Open Access Article
Open Access ArticlePerformance of single-cell ICP-MS for quantitative biodistribution studies of silver interactions with bacteria†
Ana C.
Gimenez-Ingalaturre
 a,
Isabel
Abad-Álvaro
a,
Isabel
Abad-Álvaro
 *a,
Pilar
Goñi
*a,
Pilar
Goñi
 b,
Kharmen
Billimoria
b,
Kharmen
Billimoria
 c,
Heidi
Goenaga-Infante
c,
Heidi
Goenaga-Infante
 c and
Francisco
Laborda
c and
Francisco
Laborda
 a
a
aGroup of Analytical Spectroscopy and Sensors (GEAS), Institute of Environmental Sciences (IUCA), University of Zaragoza, Pedro Cerbuna 12, 50009 Zaragoza, Spain. E-mail: iabad@unizar.es
bGroup of Water and Environmental Health, Institute of Environmental Sciences (IUCA), Domingo Miral s/n, 50009 Zaragoza, Spain
cNational Measurement Laboratory, LGC, Queens Road, Teddington, TW11 0LY, UK
First published on 23rd January 2024
Abstract
Single-cell analysis using inductively coupled plasma mass spectrometry (SC-ICP-MS) is an analytical methodology that allows to obtain qualitative and quantitative information of the element content of bioparticles (cells, bacteria, unicellular algae, yeasts…) on a cell-by-cell basis. In this study, two commercially available nebulisation systems for introduction of intact bioparticles were evaluated, showing similar performance. Bacteria (E. coli) exposed to Ag(I) and 10 nm Ag nanoparticles were detected and their complete silver content distributions successfully recorded. Analysis of the corresponding spheroplasts, obtained after enzymatic digestion of the bacterial outer membrane with lysozyme, allowed information about the intracellular silver to be obtained, providing an insight into the biodistribution of silver in the bacteria. Likewise, validity of the quantitative information obtained by SC-ICP-MS was evaluated by applying methodology based on the estimation of silver mass per bioparticle critical values and successive dilutions. The quantitative information obtained by SC-ICP-MS for the total content of silver in bacteria and spheroplasts was confirmed by acid digestion followed by conventional ICP-MS analysis. Bacteria exposed to silver nanoparticles accumulated lower amounts of silver than those exposed to ionic silver, the latter being totally internalized, showing the higher bioavailability of ionic silver. This study proves for the first time the suitability of SC-ICP-MS for the quantitative determination of silver in bacteria, as well as in spheroplasts when combined with lysozyme digestion. Limits of detection of 7 ag of Ag per bioparticle, 500 bioparticles per mL and 38 ng of total Ag per litre of bacteria/spheroplast suspension were achieved.
Introduction
Cells are the morphological and functional unit of all living organisms. Due to their associated structural complexity, cells of the same type that have been exposed to the same external stimulus, can exhibit significant differences in the intracellular expression of biomolecules and elements. These cell-to-cell variations can be the cause of different diseases, neurological and developmental disorders.1 Thus, methods that allow cell-by-cell analysis to obtain information about their variability are highly relevant. On the other hand, metals are important elements in a number of biological processes.1,2 Hence, the study of cell variability in terms of metallomics is of critical importance.1,3,4 Traditional methods used for determination of the metal content in cells provide average mass concentrations, where the homogeneous distribution of the metals in the cells is assumed.4 However, quantification of metals content in individual cells can help to understand their mechanisms of uptake and elimination, the mechanisms of interactions between metal-containing nanoparticles and metallo-drugs with cells, or the distribution of nutrients among a population of cells.5When inductively coupled plasma mass spectrometry (ICP-MS) is operated in single particle mode to analyse bioparticles, such as cells, bacteria, microalgae or yeasts, elemental information of individual bioparticles can be obtained, which is commonly known as single cell (SC) ICP-MS.6 SC-ICP-MS can be used to quantify both endogenous elements in individual bioparticles, as well as the uptake and bioaccumulation of dissolved elements and nanoparticles. SC-ICP-MS is able to provide information about the mass of element per cell, the mass distribution within a cell population and the concentration of cells containing specific elements.5,6 Conceptually, single particle and single cell analysis by ICP-MS are based on the same fundamentals, so the majority of the concepts and procedures related to SP-ICP-MS can be directly applied to SC-ICP-MS. When a sufficiently diluted suspension of cells is introduced into the plasma, each cell will be vaporised, atomised and its atoms ionised as a single pack of ions that will be detected as a discrete event. A very diluted suspension (<109 cells per L) and fast data acquisition are required so that each detected event corresponds only to an individual cell.6 Likewise, the calculations for SC-ICP-MS are also similar to those for SP-ICP-MS.4
The first work devoted to SC-ICP-MS was published by Li et al.7 in 2005. These authors analysed samples of bacteria incubated in uranium-rich culture medium and demonstrated that the uranium signals came from uranium compounds incorporated into individual bacteria, which behaved like large particles in the ICP.7 From then on, SC-ICP-MS has been used for the analysis of different types of unicellular systems from different origins, such as human and other mammalian cell lines, bacteria, algae or yeasts.1,4 Some parameters such as the size, morphology or stability of these bioparticles, determined by their composition or structure, have an influence on the analysis by SC-ICP-MS, and it is in this context where the differences between SP-ICP-MS and SC-ICP-MS become more evident because the stability of bioparticles and other organic or inorganic particles is not the same.
A problem associated with SC-ICP-MS is related to the introduction of bioparticles into the plasma. Conventional sample introduction systems usually consist of cyclonic or double pass spray chamber equipped with pneumatic nebulisers, which are designed to filter larger particles (up to 3–5 μm) preventing them from entering the plasma.1,4,5 However, some bioparticles are larger than the droplet size allowed by these conventional sample introduction systems, because the size of some human cells can range from 7–150 μm.1 Nomizu et al. used a concentric nebuliser for the analysis of 10–20 μm cells to quantify calcium content by ICP-OES, and the authors obtained a transport efficiency lower than 0.1%.8 Likewise, in more recent publications, transport efficiencies between 2–3% have been obtained using conventional sample introduction systems for the determination of cobalt and silver in different cells.9,10 In order to overcome these limitations, new sample introduction systems have been developed and commercialised. These systems have been designed to provide higher transport efficiencies using sample flow rates in the range of μL min−1. Some authors have used these sample introduction systems for the analysis of cells, algae and yeasts, obtaining transport efficiencies from 8 up to 30%.11–13 Moreover, these systems allow the introduction of intact bioparticles into the plasma keeping their integrity, which is especially relevant for fragile cells and prone to rupture.2 As mentioned above, continuous advances in SC-ICP-MS over the last years have made it possible to apply this technique in different scientific fields. The most common applications are the determination of endogenous elements in cells (Cu, Fe, K, Na, Zn, P or S) and the evaluation of uptake and distribution of exogenous elements. Cells can be exposed to metals in the context of different applications, such as metallodrug studies, exposition to nanoparticles or the use of metallic labels.2,4,14 It should be noted that most of the studies reported in the literature4 are based on the analysis of cells, algae or yeast, whereas studies on bacteria are very limited.7,15,16
In relation to the information obtained by SC-ICP-MS, it can be limited to get qualitative information, as in the monitoring of essential elements (Mg, P, Mn, Cu, Se or Zn) in single yeast cells (Saccharomyces cerevisiae)17,18 or to the detection of endogenous and exogenous elements (Mg, P, K, Cr, Zn and Pt) in single sperm cells.19 However, many studies provide quantitative information derived from SC-ICP-MS whose validity should be evaluated conveniently, which is not always the case.20–22 Comparison of the SC-ICP-MS results with those from the acid digested cells is the most frequent strategy, although significant differences have been reported for the analysis of a number of cells, algae, protozoa and yeasts.9,10,23–30 Different reasons have been proposed to explain these differences: differences in the transport efficiency of nanoparticles and cells,25 differences in the atomization and ionisation for nanoparticles and cells,27 saturation of the detector when the cells contain a high number of nanoparticles,10,28 lower number of cells involved in the SC-ICP-MS measurements,24 on line correction with internal standards was not possible,26 dwell time affected the detection limits in single cell analysis.29 In any case, robust strategies to obtain comparable results between different methods to validate the quantitative results obtained by SC-ICP-MS are required.
On the other hand, SC-ICP-MS is limited to determine the total content of specific elements in individual bioparticles. Thus, in studies where microorganisms are incubated with exogenous elements, SC-ICP-MS is unable to distinguish whether the accumulated element has been internalised by the microorganism or retained at the membrane. In this regard, cell lysis31 and enzymatic digestion32 procedures have proven to be useful for determining the average silver internalised in bacteria when used in combination with ICP-MS or size exclusion chromatography ICP-MS, respectively. Hence, it is feasible the combination of SC-ICP-MS with these methods to determine the element distribution in bioparticles in a cell-by-cell basis.
The aim of this work is the development and application for the first time of an analytical methodology based on SC-ICP-MS for the analysis of bacterial cultures exposed to silver species with two main objectives: (i) the identification of bacteria containing silver and the quantification of their individual total content, (ii) to study the biodistribution of silver by quantification of intracellular silver, through the enzymatic removal of the bacterial cell wall. In addition, the validity of the quantitative information obtained by SC-ICP-MS has been evaluated by applying a method based on the estimation of the element mass per particle critical values and by comparison with the information obtained by conventional ICP-MS analysis.
Experimental
Instrumentation
An Agilent 8900 ICP-MS/MS (Agilent Technologies, Tokyo, Japan) and a PerkinElmer NexION 2000B Inductively Coupled Plasma Mass Spectrometer (Toronto, Canada) were used throughout the study. When working in conventional mode, the sample introduction systems consisted of a conventional MicroMist nebuliser and cooled (2 °C) quartz Scott double pass spray chamber (Glass Expansion, Melbourne, Australia) for the Agilent 8900, and of a glass concentric nebuliser and a baffled cyclonic spray chamber (Meinhard, Colorado, USA) for the NexION 2000B. Data acquisition was performed using a dwell time of 0.3 s, 100 sweeps and 20 replicates per measurement in the first case, and with a dwell time of 50 ms, 20 sweeps and 10 replicates per measurement for the second one. Both mass spectrometers were also used for measurements in single cell mode (SC-ICP-MS). In this case, sample introduction systems consisted of a high efficiency glass concentric CytoNeb micronebuliser (Meinhard, Colorado, USA) with a CytoSpray linear pass spray chamber (Elemental Scientific Inc., Nebraska, USA) and a high efficiency glass concentric micronebuliser (Meinhard, Colorado, USA) and an Asperon™ linear pass spray chamber (PerkinElmer, Toronto, Canada), for the Agilent and PerkinElmer instruments, respectively. Default instrumental and data acquisition parameters are listed in Table 1.| ICP-MS | Perkin Elmer NexION 2000B | Agilent 8900 |
|---|---|---|
| Instrumental parameters | ||
| RF power | 1600 W | 1550 W |
| Argon gas flow rate | ||
| Plasma | 15 L min−1 | 15 L min−1 |
| Auxiliary | 1.2 L min−1 | 0.9 L min−1 |
| Nebuliser | 0.3 L min−1 | 0.3 L min−1 |
| Make-up | 0.9 L min−1 | 0.9 L min−1 |
| Sample flow rate | 12–15 μL min−1 | 15–18 μL min−1 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Data acquisition parameters | ||
| Dwell time | 100 μs | 100 μs |
| Total acquisition time | 60 s | 60 s |
| Isotopes monitored | 107Ag, 197Au | 107Ag, 151Eu |
Standards
Diluted suspensions of gold and silver nanoparticles were prepared from commercially available suspensions. Ultra-uniform gold nanoparticle (PEG-carboxil 0.8 kDa surface) suspension of 47.8 ± 1.8 nm, silver nanoparticle suspension (mPEG 5 kDa surface) of 51 ± 4 nm diameter and monodisperse citrate-stabilised silver nanoparticles of nominal diameters 60 ± 7 nm and 10.3 ± 2.1 nm were obtained from NanoComposix (San Diego, CA, USA). A europium microsphere suspension of nominal diameter 2.36 ± 0.22 μm was purchased from Bang Laboratories (Indiana, USA). Dilutions were prepared in ultrapure water (Milli-Q Advantage, Molsheim, France or Elga, Marlow, Buckinghamshire, UK) by accurately weighing (±0.1 mg) aliquots of the stock suspensions after 1 min sonication. After dilution and before each analysis, the suspensions were bath sonicated for 1 min. Longer sonication times were not used to avoid excessive heating of the suspensions.Aqueous gold, silver and multi-element (VHG-SM68-1, VHG-SM68-2 and VHG-SM68-3) solutions were prepared from standard stock solutions of 100–1000 mg L−1 (Sigma Aldrich, Saint Louis, MO, USA and Romil, Cambridge, UK) by dilution in ultrapure water or 1% or 25% (v/v) HNO3. Nitric acid (Baker Instra Analyzed for Trace Metals Analysis, J. T. Baker, Holland and Romil, Cambridge, UK), hydrogen peroxide (Scharlau, Scharlab. S.L., Barcelona, Spain and Romil, Cambridge, UK), physiological saline solution (NaCl 0.9%) (Panreac, Barcelona, Spain), phosphate buffered saline (PBS) (Sigma-Aldrich, Saint Louis, MO, USA), Mueller-Hinton broth (MHB) (Scharlau, Scharlab S.L., Barcelona, Spain), Mueller-Hinton (MH) agar (Bio-Rad, La Coquette, France), Tween 80 (Sigma Aldrich, Saint Louis, MO, USA), lysozyme from chicken egg white (90%) (Sigma Aldrich, Saint Louis, MO, USA), ethylenediaminetetraacetic acid (EDTA) (98%) (Scharlau, Scharlab. S.L., Barcelona, Spain), Tris–Hydrochloride (Tris–HCl) (99%) (Sigma Aldrich, Saint Louis, MO, USA), glycerol (Panreac, Barcelona, Spain) and paraformaldehyde (Fisher Scientific, Thermo Fisher Scientific, Waltham, MA, USA) were also used. The solutions of physiological saline solution, PBS, Tris–HCl and culture media were autoclaved for 20 min at 121 °C at 1 atm.
Bacterial cell culture
Escherichia coli ATCC 25922 strain was used in all experiments. The bacteria were cultured in MH agar and grown overnight at 37 °C.Procedures
A part of the bacterial samples was subjected to lyophilisation and fixation with paraformaldehyde.34 Samples exposed to 1 mg L−1 of silver(I) or 2 mg L−1 of 10 nm silver nanoparticles were centrifuged at 6000g during 3 min to remove the PBS supernatant. A 5% (w/v) glycerol solution in PBS was added to the pellet and let it work for 2 hours. After this time, samples were centrifuged under the same conditions to remove the supernatant. Bacterial pellets were frozen overnight at −80 °C and lyophilised for 24 h at −48 °C and 4.36 × 10−6 atm (Tesla Lioalfa-6). Lyophilised samples were stored at 4 °C. For the fixation process, the lyophilised pellets were resuspended in 1 mL of sterile PBS and centrifuged at 5000 rpm during 3 min. Supernatant was discarded and a 4% paraformaldehyde solution in PBS was added to the bacterial pellets. After 15 min, the samples were centrifuged under the last same conditions to remove the supernatant. The fixed pellets were resuspended in 1 mL of ultrapure water. Controls were also treated in the same way. To estimate the concentration of bacteria in samples, optical density of bacterial cultures was measured with an UV-vis spectrophotometer (UV-2004 Lan Optics) at 600 nm. Bacteria concentrations ranged 7–9 x 109 mL−1 for non post-treated bacteria, and 5 × 1010 mL−1 for the lyophilised and fixed ones.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g during 20 min to remove the PBS medium. Volumes of 500 μL of HNO3 (69–70% w/v) and 100 μL of H2O2 (30% v/v) were added to the pellets and samples were shaken and digested for 24 h at room temperature and shaken at 124 rpm. After digestion, the volume was made up to 10 mL with 1% HNO3 (v/v) and the content of total silver was quantified by ICP-MS. Five replicates of each sample were analysed. Bacterial control and bacterial control spiked with 5 μg L−1 of silver(I) were treated in the same way. In addition, the acid digestion procedure was applied to the enzymatically digested samples (final supernatants and pellets) and enzymatically digested bacterial control sample spiked with 5 μg L−1 of silver(I).
000g during 20 min to remove the PBS medium. Volumes of 500 μL of HNO3 (69–70% w/v) and 100 μL of H2O2 (30% v/v) were added to the pellets and samples were shaken and digested for 24 h at room temperature and shaken at 124 rpm. After digestion, the volume was made up to 10 mL with 1% HNO3 (v/v) and the content of total silver was quantified by ICP-MS. Five replicates of each sample were analysed. Bacterial control and bacterial control spiked with 5 μg L−1 of silver(I) were treated in the same way. In addition, the acid digestion procedure was applied to the enzymatically digested samples (final supernatants and pellets) and enzymatically digested bacterial control sample spiked with 5 μg L−1 of silver(I).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 4 °C for 20 min to remove the PBS medium. The pellets obtained were washed twice with 0.01 M Tris–HCl buffer (pH = 7.8) by centrifugation at 3000g at 4 °C for 20 min. The supernatants were removed and the bacterial cell pellets were resuspended in 500 μL of 0.01 M Tris–HCl buffer. A volume of 10 μL of 0.5 M EDTA (pH = 8) was added slowly to the bacteria, until its concentration reached 10 mM and samples were shaken and incubated for 30 min at 37 °C and 100 rpm. Afterwards, samples were centrifuged at 3000g for 20 min. The supernatants were removed and 500 μL of lysozyme in 0.01 M Tris–HCl buffer (2 mg mL−1) were added to the pellets. Samples were incubated for 1 h at 37 °C shaking at 100 rpm. Afterwards, samples were centrifuged at 4000g for 20 min at 4 °C. The supernatants were stored and analysed by ICP-MS after acid digestion. The pellets were washed twice with 0.01 M Tris–HCl buffer (pH = 7.8) by centrifugation at 3000g at 4 °C for 20 min. The final pellets were subjected to analysis by ICP-MS after acid digestion and by SC-ICP-MS. Bacterial and culture medium controls were treated in the same way.
000g and 4 °C for 20 min to remove the PBS medium. The pellets obtained were washed twice with 0.01 M Tris–HCl buffer (pH = 7.8) by centrifugation at 3000g at 4 °C for 20 min. The supernatants were removed and the bacterial cell pellets were resuspended in 500 μL of 0.01 M Tris–HCl buffer. A volume of 10 μL of 0.5 M EDTA (pH = 8) was added slowly to the bacteria, until its concentration reached 10 mM and samples were shaken and incubated for 30 min at 37 °C and 100 rpm. Afterwards, samples were centrifuged at 3000g for 20 min. The supernatants were removed and 500 μL of lysozyme in 0.01 M Tris–HCl buffer (2 mg mL−1) were added to the pellets. Samples were incubated for 1 h at 37 °C shaking at 100 rpm. Afterwards, samples were centrifuged at 4000g for 20 min at 4 °C. The supernatants were stored and analysed by ICP-MS after acid digestion. The pellets were washed twice with 0.01 M Tris–HCl buffer (pH = 7.8) by centrifugation at 3000g at 4 °C for 20 min. The final pellets were subjected to analysis by ICP-MS after acid digestion and by SC-ICP-MS. Bacterial and culture medium controls were treated in the same way.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g during 20 min to remove the PBS medium and the bacterial pellets were resuspended in ultrapure water and diluted to a cell number concentration of approximately 4 × 108 L−1. On the other hand, enzymatically digested samples were also analysed using the same ICP-MS instrument. The final pellets were resuspended and diluted in 0.01 M Tris–HCl buffer. All suspensions were measured in single cell mode using the Syngistix Single Cell-Application module version 2.5 (PerkinElmer Inc.). Recorded signals were processed by applying a 5-sigma threshold criterion calculated as five times the square root of the mean baseline intensity of the time scan.35,36 Analyte transport efficiency was calculated according to the frequency method developed by Pace et al.37 by using the ultra-uniform gold nanoparticle standard described above. Sample flow rate was measured gravimetrically.
000g during 20 min to remove the PBS medium and the bacterial pellets were resuspended in ultrapure water and diluted to a cell number concentration of approximately 4 × 108 L−1. On the other hand, enzymatically digested samples were also analysed using the same ICP-MS instrument. The final pellets were resuspended and diluted in 0.01 M Tris–HCl buffer. All suspensions were measured in single cell mode using the Syngistix Single Cell-Application module version 2.5 (PerkinElmer Inc.). Recorded signals were processed by applying a 5-sigma threshold criterion calculated as five times the square root of the mean baseline intensity of the time scan.35,36 Analyte transport efficiency was calculated according to the frequency method developed by Pace et al.37 by using the ultra-uniform gold nanoparticle standard described above. Sample flow rate was measured gravimetrically.
The samples of lyophilised and fixed bacteria were analysed using the Agilent 8900 ICP-MS. Before analysis, bacterial samples were diluted in ultrapure water to a cell number concentration of approximately 5 × 107 L−1. Suspensions were measured in single particle mode using the single particle application module for ICP-MS MassHunter 4.3 (version: G72Dc.c.01.03) software. Recorded time scans were exported and processed with the SPCal38 software by using the Poisson filter option. Analyte transport efficiency was calculated according to the frequency method developed by Pace et al.37 by using the ultra-uniform silver nanoparticle standard described above. Dynamic mass flow method (DMF), which does not require reference material for the calculation of transport efficiency, was used for the in-house characterisation on particle number concentration of this material,39 obtaining a particle number concentration of 8.4 × 1014 kg−1, which was in good agreement with the value indicated by the manufacturer. Sample flow rate was measured gravimetrically.
Results and discussion
Evaluation of nebulisation systems for SC-ICP-MS
Two nebulisation systems specifically designed for introduction of intact cells in ICP-MS were evaluated in order to determine their optimal working conditions and performance. Both systems are commercially available and consist of linear pass and laminar flow spray chambers with proprietary designs (Asperon™ by PerkinElmer and CytoSpray by Elemental Scientific Inc. (ESI)), equipped with high efficiency concentric micronebulisers (Meinhard). The behaviour of both systems was initially studied with silver nanoparticles, with the ultimate aim of their application to the analysis of bacteria by SC-ICP-MS.Both spray chambers use a make-up gas in order to create a tangential flow to the spray chamber walls, preventing the loss of cells by collision with the walls. Additionally, this make-up gas allows to control the residence time of the bioparticles within the plasma, and thus, the different processes that cells and analytes undergo, regardless of the nebuliser gas flow rate. On the other hand, the laminar gas flow within the spray chamber ensures a maximum aerosol transport at the flow rates applied.
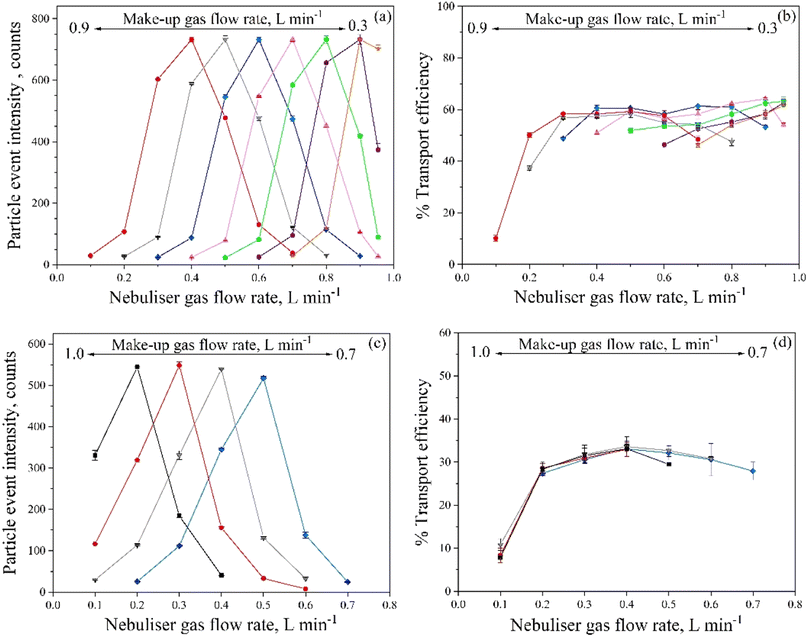
Fig. 1a and b show the effect of the nebuliser gas flow rate at different make-up gas flow rates on the mean intensity of particles detected and the transport efficiency for a suspension of citrate-stabilised silver nanoparticles of 60 nm, using the PerkinElmer nebulisation system. Whereas the transport efficiency depends on the number of particles detected, the intensity observed is related to the atomisation efficiency of the particles and the ionisation efficiency of the target element. Similar behaviours related to the intensity of the events were observed for the different combinations of make-up and nebuliser gas flow rates, with the highest intensity per particle at total gas flow rates (make-up + nebuliser gas flow rate) of 1.2–1.3 L min−1. Furthermore, for a fixed total gas flow rate, the intensity was constant in the curves studied. For example, at a total gas flow of 1.2 L min−1 (combination of nebuliser and make-up gas flow rate of 0.6 + 0.6, 0.5 + 0.7, 0.4 + 0.8 and 0.3 + 0.9 L min−1, respectively), the intensity of the events detected was around 550–600 counts. In relation to the transport efficiency, a similar efficiency was observed in those situations where the maximum event intensity was obtained (total gas flow rate of 1.2–1.3 L min−1). This transport efficiency varied between 55–60%, indicating that, for a fixed total gas flow rate, it was fairly constant at the conditions studied.
According to the results obtained, it was determined that the best conditions with the highest event intensities and the highest transport efficiency, were those with total gas flow rates (make-up + nebuliser gas flow rate) of 1.2–1.3 L min−1. The nebuliser gas flow rates typically used for cell introduction are in the range of 0.3–0.5 L min−1, whereas the make-up gas flow rates are around 0.7–0.9 L min−1.5,11,13,22,40,41 These combinations of high make-up and low nebuliser gas flow rates allow the introduction of intact cells into the plasma.5,11,13,22,24 Hence, the combination of a nebuliser gas flow rate of 0.3 L min−1 and a make-up gas flow rate of 0.9 L min−1 was chosen as optimal.
Fig. 1c and d show the behaviour observed for the CytoSpray ESI nebulisation system in an Agilent 8900 instrument. As for the PerkinElmer system, the maximum event intensity was obtained at a total gas flow rate (make-up + nebuliser gas flow rate) of 1.2 L min−1 with the mean intensity of the events detected between 520–550 counts. In relation to the transport efficiency, the highest efficiency was obtained at a nebuliser gas flow rate of 0.4 L min−1 for all make-up gas flow rates studied. This indicated that the transport efficiency depended only on the nebuliser gas flow rate and it was independent of the make-up gas flow rate. Therefore, based on the results obtained, for the sample introduction system for the Agilent 8900, the best conditions of particle intensity and transport efficiency would be obtained at a total gas flow rate of 1.2 L min−1, being the best combination a nebuliser gas flow rate of 0.4 L min−1, and thus, a make-up gas flow rate of 0.8 L min−1.
Even though both systems showed the maximum event intensity at a total gas flow rate of 1.2–1.3 L min−1, as well as this intensity was constant for a fixed total gas flow rate, the two systems did not show the same behaviour in relation with the transport efficiency (Fig. 1b and d). In the case of the PerkinElmer system, the analyte transport efficiency remained constant for a fixed total gas flow rate, whereas in the case of the CytoSpray ESI system, it was constant for a fixed nebuliser gas flow rate. In any case, the nebulisation conditions finally chosen for subsequent analysis in both systems were the same (0.3 L min−1 nebuliser gas flow rate + 0.9 L min−1 make-up gas flow rate). Under such conditions, the PerkinElmer and the CytoSpray ESI systems provided analyte transport efficiencies of 58.4 ± 1.2 and 30.8 ± 0.6%, respectively (sample flow rate: 12 and 16 μL min−1, respectively).
Bioparticle transport efficiency
A critical issue in SC-ICP-MS when compared to SP-ICP-MS is the nature and size of the particles involved. The stability of cell suspensions depends on the type of cell; bacteria is one of the most robust cell types, therefore it is possible to work with them in simple aqueous solutions.4 Another important factor is the size of the bioparticles. In the case of cells, their sizes are in the micrometre range, which can affect their nebulisation compared to nanoparticles and dissolved species.Escherichia coli bacteria used in this study had a mean length of 1.94 ± 0.43 μm (Fig. S2†). In order to assess if the difference in the size of these bacteria in comparison with nanoparticles could affect the sample introduction, and assuming that bacteria behave similarly to europium-labelled polystyrene microspheres, a suspension of these microspheres (2.36 μm) was measured and transport efficiency determined using the CytoSpray ESI system. The transport efficiencies measured using the europium-labelled microspheres and 50 nm PEG-stabilised silver nanoparticles were 30.3 ± 1.0% and 32.6 ± 1.6%, respectively. By applying a t-test (95% confidence level), it can be affirmed that both results were in agreement. Likewise, this behaviour is in agreement with that determined by Laborda et al.42 using the PerkinElmer nebulisation system, where the authors demonstrated that polystyrene microparticles up to ca. 3 μm were nebulised with the same efficiency than Au nanoparticles, whereas the transport efficiency dropped by half for 5 μm microparticles. Therefore, for both sample introduction systems used throughout this study, the transport efficiency required for characterisation of individual silver-containing bacteria can be determined by nanoparticle standard suspensions.
Detection of bacteria exposed to silver by SC-ICP-MS
The use of SC-ICP-MS as qualitative technique for the assessment of the presence or absence of (bio)particles containing specific elements in a sample relies, as in single particle ICP-MS,35,36 on the application of element mass per particle (XmassC) and number concentration critical values (XnumberC) as limits of decision.The element mass per particle critical value can be based on a 5-sigma criterion and it can be calculated in the signal intensity domain as:
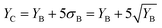 | (1) |
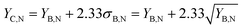 | (2) |
E. coli bacteria exposed to silver(I) and silver nanoparticles were analysed by SC-ICP-MS using the two instrumental configurations studied above. Samples of bacteria were directly analysed after isolation and washing using the PerkinElmer configuration, whereas the Agilent configuration was used for lyophilised and fixed samples. Fig. S3 and S4† show the time scans obtained by SC-ICP-MS for the different samples and instruments. Application of the critical values described above allowed to confirm the presence of silver-containing bioparticles in the samples of bacteria both exposed to Ag(I) or AgNPs.
Validity of the quantitative information obtained by SC-ICP-MS
Apart from qualitative information, SC-ICP-MS can provide quantitative information about the mass of element per bioparticle, the number of bioparticles containing the monitored element over the corresponding critical value and the corresponding mass per bioparticle distribution. Since bioparticle standards are not available, SP-ICP-MS strategies based on the use of dissolved element standards in combination with calculations involving the transport efficiency and the sample flow rate are used,37 assuming that the bioparticles nebulise like dissolved species and the element behaves in the same way both dissolved and in the bioparticles. In any case, the validity of the quantitative information provided by SP and SC-ICP-MS relies on recording complete element mass per particle distributions, which is not straightforward to prove when the particle distribution is unknown or it has not been measured by an alternative technique. In this regard, an approach for the assessment of the validity of the quantitative information obtained by SP-ICP-MS has been recently reported43 that can also be applied to SC-ICP-MS. The approach is based on the estimation of the element mass per particle critical values (XmassC), that must be below the lower end of the measured elemental mass per particle distribution, and the analysis of successive dilutions of the samples, which should provide comparable concentrations.Suspensions from bacteria isolated and washed after culturing with silver contained negligible amounts of dissolved silver, which means that baseline intensities were close to those from instrumental blanks. In any case, if the lower end of the measured mass per particle distribution is larger than the XmassC calculated from the baseline of the sample, the suspension can be diluted to make a second confirmatory measurement. In contrast, if the lower end of the measured mass distribution is next to the critical value and it cannot be improved by dilution, no additional action can be taken and the validity of the distribution and the quantitative information cannot be confirmed. If dilution involves the recording of a low number of particle events, the total acquisition time can be increased conveniently. The detailed flow chart of the procedure is available in ref. 43.
Silver mass per particle distributions from E. coli bacteria exposed to silver(I) and 10 nm silver nanoparticles are shown in Fig. 2. Bacteria exposed to silver(I) showed a broader distribution than when exposed to silver nanoparticles. In the first case, the mass per particle distribution range was 23–800 ag, while in the latter, the range was 13–350 ag for bacteria measured directly without any treatment with the PerkinElmer configuration, whereas lyophilised and paraformaldehyde fixed bacteria measured with the Agilent configuration showed ranges of 5–350 and 5–100 ag of silver, respectively. Under the corresponding measurement conditions, the XmassC values were 6.2 and 5.8 ag for bacteria exposed to silver(I) and silver nanoparticles, respectively, with the PerkinElmer configuration and 3.4 ag for both types of samples with the Agilent configuration. For all these samples, the baseline intensities were close to the blank level and the lower ends of the measured distributions were larger than the critical values, so the suspensions were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 to confirm the results obtained. The results from both dilutions showed good agreement for all the samples, obtaining quantitative recoveries over 90% for both samples of bacteria exposed to silver(I) and silver nanoparticles. This fact confirms that the mass per particle distributions obtained in Fig. 2 were complete and the obtained information could be considered quantitative. Table 2 summarises the steps followed for the assessment of the information obtained for these samples and the obtained results.
2 to confirm the results obtained. The results from both dilutions showed good agreement for all the samples, obtaining quantitative recoveries over 90% for both samples of bacteria exposed to silver(I) and silver nanoparticles. This fact confirms that the mass per particle distributions obtained in Fig. 2 were complete and the obtained information could be considered quantitative. Table 2 summarises the steps followed for the assessment of the information obtained for these samples and the obtained results.
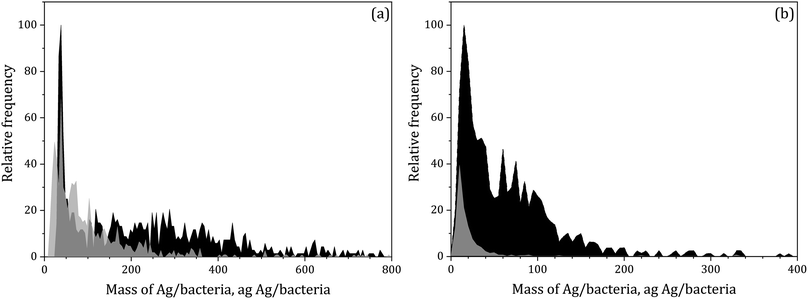 | ||
| Fig. 2 Silver mass per particle distributions for samples of E. coli bacteria exposed to Ag(I) (black) and 10 nm AgNPs (grey) analysed by SC-ICP-MS. (a) PerkinElmer NexION 2000 B (b) Agilent 8900. | ||
| Sample | Dilution | Mean baseline intensity counts | X massC (ag) | Mean mass per particle (ag) | Mass range (ag) | Number of events | Number concentration (L−1) |
|---|---|---|---|---|---|---|---|
| Perkin Elmer NexION 2000 B | |||||||
| Bacteria + Ag(I) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 80 |
0.040 ± 0.001 | 6.2 | 268.7 ± 7.8 | 23–800 | 1008 ± 49 | 2.82 ± 0.14 × 1011 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 160 160 |
0.030 ± 0.001 | 6.2 | 265.4 ± 6.4 | 23–800 | 481 ± 30 | 2.80 ± 0.18 × 1011 | |
| Bacteria + 10 nm AgNPs | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 40 |
0.010 ± 0.001 | 5.8 | 121.6 ± 4.7 | 13–350 | 1831 ± 78 | 2.46 ± 0.10 × 1011 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 80 |
0.010 ± 0.001 | 5.8 | 129.4 ± 7.2 | 13–350 | 950 ± 47 | 2.51 ± 0.11 × 1011 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Agilent 8900 | |||||||
| Bacteria + Ag(I) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
0.027 ± 0.002 | 3.4 | 66.6 ± 2.4 | 5–350 | 839 ± 7 | 1.56 ± 0.04 × 1013 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
0.016 ± 0.002 | 2.9 | 67.7 ± 4.4 | 4–350 | 368 ± 16 | 1.51 ± 0.07 × 1013 | |
| Bacteria + 10 nm AgNPs | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
0.029 ± 0.002 | 3.4 | 18.6 ± 3.0 | 5–100 | 384 ± 32 | 1.58 ± 0.10 × 1012 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
0.018 ± 0.001 | 2.9 | 16.5 ± 1.1 | 4–100 | 175 ± 8 | 1.45 ± 0.04 × 1012 | |
Quantification of the bacteria total silver uptake by SC-ICP-MS
SC-ICP-MS can be used to detect the occurrence of bacteria containing silver, to obtain the corresponding mass per particle distributions and hence to determine the total concentration of silver accumulated by bacteria during the incubation process. Table 3 shows the amount of total silver determined by this technique in samples from bacteria suspension exposed to Ag(I) and 10 nm AgNPs, following the treatments described in the Experimental section. Results are expressed as silver concentration in the original bacteria suspension. In both cases, bacteria exposed to ionic silver accumulated higher amounts of silver despite the initial amount of silver(I) incubated was lower than that of silver nanoparticles, showing the higher bioavailability of ionic silver. In the case of lyophilised and fixed samples, bacteria were exposed to a higher concentration of Ag(I) showing higher accumulation, whereas similar results were obtained for bacteria exposed to AgNPs. Whereas a part of the bacteria was directly analysed after isolation and washing, another part of bacteria was also subjected to lyophilisation and paraformaldehyde fixation. These additional steps justify the higher silver content of the control samples due to manipulation and contamination issues.| Sample | Silver mass concentration (μg L−1) | |
|---|---|---|
| SC-ICP-MS | Acid digestion-ICP-MS | |
| Non post-treated bacteria | ||
| Control | <0.1 | 0.3 ± 0.1 |
| +0.5 mg L−1 Ag(I) | 72.0 ± 3.0 | 73.3 ± 6.5 |
| +2 mg L−1 10 nm AgNPs | 26.5 ± 1.4 | 25.3 ± 0.9 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Lyophilised and fixed bacteria | ||
| Control | 9.4 ± 0.2 | 11.3 ± 0.4 |
| +1 mg L−1 Ag(I) | 921.7 ± 51.7 | 898.2 ± 31.7 |
| +2 mg L−1 10 nm AgNPs | 30.8 ± 3.5 | 30.1 ± 2.3 |
The results obtained by SC-ICP-MS were compared with those obtained by ICP-MS after acid digestion (Table 3). In some cases, the relative standard deviations were relatively high (>10%) since the amount of silver taken up by each bacteria had a high variability. Application of a t-test (95% confidence level) showed that the direct analysis by SC-ICP-MS provided similar results to those obtained by ICP-MS after acid digestion of the samples. No significant differences were found between both methods, except for the bacterial control samples (p = 0.58 and p = 0.12 for non post-treated bacteria exposed to Ag(I) and AgNPs, respectively; p = 0.04, p = 0.16 and p = 0.69 for control lyophilised and fixed bacteria and exposed to Ag(I) or AgNPs, respectively). Bacterial control sample spiked with 5 μg L−1 of silver(I) and subjected to acid digestion led to 88 ± 2% recovery. These results confirm the conclusions of the validity approach discussed above and hence the validity of the quantitative information and mass per particle distributions obtained by SC-ICP-MS.
Determination of intracellular silver in bacteria by SC-ICP-MS
SC-ICP-MS by itself cannot distinguish whether the accumulated silver is internalised by the bacteria or retained in their membrane. A SC-ICP-MS method based on the enzymatic digestion of the membrane was developed to discriminate the intracellular silver and the silver adsorbed on the cell wall and/or accumulated in the periplasmic space of the membrane. Lysozyme has been successfully used to remove the bacterial outer membrane in the presence of EDTA,32 leaving the corresponding spheroplasts, which were analysed by SC-ICP-MS to quantify intracellular silver. On the other hand, cell wall debris, where silver could be adsorbed, and liquid from the periplasmic space, where silver could be accumulated, remained in the residual fraction of this digestion. Fig. S5† shows the bacillus shape of original bacteria and the corresponding spheroplasts after lysozyme digestion of the membrane, which adopt a round shape maintaining their integrity. Although the spheroplasts are more fragile entities than the original bacteria, they remained intact after nebulisation since an increase in the baseline level due to their breaking was not observed, when compared to the control spheroplasts.Table 4 shows the mass concentration of intracellular silver obtained by the SC-ICP-MS analysis of spheroplast and the percentage of intracellular silver in relation to the total silver taken up by bacteria. It was observed that in the case of bacteria exposed to silver(I), all the silver was internalised, whereas in the case of bacteria exposed to silver nanoparticles, this percentage was limited to about 46%. To validate these results, the spheroplasts were analysed by ICP-MS after acid digestion. LOD and LOQ values for total mass concentration determined by SC-ICP-MS were 38 and 128 ng L−1, respectively, and 150 and 510 ng L−1 for total mass concentration determined in standard mode after acid digestion, showing the higher detection capability in single particle mode (detailed information about these and other figures of merit are summarised in Table S1†). In addition, the supernatants containing cell wall residue and periplasmic space components were also analysed by ICP-MS after acid digestion. These results are also included in Table 4. By applying a t-test (95% confidence level), it was confirmed that the direct analysis by SC-ICP-MS gave similar results to those obtained by ICP-MS after acid digestion of spheroplasts, corresponding to the intracellular silver (p = 0.44, p = 0.19 for bacteria exposed to Ag(I) and AgNPs, respectively). Bacterial control spheroplasts spiked with 5 μg L−1 of silver(I) and subjected to acid digestion led to 87 ± 4% recovery. In the case of bacteria exposed to ionic silver, most silver was internalised although the occurrence of a residual fraction associated to the membrane could not be discarded. However, for silver nanoparticles, the sum of the concentrations of intracellular silver and silver associated to the membrane was not in agreement with the total silver taken up by bacteria, which might be due to loses of nanoparticles during the digestion treatment.
| Bacteria | Intracellular Ag | Membrane Ag | Total Ag | ||||
|---|---|---|---|---|---|---|---|
| SC-ICP-MS | Acid digestion + ICP-MS | Acid digestion + ICP-MS | Acid digestion + ICP-MS | ||||
| μg L−1 | % | μg L−1 | % | μg L−1 | % | % | |
| Control | <0.3 | — | 0.8 ± 0.1 | — | <0.2 | — | — |
| +Ag(I) | 73.0 ± 4.2 | 101.4 ± 5.8 | 75.1 ± 3.6 | 102.5 ± 4.9 | 2.6 ± 0.1 | 3.5 ± 0.1 | 106.0 ± 4.9 |
| +10 nm AgNPs | 12.1 ± 1.0 | 45.8 ± 3.6 | 11.1 ± 0.9 | 43.9 ± 3.5 | 0.3 ± 0.1 | 1.1 ± 0.3 | 45.0 ± 3.5 |
These results of the biodistribution of silver were compared with those obtained in other studies reported in the bibliography. Dong et al. studied the biodistribution of silver in bacteria by size-exclusion chromatography coupled with ICP-MS after enzymatic digestion with lysozyme.32 After incubation E. coli JCM 1649 bacteria with silver ions and 10 and 30 nm silver nanoparticles, silver was mainly adsorbed on the cell wall (25–64%), and a small fraction of silver was internalised (6–15%). The other silver fraction (24–60%) did not interact with bacteria and was removed during the washing steps after incubation process. In contrast, in our study it was observed that silver was internalised to a greater extent (1–20%) while less than 1% of silver was adsorbed on the bacterial wall, although in the case of silver nanoparticles exposure, the percentage of adsorbed silver could be higher, as discussed above. Likewise, Long et al. determined the silver biodistribution in E. coli MG 1655 bacteria after exposure to Ag(I) and 10 nm AgNPs.31 The authors applied a procedure based on the cell lysis and the subsequent separation of membrane and cytoplasmic fractions followed by ICP-MS quantification. Higher levels of silver were obtained in the cytoplasmic fraction, around 55 and 70% for silver ion and nanoparticle exposure, respectively. These results were not in agreement with those obtained in our study.
Conclusions
A SC-ICP-MS method has been developed and evaluated to obtain quantitative information about the individual silver content in bacteria exposed to different silver species, as well as biodistribution of silver within bacteria. For this purpose, two sample introduction systems have been studied in order to determine and compare the optimal working conditions in SC-ICP-MS, selecting the same optimal conditions for both of them (nebuliser gas flow rate of 0.3 L min−1 and make-up gas flow rate of 0.9 L min−1). Once their behaviour was known, these introduction systems were applied to the analysis of bacteria samples previously incubated with silver(I) and silver nanoparticles. After applying different criteria related to detectability, SC-ICP-MS was able to detect and confirm the presence of silver-containing bioparticles and quantify their individual content, demonstrating that the introduction systems allow to introduce efficiently bacteria into ICP-MS. In addition, SC-ICP-MS has allowed to quantify directly the total concentration of silver accumulated by E. coli bacteria during the incubation process. Bacteria exposed to ionic silver accumulated higher amounts of silver than those exposed to 10 nm silver nanoparticles, showing the higher bioavailability of ionic silver. On the other hand, the SC-ICP-MS analysis of spheroplasts obtained by enzymatic digestion of the bacterial cell wall with lysozyme, has allowed to quantify the intracellular silver and to study the silver biodistribution in individual bacteria. In the case of bacteria exposed to ionic silver, the silver taken up by bacteria was mainly internalised, while in the case of 10 nm silver nanoparticles, only 45% of the silver was internalised.In summary, SC-ICP-MS can provide accurate quantitative information at both bioparticle and population average levels for bacteria exposed to silver species, as well as for the corresponding spheroplasts obtained after enzymatic digestion. Their robustness and small size (below ca. 2 μm) contribute to their efficient nebulisation as intact entities with the sample introduction systems tested, which cannot be extended to other larger and/or fragile bioparticles. Due to the features of SC-ICP-MS, the validity of the quantitative information obtained must be evaluated conveniently to confirm that the mass per bioparticle distributions have been recorded completely.
Author contributions
A. C. G.: formal analysis, investigation, data curation, methodology, validation, visualisation, writing – original draft. I. A.: formal analysis, investigation, data curation, methodology, supervision, validation, writing – review & editing. P. G.: methodology, supervision, writing – review & editing. K. B.: investigation, writing – review & editing. H. G.: resources, supervision, writing – review & editing. F. L.: conceptualisation, data curation, funding acquisition, methodology, resources, supervision, validation, writing – review & editing. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the project PID2021-123203OB-I00 funded by MCIN/AEI/10.13039/501100011033 and by “ERDF A way of making Europe” and the Government of Aragon (E29_23R). A. C. G. thanks the Government of Aragón for a predoctoral contract. I. A. thanks the European Union-Next Generation EU and the Spanish Ministry of Universities for funding under the María Zambrano Grant (MZ-240621). Authors would like to acknowledge the use of Servicio General de Apoyo a la Investigación-SAI, Universidad de Zaragoza. Funding from the UK Department for Science, Innovation and Technology (DSIT) is also acknowledged.References
- M. Corte-Rodríguez, R. Álvarez-Fernández, P. García-Cancela, M. Montes-Bayón and J. Bettmer, TrAC, Trends Anal. Chem., 2020, 132, 116042 CrossRef.
- R. Álvarez-Fernández García, M. Corte-Rodríguez, P. García-Cancela, J. Bettmer and M. Montes-Bayón, in Analytical Nebulizers, Elsevier, 2023, pp. 197–216 Search PubMed.
- X. Wei, Y. Lu, X. Zhang, M. L. Chen and J. H. Wang, TrAC, Trends Anal. Chem., 2020, 127, 115886 CrossRef CAS.
- M. Resano, M. Aramendía, E. García-Ruiz, A. Bazo, E. Bolea-Fernandez and F. Vanhaecke, Chem. Sci., 2022, 13, 4436–4473 RSC.
- Perkin Elmer, Single Cell ICP-MS Analysis: Quantification of Metal Content at the Cellular Level White Paper, 2017, pp. 1–4 Search PubMed.
- L. Amable, C. Stephan, S. Smith and R. Merrifield, White Paper, 2017, vol. 1–5 Search PubMed.
- F. Li, D. W. Armstrong and R. S. Houk, Anal. Chem., 2005, 77, 1407–1413 CrossRef CAS PubMed.
- T. Nomizu, S. Kaneco, T. Tanaka, D. Ito, H. Kawaguchi and B. T. Vallee, Anal. Chem., 1994, 66, 3000–3004 CrossRef CAS.
- Q. X. Sun, X. Wei, S. Q. Zhang, M. L. Chen, T. Yang and J. H. Wang, Anal. Chim. Acta, 2019, 1066, 13–20 CrossRef CAS PubMed.
- A. López-Serrano Oliver, S. Baumgart, W. Bremser, S. Flemig, D. Wittke, A. Grützkau, A. Luch, A. Haase and N. Jakubowski, J. Anal. At. Spectrom., 2018, 33, 1256–1263 RSC.
- R. C. Merrifield, C. Stephan and J. R. Lead, Environ. Sci. Technol., 2018, 52, 2271–2277 CrossRef CAS PubMed.
- Y. ki Tanaka, R. Iida, S. Takada, T. Kubota, M. Yamanaka, N. Sugiyama, Y. Abdelnour and Y. Ogra, ChemBioChem, 2020, 21, 3266–3272 CrossRef PubMed.
- T. Liu, E. Bolea-Fernandez, C. Mangodt, O. De Wever and F. Vanhaecke, Anal. Chim. Acta, 2021, 1177, 338797 CrossRef CAS PubMed.
- S. Miyashita, S. Fujii, K. Shigeta and K. Inagaki, Metallomics, Springer Japan, Tokyo, 2017, pp. 107–124 Search PubMed.
- Y. Liang, Q. Liu, Y. Zhou, S. Chen, L. Yang, M. Zhu and Q. Wang, Anal. Chem., 2019, 91, 8341–8349 CrossRef CAS PubMed.
- B. Gomez-Gomez, M. Corte-Rodríguez, M. T. Perez-Corona, J. Bettmer, M. Montes-Bayón and Y. Madrid, Anal. Chim. Acta, 2020, 1128, 116–128 CrossRef CAS PubMed.
- A. S. Groombridge, S. I. Miyashita, S. I. Fujii, K. Nagasawa, T. Okahashi, M. Ohata, T. Umemura, A. Takatsu, K. Inagaki and K. Chiba, Anal. Sci., 2013, 29, 597–603 CrossRef CAS PubMed.
- K. Shigeta, G. Koellensperger, E. Rampler, H. Traub, L. Rottmann, U. Panne, A. Okino and N. Jakubowski, J. Anal. At. Spectrom., 2013, 28, 637 RSC.
- Y. Cao, Y. Wu, Z. Liu, Q. Kang, H. Wei, J. Peng and S. Li, Spectrochim. Acta, Part B, 2022, 197, 106534 CrossRef CAS.
- Y. Tao, M. He, B. Chen, G. Ruan, P. Xu, Y. Xia, G. Song, Y. Bi and B. Hu, Aquat. Toxicol., 2023, 258, 106499 CrossRef CAS PubMed.
- R. Álvarez-Fernández García, L. Gutiérrez Romero, J. Bettmer and M. Montes-Bayón, Nanomaterials, 2022, 13, 12 CrossRef PubMed.
- L. Rasmussen, H. Shi, W. Liu and K. B. Shannon, Anal. Bioanal. Chem., 2022, 414, 3077–3086 CrossRef CAS PubMed.
- X. Wei, L. L. Hu, M. L. Chen, T. Yang and J. H. Wang, Anal. Chem., 2016, 88, 12437–12444 CrossRef CAS PubMed.
- S. Meyer, A. López-Serrano, H. Mitze, N. Jakubowski and T. Schwerdtle, Metallomics, 2018, 10, 73–76 CrossRef CAS PubMed.
- E. Mavrakis, L. Mavroudakis, N. Lydakis-Simantiris and S. A. Pergantis, Anal. Chem., 2019, 91, 9590–9598 CrossRef CAS PubMed.
- J. Shi, X. Ji, Q. Wu, H. Liu, G. Qu, Y. Yin, L. Hu and G. Jiang, Anal. Chem., 2020, 92, 622–627 CrossRef CAS PubMed.
- W. Qin, H.-J. Stärk, S. Müller, T. Reemtsma and S. Wagner, Metallomics, 2021, 13, 32 CrossRef PubMed.
- C. Suárez-Oubiña, P. Herbello-Hermelo, N. Mallo, M. Vázquez, S. Cabaleiro, I. Pinheiro, L. Rodríguez-Lorenzo, B. Espiña, P. Bermejo-Barrera and A. Moreda-Piñeiro, Anal. Bioanal. Chem., 2023, 415, 3399–3413 CrossRef PubMed.
- Z. Liu, A. Xue, H. Chen and S. Li, Appl. Microbiol. Biotechnol., 2019, 103, 1475–1483 CrossRef CAS PubMed.
- X. Yu, B. Chen, M. He, H. Wang and B. Hu, Anal. Chem., 2019, 91, 2869–2875 CrossRef CAS PubMed.
- Y. M. Long, L. G. Hu, X. T. Yan, X. C. Zhao, Q. F. Zhou, Y. Cai and G. Bin Jiang, Int. J. Nanomed., 2017, 12, 3193–3206 CrossRef CAS PubMed.
- L.-J. Dong, Y.-J. Lai, S.-J. Yu and J.-F. Liu, Anal. Chem., 2019, 91, 12525–12530 CrossRef CAS PubMed.
- A. C. Gimenez-Ingalaturre, E. Rubio, P. Chueca, F. Laborda and P. Goñi, J. Microbiol. Methods, 2022, 203, 106618 CrossRef CAS PubMed.
- T. Hilberath, A. Raffaele, L. M. Windeln and V. B. Urlacher, AMB Express, 2021, 11, 162 CrossRef CAS PubMed.
- F. Laborda, A. C. Gimenez-Ingalaturre, E. Bolea and J. R. Castillo, Spectrochim. Acta, Part B, 2019, 159, 105654 CrossRef CAS.
- F. Laborda, A. C. Gimenez-Ingalaturre, E. Bolea and J. R. Castillo, Spectrochim. Acta, Part B, 2020, 169, 105883 CrossRef CAS.
- H. E. Pace, N. J. Rogers, C. Jarolimek, V. A. Coleman, C. P. Higgins and J. F. Ranville, Anal. Chem., 2011, 83, 9361–9369 CrossRef CAS PubMed.
- T. E. Lockwood, R. Gonzalez De Vega and D. Clases, J. Anal. At. Spectrom., 2021, 36, 2536–2544 RSC.
- S. Cuello-Nuñez, I. Abad-Álvaro, D. Bartczak, M. E. Del Castillo Busto, D. A. Ramsay, F. Pellegrino and H. Goenaga-Infante, J. Anal. At. Spectrom., 2020, 35, 1832–1839 RSC.
- L. Amable, S. Smith and C. Stephan, New Research Evaluating Cisplatin Uptake in Ovarian Cancer Cells by Single Cell ICP-MS, PerkinElmer, 2017, vol.1–6 Search PubMed.
- S. Wilhelm, R. C. Bensen, N. Rama Kothapalli, A. W. G. Burgett, R. Merrifield and C. Stephan, Quantification of Gold Nanoparticle Uptake into Cancer Cells using Single Cell ICP-MS, PerkinElmer, 2017, vol. 4 Search PubMed.
- F. Laborda, C. Trujillo and R. Lobinski, Talanta, 2021, 221, 121486 CrossRef CAS PubMed.
- A. C. Gimenez-Ingalaturre, K. Ben-Jeddou, J. Perez-Arantegui, M. S. Jimenez, E. Bolea and F. Laborda, Anal. Bioanal. Chem., 2023, 415, 2101–2112 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ja00378g |
| This journal is © The Royal Society of Chemistry 2024 |