objectives. |
1. Introduction
Enzyme biocatalysis, encompassing the utilization of microorganisms or enzyme preparations to catalyze chemical transformations, has arisen as a pivotal force in the field of pharmaceutical synthesis, providing a sustainable and efficient pathway for drug discovery and production.1–5 In an era characterized by unprecedented demands for speed, sustainability, cost-efficiency, and precision in drug development, the pharmaceutical industry is undergoing a profound shift catalyzed by the rise of enzyme biocatalysis.6,7 This paradigm shift toward environmentally friendly and more precise methodologies addresses both environmental concerns and efficacy issues associated with conventional chemical synthesis approaches.8,9 Historically, the pharmaceutical industry heavily relied on chemical synthesis methods to produce therapeutic compounds.10 While these approaches undeniably accelerated drug development and production, they also introduced inherent challenges. For example, the employment of hazardous reagents, significant energy consumption, and the generation of substantial waste posed significant environmental and economic concerns.11 The call for more sustainable alternatives has grown louder in recent years, and in response, enzyme biocatalysis has emerged as a compelling solution.Enzymes, often referred to as the catalysts of life, have evolved over millions of years to orchestrate precise chemical transformations within living organisms.12 Their intrinsic specificity, combined with compatibility with mild reaction conditions, positions enzymes as ideal candidates for pharmaceutical applications.5,12 Harnessing enzymes in drug development and synthesis not only enhances the selectivity of reactions but also aligns pharmaceutical processes with the principles of green chemistry, mitigating their environmental footprint.13 Enzyme biocatalysis in pharmaceuticals is no longer confined to the realm of theoretical promise, it has evolved into a practical, advantageous reality with discernible benefits. Globally, researchers have harnessed enzymes' catalytic potency to execute a diverse range of transformations, encompassing small-molecule drug synthesis and the production of intricate biological therapeutics.14 For example, enzymes such as lipases and hydrolases have played a fundamental role in optimizing the synthesis of enantiopure chiral intermediates—a formidable challenge frequently confronted in the pharmaceutical chemistry.15 This progress underscores the central role of enzyme biocatalysis in modern pharmaceutical research and manufacturing.
Indeed, enzyme biocatalysis in pharmaceuticals has offered a multitude of advantages that have an impact on various facets of drug development and production. Firstly, enzymes have played a crucial role in the assembly of complex peptide and oligonucleotide therapeutics, elevating the precision of drug design.16–19 By selectively and efficiently catalyzing specific chemical reactions, enzymes enable pharmaceutical scientists to fine-tune the molecular structures of drugs, optimizing their efficacy and safety profiles. This newfound precision in drug synthesis carries profound implications, extending beyond the creation of novel therapeutic agents to encompass the optimization of existing medications.20 The ability to finely manipulate molecular structures through enzymatic catalysis not only facilitates the design of innovative drugs but also opens avenues for enhancing the efficacy, safety, and targeted delivery of established pharmaceuticals.21
Additionally, one of the outstanding features that distinguishes enzyme biocatalysis in pharmaceuticals is its inherent compatibility with aqueous environments and mild operational conditions.7,12,22,23 Unlike conventional chemical catalysts, enzymes thrive in water-based solutions and typically function at relatively low temperatures. These favorable conditions not only diminish the energy requirements of pharmaceutical processes but also elevate safety standards by minimizing potential hazards linked to high-temperature reactions and the use of hazardous reagents.9,24 Furthermore, enzymes frequently showcase an exceptional level of substrate versatility, demonstrating the capacity to accommodate a wide range of substrates.2,24,25 This adaptability paves the way for innovative approaches in synthesizing a wide spectrum of pharmaceutical compounds. Such versatility has notable importance for addressing the dynamic requirements of the pharmaceutical sector, characterized by a continual demand for complex and highly targeted drug molecules.
The evolutionary journey of enzyme biocatalysis from a promising concept to an indispensable tool in pharmaceutical research and production can further be driven by multidisciplinary approaches. This convergence of disciplines encompasses three central pillars: protein engineering, computational biology, and nanoarchitectonics. Protein engineering, through directed evolution and rational design, has empowered researchers to tailor enzymes for specific reactions and enhance their stability, paving the way to novel applications in pharmaceutical synthesis.12,22,26,27 Computational biology has augmented our understanding of enzymatic mechanisms and facilitated the rational design of enzymes with enhanced catalytic activity.28–32 Additionally, computational tools have streamlined the prediction of enzyme–substrate interactions, expediting the development and optimization of enzymatic processes.33,34 Meanwhile, nanoarchitectonics has introduced innovative strategies for enzyme immobilization and encapsulation within custom-designed nanomaterials.35–37 These advances not only enhance enzyme stability and reusability but also enable their seamless integration into continuous flow processes, a crucial requirement for large-scale pharmaceutical production.
This review paper presents a comprehensive examination of enzyme biocatalysis within the realm of pharmaceutical applications, with a primary focus on introducing multidisciplinary approaches to advance the field of enzyme biocatalysis (Fig. 1). We begin by providing a concise overview of the background and advantages of utilizing enzyme biocatalysis in pharmaceutical contexts. Following this, we introduce the key factors enabling the development of advanced enzymes for pharmaceutical applications. Within the context of multidisciplinary approaches, we extensively explore the significant contributions made by disciplines such as protein engineering, computational biology, and nanoarchitectonics in advancing the field of enzyme biocatalysis. Additionally, we address the persistent challenges that remain and offer insights into the prospects for these multidisciplinary strategies. As we explore this transformative landscape, it becomes evident that the synergy among these multidisciplinary facets holds great potential to have a significant impact on the evolution of pharmaceutical research and production. This potential contribution revolves around the core principles of speed, sustainability, efficiency, and precision, which are increasingly important in today's pharmaceutical landscape.
 | ||
| Fig. 1 Outline of the multidisciplinary approaches to advanced enzyme biocatalysis for pharmaceutical applications. | ||
2. Enzyme biocatalysis as a potential solution for pharmaceutical applications
2.1 Definition of enzyme biocatalysis
Enzyme biocatalysis, situated at the crossroads of biology and chemistry, represents a cutting-edge discipline with the power to reshape how we approach chemical reactions and synthesis processes.12,38 At its core, this transformative field harnesses the remarkable catalytic properties of enzymes—biological macromolecules finely honed by evolution over millions of years to function as molecular machinery within living organisms.8,12 These enzymes exhibit an exceptional blend of specificity and efficiency, unrivalled in their capacity to orchestrate complex chemical transformations.Enzyme biocatalysis, in essence, can be defined as the application of enzymes as catalysts to drive and accelerate chemical reactions.1 These biological catalysts possess the unique ability to enhance the speed and precision of chemical transformations under mild conditions, making them invaluable tools in various scientific and industrial applications.2,12 Central to the concept of enzyme biocatalysis is the idea of catalysis itself. In chemical reactions, catalysts are substances that facilitate the conversion of reactants into products without undergoing any permanent change themselves.39 Enzymes excel in this role, lowering the activation energy required for reactions and thus effectively expediting chemical transformations. Their catalytic action is highly specific, with each enzyme tailored to interact with a particular substrate or class of molecules, ensuring precision in the reactions that they catalyze.2,40
Furthermore, enzyme biocatalysis offers a sustainable and environmentally friendly approach to chemical processes.5,9 Enzymes typically operate in aqueous environments and under relatively mild temperatures and pH conditions, which reduces energy consumption and minimizes the production of hazardous byproducts.23 These characteristics align with the principles of green chemistry, making enzyme biocatalysis an attractive choice for industries seeking eco-conscious and economically viable solutions.41,42 Ultimately, enzyme biocatalysis represents a convergence of nature's biochemical marvels with human ingenuity, opening doors to a wide range of applications across fields as diverse as pharmaceuticals, biotechnology, food production, and more. It holds the potential to transform the way we approach chemical synthesis and catalysis, offering a greener, more efficient, and more precise alternative to traditional chemical methods.
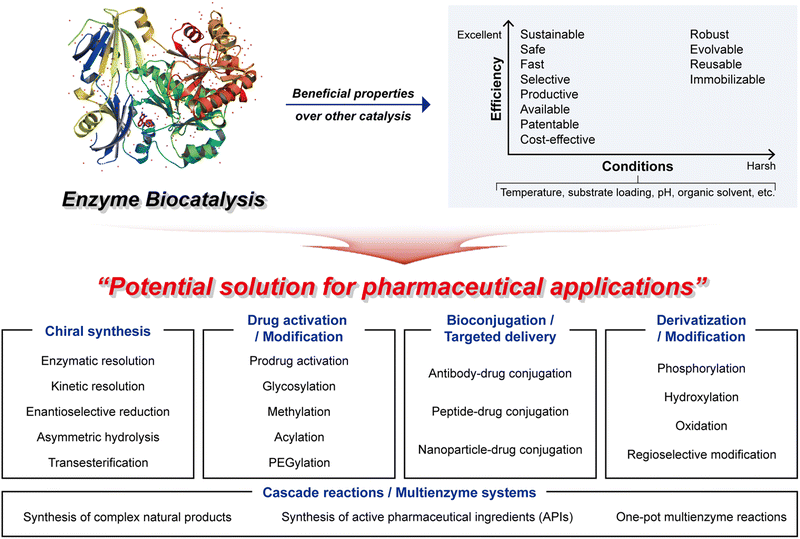
2.2 Beneficial properties of enzyme biocatalysis in pharmaceutical applications
Enzyme biocatalysis has emerged as a game-changer in the realm of pharmaceuticals, offering a plethora of highly advantageous properties that are transforming the drug development and manufacturing landscape. Within this section, we will explore several key advantageous properties that define the potential of enzyme biocatalysis, with a specific focus on its applications in the pharmaceutical sector. These properties encapsulate the versatility and sustainability that make enzyme biocatalysis an indispensable tool in modern pharmaceutical research and production.In conclusion, enzyme biocatalysis in pharmaceuticals embodies a host of advantageous properties that have positioned it at the forefront of drug discovery and production (Fig. 2). The plot (top right) highlights the most desirable characteristics of enzyme biocatalysis. In harsh industrial settings, attributes such as robustness and tolerance to mutations, which enhance stability, often become imperative. Conversely, whether operating under mild or more challenging conditions, the pharmaceutical industry consistently seeks biocatalyst properties that offer flexibility and efficiency.12 These advantageous attributes can be synergistically harnessed to yield high-value pharmaceutical compounds, even in the most taxing industrial scenarios, while concurrently achieving commendable economic and environmental performance.
2.3 Main applications of enzyme biocatalysis in pharmaceutical applications
Enzyme biocatalysis has become a significant factor behind pharmaceutical advances, offering a diverse set of tools and techniques that enhance speed, precision, efficiency, and sustainability. As a result, it finds widespread utility in the pharmaceutical industry, covering various applications, from chiral synthesis to targeted drug delivery and cascade reactions (Fig. 2). These applications leverage the exceptional catalytic capabilities of enzymes, allowing pharmaceutical researchers to address challenges, refine drug development processes, and create innovative therapeutic solutions. In this section, we explore the primary applications of enzyme biocatalysis in pharmaceuticals, each representing a foundational step towards greener, more effective, and more precisely tailored pharmaceuticals.This capability eliminates the need for cumbersome and expensive separation methods, which are traditionally required to isolate desired enantiomers from racemic mixtures.53 By harnessing the power of enzyme biocatalysis, pharmaceutical scientists can confidently synthesize single enantiomers, thus ensuring both the safety and the efficacy of pharmaceutical compounds. This transformative aspect of enzyme biocatalysis has led to the development of safer and more effective chiral pharmaceuticals, offering enhanced therapeutic outcomes.
By harnessing enzyme biocatalysis for bioconjugation, pharmaceutical scientists can tailor drug delivery systems to suit the unique requirements of diverse diseases and patient populations, thus advancing the field of personalized medicine.
By harnessing enzyme biocatalysis in these manufacturing procedures, pharmaceutical manufacturers can attain a heightened level of control over drug production, yielding pharmaceuticals that are more dependable, uniform, and ultimately beneficial for patients.
Concurrently, multienzyme systems, often featuring engineered enzymes, unlock the potential for highly customized and efficient reactions that would present formidable challenges or even impossibilities, using traditional chemical methods.75,76 These intricate enzymatic networks bestow upon pharmaceutical scientists an unprecedented level of control and precision. Consequently, they elevate the versatility and efficiency of drug synthesis, ultimately culminating in more streamlined and cost-effective pharmaceutical production processes.
In summary, enzyme biocatalysis in pharmaceutical applications encompasses a broad range of vital functions, extending from chiral synthesis to drug activation, bioconjugation, derivation, and cascade reactions. These applications collectively empower pharmaceutical scientists to craft safer, more effective, and more precisely targeted therapies, all while streamlining drug synthesis and production processes. Enzyme biocatalysis remains a driving force in the evolution of the pharmaceutical industry, propelling innovation and extending the boundaries of drug discovery and development. For a detailed overview of various enzyme biocatalysis applications, please refer to Table 1.51,52,58,59,61,62,65,66,71–74,77–93
| Applications | Used enzyme | Biocatalysis | Key features | Ref. |
|---|---|---|---|---|
| Chiral synthesis | Lipase from Candida antarctica | Kinetic resolution for the synthesis of enantiomerically pure compounds | • Stereoselective hydrolysis of racemic mixtures | 51 |
| • High enantioselectivity | ||||
| • Broad substrate scope | ||||
| Transaminase | Asymmetric amination for chiral amine synthesis | • Enantioselective conversion of ketones/aldehydes to chiral amines | 77 and 78 | |
| • Mild reaction conditions | ||||
| • Compatibility with diverse substrates | ||||
| Hydantoinase | Enantioselective hydrolysis of hydantoins for chiral amino acid synthesis | • Regio- and stereoselective hydrolysis | 79 | |
| • High enantioselectivity | ||||
| • Biocatalytic resolution of racemic hydantoins | ||||
| Ketoreductase | Asymmetric reduction for the synthesis of chiral alcohols | • Stereoselective reduction of ketones | 52, 80 and 81 | |
| • High enantioselectivity | ||||
| • Broad substrate compatibility | ||||
| Drug activation and modification | Glucuronosyltransferase | Glucuronidation for drug metabolism and detoxification | • Selective conjugation of glucuronic acid to drugs, facilitating elimination and enhancing water solubility | 58 and 82 |
| Sulfotransferase | Sulfation for drug metabolism and bioactivation | • Selective sulfonation of xenobiotics, modulating pharmacokinetics and biological activity | 59 | |
| Thiopurine S-methyltransferase | Methylation of thiopurine drugs for pharmacogenetic applications | • Methylation of thiopurine drugs, influencing efficacy and toxicity in personalized medicine | 83 and 84 | |
| Cytochrome P450 3A4 | Drug metabolism and activation | • Oxidative metabolism of diverse drugs, influencing efficacy and toxicity profiles | 85 and 86 | |
| Bioconjugation and targeted drug delivery | Sortase | Site-specific protein labeling and bioconjugation | • Selective ligation of peptides or proteins, facilitating protein engineering and bioconjugate synthesis | 61 and 87 |
| Transglutaminase | Protein cross-linking for drug delivery systems | • Site-specific conjugation of proteins, forming stable and controlled drug delivery vehicles | 88 | |
| Lipoic acid ligase | Protein modification for targeted drug delivery | • Site-specific attachment of lipoic acid to proteins, enabling targeted drug delivery and imaging | 62 and 89 | |
| Protein kinase | Phosphorylation for targeted drug delivery and protein engineering | • Site-specific phosphorylation of proteins, modulating activity and interactions | 90 and 91 | |
| Derivatization and modification | Glycosyltransferase | Enzymatic glycosylation for the synthesis of glycosylated compounds | • Regio- and stereoselective glycosylation | 65 |
| • Versatile donor and acceptor substrate specificity | ||||
| Methyltransferase | Methylation for the synthesis of methylated compounds | • Selective transfer of methyl groups to substrates, enabling site-specific modification | 92 | |
| Halogenase | Halogenation for the synthesis of halogenated compounds | • Regio- and stereoselective halogenation, introducing halogen atoms into organic molecules | 66 | |
| Decarboxylase | Decarboxylation for the synthesis of bioactive compounds | • Conversion of carboxylic acids to corresponding amines or aldehydes, enabling diversification of chemical space | 93 | |
| Cascade reactions and multienzyme systems | Taxadiene synthase, taxadiene-5-α-hydroxylase, and acetyltransferase | Taxol biosynthesis cascade | • Sustainable production of the anticancer drug taxol from simple precursors, showcasing the potential of enzymatic synthesis for pharmaceutical compounds | 71 |
| Amorphadiene synthase, CYP71AV1, and ALDH1 | Artemisinin biosynthesis multienzyme system | • Multienzyme system for artemisinin biosynthesis illustrating the power of enzymatic pathways in producing vital antimalarial drugs efficiently | 72 | |
| ACVS, IPNS, and IAT | Penicillin G biosynthesis cascade | • Penicillin G biosynthesis cascade that can efficiently produce antibiotics, revolutionizing pharmaceutical manufacturing | 73 | |
| HMG-CoA synthase, HMG-CoA reductase, and mevalonate kinase | Statin synthesis multienzyme network | • Multienzyme system for statin synthesis to produce cholesterol-lowering drugs with precision | 74 | |
3. Key factors enabling enzyme biocatalysis in pharmaceutical applications
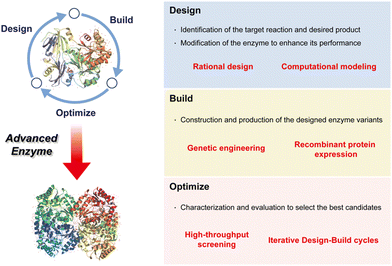
Enzyme biocatalysis has emerged as a vital facet of contemporary organic synthesis, exerting substantial influence across academic research and the chemical and pharmaceutical industries. Originally, during the early 2000s, its primary application lay in the production of optically active intermediates.94 The field of biocatalysis has undergone a transformative evolution, however, expanding its domain to encompass a diverse array of applications, with profound implications for the synthesis of chiral compounds integral to pharmaceuticals. This transformation has engendered the imperative adoption of a cyclical and iterative approach, characterized by three pivotal phases: design, build, and optimize (Fig. 3).4 These phases have become indispensable in harnessing the immense potential of enzyme biocatalysis, enabling pharmaceutical scientists to judiciously engineer enzymes, fabricate novel biocatalysts, and systematically refine their performance.95 In this manner, they drive innovation and enhance efficiency in both drug discovery and production.Design: the process commences with a rigorous design phase, marked by the pursuit of high efficacy, enantioselectivity, and specificity in enzyme biocatalysis tailored for pharmaceutical applications. It necessitates the meticulous selection of target substrates and the systematic delineation of requisite enzyme engineering strategies essential for achieving precise substrate binding.
Build: following the design phase, the spotlight shifts to the efficient production of biocatalysts. The fabrication of biocatalysts for industrial pharmaceutical use demands productivity, practicality, and cost-effectiveness. This pivotal phase serves as the bridge between conceptual enzyme design and its tangible application.
Optimize: once biocatalysts are assembled, they undergo comprehensive testing to ascertain their capacity for effective biocatalysis. The development of a rigorous examination system assumes paramount importance in the progression of pharmaceutical biocatalysis, ensuring reliability and efficiency in the synthesis of pharmaceutical compounds.
In this section, we embark on a comprehensive exploration of these three critical phases that underpin the success of enzyme biocatalysis in pharmaceutical applications. By exploring the intricacies of design, production, and optimization, we aim to provide a thorough scientific elucidation of the dynamic field of enzymatic biocatalysis and its seminal role in shaping the future of pharmaceutical science.
3.1 Design: precision at the molecular level
Protein design in the context of enzymes involves creating or modifying proteins to enhance their catalytic activity, substrate specificity, stability, and other properties for various biocatalytic applications.96 A comprehensive understanding of protein structure and function to engineer enzymes makes it possible to improve their performance. Designing enzymes for high efficacy, enantioselectivity, and specificity is the starting point in developing enzyme biocatalysis for pharmaceutical applications.97 Selecting a target substrate and uncovering which enzyme engineering is required for specific substrate binding are fundamental aspects of this process.Rational design particularly involves making specific changes to an enzyme's amino acid sequence or structure based on our understanding of its catalytic mechanism. Unlike random or empirical methods, rational design relies on scientific principles, computational tools, and biochemical knowledge to make precise changes to a protein's amino acid sequence or structure, with the goal of improving or customizing its properties. Because rational design starts with knowledge of the protein's three-dimensional structure, technology such as X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, or cryogenic electron microscopy (cryo-EM) is important for understanding the structure of the desired enzyme and provides insights into its active site, substrate-binding regions, and overall conformation.
Rational design focuses on the enzyme's active site, where chemical reactions take place. Researchers can introduce mutations or structural alterations to optimize the active site's geometry, electrostatics, or binding affinity towards substrates. Additionally, by altering key residues in the active site, researchers can make the enzyme preferentially recognize different substrates. Rational design can also enhance protein stability and solubility, making the enzyme more robust under varying conditions, and create enzymes with high specificity for a particular substrate or to improve selectivity.
Furthermore, computational molecular modeling of enzymes is a valuable tool for gaining insights into enzyme structure and function, elucidating reaction mechanisms, and facilitating the design of novel enzymes or engineered enzymes for various applications in pharmaceuticals. Computational tools and molecular modeling techniques can predict how changes to the protein's structure, such as mutations or structural modifications, will impact its function. Among these tools, molecular dynamics (MD) simulations and energy calculations provide dynamic information on enzyme behavior, including conformational changes, flexibility, and the simulation of enzyme–substrate interactions in a realistic environment.
The integration of rational design principles and computational techniques has revolutionized the field of enzyme biocatalysis, enabling the precise engineering of enzymes for pharmaceutical applications. In the subsequent sections of this review, we will explore the crucial phases of building and optimizing these designed enzymes, culminating in their impactful role in pharmaceutical synthesis.
3.2 Build: crafting custom biocatalysis
Following the meticulous design phase, the focus shifts to the efficient production of biocatalysts—a phase that stands as a critical bridge between conceptual enzyme design and its tangible application. While some well-known enzymes such as oxidoreductases, transferases, and lyases are readily available through chemical vendors, the development of designed enzymes requires a more tailored approach.To obtain a designed enzyme with a known amino acid sequence, researchers typically turn to gene synthesis. This process entails the creation of a synthetic gene encoding the enzyme engineered through rational design and computational molecular modeling to enhance its catalytic activity. Once the synthetic gene is prepared, it undergoes cloning using techniques such as the polymerase chain reaction (PCR) to amplify the gene. The cloned gene is subsequently inserted into a suitable host organism or expressed using cell-free protein synthesis systems.
The choice of expression system depends on various factors, including the nature of the enzyme and the intended application. Common expression hosts encompass bacteria, yeast, mammalian cells, or even cell-free systems. For living organisms, optimizing cultivation conditions, such as temperature and nutrient supply, becomes essential to ensure efficient enzyme production.
Upon successful expression, the next crucial step involves the harvesting and purification of the protein from the expression system. This purification process typically commences with cell lysis to release the protein, followed by various purification techniques such as chromatography or filtration. Importantly, the purified protein undergoes rigorous characterization to ensure it aligns with the desired specifications. Techniques such as mass spectrometry and gel electrophoresis are important in verifying the protein's identity and purity.
Moreover, if necessary, the production process can be further optimized to achieve higher yields and can be scaled up to generate larger quantities of the protein suitable for pharmaceutical applications. The specific details of each step may vary based on the protein, the chosen host organism, and the intended application. Genetic engineering, through the precise control and manipulation of genes, offers the versatility needed to produce a wide range of proteins for diverse purposes.
In essence, the efficient production of biocatalysts represents a pivotal phase that culminates in the transformation of conceptual enzyme design into tangible solutions for pharmaceutical applications. It is at this juncture that the engineered enzymes become the tools of choice for driving innovation and efficiency in drug discovery and production.
3.3 Optimize: continuous refinement and enhancement
Optimizing the engineered enzyme is a systematic approach that involves testing numerous enzyme variants and making incremental improvements through high-throughput screening (HTS) and iterative design-build cycles. The goal of optimization can encompass various aspects, such as improved activity, substrate specificity, and improved stability, depending on the defined purpose of enzyme enhancement. To assess enzymatic activity efficiently and accurately, a robust screening method is crucial.HTS, a good example, serves as a valuable tool in this regard, enabling the rapid and systematic testing of a large library of enzyme variants.98 HTS, a method widely utilized across various scientific and industrial disciplines, including drug discovery, materials science, and enzyme engineering, automates the screening process for enhanced efficiency. HTS allows researchers to identify enzyme variants with improved properties, whether they are enhanced catalytic activity, altered substrate specificity, or increased stability. This method accelerates the identification of promising enzyme candidates from the vast pool of variants. Analysis of HTS screening data further refines the selection process, pinpointing the most promising enzyme variants. These selected variants often become the foundation for the next generation of enzyme enhancements, where targeted mutations or modifications are introduced to fine-tune their performance to meet specific application requirements.
The design–build–optimize process will not reach its conclusion in just one iteration. With each successive cycle, enzymes become increasingly customized, more efficient, and more sustainable. In practice, the iterative process described here need not rigidly adhere to a predetermined sequence and exhaustively cover all three phases. Researchers, particularly those with potent design strategies, can have the flexibility to focus their efforts primarily on the build and optimize phases, persisting until the most favourable candidate is identified. However, it is worth noting that there are instances where, prior to advancing to the optimize phase, a reassessment of the design phase may be warranted, especially when certain enzymes have not been adequately constructed or produced.
This iterative design–build–optimize methodology, as elucidated, underscores the key role of precision, adaptability, and optimization in the quest for innovative pharmaceuticals and therapeutic solutions. It emphasizes the dynamic nature of the biocatalyst development process, acknowledging that the sequence of phases may vary based on the unique requirements and challenges of each biocatalyst design.
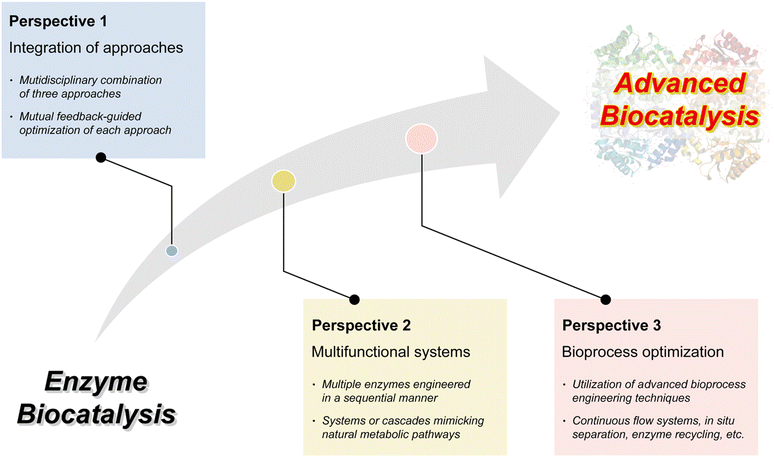
4. Multidisciplinary approaches for advanced enzyme biocatalysis
The necessity of introducing multidisciplinary approaches in the realm of enzyme biocatalysis for pharmaceutical applications is intimately tied to the field's continuous quest for optimization and advancement (Fig. 4). Enzyme biocatalysis, while already a transformative force in drug development and manufacturing, is far from reaching its full potential. To harness the full power of enzyme biocatalysis and overcome the multifaceted challenges presented by modern pharmaceutical demands, a more defined and advanced approach is required. Enzymes, as Nature's molecular machines, possess remarkable catalytic capabilities, yet there is room for improvement.6,99,100 To tailor enzymes for specific pharmaceutical reactions, enhance their stability, and broaden their substrate scope, protein engineering emerges as a pivotal multidisciplinary approach.22,25,26,101,102 Directed evolution, semi-rational design, and rational design techniques offer sophisticated strategies for fine-tuning enzymes. These advances are not only driven by biology, but are heavily reliant on computational biology.103–109 The marriage of computational modeling and biological understanding empowers researchers to predict, manipulate, and optimize enzyme behavior at a molecular level. Furthermore, the integration of nanoarchitectonics into enzyme biocatalysis brings novel strategies for enzyme immobilization, encapsulation, and deployment within custom-designed nanomaterials.35–37,110 These innovations enhance enzyme stability, reusability, and their seamless integration into continuous flow processes—critical requirements for large-scale pharmaceutical production.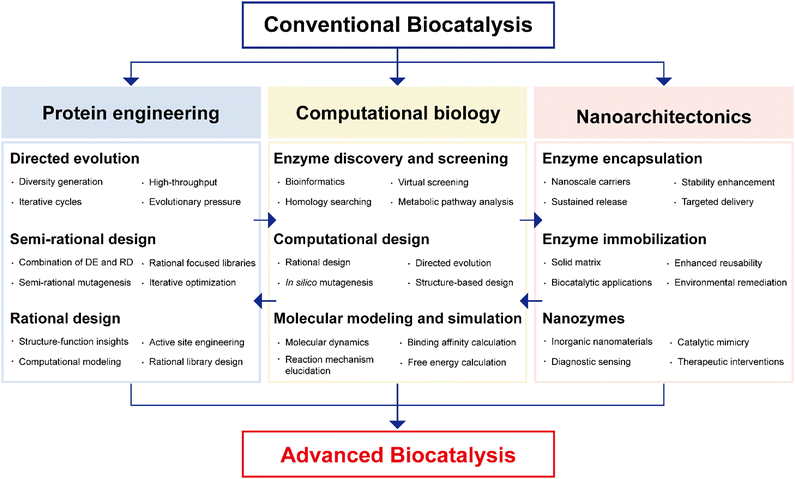 | ||
| Fig. 4 Multidisciplinary approaches to advanced enzyme biocatalysis in the pharmaceutical industry. DE: directed evolution; RD: rational design. | ||
It will be the convergence of multidisciplinary approaches: protein engineering, computational biology, and nanoarchitectonics that promises to unlock the full potential of enzyme biocatalysis, making it an indispensable tool in pharmaceutical research and production (Fig. 4). This synergy not only enhances efficiency but also facilitates more sustainable and precise pharmaceutical processes, solidifying the significance of enzyme biocatalysis in shaping the future of the pharmaceutical industry.
4.1 Protein engineering for advanced enzyme biocatalysis
In recent decades, the field of protein engineering has witnessed significant advances, driven by breakthroughs in molecular biology, crystallography, and computational methods.111 These innovations have equipped researchers with powerful tools to enhance existing biocatalysts and tailor them for novel substrates. Beginning with the emergence of molecular biology in the late 1970s, the discipline of protein engineering has seen remarkable growth.112 In the context of biotechnology and industrial applications, the exploitation of enzymes as biocatalysts holds immense promise. Enzymes, with their exquisite specificity and catalytic efficiency, are the biological marvels at the heart of this endeavor. To unlock their full potential, scientists have developed a repertoire of protein engineering techniques, prominently including directed evolution, semi-rational design, and rational design (Fig. 5).38 Directed evolution emulates the forces of natural selection in the laboratory, enabling the creation of enzyme variants with enhanced traits through controlled mutation and selection processes. Semi-rational design integrates empirical methods with structural insights, allowing researchers to strategically introduce mutations to improve enzyme properties. Rational design leverages a deep understanding of enzyme structure–activity relationships, enabling precise modification for targeted applications. These methodologies have ushered in a new era, opening exciting possibilities for tailoring enzymes with augmented catalytic activity, stability, and substrate specificity. These properties are of paramount significance, serving as the bedrock for advances in enzyme biocatalysis, underpinning achievements in biotechnology, pharmaceuticals, and green chemistry.113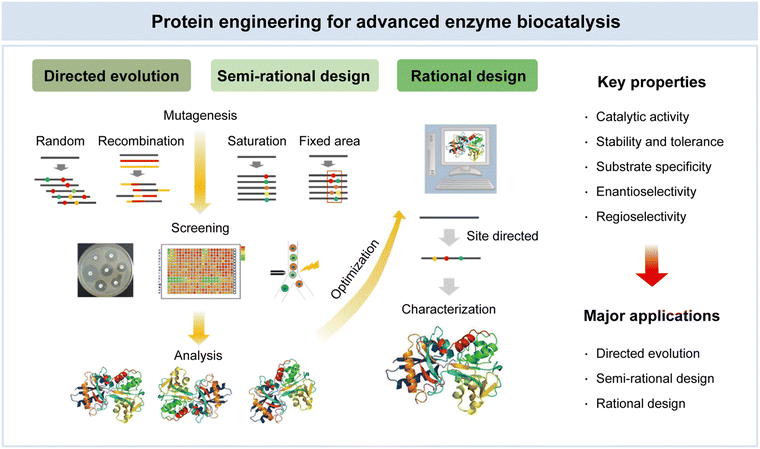 | ||
| Fig. 5 Protein engineering for advanced enzyme biocatalysis in pharmaceuticals. Reproduced with permission from ref. 95. Copyright 2023, Springer Nature. | ||
In a notable example, Brandenberg et al. directed their attention towards cytochrome P450 variants, particularly P411-CIS, employing the formidable tools of directed evolution to enhance the selective functionalization of cyclic compounds (Fig. 6).116 The outcome was nothing short of astonishing, as the engineered enzyme variant exhibited remarkable prowess, catalyzing the C3-alkylation of 1-methylindole with a remarkable 14% yield and an astonishing turnover number (TTN) of 690. The intrigue deepened as the resulting alkylated product showcased a pronounced red-shift in ultraviolet (UV) absorbance compared to its parent compound (Fig. 6A). P411-CIS, an intricate 1048-residue protein, comprises a heme domain encompassing the active site (residues 1–470) and a reductase domain responsible for electron transport from nicotinamide adenine dinucleotide phosphate (NADPH) to the heme domain. This electron-shuttling mechanism is essential for the native oxo-transfer chemistry of P450-BM3, with residues 471–1048 subdivided into flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) domains (Fig. 6B). Through an iterative sequence of directed evolution rounds, the research team culminated in the emergence of P411-HF (where HF stands for heterocycle functionalization; Fig. 6C). The quest continued with three compelling ‘case studies’ on P411-HF, delving into regioselective pyrrole alkylation, enantioselective indole alkylation employing an α-disubstituted carbene precursor, and the complex domain of cyclopropanetrione involving cyclic alkenes (Fig. 6D). As the research unfolded, it became evident that P411-HF bore more than its designated function. It exhibited additional ‘promiscuous’ activities, thus revealing a gateway to further evolutionary adaptations. This versatility was harnessed in the context of regioselective 1-methylpyrrole alkylation, enantioselective 1-methylindole alkylation with ethyl 2-diazopropanoate, and stereoselective cyclic alkene cyclopropanation. The significance of this study lies in its profound demonstration of precise control over regioselectivity, enantioselectivity, and catalytic activity. The remarkable adaptability and tunability of P411-HF underscore its potential as a catalyst in a diverse array of challenging reactions.
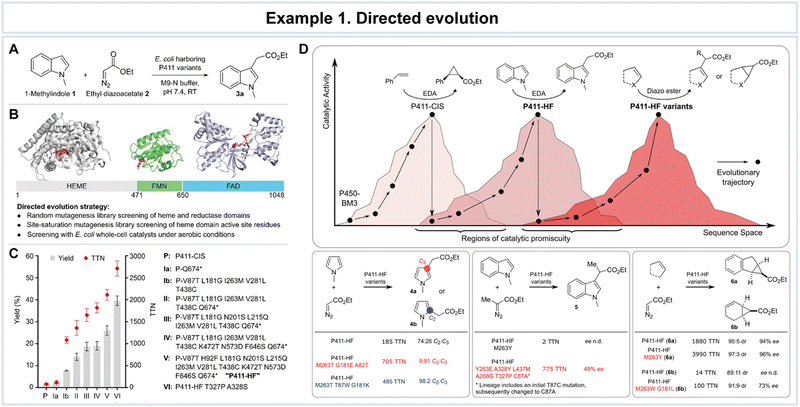 | ||
| Fig. 6 Representative example of directed evolution for advanced enzyme biocatalysis. Directed evolution of P411-CIS for 1-methylindole C3-alkylation. (A) Screening reaction of P411 variants for higher indole alkylation activity. (B) Domain structure of P450-BM3. Individual domain structures with approximate domain boundaries were used to construct the P450-BM3 protein structure. (C) Evolutionary trajectory from P411-CIS to P411-HF T327P A328S. (D) Evolving P411-HF for selective (hetero)cycle functionalization reactions. Schematic representation of the relation between protein sequence and catalytic function. A given enzyme sequence may exhibit additional promiscuous functions that can be optimized by directed evolution. Reproduced with permission from ref. 116. Copyright 2019, American Chemical Society. | ||
In the work by Chen et al., the application of semi-rational design to the Ralstonia sp. alcohol dehydrogenase (RasADH) was aimed at overcoming challenges associated with the reductive desymmetrization of cyclic diketones by engineering an enzyme, designated as F12, which exhibited exceptional efficiency in catalyzing the reductive desymmetrization of 1a, as well as other 2,2-disubstituted-1,3-cyclopentanediones (Fig. 7).119 To gain a comprehensive understanding of the molecular basis underlying the remarkable enhancements in catalytic activity and stereocontrol achieved through semi-rational design, the researchers conducted a combination of crystal structural studies and MD simulations. These investigations involved a detailed comparison of the wild-type and mutant enzymes in the presence of the co-factor NADPH and the substrate, (13R,17S)-ethyl secol (2a). While an overall structural similarity was observed in the presence of NADPH for both the wild-type and mutant enzymes, subtle but significant differences were noted in the conformation of the α6-helix of the mutant enzyme when bound to 2a (Fig. 7A).
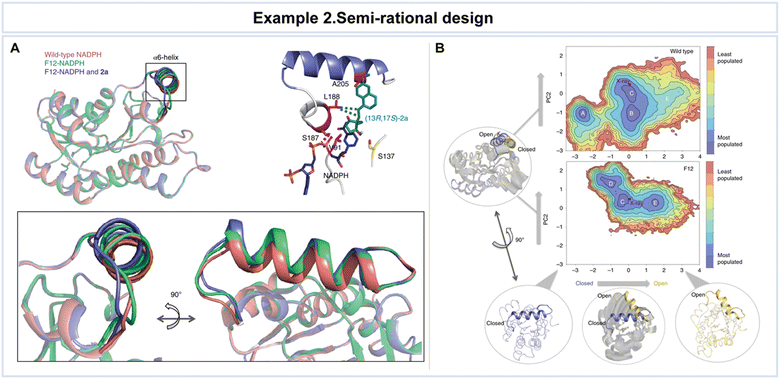 | ||
| Fig. 7 Representative example of semi-rational design for advanced enzyme biocatalysis. (A) Crystal structures of wild-type NADPH and mutant F12-NADPH with or without 2a. (B) Conformational population analyses. The two most important principal components (PC1 and PC2) that are based on Cα contacts for the RasADH wild-type enzyme and the F12 variant were analyzed. In the conformational landscape represented, the most populated conformations are colored in blue, whereas the least populated are in red. Reproduced with permission from ref. 119. Copyright 2019, Springer Nature. | ||
A meticulous analysis of these structural findings unveiled critical interactions that take a lead in enhancing the enzyme's activity and stereocontrol. For example, amino acid S187 was found to interact indirectly with (13R,17S)-2a, forming a crucial hydrogen bond with the phosphate group of NADPH. While the precise influence of the α6-helix and the factors responsible for the improved activity and stereoselectivity remained elusive solely through structural analyses, computational assessments were undertaken to provide valuable insights. MD simulations unveiled dynamic transitions of the α6-helix, with the enzyme switching between open, partially closed, and closed conformations at the entrance of the active site (Fig. 7B). Notably, these conformational changes, especially the transition to an open state, were notably more pronounced in the F12 variant compared to the wild type. The combination of structural insights and computational analyses clarifies the intricate conformational dynamics that govern enzyme–substrate interactions and catalytic outcomes, providing valuable guidance for future enzyme engineering endeavors. This resulted in high yields of the desired (13R,17S)-stereoisomer of ketol products, along with favorable diastereomeric ratio values.
In a study conducted by Son et al., rational design was employed to enhance the thermal stability of IsPETase (Fig. 8).121 Key structural issues affecting thermal stability were identified, including a disrupted central β-sheet caused by an abnormal β6 strand conformation and the flexibility of the β6–β7 connecting loop in the enzyme (Fig. 8A). To address these challenges, the researchers developed the IsPETaseP181A variant to restore the disrupted β-sheet. Furthermore, the flexibility of the β6–β7 connecting loop in IsPETase was found to be critical for stability, as revealed through a comparison with the more stable TfCUT2 enzyme (Fig. 8B). To improve loop stability, the IsPETaseS121D/D186H variant was created by introducing a hydrogen bond.
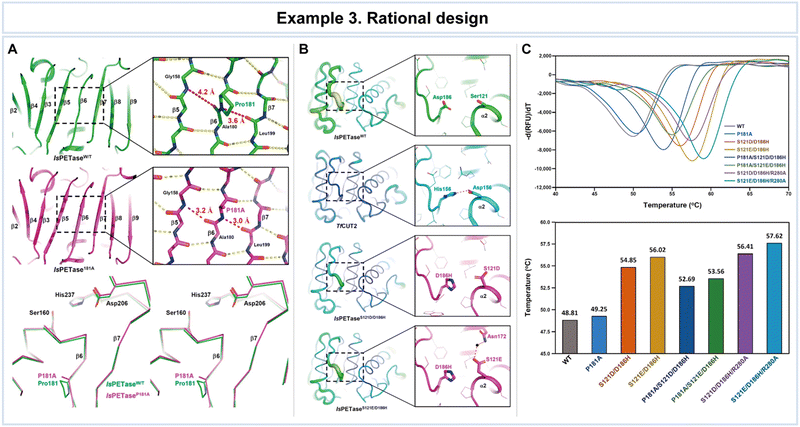 | ||
| Fig. 8 Representative example of rational design for advanced enzyme biocatalysis. (A) Structural comparison of IsPETaseWT with IsPETaseP181A variant: the central mixed β-sheets of IsPETaseWT and IsPETaseP181A variant; stereoview of superimposed structures of IsPETaseWT and IsPETaseP181A variant. (B) The β6–β7 connecting loops of IsPETaseWT, TfCUT2, IsPETaseS121D/D186H, and IsPETaseS121E/D186H. (C) Thermal stability comparison of IsPETaseWT and variants. Reproduced with permission from ref. 121. Copyright 2019, American Chemical Society. | ||
The impact of these mutations on thermal stability was assessed by comparing the melting temperatures (Tm) that IsPETaseP181A and IsPETaseS121D/D186H exhibited Tm values of 49.25 °C and 54.85 °C, respectively, representing increases of 0.5 °C and 6 °C compared to IsPETaseWT at 48.81 °C (Fig. 8C). The IsPETaseP181A variant successfully restored the disrupted β-sheet, while the IsPETaseS121D/D186H variant mitigated flexibility in the β6–β7 connecting loop (Fig. 8B). Remarkably, the IsPETaseS121E/D186H variant displayed an impressive Tm of 56.02 °C, surpassing IsPETaseWT by 7.21 °C (Fig. 8C). Unexpectedly, enhanced thermal stability in this variant was attributed to a water-mediated hydrogen bond between Glu121 and Asn172. IsPETaseS121D and IsPETaseS121E variants exhibited slightly increased polyethylene terephthalate (PET) degradation ability at 30 °C, however, but demonstrated lower Tm values and decreased activity at 40 °C.
In summary, rational design has proven to be a potent approach for enhancing enzyme properties, as demonstrated in the context of improving thermal stability. Structural insights and targeted mutations guided the development of variants with enhanced thermal resilience, albeit with some trade-offs in activity at higher temperatures.
The combination of directed evolution, semi-rational design, and rational design represents an impact shift in the field of enzyme biocatalysis. These methodologies have emerged as indispensable tools for the precise tailoring of enzymes, conferring upon them exceptional properties that are paramount for pushing the boundaries of biocatalysis. Further exemplification of enzyme engineering can be found in Table 2, where additional instances are provided.122–140 Enhanced catalytic activity accelerates reactions, stability ensures that enzymes can endure challenging environments, and refined substrate specificity opens doors to a plethora of novel applications. However, these methods still have several limitations, for example, directed evolution can be time-consuming and resource-intensive and may not always yield enzymes with the desired properties. The accuracy of semi-rational design heavily depends on the available structural data and the quality of computational algorithms. Rational design requires in-depth knowledge of protein structure and biochemistry, which may not be available for all enzymes. While these methods have undeniably pushed the boundaries of enzyme biocatalysis, their full potential is yet to be realized. Continued research to address their limitations will be crucial in maximizing the practical benefits of directed evolution, semi-rational design, and rational design in enzyme engineering.
| Example | Applications | Enzyme | Key features | Detailed information | Ref. |
|---|---|---|---|---|---|
| (a) Directed evolution | |||||
| Fucosyltransferase | Fucosylated glycoconjugates | α-1,3-Fucosyltransferase | • Improved activity | • Engineered variants with improved activity and substrate specificity | 122 |
| • Enhanced substrate specificity | • Enabling the efficient synthesis of key intermediates for the production of pharmaceutical compounds | ||||
| • Synthesis of key intermediates | |||||
| Transaminases | Synthesis of chiral amines | ω-Transaminase (ω-TA) | • Expanded substrate scope | • Engineered variants with expanded substrate scope and improved activity | 123 |
| • Improved activity | • Enabling efficient synthesis of diverse chiral amines | ||||
| • Synthesis of diverse chiral amines | |||||
| Esterases | Enantioselective synthesis of chiral compounds | Bacillus subtilis esterase | • Enhanced stability | • Engineered variants with enhanced stability and activity towards specific ester substrates | 124 |
| • Improved activity | • Efficient hydrolysis of ester prodrugs, contributing to drug activation and improving drug delivery | ||||
| • Hydrolysis of ester prodrugs | |||||
| • Drug activation | |||||
| Nitrilases | Nitrile hydrolysis, synthesis | Rhodococcus rhodochrous nitrilase | • Improved substrate specificity | • Engineered nitrilase variants with improved substrate specificity and resistance to inhibitory compounds | 125 |
| • Resistance to inhibitory compounds | • Conversion of nitrile-containing precursors into valuable pharmaceutical intermediates, contributing to the synthesis of bioactive compounds | ||||
| • Conversion of nitrile precursors | |||||
| • Synthesis of drug intermediates | |||||
| Glycosidases | Glycosylation, glycoside synthesis | β-Glucosidase | • Enhanced thermostability | • Engineered variants with enhanced thermostability and substrate tolerance | 126 |
| • Substrate tolerance | • Efficient enzymatic hydrolysis of glycosidic bonds in drug glycosides, facilitating drug activation and improving bioavailability | ||||
| • Enzymatic hydrolysis | |||||
| • Drug activation | |||||
| (b) Semi-rational design | |||||
| Proteases | Drug synthesis, enzymatic modifications | Engineered subtilisin E | • Altered substrate specificity | • Semi-rational design strategies to modify specific amino acid residues in the active site for broadened substrate specificity towards non-natural amino acids | 127 |
| • Enhanced catalytic efficiency | |||||
| • Improved selectivity | |||||
| Kinases | Drug discovery, screening, activation | Engineered protein kinase | • Enhanced substrate specificity | • Semi-rational design approaches to modify the active site of a protein kinase, enhancing its substrate specificity towards specific peptide sequences relevant to drug targets | 128 |
| • Improved binding affinity | |||||
| • Improved thermostability | |||||
| Isomerases | Drug synthesis, pharmaceutical production, bioconversion | Engineered xylose isomerase | • Enhanced catalytic activity | • Modification of active site to enhance catalytic activity and stability during bioconversion through semi-rational design | 129 |
| • Improved stability | |||||
| • Increased conversion efficiency | |||||
| Ligases | DNA manipulation, protein production, diagnostics | Engineered DNA ligase | • Enhanced fidelity in DNA ligation | • Semi-rational design to enhance the fidelity and efficiency for precise DNA manipulation in pharmaceutical research and development | 130 |
| • Improved efficiency | |||||
| • Reduced side reactions | |||||
| Dehalogenases | Detoxification, remediation, drug production | Engineered haloalkane dehalogenase | • Altered substrate specificity | • Semi-rational design techniques to modify the active site, enhancing its substrate specificity towards specific halogenated compounds encountered in pharmaceutical manufacturing processes | 131 and 140 |
| • Enhanced catalytic efficiency | |||||
| • Increased detoxification efficacy | |||||
| (c) Rational design | |||||
| Oxidases | Stereoselective oxidation, drug synthesis | Engineered laccase variant T2/T3 site mutants | • Enhanced activity towards specific substrates | • Rational design to introduce mutations at the T2 and T3 copper-binding sites in laccase | 132 and 133 |
| • Improved pH stability | • Computational analysis and MD simulations to identify key residues for targeted mutagenesis | ||||
| Transferases | Drug synthesis, enzymatic cascade | Engineered transaminase variant with optimized active site residues | • Altered substrate specificity | • Rational design to optimize active site residues | 134 |
| • Increased thermostability | • Computational analysis and protein structure modeling to identify key amino acids for modification | ||||
| Esterases | Drug synthesis, chiral resolution | Engineered esterase variant with altered binding pocket residues | • Enhanced AHL-degrading activity | • Rational design to modify binding pocket residues | 135 and 136 |
| • Improved binding affinity with substrate | • Computational modeling and substrate docking studies to select key amino acids for mutation | ||||
| Glycosidases | Glycosylation, drug synthesis | Engineered glycosidase variant with altered active site residues | • Increased thermostability | • Rational design strategies to modify active site residues in glycosidases | 137 |
| • Improved glycosylation efficiency | • Computational modeling and MD simulations to select key amino acids for mutation | ||||
| Oxidoreductases | Precursor synthesis, oxidation | Engineered oxidoreductase variant with expanded substrate | • Enhanced substrate promiscuity | • Rational design to expand the substrate binding pocket in oxidoreductases | 138 and 139 |
| • Catalytic efficiency | • Computational analysis and protein–ligand docking studies to select key amino acids for modification | ||||
4.2 Computational biology for advanced enzyme biocatalysis
Within the domain of advanced enzyme biocatalysis, computational biology emerges as a significant discipline, catalyzing groundbreaking advances at the intersection of biology and computation (Fig. 9).12,141,142 Computational biology, broadly defined, is the application of computational techniques and mathematical modeling to solve complex biological problems.143–145 Its significance lies in its ability to harness the power of algorithms and high-performance computing to decipher intricate biological processes, predict molecular interactions, and optimize enzymatic properties.146,147 Key among these properties are the prediction of enzyme properties, including catalytic activity and stability, substrate specificity, docking and elucidation of binding mechanisms, and the mapping of intricate metabolic pathways.28,30,34,106,141 These predictive capabilities are indispensable in the rational design and engineering of enzymes, ultimately driving the evolution of enzyme biocatalysis. | ||
| Fig. 9 Computational biology for advanced enzyme biocatalysis in pharmaceuticals. Reproduced with permission from ref. 173. Copyright 2023, Springer Nature. | ||
Foremost among these applications is enzyme discovery and screening.108,148 By employing computational tools, researchers can efficiently sift through vast databases of potential enzymes, identifying candidates with the desired catalytic properties for specific reactions. This accelerates the process of enzyme selection, streamlining the search for novel biocatalysts. Another notable application lies in computational enzyme design.28,34,104,108,109,149 Here, computational biology enables the tailored engineering of enzymes with enhanced catalytic activity and selectivity. Researchers can iteratively model enzyme structures and interactions to predict the effects of mutations, ultimately guiding the development of enzymes optimized for specific reactions. This approach revolutionizes the field by expediting the creation of tailor-made biocatalysts for pharmaceutical applications. Molecular modelling and simulation constitute yet another critical facet of computational biology in enzyme biocatalysis.140,150,151 These techniques enable researchers to simulate enzyme–substrate interactions, providing essential insights into reaction mechanisms, binding affinities, and conformational changes. By unraveling these intricacies at the molecular level, computational biology empowers scientists to fine-tune enzymatic reactions with unparalleled precision, ensuring the development of efficient and sustainable bioprocesses.
In summary, computational biology serves as an indispensable partner in the pursuit of advanced enzyme biocatalysis. Its predictive prowess in property assessment, combined with its major applications in enzyme discovery, design, and molecular modeling, shapes a landscape of innovation that continues to redefine the boundaries of biocatalytic research and applications (Fig. 9).
In a study conducted by Vanacek et al., computational methodologies proved highly effective in facilitating the exploration of a vast pool of protein sequences (Fig. 10).154 This capability addressed a critical need in the postgenomic era, characterized by an exponential surge in available protein sequences, representing an untapped source of diverse enzyme catalysts. Despite the immense potential for biological and biotechnological discoveries, only a small fraction of these sequences has undergone experimental characterization.155 To efficiently harness this genomic wealth stored in public databases, the research team devised an integrated system comprising an automated in silico screening protocol and experimental procedures. This system aimed to unlock the structural and functional diversity of entire enzyme families (Fig. 10A). As a proof of concept, the study focused on exploring the diversity within the microbial enzyme group known as haloalkane dehalogenases (EC 3.8.1.5, HLDs).
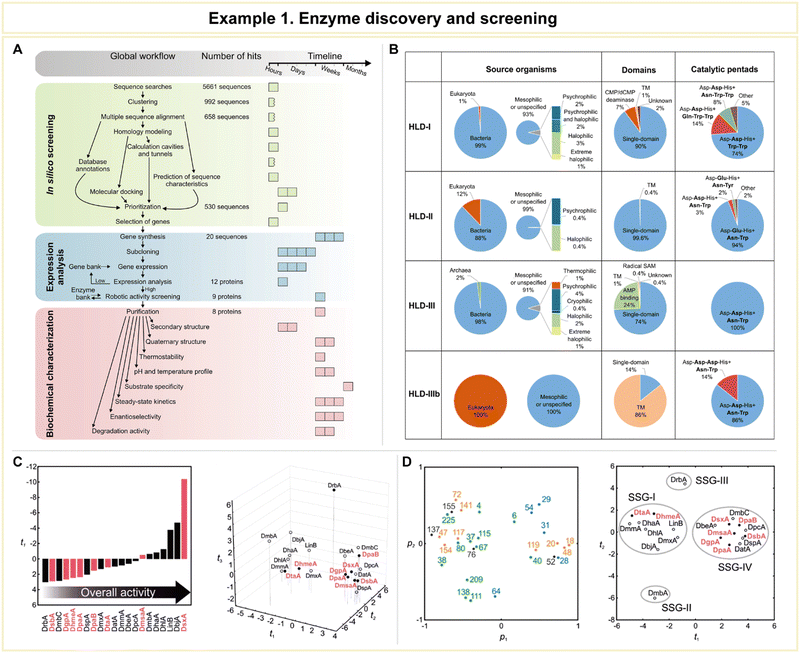 | ||
| Fig. 10 Representative example of enzyme discovery and screening for advanced enzyme biocatalysis. (A) Workflow of an integrated system for the exploitation of enzyme structural and functional diversity. (B) Overview of putative haloalkane dehalogenases (HLDs) identified. (C) Comparison of the substrate specificities and overall catalytic activities of novel HLDs with previously characterized enzymes using multivariate statistics. (D) The loading plot p1/p2 and the corresponding score plot t1/t2 from principal component analysis (PCA) of the transformed data set. Reproduced with permission from ref. 154. Copyright 2023, Springer Nature. | ||
The workflow of this integrated system encompassed three distinct phases: (1) automated sequence and structural bioinformatics, (2) protein production and robotic activity screening, and (3) biochemical characterization. Beginning with a staggering 5661 sequences, the researchers successfully entified 20 sequences corresponding to putative HLDs. These sequences provided valuable information about their source organisms, catalytic pentads, and domain compositions (Fig. 10B). Following expression analysis and robotic activity screening, a mere eight proteins emerged as candidates for comprehensive biochemical and biophysical characterization. By comparing the substrate specificities and overall catalytic activities of these novel HLDs with previously characterized enzymes using advanced multivariate statistics (Fig. 10C and D), the team achieved significant milestones. These included the discovery of the most catalytically proficient native HLD to date (kcat/K0.5 = 96.8 mM−1 s−1, where kcat is the catalytic constant and K0.5 is the substrate concentration that leads to half of the maximal velocity), the identification of the most thermostable enzyme boasting a melting temperature of 71 °C, the characterization of three distinct cold-adapted enzymes exhibiting dehalogenase activity at near-zero temperatures, and the development of a biocatalyst capable of degrading the warfare chemical sulfur mustard. This study exemplifies the vast potential of computational approaches in addressing the big data challenge presented by the postgenomics era.
In a groundbreaking study led by Lu et al., computational methodologies emerged as a potent tool for the engineering of PET hydrolases, enabling them to exhibit robustness across a wide pH and temperature range, accelerated reaction rates, and the unique ability to directly degrade untreated postconsumer plastics (Fig. 11).158 To achieve this remarkable feat, the research team harnessed the power of a structure-based machine learning algorithm, applying it to engineer a highly robust and active PET hydrolase. Specifically, the researchers employed a cutting-edge three-dimensional self-supervised convolutional neural network, known as MutCompute,159 to pinpoint stabilizing mutations within the enzyme (Fig. 11A). This innovative approach allowed them to identify positions within the enzyme's crystal structure where the original amino acid residues were suboptimal compared to potential substitutions, and these positions were ranked based on predicted probabilities (Fig. 11B). Subsequently, the team uncovered four pivotal mutations (S121E, T140D, R224Q, and N233K) that held the key to enhancing the PET-hydrolytic activity and expanding its operational temperature range. Notably, the machine-learning-guided predictions led to a significant enhancement in enzymatic activity across all tested conditions, spanning temperatures from 30 to 60 °C (Fig. 11C).
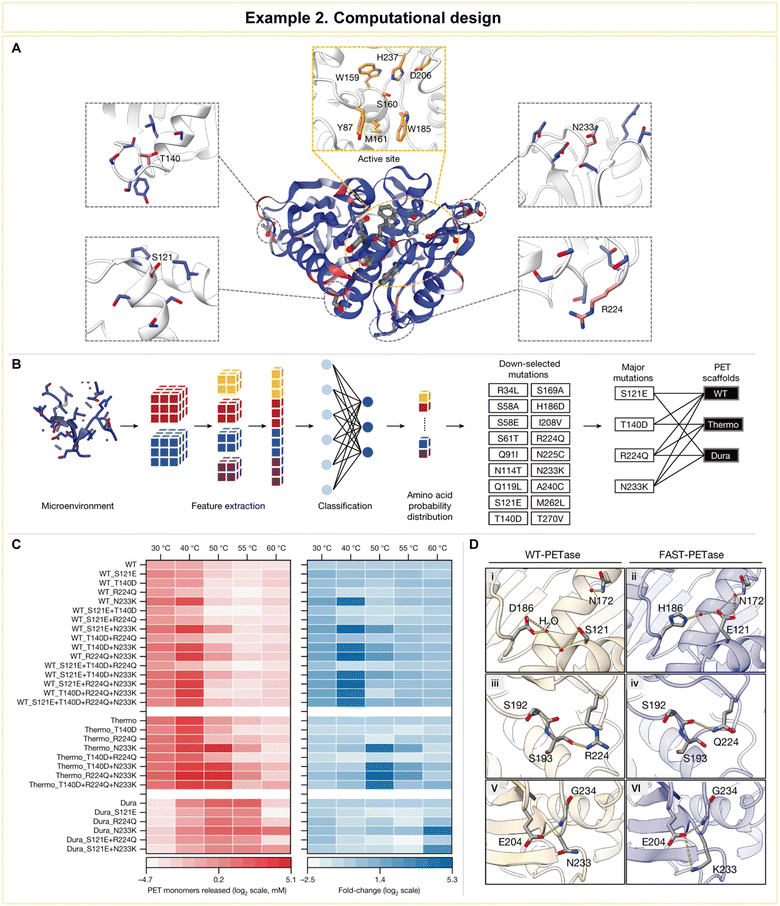 | ||
| Fig. 11 Representative example of computational design for advanced enzyme biocatalysis. (A) WT PETase protein structure rendered by the output of MutCompute. (B) Predictions based on both WT PETase and ThermoPETase were ranked by the fold change in the probabilities between the predicted and the WT amino acid. (C) The red heatmap shows the PET-hydrolytic activity of the resulting variants, and the blue heatmap shows the fold change of activity over their respective scaffolds based on the total PET monomers from the hydrolysis of circular gf-PET films by the PETase variants. (D) Predicted mutations from the neural network algorithm to stabilize FAST-PETase. Reproduced with permission from ref. 158. Copyright 2022, Springer Nature. | ||
To gain further insights into the enhanced stability and catalytic prowess of the engineered enzyme, crystal structure analysis was conducted on the top-performing variant, aptly named FAST-PETase (PDB 7SH6, where PDB stands for Protein Data Bank), which contained a total of five mutations, including those identified through predictions (N233K/R224Q/S121E) and scaffold-derived mutations (D186H/R280A). The structural analysis revealed the formation of favorable residue interactions that contributed to FAST-PETase's superior PET-hydrolytic activity compared to both the wild-type enzyme and previously engineered variants (Fig. 11D).
This groundbreaking study illustrates how the integration of computational biology can revolutionize enzyme engineering, resulting in enzymes with enhanced functionalities that were previously unattainable through traditional experimental methods. Such an approach holds immense promise not only in the realm of sustainable plastic degradation but also in diverse practical applications, including the pharmaceutical industry. The ability to tailor enzymes with precision and efficiency opens new avenues for addressing complex challenges in drug discovery, development, and manufacturing.
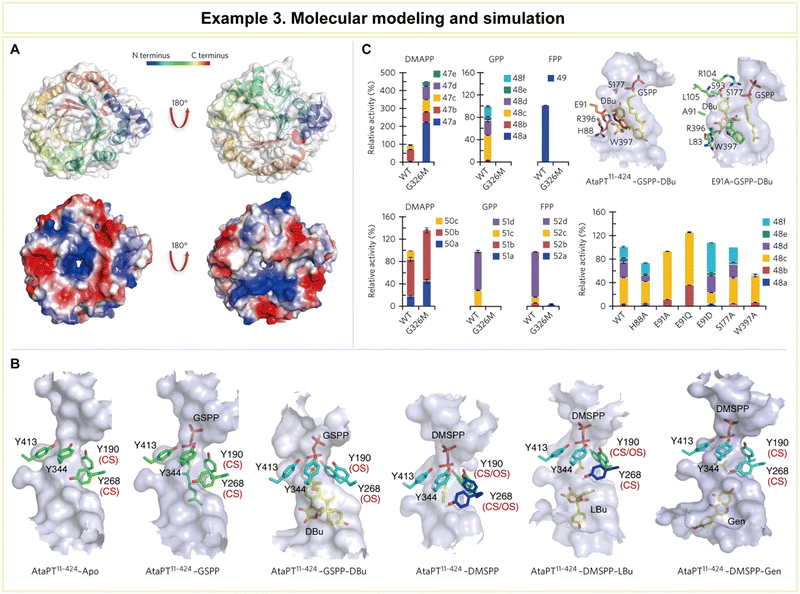 | ||
| Fig. 12 Representative example of molecular modeling and simulation for advanced enzyme biocatalysis. (A) Crystal structure of AtaPT and its catalytic chamber. (B) Conformational dynamics of the tyrosine shield and various acceptor binding sites of AtaPT. (C) Structure-guided mutagenesis of AtaPT to alter its substrate promiscuity. Reproduced with permission from ref. 164. Copyright 2017, Springer Nature. | ||
Furthermore, the research team conducted mutagenesis experiments to pinpoint the residues responsible for prenylation promiscuity (Fig. 12C). Armed with a profound understanding of AtaPT's molecular mechanisms governing prenylations, it became feasible to employ structure-guided mutagenesis to fine-tune both the prenyl donor and the prenylation selectivity of this enzyme. Such precision engineering has the potential to enhance the utility of AtaPT by synthesizing novel prenylated derivatives, opening new avenues in drug discovery programs and beyond. This study exemplifies how computational methodologies can provide invaluable insights into enzyme behavior, offering a strategic advantage in various scientific and industrial applications.
In summary, computational biology has revolutionized enzyme biocatalysis in pharmaceutical applications by enabling efficient enzyme discovery and screening, precise computational design, and insightful molecular modeling and simulation. These computational approaches have accelerated the identification of novel enzymes, facilitated tailored biocatalyst engineering, and deepened our understanding of enzymatic behaviour. Further exemplification of enzyme engineering can be found in Table 3, where additional instances are provided.52,103,165–180 As the pharmaceutical industry seeks sustainable, efficient, and precise solutions, computational biology emerges as a transformative tool, bridging the gap between theory and experiment and propelling innovation in drug development and production.
| Example | Applications | Enzyme | Key features | Detailed information | Ref. |
|---|---|---|---|---|---|
| (a) Enzyme discovery and screening | |||||
| Substrate-specific enzyme design | Peptide synthesis, drug discovery, protein engineering | Subtilisin Carlsberg | • Improved enantioselectivity and stability | • In silico screening methods to design enzymes with improved substrate specificity | 165 |
| • Virtual screening and molecular docking | • Specific mutations to enhance affinity and selectivity through substrate screening and computational analysis of binding interactions | ||||
| Substrate binding pocket engineering | Lipid modification, drug synthesis | Candida rugosa lipase | • Expanded substrate specificity and catalytic efficiency | • In silico screening and molecular docking to engineer the substrate binding pocket | 166 |
| • In silico screening and molecular docking | • Mutations to enhance substrate specificity and catalytic efficiency for drug synthesis through virtual screening and computational analysis | ||||
| Enzyme inhibitor design | Cancer treatment, drug discovery | Dihydrofolate reductase (DHFR) | • High affinity and specificity | • In silico screening and molecular docking to design DHFR inhibitors for pharmaceutical applications | 167 |
| • In silico screening and molecular docking | • Identification of potential inhibitors through virtual screening against target enzyme and binding interaction analysis | ||||
| Substrate promiscuity analysis | Drug synthesis, biotransformation | Alcaligenes faecalis nitrilase | • Understanding substrate preferences and promiscuity | • In silico screening and molecular docking to analyze the substrate promiscuity of enzymes | 168 |
| • In silico screening and molecular docking | • Substrate preferences and promiscuity study by virtual screening and computational analysis of binding interactions for pharmaceutical synthesis | ||||
| Substrate docking and reactivity analysis | Drug metabolism, pharmacokinetics, personalized medicine | Human cytochrome P450 2D6 (CYP2D6) | • Understanding substrate binding and metabolism | • In silico docking and reactivity analysis to study the binding and metabolism of substrates by enzymes | 169 and 170 |
| • In silico docking simulations and reactivity analysis | • Investigation of binding affinity and regioselectivity through virtual docking simulations and computational analysis for insights into drug metabolism and drug-drug interactions | ||||
| (b) Computational design | |||||
| Engineered lipase | Chiral drug synthesis, enzymatic resolution | Candida antarctica lipase B | • Improved substrate specificity and catalytic activity | • Prediction and introduction of specific amino acid substitutions in the active site of the enzyme using computational modeling and MD simulations | 171 and 172 |
| • Enhanced activity and selectivity towards a range of pharmaceutical substrates, enabling efficient synthesis of enantiopure drug intermediates | |||||
| Engineered oxidoreductase | Chiral synthesis, pharmaceutical intermediate production | Ketoreductase ChKRED12 | • Altered co-factor specificity | • Computational analysis of active site and molecular docking simulations to modify co-factor binding and enhance catalytic efficiency | 52 |
| • Improved catalytic efficiency | • Improved activity and selectivity of chiral alcohol synthesis for pharmaceutical applications, facilitating more efficient and sustainable routes | ||||
| Engineered esterase | Ester hydrolysis for drug synthesis | Pseudomonas fluorescens esterase | • Enhanced substrate specificity and stability | • Computational methods, including homology modeling and MD simulations, to explore the active site and predict mutations | 173 |
| • Increased enantioselectivity | • Enhanced catalytic activity and selectivity towards specific substrates, enabling efficient enzymatic synthesis of pharmaceutical intermediates | ||||
| Engineered halogenase | Halogenated compound synthesis, drug lead optimization | CYP450 | • Altered substrate specificity and regioselectivity | • Computational modeling and MD simulations to study the active site and predict mutations for altering substrate specificity and regioselectivity | 103 |
| • Improved activity and selectivity in halogenation reactions, enabling the synthesis of halogenated compounds for drug development | |||||
| Engineered decarboxylase | Aromatic compound, drug precursor production | O-Succinylbenzoate synthase | • Altered substrate specificity | • Computational methods to study active site and identify key residues in substrate binding and catalysis | 174 |
| • Improved catalytic efficiency | • Improved activity in synthesis of valuable aromatic compounds for pharmaceutical applications | ||||
| (c) Molecular modeling and simulation | |||||
| Molecular modeling for antibiotic design | Antibiotic resistance studies, antibiotic design, optimization | Beta-lactamase | • Molecular modeling and simulation for substrate specificity analysis | • Computational modeling and simulation methods to investigate the binding interactions with different beta-lactam antibiotics | 175 and 176 |
| • Analysis for insights into the enzyme's substrate specificity and molecular determinants governing substrate recognition and binding | |||||
| Understanding chitin substrate interactions in drug delivery | Chitin degradation, drug delivery systems | Chitinase | • MD simulations for substrate binding and catalytic mechanism analysis | • MD simulations to study the substrate binding process and catalytic mechanism | 177 |
| • Insights into dynamics of substrate binding, role of key residues in catalysis, and binding energy landscape of different chitin substrates | |||||
| Computational analysis for neurological disorder treatments | Alzheimer's disease treatment, drug discovery for neurological disorders | Acetylcholinesterase | • Molecular docking and binding free energy calculations for substrate binding analysis | • Computational methods, including molecular docking and binding free energy calculations, to investigate binding with substrates or inhibitors | 178 |
| • Analysis for insights into enzyme–substrate/inhibitor binding modes and key residues in binding affinity | |||||
| Enzyme design for pharmaceutical synthesis | Glycosylation reactions, drug conjugation | Glycosyltransferase | • Homology modeling and MD simulations for substrate binding analysis | • Homology modeling and MD simulations to investigate substrate binding process | 179 |
| • Analysis for insights into binding with various donor and acceptor substrates, elucidating the structural basis for substrate recognition and specificity | |||||
| Virtual screening for drug metabolism studies | Drug metabolism, pharmacokinetics studies, drug design and optimization | Cytochrome P450 | • Virtual screening and molecular docking for substrate identification and binding analysis | • Virtual screening techniques and molecular docking simulations to identify potential substrates for enzymes and predict their binding modes | 180 |
| • Analysis for insights into substrate preference and binding interactions, enabling the identification of novel drug metabolites or optimization of activities | |||||
The critical point for computational biology is that the validation of computational predictions through rigorous experimental testing is imperative to establish the trustworthiness of designed enzymes. Reliable experimental validation not only verifies the accuracy of computational models but also provides valuable feedback loops for refining these models. As the complexity of biological systems increases, it becomes crucial to address the limitations of current computational methodologies. Factors such as protein dynamics, solvent effects, and long-range interactions pose significant challenges to accurate predictions. Understanding these complexities and developing computational approaches that can capture them will be essential for advancing the field.
4.3 Nanoarchitectonics for advanced enzyme biocatalysis
Nanoarchitectonics is an interdisciplinary technological paradigm, initially proposed by Masakazu Aono.181 This innovative concept revolves around the meticulous design and construction of structures and materials on the nanoscale, where precision and organization are important. Nanoarchitectonics is driven by synthesis techniques, such as chemical synthesis, molecule manipulation, chemical nano-manipulation, self-assembly, and self-organization, to craft functional materials from fundamental building blocks at the nanoscale.182,183This versatile concept extends its range into various fields and applications. Notably, it has been employed in the design and synthesis of catalysts spanning a diverse spectrum, including photocatalysts, environmental catalysts, electrochemical catalysts, and biocatalysts. Beyond catalysis, the applications of nanoarchitectonics extend to material production, structural regulation, device fabrication, sensor development, energy harnessing, environmental science, and even the biological and biomedical realms.35
In particular, nanoarchitectonics is widely considered for sensing various biomolecules. Recently, microRNA (miRNA) has emerged as prominent biomolecule for next generation of diagnostic and prognostic biomarkers. Numerous RNA detection techniques based on engineered nanomaterials-based electrochemical biosensing strategies, such as gold nanoparticle, carbon-based nanomaterials, quantum dots (QDs), metal–organic frameworks (MOF) show huge potential for cancer management.184
One of the distinguishing facets of the nanoarchitectonics concept is its dynamic and harmonized nature.185,186 At the nanoscale, there are uncertainties, including thermal fluctuations, static distributions, and quantum effects, so a balanced harmonization incorporating dynamism becomes crucial within the framework of nanoarchitectonics. The dynamics bears a striking similarity to the intricacies observed in biological systems, where functional components dynamically cooperate and harmonize their operations.187 Consequently, nanoarchitectonics appears exceptionally promising in catalyst design that is tailored for biological functions.
For instance, in nanoarchitectonics, integration of hydrogel into common biosensing platforms creates soft structures that are both physically and chemically regulated. These structures boast superior biocompatibility, improved immobilization of biomolecules, and the ability to design biosensors that are specific and highly sensitive. The physical and chemical properties of 3D hydrogel structures can be modified by integrating with nanostructures, increasing their sensitivity to various stimuli like mechanical, optical, thermal, magnetic, and electric forces.188
Porous materials derived from coordination compounds like metal–organic frameworks (MOFs) and porous coordination polymers (PCPs) possess well-defined pore structures and promising properties. As these coordination compounds, Prussian blue (PB) and its analogues (PBA) exhibit excellent physical and chemical properties, making them versatile nanoarchitectonics for a wide range of applications including sensing, batteries, biomedicine, and imaging.189
In the context of enhancing enzyme biocatalysis, nanoarchitectonics has emerged as a dynamic fusion of nanotechnology, materials science, and enzymology (Fig. 13).35 It offers a compelling route to not only amplify catalytic activity but also strengthen enzyme stability, bolster reusability, and enhance versatility. In this part, we will describe in more detail the three central strategies encompassed by nanoarchitectonics for advanced enzyme biocatalysis: enzyme encapsulation, enzyme immobilization, and nanozymes. These strategies harness the principles of nanoarchitectonics to revolutionize the world of biocatalysis and hold immense promise for a multitude of applications across diverse industries.
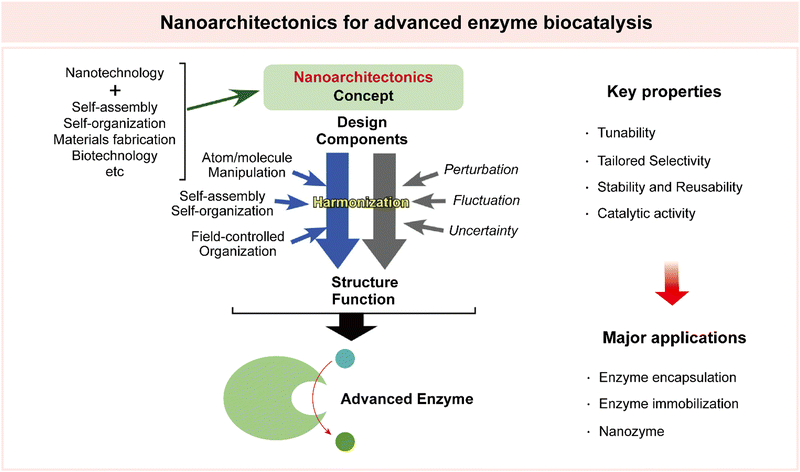 | ||
| Fig. 13 Illustration of the nanoarchitectonics concept for advanced enzyme biocatalysis. Reproduced with permission from ref. 35. Copyright 2019, Elsevier. | ||
Chen et al. designed an external hydrogen-bonded organic framework that could modulate the shape of the enzyme cytochrome c (Cyt c) (Fig. 14).190 This structural alteration enables the enzyme to exhibit functions that it could not naturally perform. A hydrogen-bonded organic framework, which is sturdy and has carboxylate molecules arranged in a specific way, is introduced onto the native Cyt c (Fig. 14A). The resultant hydrogen bonded nano-biointerface causes Cyt c to adopt a catalase-like conformation, which it typically could not take on when interacting with porous organic frameworks. The heme macrocycle of the Cyt c nanosystem in this paper is more structurally stable than those in other heme enzymes such as catalase (CAT). Also, this hydrogen-bonded organic framework continues to stabilize the enzyme, ensuring its structural integrity and activity (Fig. 14B).
 | ||
| Fig. 14 Representative example of enzyme encapsulation for advanced enzyme biocatalysis. (A) Schematic illustration of the technique for modulating the conformation of an enzyme by H-bonded cage encapsulation. (B) Structure of the heme macrocycle of Cyt c@HOF-101 based on MD simulation and its catalase-like biocatalysis performance. Reproduced with permission from ref. 190. Copyright 2022, Springer Nature. | ||
This research introduces a novel nanotechnological concept, which involves controlling the flexible shapes of enzymes. It demonstrates how artificial hydrogen-bonded frameworks can be used to modify enzyme activity, highlighting the benefits of using such scaffolds to modulate enzyme behavior. In essence, the study explores a new way to manipulate the structure and function of enzymes for various practical applications.
In an important example, Chen et al. presented a feasible approach that leverages the combined action of triazoles and metal ions to trigger the creation of porous hydrogels assembled with enzymes (Fig. 15).191 They developed an enzyme-assembled hydrogel (EAG) by adding 1H-3-methyl-1,2,4-triazole (Hmtz) and Mg2+ ions to the enzyme solution before the pre-gelation and gelation processes, respectively (Fig. 15A). Specifically, when they tested the catalytic efficiency of EAG in reducing acetophenone, it was an impressive 6.3 times higher than that of the free enzyme. Furthermore, they showed the reusability of this hydrogel by demonstrating its sustained high catalytic activity even after undergoing 12 cycles of use. To gain a deeper understanding of the structure of the enzyme within the hydrogel, they employed cryo-EM, achieving a near-atomic resolution of 2.1 Å (PDB: 7XY9). With these results, they analyzed the differences in enzyme activity and stability, before (1YKF) and after (7XY9) gelation. An overlay of the 7XY9 (salmon) and 1YKF (gray) structures shows the coordination of the Mg2+ ion with the adjacent residues (Fig. 15B). Also, EAG exhibited a higher specific activity than that of the free enzyme in various concentrations of acetophenone, and its enantioselectivity remained above 99% without any decrease (Fig. 15C).
 | ||
| Fig. 15 Representative example of enzyme immobilization for advanced biocatalysis. (A) Schematic illustration of enzyme assembled hydrogel (EAG) preparation using 1H-3-methyl-1,2,4-triazole (Hmtz) and Mg2+. (B) Overlay of the 7XY9 (salmon) and 1YKF (gray) structure showing the coordination of the Mg2+ ion with the adjacent residues. (C) Specific activity and chiral selectivity values of the free enzyme and EAG when catalyzing the conversion of acetophenone with concentrations of 5–20 nM. Reproduced with permission from ref. 191. Copyright 2019, Springer Nature. | ||
Additionally, this paper was able to shed light on the mechanism behind the formation of these hydrogels.191 The team discovered that both triazoles and metal ions play essential roles in this process. Armed with this knowledge, they extended our approach to utilize two other enzymes to create enzyme-assembled hydrogels, ensuring their excellent reusability. In conclusion, the strategy that they have described holds significant promise for advancing the development of practical catalytic biomaterials and immobilized biocatalysts. This approach represents a notable step forward in creating efficient and reusable biocatalytic materials with potential applications in various industries and processes.
In 2021, Ji et al. published their results on their single-atom iron nanozyme (Fig. 16).192 They prepared their FeN3P-single-atom nanozyme (SAzyme) based on zeolitic imidazolate framework-8 (ZIF-8) as a raw material followed by a coating process that involved polymerization and pyrolysis at 950 °C for 3 hours under flowing nitrogen (Fig. 16A). The engineered FeN3P-SAzyme exhibited peroxidase-like catalytic activity that is comparable to that of natural enzymes. In this study, they quantitatively determined the specific activity values by measuring the absorption intensity of the nanozyme-catalyzed colorimetric reactions. FeN3P-SAzyme showed much higher catalytic activity compared to the most widely used Fe3O4 nanozyme or FeN4-SAzyme without P coordination. The measured specific activity of the FeN3P-SAzyme was more than 30-fold higher than that of the Fe3O4 nanozyme and almost 10-fold higher than that of the FeN4-SAzyme (Fig. 16B). Among these three catalysts, adsorbed H2O2 will be easily dissociated into two surface OH species (H2O2 → 2OH) with very low barriers of 0.07, 0.17, and 0.10 eV on the three models of FeN3P-SAzyme, FeN4-SAzyme, and Fe3O4 (Fig. 16C).
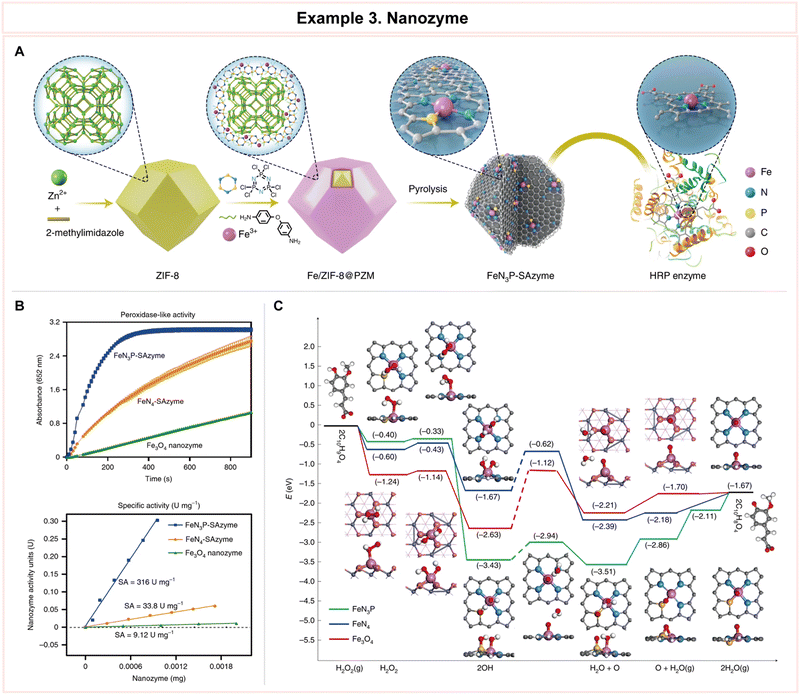 | ||
| Fig. 16 Representative example of nanozyme for advanced enzyme biocatalysis. (A) Schematic illustration of the preparation process for FeN3P-SAzyme. (B) Reaction–time curves of the colorimetric reaction and comparison of the specific activities of FeN3P-SAzyme, FeN3-SAzyme, and Fe3O4 nanozyme. (C) Density functional theory (DFT) studies and energy profile of FeN3P-SAzyme, FeN4-SAzyme and Fe3O4 nanozyme. Reproduced with permission from ref. 192. Copyright 2021, Springer Nature. | ||
To understand the basis for its high enzyme-like activity, they conducted density functional theory (DFT) calculations, providing insights into the underlying mechanisms. Importantly, they demonstrated that FeN3P-SAzyme with its superior peroxidase-like activity can serve as an effective therapeutic strategy for inhibiting the growth of tumor cells both in laboratory experiments and within living organisms. This shows the promising potential of SAzymes, such as FeN3P-SAzyme, for creating artificial enzymes that replicate the catalytic kinetics of natural enzymes. In summary, this research presents a significant step forward in the quest to develop artificial enzymes that rival the catalytic capabilities of their natural counterparts. The precise coordination of single atoms, as demonstrated by FeN3P-SAzyme, offers exciting prospects for future applications in biomedicine.
In summary, nanoarchitectonics for advanced enzyme biocatalysis is a cutting-edge field that harnesses nanotechnology to optimize the performance of enzymes in various applications. This multidisciplinary approach involves the precise design and construction of nanoscale structures and materials to enhance catalytic activity, enzyme stability, reusability, and versatility. Three primary strategies within this field are enzyme encapsulation, enzyme immobilization, and the development of nanozymes. However, these three strategies are not fully separated, for example, enzyme encapsulation and enzyme immobilization within hydrogel could be considered as both strategies. Also, when nanozymes, nanomaterials with intrinsic enzyme-like catalytic activity, are immobilized or encapsulated, several challenges such as loss of activity and specificity could be solved.193 So, by leveraging nanoscale structures and materials, researchers can optimize enzymes for a wide range of applications, including pharmaceuticals, biocatalysis, environmental remediation, and diagnostics. More research examples of nanoarchitectonics for advanced enzyme biocatalysis are described in Table 4.194–221
| Example | Applications | Enzyme | Key features | Detailed information | Ref. |
|---|---|---|---|---|---|
| (a) Enzyme encapsulation | |||||
| Metal–organic framework (MOF) | Pharmaceuticals, gas storage | Cytochrome c (Cyt c) | • A porous, crystalline material composed of inorganic nodes and organic linkers | • Common approaches: surface adsorption, covalent attachment, co-precipitation & biomineralization, diffusion into MoF pores/channels | 194 and 195 |
| • MOF could be also used as solid supports for enzyme immobilization | • Extremely high Brunauer–Emmett–Teller (BET) surface areas compared with other porous materials up to 7000 m2 g−1 | ||||
| Liposomal vesicle | Pharmaceuticals, food industry | α-Amylase | • Encapsulation efficiency is influenced by liposome number and size | • Glucose oxide (GOx) loaded liposome for starvation therapy shows high enzymatic activity | 196 and 197 |
| • Hydrophilic, amphipathic, and hydrophobic enzymes can all be loaded into liposomes | • Cyclic RCD (cRGD) functionalized liposome encapsulated with urokinase for thrombosis | ||||
| Hydrogel | Diagnostics, biomedical engineering | Horseradish peroxidase (HRP) | • In non-aqueous environments, hydrogels are essential for creating the favorable environment required by most enzymes | • Insulinase entrapped in calcium alginate–gelatin beads exhibits better thermal stability and reusability | 198 and 199 |
| • Most of natural polymers for hydrogels, are biocompatible, biodegradable, mechanically flexible, and renewable | • Laccase with synthetic polymer hydrogel composed of HEMA, ITA, and ECOA for wastewater treatment | ||||
| Viral capsid/virus like particle (VLP) | Pharmaceuticals, diagnostics | Candida antarctica lipase B (CALB) | • Various strategies inspired by viral capsid assembly and genome loading mechanisms have been developed to achieve greater and more controlled enzyme loading | • GFP fused with positively charged tag is directed to inside of the AaLS-neg cage via electrostatic interaction | 200 and 201 |
| • Encapsulation method: passive, electrostatic interaction, RNA packing signal, scaffold protein (SP), covalent conjugation | • Co-encapsulation of SP-tagged alcohol dehydrogenase (AdhD) and wild-type SP, with controlled stoichiometry is achieved by modulating the input ratio | ||||
| Biomimetic silica | Pharmaceuticals, diagnostics | Protease | • Biomimetic synthesis of silica nanoparticles is inspired by bio mineralization in marine organisms, such as diatoms | • Cargo molecules fused to the Cys-R5 peptide via a disulfide bond are incorporated into silica particles. | 202 and 203 |
| • Size and morphology of the particle could be tuned by controlling the initiator molecular weight and the precipitation conditions | • Co-precipitation by means of physically mixing the protein with the R5 peptide | ||||
| (b) Enzyme immobilization | |||||
| Immobilization of lipase | Pharmaceuticals, food industry | Candida antarctica lipase B (CALB) | • Enhanced stability | • Physical adsorption on hydrophobic media is most often used to immobilize lipase | 204 and 205 |
| • Reusability | • Lipase immobilization for biofuel production in mesoporous silica nanoparticles increases thermal and mechanical stability | ||||
| • Regeneration | |||||
| Immobilization of protease | Pharmaceuticals, detergents, food industry | Protease from Bacillus subtilis | • Enhanced stability | • Various proteases immobilized in alginate–chitosan beads exhibits reasonable stability and good activity | 206 |
| • Reduced autolysis | • Catalase embedded into single crystalline ZIF-90 crystals shows activity in hydrogen peroxide | ||||
| • Tailored selectivity | |||||
| Immobilization of kinase | Pharmaceuticals, diagnostics | Protein kinase A (PKA) | • Enhanced stability | • Immobilization of deoxyadenosine kinase from dictyostelium discoideum (DddAK) by ionic interaction on an aminated epoxy functionalized support enhances stability under high pH and temperature conditions | 207 and 208 |
| • Reusability | • A gold-binding polypeptide (GBP)-linked kinase immobilized on gold surface shows kinase assay sensitivity | ||||
| • High selectivity | |||||
| Immobilization of esterase | Pharmaceuticals, bioremediation, paper industry | Carboxylesterase (CEs) | • Enhanced stability | • Recombinant NStcI esterase adsorbed on accurel MP1000 exhibits high immobilization efficiency and storage stability under various conditions | 209 and 210 |
| • Reusability | • Est684, a cold adapted esterase, immobilization enhances thermostability and reusability for bioremediation | ||||
| • High activity | |||||
| Immobilization of aldolases | Bioremediation, diagnostics, organic synthesis | 2-Deoxy-D-ribose-5-phosphate aldolase (DERA) | • Enhanced stability | • Threonine aldolase immobilization on different supports for productive, cost-efficient enzymatic microreactors | 211 and 212 |
| • Longer lifespan | • Fixation of DERA-bearing thin film on a polymeric membrane support to gain atorvastatin efficiently | ||||
| • Cost effective | |||||
| (c) Nanozymes | |||||
| Gold nanozymes | Pharmaceuticals, diagnostics | Peroxidase, catalase, reductase | • Tunability | • A simple, sensitive and selective label-free colorimetric sensing of malathion using palladium–gold nanorod as nanozyme | 213 and 214 |
| • Biocompatibility | • A mesoporous carbon–gold hybrid nanoprobes for real-time imaging, photothermal/photodynamic and nanozyme oxidative therapy | ||||
| • Cost effectiveness | |||||
| Iron oxide nanozymes | Pharmaceuticals, diagnostics | Peroxidase, catalase | • Non-toxic and allergenic | • Yan's group discovered that iron oxide nanoparticles have an intrinsic peroxidase-like activity and could be used as a natural enzyme substitute | 215 |
| • High stability | • Gu's group showed that iron oxide nanozymes perform peroxidase-like activity under acidic pH and catalase-like activity under neutral pH | ||||
| • Tunability | |||||
| Magnetic nanozymes | Pharmaceuticals, diagnostics | Peroxidase, catalase, oxidase | • High stability | • Cobalt(II,III) oxide (Co3O4) nanotubes, prepared by electrospinning, show oxidase-like activity, peroxidase-like activity, and catalase-like activity | 216 and 217 |
| • Cost-effective | • An aptamer conjugated PtCo bimetallic nanoparticle has improved oxidase-like catalytic activity for cancer-cell detection without the destructive H2O2 | ||||
| • High activity | |||||
| Quantum dot (QD) nanozymes | Pharmaceuticals, diagnostics | Peroxidase, catalase, oxidase | • High stability | • Mo, S co-doped carbon quantum dots (Mo-CQDs) as a peroxidase mimic detects cholesterol with excellent selectivity and high sensitivity in the range of 0.01–1.0 mM | 218 and 219 |
| • Low toxicity | • Graphene oxide (GO) nanosheets, GO quantum dots (GOQDs), as nanozymes, efficiently reduce reactive oxygen species (ROS) and H2O2 in 1-methyl-4-phenyl-pyridinium ion (MPP+)-induced PC12 cells | ||||
| • High activity | |||||
| MOF nanozymes | Pharmaceuticals, diagnostics | Peroxidase, catalase, oxidase, hydrolases | • High activity | • Hierarchically structured and highly dispersible MOF nanozymes showed enhanced peroxidase-like activity for persistent chemiluminescence immunoassay | 220 and 221 |
| • High stability | • Ultrasensitive detection of pathogenic bacteria using a 2D MOF nanozyme with peroxidase-like activity | ||||
| • Low toxicity | |||||
In conclusion, multidisciplinary approaches in enzyme biocatalysis mark a critical moment in the evolution of pharmaceutical research and production. By the precision of protein engineering, the computational prowess of computational biology, and the innovative strategies of nanoarchitectonics, scientists are poised to unlock the full potential of enzymes as molecular tools. These methods not only promise to address the challenges posed by modern pharmaceutical demands but also lay the groundwork for a more sustainable and efficient future in drug development and manufacturing. As stated in the following sections, the intricate interplay of these multidisciplinary techniques will shed light on the transformative impact they have on enzyme biocatalysis. Through collaborative efforts and continuous advancements, the pharmaceutical industry is on the brink of a new era, one where enzyme biocatalysis stands as an indispensable cornerstone, shaping the landscape of pharmaceutical innovation for years to come. Looking ahead, for industrial applications, further exploration, and consideration of additional modifications in biocatalysis for long-term use, such as enzyme cross-linking,222 chemical modification,223 and formulation environment control,224 hold the potential to revolutionize the efficiency and scalability of enzyme-driven processes.
5. Conclusions
5.1 Summary
In the ever-evolving realm of pharmaceutical research and production, the utilization of enzyme biocatalysis stands as a beacon of innovation and sustainability. Throughout this comprehensive exploration, we have emphasized the significance of enzyme biocatalysis in advancing pharmaceutical applications, with particular attention to the introduction of multidisciplinary approaches as catalysts for progress. The convergence of multidisciplinary approaches, encompassing protein engineering, computational biology, and nanoarchitectonics, has heralded a new era for enzyme biocatalysis. This interdisciplinary synergy has proven to be greater than the sum of its parts, unlocking novel opportunities and redefining the boundaries of what is achievable in pharmaceutical research and production.This transformative technology offers a wealth of advantages, from the precision and selectivity of enzyme-catalyzed reactions to their eco-friendly and sustainable attributes. Significant progress lies in the field of computational biology. This burgeoning field has emerged as a groundbreaking approach for optimizing enzyme biocatalysis, and its computational prowess empowers pharmaceutical scientists to optimize drug candidates, improve pharmacokinetics, and reduce toxicity, all with a level of precision that was once unimaginable. In the field of nanoarchitectonics, innovative strategies for enzyme encapsulation and immobilization within tailor-designed nanomaterials have enhanced enzyme stability and reusability.
5.2 Perspectives
As we conclude this comprehensive review, it is crucial to cast our gaze towards the future of advanced enzyme biocatalysis in pharmaceutical applications. There are three key perspectives that hold immense promise (Fig. 17):In summary, the evolution of enzyme biocatalysis in pharmaceutical applications continues to unfold. The integration of multidisciplinary approaches, the advancement of multifunctional systems, and the continued pursuit of bioprocess optimization beckon as uncharted territories brimming with untapped potential. As researchers embark on this next phase, their vision encompasses a pharmaceutical domain marked by heightened speed, sustainability, efficiency, and precision. Through collaborative efforts and innovative strides, researchers are poised to expand the boundaries of what can be accomplished, confirming enzyme biocatalysis as an indispensable cornerstone of pharmaceutical research and production for the foreseeable future.
Author contributions
S. Kim: conceptualization, visualization, writing – original draft, writing – review & editing. S. Ga: writing – original draft. H. Bae: writing – original draft. R. Sluyter: writing – review & editing. K. Konstantinov: writing – review & editing. L. K. Shrestha: writing – review & editing. Y. H. Kim: supervision, funding acquisition, writing – review & editing. J. H. Kim: supervision, project administration, writing – review & editing. K. Ariga: supervision, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2023R1A2C3005731). This work was also supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government Ministry of Science and ICT (MSIT) (No. 2021R1A4A1033424).References
- U. T. Bornscheuer, G. W. Huisman, R. J. Kazlauskas, S. Lutz, J. C. Moore and K. Robins, Nature, 2012, 485, 185–194 CrossRef CAS
.
- E. M. M. Abdelraheem, H. Busch, U. Hanefeld and F. Tonin, React. Chem. Eng., 2019, 4, 1878–1894 RSC
.
- A. Fryszkowska and P. N. Devine, Curr. Opin. Chem. Biol., 2020, 55, 151–160 CrossRef CAS
.
- M. D. Truppo, ACS Med. Chem. Lett., 2017, 8, 476–480 CrossRef CAS PubMed
.
- D. Yi, T. Bayer, C. P. S. Badenhorst, S. Wu, M. Doerr, M. Höhne and U. T. Bornscheuer, Chem. Soc. Rev., 2021, 50, 8003–8049 RSC
.
- P. N. Devine, R. M. Howard, R. Kumar, M. P. Thompson, M. D. Truppo and N. J. Turner, Nat. Rev. Chem., 2018, 2, 409–421 CrossRef
.
- A. Kinner, P. Nerke, R. Siedentop, T. Steinmetz, T. Classen, K. Rosenthal, M. Nett, J. Pietruszka and S. Lütz, Biomedicines, 2022, 10, 964 CrossRef CAS
.
- N. C. Goodwin, J. P. Morrison, D. E. Fuerst and T. Hadi, ACS Med. Chem. Lett., 2019, 10, 1363–1366 CrossRef CAS PubMed
.
- J.-M. Choi, S.-S. Han and H.-S. Kim, Biotechnol. Adv., 2015, 33, 1443–1454 CrossRef CAS
.
- C. A. Busacca, D. R. Fandrick, J. J. Song and C. H. Senanayake, Adv. Synth. Catal., 2011, 353, 1825–1864 CrossRef CAS
.
- J. D. Hayler, D. K. Leahy and E. M. Simmons, Organometallics, 2019, 38, 36–46 CrossRef CAS
.
- E. L. Bell, W. Finnigan, S. P. France, A. P. Green, M. A. Hayes, L. J. Hepworth, S. L. Lovelock, H. Niikura, S. Osuna, E. Romero, K. S. Ryan, N. J. Turner and S. L. Flitsch, Nat. Rev. Methods Primers, 2021, 1, 46 CrossRef CAS
.
- S. P. France, R. D. Lewis and C. A. Martinez, JACS Au, 2023, 3, 715–735 CrossRef CAS PubMed
.
- B. Wiltschi, T. Cernava, A. Dennig, M. Galindo Casas, M. Geier, S. Gruber, M. Haberbauer, P. Heidinger, E. H. Acero, R. Kratzer, C. Luley-Goedl, C. A. Müller, J. Pitzer, D. Ribitsch, M. Sauer, K. Schmölzer, W. Schnitzhofer, C. W. Sensen, J. Soh, K. Steiner, C. K. Winkler, M. Winkler and T. Wriessnegger, Biotechnol. Adv., 2020, 40, 107520 CrossRef CAS
.
- B.-S. Chen and F. Z. Ribeiro de Souza, RSC Adv., 2019, 9, 2102–2115 RSC
.
- Y. Ding, D. Zheng, L. Xie, X. Zhang, Z. Zhang, L. Wang, Z.-W. Hu and Z. Yang, J. Am. Chem. Soc., 2023, 145, 4366–4371 CrossRef CAS PubMed
.
- F. Lin, C. Jia and F.-G. Wu, Molecules, 2022, 27, 6557 CrossRef CAS PubMed
.
- S. Palluk, D. H. Arlow, T. de Rond, S. Barthel, J. S. Kang, R. Bector, H. M. Baghdassarian, A. N. Truong, P. W. Kim, A. K. Singh, N. J. Hillson and J. D. Keasling, Nat. Biotechnol., 2018, 36, 645–650 CrossRef CAS PubMed
.
- S. Agarwal and E. Franco, J. Am. Chem. Soc., 2019, 141, 7831–7841 CrossRef CAS PubMed
.
- T. T. Ashburn and K. B. Thor, Nat. Rev. Drug Discovery, 2004, 3, 673–683 CrossRef CAS PubMed
.
- A. Debon, E. Siirola and R. Snajdrova, JACS Au, 2023, 3, 1267–1283 CrossRef CAS
.
- R. A. Sheldon and P. C. Pereira, Chem. Soc. Rev., 2017, 46, 2678–2691 RSC
.
- J. B. Pyser, S. Chakrabarty, E. O. Romero and A. R. H. Narayan, ACS Cent. Sci., 2021, 7, 1105–1116 CrossRef CAS PubMed
.
- J. P. Adams, M. J. B. Brown, A. Diaz-Rodriguez, R. C. Lloyd and G.-D. Roiban, Adv. Synth. Catal., 2019, 361, 2421–2432 CrossRef CAS
.
- W. Li, M. Bilal, A. K. Singh, F. Sher, S. S. Ashraf, M. Franco, J. H. P. Américo-Pinheiro and H. M. N. Iqbal, Catal. Lett., 2023, 153, 1227–1239 CrossRef CAS
.
- K. Chen and F. H. Arnold, Nat. Catal., 2020, 3, 1–11 CrossRef
.
- J. A. McIntosh and A. E. Owens, Curr. Opin. Green Sustainable, 2021, 29, 100448 CrossRef CAS
.
- A. H.-W. Yeh, C. Norn, Y. Kipnis, D. Tischer, S. J. Pellock, D. Evans, P. Ma, G. R. Lee, J. Z. Zhang, I. Anishchenko, B. Coventry, L. Cao, J. Dauparas, S. Halabiya, M. DeWitt, L. Carter, K. N. Houk and D. Baker, Nature, 2023, 614, 774–780 CrossRef CAS
.
- L. Tallorin, J. Wang, W. E. Kim, S. Sahu, N. M. Kosa, P. Yang, M. Thompson, M. K. Gilson, P. I. Frazier, M. D. Burkart and N. C. Gianneschi, Nat. Commun., 2018, 9, 5253 CrossRef CAS PubMed
.
- S. Goldman, R. Das, K. K. Yang and C. W. Coley, PLoS Comput. Biol., 2022, 18, e1009853 CrossRef CAS PubMed
.
- Y. Qiu, J. Hu and G.-W. Wei, Nat. Comput. Sci., 2021, 1, 809–818 CrossRef
.
- K. K. Yang, Z. Wu and F. H. Arnold, Nat. Methods, 2019, 16, 687–694 CrossRef CAS PubMed
.
- P. Schneider and G. Schneider, Angew. Chem., Int. Ed., 2017, 56, 11520–11524 CrossRef CAS PubMed
.
- S. Roda, L. Fernandez-Lopez, R. Cañadas, G. Santiago, M. Ferrer and V. Guallar, ACS Catal., 2021, 11, 3590–3601 CrossRef CAS
.
- M. Komiyama and K. Ariga, Mol. Catal., 2019, 475, 110492 CrossRef CAS
.
- K. Ariga, Q. Ji, T. Mori, M. Naito, Y. Yamauchi, H. Abe and J. P. Hill, Chem. Soc. Rev., 2013, 42, 6322–6345 RSC
.
- H. Abe, J. Liu and K. Ariga, Mater. Today, 2016, 19, 12–18 CrossRef CAS
.
- R. A. Sheldon, D. Brady and M. L. Bode, Chem. Sci., 2020, 11, 2587–2605 RSC
.
- B. Hauer, ACS Catal., 2020, 10, 8418–8427 CrossRef CAS
.
- D. J. Pollard and J. M. Woodley, Trends Biotechnol., 2007, 25, 66–73 CrossRef CAS
.
- S.-i Shoda, H. Uyama, J.-i Kadokawa, S. Kimura and S. Kobayashi, Chem. Rev., 2016, 116, 2307–2413 CrossRef CAS PubMed
.
- Nat. Catal., 2020, 3, 179–180, DOI:10.1038/s41929-020-0447-8.
- R. A. Sheldon and J. M. Woodley, Chem. Rev., 2018, 118, 801–838 CrossRef CAS
.
- R. G. Araújo, R. B. González-González, M. Martinez-Ruiz, K. G. Coronado-Apodaca, H. Reyes-Pardo, Z. P. Morreeuw, M. A. Oyervides-Muñoz, J. E. Sosa-Hernández, D. Barceló, R. Parra-Saldívar and H. M. N. Iqbal, ACS Omega, 2022, 7, 32863–32876 CrossRef
.
- Y. Miao, P. G. Tepper, E. M. Geertsema and G. J. Poelarends, Eur. J. Org. Chem., 2016, 5350–5354 CrossRef CAS
.
- S. Wu, R. Snajdrova, J. C. Moore, K. Baldenius and U. T. Bornscheuer, Angew. Chem., Int. Ed., 2021, 60, 88–119 CrossRef CAS
.
- M. Schober, C. MacDermaid, A. A. Ollis, S. Chang, D. Khan, J. Hosford, J. Latham, L. A. F. Ihnken, M. J. B. Brown, D. Fuerst, M. J. Sanganee and G.-D. Roiban, Nat. Catal., 2019, 2, 909–915 CrossRef CAS
.
- R. N. Patel, Coord. Chem. Rev., 2008, 252, 659–701 CrossRef CAS
.
- W. H. Brooks, W. C. Guida and K. G. Daniel, Curr. Top. Med. Chem., 2011, 11, 760–770 CrossRef CAS PubMed
.
- Z. S. Seddigi, M. S. Malik, S. A. Ahmed, A. O. Babalghith and A. Kamal, Coord. Chem. Rev., 2017, 348, 54–70 CrossRef CAS
.
- Z. Wu, W. Shi, M. Jin and W. Zhou, RSC Adv., 2023, 13, 18953–18959 RSC
.
- L. Qiao, Z. Luo, H. Chen, P. Zhang, A. Wang and R. A. Sheldon, Chem. Commun., 2023, 59, 7518–7533 RSC
.
- M. Furegati and S. Nocito, Org. Process Res. Dev., 2017, 21, 1822–1827 CrossRef CAS
.
- I. Nasibullin, I. Smirnov, P. Ahmadi, K. Vong, A. Kurbangalieva and K. Tanaka, Nat. Commun., 2022, 13, 39 CrossRef CAS
.
- Y.-h Yang, H. Aloysius, D. Inoyama, Y. Chen and L.-q Hu, Acta Pharm. Sin. B, 2011, 1, 143–159 CrossRef CAS
.
- G. A. Holdgate, T. D. Meek and R. L. Grimley, Nat. Rev. Drug Discovery, 2018, 17, 115–132 CrossRef CAS PubMed
.
- A. S. Sheweita, Curr. Drug Metab., 2000, 1, 107–132 CrossRef
.
- Y. Jarrar and S.-J. Lee, J. Pers. Med., 2021, 11, 554 CrossRef PubMed
.
- N. Gamage, A. Barnett, N. Hempel, R. G. Duggleby, K. F. Windmill, J. L. Martin and M. E. McManus, Toxicol. Sci., 2006, 90, 5–22 CrossRef CAS
.
- Q. Hu, P. S. Katti and Z. Gu, Nanoscale, 2014, 6, 12273–12286 RSC
.
- R. M. Lieser, D. Yur, M. O. Sullivan and W. Chen, Bioconjugate Chem., 2020, 31, 2272–2282 CrossRef CAS
.
- J. G. Plaks, R. Falatach, M. Kastantin, J. A. Berberich and J. L. Kaar, Bioconjugate Chem., 2015, 26, 1104–1112 CrossRef CAS PubMed
.
- Z. Zhang and W. Tang, Acta Pharm. Sin. B, 2018, 8, 721–732 CrossRef
.
- J. Shanu-Wilson, L. Evans, S. Wrigley, J. Steele, J. Atherton and J. Boer, ACS Med. Chem. Lett., 2020, 11, 2087–2107 CrossRef CAS
.
- J. Li, J. Yang, S. Mu, N. Shang, C. Liu, Y. Zhu, Y. Cai, P. Liu, J. Lin, W. Liu, Y. Sun and Y. Ma, ACS Catal., 2020, 10, 3629–3639 CrossRef CAS
.
- J. Latham, E. Brandenburger, S. A. Shepherd, B. R. K. Menon and J. Micklefield, Chem. Rev., 2018, 118, 232–269 CrossRef CAS PubMed
.
- E. T. Hwang and S. Lee, ACS Catal., 2019, 9, 4402–4425 CrossRef CAS
.
- Y. Wu, R. Xu, Y. Feng and H. Song, ACS Catal., 2022, 12, 3767–3775 CrossRef CAS
.
- R. Cutlan, S. De Rose, M. N. Isupov, J. A. Littlechild and N. J. Harmer, Biochim. Biophys. Acta, Proteins Proteomics, 2020, 1868, 140322 CrossRef CAS PubMed
.
- A. I. Benítez-Mateos, D. Roura Padrosa and F. Paradisi, Nat. Chem., 2022, 14, 489–499 CrossRef PubMed
.
- R. Croteau, R. E. B. Ketchum, R. M. Long, R. Kaspera and M. R. Wildung, Phytochem. Rev., 2006, 5, 75–97 CrossRef CAS
.
- X. Chen, C. Zhang and H.-P. Too, Molecules, 2017, 22, 1422 CrossRef
.
- J. K. Blum, A. L. Deaguero, C. V. Perez and A. S. Bommarius, ChemCatChem, 2010, 2, 987–991 CrossRef CAS
.
- A. Švarc, Z. Findrik Blažević, Đ. Vasić-Rački, S. J. Charnock and A. Vrsalović Presečki, Chem. Eng. Res. Des., 2020, 164, 35–45 CrossRef
.
- J. M. Sperl and V. Sieber, ACS Catal., 2018, 8, 2385–2396 CrossRef CAS
.
- S. Ren, C. Li, X. Jiao, S. Jia, Y. Jiang, M. Bilal and J. Cui, Chem. Eng. J., 2019, 373, 1254–1278 CrossRef CAS
.
- I. Slabu, J. L. Galman, R. C. Lloyd and N. J. Turner, ACS Catal., 2017, 7, 8263–8284 CrossRef CAS
.
- A. Gomm and E. O’Reilly, Curr. Opin. Chem. Biol., 2018, 43, 106–112 CrossRef CAS
.
- J. Altenbuchner, M. Siemann-Herzberg and C. Syldatk, Curr. Opin. Biotechnol, 2001, 12, 559–563 CrossRef CAS PubMed
.
- G.-W. Zheng, Y.-Y. Liu, Q. Chen, L. Huang, H.-L. Yu, W.-Y. Lou, C.-X. Li, Y.-P. Bai, A.-T. Li and J.-H. Xu, ACS Catal., 2017, 7, 7174–7181 CrossRef CAS
.
- C. Huang, J. Liu, J. Fang, X. Jia, Z. Zheng, S. You and B. Qin, Front. Bioeng. Biotechnol., 2022, 10, 929784 CrossRef PubMed
.
- E. P. Allain, M. Rouleau, E. Lévesque and C. Guillemette, Br. J. Cancer, 2020, 122, 1277–1287 CrossRef
.
- C. Asadov, G. Aliyeva and K. Mustafayeva, Cardiovasc. Hematol. Agents Med. Chem., 2017, 15, 23–30 CAS
.
- W. E. Evans, Ther. Drug Monit., 2004, 26, 186–191 CrossRef CAS
.
- N. D. Fessner, C. Grimm, M. Srdič, H. Weber, W. Kroutil, U. Schwaneberg and A. Glieder, ChemCatChem, 2022, 14, e202101564 CrossRef CAS
.
- J. H. Park, S. H. Lee, G. S. Cha, D. S. Choi, D. H. Nam, J. H. Lee, J.-K. Lee, C.-H. Yun, K. J. Jeong and C. B. Park, Angew. Chem., Int. Ed., 2015, 54, 969–973 CrossRef CAS PubMed
.
- L. Schmohl and D. Schwarzer, Curr. Opin. Chem. Biol., 2014, 22, 122–128 CrossRef CAS PubMed
.
- P. Dennler, A. Chiotellis, E. Fischer, D. Brégeon, C. Belmant, L. Gauthier, F. Lhospice, F. Romagne and R. Schibli, Bioconjugate Chem., 2014, 25, 569–578 CrossRef CAS PubMed
.
- J. A. Shadish and C. A. DeForest, Matter, 2020, 2, 50–77 CrossRef
.
- M. T. Manzari, Y. Shamay, H. Kiguchi, N. Rosen, M. Scaltriti and D. A. Heller, Nat. Rev. Mater., 2021, 6, 351–370 CrossRef CAS
.
- J. Hines, J. D. Gough, T. W. Corson and C. M. Crews, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 8942–8947 CrossRef CAS
.
- M. R. Bennett, S. A. Shepherd, V. A. Cronin and J. Micklefield, Curr. Opin. Chem. Biol., 2017, 37, 97–106 CrossRef CAS PubMed
.
- S. E. Payer, S. A. Marshall, N. Bärland, X. Sheng, T. Reiter, A. Dordic, G. Steinkellner, C. Wuensch, S. Kaltwasser, K. Fisher, S. E. J. Rigby, P. Macheroux, J. Vonck, K. Gruber, K. Faber, F. Himo, D. Leys, T. Pavkov-Keller and S. M. Glueck, Angew. Chem., Int. Ed., 2017, 56, 13893–13897 CrossRef CAS PubMed
.
- M. Breuer, K. Ditrich, T. Habicher, B. Hauer, M. Keßeler, R. Stürmer and T. Zelinski, Angew. Chem., Int. Ed., 2004, 43, 788–824 CrossRef CAS PubMed
.
-
G. Zhang, Y. Chen and G. Du, in Biomass, Biofuels, Biochemicals, ed. S. P. Singh, A. Pandey, R. R. Singhania, C. Larroche and Z. Li, Elsevier, Amsterdam, The Netherlands, 2020, ch. 7, pp. 103–117 Search PubMed
.
- R. A. Sheldon and S. van Pelt, Chem. Soc. Rev., 2013, 42, 6223–6235 RSC
.
- A. I. Benítez-Mateos, M. L. Contente, D. Roura Padrosa and F. Paradisi, React. Chem. Eng., 2021, 6, 599–611 RSC
.
- R. Macarron, M. N. Banks, D. Bojanic, D. J. Burns, D. A. Cirovic, T. Garyantes, D. V. S. Green, R. P. Hertzberg, W. P. Janzen, J. W. Paslay, U. Schopfer and G. S. Sittampalam, Nat. Rev. Drug Discovery, 2011, 10, 188–195 CrossRef CAS
.
- M. Baweja, L. Nain, Y. Kawarabayasi and P. Shukla, Front. Microbiol., 2016, 7, 965 Search PubMed
.
- R. A. Rocha, R. E. Speight and C. Scott, Org. Process Res. Dev., 2022, 26, 1914–1924 CrossRef CAS
.
- M. Wang, T. Si and H. Zhao, Bioresour. Technol., 2012, 115, 117–125 CrossRef CAS
.
- L. Lancaster, W. Abdallah, S. Banta and I. Wheeldon, Chem. Soc. Rev., 2018, 47, 5177–5186 RSC
.
- M. A. Ashworth, E. Bombino, R. M. de Jong, H. J. Wijma, D. B. Janssen, K. J. McLean and A. W. Munro, ACS Catal., 2022, 12, 15028–15044 CrossRef CAS
.
- V. M. de Oliveira, M. N. da Rocha, E. P. Magalhães, F. R. da Silva Mendes, M. M. Marinho, R. R. P. P. B. de Menezes, T. L. Sampaio, H. S. dos Santos, A. M. C. Martins and E. S. Marinho, Future J. Pharm. Sci., 2021, 7, 185 CrossRef
.
- M. Musil, H. Konegger, J. Hon, D. Bednar and J. Damborsky, ACS Catal., 2019, 9, 1033–1054 CrossRef CAS
.
- G. Kiss, N. Çelebi-Ölçüm, R. Moretti, D. Baker and K. N. Houk, Angew. Chem., Int. Ed., 2013, 52, 5700–5725 CrossRef CAS
.
- W. S. Mak and J. B. Siegel, Curr. Opin. Struct. Biol., 2014, 27, 87–94 CrossRef CAS PubMed
.
- M. Scherer, S. J. Fleishman, P. R. Jones, T. Dandekar and E. Bencurova, Front. Bioeng. Biotechnol., 2021, 9, 673005 CrossRef PubMed
.
- Y. Cui, J. Sun and B. Wu, Trends Chem., 2022, 4, 409–419 CrossRef CAS
.
- S. Dutta, J. Kim, P.-H. Hsieh, Y.-S. Hsu, Y. V. Kaneti, F.-K. Shieh, Y. Yamauchi and K. C.-W. Wu, Small Methods, 2019, 3, 1900213 CrossRef CAS
.
- A. S. Bommarius, J. K. Blum and M. J. Abrahamson, Curr. Opin. Chem. Biol., 2011, 15, 194–200 CrossRef CAS PubMed
.
- T. Dinmukhamed, Z. Huang, Y. Liu, X. Lv, J. Li, G. Du and L. Liu, Syst. Microbiol. Biomanuf., 2021, 1, 15–23 CrossRef CAS
.
- M.-J. Seo and C. Schmidt-Dannert, Catalysts, 2021, 11, 409 CrossRef CAS
.
- R. E. Cobb, R. Chao and H. Zhao, AIChE J., 2013, 59, 1432–1440 CrossRef CAS
.
- O. Khersonsky, D. Röthlisberger, O. Dym, S. Albeck, C. J. Jackson, D. Baker and D. S. Tawfik, J. Mol. Biol., 2010, 396, 1025–1042 CrossRef CAS
.
- O. F. Brandenberg, K. Chen and F. H. Arnold, J. Am. Chem. Soc., 2019, 141, 8989–8995 CrossRef CAS PubMed
.
- U. Markel, K. D. Essani, V. Besirlioglu, J. Schiffels, W. R. Streit and U. Schwaneberg, Chem. Soc. Rev., 2020, 49, 233–262 RSC
.
- S. Lutz, Curr. Opin. Biotechnol, 2010, 21, 734–743 CrossRef CAS PubMed
.
- X. Chen, H. Zhang, M. A. Maria-Solano, W. Liu, J. Li, J. Feng, X. Liu, S. Osuna, R.-T. Guo, Q. Wu, D. Zhu and Y. Ma, Nat. Catal., 2019, 2, 931–941 CrossRef CAS
.
- C. J. Wilson, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2015, 7, 330–341 CAS
.
- H. F. Son, I. J. Cho, S. Joo, H. Seo, H.-Y. Sagong, S. Y. Choi, S. Y. Lee and K.-J. Kim, ACS Catal., 2019, 9, 3519–3526 CrossRef CAS
.
- Y. Tan, Y. Zhang, Y. Han, H. Liu, H. Chen, F. Ma, S. G. Withers, Y. Feng and G. Yang, Sci. Adv., 2019, 5, eaaw8451 CrossRef CAS PubMed
.
- F. Cheng, X.-L. Chen, C. Xiang, Z.-Q. Liu, Y.-J. Wang and Y.-G. Zheng, Appl. Microbiol. Biotechnol., 2020, 104, 2999–3009 CrossRef CAS PubMed
.
- Y. Gong, G.-C. Xu, Q. Chen, J.-G. Yin, C.-X. Li and J.-H. Xu, Catal. Sci. Technol., 2016, 6, 2370–2376 RSC
.
- M. G. Gall, A. Nobili, I. V. Pavlidis and U. T. Bornscheuer, Appl. Microbiol. Biotechnol., 2014, 98, 1719–1726 CrossRef CAS
.
- L.-c Cao, Z.-j Wang, G.-h Ren, W. Kong, L. Li, W. Xie and Y.-h Liu, Biotechnol. Biofuels, 2015, 8, 202 CrossRef
.
- H. Takagi, I. Ohtsu and S. Nakamori, Protein Eng., 1997, 10, 985–989 CrossRef CAS PubMed
.
- Y.-H. Huang, S. T. Henriques, C. K. Wang, L. Thorstholm, N. L. Daly, Q. Kaas and D. J. Craik, Sci. Rep., 2015, 5, 12974 CrossRef CAS
.
- D. H. Patel, E. J. Cho, H. M. Kim, I. S. Choi and H. J. Bae, Protein Eng., Des. Sel., 2012, 25, 331–336 CrossRef CAS PubMed
.
- L. Zhai, Z. Wang, L. Fen, C. Xu, J. Wang, H. Han, Q. Xie, W. Zhang, Y. Zheng, A. K. Buell and Y. Dong, bioRxiv, 2023, preprint DOI:10.1101/2023.03.20.533374.
- N. H. Daud, T. C. Leow, S. N. Oslan and A. B. Salleh, Mol. Biotechnol., 2019, 61, 477–488 CrossRef CAS PubMed
.
- I. Pardo and S. Camarero, Cell. Mol. Life Sci., 2015, 72, 897–910 CrossRef CAS PubMed
.
- F. Autore, C. Del Vecchio, F. Fraternali, P. Giardina, G. Sannia and V. Faraco, Enzyme Microb. Technol., 2009, 45, 507–513 CrossRef CAS
.
- H. Land, F. Ruggieri, A. Szekrenyi, W.-D. Fessner and P. Berglund, Adv. Synth. Catal., 2020, 362, 812–821 CrossRef CAS
.
- X. Liu, L.-C. Cao, X.-J. Fan, Y.-H. Liu and W. Xie, Sci. Rep., 2016, 6, 38137 CrossRef CAS
.
- X. Yan, J. Wang, Y. Sun, J. Zhu and S. Wu, Appl. Environ. Microbiol., 2016, 82, 6748–6756 CrossRef CAS
.
- D. Mariano, N. Pantuza, L. H. Santos, R. E. O. Rocha, L. H. F. de Lima, L. Bleicher and R. C. de Melo-Minardi, BMC Mol. Cell Biol., 2020, 21, 50 CrossRef CAS PubMed
.
- H. Wulf, H. Mallin and U. T. Bornscheuer, Enzyme Microb., 2012, 51, 217–224 CrossRef CAS
.
- L. Sellés Vidal, C. L. Kelly, P. M. Mordaka and J. T. Heap, Biochim. Biophys. Acta, Proteins Proteomics, 2018, 1866, 327–347 CrossRef PubMed
.
- A. Schenkmayerova, G. P. Pinto, M. Toul, M. Marek, L. Hernychova, J. Planas-Iglesias, V. Daniel Liskova, D. Pluskal, M. Vasina, S. Emond, M. Dörr, R. Chaloupkova, D. Bednar, Z. Prokop, F. Hollfelder, U. T. Bornscheuer and J. Damborsky, Nat. Commun., 2021, 12, 3616 CrossRef CAS PubMed
.
- S. L. Lovelock, R. Crawshaw, S. Basler, C. Levy, D. Baker, D. Hilvert and A. P. Green, Nature, 2022, 606, 49–58 CrossRef CAS PubMed
.
- T. Yu, A. G. Boob, M. J. Volk, X. Liu, H. Cui and H. Zhao, Nat. Catal., 2023, 6, 137–151 CrossRef CAS
.
- C. Angermueller, T. Pärnamaa, L. Parts and O. Stegle, Mol. Syst. Biol., 2016, 12, 878 CrossRef PubMed
.
- G. Sliwoski, S. Kothiwale, J. Meiler, J. Edward and W. Lowe, Pharmacol. Rev., 2014, 66, 334–395 CrossRef PubMed
.
- M. R. Hurle, L. Yang, Q. Xie, D. K. Rajpal, P. Sanseau and P. Agarwal, Clin. Pharmacol. Ther., 2013, 93, 335–341 CrossRef CAS PubMed
.
- G. Eraslan, Ž. Avsec, J. Gagneur and F. J. Theis, Nat. Rev. Genet., 2019, 20, 389–403 CrossRef CAS
.
- S. J. Y. Macalino, V. Gosu, S. Hong and S. Choi, Arch. Pharmacal Res., 2015, 38, 1686–1701 CrossRef CAS
.
- A. V. Sadybekov and V. Katritch, Nature, 2023, 616, 673–685 CrossRef CAS
.
- R. Li, H. J. Wijma, L. Song, Y. Cui, M. Otzen, Y. E. Tian, J. Du, T. Li, D. Niu, Y. Chen, J. Feng, J. Han, H. Chen, Y. Tao, D. B. Janssen and B. Wu, Nat. Chem. Biol., 2018, 14, 664–670 CrossRef CAS PubMed
.
- H. A. Bunzel, J. L. R. Anderson, D. Hilvert, V. L. Arcus, M. W. van der Kamp and A. J. Mulholland, Nat. Chem., 2021, 13, 1017–1022 CrossRef CAS
.
- P. Wang, J. Zhang, S. Zhang, D. Lu and Y. Zhu, J. Chem. Inf. Model., 2023, 63, 1323–1337 CrossRef CAS PubMed
.
- S. L. Robinson, J. Piel and S. Sunagawa, Nat. Prod. Rep., 2021, 38, 1994–2023 RSC
.
- R. Vanella, G. Kovacevic, V. Doffini, J. Fernández de Santaella and M. A. Nash, Chem. Commun., 2022, 58, 2455–2467 RSC
.
- P. Vanacek, E. Sebestova, P. Babkova, S. Bidmanova, L. Daniel, P. Dvorak, V. Stepankova, R. Chaloupkova, J. Brezovsky, Z. Prokop and J. Damborsky, ACS Catal., 2018, 8, 2402–2412 CrossRef CAS
.
- A. M. Schnoes, S. D. Brown, I. Dodevski and P. C. Babbitt, PLoS Comput. Biol., 2009, 5, e1000605 CrossRef
.
- Y. Ding, G. Perez-Ortiz, J. Peate and S. M. Barry, Front. Mol. Biosci., 2022, 9, 908285 CrossRef CAS
.
- M. B. Quin and C. Schmidt-Dannert, ACS Catal., 2011, 1, 1017–1021 CrossRef CAS
.
- H. Lu, D. J. Diaz, N. J. Czarnecki, C. Zhu, W. Kim, R. Shroff, D. J. Acosta, B. R. Alexander, H. O. Cole, Y. Zhang, N. A. Lynd, A. D. Ellington and H. S. Alper, Nature, 2022, 604, 662–667 CrossRef CAS
.
- R. Shroff, A. W. Cole, D. J. Diaz, B. R. Morrow, I. Donnell, A. Annapareddy, J. Gollihar, A. D. Ellington and R. Thyer, ACS Synth. Biol., 2020, 9, 2927–2935 CrossRef CAS
.
- S. Vardhan and S. K. Sahoo, Comput. Biol. Med., 2020, 124, 103936 CrossRef CAS PubMed
.
- Y. Kumar, H. Singh and C. N. Patel, J. Infect. Public Health, 2020, 13, 1210–1223 CrossRef PubMed
.
- G. Oanca, M. Purg, J. Mavri, J. C. Shih and J. Stare, Phys. Chem. Chem. Phys., 2016, 18, 13346–13356 RSC
.
- L. Fang, M. Geng, C. Liu, J. Wang, W. Min and J. Liu, PLoS One, 2019, 14, e0215609 CrossRef CAS PubMed
.
- R. Chen, B. Gao, X. Liu, F. Ruan, Y. Zhang, J. Lou, K. Feng, C. Wunsch, S.-M. Li, J. Dai and F. Sun, Nat. Chem. Biol., 2017, 13, 226–234 CrossRef CAS
.
- R. Dorau, T. Görbe and M. Svedendahl Humble, ChemBioChem, 2018, 19, 338–346 CrossRef CAS
.
- G. Sawant, S. Ghosh, S. Banesh, J. Bhaumik and U. Chand Banerjee, RSC Adv., 2016, 6, 49150–49157 RSC
.
- R. M. Rana, S. Rampogu, A. Zeb, M. Son, C. Park, G. Lee, S. Yoon, A. Baek, S. Parameswaran, S. J. Park and K. W. Lee, J. Clin. Med., 2019, 8, 233 CrossRef CAS
.
- Z.-Q. Liu, P. J. Baker, F. Cheng, Y.-P. Xue, Y.-G. Zheng and Y.-C. Shen, PLoS One, 2013, 8, e67197 CrossRef CAS PubMed
.
- J. M. Kriegl, T. Arnhold, B. Beck and T. Fox, QSAR Comb. Sci., 2005, 24, 491–502 CrossRef CAS
.
- R. G. Susnow and S. L. Dixon, J. Chem. Inf. Comput. Sci., 2003, 43, 1308–1315 CrossRef CAS PubMed
.
- H. J. Park, K. Park, Y. H. Kim and Y. J. Yoo, J. Biotechnol., 2014, 192, 66–70 CrossRef CAS PubMed
.
- P. Trodler and J. Pleiss, BMC Struct. Biol., 2008, 8, 9 CrossRef PubMed
.
- A. Schließmann, A. Hidalgo, J. Berenguer and U. T. Bornscheuer, ChemBioChem, 2009, 10, 2920–2923 CrossRef PubMed
.
- W. W. Zhu, C. Wang, J. Jipp, L. Ferguson, S. N. Lucas, M. A. Hicks and M. E. Glasner, Biochemistry, 2012, 51, 6171–6181 CrossRef CAS PubMed
.
- C. Fröhlich, J. Z. Chen, S. Gholipour, A. N. Erdogan and N. Tokuriki, Protein Eng., Des. Sel., 2021, 34, gzab013 CrossRef
.
- J.-F. Wang and K.-C. Chou, PLoS One, 2011, 6, e18414 CrossRef CAS
.
- E. Jiménez-Ortega, P. E. Kidibule, M. Fernández-Lobato and J. Sanz-Aparicio, Int. J. Mol. Sci., 2022, 23, 7599 CrossRef
.
- M. Makarian, M. Gonzalez, S. M. Salvador, S. Lorzadeh, P. K. Hudson and S. Pecic, J. Mol. Struct., 2022, 1247, 131425 CrossRef CAS PubMed
.
- S. A. Osmani, S. Bak and B. L. Møller, Phytochemistry, 2009, 70, 325–347 CrossRef CAS PubMed
.
- M. Pickl, S. Kurakin, F. G. Cantú Reinhard, P. Schmid, A. Pöcheim, C. K. Winkler, W. Kroutil, S. P. de Visser and K. Faber, ACS Catal., 2019, 9, 565–577 CrossRef CAS PubMed
.
- K. Ariga, Q. Ji, J. P. Hill, Y. Bando and M. Aono, NPG Asia Mater., 2012, 4, e17 CrossRef
.
- K. Ariga, M. Li, G. J. Richards and J. P. Hill, J. Nanosci. Nanotechnol., 2011, 11, 1–13 CrossRef CAS PubMed
.
- K. Ariga, M. Nishikawa, T. Mori, J. Takeya, L. K. Shrestha and J. P. Hill, Sci. Technol. Adv. Mater., 2019, 20, 51–95 CrossRef CAS PubMed
.
- M. K. Masud, M. Umer, M. S. A. Hossain, Y. Yamauchi, N.-T. Nguyen and M. J. A. Shiddiky, Trends Biochem. Sci., 2019, 44, 433–452 CrossRef CAS PubMed
.
- K. Ariga, ChemNanoMat, 2016, 2, 333–343 CrossRef CAS
.
- K. Ariga, J. Li, J. Fei, Q. Ji and J. P. Hill, Adv. Mater., 2016, 28, 1251–1286 CrossRef CAS PubMed
.
- K. Ariga, Mater. Chem. Front., 2017, 1, 208–211 RSC
.
- Z. S. Nishat, T. Hossain, M. N. Islam, H.-P. Phan, M. A. Wahab, M. A. Moni, C. Salomon, M. A. Amin, A. A. I. Sina, M. S. A. Hossain, Y. V. Kaneti, Y. Yamauchi and M. K. Masud, Small, 2022, 18, 2107571 CrossRef CAS PubMed
.
- A. Azhar, Y. Li, Z. Cai, M. B. Zakaria, M. K. Masud, M. S. A. Hossain, J. Kim, W. Zhang, J. Na, Y. Yamauchi and M. Hu, Bull. Chem. Soc. Jpn., 2019, 92, 875–904 CrossRef CAS
.
- G. Chen, L. Tong, S. Huang, S. Huang, F. Zhu and G. Ouyang, Nat. Commun., 2022, 13, 4816 CrossRef CAS PubMed
.
- Q. Chen, G. Qu, X. Li, M. Feng, F. Yang, Y. Li, J. Li, F. Tong, S. Song, Y. Wang, Z. Sun and G. Luo, Nat. Commun., 2023, 14, 2117 CrossRef CAS PubMed
.
- S. Ji, B. Jiang, H. Hao, Y. Chen, J. Dong, Y. Mao, Z. Zhang, R. Gao, W. Chen, R. Zhang, Q. Liang, H. Li, S. Liu, Y. Wang, Q. Zhang, L. Gu, D. Duan, M. Liang, D. Wang, X. Yan and Y. Li, Nat. Catal., 2021, 4, 407–417 CrossRef CAS
.
- N. Feng, Q. Li, Q. Bai, S. Xu, J. Shi, B. Liu and J. Guo, J. Colloid Interface Sci., 2022, 618, 68–77 CrossRef CAS PubMed
.
- R. J. Drout, L. Robison and O. K. Farha, Coord. Chem. Rev., 2019, 381, 151–160 CrossRef CAS
.
- O. K. Farha, I. Eryazici, N. C. Jeong, B. G. Hauser, C. E. Wilmer, A. A. Sarjeant, R. Q. Snurr, S. T. Nguyen, A. Ö. Yazaydın and J. T. Hupp, J. Am. Chem. Soc., 2012, 134, 15016–15021 CrossRef CAS PubMed
.
- Y. Xia, Y. Wu, J. Cao, J. Wang, Z. Chen, C. Li and X. Zhang, ACS Biomater. Sci. Eng., 2022, 8, 1892–1906 CrossRef CAS PubMed
.
- N. Zhang, C. Li, D. Zhou, C. Ding, Y. Jin, Q. Tian, X. Meng, K. Pu and Y. Zhu, Acta Biomater., 2018, 70, 227–236 CrossRef CAS PubMed
.
- H. Hang, C. Wang, Y. Cheng, N. Li and L. Song, Appl. Biochem., 2018, 184, 453–470 CrossRef CAS PubMed
.
- C. Horn, D. Pospiech, P. J. Allertz, M. Müller, K. Salchert and R. Hommel, ACS Appl. Polym. Mater., 2021, 3, 2823–2834 CrossRef CAS
.
- F. P. Seebeck, K. J. Woycechowsky, W. Zhuang, J. P. Rabe and D. Hilvert, J. Am. Chem. Soc., 2006, 128, 4516–4517 CrossRef CAS PubMed
.
- J. Sharma and T. Douglas, Nanoscale, 2020, 12, 336–346 RSC
.
- C. C. Lechner and C. F. W. Becker, Bioorg. Med. Chem., 2013, 21, 3533–3541 CrossRef CAS PubMed
.
- H. R. Luckarift, J. C. Spain, R. R. Naik and M. O. Stone, Nat. Biotechnol., 2004, 22, 211–213 CrossRef CAS PubMed
.
- D. Remonatto, R. H. Miotti Jr, R. Monti, J. C. Bassan and A. V. de Paula, Process Biochem., 2022, 114, 1–20 CrossRef CAS
.
- A. Costantini and V. Califano, Catalysts, 2021, 11, 629 CrossRef CAS
.
- F.-K. Shieh, S.-C. Wang, C.-I. Yen, C.-C. Wu, S. Dutta, L.-Y. Chou, J. V. Morabito, P. Hu, M.-H. Hsu, K. C. W. Wu and C.-K. Tsung, J. Am. Chem. Soc., 2015, 137, 4276–4279 CrossRef CAS PubMed
.
- I. Serra, D. Ubiali, J. Piškur, B. Munch-Petersen, T. Bavaro and M. Terreni, ChemistrySelect, 2017, 2, 5403–5408 CrossRef CAS
.
- J. K. Rho, J. A. Kang, M. H. Choi, K.-C. Ko and S. H. Park, Sens. Actuators, B, 2011, 160, 632–636 CrossRef
.
- C. Peña-Montes, M. E. Mondragón-Tintor, J. A. Castro-Rodríguez, I. Bustos-Jaimes, A. Navarro-Ocaña and A. Farrés, Enzyme. Res., 2013, 2013, 928913 Search PubMed
.
- X. Fan, W. Liang, Y. Li, H. Li and X. Liu, Microb. Cell Fact., 2017, 16, 149 CrossRef PubMed
.
- H. Fu, I. Dencic, J. Tibhe, C. A. Sanchez Pedraza, Q. Wang, T. Noel, J. Meuldijk, M. de Croon, V. Hessel, N. Weizenmann, T. Oeser, T. Kinkeade, D. Hyatt, S. Van Roy, W. Dejonghe and L. Diels, Chem. Eng. J., 2012, 207–208, 564–576 CrossRef CAS
.
- S. Zhang, J. Bramski, M. Tutus, J. Pietruszka, A. Böker and S. Reinicke, ACS Appl. Mater. Interfaces, 2019, 11, 34441–34453 CrossRef CAS PubMed
.
- S. Singh, P. Tripathi, N. Kumar and S. Nara, Biosens. Bioelectron., 2017, 92, 280–286 CrossRef CAS PubMed
.
- A. Zhang, S. Pan, Y. Zhang, J. Chang, J. Cheng, Z. Huang, T. Li, C. Zhang, J. M. D. L. Fuentea, Q. Zhang and D. Cui, Theranostics, 2019, 9, 3443–3458 CrossRef CAS PubMed
.
- Z. Chen, J.-J. Yin, Y.-T. Zhou, Y. Zhang, L. Song, M. Song, S. Hu and N. Gu, ACS Nano, 2012, 6, 4001–4012 CrossRef CAS PubMed
.
- T. Wang, P. Su, H. Li, Y. Yang and Y. Yang, New J. Chem., 2016, 40, 10056–10063 RSC
.
- S. Cai, C. Qi, Y. Li, Q. Han, R. Yang and C. Wang, J. Mater. Chem. B, 2016, 4, 1869–1877 RSC
.
- L. Zhao, Z. Wu, G. Liu, H. Lu, Y. Gao, F. Liu, C. Wang, J. Cui and G. Lu, J. Mater. Chem. B, 2019, 7, 7042–7051 RSC
.
- C. Ren, X. Hu and Q. Zhou, Adv. Sci., 2018, 5, 1700595 CrossRef PubMed
.
- T. Jiang, L. Dai, Y. Lou, H. Wang, Z. Gao, D. Wu, H. Ma and Q. Wei, ACS Appl. Mater. Interfaces, 2023, 15, 42404–42412 CrossRef CAS PubMed
.
- W.-C. Hu, J. Pang, S. Biswas, K. Wang, C. Wang and X.-H. Xia, Anal. Chem., 2021, 93, 8544–8552 CrossRef CAS PubMed
.
- A. Madhavan, R. Sindhu, P. Binod, R. K. Sukumaran and A. Pandey, Bioresour. Technol., 2017, 245, 1304–1313 CrossRef CAS PubMed
.
- D. A. Cowan and R. Fernandez-Lafuente, Enzyme Microb. Technol., 2011, 49, 326–346 CrossRef CAS PubMed
.
- E. P. Hudson, R. K. Eppler, J. M. Beaudoin, J. S. Dordick, J. A. Reimer and D. S. Clark, J. Am. Chem. Soc., 2009, 131, 4294–4300 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2024 |