Open Access Article
Open Access ArticleEncapsulating bacteria in alginate-based electrospun nanofibers†
Emily
Diep
 and
Jessica D.
Schiffman
and
Jessica D.
Schiffman
 *
*
Department of Chemical Engineering, University of Massachusetts, Amherst, Amherst, Massachusetts 01003-9303, USA. E-mail: schiffman@ecs.umass.edu
First published on 13th April 2021
Abstract
Encapsulation technologies are imperative for the safe delivery of live bacteria into the gut where they regulate bodily functions and human health. In this study, we develop alginate-based nanofibers that could potentially serve as a biocompatible, edible probiotic delivery system. By systematically exploring the ratio of three components, the biopolymer alginate (SA), the carrier polymer poly(ethylene oxide) (PEO), and the FDA approved surfactant polysorbate 80 (PS80), the surface tension and conductivity of the precursor solutions were optimized to electrospin bead-free fibers with an average diameter of 167 ± 23 nm. Next, the optimized precursor solution (2.8/1.2/3 wt% of SA/PEO/PS80) was loaded with Escherichia coli (E. coli, 108 CFU mL−1), which served as our model bacterium. We determined that the bacteria in the precursor solution remained viable after passing through a typical electric field (∼1 kV cm−1) employed during electrospinning. This is because the microbes are pulled into a sink-like flow, which encapsulates them into the polymer nanofibers. Upon electrospinning the E. coli-loaded solutions, beads that were much smaller than the size of an E. coli were initially observed. To compensate for the addition of bacteria, the SA/PEO/PS80 weight ratio was reoptimized to be 2.5/1.5/3. Smooth fibers with bulges around the live microbes were formed, as confirmed using fluorescence and scanning electron microscopy. By dissolving and plating the nanofibers, we found that 2.74 × 105 CFU g−1 of live E. coli cells were contained within the alginate-based fibers. This work demonstrates the use of electrospinning to encapsulate live bacteria in alginate-based nanofibers for the potential delivery of probiotics to the gut.
Introduction
Exploration into the gut microbiota has shed light on the influence that commensal bacteria have on human health and disease.1 As most of the immune system is found in the gut,1,2 these bacteria help to metabolize nutrients3 and also influence immune responses.4 Studies have linked the gut microbiota with ailments including allergies,2 inflammatory bowel disease,5 multiple sclerosis,6 depression,7 diabetes,8 obesity,9 autism,10 and cancer.3 According to the Food and Agriculture Organization (FAO) and the World Health Organization (WHO), probiotic bacteria must be administered to the gut in high amounts in order to provide therapeutic effects.11–13The current approach to deliver “probiotics” adopts pharmaceutical processing and formulations. Probiotic bacteria are often freeze-dried, mixed with excipient, and packed into tablets.14 In terms of processing, freeze drying is a common method used for the long-term storage of bacteria. Freezing causes bacteria to enter a hibernation-like state extending their shelf-life. However, ice formation during freezing can kill bacteria by rupturing their cell membrane. Drying removes excess water, which causes degradation of the cell and various molecules within the cell, but it also decreases the likelihood of ice formation. To counter these shortcomings, chemical additives like cryoprotectants are added to the bacterial suspension to protect the cells from damage. These additives include a variety of polymers like lactose,15 fructooligosaccharides,16,17 carboxymethyl high amylose starch,14 and calcium alginate1,18,19 that effectively encapsulate the bacteria during the freeze drying process. Additionally, optimizing the formulation by co-encapsulating probiotics with prebiotics,17 antacids,20 and fats21 may help bacteria maintain their viability as they travel through the gastrointestinal tract. Other studies22–24 have shown that the food delivery matrix of probiotics (e.g. chocolate,25 kimchi,26 yogurt23) affect their probiotic efficacy in the body.24 The goal of the full formulation is to protect the bacteria against environmental factors and cell damage during processing and later during travel through the gastrointestinal tract where the bacteria can confer beneficial effects.1,14,22
Here, we suggest an alternative approach: to encapsulate the live microbes into alginate-based nanofiber mats, which could be ingested. Alginate is a polysaccharide that is compatible with both the human body and bacterial biofilms making it an ideal material for the encapsulation of bacteria for delivery into the gut.27–30 Nanofiber-based technologies are of growing interest in the drug delivery industry for their high surface area to volume ratio which allows for controllable diffusion of active ingredients out of the material.31 During the electrospinning process, fibers are formed by applying an electric field onto a polymeric solution to pull the polymers into solid nanofibers. Fiber formation, diameter, and morphology depend on both solution properties, like concentration, density, conductivity, and surface tension, as well as experimental parameters, including solution flow rate, tip-to-collector distance, and applied voltage.32–40 The high surface area to volume ratios and high porosities of these nanofiber mats make them suitable for many biomedical applications, including protective clothing,41 wound dressing,32,42 vascular grafts,43 and drug delivery.31 Previously, it has been demonstrated that electrospinning can be used to encapsulate a wide variety of viable bacteria, including Gram-negative and Gram-positive cells into dry nanofiber mats.17,39,42,44–47 By adding glycerol, a cryoprotectant, into the electrospinning precursor solutions, Escherichia coli (E. coli) and Staphylococcus albus were encapsulated in polyvinyl alcohol (PVA) fibers with viabilities of 48% and 100% viability, respectively.39 Other bacteria have also been electrospun into polymers for a range of biomedical applications. For example, Staphylococcus epidermidis has been loaded into carboxymethyl cellulose/polyethylene oxide (PEO) fibers to treat diabetic foot ulcers,42 and Lactobacillus acidophilus has been loaded into PVA and polyvinyl pyrrolidone fibers to treat bacterial vaginosis.44 The encapsulation and delivery of gut probiotic bacteria from PVA and PEO fibers has also been demonstrated.17,45,47 However, research on the encapsulation of microbes into nanofibers fabricated from the generally regarded as safe (GRAS) biopolymer alginate is scarce; to date the authors are only aware of one report48 (that was published while we were writing this manuscript).
Bacteria produce polysaccharides, proteins, and other biopolymers which make up their protective biofilm while encouraging community growth. Polysaccharides, such as alginate, can be used to encapsulate bacteria within a matrix that mimics their natural environment.27–30 Alginate – linear copolymers of β-(1–4) linked D-mannuronic acid and β-(1–4)-linked L-guluronic acid units – is well known for its biocompatibility, sustainability, low cost, and ease of use.29,49 The carboxylic groups on the alginate chain can be crosslinked with multivalent ions, like calcium. Crosslinked alginate materials are stable in the low pH environment of the stomach, and the crosslinks are reversed in high pH environments like that of the intestines. The controllable and reversible nature of these crosslinks make alginate a promising encapsulation polymer for the targeted delivery of probiotics to the gut and its microbiota.22,29,49,50 Alginate hydrogel microbeads have been previously explored for the encapsulation of drugs,19 proteins,51 and bacteria.20 Unfortunately, the size of these microbeads, which range from a few microns to several hundred microns, limits their use in food-based products as mastication would immediately break down the beads and release the encapsulated cargo.51 Electrospinning can be used to form nanofibers that are much smaller than these microbeads. The rigid structure of the alginate coupled with its high surface tension and conductivity in solution has made it a challenge to electrospin. Some studies have utilized various surfactant and solvent mixtures to overcome these challenges (e.g., 3.6/0.4/0.5 wt% SA/PEO/Triton X-100 in water and DMSO,52 2.7/0.3/0.8 wt% SA/PEO/Triton X-100 in water and ethanol,53 3.2/0.8/1.5 wt% SA/PEO/Pluronic F127 in water and DMF,54 and 8.0/1.6/2.0 wt% SA/PEO/Pluronic F127 in water).34 While these published studies have pioneered key parameters for electrospinning alginate into nanofibers, none of them have demonstrated their ability to encapsulate living cells and most featured cytotoxic solvents.
In this study, we systematically investigated precursor solutions that enabled the electrospinning of smooth alginate-based nanofibers featuring viable E. coli. Due to its bulky uronic groups, alginate has a rigid structure that limits chain entanglements and subsequent electrospinning.34–36 Thus, previous studies have mixed alginate with “carrier polymers”, including PEO34–36 and PVA,55,56 so that sufficient chain entanglements could be achieved without greatly increasing the viscosity of the precursor solution.34–36 Another precursor additive, the nonionic surfactant, Triton X-100,34 was previously reported to enable the formation of bead-free fibers by reducing the surface tension of precursor solution. Here, we focused on using only GRAS solvents and additives to manufacture smooth fibers. PEO was selected as a carrier polymer because it is biocompatible and can also act as a mucoadhesive for prolonged probiotic presence in the gut.46 We also investigated the effect of adding polysorbate 80 (PS80), an FDA approved surfactant, to enable smooth fiber formation during electrospinning. After optimizing the electrospinning of alginate/PEO/PS80 fibers, we encapsulated bacteria in these alginate-based fibers. We suggest that alginate-based nanofibers have the potential to protect and deliver a sufficient concentration of live bacteria into the gut where they will provide therapeutic effects.
Materials & methods
Materials
All chemicals were used as received without further purification. Low viscosity alginic acid sodium salt from brown algae (SA, ν = 4–12 cP, 1 wt% in H2O at 25 °C), poly(ethylene oxide) (PEO, Mw = 600 kDa), polysorbate 80 (PS80), tryptone, sodium chloride (NaCl), yeast extract, agar, potassium chloride (KCl), sodium phosphate dibasic (Na2HPO4), potassium phosphate (KH2PO4), hydrochloric acid (HCl), calcium chloride (CaCl2), carbenicillin, and propidium iodide (PI) were purchased from Sigma Aldrich (St. Louis, MO). Deionized (DI) water was obtained from a Barnstead Nanopure Infinity water purification system (Thermo Fisher Scientific, Waltham, MA).E. coli preparation
E. coli K12 MG1655, engineered with GFP plasmid, was used throughout our studies as a model microorganism. Overnight cultures of E. coli were grown in Luria Bertani (LB) broth (10 g L−1 tryptone, 10 g L−1 NaCl, 5 g L−1 yeast extract in DI water) with carbenicillin (1 μL mL−1) for 16 h at 37 °C. Cells were harvested by centrifugation at 2500g for 5 min at 25 °C (Sorvall™ ST 40R, ThermoFisher Scientific, Waltham, MA). Bacteria pellets were washed three times with phosphate-buffered saline (PBS, 8 g L−1 NaCl, 0.2 g L−1 KCl, 1.44 g L−1 Na2HPO4, 0.24 g L−1 KH2PO4 in DI water, adjusted to a pH of 7.4 using HCl) before being resuspended in sterile DI water.Electrospinning alginate-based nanofibers loaded with bacteria
Separate solutions of SA and PEO in sterile DI water were prepared. The two polymeric solutions were mixed with PS80 in predetermined weight ratios, see the Results and Discussion section for ratios. SA/PEO/PS80 solutions were mixed for 24 h at 20 rpm using a Rotator Genie Lab Rotator (Scientific Industries, Bohemia, NY). To create the bacteria-loaded electrospinning solutions, a suspension of E. coli was added to the SA/PEO/PS80 solutions to make a concentration of 108 CFU mL−1. The solutions were mixed by rotating for at least 1 h at room temperature (23 °C) before electrospinning.Electrospinning solutions were loaded into 5 mL Luer-Lock syringes (Norm-Ject, Tuttlingen, Germany) capped with an 18-gauge hypodermic needle (Exelint, Redondo Beach, CA). Syringes were secured to a horizontal syringe pump (Cole Parmer, Vernon Hills, IL) that advanced the solution at a constant volumetric flow rate of 2 mL h−1. The high voltage supply (Gamma High Voltage Research, Ormond Beach, FL) applied 17.5 kV between the syringe needle and collector plate which were attached to the supply via alligator clips. The 15 cm × 15 cm × 0.32 cm copper collector plate covered with aluminum foil was set at a horizontal separation distance of 17 cm away from the tip of the needle. Temperature and relative humidity were kept constant at 23 °C and 20–30% using an environmental box.
Characterization of electrospinning solutions
Surface tension measurements were made using a tensiometer (Optical Contact Angle 20, Dataphysics Inc., GmbH, Germany) that utilized the drop pendant method. A droplet of solution was suspended in air from an 18-gauge needle. The needle gauge was selected because it matches that used for electrospinning. The droplet was recorded until reaching equilibrium; analysis of surface tension was conducted using the integrated system software (SCA20, Dataphysics Inc., GmbH, Germany). Conductivity was measured using a conductivity probe (Tetracon 325, Vernon Hills, IL). Surface tension and conductivity experiments were conducted in triplicate.Characterization of alginate-based nanofibers
Visualization of the GFP produced by the encapsulated bacteria was captured using a Zeiss Axio Imager A2 M Microscope (Carl Zeiss Microscopy, White Plains, NY). Micrographs of the nanofiber mats were taken using scanning electron microscopy (SEM). Samples were sputter-coated (Cressington208 Sputter Coater, Watford, UK) with 3 mm of platinum before imaging using an FEI Magellan 400 XHR-SEM (ThermoFisher Scientific, Hillsboro, OR). Average nanofiber diameter distribution was determined by measuring the diameters of 50 random fibers from five micrographs using ImageJ 1.52a software (National Institutes of Health, Bethesda, MD).35 Fiber diameter was measured excluding beads or bacterial cells. Chemical composition was analyzed using Fourier-transform infrared spectroscopy (Bruker Alpha, Bruker Optics, Billerica, MA). Spectra of electrospun fibers were compared to spectra acquired from dried SA films, dried PEO dry films, liquid PS80, and dried SA/PEO/PS80 (2.8/1.2/3 wt%) mixture films.Bacterial loading in alginate-based solution and nanofibers
The single plate-serial dilution spotting (SP-SDS) drop plate method51 was used to determine cell viability within the precursor solutions and fiber mats. To determine bacterial loading of a solution, solutions were serial diluted. Samples of 20 μL from dilutions were plated on agar plates (10 g L−1 tryptone, 10 g L−1 sodium chloride, 5 g L−1 yeast extract, 15 g L−1 agar, 1 μL mL−1 carbenicillin in DI water).52 Plates were incubated for 16 h at 37 °C. Each bacterium that grew into a distinct, visible colony was counted as a colony forming unit (CFU). The quantity of viable bacteria in the electrospun fibers was determined using a similar method. Instead of a serial dilution, fiber mats were weighed (Mettler Toldeo XP204, Columbus, OH) and dissolved in PBS prior to being plated and counted.Statistics
Surface tension, conductivity, and bacterial loading are reported as average values ± standard error, whereas fiber diameter, bead size, and E. coli size were reported as average values ± standard deviation. A two-sided unpaired Student's t-test was used to determine statistical significance reports in figures, consistent with previous work from our group.57–59Results and discussion
Effect of surfactant on alginate-based nanofibers
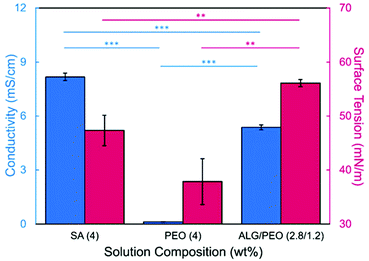
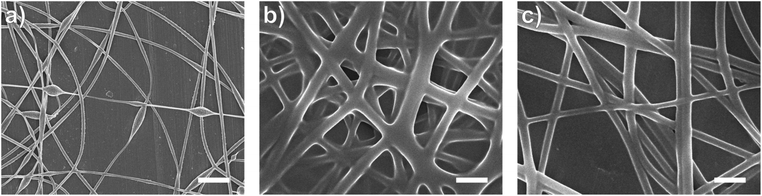
In this study, bead-free alginate-based nanofibers were electrospun from a solution of alginate (SA), poly(ethylene oxide) (PEO), polysorbate 80 (PS80). Each component was optimized to reduce bead formation along the fiber to create a smooth, consistent fiber product. Attempts were first made to electrospin pure solutions of SA; however, as expected and consistent with previous works,34,35,60,61 the rigid structure, high surface tension (47.35 N m−1 for 4 wt% SA), and high conductivity (8.17 mS cm−1 for 4 wt%) of a pure aqueous SA solution prevented fiber formation (Table 1, Fig. 1). To enable electrospinning, 4 wt% of the linear polymer PEO was added to the 4 wt% SA solution in a 3 to 7 ratio to create a 2.8/1.2 wt% SA/PEO solution. This concentration of SA/PEO was used as a starting point as it has been electrospun in other studies.34,35 Large beads interconnected by thin fibers or heavily beaded fibers were produced from this SA/PEO solution (Fig. 2a). Beading can be caused by factors including low polymer concentration,62 slow solution flow rate,63 low applied voltage,64 and low surface tension.34,35,65 In our case, we hypothesized that the reason for beading was due to high surface tension which has been shown by other studies to be common for alginate systems.34,35,52,60,61 As the electric field pulls the solution into a conical jet, surface tension forces attempt to decrease the surface area per unit of mass turning the jet into spherical shapes that create the beaded fiber morphology.66| Precursor solution composition | Solution properties | Fiber characteristics | |||||
|---|---|---|---|---|---|---|---|
| Polymer (wt%) | E. coli concentration | Conductivitya | Surface tensiona | Fiber diameterb | Bead formation | ||
| SA | PEO | PS80 | (CFU mL−1) | (mS cm−1) | (mN m−1) | (nm) | (yes/no) |
| a Values are reported as mean ± standard error. b Values are reported as mean ± standard deviation. | |||||||
| 4.0 | n/a | n/a | n/a | 8.17 ± 0.21 | 47.35 ± 2.82 | n/a | n/a |
| 2.8 | 1.2 | n/a | n/a | 5.38 ± 0.14 | 51.70 ± 0.66 | n/a | Yes |
| 2.8 | 1.2 | 0.5 | n/a | 5.58 ± 0.04 | 41.92 ± 0.91 | 125 ± 37 | Yes |
| 2.8 | 1.2 | 1.0 | n/a | 5.44 ± 0.05 | 40.37 ± 0.58 | 127 ± 38 | Yes |
| 2.8 | 1.2 | 3.0 | n/a | 5.04 ± 0.10 | 39.65 ± 0.63 | 167 ± 23 | No |
| 2.8 | 1.2 | 3.0 | 108 | 5.28 ± 0.03 | 41.25 ± 0.24 | 100 ± 23 | Yes |
| 2.8 | 1.2 | 5.0 | 108 | 5.09 ± 0.05 | 39.50 ± 0.34 | 305 ± 110 | Yes |
| 2.5 | 1.5 | 3.0 | 108 | 4.74 ± 0.04 | 40.43 ± 0.56 | 273 ± 64 | No |
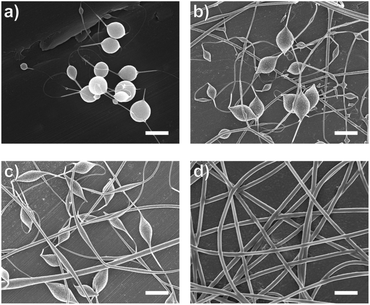
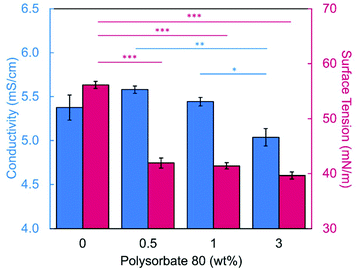
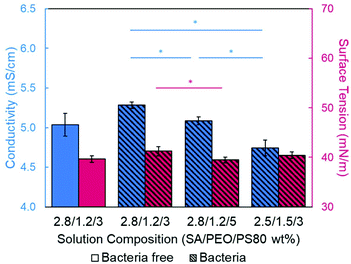
Introducing surfactant into the precursor solution decreased the surface tension and allowed for the formation of smooth fibers. We explored the use of a nonionic surfactant, PS80 because it is an FDA approved surfactant already used in oral delivery.67 The addition of 0.5 wt% PS80 into the 2.8/1.2 wt% SA/PEO solution reduced the surface tension by 19%, to ∼41.93 mN m−1, compared to the surfactant-free solution. Continuous fibers that were 125 ± 37 nm in diameter were formed at the smallest incorporation of surfactant (0.5 wt% PS80), but elongated beads were still present (Fig. 2b). Increasing the surfactant concentration to 1.0 wt% produced more spindle-like beads, as well as some smooth fibers (Fig. 2c). A mixture of beaded and bead-free fibers would alter the delivery rates of drug loaded fibers.31,68,69 The average diameter was not statistically different compared to the previous fibers made from 0.5 wt% PS80. As the surface tension decreased, the fiber diameter (and consequently the fiber mass) is more consistent along the length of the fiber leading to smaller, less spherical beads and thicker fiber diameters.66 Smooth fibers with an average fiber diameter of 167 ± 24 nm were produced by further increasing the surfactant concentration to 3.0 wt% PS80 (Fig. 2d). The presence of surfactant, in any amount, was able to significantly decrease the surface tension of the precursor solution compared to the solution without surfactant. However, a two tailed t-test proved that the surface tension did not significantly change between the varying amounts of surfactant, from 0.5–3.0 wt% (Fig. 3).
Similar trends of increasing the surfactant concentration to reduce beading in SA/PEO electrospun fibers have been reported in other studies. Saquing et al.35 found that lower concentrations of Triton X-100 (0.1 wt% and 0.3 wt%) produced beaded fibers with an 80 to 20 ratio of SA to PEO, whereas higher concentrations (0.5 wt%, 1 wt%, 1.5 wt%) produced bead-free fibers. Similar to our study, all these concentrations were well above critical micelle concentration (CMC). At this concentration, surfactant molecules have saturated the surface of the liquid allowing excess molecules in the bulk solution to form micelles. Therefore, increasing the surfactant concentration should not significantly affect surface tension values after reaching the CMC.70 Yet, changes in morphology were reported. Notably, different sources of alginate and varying molecular weights of PEO were used for each of these studies leading to different weight ratios and concentrations. It is interesting that various concentrations of surfactant can be used to form smooth fibers despite the fact that the surface tension should not change after reaching CMC; this led us to hypothesize that the surfactant concentration might have a compounding effect on solution properties, which would affect fiber formation.
Effect of conductivity on alginate-based nanofibers
We next determined the conductivity of the precursor solutions because sodium alginate dissolved in water has a high conductivity due to its ionic nature. Notably, high conductivity can prevent fiber formation by causing strong repulsive forces between polymer chains that prevent entanglements during electrospinning.34–36,61,71 One of the reasons that a linear copolymer, like PEO, is used to electrospin alginate is because the PEO chains wrap around the rigid alginate chains to interrupt interactions between alginate and sodium ions lowering the conductivity of the solution.36 We observed this effect, too. Mixing the SA solution with the PEO solution decreased its conductivity from 8.17 mS cm−1 (4 wt% SA) to 5.38 mS cm−1 (2.8/1.2 wt% SA/PEO) (Table 1, Fig. 1). Though this change was statistically significant, bead-free fibers were not formed upon the addition of PEO into the solution. While the lowest addition of 0.5 wt% PS80 did not have a statistical effect on the conductivity of the solution, additional amounts of PS80 significantly decreased the solution conductivity from 5.58 mS cm−1 (0.5 wt% PS80) to 5.44 mS cm−1 (1.0 wt% PS80) and to 5.04 mS cm−1 (3.0 wt% PS80) (Fig. 3). Increasing the concentration of surfactant above the CMC leads to the formation of micelles in the bulk solution. These micelles tend to have lower conductivity than their single-molecule counterparts thereby decreasing the conductivity of the solution as surfactant concentration increases.72,73Decreasing conductivity has been previously shown to be able to supress beading. While electrospinning relies on charges within the polymer solution to form the Taylor cone, a highly conductive solution can also be detrimental to the formation of fibers. For example, Zhang et al.63 reported that when they added protein to their precursor solution, it increased solution conductivity and was a source of instability that caused fiber beading. Furthermore, increases in conductivity decreases the tangential electrostatic force within the polymer droplet which hinders Taylor cone and subsequent fiber formation.38,74 In a SA/PEO/Triton X-100 solution, Saquing et al.35 found that even a low concentration (1 wt%) of sodium chloride salt which increases solution conductivity began to decrease fiber formation. A multivalent salt was used by Fang et al.61 to form calcium alginate fibers. However, their fibers formed due to the crosslinking of calcium ions with alginate chains, which allowed for entanglement-like interactions that promoted electrospinning. Conductivities of solutions used in the aforementioned study remained relatively low due to the mixed solvent system used (i.e., a combination of DI water, ethanol, and DMF). Solvents like glycerol60 and DMSO52 also alter interactions between alginate molecules in solution and aid in fiber formation. In our system, we used an aqueous solution to ensure that the final product would be safe for consumption as a delivery vehicle for probiotics. In sum, both the lower surface tension and lower conductivity of the precursor solution aided in the suppression of the beading in our system allowing us to form smooth fibers using 2.8/1.2/3 wt% SA/PEO/PS80 solution (Fig. 2d).
Chemical confirmation of alginate-based nanofibers
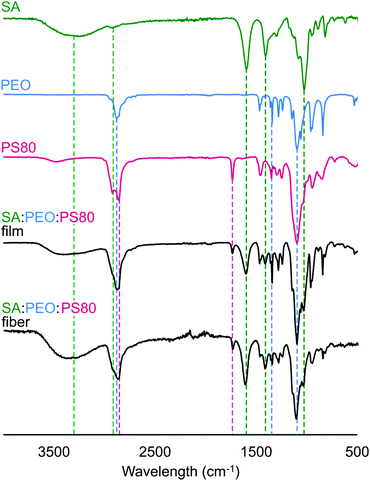
In Fig. 4, we confirmed that all components in the precursor solution were included in the final electrospun nanofiber product by acquiring Fourier-transform infrared (FTIR) spectra and comparing them to control spectra of SA, PEO, PS80, and SA/PEO/PS80. Characteristic peaks of each compound also appeared in the FTIR spectra of the SA/PEO/PS80 fibers and control films, thus indicating that all three components were present. Characteristic peaks in the pure SA spectra included hydroxyl groups (3500–3000 cm−1 broad peak followed by a sharp peak at 2800 cm−1), asymmetric stretches of carboxylate anions (1566 cm−1 and 1408 cm−1 peaks), and ester bonds (1026 cm−1 strong peak).75 Similarly, the PEO spectra exhibited strong peaks at 2878 cm−1 and 1098 cm−1 indicating terminal hydrogens and ester groups, respectively. The series of peaks in the 1460–1180 cm−1 range of the PEO spectra were attributed to secondary hydrogens along the PEO backbone.76 Different from SA and PEO, the PS80 spectra showed a peak at 1700 cm−1 representing a carbonyl group.77 Indeed, fibers were comprised of alginate, PEO, and PS80.Effect of E. coli suspension on fiber morphology
Next, the effect of adding bacteria into the precursor solution was explored using E. coli K12 MG1655 as a model bacterium. The previously described optimized 2.8/1.2/3 wt% SA/PEO/PS80 solution was loaded with 108 CFU mL−1 of E. coli and electrospun into beaded fibers with an average diameter of 100 ± 23 nm (Table 1). The beads present along these bacteria loaded fibers were 0.81 ± 0.25 μm in length and 0.23 ± 0.04 μm in width (Fig. 5a). We measured the rod-shaped E. coli cells using SEM (Fig. S1†), and they were 2.44 ± 0.57 μm in length by 0.65 ± 0.07 μm in diameter; therefore, we did not suspect that these beads were bacteria. To be able to differentiate between these polymer beads and the bacteria, as well as provide a consistent product, we aimed to once again produce smooth fibers by adjusting our formulation.Conductivity and surface tension of bacteria-loaded solutions were determined to explore their role in fiber beading. These results were compared to the solution properties of the previously optimized bacteria-free solutions, as shown in Fig. 6. The change in conductivity due to the addition of bacteria in the 2.8/1.2/3 wt% SA/PEO/PS80 solution, though statistically insignificant, was likely due to the negative charge carried on the surface of the E. coli.78 Škrlec et al.46 saw a similar increase in conductivity due to the addition of L. plantarum in their PEO precursor solution and attributed it to extracellular proteins and ions. Increases in solution conductivity can cause thinner fibers as additional repulsive charges stretch the electrospinning jet.63 This would explain the decrease in average fiber diameter due to the addition of the bacteria. As previously mentioned, increased conductivity also causes beading along electrospun nanofibers due to Taylor Cone instability.66 The surface tension of the bacteria-loaded solution was found to be 41.25 mN m−1. From our previous studies, high surface tension could also have caused the beading. Though neither the change in solution conductivity nor surface tension was statistically significant, we propose changing these solution properties could compensate for the addition of E. coli and produce smooth fibers due to previous indications that these properties influence fiber morphology.
Increasing the PS80 concentration would lower both surface tension and conductivity as proven by the previous studies. By increasing the surfactant from 3 to 5 wt% PS80, the 2.8/1.2 wt% SA/PEO solution with bacteria electrospun into flattened and webbed structures indicative of wet fibers (Fig. 5b). These fibers were 305 ± 110 nm in diameter, which was much larger than any of our previously spun fibers. Surface tension was reduced to 39.50 mN m−1 compared to the 3 wt% PS80 with bacteria, which could explain the production of wet fibers. The force of the electric field that aims to pull entangled polymers out from the electrospinning droplet to form nanofibers is countered by the force of surface tension which stabilizes the electrospinning jet. High surface tension may cause beading, but the low surface tension cannot hold the jet together.63–65 Therefore, excess amounts of solution, both polymer and solvent, is removed from the electrospinning droplet and lands on the collector. In this case, the produced fibers would be larger due to increased mass flow rate and can be wet due to increased amounts of solvent being pulled into fibers.64
Alternatively, conductivity and surface tension can be altered by changing the SA to PEO ratio. PEO chains can interfere with the interaction between sodium ions and alginate chains which are the major contributors to the conductivity in this system.34–36,61,71 Therefore, decreasing the concentration of alginate while increasing the PEO concentration will decrease the solution conductivity. When the ratio of SA to PEO was changed from 2.5 to 1.5, the resulting conductivity of the 2.5/1.5/3 wt% SA/PEO/PS80 solution with 108 CFU mL−1 of E. coli was 4.74 ± 0.08 mS cm−1, which was significantly lower than all other bacteria loaded solutions. Consequently, the surface tension was also altered to a value of 40.43 mN m−1 though this was not significantly different from the 2.8/1.2/3 wt% SA/PEO/PS80 solution with or without bacteria. We suspect lower solution conductivity stabilizes the electrospinning jet while surface tension allows a moderate amount of polymer and solvent to be drawn into dry fibers enabling this solution to produce smooth, bead-free fibers with encapsulated E. coli. The optimized fibers containing bacteria were 273.40 ± 62.53 nm in diameter (Fig. 5c).
Viability of bacteria during electrospinning and after encapsulation in alginate-based fibers
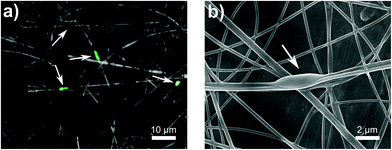
We further analyzed the electrospun fibers with encapsulated E. coli via fluorescence microscopy and SEM. The GFP which are produced by metabolically active bacteria can be monitored using fluorescence microscopy to locate encapsulated bacteria and confirm their viability. In these micrographs (Fig. 7a, and S2†), fluorescent bacteria cells appeared to be encapsulated within the nanofibers. The distinct rod shape of E. coli was oriented length-wise along the fiber due to a sink-like flow in the electrospinning jet.39 SEM micrographs were also able to confirm the encapsulation of bacteria. Bulges appeared along the length of the fiber which were 2.52 ± 1.14 μm in length and 0.86 ± 0.05 μm in width (Fig. 7b). These dimensions closely resemble that of our E. coli (2.44 μm × 0.65 μm), Fig. S1.†Previous work on electrospinning live cells (including mammalian,79,80 yeast,81 and bacterial cells30,39,44–46) have shown variable results on the viability of encapsulated cells. Specifically for E. coli strains encapsulated via electrospinning, viability from 0.1%82 to 48%39,83 has been reported. Severe declines in viability were attributed dehydration,46,47 cell rupture,80 and hydrophobicity,45 whereas viability could be improved with the addition of nutrients, like glycerol39 or prebiotics.17 To our knowledge, only one other paper has encapsulated bacteria in alginate-based fibers which was published while these results were being written. Yilmaz et al.48 published a paper on encapsulating Lactobacillus paracasei in electrospun alginate nanofibers. They were able to achieve a bacterial loading of 8.57 log (CFU g−1) in their fibers made of 7 to 1 ratio of 20 wt% PVA to 3 wt% SA.
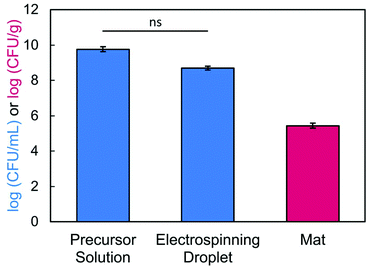
To confirm that we achieved high viability, we determined how many bacteria were loaded into the precursor solution versus how many survived encapsulation within the electrospun fibers. After conducting our typical mixing protocol, a sample of the bacteria-loaded precursor solution (2.5/1.5/3 wt% of SA/PEO/PS80) was serial diluted and plated. The concentration of bacteria in solution before electrospinning was determined to be 4.03 × 109 CFU mL−1 or 9.61 log (CFU mL−1). Next, we confirmed bacterial viability post exposure to the electric field.79 During electrospinning, excess precursor solution falls from the needle tip as the volumetric flow rate exceeds the rate at which solution is drawn into fibers. These droplets contain bacteria that have been subjected to the electric field. In our study, 17.5 kV was applied across a tip-to-collector distance of 17 cm. After passing through the electric field, 4.94 × 108 CFU mL−1 or 8.69 log (CFU mL−1) of E. coli cells were viable, which was statistically equivalent to the concentration that was loaded in the precursor solution (Fig. 8). Bacteria viability and loading is usually self-defined within an individual study.17,39,42,44–48 Some studies39,44,48 have reported bacterial retention as a percentage of the quantity of encapsulated/delivered viable bacteria divided by a measured or theoretical total. We chose not to report bacterial retention as a percentage because we are comparing bacterial concentrations between a liquid phase (CFU mL−1 of precursor) and a solid phase (CFU/gram of nanofiber mat). Notably, this experiment demonstrates that bacteria remain viable after electrospinning. Despite the applied voltage,84 the amount of charge that travels through the bacteria remains low and does not lead to cell inactivation.64,85 Finally, we determined the number of viable bacteria encapsulated in our nanofibers by dissolving and plating the fibers. Bacteria loading in our fibers was determined to be 2.74 × 105 CFU g−1. Importantly, our system is made of green chemistries, an FDA approved surfactant, and aqueous solvent to support biocompatibility with the encapsulated bacteria and potentially to enable delivery to the gut.
The purpose of this paper was to systematically study the precursor solution properties as they relate to the production of smooth, alginate-based fibers with bacteria encapsulants; therefore, techniques used to increase bacterial viability were beyond the scope of this study. As previously mentioned, researchers17,39,42,44–47 have also encapsulated high loadings of bacteria in other polymer nanofibers with the help of co-encapsulates. While we are encouraged by all of these reports, more research is needed to understand the correlation between precursor solution properties, fiber formation, and the distribution of bacteria in these systems. We suggest that electrospun fibers that contain a high concentration of alginate hold potential as delivery vehicles to protect and deliver probiotics to the gut.
Conclusions
This study aimed to encapsulate live bacteria into alginate-based nanofibers to demonstrate the potential of using these materials to deliver probiotics to the gut. First, we optimized the morphology of SA/PEO fibers using the surfactant PS80. The addition of surfactant decreased both the surface tension and the conductivity of the precursor solution to form smooth fibers from a SA/PEO/PS80 (2.8/1.2/3 wt%) solution. Next, a suspension of E. coli was blended into this polymer solution to be encapsulated during electrospinning. However, fibers with a beaded morphology were electrospun from this bacteria-loaded solution. The polymer concentration was modified to compensate for the addition of bacteria into the solution, and smooth fibers were once again produced. The electrospun solution of 2.5/1.5/3 wt% SA/PEO/PS80 encapsulated 2.74 × 105 CFU g−1 of viable E. coli. These bacteria-loaded and alginate-based nanofibers could be useful for the delivery of bacteria into the gut, but more work needs to be conducted to understand the release kinetics. The goal of encapsulating probiotics is to ensure the delivery of a sufficient concentration of bacteria into the gut where they can provide therapeutic effects. We suggest that by thoroughly understanding how surfactant and cargo loading impacts precursor solution properties and subsequently biopolymer fiber formation, that the ease of encapsulating living active agents into biopolymer fibers can be facilitated, thus broadening their application space. The need to deliver living microorganisms using a flexible, porous, and biocompatible textile will continue to expand in years to come.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Sara M. Koprek for her laboratory assistance. E.D. acknowledges the support of the Spaulding Smith Fellowship Program and the National Science Foundation NRT-SMLS program (DGE-1545399). We acknowledge the facilities at the W.M. Keck Center for Electron Microscopy.References
- H. Tlaskalová-Hogenová, R. Štěpánková, H. Kozáková, T. Hudcovic, L. Vannucci, L. Tučková, P. Rossmann, T. Hrnčíř, M. Kverka, Z. Zákostelská, K. Klimešová, J. Přibylová, J. Bártová, D. Sanchez, P. Fundová, D. Borovská, D. Šrůtková, Z. Zídek, M. Schwarzer, P. Drastich and D. P. Funda, Cell. Mol. Immunol., 2011, 8, 110–120 CrossRef PubMed
.
- G. Vighi, F. Marcucci, L. Sensi, G. Di Cara and F. Frati, Clin. Exp. Immunol., 2008, 153, 3–6 CrossRef CAS PubMed
.
- C.-Z. Wang, X.-Q. Ma, D.-H. Yang, Z.-R. Guo, G.-R. Liu, G.-X. Zhao, J. Tang, Y.-N. Zhang, M. Ma, S.-Q. Cai, B.-S. Ku and S.-L. Liu, BMC Microbiol., 2010, 10, 115 CrossRef PubMed
.
- R. Medzhitov and C. J. Janeway, Immunol. Rev., 2000, 173, 89–97 CrossRef CAS PubMed
.
- J. L. Round and S. K. Mazmanian, Nat. Rev. Immunol., 2009, 9, 313–323 CrossRef CAS PubMed
.
- Y. K. Lee, J. S. Menezes, Y. Umesaki and S. K. Mazmanian, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4615–4622 CrossRef CAS PubMed
.
- M. Valles-Colomer, G. Falony, Y. Darzi, E. F. Tigchelaar, J. Wang, R. Y. Tito, C. Schiweck, A. Kurilshikov, M. Joossens, C. Wijmenga, S. Claes, L. Van Oudenhove, A. Zhernakova, S. Vieira-Silva and J. Raes, Nat. Microbiol., 2019, 4, 623–632 CrossRef CAS PubMed
.
- L. Wen, R. E. Ley, P. Yu. Volchkov, P. B. Stranges, L. Avanesyan, A. C. Stonebraker, C. Hu, F. S. Wong, G. L. Szot, J. A. Bluestone, J. I. Gordon and A. V. Chervonsky, Nature, 2008, 455, 1109–1113 CrossRef CAS PubMed
.
- F. Bäckhed, J. K. Manchester, C. F. Semenkovich and J. I. Gordon, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 979–984 CrossRef PubMed
.
- L. de Magistris, V. Familiari, A. Pascotto, A. Sapone, A. Frolli, P. Iardino, M. Carteni, M. De Rosa, R. Francavilla, G. Riegler, R. Militerni and C. Bravaccio, J. Pediatr. Gastroenterol. Nutr., 2010, 51, 418–424 CrossRef PubMed
.
- Z. Li, A. M. Behrens, N. Ginat, S. Y. Tzeng, X. Lu, S. Sivan, R. Langer and A. Jaklenec, Adv. Mater., 2018, 30, 1803925 CrossRef PubMed
.
- S. Stojanov and A. Berlec, Front. Bioeng. Biotechnol., 2020, 8, 130 CrossRef PubMed
.
-
FAO/WHO, Health and Nutritional Properties of Probiotics in Food including Powder Milk with Live Lactic Acid Bacteria, Cordoba, Argentina, 2001 Search PubMed
.
- C. Calinescu, J. Mulhbacher, É. Nadeau, J. M. Fairbrother and M. A. Mateescu, Eur. J. Pharm. Biopharm., 2005, 60, 53–60 CrossRef CAS PubMed
.
- G. Zárate and M. E. Nader-Macias, Process Biochem., 2006, 41, 1779–1785 CrossRef
.
- R. Rajam and C. Anandharamakrishnan, LWT – Food Sci. Technol., 2015, 60, 773–780 CrossRef CAS
.
- K. Feng, M.-Y. Zhai, Y. Zhang, R. J. Linhardt, M.-H. Zong, L. Li and H. Wu, J. Agric. Food Chem., 2018, 66, 10890–10897 CrossRef CAS PubMed
.
- M. J. Lemay, C. P. Champagne, C. Gariépy and L. Saucier, J. Food Sci., 2002, 67, 3428–3434 CrossRef CAS
.
- G. Pasparakis and N. Bouropoulos, Int. J. Pharm., 2006, 323, 34–42 CrossRef CAS PubMed
.
- M. Gu, Z. Zhang, C. Pan, T. R. Goulette, R. Zhang, G. Hendricks, D. J. McClements and H. Xiao, Food Hydrocolloids, 2019, 91, 283–289 CrossRef CAS
.
- F. Weinbreck, I. Bodnár and M. L. Marco, Int. J. Food Microbiol., 2010, 136, 364–367 CrossRef CAS PubMed
.
- M. Govender, Y. E. Choonara, P. Kumar, L. C. du Toit, S. van Vuuren and V. Pillay, AAPS PharmSciTech, 2014, 15, 29–43 CrossRef CAS PubMed
.
- E. Isolauri, M. Juntunen, T. Rautanen, P. Sillanaukee and T. Koivula, Pediatrics, 1991, 88, 90–97 CAS
.
- M. E. Sanders and M. L. Marco, Annu. Rev. Food Sci. Technol., 2010, 1, 65–85 CrossRef PubMed
.
- S. Possemiers, M. Marzorati, W. Verstraete and T. Van de Wiele, Int. J. Food Microbiol., 2010, 141, 97–103 CrossRef CAS PubMed
.
- I. Khan and S. C. Kang, Food Control, 2016, 60, 88–94 CrossRef CAS
.
- L. A. Anderson, M. A. Islam and K. L. J. Prather, J. Biol. Chem., 2018, 293, 5053–5061 CrossRef CAS PubMed
.
- M. Magana, C. Sereti, A. Ioannidis, C. A. Mitchell, A. R. Ball, E. Magiorkinis, S. Chatzipanagiotou, M. R. Hamblin, M. Hadjifrangiskou and G. P. Tegos, Clin. Microbiol. Rev., 2018, 31, 3 CrossRef PubMed
.
- E. Ning, G. Turnbull, J. Clarke, F. Picard, P. Riches, M. Vendrell, D. Graham, A. W. Wark, K. Faulds and W. Shu, Biofabrication, 2019, 11, 045018 CrossRef CAS PubMed
.
- Y. Liu, M. H. Rafailovich, R. Malal, D. Cohn and D. Chidambaram, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 14201–14206 CrossRef CAS PubMed
.
- P. Taepaiboon, U. Rungsardthong and P. Supaphol, Nanotechnology, 2006, 17, 2317–2329 CrossRef CAS
.
- K. A. Rieger, N. P. Birch and J. D. Schiffman, J. Mater. Chem. B, 2013, 1, 4531–4541 RSC
.
-
S. Ramakrishna, K. Fujihara, W.-E. Teo, T.-C. Lim and Z. Ma, An Introduction to Electrospinning and Nanofibers, World Scientific Publishing Co. Pte. Ltd., 2005 Search PubMed
.
- C. A. Bonino, M. D. Krebs, C. D. Saquing, S. I. Jeong, K. L. Shearer, E. Alsberg and S. A. Khan, Carbohydr. Polym., 2011, 85, 111–119 CrossRef CAS
.
- C. D. Saquing, C. Tang, B. Monian, C. A. Bonino, J. L. Manasco, E. Alsberg and S. A. Khan, Ind. Eng. Chem. Res., 2013, 52, 8692–8704 CrossRef CAS
.
- J. Mirtič, H. Balažic, Š. Zupančič and J. Kristl, Polymers, 2019, 11, 4 CrossRef PubMed
.
- D. H. Reneker and A. L. Yarin, Polymer, 2008, 49, 2387–2425 CrossRef CAS
.
- A. Haider, S. Haider and I.-K. Kang, Arabian J. Chem., 2018, 11, 1165–1188 CrossRef CAS
.
- W. Salalha, J. Kuhn, Y. Dror and E. Zussman, Nanotechnology, 2006, 17, 4675–4681 CrossRef CAS PubMed
.
- S. Zargham, S. Bazgir, A. Tavakoli, A. S. Rashidi and R. Damerchely, J. Eng. Fibers Fabr., 2012, 7, 155892501200700 Search PubMed
.
- M. Faccini, C. Vaquero and D. Amantia, J. Nanomater., 2012, 2012, 1–9 CrossRef
.
- M. Kurečič, T. Rijavec, S. Hribernik, A. Lapanje, K. S. Kleinschek and U. Maver, Nanomed, 2018, 13, 1583–1594 CrossRef PubMed
.
- P. Uttayarat, A. Perets, M. Li, P. Pimton, S. J. Stachelek, I. Alferiev, R. J. Composto, R. J. Levy and P. I. Lelkes, Acta Biomater., 2010, 6, 4229–4237 CrossRef CAS PubMed
.
- Zs. K. Nagy, I. Wagner, A. Suhajda, T. Tobak, A. H. Harasztos, T. Vigh, P. L. Soti, H. Pataki, K. Molnar and Gy. Marosi, eXPRESS Polym. Lett., 2014, 8, 352–361 CrossRef
.
- Š. Zupančič, K. Škrlec, P. Kocbek, J. Kristl and A. Berlec, Pharmaceutics, 2019, 11, 483 CrossRef PubMed
.
- K. Škrlec, Š. Zupančič, S. Prpar Mihevc, P. Kocbek, J. Kristl and A. Berlec, Eur. J. Pharm. Biopharm., 2019, 136, 108–119 CrossRef PubMed
.
- A. López-Rubio, E. Sanchez, Y. Sanz and J. M. Lagaron, Biomacromolecules, 2009, 10, 2823–2829 CrossRef PubMed
.
- M. T. Yilmaz, O. Taylan, C. Y. Karakas and E. Dertli, Carbohydr. Polym., 2020, 244, 116447 CrossRef CAS PubMed
.
- S. Nualkaekul, M. T. Cook, V. V. Khutoryanskiy and D. Charalampopoulos, Food Res. Int., 2013, 53, 304–311 CrossRef CAS
.
- M. Bruchet and A. Melman, Carbohydr. Polym., 2015, 131, 57–64 CrossRef CAS PubMed
.
- Y. Su, L. Gu, Z. Zhang, C. Chang, J. Li, D. J. McClements and Y. Yang, Food Res. Int., 2019, 120, 305–311 CrossRef CAS PubMed
.
- N. Bhattarai and M. Zhang, Nanotechnology, 2007, 18, 455601 CrossRef
.
- Y. Ma, P. Qi, J. Ju, Q. Wang, L. Hao, R. Wang, K. Sui and Y. Tan, Composites, Part B, 2019, 162, 671–677 CrossRef CAS
.
- H. Hajiali, M. Summa, D. Russo, A. Armirotti, V. Brunetti, R. Bertorelli, A. Athanassiou and E. Mele, J. Mater. Chem. B, 2016, 4, 1686–1695 RSC
.
- Y. Tang, X. Lan, C. Liang, Z. Zhong, R. Xie, Y. Zhou, X. Miao, H. Wang and W. Wang, Carbohydr. Polym., 2019, 219, 113–120 CrossRef CAS PubMed
.
- J. M. Yang, J. H. Yang, S. C. Tsou, C. H. Ding, C. C. Hsu, K. C. Yang, C. C. Yang, K. S. Chen, S. W. Chen and J. S. Wang, Mater. Sci. Eng., C, 2016, 66, 170–177 CrossRef CAS PubMed
.
- K. A. Rieger, M. Porter and J. D. Schiffman, Materials, 2016, 9, 297 CrossRef PubMed
.
- K. M. Dobosz, C. A. Kuo-Leblanc, T. J. Martin and J. D. Schiffman, Ind. Eng. Chem. Res., 2017, 56, 5724–5733 CrossRef CAS PubMed
.
- T. S. Heckmann and J. D. Schiffman, ACS Appl. Nano Mater., 2020, 3, 977–984 CrossRef CAS
.
- H. Nie, A. He, J. Zheng, S. Xu, J. Li and C. C. Han, Biomacromolecules, 2008, 9, 1362–1365 CrossRef CAS PubMed
.
- D. Fang, Y. Liu, S. Jiang, J. Nie and G. Ma, Carbohydr. Polym., 2011, 85, 276–279 CrossRef CAS
.
- L. A. Bosworth and S. Downes, J. Polym. Environ., 2012, 20, 879–886 CrossRef CAS
.
- C. Zhang, X. Yuan, L. Wu, Y. Han and J. Sheng, Eur. Polym. J., 2005, 41, 423–432 CrossRef CAS
.
- J. M. Deitzel, J. Kleinmeyer, D. Harris and N. C. Beck Tan, Polymer, 2001, 42, 261–272 CrossRef CAS
.
- S. Chung, A. K. Moghe, G. A. Montero, S. H. Kim and M. W. King, Biomed. Mater., 2009, 4, 015019 CrossRef PubMed
.
- H. Fong, I. Chun and D. H. Reneker, Polymer, 1999, 40, 4585–4592 CrossRef CAS
.
- A. Kazsoki, P. Szabó, A. Domján, A. Balázs, T. Bozó, M. Kellermayer, A. Farkas, D. Balogh-Weiser, B. Pinke, A. Darcsi, S. Béni, J. Madarász, L. Szente and R. Zelkó, Mol. Pharm., 2018, 15, 4214–4225 CrossRef CAS PubMed
.
-
H. Zhao and H. Chi, Chapter 5: bead-on-string nanofibers: useless or something of value?, in Novel aspects of nanofibers, ed. T. Lin, IntechOpen Limited, 201815, pp. 87–102 Search PubMed
.
- Y. Sun, S. Cheng, W. Lu, Y. Wang, P. Zhang and Q. Yao, RSC Adv., 2019, 9, 25712–25729 RSC
.
- D. M. Ćirin, M. M. Poša and V. S. Krstonošić, Ind. Eng. Chem. Res., 2012, 51, 3670–3676 CrossRef
.
- T. C. Mokhena, M. J. Mochane, A. Mtibe, M. J. John, E. R. Sadiku and J. S. Sefadi, Materials, 2020, 13, 934 CrossRef CAS PubMed
.
- M. Pérez-Rodríguez, G. Prieto, C. Rega, L. M. Varela, F. Sarmiento and V. Mosquera, Langmuir, 1998, 14, 4422–4426 CrossRef
.
- S. Durand-Vidal, O. Bernard, Ž. Medoš and M. Bešter-Rogač, J. Mol. Liq., 2020, 309, 112968 CrossRef CAS
.
- C. J. Angammana and S. H. Jayaram, IEEE Trans. Ind. Appl., 2011, 47, 1109–1117 CAS
.
- Md. S. Islam and M. R. Karim, Colloids Surf., A, 2010, 366, 135–140 CrossRef CAS
.
- M. Sethupathy, V. Sethuraman, A. R. Jeyaraman, P. Muthuraja and P. Manisankar, AIP Conf. Proc., 2014, 1620, 253 CrossRef CAS
.
- B. Kim, J. Yang, M. Hwang, J. Choi, H.-O. Kim, E. Jang, J. Lee, S. Ryu, J. Suh, Y.-M. Huh and S. Haam, Nanoscale Res. Lett., 2013, 8, 399 CrossRef PubMed
.
- J. S. Dickson and M. Koohmaraie, Appl. Environ. Microbiol., 1989, 55, 832–836 CrossRef CAS PubMed
.
- A. Townsend-Nicholson and S. N. Jayasinghe, Biomacromolecules, 2006, 7, 3364–3369 CrossRef CAS PubMed
.
- M. F. Canbolat, C. Tang, S. H. Bernacki, B. Pourdeyhimi and S. Khan, Macromol. Biosci., 2011, 11 Search PubMed
.
- M. F. Canbolat, N. Gera, C. Tang, B. Monian, B. M. Rao, B. Pourdeyhimi and S. A. Khan, ACS Appl. Mater. Interfaces, 2013, 5, 9349–9354 CrossRef CAS PubMed
.
- M. Gensheimer, M. Becker, A. Brandis,–Heep, J. H. Wendorff, R. K. Thauer and A. Greiner, Adv. Mater., 2007, 19, 2480–2482 CrossRef CAS
.
- S. Xie, S. Tai, H. Song, X. Luo, H. Zhang and X. Li, J. Mater. Chem. B, 2016, 4, 6820–6829 RSC
.
- H. Hülsheger and E.-G. Niemann, Radiat. Environ. Biophys., 1980, 18, 281–288 CrossRef PubMed
.
- P. K. Baumgarten, J. Colloid Interface Sci., 1971, 36, 71–79 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0bm02205e |
| This journal is © The Royal Society of Chemistry 2021 |