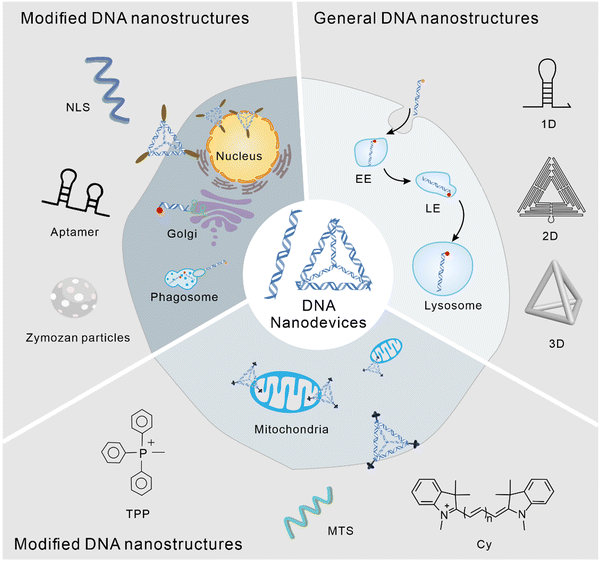
Subcellular localization of DNA nanodevices and their applications
Xia
Liu†
ad,
Shuting
Cao†
ad,
Yue
Gao
ad,
Shihua
Luo
c,
Ying
Zhu
abd and
Lihua
Wang
 *abd
*abd
aDivision of Physical Biology, CAS Key Laboratory of Interfacial Physics and Technology, Shanghai Institute of Applied Physics, Chinese Academy of Sciences, Shanghai 201800, China. E-mail: wanglh@sari.ac.cn
bThe Interdisciplinary Research Center, Shanghai Synchrotron Radiation Facility, Zhangjiang Laboratory, Shanghai Advanced Research Institute, Chinese Academy of Sciences, Shanghai 201210, China
cDepartment of Traumatology, Rui Jin Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, 200025, China
dUniversity of Chinese Academy of Sciences, Beijing 100049, China
First published on 6th March 2023
Abstract
The application of nanodevices based on DNA self-assembly in the field of cell biology has made significant progress in the past decade. In this study, the development of DNA nanotechnology is briefly reviewed. The subcellular localization of DNA nanodevices, and their new progress and applications in the fields of biological detection, subcellular and organ pathology, biological imaging, and other fields are reviewed. The future of subcellular localization and biological applications of DNA nanodevices is also discussed.
Introduction
Deoxyribonucleic acid (DNA), as the genetic material in life, has natural biocompatibility. In 1953, Watson of the United States and Crick of the United Kingdom successfully predicted the molecular model of the double helix structure of DNA using X-ray crystal diffraction.1 The development of this model, arguably one of the greatest discoveries of the 20th century, made the subsequent development of DNA nanotechnology possible. Later, the Watson-Crick base-complementary pairing principle that DNA molecules follow was proposed, making the double-stranded DNA (dsDNA) structure somewhat rigid and predictable at the nanoscale. In 1982, Professor Seeman of New York University first proposed that DNA can form specific structures by the principle of base complementary pairing, and that individual structures can form complex two-dimensional (2D) or three-dimensional (3D) structures through sticky ends.2 This principle is at the heart of DNA nanotechnology. Since then, DNA molecules have attracted considerable attention in the field of nanoscience, and researchers have designed and synthesized various functional DNA structures. DNA nanotechnology has been applied in various life science fields, such as tissue regeneration, disease prevention, inflammation inhibition, bio-imaging, biosensors, diagnosis, and anti-tumor drug delivery and treatment.3–7Among DNA nanostructures, functional nucleic acids are a class of DNA structures that can undergo conformational changes after binding or interacting with target analytes, which are often used in biosensor design.8,9 For example, in 1990, Gold and Szostak introduced the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) in vitro screening.10 This technique can be used to screen for aptamers,11 which are single-stranded DNA (ssDNA) structures that specifically bind to metal ions, adenosine triphosphate, nucleic acids, and biomacromolecules.12–15 DNAzymes are another class of functional nucleic acids capable of specifically cutting DNA/RNA substrates in the presence of cofactors. To date, Pb2+, Zn2+, Ca2+, and histidine-dependent DNAzymes have been obtained through advanced in vitro screening.16,17 In addition, structures containing T–Hg2+–T or C–Ag+–C are also commonly used for Hg2+ and Ag+ sensor detection.18,19 In addition to functional nucleic acids that can directly and specifically bind to the target analyte, there is also a category of DNA-responsive devices with environmental specificity. Such DNA nanodevices tend to undergo conformational transformation under specific circumstances, for example, DNA nanostructures with specific sequences can form triplex20–23 or i-motif24,25 for pH sensing under an acidic environment. G-quadruplex can be used for K+ or Na+ sensing.26 DNA nanostructures show great promise in biomedical applications.
Compared to that of traditional biological macromolecules, DNA has excellent programmability and can be designed more accurately according to the principle of base complementary pairing. Diverse DNA nanostructures, including 2D and complex 3D structures, can be constructed using bottom-up or top-down methods.27–29 Both DNA and cell membranes are electronegative. When they come into contact, it is difficult for DNA to cross the cell membrane and enter the cell matrix due to electrostatic repulsion. In contrast to single-stranded DNA (ssDNA), a variety of 2D and 3D DNA nanostructures can be actively taken up by cells and localized into organelles without the help of transfection reagents. For example, a previous study prepared ssDNA of 55 nt in length by simple thermal annealing in order to produce DNA tetrahedra. After cell incubation, it was transported into lysosomes via the caveolin-mediated endocytic pathway30 and could be used for the detection of the lysosomal microenvironment and lysosomal pathological studies.31–33 In addition, DNA nanostructures are addressable and can be modified with various groups possessing regular positions and valences, further endowing DNA nanostructures with new functions. Moreover, DNA nanostructures modified with targeting molecules can alter their fate in cells. For example, DNA tetrahedra modified with triphenylphosphine (TPP) can be localized in the mitochondria, and can be conveniently used for mitochondrial imaging and mitochondrial-related functional interventions, such as aerobic respiration and glycolysis.34
The application of DNA nanostructures in the field of cell biology has developed rapidly over the past decade. In this study, we reviewed the subcellular localization of DNA nanodevices, that is how DNA nanostructures can target specific organelles and achieve specific physiological functions within them. In DNA nanodevices, organelle targeting, normalization, and sensing modules can be combined in a single structure with high stoichiometric purity and high yields, enabling precise imaging and subcellular targeting in different living systems.35 The modularity and control of chemometrics provided by DNA can lead to precision medicine, enabling the overlay of diagnosis and personalized treatment.36 The following is a brief overview of the biological application of the DNA nanostructure according to its different subcellular localization (Fig. 1).
Lysosome
Lysosomes, which act as the garbage-disposal system of cells,37 play a crucial role in regulating the homeostasis of cells and organisms.38 Changes in the components of the lysosomal microenvironment (such as H+, Cl−, Ca2+, and various enzyme activities) are mostly related to the occurrence and development of many diseases, including autoimmune diseases, neurodegenerative diseases, cancer, and metabolic diseases.39 Studies have shown that most DNA nanodevices are localized to lysosomes through specific endocytic pathways after being ingested by cells (Fig. 2(a)).35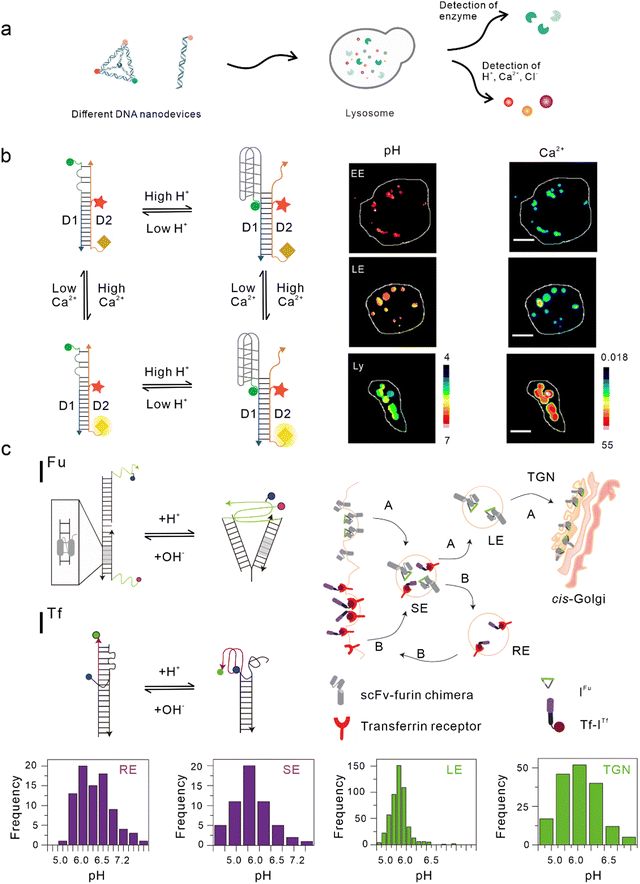 | ||
| Fig. 2 Bio-application of DNA nanodevices within lysosomes. (a) Schematic of the design principle of the DNA nanodevice in lysosomes. (b) Two-ion measurement (2-IM) using combination ion sensors and representative pseudocolor pH and Ca2+ maps of early endosomes (EE), late endosomes (LE), and lysosomes (Ly) labeled with CalipHluorLy. Reproduced from ref. 42 with permission from Springer Nature, copyright 2018. (c) Schematic of the DNA nanomachines that map the furin (Fu) and transferrin (Tf) pathways. Recycling endosomes (REs), LEs, EE/SEs, and Trans-Golgi network (TGN) are marked with Tf-ITf and IFu and their corresponding pH distributions. Reproduced from ref. 51 with permission from Springer Nature, copyright 2013. | ||
Double-stranded DNA nanodevices
One of the simplest DNA nanodevices is the dsDNA structure. Compared with the structure of ssDNA, dsDNA structures are more rigid, easier to be taken up by cells,40 and can provide more functional binding sites. Under physiological conditions, dsDNA structures are also more resistant to nuclease degradation. Krishnan et al. designed a series of dsDNA nanodevices, which can be localized to lysosomes through endocytosis mediated by the anion ligand-binding receptor (ALBR)41–44 or scavenger receptor.45,46 Modular modification can be used to accurately and directly integrate these modules with different functions into the same dsDNA nanostructure, thus attaining an accurate detection of chemical components in endocytotic organelles during lysosome maturation. In 2009, Modi et al. designed a dsDNA nanomachine that could independently respond to pH changes in living cells and that is also localized in lysosomes through the endocytic pathway mediated by ALBR. Fluorescence resonance energy transfer (FRET) facilitated by the i-motif in the dsDNA nanomachine was used to detect the pH range (5.8–7) in lysosomes and map the change in pH with a temporal and spatial resolution during lysosome maturation.41 Then, they modified the Cl− sensitive small molecule BAC at the end of the dsDNA nanostructure to construct a Cl− sensor, which is referred to as Clensor, in order to detect Cl− in lysosomes.43,47 In this study, Cl− was detected in endosomes and it was revealed that chloride plays an essential role in lysosomal functions.44 Leung et al. developed a two-ion measurement (2-IM) method based on dsDNA nanostructures.45 In this study, a Clensor module was combined with an i-motif module in order to simultaneously measure H+ and Cl− in lysosomes. This technology was able to identify lysosomal subgroups according to the correlation between H+ and Cl− concentrations by locating a single lysosome, and is of great significance to the study of lysosomal physiological function.48 In addition, Narayanaswamy et al. reported another 2-IM nanomachine that can detect Ca2+ and H+ simultaneously. The nanomachine was combined with a Ca2+ reporter, referred to as CalipHluor, with a built-in pH reporter, which was able to monitor the change in the affinity of Ca2+ fluorophore in the endocytic pathway and its dissociation constant (Kd) with the pH value, so as to accurately calculate the true value of Ca2+ in the endocytic cavity (Fig. 2(b)).42Modified double-stranded DNA nanodevices
In the lysosomal acid chamber, more than 60 types of acid hydrolases have been found to be responsible for the degradation of proteins, nucleic acids, and other macromolecules.49 By integrating fluorophores that are sensitive to thiol-disulfide exchange into lysosome-targeted DNA nanodevices, Dan et al. mapped the disulfide reduction in endocytic organelles of Caenorhabditis elegans.50 This strategy provides a new method that can be used to measure a series of enzyme cleavage reactions in organelles. Compared with simple dsDNA nanodevices, modified dsDNA nanodevices can achieve lysosomal targeting of other endocytic pathways excluding ALBR and scavenger receptors. In another study, Modi et al. reported a strategy in which cells can take in two kinds of dsDNA nanodevices that are able to detect pH through different endocytic pathways at the same time. One of the nanodevices contained an 8 bp dsDNA sequence, which acted as the binding site of the engineered protein, thus enabling it to be located in the furin retrograde endocytic pathway.51–53 The other was modified with transferrin so that it could be located in the endocytic/circulation pathway of transferrin.51,54 Both dsDNA nanodevices could be located in endocytic organelles and could independently detect the corresponding pH (Fig. 2(c)).51 Similarly, by modifying RNA aptamers at one end of the Clensor, Saha et al. achieved Cl− imaging and detection during transferrin receptor-mediated endocytosis.43 In addition, Saminathan et al. modified 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine for lipid anchoring and voltage-sensitive fluorophores on dsDNA nanodevices, obtaining a change map of the membrane potential during lysosome maturation.463D DNA nanodevices
Research has shown that in contrast to dsDNA nanodevices, 3D DNA nanostructures with precise morphology and size are more stable in complex biological systems and can also enter lysosomes through membrane receptor-mediated endocytosis.30,55,56 For example, Fan et al. found that weakened bands of a DNA tetrahedron could still be seen in gel electrophoresis even after 24 h of co-culturing with serum, whereas dsDNA had rapidly degraded.57 Single-particle tracing technology has revealed that DNA tetrahedra are localized to lysosomes via caveolin-mediated endocytosis in HeLa cells.30 Tetrahedrals combined with i-motifs can be used to detect the pH in lysosomes.58–60Tumor therapeutic
In addition to lysosomal microenvironment detection, DNA nanostructures have been used in lysosomal-related tumor therapy. Through proteomics, Cui et al. found that the low antigen cross-presentation efficiency of M2 tumor-associated macrophages (TAMs), which promote tumor angiogenesis and metastasis and inhibit immunity, thus promoting tumor growth, was related to the high lysosomal cysteine protease activity of TAMs.61 Given that DNA nanodevices are easily located in and easily degraded by lysosomes, can DNA nanodevices be reasonably designed to directly inhibit lysosomal activity after being endocytosed to lysosomes, so as to avoid DNA degradation? Utilizing the inherent lysosome localization properties of DNA nanodevices and exploiting their potential in lysosomes may be a better strategy for biomedical applications. Based on this, they anchored the cysteine protease inhibitor, E64, on a DNA nanodevice and injected the nanodevice into mice through the tail vein. The DNA nanodevice was preferentially internalized and located in the lysosome by TAMs through the scavenger receptor-mediated pathway, after which the cysteine protease inhibitor E64 was released, effectively inhibiting the activity of cysteine protease in the lysosome of TAMs, and improving the antigen presentation ability of TAMs. Subsequently, the DNA nanodevice was combined with the anti-cancer drug, cyclophosphamide (CTX), and displayed favorable effects in the treatment of mice with triple-negative breast cancer.40,61 Liu et al. designed a nanomachine based on rectangular DNA origami extending three different capture strands at a predetermined position. Through DNA hybridization, antigens (peptides), toll-like receptor (TLR) agonists, double-stranded RNA (dsRNA), and CpG DNA were precisely arranged on the DNA nanodevice. To protect the antigen/adjuvant from interference by extracellular ribonucleic acid, a DNA strand was added to the edge of the rectangle to lock it and the latter was only unlocked within a specific pH range. When DNA nanovaccines enter antigen-presenting cells, they preferentially enter the lysosomes. In the acidic environment of lysosomes, the locks on the sides of the rectangle were opened, and adjuvants and antigenic peptides were exposed, which activated a strong immune response, effectively eliminating the tumor in mice and protecting mice from subsequent tumors.62 Gu et al. designed tetrahedral DNA nanostructures (TDNs) loaded with gold nanoparticles (GNPs) and doxorubicin (DOX) to treat tumors. Initially, large nanoparticles accumulated in tumors. When the pH of the tumor microenvironment was approximately 6.5, the junction of TDNs and GNP was transformed into a triple structure, resulting in the separation of TDNs and GNPs. When endocytosed into cells, TDNs formed an i-motif in order to release DOX into lysosomes at pH 5.0, thus displaying a favored anti-tumor effect.63 Dong et al. designed a proton-driven DNA framework using the i-motif. Under the acidic environment of the lysosome, the DNA framework dynamically assembled into aggregates and remained in the lysosome. Meanwhile, the protonation effect led to a decrease in lysosomal acidity and hydrolase activity, which hindered the degradation of nucleic acid drugs in the lysosome and improved the efficiency of gene silencing. This lysosomal interference method may play a prominent role in protecting nucleic acid cargo.64In summary, dsDNA nanodevices can directly locate lysosomes through ALBR or scavenger receptor-mediated endocytosis pathways, and the sensor detection of these two pathways can be realized by designing suitable dsDNA nanodevices. Lysosomal sensing of other endocytic pathways, such as transferrin-mediated endocytic pathways, can be achieved by modifying other targeted groups on dsDNA. 3D DNA nanodevices, such as DNA tetrahedra, are increasingly used to intervene in lysosome microenvironments because of their more accurate morphology and size, and more stable structure under physiological conditions. Based on the characteristic that DNA nanodevices are usually endocytosed into lysosomes, the functional intervention of lysosomes is realized. By designing pH-responsive DNA nanodevices through the low pH in lysosomes, the controlled release of anti-tumor drugs is accomplished and the powerful tumor therapeutic effect is also demonstrated.
Mitochondria
Mitochondria play important roles in the production of adenosine triphosphate (ATP), reactive oxygen species (ROS), and cell apoptosis.65 Mitochondrial abnormalities often contribute to a variety of diseases, including cancer, neurodegenerative diseases, and cardiovascular diseases.66–70 Therefore, mitochondria have long been considered ideal subcellular targets for cancer treatment. Because of the proton pump in the process of mitochondrial oxidative respiration, a large amount of negative charge is generated in the mitochondrial matrix. After lipophilic cations enter the cell, they can actively target the mitochondria by taking advantage of its high membrane potential. Therefore, most DNA nanodevices targeted by mitochondria are achieved by modifying lipophilic cationic ligands (Fig. 3(a)).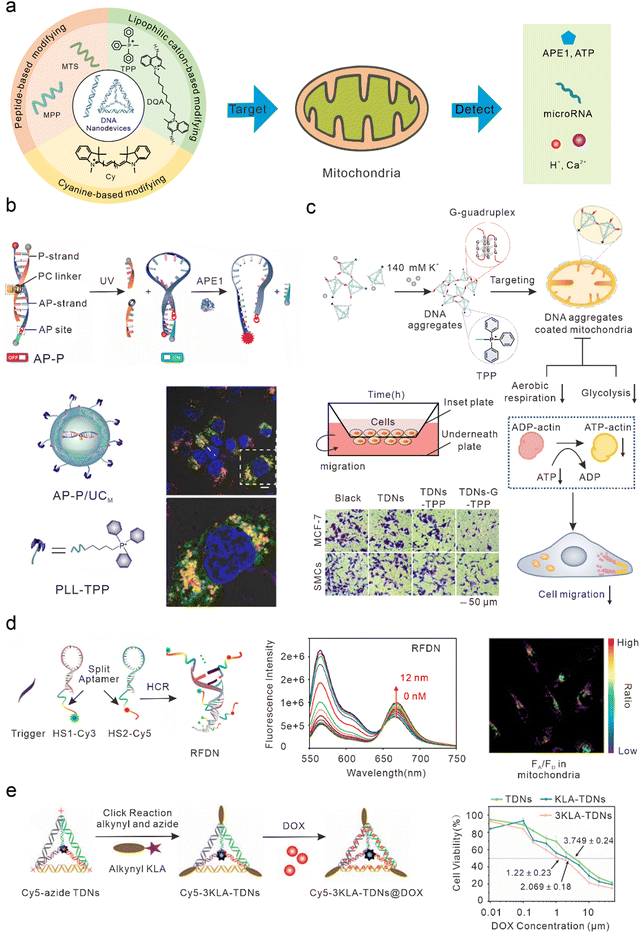 | ||
| Fig. 3 Bio-application of DNA nanodevices within mitochondria. (a) Schematic of the design principle of the DNA nanodevice in mitochondria. (b) Design of the DNA nanosensor for in situ localization and near-infrared (NIR)-light-activatable imaging of apurinic/apyrimidinic endonuclease 1 (APE1) in mitochondria. Reproduced from ref. 76 with permission from Wiley, copyright 2021. (c) Dynamic assembly of TDNs-G-TPP in living cells for mitochondrial interference and the consequent regulations on cellular behaviors. Reproduced from ref. 34 with permission from American Chemical Society, copyright 2022. (d) Illustration of the RFDN assembled by hybridization chain reaction. and ATP detection in vitro and in vivo. Reproduced from ref. 83 with permission from American Chemical Society, copyright 2021. (e) Schematic design of mitochondria-targeted 3KLA-TDNs/DOX for the enhancement of anti-breast cancer therapy. Reproduced from ref. 89 with permission from The Royal Society of Chemistry, copyright 2020. | ||
Mitochondrial targeting with TPP modification
DNA nanostructures can be selectively localized to mitochondria after modification with triphenylphosphine (TPP). TPP, which contains lipophilic cations, is the most widely used mitochondrial localization group.71–75 TPP contains three lipophilic phenyl groups with a positive charge, which can quickly penetrate the mitochondrial membrane through to the mitochondrial matrix. Recently, Shao et al. designed organelle-specific, light-activated DNA nanosensors by assembling mitochondria-targeted TPP on lanthanide-doped up-conversion nanoparticles modified with DNA probes.76 Under ultraviolet light stimulation, the P-strand, which contained a photocleavable (PC) linker of dsDNA, underwent photolysis to form the loop structure of the molecular beacon. At this time, the fluorophore and quencher modified at the adjacent ends of DNA were in proximity, resulting in quenched fluorescence. In the presence of human apurinic/apyrimidinic endonuclease 1 (APE1, a marker of malignant tumors, inhibition of which can significantly kill tumor cells),77 the apurinic/apyrimidinic (AP) site of the molecular beacon was enzymatically hydrolyzed and the quencher was cut off, resulting in the recovery of the fluorescence signal. This sensor could achieve accurate APE1 imaging activated by “on-demand” remote light in mitochondria and improve subcellular resolution (Fig. 3(b)). Using similar DNA nanostructures, Zhao et al. also developed a near-infrared (NIR) light-controlled DNA strand displacement reaction to image MicroRNAs (miRNAs) in the mitochondria of living cells.78,79 This method is expected to be further used in the study of physiological events related to miRNAs in the mitochondria. In addition, Chai et al. designed redox-activated aptamer sensors that assembled redox-responsive ATP aptamer probes on cationic polymer-coated nanoparticles, which were further modified with TPP to achieve spatially controlled and gated imaging of ATP and glutathione (GSH) in the mitochondria.80 Liu et al. modified TPP at one vertex of a DNA tetrahedron, and a Ca2+ fluorescence probe, a pH fluorescence probe, and an internal reference probe at the other three vertices to construct a mitochondrially targeted DNA nanodevice, achieving simultaneous imaging as well as biological sensing of pH and Ca2+ in mitochondria.81 Li et al. modified TPP at one vertex of a DNA tetrahedron, and designed a guanine-rich DNA sequence at the remaining three vertices to form a stable G-quadruplex structure among multiple tetrahedral frames under the trigger of a high concentration of potassium ions (∼140 mM) in the cytoplasm, to allow the DNA tetrahedron to aggregate on the mitochondrial surface and form an anion barrier. Thus, such a nanodevice significantly inhibits aerobic respiration and glycolysis in mitochondria, reduces the production of intracellular ATP, and inhibits the migration of cancer cells (Fig. 3(c)).34Mitochondrial targeting with Cy modification
In addition to TPP modification, DNA nanodevices that modify themselves to be fat-soluble and positively charged fluorescent dyes (such as cyanine) can also efficiently accumulate in mitochondria. Such modified DNA nanodevices which can not only serve as reporting ligands, but can also achieve mitochondrial targeting, have attracted a significant amount of attention. Liu et al. combined cyanine dyes with a Y-shaped acid aptamer to achieve sensing and imaging of ATP in the mitochondria of living cells.82 Luo et al. hybridized two hairpin DNA strands (HS1-Cy3 and HS2-Cy5) containing multiple ATP aptamer fragments. When ATP combined with the aptamer fragments on the DNA double-strand, the distance between Cy3 and Cy5 was shortened and a FRET signal was generated, resulting in ratio imaging of ATP in the mitochondria (Fig. 3(d)).83 Jiang et al. designed a DNA-based “nanotrain” drug delivery system with DNA as the basic unit, cyanine dye (self-luminescence and mitochondrial targeting) as the locomotive, and the chemotherapeutic drug DOX, which could be inserted into the CG sequence of the DNA. Cy5.5 produces weak ROS under NIR light irradiation conditions, thus enhancing the sensitivity of tumor cells to traditional chemotherapy drugs and enhancing the effect of chemotherapy drugs to a certain extent.84Mitochondrial targeting with MTS modification
Mitochondrial targeting sequence (MTS) peptides are recognized by receptors on the mitochondrial surface, which are composed of 20–40 amino acids.85 D-(KLAKLAK)2 (KLA) is a positively charged specific mitochondrial-targeting sequence that can destroy the mitochondrial membrane and initiate apoptosis.86–88 Efficient mitochondrial targeting is achieved by modifying KLA on DNA nanodevices. Yan et al. modified the precise amount of KLA peptide in a DNA tetrahedron carrying DOX, which enhanced the anti-cancer effect in vitro by activating the mitochondrial-mediated programmed apoptotic pathway (Fig. 3(e)).89 Abnous et al. designed a simple DNA nanostructure composed of two sequences: one containing ATP and the AS1411 aptamer and the other containing antimiR-21. By modifying the KLA peptide, the structure achieved the delivery of antimiR-21 and effectively inhibited tumor growth in vitro and in vivo.90DNA nanodevices cannot usually be used to directly target mitochondria. Specific mitochondrial targeting molecules need to be modified on DNA nanodevices, such as the lipophilic cationic ligands or mitochondrial targeting sequences mentioned earlier. Compared with other nanomaterials modifying targeted molecules, DNA nanostructures have higher biocompatibility and safety. Moreover, DNA nanostructures are programmable, and can precisely control modification sites and quantities of targeted molecules, making DNA nanodevices convenient for scientists to manipulate.
Other organelles
Nuclear targeting
The nucleus is the control center of the cell and contains the majority of the cell's genetic material, playing an important role in cell metabolism, growth, and differentiation. Targeted intervention in the nucleus can directly induce apoptosis and effectively treat various diseases. Nucleolin is highly expressed on the surface of many types of tumor cells and is associated with the neovascularization of endothelial cells. The nucleic acid aptamer, AS1411, can specifically bind to nucleolin, preventing DNA damage repair, promoting cell apoptosis, and delivering drugs from the cell surface to the nucleus in order to actively target the nucleus for therapy.91,92 However, AS1411 is ssDNA, which has a low uptake efficiency in cells and poor stability in vivo. Therefore, AS1411 is often modified on DNA nanodevices to improve stability and achieve nuclear targeting and therapy. Li et al. modified AS1411 on DNA tetrahedra and were able to specifically inhibit tumor cell growth compared to DNA tetrahedra alone.93 Modification of the nuclear localization signal (NLS) on DNA nanostructures is also a commonly used method to guide foreign materials into the nucleus.94 Liang et al. modified the NLS on DNA tetrahedra, which initially targeted lysosomes, to target the nuclei (Fig. 4(b)).30 Furthermore, Zhao et al. studied the efficiency of the connection between the NLS of different lengths and DNA tetrahedra. They found that both the long NLS (NLS29) and the original NLS12 could target the nucleus; however, the connection efficiency between the lengthened NLS and DNA tetrahedra was higher.95 Yang et al. modified the DNA tetrahedron with NLS in order to assist antisense oligonucleotides with the transfer to the nucleus after interacting with A549 cells, thus downregulating target mRNA in the nucleus and cytoplasm.96 Similar to the function of NLS, TAT is a nuclear-localized polypeptide that promotes the aggregation of nanomaterials in the nucleus by binding to the import receptors, importin α and β, and targeting the nuclear pore complex.97 Shao et al. introduced a TAT peptide on the surface of a DNA nanosensor in order to obtain APE1 imaging in the nucleus.76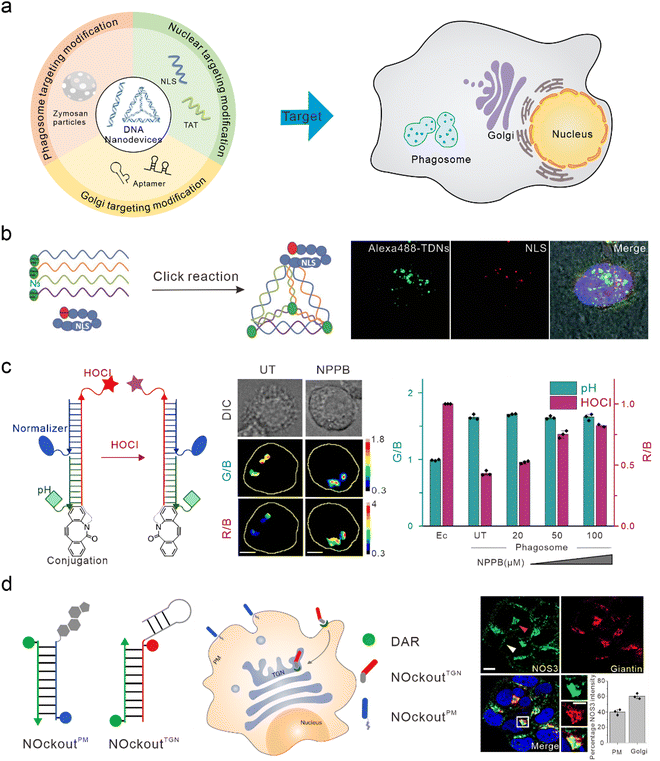 | ||
| Fig. 4 Bio-application of DNA nanodevices within other organelles. (a) Schematic of the design principle of the DNA nanodevice in other organelles. (b) The structure of the DNA tetrahedron modified by NLS is used to target the nucleus. Reproduced from ref. 30 with permission from Wiley, copyright 2014. (c) Design of HOCl sensors that target phagosomes. Reproduced from ref. 107 with permission from Springer Nature, copyright 2018. (d) Two DNA nanodevices for targeting the TGN and the plasma membrane. Reproduced from ref. 115 with permission from Springer Nature, copyright 2020. | ||
Phagosomes targeting
Phagosomes are common subcellular structures of the immune system, and pathogens that invade host cells are captured by phagosomes.98,99 Lysosomes can combine with phagosomes to form phagolysosomes, which transport destructive hydrolases in lysosomes to phagosomes,100 and produce large amounts of ROS and reactive nitrogen in order to induce the elimination and degradation of pathogens.101 Phagosomes can be marked by swallowing zymosan particles. Zymosan is a boiled, trypsin-treated preparation of the cell wall of Saccharomyces cerevisiae.102 DNA nanodevices can be modified onto zymosan particles to target phagosomes. Myeloperoxidase (MPO) is a destructive enzyme transported to the phagosome and is the only enzyme that catalyzes the reaction of hydrogen peroxide and chloride ions to produce HOCl (a bactericide).103,104 Therefore, MPO activity can be determined through the detection of HOCl. When MPO activity is unbalanced, it leads to various immune-mediated inflammatory diseases.105,106 Thekkan et al.107 developed a DNA-based reporter group to simultaneously quantify HOCl and pH in phagosomes by integrating HOCl-sensitive fluorophores, pH-sensitive fluorophores, reference fluorophores, and phagosome targeting groups, thus helping us to better understand the role of MPO in immune-mediated inflammatory diseases (Fig. 4(c)).108 Nitric oxide synthase 2 (NOS2) is primarily found in immune cells. When pathogens are phagocytosed into the phagosomes of immune cells, the pathogen-associated molecular patterns (PAMPs) of some pathogens are recognized by TLRs that activate NOS2 and produce a large amount of nitric oxide (NO) to clear pathogens.109–111 Veetil et al. quantified NO in phagosomes and endosomes by modifying the targeting group “Fn” sequence (a functional DNA or RNA sequence that recognizes PAMP) on DNA nanodevices. Altered PAMPs can also be used to detect NO in the phagosomes of different species (live zebrafish brain microglia), and reflects NOS2 activity.112Golgi apparatus targeting
The Golgi apparatus consists of many flattened vesicles, which are mainly responsible for biosynthesis, secretion, and intracellular signal transduction.113 Recent studies have shown that the Golgi apparatus plays an important role in the cytoskeleton, and in the regulation of calcium homeostasis and mitosis.114 However, at present, there are few researches conducted on DNA nanodevices for Golgi apparatus targeting. Jani et al. designed a DNA nanodevice called NOckout, which integrated NO-sensitive fluorophores, internal reference fluorophores, and plasma membrane targeting (modified with cholesterol) or Trans-Golgi network (TGN)-targeting groups (modified with MUC1 aptamer, which was combined with the low glycosylated MUC1 protein on the cancer cell cytoplasm membrane and could label the lumen of TGN through retrograde endocytosis) (Fig. 4(d)). By monitoring the production of NO in these two subcellular structures, they found that although NOS3 activity on the TGN was ten times lower than that on the plasma membrane, it was essential in maintaining the structural integrity of the Golgi apparatus.115To summarize, when DNA nanodevices are used for targeting other subcellular structures, the corresponding targeting module also needs to be modified. However, there are few studies on DNA nanodevices used for targeting phagosomes and Golgi apparatus at present. This may be because the localization mechanism for these organelles is not well understood. On the other hand, there may be technical difficulties in binding existing targeted groups to DNA nanodevices. Further enriching the DNA nanodevices targeted by various organelles will provide a powerful tool for basic research and clinical diagnosis at the subcellular level.
Conclusions and perspectives
The morphology, activity, and chemical composition of subcellular structures reflect the metabolic state of cells. Subcellular structures have been found to play a significant role in regulating the disease microenvironment, and early therapeutic intervention at the organelle level is very important.37,116,117 Modulating organelle activity through precisely targeted therapies at the subcellular level provides one more option for studying and understanding disease. The precision of targeted organelles in vivo depends on the physiological mechanism of intracellular transport and the unique properties of the subcellular structures. Precise targeting at the organelle level using fluorescently labeled DNA nanodevices has established the value of DNA nanostructures for imaging organelles for diagnostic applications, and the same can be explored for therapeutic applications. Cracking the rules of appropriate transmission between organelles, cells, and tissues could advance the use of DNA nanodevices as diagnostic and therapeutic agents.Herein, we have reviewed recent advances in organelle targeting based on DNA nanodevices, highlighting the biological applications of the same. Cell biology research has gone through a remarkable development path from simply monitoring and analyzing the lysosomal microenvironment using the DNA nanostructures, which are naturally localized to lysosomes, to controlling its spatial distribution and biological functions in cells through modification, and further to building intelligent nano-reactors for organelle pathology and molecular diagnosis. In contrast, although liposomes or nanoparticles can now also be designed with uniform size, composition, and surface chemical modifications, they are not molecularly the same, and this variability can limit their biocompatibility, specificity, and performance, among other things.
The programmability, modularity, and biocompatibility of DNA nanodevices make them ideal for targeting organelle-level resolution in living organisms. Although some progress has been made, there are still several problems with its wide application. Firstly, research on DNA nanodevices at the subcellular level mainly focuses on lysosomes, mitochondria, and nuclei. The study of other organelles, such as the endoplasmic reticulum and Golgi apparatus, which are responsible for protein synthesis and processing, is still insufficient. Secondly, the detection of other components of the organelle, such as other small molecules and enzyme activity in the lysozyme, depends on chemists synthesizing novel chemical molecules corresponding to the response. Finally, although the studies reviewed in this paper show that organelle function is closely related to multiple pathways that control cell and body homeostasis, the structure and function of organelles have only been understood to some extent. The understanding of how each subcellular structure is related may merely be the tip of the iceberg, and it is necessary to ultimately investigate the function of each organelle in the context of the whole cell and organism.
In the foreseeable future, we believe that DNA nanotechnology itself will be more closely integrated with cell biology and synthetic biology, so that scientists can participate more deeply in the basic levels of cell biosensors and cell life activities through the design of DNA structures, thus revealing more details of such activities and better understanding the mechanisms of life activities.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 22022410, No. 82050005), 2022 Shanghai “Science and Technology Innovation Action Plan” Fundamental Research Project (22JC1401203), and the Youth Innovation Promotion Association of CAS (grant no. 2016236).Notes and references
- J. D. Watson and F. H. Crick, Cold Spring Harbor Symp. Quant. Biol., 1953, 18, 123–131 CrossRef CAS PubMed.
- N. C. Seeman, J. Theor. Biol., 1982, 99, 237–247 CrossRef CAS PubMed.
- J. Li, C. Zheng, S. Cansiz, C. Wu, J. Xu, C. Cui, Y. Liu, W. Hou, Y. Wang, L. Zhang, I. T. Teng, H. H. Yang and W. Tan, J. Am. Chem. Soc., 2015, 137, 1412–1415 CrossRef CAS PubMed.
- S. Li, Q. Jiang, S. Liu, Y. Zhang, Y. Tian, C. Song, J. Wang, Y. Zou, G. J. Anderson, J. Y. Han, Y. Chang, Y. Liu, C. Zhang, L. Chen, G. Zhou, G. Nie, H. Yan, B. Ding and Y. Zhao, Nat. Biotechnol., 2018, 36, 258–264 CrossRef CAS PubMed.
- J. Song, K. Im, S. Hwang, J. Hur, J. Nam, G. O. Ahn, S. Hwang, S. Kim and N. Park, Nanoscale, 2015, 7, 9433–9437 RSC.
- Q. Yuan, Y. Wu, J. Wang, D. Q. Lu, Z. L. Zhao, T. Liu, X. B. Zhang and W. H. Tan, Angew. Chem., Int. Ed., 2013, 52, 13965–13969 CrossRef CAS PubMed.
- H. M. Zhang, Y. L. Ma, Y. Xie, Y. An, Y. S. Huang, Z. Zhu and C. Y. J. Yang, Sci. Rep., 2015, 5, 10099 CrossRef CAS PubMed.
- Y. Peng, X. Wang, Y. Xiao, L. Feng, C. Zhao, J. Ren and X. Qu, J. Am. Chem. Soc., 2009, 131, 13813–13818 CrossRef CAS PubMed.
- F. Li, X. Han and S. Liu, Biosens. Bioelectron., 2011, 26, 2619–2625 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- B. Billet, B. Chovelon, E. Fiore, F. Oukacine, M. A. Petrillo, P. Faure, C. Ravelet and E. Peyrin, Angew. Chem., Int. Ed., 2021, 60, 12346–12350 CrossRef CAS PubMed.
- E. Del Grosso, G. Ragazzon, L. J. Prins and F. Ricci, Angew. Chem., Int. Ed., 2019, 58, 5582–5586 CrossRef CAS PubMed.
- M. Liu, J. Wang, Y. Chang, Q. Zhang, D. Chang, C. Y. Hui, J. D. Brennan and Y. Li, Angew. Chem., Int. Ed., 2020, 59, 7706–7710 CrossRef CAS PubMed.
- H. Qu, A. T. Csordas, J. Wang, S. S. Oh, M. S. Eisenstein and H. T. Soh, ACS Nano, 2016, 10, 7558–7565 CrossRef CAS PubMed.
- X. B. Zhang, R. M. Kong and Y. Lu, Annu. Rev. Anal. Chem., 2011, 4, 105–128 CrossRef CAS PubMed.
- W. Zhou, R. Saran and J. Liu, Chem. Rev., 2017, 117, 8272–8325 CrossRef CAS PubMed.
- P. Chen, Y. Wang, Y. He, K. Huang, X. Wang, R. Zhou, T. Liu, R. Qu, J. Zhou, W. Peng, M. Li, Y. Bai, J. Chen, J. Huang, J. Geng, Y. Xie, W. Hu and B. Ying, ACS Nano, 2021, 15, 11634–11643 CrossRef CAS PubMed.
- F. Gao, F. Zhan, S. Li, P. Antwi-Mensah, L. Niu and Q. Wang, Biosens. Bioelectron., 2022, 209, 114280 CrossRef CAS PubMed.
- Y. Zhou, E. Kierzek, Z. P. Loo, M. Antonio, Y. H. Yau, Y. W. Chuah, S. Geifman-Shochat, R. Kierzek and G. Chen, Nucleic Acids Res., 2013, 41, 6664–6673 CrossRef CAS PubMed.
- L. Liu, J. W. Liu, Z. M. Huang, H. Wu, N. Li, L. J. Tang and J. H. Jiang, Anal. Chem., 2017, 89, 6944–6947 CrossRef CAS PubMed.
- H. Li, X. Zhou, D. Yao and H. Liang, Chem. Commun., 2018, 54, 3520–3523 RSC.
- Q. Tang, W. Lai, P. P. Wang, X. W. Xiong, M. S. Xiao, L. Li, C. H. Fan and H. Pei, Angew. Chem., Int. Ed., 2021, 60, 15013–15019 CrossRef CAS PubMed.
- H. L. Liu, Q. C. Jiang, J. Pang, Z. Y. Jiang, J. Cao, L. N. Ji, X. H. Xia and K. Wang, Adv. Funct. Mater., 2018, 28, 1703847 CrossRef.
- M. Debnath, K. Fatma and J. Dash, Angew. Chem., Int. Ed., 2019, 58, 2942–2957 CrossRef CAS PubMed.
- M. Debnath, S. Chakraborty, Y. P. Kumar, R. Chaudhuri, B. Jana and J. Dash, Nat. Commun., 2020, 11, 469 CrossRef CAS PubMed.
- F. Zhang, J. Nangreave, Y. Liu and H. Yan, J. Am. Chem. Soc., 2014, 136, 11198–11211 CrossRef CAS PubMed.
- X. Lim, Nature, 2017, 546, 687–689 CrossRef CAS PubMed.
- N. C. Seeman and H. F. Sleiman, Nat. Rev. Mater., 2018, 3, 17068 CrossRef CAS.
- L. Liang, J. Li, Q. Li, Q. Huang, J. Shi, H. Yan and C. Fan, Angew. Chem., Int. Ed., 2014, 53, 7745–7750 CrossRef CAS PubMed.
- L. He, D. Lu, H. Liang, S. Xie, X. Zhang, Q. Liu, Q. Yuan and W. Tan, J. Am. Chem. Soc., 2018, 140, 258–263 CrossRef CAS PubMed.
- A. Lacroix and H. F. Sleiman, ACS Nano, 2021, 15, 3631–3645 CrossRef CAS PubMed.
- P. Peng, Y. Du, J. Zheng, H. Wang and T. Li, Angew. Chem., Int. Ed., 2019, 58, 1648–1653 CrossRef CAS PubMed.
- F. Li, Y. Liu, Y. Dong, Y. Chu, N. Song and D. Yang, J. Am. Chem. Soc., 2022, 144, 4667–4677 CrossRef CAS PubMed.
- A. Saminathan, M. Zajac, P. Anees and Y. Krishnan, Nat. Rev. Mater., 2022, 7, 355–371 CrossRef.
- Y. Krishnan, J. Zou and M. S. Jani, ACS Cent. Sci., 2020, 6, 1938–1954 CrossRef CAS PubMed.
- A. Ballabio and J. S. Bonifacino, Nat. Rev. Mol. Cell Biol., 2020, 21, 101–118 CrossRef CAS PubMed.
- M. Cao, X. Luo, K. Wu and X. He, Signal Transduction Targeted Ther., 2021, 6, 379 CrossRef CAS PubMed.
- S. R. Bonam, F. Wang and S. Muller, Nat. Rev. Drug Discovery, 2019, 18, 923–948 CrossRef CAS PubMed.
- J. Li and C. Fan, Nat. Nanotechnol., 2021, 16, 1306–1307 CrossRef CAS PubMed.
- S. Modi, M. G. Swetha, D. Goswami, G. D. Gupta, S. Mayor and Y. Krishnan, Nat. Nanotechnol., 2009, 4, 325–330 CrossRef CAS PubMed.
- N. Narayanaswamy, K. Chakraborty, A. Saminathan, E. Zeichner, K. Leung, J. Devany and Y. Krishnan, Nat. Meth., 2019, 16, 95–102 CAS.
- S. Saha, V. Prakash, S. Halder, K. Chakraborty and Y. Krishnan, Nat. Nanotechnol., 2015, 10, 645–651 CrossRef CAS PubMed.
- K. Chakraborty, K. Leung and Y. Krishnan, eLife, 2017, 6, e28862 CrossRef PubMed.
- K. Leung, K. Chakraborty, A. Saminathan and Y. Krishnan, Nat. Nanotechnol., 2019, 14, 176–183 CrossRef CAS PubMed.
- A. Saminathan, J. Devany, A. T. Veetil, B. Suresh, K. S. Pillai, M. Schwake and Y. Krishnan, Nat. Nanotechnol., 2021, 16, 96–103 CrossRef CAS PubMed.
- V. Prakash, S. Saha, K. Chakraborty and Y. Krishnan, Chem. Sci., 2016, 7, 1946–1953 RSC.
- A. Saminathan, V. S. Noyola and Y. Krishnan, Trends Biochem. Sci., 2020, 45, 365–366 CrossRef CAS PubMed.
- P. Saftig and J. Klumperman, Nat. Rev. Mol. Cell Biol., 2009, 10, 623–635 CrossRef CAS PubMed.
- K. Dan, A. T. Veetil, K. Chakraborty and Y. Krishnan, Nat. Nanotechnol., 2019, 14, 252–259 CrossRef CAS PubMed.
- S. Modi, C. Nizak, S. Surana, S. Halder and Y. Krishnan, Nat. Nanotechnol., 2013, 8, 459–467 CrossRef CAS PubMed.
- S. Modi, S. Halder, C. Nizak and Y. Krishnan, Nanoscale, 2014, 6, 1144–1152 RSC.
- W. G. Mallet and F. R. Maxfield, J. Cell Biol., 1999, 146, 345–359 CrossRef CAS PubMed.
- J. F. Presley, S. Mayor, K. W. Dunn, L. S. Johnson, T. E. McGraw and F. R. Maxfield, J. Cell Biol., 1993, 122, 1231–1241 CrossRef CAS PubMed.
- L. He, D. Q. Lu, H. Liang, S. Xie, C. Luo, M. Hu, L. Xu, X. Zhang and W. Tan, ACS Nano, 2017, 11, 4060–4066 CrossRef CAS PubMed.
- F. Hong, F. Zhang, Y. Liu and H. Yan, Chem. Rev., 2017, 117, 12584–12640 CrossRef CAS PubMed.
- J. Li, H. Pei, B. Zhu, L. Liang, M. Wei, Y. He, N. Chen, D. Li, Q. Huang and C. Fan, ACS Nano, 2011, 5, 8783–8789 CrossRef CAS PubMed.
- X. Yue, Y. Qiao, D. Gu, R. Qi, H. Zhao, Y. Yin, W. Zhao, R. Xi and M. Meng, Anal. Chem., 2021, 93, 7250–7257 CrossRef CAS PubMed.
- S. He, M. Liu, F. Yin, J. Liu, Z. Ge, F. Li, M. Li, J. Shi, L. Wang, X. Mao, X. Zuo and Q. Li, Chem. Commun., 2021, 57, 3247–3250 RSC.
- A. Rajwar, S. R. Shetty, P. Vaswani, V. Morya, A. Barai, S. Sen, M. Sonawane and D. Bhatia, ACS Nano, 2022, 16, 10496–10508 CrossRef CAS PubMed.
- C. Cui, K. Chakraborty, X. A. Tang, K. Q. Schoenfelt, A. Hoffman, A. Blank, B. McBeth, N. Pulliam, C. A. Reardon, S. A. Kulkarni, T. Vaisar, A. Ballabio, Y. Krishnan and L. Becker, Nat. Nanotechnol., 2021, 16, 1394–1402 CrossRef CAS PubMed.
- S. Liu, Q. Jiang, X. Zhao, R. Zhao, Y. Wang, Y. Wang, J. Liu, Y. Shang, S. Zhao, T. Wu, Y. Zhang, G. Nie and B. Ding, Nat. Mater., 2021, 20, 421–430 CrossRef CAS PubMed.
- D. Gu, Y. Qiao, H. Fu, H. Zhao, X. Yue, S. Wang, Y. Yin, R. Xi, X. Fu, X. Zhao and M. Meng, ACS Appl. Mater. Interfaces, 2022, 14, 38048–38055 CrossRef CAS PubMed.
- Y. Dong, F. Li, Z. Lv, S. Li, M. Yuan, N. Song, J. Liu and D. Yang, Angew. Chem., Int. Ed., 2022, 61, e202207770 CAS.
- A. T. Hoye, J. E. Davoren, P. Wipf, M. P. Fink and V. E. Kagan, Acc. Chem. Res., 2008, 41, 87–97 CrossRef CAS PubMed.
- S. S. Liew, X. Qin, J. Zhou, L. Li, W. Huang and S. Q. Yao, Angew. Chem., Int. Ed., 2021, 60, 2232–2256 CrossRef CAS PubMed.
- H. Singh, D. Sareen, J. M. George, V. Bhardwaj, S. Rha, S. J. Lee, S. Sharma, A. Sharma and J. S. Kim, Coord. Chem. Rev., 2022, 452, 214283 CrossRef CAS.
- M. J. Devine and J. T. Kittler, Nat. Rev. Neurosci., 2018, 19, 63–80 CrossRef CAS PubMed.
- F. J. Bock and S. W. G. Tait, Nat. Rev. Mol. Cell Biol., 2020, 21, 85–100 CrossRef CAS PubMed.
- M. Khacho, R. Harris and R. S. Slack, Nat. Rev. Neurosci., 2019, 20, 34–48 CrossRef CAS PubMed.
- J. Zielonka, J. Joseph, A. Sikora, M. Hardy, O. Ouari, J. Vasquez-Vivar, G. Cheng, M. Lopez and B. Kalyanaraman, Chem. Rev., 2017, 117, 10043–10120 CrossRef CAS PubMed.
- R. A. Smith, R. C. Hartley and M. P. Murphy, Antioxid. Redox Signaling, 2011, 15, 3021–3038 CrossRef CAS PubMed.
- R. A. Smith, R. C. Hartley, H. M. Cocheme and M. P. Murphy, Trends Pharmacol. Sci., 2012, 33, 341–352 CrossRef CAS PubMed.
- W. Xu, Z. Zeng, J. H. Jiang, Y. T. Chang and L. Yuan, Angew. Chem., Int. Ed., 2016, 55, 13658–13699 CrossRef CAS PubMed.
- P. Lu, B. J. Bruno, M. Rabenau and C. S. Lim, J. Controlled Release, 2016, 240, 38–51 CrossRef CAS PubMed.
- Y. L. Shao, J. Zhao, J. Y. Yuan, Y. L. Zhao and L. L. Li, Angew. Chem., Int. Ed., 2021, 60, 8923–8931 CrossRef CAS PubMed.
- F. Shah, D. Logsdon, R. A. Messmann, J. C. Fehrenbacher, M. L. Fishel and M. R. Kelley, NPJ Precis. Oncol., 2017, 1, 19 CrossRef PubMed.
- F. Z. Yu, Y. L. Shao, X. Chai, Y. L. Zhao and L. L. Li, Angew. Chem., Int. Ed., 2022, 61, e202203238 CAS.
- J. Zhao, Z. Li, Y. Shao, W. Hu and L. Li, Angew. Chem., Int. Ed., 2021, 60, 17937–17941 CrossRef CAS PubMed.
- X. Chai, Z. Fan, M. M. Yu, J. Zhao and L. Li, Nano Lett., 2021, 21, 10047–10053 CrossRef CAS PubMed.
- Z. Liu, H. Pei, L. Zhang and Y. Tian, ACS Nano, 2018, 12, 12357–12368 CrossRef CAS PubMed.
- J. Liu, L. Yang, C. Xue, G. Huang, S. Chen, J. Zheng and R. Yang, ACS Appl. Mater. Interfaces, 2021, 13, 33894–33904 CrossRef CAS PubMed.
- L. Luo, M. Wang, Y. Zhou, D. Xiang, Q. Wang, J. Huang, J. Liu, X. Yang and K. Wang, Anal. Chem., 2021, 93, 6715–6722 CrossRef CAS PubMed.
- T. Jiang, L. Zhou, H. Liu, P. Zhang, G. Liu, P. Gong, C. Li, W. Tan, J. Chen and L. Cai, Anal. Chem., 2019, 91, 6996–7000 CrossRef CAS PubMed.
- L. F. Yousif, K. M. Stewart, K. L. Horton and S. O. Kelley, ChemBioChem, 2009, 10, 2081–2088 CrossRef CAS PubMed.
- L. Agemy, D. Friedmann-Morvinski, V. R. Kotamraju, L. Roth, K. N. Sugahara, O. M. Girard, R. F. Mattrey, I. M. Verma and E. Ruoslahti, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 17450–17455 CrossRef CAS PubMed.
- J. W. Sun, L. Jiang, Y. Lin, E. M. Gerhard, X. H. Jiang, L. Li, J. Yang and Z. W. Gu, Int. J. Nanomed., 2017, 12, 1517–1537 CrossRef CAS PubMed.
- H. M. Ellerby, W. Arap, L. M. Ellerby, R. Kain, R. Andrusiak, G. D. Rio, S. Krajewski, C. R. Lombardo, R. Rao, E. Ruoslahti, D. E. Bredesen and R. Pasqualini, Nat. Med., 1999, 5, 1032–1038 CrossRef CAS PubMed.
- J. Yan, J. Chen, N. Zhang, Y. Yang, W. Zhu, L. Li and B. He, J. Mater. Chem. B, 2020, 8, 492–503 RSC.
- K. Abnous, N. M. Danesh, M. Ramezani, M. Alibolandi, A. Bahreyni, P. Lavaee, S. A. Moosavian and S. M. Taghdisi, J. Drug Targeting, 2020, 28, 852–859 CrossRef CAS PubMed.
- D. H. Dam, J. H. Lee, P. N. Sisco, D. T. Co, M. Zhang, M. R. Wasielewski and T. W. Odom, ACS Nano, 2012, 6, 3318–3326 CrossRef CAS PubMed.
- D. F. Wang, M. Liu, Y. S. Wu, T. X. Weng, L. Wang, Y. F. Zhang, Y. N. Zhao and J. Han, J. Mol. Liq., 2022, 355, 118947 CrossRef CAS.
- Q. Li, D. Zhao, X. Shao, S. Lin, X. Xie, M. Liu, W. Ma, S. Shi and Y. Lin, ACS Appl. Mater. Interfaces, 2017, 9, 36695–36701 CrossRef CAS PubMed.
- Y. C. Pan, R. Weng, L. H. Zhang, J. Qiu, X. L. Wang, G. Q. Liao, Z. H. Qin, L. P. Zhang, H. H. Xiao, Y. Z. Qian and X. Su, Nano Today, 2022, 46, 101573 CrossRef CAS.
- Y. Zhao, F. Li, L. J. Guo, J. B. Dai, S. Xing and L. H. Wang, Nucl. Technol., 2017, 40, 48–53 Search PubMed.
- J. Yang, Q. Jiang, L. He, P. Zhan, Q. Liu, S. Liu, M. Fu, J. Liu, C. Li and B. Ding, ACS Appl. Mater. Interfaces, 2018, 10, 23693–23699 CrossRef CAS PubMed.
- L. Pan, Q. He, J. Liu, Y. Chen, M. Ma, L. Zhang and J. Shi, J. Am. Chem. Soc., 2012, 134, 5722–5725 CrossRef CAS PubMed.
- L. M. Stuart and R. A. Ezekowitz, Immunity, 2005, 22, 539–550 CrossRef CAS PubMed.
- L. Chernyak and A. I. Tauber, Cell. Immunol., 1988, 117, 218–233 CrossRef CAS PubMed.
- A. W. Segal, Annu. Rev. Immunol., 2005, 23, 197–223 CrossRef CAS PubMed.
- C. C. Winterbourn and A. J. Kettle, Antioxid. Redox Signaling, 2013, 18, 642–660 CrossRef CAS PubMed.
- D. M. Underhill, J. Endotoxin Res., 2003, 9, 176–180 CrossRef CAS PubMed.
- S. J. Klebanoff, A. J. Kettle, H. Rosen, C. C. Winterbourn and W. M. Nauseef, J. Leukoc. Biol., 2013, 93, 185–198 CrossRef CAS PubMed.
- M. B. Hampton, A. J. Kettle and C. C. Winterbourn, Blood, 1998, 92, 3007–3017 CrossRef CAS PubMed.
- P. Valadez-Cosmes, S. Raftopoulou, Z. N. Mihalic, G. Marsche and J. Kargl, Pharmacol. Ther., 2022, 236, 108052 CrossRef CAS PubMed.
- S. E. Janus, J. Hajjari, T. Chami, M. Karnib, S. G. Al-Kindi and I. Rashid, Curr. Probl. Cardiol., 2021, 47, 101080 CrossRef PubMed.
- S. Thekkan, M. S. Jani, C. Cui, K. Dan, G. Zhou, L. Becker and Y. Krishnan, Nat. Chem. Biol., 2019, 15, 1165–1172 CrossRef CAS PubMed.
- P. Anees, M. Zajac and Y. Krishnan, Methods Cell Biol., 2021, 164, 119–136 CAS.
- A. P. West, A. A. Koblansky and S. Ghosh, Annu. Rev. Cell Dev. Biol., 2006, 22, 409–437 CrossRef CAS PubMed.
- O. Takeuchi and S. Akira, Cell, 2010, 140, 805–820 CrossRef CAS PubMed.
- T. H. Mogensen, Clin. Microbiol. Rev., 2009, 22, 240–273 CrossRef CAS PubMed.
- A. T. Veetil, J. Zou, K. W. Henderson, M. S. Jani, S. M. Shaik, S. S. Sisodia, M. E. Hale and Y. Krishnan, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 14694–14702 CrossRef CAS PubMed.
- R. S. Li, C. Wen, C. Z. Huang and N. Li, Trends Anal. Chem., 2022, 156, 116714 CrossRef CAS.
- G. Guizzunti and J. Seemann, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E6590–E6599 CrossRef CAS PubMed.
- M. S. Jani, J. Zou, A. T. Veetil and Y. Krishnan, Nat. Chem. Biol., 2020, 16, 660–666 CrossRef CAS PubMed.
- N. Zheng, Q. Wang, S. Zhang, C. Mao, L. He and S. Liu, J. Mater. Chem. B, 2022, 10, 7450–7459 RSC.
- M. Zhang, N. Xu, W. Xu, G. Ling and P. Zhang, Pharmacol. Res., 2022, 175, 105861 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |