Open Access Article
Open Access ArticleLearning from nature: recovery of rare earth elements by the extremophilic bacterium Methylacidiphilum fumariolicum†
Helena
Singer
a,
Robin
Steudtner
 b,
Ignacio
Sottorff
a,
Björn
Drobot
b,
Ignacio
Sottorff
a,
Björn
Drobot
 b,
Arjan
Pol
c,
Huub J. M.
Op den Camp
b,
Arjan
Pol
c,
Huub J. M.
Op den Camp
 c and
Lena J.
Daumann
c and
Lena J.
Daumann
 *a
*a
aDepartment of Chemistry, Ludwig-Maximilians-University Munich, 81377 München, Germany. E-mail: lena.daumann@lmu.de
bInstitute of Resource Ecology, Helmholtz-Zentrum Dresden-Rossendorf e.V., 01328 Dresden, Germany
cDepartment of Microbiology, Research Institute for Biological and Environmental Sciences, Radboud University Nijmegen, 6525 AJ Nijmegen, The Netherlands
First published on 29th June 2023
Abstract
We present the extremophilic bacterium Methylacidiphilum fumariolicum SolV as a platform for the recovery of rare earth elements (REE). Strain SolV is able to selectively extract the light REE from artificial industrial waste sources, natural REE-containing and post-mining waters. Upscaling, different media composition and accumulation over several cycles were successfully implemented, underlining the potential for bio-recovery of REE.
Modern life without rare earth elements (REE) can hardly be imagined today. The elements Sc, Y, La and the following 14 lanthanides (Ln) Ce to Lu have made their way into every aspect of our lives. REE are mainly obtained from primary sources such as the ore bastnaesite which also releases the radioactive elements thorium and uranium as by-products.1 Secondary sources are hardly used to recycle REE (less than 2%) despite their large potential,2,3 focusing end-of-life products (EOL)4,5 such as fluorescent lamps,6 or industrial by-products and residues like mining waters.7,8 REE are crucial for new energy technologies, however their mining and refining poses environmental problems, since these processes are energy and material consuming. So, it appears indispensable to develop ecologically and economically compatible methods to recycle REE to extend the life cycle and reduce the environmental footprint of these elements.9 Nowadays, recovery of REE from EOL is based on hydro- and pyrometallurgical strategies, electrochemical treatments, biometallurgical technologies, nanomaterials, siderophores as chelators, and physical methods.4,10 All of them have in common that they have to face complex mixtures with non-REE in combination with low REE amounts.10
Not only mankind relies on REE, but in nature bacteria can be found that use these elements for their metabolism such as the methanotroph Methylacidiphilum fumariolicum SolV11 or Methylorubrum extorquens AM1.12 While strain SolV is strictly dependent on Ln, strain AM1 is able to switch between a Ln and Ca-based pathway.12 Ongoing, proteins are identified which include the essential lanthanides (Ln) as Lewis acid in the active site (methanol dehydrogenase XoxF)13,14 or can bind them such as LanModulin15–17 or LanPepsy.18 As a consequence, these biological systems have evolved a highly-efficient, selective Ln-uptake machinery that can be used to develop alternative approaches for bio-recovery of REE.19 Connecting hydrometallurgy and biochemistry was already shown for various metals, mainly focusing on d-block elements.20,21 Recently, copper-binding peptides were functionalized onto the surface of fungal mycelia,22 and microorganisms can be genetically engineered to bioaccumulate heavy metals.20 In addition, the field of biosorption and accumulation for REE, using bacteria, fungi or algae is progressing.10,23,24 High adsorption capacities of REE were lately discovered for 12 cyanobacterial strains.25 The Ln-utilizing bacterium strain AM1 was genetically modified for a selective uptake of heavier Ln, such as Gd.26
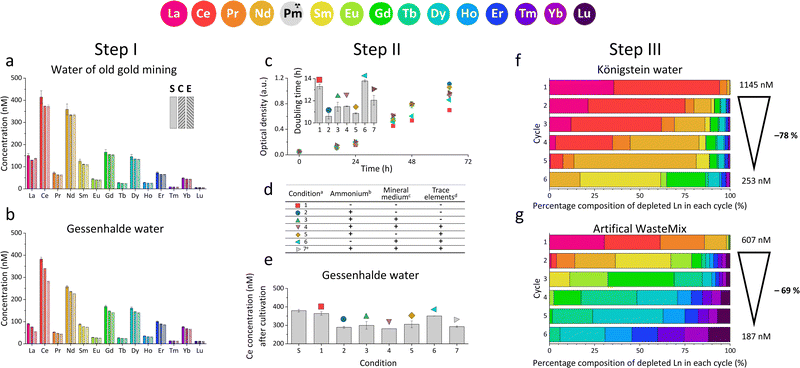
For our studies, we used the native Ln-dependent strain SolV11 to accumulate and enrich certain Ln from a variety of natural and artificial REE sources. As an extremophile, strain SolV needs high temperatures up to 60 °C and an acidic environment of pH 2–3 for optimal growth.11 Since metal ions display a good solubility in acids (H2SO4, HNO3 or HCl), this is also used by various hydrometallurgical metal recovery strategies.10,27 Consequentially, many industrial waste waters are in an acidic matrix, with a high concentration of other elements e.g. Mg, Ca, Sr, Se, As, Fe, Al, Hg and U.5 Since strain SolV is strictly Ln-dependent, and can tolerate these conditions, it is an ideal candidate for recovering REE from different low-grade sources. It has been previously shown that especially larger Ln such as La enhance growth of strain SolV, and in this regard, it is also interesting to examine the system for potential selectivity.11,14 To ensure a complex mixture of elements with high composition variability, we tested various REE sources – ranging from flooding waters, mining leachates, primary sources and EOL industrial waste (ESI† 1.1). Königstein and Gessenhalde water are redevelopment surface flooding waters from former uranium ore mining by sulfuric acid leaching. Other low-grade sources discussed and tested are post-mining waters, more specifically industrial decomposition waters from leaching of different ores (zinc blende, Li mica, quartz) which contain REE in low concentrations as by-products. To include a primary source with a high REE concentration in contrast, two digestion samples of bastnaesite were added. Finally, an industrial sample of an EOL product was screened. To analyse the potential of the bacterial strain SolV to selectively deplete REE from the different sources, ICP-MS analysis of the supernatants as well as of the bacterial cells were performed. Growth was monitored through optical density measurement at 600 nm (OD600). The amount of REE depleted by strain SolV, growth curves and thereof calculated doubling times helped us to understand how strain SolV can be used as bio-recovery system for Ln from a small-scale approach (Step I), to optimization (Step II and III) to larger-scale application (Step IV).
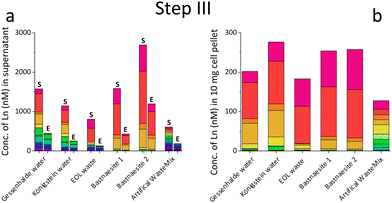
We initially investigated, whether SolV showed depletion of REE in the 10 different REE sources (ESI† Table S1, Fig. S20, S21, Step I). We analysed the REE concentration in the supernatant before (S) and after cultivation (E) with strain SolV as well as in the control (C) without added biomass to investigate precipitation formation that could hamper with the OD600 readout (Fig. 1a, b and Fig. S1, ESI†), in combination with OD600 measurements to monitor the bacterial growth (Fig. S2, ESI†). Fig. 1a shows the amount of REE in waste water from an old gold mine, where the Ln concentration does not significantly decrease in the presence of strain SolV. The slight reduction observed is comparable to the control which it is due to precipitate formation by the complex media matrix. Similar observations were made for Lithium mica (with strong precipitate formation in the control), water of ore leaching, and quartz leaching. In these samples some metal ions may be in too high concentration and thus toxic (e.g. As, Cr, Table S1, ESI†). In contrast, a significant decrease of the Ln amount in the presence of strain SolV was observed in the water coming from Königstein, Gessenhalde (Fig. 1b) and EOL waste. Leaching of zinc blende also showed a decrease of Ln concentration in the medium, but strong precipitate formation was present in the strain SolV and control, which hampered OD600 readout for growth experiments. Bastnaesite samples did not show a clear reduction of REE during the course of a three-day cultivation, very likely due to the high REE concentration (around 10 μM). As bacterial growth was observed, this REE source was investigated further in a diluted fashion. To confirm that depletion of Ln is selective within the series and over other elements and due to active uptake of Ln by SolV (and not a result of simple cell-wall adsorption), a control experiment without CH4 and CO2 was conducted (including a measurement of all elements present). Without these gases, SolV cells are alive and intact but not growing. Fig. S3–S6 (ESI†) show that from a bastnaesite solution only P, Mg, Fe, Cu, Zn, Mn and La/Ce concentrations are decreasing when SolV is actively growing. More importantly, other metals such as U or Pb are not taken up.
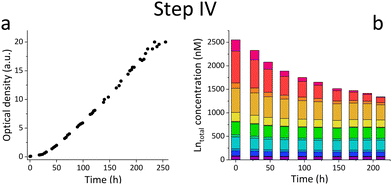
Based on the results of the first screening for potential candidates, we tested different media compositions (omitting/limiting N-source, salts for buffering, and metals for cofactors) to optimize the growth and Ln uptake by strain SolV (Step II). Ammonium (nitrogen source),28 mineral medium (MM, minimal salts without additional nutritional supplements, mainly for buffering) and trace element solution (TE, providing cofactors for essential enzymes) were added to Gessenhalde and Königstein water in seven different combinations (Fig. 1d). The required gas supply, consistent of CH4 and CO2 was indispensable for growth of the methanotrophic strain SolV, and therefore couldn’t be omitted. CO2 is required for the carbon fixation pathway,29 while CH4 is an energy source, as it is first oxidized by methane monooxygenases to methanol, and finally to CO2.30Fig. 1c shows the growth curves of strain SolV in Gessenhalde water under varying conditions, and calculated doubling times, which both indicated a reduced and limited growth behavior under condition 1 and 6 compared to the other tested medium compositions. This was also supported by the ICP-MS analysis (Fig. 1c), shown here with Ce (other elements in Fig. S7, ESI†). Under condition 1 and 6, almost no Ce was depleted by strain SolV within one incubation cycle whereas under better conditions 2–5 and 7 up to 25% of the initial Ce was removed. Similar observations were made for Königstein water (Fig. S8, S9, ESI†), EOL waste and bastnaesite samples (Fig. S10, condition 1,2,7, ESI†). Thus, ammonium, acting as a nitrogen source for growth, is the most crucial part of the media composition. It was shown that strain SolV is able to replace ammonium with atmospheric N2 (under reduced oxygen concentrations <0.5%), however, a 2.8 times slower growth was observed,31 which could also explain the higher doubling times, and reduced Ln depletion from the growth medium observed, when no ammonium was added. The addition of the components TE and MM was not necessary, as the ICP-MS/OES analyses revealed a sufficiently high concentration of these elements in the waste waters themselves. These results are particularly important for a potential industrial application, where processes with the least amount of (potentially costly) additives are economically feasible. Thus, recovery in a process that can be repeated the same way several times to remove the maximum amount of Ln from a single sample would be beneficial. Consequentially, we tested whether SolV bacteria can be added for several cycles to REE sources until maximum OD600 was reached. The experimental setup was upscaled from 10 mL in Step I and II to a volume of 50 mL and the accumulation process was run up to six 72 h cycles while the biomass was removed by centrifugation after every cycle, and fresh bacterial cells were added for standardization of all samples (Step III). However, it is also possible to use the harvested biomass for starting a new growth cycle. Independent of the used REE source, the multi-cycle upscaling experiment (Step III) revealed that about 70% of the initial Ln amount was removed from the sources by strain SolV constantly in the 6 cycles (Fig. 1e, f and Fig. S11–15, ESI†). By ongoing cycling, it seems achievable to deplete 100% of the Ln. Matching, the depleted Ln can be found in the lyophilized and subsequently disrupted cell pellet (Fig. 2b). Particularly noteworthy is that Ln are not removed randomly by strain SolV, but a clear trend is present, showing a selective uptake of Ln. First, the light Ln are completely removed, then the heavier Ln are depleted from the source. We already showed that when strain SolV is presented to equal amounts of Ln and selected actinides, the strain will deplete preferentially the light Ln over the heavier ones.11,14 We proposed that this is mainly based on the size of these elements. While Ln have very similar coordination and chemical properties, they display a decreasing size with increasing atomic number along the series (Ln contraction). In addition, we assume that strain SolV evolved for a preferential uptake of the more abundant Ln, which are the larger ones in the series. Despite the versatile REE source composition, a trend for light Ln-preference is clearly observed. Following the up-scaling experiments, we performed a large-scale approach in a self-constructed batch reactor system with a continuous gas feeding to outline a possible industrial approach. 3.7 liters of Königstein water were mixed with medium, and bacterial cells of strain SolV were added to start the cultivation while a continuous supply of air, CH4 and CO2 was provided. Samples for OD600 measurements were collected at different time intervals (Fig. 3a), and supernatant samples from selected time points were analysed with ICP-MS, as well as the final biomass. As expected, a clear decrease in the total Ln concentration (Fig. 3b) occurred as the cultivation progressed, and again the light Ln were depleted first. In total, 47.4% of present Ln were removed from Königstein water by strain SolV, however, here a maximum in bacterial growth seemed to be reached due to flattening of the OD600 values around 20. For further uptake of Ln, biomass might be removed while the supernatant should be fed to the bioreactor again. In total, 15 g of lyophilized bacterial cells were obtained from 3.7 L of medium, with mainly incorporated La, Ce, Pr and Nd (Fig. S18 and S19, ESI†), while within the cycling experiments around 50–70 mg of lyophilized biomass were obtained over 6 cycles with 50 mL of medium. A direct comparison to the cyclic experiments (Step III) is not possible, since different limiting factors apply to a closed system such as bottles compared to a continuous gas-flow system. However, it was shown that under both setups, there is a significant as well as a selective removal of Ln from the REE sources by strain SolV.
The reported results are promising, the extremophilic strain Methylacidiphilum fumariolicum can be used to recover REE from a variety of low-grade sources. Other present elements play a subordinate role, since strain SolV shows sufficient growth even under minimal conditions, in combination with an acidic pH and high temperatures. Even the presence of toxic heavy metals such as Th and U in non-negligible concentrations did not impair the growth of strain SolV. Only the addition of ammonium, CH4, O2 and CO2 are essential for sufficient bacterial growth and Ln uptake. Also, the flexible procedure, as small-scale-, up-scale cycling, or large-scale approach shows the promising potential application. By carefully adjusting the REE concentration in the system, it is possible to control which Ln are absorbed at which time. At high concentrations (μM range), mainly the light ones are depleted, whereas at low concentrations (up to 100 nM), the heavier ones can also be removed in selected fractions. This study shows that using bacteria for the recovery of Ln hold great potential, since already existing biological systems which evolved for a selective Ln uptake can be used to pave new ways for, ultimately, the recycling of critical elements.
The authors are grateful to the Wismut GmbH, G. E. O. S. GmbH, Amberger Kaolinwerke GmbH and to the Applied Geology and Microbiology groups of Friedrich Schiller University Jena for the post-mining waters. We thank Falk Lehmann and Dr Manja Vogel for kindly providing EOL and bastnaesite samples. LJD greatly acknowledges the ERC Starting Grant Lanthanophor (945846), BD thanks the PepTight Projekt (BMBF 031B1122).
Conflicts of interest
There are no conflicts to declare.References
- P. Fröhlich, T. Lorenz, G. Martin, B. Brett and M. Bertau, Angew. Chem., Int. Ed., 2017, 56, 2544–2580 CrossRef PubMed.
- A. B. Patil, V. Paetzel, R. P. W. J. Struis and C. Ludwig, Separations, 2022, 9, 56 CrossRef CAS.
- K. Binnemans, P. McGuiness and P. T. Jones, Nat. Rev. Mater., 2021, 6, 459–461 CrossRef CAS.
- C. Ramprasad, W. Gwenzi, N. Chaukura, N. Izyan Wan Azelee, A. Upamali Rajapaksha, M. Naushad and S. Rangabhashiyam, Chem. Eng. J., 2022, 442, 135992 CrossRef CAS.
- R. K. Jyothi, T. Thenepalli, J. W. Ahn, P. K. Parhi, K. W. Chung and J.-Y. Lee, J. Clean. Prod., 2020, 267, 122048 CrossRef CAS.
- C. Tunsu, M. Petranikova, C. Ekberg and T. Retegan, Sep. Purif. Technol., 2016, 161, 172–186 CrossRef CAS.
- M. Hermassi, M. Granados, C. Valderrama, C. Ayora and J. L. Cortina, Sci. Total Environ, 2022, 810, 152258 CrossRef CAS PubMed.
- S. Costis, K. K. Mueller, L. Coudert, C. M. Neculita, N. Reynier and J.-F. Blais, J. Geochem. Explor., 2021, 221, 106699 CrossRef CAS.
- P. S. Arshi, E. Vahidi and F. Zhao, ACS Sustainable Chem. Eng., 2018, 6, 3311–3320 CrossRef CAS.
- T. G. Ambaye, M. Vaccari, F. D. Castro, S. Prasad and S. Rtimi, Environ. Sci. Pollut. R., 2020, 27, 36052–36074 CrossRef CAS PubMed.
- A. Pol, T. R. M. Barends, A. Dietl, A. F. Khadem, J. Eygensteyn, M. S. M. Jetten and H. J. M. Op den Camp, Environ. Microbiol., 2014, 16, 255–264 CrossRef CAS PubMed.
- H. N. Vu, G. A. Subuyuj, S. Vijayakumar, N. M. Good, N. C. Martinez-Gomez and E. Skovran, J. Bacteriol., 2016, 198, 1250–1259 CrossRef CAS PubMed.
- H. Lumpe, A. Pol, H. J. M. Op den Camp and L. J. Daumann, Dalton Trans., 2018, 47, 10463–10472 RSC.
- H. Singer, R. Steudtner, A. S. Klein, C. Rulofs, C. Zeymer, B. Drobot, A. Pol, N. C. Martinez-Gomez, H. J. M. Op den Camp and L. J. Daumann, Angew. Chem., Int. Ed., 2023, e202303669 Search PubMed.
- G. J. P. Deblonde, J. A. Mattocks, D. M. Park, D. W. Reed, J. A. Cotruvo and Y. Jiao, Inorg. Chem., 2020, 59, 11855–11867 CrossRef CAS PubMed.
- H. Singer, B. Drobot, C. Zeymer, R. Steudtner and L. J. Daumann, Chem. Sci., 2021, 12, 15581–15587 RSC.
- C.-E. Wegner, M. Westermann, F. Steiniger, L. Gorniak, R. Budhraja, L. Adrian and K. Küsel, Appl. Environ. Microbiol., 2021, 87, e03144–03120 CrossRef CAS PubMed.
- J. L. Hemmann, P. Keller, L. Hemmerle, T. Vonderach, A. M. Ochsner, M. Bortfeld-Miller, D. Gunther and J. A. Vorholt, J. Biol. Chem., 2023, 299, 102940 CrossRef CAS PubMed.
- Z. Dong, J. A. Mattocks, G. J. Deblonde, D. Hu, Y. Jiao, J. A. Cotruvo, Jr. and D. M. Park, ACS Cent. Sci., 2021, 7, 1798–1808 CrossRef CAS PubMed.
- P. Diep, R. Mahadevan and A. F. Yakunin, Front. Bioeng. Biotechnol., 2018, 6, 157 CrossRef PubMed.
- M. J. Capeness and L. E. Horsfall, Biochem. Soc. Trans., 2020, 48, 1367–1378 CrossRef CAS PubMed.
- J. Urbina, A. Patil, K. Fujishima, I. G. Paulino-Lima, C. Saltikov and L. J. Rothschild, Sci. Rep., 2019, 9, 16422 CrossRef PubMed.
- F. Barmettler, C. Castelberg, C. Fabbri and H. Brandl, AIMS Microbiol., 2016, 2, 190–204 CAS.
- R. M. Brown, A. Mirkouei, D. Reed and V. Thompson, Renew. Sust. Energ. Rev., 2023, 173, 113099 CrossRef CAS.
- M. Paper, M. Koch, P. Jung, M. Lakatos, T. Nilges and T. B. Brück, Front. Bioeng. Biotechnol., 2023, 11, 1130939 CrossRef PubMed.
- N. M. Good, H. D. Lee, E. R. Hawker, M. Z. Su, A. A. Gilad and N. C. Martinez-Gomez, Front. Microbiol., 2022, 13, 820327 CrossRef PubMed.
- E. O. Opare, E. Struhs and A. Mirkouei, Renew. Sust. Energ. Rev., 2021, 143, 110917 CrossRef CAS.
- S. S. Mohammadi, A. Pol, T. van Alen, M. S. M. Jetten and H. J. M. Op den Camp, Front. Microbiol., 2017, 8, 1901 CrossRef PubMed.
- A. F. Khadem, A. Pol, A. Wieczorek, S. S. Mohammadi, K.-J. Francoijs, H. G. Stunnenberg, M. S. M. Jetten and H. J. M. Op den Camp, J. Bacteriol., 2011, 193, 4438–4446 CrossRef CAS PubMed.
- N. Picone, S. S. Mohammadi, A. C. Waajen, T. A. van Alen, M. S. M. Jetten, A. Pol and H. J. M. Op den Camp, Front. Microbiol., 2020, 11, 604485 CrossRef PubMed.
- A. F. Khadem, A. Pol, M. S. M. Jetten and H. J. M. Op den Camp, Microbiology, 2010, 156, 1052–1059 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc01341c |
| This journal is © The Royal Society of Chemistry 2023 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pH adjusted to 2.7 with H2SO4 and 10% CH4 and 5% CO2 added; b
pH adjusted to 2.7 with H2SO4 and 10% CH4 and 5% CO2 added; b