Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Pentafluorophenyl-based single-chain polymer nanoparticles as a versatile platform towards protein mimicry†
A. Pia P.
Kröger‡
 a,
Jan-Willem D.
Paats‡
a,
Jan-Willem D.
Paats‡
 a,
Roy J. E. A.
Boonen
a,
Naomi M.
Hamelmann
a,
Roy J. E. A.
Boonen
a,
Naomi M.
Hamelmann
 a and
Jos M. J.
Paulusse
a and
Jos M. J.
Paulusse
 *ab
*ab
aDepartment of Biomolecular Nanotechnology, MESA+ Institute for Nanotechnology and TechMed Institute for Health and Biomedical Technologies, Faculty of Science and Technology, University of Twente, P.O. Box 217, 7500 AE Enschede, The Netherlands. E-mail: J.M.J.Paulusse@utwente.nl
bDepartment of Nuclear Medicine and Molecular Imaging, University Medical Center Groningen, P.O. Box 30.001, 9700 RB Groningen, The Netherlands
First published on 26th August 2020
Abstract
Proteins are biopolymers folded into 3D-structures and are omnipresent in biological systems, where they fulfil a wide array of complex functions. Mimicking the exceptional characteristics of proteins with synthetic analogues may likewise give unprecedented control over a nanomaterial's pharmacokinetic behaviour, enabling controlled delivery of therapeutics or imaging agents. Recent advances in polymer science have enabled the formation of bio-inspired single-chain polymer nanoparticles (SCNPs), which are formed by intramolecular collapse of individual polymer chains, and display sizes ranging from 5–20 nm. Here, we describe the preparation of SCNPs containing activated ester moieties, facilitating SCNP functionalization without altering its backbone structure. Pentafluorophenyl-functional SCNPs were prepared through intramolecular thiol-Michael addition crosslinking of thiol-functional precursor copolymers. Post-formation functionalization of the resulting SCNPs through substitution of the activated pentafluorophenyl esters with a variety of amines resulted in a series of water-soluble SCNPs with fluorescent labels, ‘click’ functionality, amino acids and even peptides. This synthetic strategy offers a straightforward method towards SCNP modification and SCNP-protein hybrids, giving access to easily adjustable physicochemical properties and protein mimicry.
Introduction
Protein-like synthetic polymers aim at combining the modularity and scalability of polymer synthesis1 with the specific functions of proteins, such as catalytic activity, signalling, transport and storage.2–5 Since proteins are able to perform a remarkable number of intricate functions under physiological conditions based on their structural conformation, replication of protein function and shape has become a primary goal for researchers worldwide. Mimicking natural proteins by synthetic polymer chemistry may be approached by several different routes. A promising strategy is the bioconjugation of peptides and proteins with polymers.6,7 By combining the structural properties of the synthetic backbone with highly specific functions of a protein domain, hybrid materials are obtained with a range of versatile properties.8 One of the first striking examples of a protein–polymer conjugate is the attachment of (poly)ethylene glycol chains onto bovine serine albumin, reducing toxicity and improving pharmacokinetic profile.9 Since then, a wide variety of chemistries has been used to create polymer–protein hybrids,10 introducing functionalities such as catalysis4 and biosensing11 into synthetic polymeric materials. Although protein–polymer conjugates can mimic specific characteristics, there is little resemblance to the structural features and shape of a protein. Therefore, the folding of synthetic polymers, like the spatial arrangement of a protein, has become a primary goal towards protein mimicry.Single-chain polymer nanoparticles (SCNPs) are prepared through exclusive intramolecular crosslinking of individual polymer chains.12–23 Size and dispersity of SCNPs are directly related to the size and size distribution of the polymers from which they are prepared, and can therefore be controlled very accurately, yielding nanoparticles in the 5–20 nm size range, even resembling size and structure of proteins.24–26 In order to approximate specific protein characteristics, control over the self-recognizing motifs determining secondary and tertiary structure,27–31 as well as catalytic activity have been studied.32–34 Current applications of protein-like SCNPs have focused on the development of enzyme-like materials,33–36 and applications in the field of drug delivery, for example aiming at biocompatibility and targeted therapeutics delivery.37–41
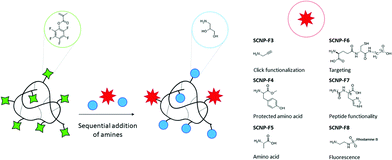
In order to achieve protein mimicry, besides controlling monomer sequence and chain folding, equipping SCNPs with different amino acids or peptide moieties further approaches the composition of natural proteins. SCNPs with protein-like characteristics can either be prepared through functional precursor polymers, as is the case with poly(γ-glutamic acid) precursors for example,42–44 or by post-formation functionalization of SCNPs.45–47 Post-formation functionalization enables modification of polymers,48 nanoparticles49,50 and nanogels51,52 without markedly altering their size and structure. This strategy has been utilized to conjugate peptides onto SCNPs for targeting purposes, by covalently attaching the peptide via amidation to carboxylic acid-functional SCNPs,38 as well as for peptide delivery by reversibly binding the peptide via disulfide linkages.38,45 Even proteins have been attached onto SCNPs via disulfide linkages or NHS (N-hydroxysuccinimide) ester chemistry.46 Exploiting post-formation functionalization of SCNPs to mimic proteins would allow careful tuning of the surface properties, while maintaining the SCNP structure intact.
Pentafluorophenyl (PFP) esters have found widespread application in bioconjugation chemistry.53 These activated esters are soluble in a range of organic solvents and less susceptible to hydrolysis than NHS esters.54,55 PFP-functional polymers have been successfully used in post-polymerization functionalization reactions with both primary and secondary amines.55 Conjugation of antibodies to PFP-acrylate polymer brushes was achieved by Son et al., enabling capture and purification of target proteins.56 In the stepwise compaction of N-substituted maleimide copolymers, the PFP-moiety was used as intramolecular crosslinking reaction to prepare SCNPs.30,57 Furthermore, Palmans and co-workers used poly(PFP-acrylate) to prepare polymer precursors for intramolecular self-assembly of SCNPs.58 Recently, PFP-polymers were also used to synthesize SCNPs with increased catalytic activity, even working inside HeLa cells.59,60
In this study, we report the preparation and characterization of PFP-functional SCNPs via thiol-Michael addition crosslinking, and their use as a flexible platform to easily modify and functionalize SCNPs, while maintaining the SCNP backbone structure unaltered. Various functional amines, including amino acids and peptides, are conjugated onto the SCNPs to yield protein-like materials, while at the same time allowing for facile conjugation of fluorescent labels, click-functionality, and affording control over polarity, hydrophilicity and biocompatibility.
Experimental section
Materials & methods
Potassium ethyl xanthogenate (96%), 2-bromoethanol (95%), methacryloyl chloride (98%), methyl acrylate (99%), pentafluorophenol (99%), 1,4-butanediol diacrylate (90%), tributylphosphane (93.5%), hydrazine 1.0 M in THF, triethylamine (TEA, 99%), ethanolamine, propargylamine (98%), 3-amino-1,2-propanediol (1-aminoglycerol, 97%), di-isopropylethylamine (99.5%), CuI (99.9%), HIS-Select® Nickel Affinity Gel, L-gluthathione reduced (>99%), L-tyrosine methyl ester (>98%), glutathione-(glycine-13C2,15N) trifluoroacetate salt and azide Alexa Fluor 488 (≥90%) were purchased from Sigma Aldrich. Hexa His tag peptide (99%) was purchased from APExBIO and L-(+)-α-alanine (>99%) from J.T. Baker. Lissamine™ rhodamine B ethylenediamine was obtained from Invitrogen, ThermoFischer Scientific. Dichloromethane (100%), hexane (100%), methanol (100%) and tetrahydrofuran (THF, 100%) were purchased from LPS B.V. Acetone (100%), 1,4-dioxane (99+%) and dimethylformamide (DMF, 100%) were purchased from VWR and chloroform (100%) was purchased from Merck. Dichloromethane, 1,4-dioxane, THF and DMF were dried were dried over 3 Å molecular sieves for at least 24 hours prior to use. 4-Cyanopentanoic acid dithiobenzoate (CPADB) was synthesized following a literature procedure, purified using silica chromatography (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 v/v ethyl acetate
3 v/v ethyl acetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) hexane) and stored at −20 °C.61 2-(Ethyl xanthate) ethyl methacrylate (XMA) was synthesized as previously reported.62,63 SnakeSkin™ dialysis tubing (10 K MWCO) from ThermoFisher was employed for dialysis and Macrosep® Advance centrifugal devices from Pall were used for desalting after affinity chromatography.
hexane) and stored at −20 °C.61 2-(Ethyl xanthate) ethyl methacrylate (XMA) was synthesized as previously reported.62,63 SnakeSkin™ dialysis tubing (10 K MWCO) from ThermoFisher was employed for dialysis and Macrosep® Advance centrifugal devices from Pall were used for desalting after affinity chromatography.
Phosphate buffered saline (PBS, pH 7.4), trypsin and 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI, 98%), were purchased from Sigma Aldrich. EndoGro-11 LS Complete Culture Media Kit (SCME-BM and SCME 001) containing EndoGro™ Basal Medium, 5 ng mL−1 rh EGF, 10 mM L-glutamine, 1.0 μg mL−1 hydrocortisone hemisuccinate, 0.75 U mL−1 heparin sulfate, 50 μg mL−1 ascorbic acid and 5% FBS (Fetal Bovine Serum), collagen I and hCMEC/D3 cells were purchased from Merck Millipore. Live Cell Imaging solution (HEPES buffered physiological saline pH 7.4 by life technologies), was purchased from Thermo fisher.
1H-NMR (400 MHz), 13C NMR (101 MHz) and 19F-NMR (376 MHz) spectra were recorded on a Bruker 400 spectrometer. Size exclusion chromatography (SEC) analysis was performed on a Waters e2695 Separations Module equipped with an Agilent PLgel 5 μm MIXED-D 300 × 7.5 mm column and Waters photodiode array detector (PDA 2998), fluorescence detector (FLR 2475) and refractive index detector (RI 2414). Chloroform and DMF were employed as eluent with molecular weights calibrated against linear polystyrene and PEO/PEG, respectively. Dynamic light scattering (DLS) and zeta potential measurements were performed on a Malvern Instruments Zetasizer ZS in chloroform, DMSO, DMF and milliQ water. Samples for SEC and DLS were filtered using a GE Healthcare Whatman SPARTAN 13/0.2 RC 0.2 μm syringe filter prior to measurements. Transmission Electron Microscopy (TEM) was performed on a Philips CM300ST-FEG Transmission Electron Microscope 300 kV equipped with GATAN Ultrascan1000 (2k × 2k CCD camera) and GATAN Tridiem energy filter (2k × 2k CCD camera). For the sample preparation, 5 μL of a 0.5 mg mL−1 solution in milliQ was casted on copper grids and incubated for 30 seconds. Excess solution was removed by filter paper. Subsequently, the sample was stained by adding 5 μL of a 1% (w/v) uranyl acetate solution for 30 seconds, followed by removal of the excess solution by filter paper. For nickel affinity chromatography, a column loaded with 7.5 mL HIS-select Ni affinity gel was washed with 15 mL demi water and 15 mL washing buffer prior to usage. Washing buffer: 50 mM Na2HPO4, 150 mM NaCl (pH 8); elution buffer: 50 mM Na2HPO4, 300 mM NaCl, 250 mM imidazole (pH 8).
Monomers
Pentafluorophenyl methacrylate (PFPMA) was prepared following a modified literature procedure.55 Pentafluorophenol (15.0 g, 81.5 mmol) was dissolved in dry dichloromethane (150 mL) under nitrogen atmosphere. The solution was cooled to 0 °C, followed by addition of TEA (12.5 mL, 89.6 mmol, 1.1 eq.), and methacryloyl chloride (7.5 mL, 77.0 mmol, 0.95 equivalents) was added dropwise to the mixture over 5 minutes. The mixture was stirred overnight, while left to warm up to room temperature. After removal of salts by filtration, the reaction mixture was washed with demineralized water (3 × 50 mL), and the combined organic phase was dried over MgSO4 and concentrated under reduced pressure. PFPMA (11.6 g, 46.0 mmol, 57% isolated yield) was obtained as a clear liquid by distillation under reduced pressure in the presence of hydroquinone (6 × 10−2 mbar, 37 °C). The product was stored with hydroquinone as inhibitor at 4 °C.1H NMR (400 MHz, CDCl3) δH: 6.45 (m, 1H), 5.91 (m, 1H), 2.09 (s, 3H). 19F NMR (400 MHz, CDCl3) δF: −152.7 (m, 2F), −158.2 (td, 1F), −162.5 (tt, 2F).
PFP-polymer
Prior to polymerization, PFPMA was filtered over neutral alumina to remove inhibitor. PFPMA and XMA were copolymerized in molar ratios of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. In a typical polymerization experiment, PFPMA (3.38 g, 13.4 mmol), XMA (0.35 g, 1.52 mmol), RAFT agent CPADB (12.5 mg, 0.045 mmol) and AIBN (1.47 mg, 0.009 mmol) were combined with 1,4-dioxane (6 mL) in a polymerization flask fitted with a rubber septum. After purging for 15 minutes with nitrogen, the flask was placed in a preheated oil bath at 80 °C. After reaching 60–70% conversion, dichloromethane (2 mL) was added to the cooled flask and the polymer was precipitated twice in cold n-hexane.
1. In a typical polymerization experiment, PFPMA (3.38 g, 13.4 mmol), XMA (0.35 g, 1.52 mmol), RAFT agent CPADB (12.5 mg, 0.045 mmol) and AIBN (1.47 mg, 0.009 mmol) were combined with 1,4-dioxane (6 mL) in a polymerization flask fitted with a rubber septum. After purging for 15 minutes with nitrogen, the flask was placed in a preheated oil bath at 80 °C. After reaching 60–70% conversion, dichloromethane (2 mL) was added to the cooled flask and the polymer was precipitated twice in cold n-hexane.
1H NMR (400 MHz, acetone-d6) δH: 4.71–4.50 (br), 4.41–4.19 (br), 3.65–3.37 (br), 2.78–2.18 (br), 1.77–1.06 (br). 19F NMR (400 MHz, acetone-d6) δF: −152.1 (br), −159.6 (br), −164.5 (br).
PFP-SCNP formation
In a typical experiment, PFP-Polymer (600 mg, 0.24 mmol XMA-units, Mw = 42 kDa) was dissolved in THF (12 mL) and purged with nitrogen for 15 minutes, followed by addition of ethanolamine (87.0 μL, 1.92 mmol, 6 eq.), or alternatively hydrazine (480.0 μL, 0.48 mmol, 1.0 M in THF, 2 eq.), resulting in immediate loss of pink color. The solution was stirred for 150 minutes, followed by precipitation in cold methanol. After centrifugation (10 minutes × 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm), the deprotected polymer was redissolved in 12 mL of THF.
000 rpm), the deprotected polymer was redissolved in 12 mL of THF.
1H NMR (400 MHz, acetone-d6) δH: 4.71–4.50 (br), 4.41–4.19 (br), 3.65–3.37 (br), 2.78–2.18 (br), 1.77–1.06 (br).
The deprotected polymer was filtered and under vigorous stirring slowly dropped into a nitrogen purged dichloromethane solution (120 mL), containing 1,4-butanediol diacrylate (50.0 μL, 0.24 mmol, 1 eq.) and tributylphosphine (11.8 μL, 0.05 mmol, 0.2 eq.). The solution was stirred for another 150 minutes, after which methyl acrylate (0.5 mL, 5.52 mmol, 23 eq.) was added. The solution was stirred overnight and concentrated under reduced pressure. PFP-SCNPs were isolated by precipitation twice into cold methanol as a white solid.
1H NMR (400 MHz, acetone-d6) δH: 7.66–7.04 (br), 4.53–3.99 (br), 3.75–3.48 (br), 3.45–3.15 (br), 2.70–2.25 (br), 1.79–1.00 (br). 19F NMR (400 MHz, acetone-d6) δF: −152.2 (br), −159.5 (br), −164.4 (br).
PFP-SCNP functionalization
In all functionalization experiments, PFP-SCNPs were dissolved in a solvent compatible with the reagents and products under nitrogen atmosphere, followed by addition of TEA and a primary amine (see Table 1). The solution was heated to 45 °C and stirred until full conversion was observed by 19F NMR measurements.| Name | Functional amine | Aminoglycerol modification (mol%) | Reaction solvent | r H (nm) | Zeta potential (mV) |
|---|---|---|---|---|---|
| DLS measurements were performed in DMF, zeta potential measurements in water with 10 wt% NaCl.a Measured in chloroform.b Measured in DMSO. | |||||
| SCNP-F1 | Butylamine | 0 | THF | 10.0a | n.d. |
| SCNP-F2 | Aminoglycerol | 100 | THF/DMSO | 7.2b | −13.3 |
| SCNP-F3 | Propargylamine | 90 | THF | 7.3 | n.d. |
| SCNP-F4 | Tyrosine methyl ester | 95 | THF | 9.2 | −10.9 |
| SCNP-F5 | L-Alanine | 70 | DMSO/PBS | 9.4 | −23.1 |
| SCNP-F6 | L-Glutathione | <100 | DMSO/PBS | n.d. | n.d. |
| SCNP-F7 | His6-peptide | <100 | DMSO/PBS | 7.7 | −19.9 |
| SCNP-F8 | Rhodamine ethylenediamine | >70 | THF/DMSO | 8.8 | −16.1 |
For example, for SCNP-F2, 175 mg of PFP-SCNP (0.63 mmol PFPMA units) was dissolved in 4 mL THF. Under nitrogen atmosphere, TEA (265 mg, 2.52 mmol, 4 eq.) in 3 mL DMSO was added to the solution, followed by 1-aminoglycerol (575 mg, 6.30 mmol, 10 eq.) in 6 mL DMSO. The reaction was stirred overnight at 45 °C, followed by dialysis against H2O. Functionalized SCNPs were isolated by lyophilization.
For SCNP-F5–F7, PFP-SCNPs were first partially reacted with 1-aminoglycerol. For example, for SCNP-F5, a solution of 75 mg PFP-SCNP (0.25 mmol PFP) and 140 μL TEA in 7 mL THF was nitrogen purged and heated to 45 °C. 1-Aminoglycerol (20 mg, 0.85 eq.) in 6 mL DMSO was added and the heated solution was stirred overnight. Full amine conversion of the reaction mixture was determined by 19F NMR measurements. Subsequently, L-alanine (H NMR and 19F NMR measurements and 221 mg, 2.5 mmol, 10 eq.) in 3 mL PBS were added and the mixture was stirred overnight. Subsequently, 1-aminoglycerol (107 mg, 4.5 eq., 1.12 mmol) in 2 mL DMSO were added. After full conversion of the PFP moieties, the product was dialysed against water and lyophilized.
SCNP-F1: 1H NMR (400 MHz, CDCl3) δH: 6.96–4.90 (br), 4.46–3.94 (br), 3.85–3.58 (br), 3.25–2.85 (br), 1.81–1.63 (br), 1.55–1.17 (br), 1.01–0.76 (br).
SCNP-F2: 1H NMR (400 MHz, DMSO-d6) δH: 7.77–7.09 (br), 4.99–4.67 (br), 4.66–4.30 (br), 3.81–3.44 (br), 3.04–2.65 (br), 2.31–1.41 (br), 1.40–0.53 (br).
13C NMR (101 MHz, DMSO-d6) δC: 177.0 (br), 70.0 (br), 69.9 (br), 68.6 (br), 64.1 (br), 44.8 (br), 43.5 (br), 42.1 (br), 29.0 (br), 17.8 (br).
SCNP-F3: 1H NMR (400 MHz, DMSO-d6) δH 7.76–6.95, 5.82–5.73 (br), 5.63–5.43 (br), 5.38–5.20 (br), 5.14–4.15 (br), 4.14–3.87 (br), 3.04–2.70 (br), 2.31–2.12 (br), 2.16–0.50 (br).
SCNP-F4: 1H NMR (400 MHz, DMSO-d6) δH: 8.02–6.90 (br), 4.99–4.67 (br), 4.66–4.30 (br), 3.81–3.44 (br), 3.04–2.65 (br), 2.31–1.41 (br), 1.40–0.53 (br).
SCNP-F5: 1H NMR (400 MHz, DMSO-d6) δH: 12.82–11.89 (br), 8.38–6.94 (br), 5.50–5.18 (br), 5.14–4.22 (br), 4.18–3.94 (br), 3.05–2.86 (br), 2.81–2.62 (br), 2.29–2.10 (br), 2.10–1.83(br), 1.70–1.57 (br), 1.56–1.40 (br), 097–0.60 (br).
SCNP-F6: 1H NMR (400 MHz, DMSO-d6) δH: 13.00–11.06 (br), 8.08–6.95 (br), 6.87–6.65 (br), 5.40–5.18 (br), 5.16–4.28 (br), 4.16–3.82 (br), 3.85–3.07 (br), 3.09–2.65 (br), 2.30–2.10 (br), 2.08–1.85 (br), 1.69–1.58 (br), 1.56–1.37 (br), 1.37–0.56 (br).
13C NMR (101 MHz, DMSO-d6) δC: 177.1 (br), 171.4 (br), 69.9 (br), 68.8 (br), 64.2 (br), 44.6 (br), 43.1 (br), 41.7 (br), 28.9 (br), 17.5 (br).
SCNP-F7: 1H NMR (400 MHz, DMSO-d6) δH: 12.83–11.68 (br), 8.21–6.95 (br), 5.55–5.28 (br), 5.10–5.01 (br), 4.98–4.30 (br), 4.14–3.90 (br), 3.93–3.06 (br), 3.05–2.72 (br), 2.09–1.84 (br), 1.82–1.69 (br),1.66–1.57 (br), 1.57–1.42 (br), 1.40–0.69 (br), 0.67–0.60 (br).
Nickel affinity chromatography
The nickel affinity column was loaded with 10 mg of SCNP-F7 or SCNP-F8 in buffer solution and incubated on a roller bank overnight. The fractions were subsequently eluted with washing buffer (15 mL) and elution buffer (2 × 15 mL). Fractions were desalted using centrifugal desalting devices and lyophilized.Fluorescent labeling via click conjugation
To a solution of SCNP-F3 (10 mg, 6.4 μmol alkyne units) in 3 mL THF nitrogen purged, azide Alexa Fluor 488 (0.5 mg in 2 mL DMF), 2 μL N,N-diisopropylethylamine and 1.2 mg CuI in 1.2 mL DMF were added. The solution was stirred overnight at 60 °C. After isolation by dialysis and lyophilisation, SCNP-F3a was obtained as an orange solid.Cytotoxicity assay
hCMEC/D3 cells (passage 30) were seeded on collagen I coated 96-well plates at 10 × 103 cells per well in 100 μL EndoGro medium and incubated for 24 h at 37 °C in a humidified 5% CO2-containing atmosphere. SCNP-F2 was dissolved in EndoGro medium and filtered with a 0.2 μm filter. The stock solution was diluted to the desired concentrations (500, 200, 100, 50, 10 and 1 μg mL−1) and 100 μL was added to the cells. The measurements were performed in triplicates. For the reference and the positive control, 100 μL of EndoGro medium was added. The medium of the positive control was aspirated 30 min prior to the assay and replaced by 70% methanol. After 24 and 48 h of incubation, the cells were washed with HEPES buffer and 100 μL of resazurin sodium salt (440 mM) in PBS was added to the cells. The fluorescence intensity was analyzed after 4 h for hCMEC/D3 cells incubation at 37 °C in a humidified 5% CO2-containing atmosphere with a Tecan infinite m200 plate reader at an excitation and emission of 560/590 nm (gain of 70).Results and discussion
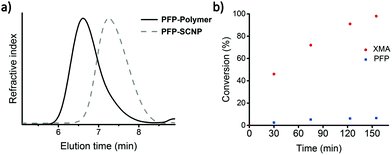
Protected thiol monomer (XMA)63 and pentafluorophenyl methacrylate monomer (PFPMA)55 were synthesized according to literature procedures and copolymerized via RAFT polymerization. The polymerization followed first order kinetics and both monomers were consumed at comparable rates as observed by 1H NMR spectroscopy, yielding copolymers (PFP-polymer) with PDI ∼ 1.4, as determined by size exclusion chromatography (SEC) measurements (Fig. 1 and Fig. S1†).PFP-SCNP formation was carried out via thiol-Michael addition crosslinking as reported previously.62 Full deprotection of the xanthate moieties in precursor polymer with ethanolamine was confirmed by 1H NMR and 19F NMR spectroscopy (Fig. 1b and Fig. S2†). The reaction proceeded more slowly than in earlier investigated XMA copolymers, but nonetheless remained much favored over amine substitution of PFP groups under the selected reaction conditions.39,62 Within 2.5 h, the xanthate moieties are fully deprotected as observed by the disappearance of signals corresponding to the xanthate moiety at 4.6 ppm and the shift of the CH2-S signal in 1H NMR spectroscopy (Fig. S2†). At the same time, the PFP moieties are functionalized only to a limited extend (∼7%), as observed by 19F NMR spectroscopy (data not shown).
Intramolecular crosslinking of the deprotected PFP-Polymer with 1,4-butanediol diacrylate, followed by endcapping of residual thiol moieties with methyl acrylate, yielded PFP-SCNPs, displaying 30–65% size reductions in comparison to the polymer precursor as determined by SEC analysis (Scheme 1 and Fig. 1a). The extent of size reduction increased with increasing precursor chain length (Fig. S3†). DLS measurements revealed particles of approximately 14 nm in diameter. Potential residual thiols were reacted with methyl acrylate. Additional signals of the crosslinker (signals d and f in Fig. 2) and of the methyl acrylate endcapper (signal e in Fig. 2) are identified in the 1H NMR spectra of PFP-SCNPs. The 19F NMR signal corresponding to the ortho-fluoro atoms displays a broadening upon crosslinking, which is not observed for the meta- and para-fluoro peaks (see Fig. S4†). This peak broadening might be assigned to increased crowding around the pentafluorophenyl rings upon chain compaction. The restricted mobility may lead to additional line-broadening of the ortho-fluorine atoms. Furthermore, increased dipole–dipole relaxation between the fluorine and hydrogen nuclei due to their increased vicinity upon compaction further contributes to the increased signal width.64
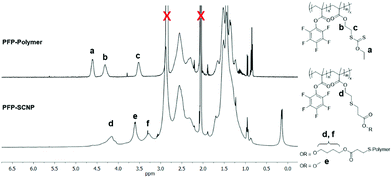 | ||
| Fig. 2 1H NMR spectra of PFP-polymer (top) and PFP-SCNP (below) in acetone-d6, indicating the appearance of crosslinker and endcap signals. | ||
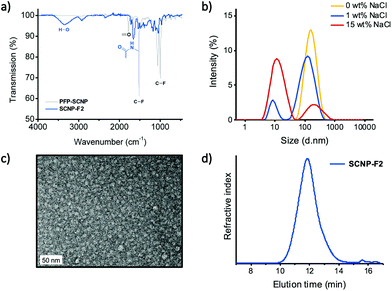
PFP-SCNPs were functionalized with n-butyl amine (SCNP-F1) to achieve acrylamides, which are known for their thermoresponsiveness.65,66 Complete functionalization was confirmed by disappearance of the signals in 19F NMR measurements (Fig. S5†), whereas in the 1H NMR spectrum n-butyl amide signals appeared (Fig. S6†). In addition, FT-IR measurements further confirmed PFP substitution by the disappearance of signals at 1000 cm−1 and 1515 cm−1, assigned to C–F bonds and the ring vibration of the PFP moiety, as well as by the emergence of signals in the amide region at 1658 cm−1.
Experiments with PFP substitution by ethanolamine proved insufficient to grant water-solubility to the nanoparticles. PFP-SCNPs were therefore reacted with 1-aminoglycerol to obtain water-soluble and biocompatible SCNPs (SCNP-F2), which was confirmed similarly by FT-IR and 1H NMR spectroscopy (Fig. 3a and Fig. S7† respectively). Surprisingly, DLS measurements indicated structures of 100 nm in diameter for SCNP-F2, as depicted in Fig. 3b, whereas the size of SCNP-F1 remained unaltered after functionalization (∼10 nm). Upon sodium chloride addition, however, two populations of 100 nm and 10 nm became apparent. With increasing amounts of salt, the 10 nm species became more pronounced, suggesting the earlier observed larger aggregates formed as a result of electrostatic interactions between the amide diols. TEM imaging of SCNP-F2 confirmed particles of 10 nm in diameter (Fig. 3c). Furthermore, SEC elution displayed a single size population for SCNP-F2 (Fig. 3d). The relative sizes of PFP-SCNP and SCNP-F2 cannot be compared by SEC as they are obtained by relative calibrations in different solvent systems with different standards.
As a first step towards future biomedical applications, cellular toxicity of SCNP-F2 was evaluated on the brain endothelial cell line hCMEC/D3. No significant decreases in metabolic activity were observed, even after 48 hours of incubation with increasing concentrations of SCNPs up to 500 μg mL−1 (Fig. S8†). These preliminary results indicate the potential of aminoglycerol-functionalized SCNPs for biological use and point to further investigations into cellular compatibility and intracellular localization.
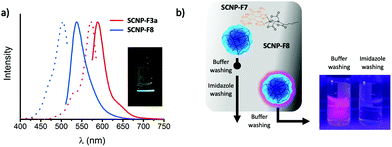
In order to utilize SCNPs in bioconjugations, propargylamine was conjugated onto the PFP-SCNPs as an orthogonal handle for efficient click-functionalization (SCNP-F3). Given that click chemistry can be performed on these SCNPs, the alkyne pendant may for example be utilized to add radioactive labels for tracking the particles in in vivo studies.671H NMR spectroscopy confirms signals of the alkyne bonds at 2.2 and 3.9 ppm, respectively (see Fig. S9†), indicating replacement of approximately 10% of the PFP moieties by propargyl amine. The presence and accessibility of the alkyne moieties were verified by a copper catalyzed azide–alkyne Huisgen click reaction with azide-functional Alexa Fluor 488. The click reaction resulted in an orange, fluorescent solid and, in accordance with rhodamine conjugation, SEC chromatography revealed UV-vis and fluorescence signals at the elution time of the click product (SCNP-F3a; Fig. 4a and Fig. S10a†).
With the aim of approaching protein-mimicry, functionalization with amino acids was pursued next, as summarized in Scheme 2. PFP-SCNPs were reacted with 0.05 molar equivalents of tyrosine methyl ester, an amino acid with a protected carboxylic acid, and accordingly, 5% conversion of PFP-esters was observed by 19F NMR spectroscopy (Fig. S11†). Subsequently, excess 1-aminoglycerol was added to yield water-soluble particles SCNP-F4.
In order to modify PFP-SCNPs with natural amino acids such as alanine, PFP-SCNPs were first partially functionalized with 1-aminoglycerol to render the particles water-soluble and then reacted with alanine in a buffered solution at pH = 7 to minimize potential hydrolysis. The sequential addition of 1-aminoglycerol and alanine was followed by 1H and 19F NMR spectroscopy revealing 30% alanine substitution, which is in agreement with the prospected degree of functionalization. Excess 1-aminoglycerol was added after conjugation of the respective amino acids to ensure full conversion of PFP moieties, yielding SCNP-F5. Successful conjugation of alanine and 1-aminoglycerol onto the SCNPs was confirmed by 1H NMR spectroscopy revealing among others new amide signals, as well as a carboxylic acid signal at 12.5 ppm (Fig. S13†). In control experiments without alanine, no significant PFP hydrolysis was detected. Interestingly, the zeta potential of SCNP-F5 decreased to −23 mV as compared to −13 mV for the fully 1-aminoglycerol functionalized SCNP-F2 (Table 1), due to the presence of carboxylate groups.
Hence, the sequential addition of functional amines may be conducted to facilitate conjugation of non-water-soluble components or to economize the use of precious substituents. Analogously, the peptides glutathione (GSH) and hexa-histidine (His6) were added onto partially modified aminoglycerol SCNPs, yielding SCNPs-F6 and SCNP-F7, respectively. His6-functionalized SCNP-F7 showed comparable size and zeta potential to the alanine-functional SCNP-F5. Carboxylic acid signals became apparent after the functionalization with peptides, but no other distinct signals corresponding to the conjugated peptides could be identified by 1H NMR spectroscopy, due to strongly overlapping amide, alkane and water signals. In order to confirm GSH conjugation to PFP-SCNPs, doubly 13C-labeled GSH was incorporated (2%) in a similar manner. 13C NMR spectroscopy revealed two additional signals at 41 and 172 ppm as compared to SCNP-F2, corresponding to the 13C-enriched carbonyl and α-carbon respectively, thereby confirming successful conjugation (Fig. S14†).
The sequential substitution of PFP-ester moieties was further utilized to directly equip SCNPs with a fluorescent label. Rhodamine ethylenediamine was added to SCNPs with 70% 1-aminoglycerol substituted PFP moieties to yield SCNP-F8. Successful addition of the dye was confirmed by new UV-vis and fluorescence signals in SEC measurements at the elution volume corresponding to the SCNPs (Fig. 4a and S10b†).
Affinity chromatography with nickel-nitrilotriacetic acid (Ni2+-NTA) columns is a well-established method for purification of recombinant proteins from cell lysate.68 Proteins modified with a polyhistidine-tag (usually His6-tag) display strong affinity to the nickel column and can thus be isolated from other proteins. With the aim of validating the integrity of peptides after coupling to the nanoparticles, SCNP-F7, which contains His6 peptides in addition to 1-aminoglycerol moieties, was tested for affinity towards Ni2+-NTA affinity columns. Rhodamine-functionalized SCNPs (SCNP-F8) served as a negative control. SCNP-F8 fully eluted from the Ni2+-NTA column already upon flushing with the washing buffer (see Fig. 4b) and no further rhodamine elution was observed when flushing the column with imidazole-containing buffer (elution buffer). On the contrary, 1H NMR analysis on the lyophilized fractions eluting from the SCNP-F7 loaded Ni2+-NTA column revealed that SCNP-F7 is only released upon eluting with an imidazole-containing buffer. Consequently, the His6-conjugated SCNP-F7 interacts well with the affinity column, confirming successful peptide conjugation.
Conclusions
In this work, we demonstrate the development of PFP-SCNPs, based on covalent intramolecular chain collapse and the subsequent modification of these nanoparticles yielding highly modular, water-soluble SCNPs. Facile amine conjugation provides an easy way towards peptide-functionalization for more advanced biomedical applications. Furthermore, conjugation of a His6-tag peptide to the SCNPs, enabled to transfer a classical protein purification method to polymers. Besides direct addition of amino acids and peptides, also modification with simple, functional amines, such as ethanolamine as chemical analogue to serine, aids in mimicking protein appearance.In principle, nearly any physicochemical function may be added to the SCNPs via amine post-formation in an easy and efficient manner. Additionally, parallel functionalization may be performed on a single type of nanoparticles, offering the possibility to alter particle functionality, without inherently changing particle size and dispersity, which would greatly facilitate comparative cell experiments, for example to study ligand density effects. Addition of alkyne groups onto the SCNPs enables click functionalization, giving way to radiolabeling of SCNPs for in vivo studies. Current work is focused on the effects of peptide conjugation on SCNP uptake and biodistribution behavior. As a protein mimic, amino acid decorated SCNPs may appropriate some of the numerous features proteins display, such as biocompatibility and receptor binding, greatly extending the potential and understanding of SCNPs in biomedical applications.
Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
Funding from the Netherlands Organization for Health Research and Development (ZonMw, project number 733050304) and through the EuroNanoMed III research program (Ref. EURO-NANOMED2017-178) is gratefully acknowledged. This research was funded by Alzheimer Netherlands and co-funded by the PPP Allowance made available by Health∼Holland, Top Sector Life Sciences & Health, to stimulate public-private partnerships. We further thank Regine van der Hee for support in cell experiments and Dr E. G. Keim for TEM measurements.Notes and references
- C. J. Hawker and K. L. Wooley, The Convergence of Synthetic Organic and Polymer Chemistries, Science, 2005, 309(5738), 1200 CrossRef CAS PubMed.
- B. E. Ramakers, J. C. van Hest and D. W. Lowik, Molecular tools for the construction of peptide-based materials, Chem. Soc. Rev., 2014, 43(8), 2743–2756 RSC.
- J. P. Cole, A. M. Hanlon, K. J. Rodriguez and E. B. Berda, Protein-like structure and activity in synthetic polymers, J. Polym. Sci., Part A: Polym. Chem., 2017, 55(2), 191–206 CrossRef CAS.
- M. A. Gauthier and H.-A. Klok, Polymer–protein conjugates: an enzymatic activity perspective, Polym. Chem., 2010, 1(9), 1352–1373 RSC.
- J.-F. Lutz, J.-M. Lehn, E. W. Meijer and K. Matyjaszewski, From precision polymers to complex materials and systems, Nat. Rev. Mater., 2016, 1(5), 16024 CrossRef CAS.
- H.-A. Klok, Peptide/Protein−Synthetic Polymer Conjugates:Quo Vadis, Macromolecules, 2009, 42(21), 7990–8000 CrossRef CAS.
- M. ler Meißig, S. Wieczorek, N. ten Brummelhuis and H. G. Börner, Synthetic Aspects of Peptide– and Protein–Polymer Conjugates in the Post-click Era. in Bio-inspired Polymers, 2016, ch. 1, pp. 1–30 Search PubMed.
- A. C. Obermeyer and B. D. Olsen, Synthesis and Application of Protein-Containing Block Copolymers, ACS Macro Lett., 2015, 4(1), 101–110 CrossRef CAS.
- A. Abuchowski, T. van Es, N. C. Palczuk and F. F. Davis, Alteration of immunological properties of bovine serum albumin by covalent attachment of polyethylene glycol, J. Biol. Chem., 1977, 252(11), 3578–3581 CAS.
- P. Wilson, Synthesis and Applications of Protein/Peptide-Polymer Conjugates, Macromol. Chem. Phys., 2017, 218(9), 1600595 CrossRef.
- Q. Liu, J. Wang and B. J. Boyd, Peptide-based biosensors, Talanta, 2015, 136, 114–127 CrossRef CAS PubMed.
- J. B. Beck, K. L. Killops, T. Kang, K. Sivanandan, A. Bayles, M. E. Mackay, K. L. Wooley and C. J. Hawker, Facile Preparation of Nanoparticles by Intramolecular Crosslinking of Isocyanate Functionalized Copolymers, Macromolecules, 2009, 42(15), 5629–5635 CrossRef CAS PubMed.
- E. Harth, B. V. Horn, V. Y. Lee, D. S. Germack, C. P. Gonzales, R. D. Miller and C. J. Hawker, A Facile Approach to Architecturally Defined Nanoparticles via Intramolecular Chain Collapse, J. Am. Chem. Soc., 2002, 124(29), 8653–8660 CrossRef CAS PubMed.
- F. Lo Verso, J. A. Pomposo, J. Colmenero and A. J. Moreno, Multi-orthogonal folding of single polymer chains into soft nanoparticles, Soft Matter, 2014, 10(27), 4813–4821 RSC.
- I. Perez-Baena, I. Asenjo-Sanz, A. Arbe, A. J. Moreno, F. Lo Verso, J. Colmenero and J. A. Pomposo, Efficient Route to Compact Single-Chain Nanoparticles: Photoactivated Synthesis via Thiol–Yne Coupling Reaction, Macromolecules, 2014, 47(23), 8270–8280 CrossRef CAS.
- A. Prasher, C. M. Loynd, B. T. Tuten, P. G. Frank, D. Chao and E. B. Berda, Efficient fabrication of polymer nanoparticles via sonogashira cross-linking of linear polymers in dilute solution, J. Polym. Sci., Part A: Polym. Chem., 2016, 54(1), 209–217 CrossRef CAS.
- A. M. Hanlon, R. Chen, K. J. Rodriguez, C. Willis, J. G. Dickinson, M. Cashman and E. B. Berda, Scalable Synthesis of Single-Chain Nanoparticles under Mild Conditions, Macromolecules, 2017, 50(7), 2996–3003 CrossRef CAS.
- H. Frisch, F. R. Bloesser and C. Barner-Kowollik, Controlling Chain Coupling and Single-Chain Ligation by Two Colours of Visible Light, Angew. Chem., Int. Ed., 2019, 58(11), 3604–3609 CrossRef CAS PubMed.
- J. Rubio-Cervilla, H. Frisch, C. Barner-Kowollik and J. A. Pomposo, Synthesis of Single-Ring Nanoparticles Mimicking Natural Cyclotides by a Stepwise Folding-Activation-Collapse Process, Macromol. Rapid Commun., 2019, 40(1), e1800491 CrossRef PubMed.
- Y. Zhou, Y. Qu, Q. Yu, H. Chen, Z. Zhang and X. Zhu, Controlled synthesis of diverse single-chain polymeric nanoparticles using polymers bearing furan-protected maleimide moieties, Polym. Chem., 2018, 9(23), 3238–3247 RSC.
- C. Song, L. Li, L. Dai and S. Thayumanavan, Responsive single-chain polymer nanoparticles with host–guest features, Polym. Chem., 2015, 6(26), 4828–4834 RSC.
- M. Artar, E. R. J. Souren, T. Terashima, E. W. Meijer and A. R. A. Palmans, Single Chain Polymeric Nanoparticles as Selective Hydrophobic Reaction Spaces in Water, ACS Macro Lett., 2015, 4(10), 1099–1103 CrossRef CAS.
- M. Seo, B. J. Beck, J. M. J. Paulusse, C. J. Hawker and S. Y. Kim, Polymeric Nanoparticles via Noncovalent Cross-Linking of Linear Chains, Macromolecules, 2008, 41(17), 6413–6418 CrossRef CAS.
- C. K. Lyon, A. Prasher, A. M. Hanlon, B. T. Tuten, C. A. Tooley, P. G. Frank and E. B. Berda, A brief user's guide to single-chain nanoparticles, Polym. Chem., 2015, 6(2), 181–197 RSC.
- M. Huo, N. Wang, T. Fang, M. Sun, Y. Wei and J. Yuan, Single-chain polymer nanoparticles: Mimic the proteins, Polymer, 2015, 66, A11–A21 CrossRef CAS.
- E. Blasco, B. T. Tuten, H. Frisch, A. Lederer and C. Barner-Kowollik, Characterizing single chain nanoparticles (SCNPs): a critical survey, Polym. Chem., 2017, 8(38), 5845–5851 RSC.
- B. V. Schmidt, N. Fechler, J. Falkenhagen and J. F. Lutz, Controlled folding of synthetic polymer chains through the formation of positionable covalent bridges, Nat. Chem., 2011, 3(3), 234–238 CrossRef CAS PubMed.
- D. Chao, X. Jia, B. Tuten, C. Wang and E. B. Berda, Controlled folding of a novel electroactive polyolefin via multiple sequential orthogonal intra-chain interactions, Chem. Commun., 2013, 49(39), 4178–4180 RSC.
- J. Lu, N. Ten Brummelhuis and M. Weck, Intramolecular folding of triblock copolymers via quadrupole interactions between poly(styrene) and poly(pentafluorostyrene) blocks, Chem. Commun., 2014, 50(47), 6225–6227 RSC.
- R. K. Roy and J. F. Lutz, Compartmentalization of single polymer chains by stepwise intramolecular cross-linking of sequence-controlled macromolecules, J. Am. Chem. Soc., 2014, 136(37), 12888–12891 CrossRef CAS PubMed.
- J. P. Cole, J. J. Lessard, K. J. Rodriguez, A. M. Hanlon, E. K. Reville, J. P. Mancinelli and E. B. Berda, Single-chain nanoparticles containing sequence-defined segments: using primary structure control to promote secondary and tertiary structures in synthetic protein mimics, Polym. Chem., 2017, 8(38), 5829–5835 RSC.
- T. Terashima, T. Mes, T. F. De Greef, M. A. Gillissen, P. Besenius, A. R. Palmans and E. W. Meijer, Single-chain folding of polymers for catalytic systems in water, J. Am. Chem. Soc., 2011, 133(13), 4742–4745 CrossRef CAS PubMed.
- I. Perez-Baena, F. Barroso-Bujans, U. Gasser, A. Arbe, A. J. Moreno, J. Colmenero and J. A. Pomposo, Endowing Single-Chain Polymer Nanoparticles with Enzyme-Mimetic Activity, ACS Macro Lett., 2013, 2(9), 775–779 CrossRef CAS.
- H. Rothfuss, N. D. Knofel, P. W. Roesky and C. Barner-Kowollik, Single-Chain Nanoparticles as Catalytic Nanoreactors, J. Am. Chem. Soc., 2018, 140(18), 5875–5881 CrossRef CAS PubMed.
- E. Huerta, P. J. Stals, E. W. Meijer and A. R. Palmans, Consequences of folding a water-soluble polymer around an organocatalyst, Angew. Chem., Int. Ed., 2013, 52(10), 2906–2910 CrossRef CAS PubMed.
- J. Rubio-Cervilla, E. Gonzalez and J. A. Pomposo, Advances in Single-Chain Nanoparticles for Catalysis Applications, Nanomaterials, 2017, 7(10), 341 CrossRef PubMed.
- Y. Bai, X. Hang, P. Wu, X. Feng, K. Hwang, J. M. Lee, X. Y. Phang, Y. Lu and S. C. Zimmerman, Chemical Control over Cellular Uptake of Organic Nanoparticles by Fine Tuning Surface Functional Groups, ACS Nano, 2015, 9(10), 10 CrossRef PubMed.
- A. B. Benito, M. K. Aiertza, M. Marradi, L. Gil-Iceta, T. Shekhter Zahavi, B. Szczupak, M. Jimenez-Gonzalez, T. Reese, E. Scanziani, L. Passoni, M. Matteoli, M. De Maglie, A. Orenstein, M. Oron-Herman, G. Kostenich, L. Buzhansky, E. Gazit, H. J. Grande, V. Gomez-Vallejo, J. Llop and I. Loinaz, Functional Single-Chain Polymer Nanoparticles: Targeting and Imaging Pancreatic Tumors in Vivo, Biomacromolecules, 2016, 17(10), 3213–3221 CrossRef CAS PubMed.
- A. P. P. Kroger, N. M. Hamelmann, A. Juan, S. Lindhoud and J. M. J. Paulusse, Biocompatible Single-Chain Polymer Nanoparticles for Drug Delivery-A Dual Approach, ACS Appl. Mater. Interfaces, 2018, 10(37), 30946–30951 CrossRef PubMed.
- A. P. P. Kroger and J. M. J. Paulusse, Single-chain polymer nanoparticles in controlled drug delivery and targeted imaging, J. Controlled Release, 2018, 286, 326–347 CrossRef PubMed.
- A. P. P. Kroger, M. I. Komil, N. M. Hamelmann, A. Juan, M. H. Stenzel and J. M. J. Paulusse, Glucose Single-Chain Polymer Nanoparticles for Cellular Targeting, ACS Macro Lett., 2019, 8(1), 95–101 CrossRef PubMed.
- J.ÉF. Radu, L. Novak, J. F. Hartmann, N. Beheshti, A.-L. Kjøniksen, B. Nyström and J. Borbély, Structural and dynamical characterization of poly-gamma-glutamic acid-based cross-linked nanoparticles, Colloid Polym. Sci., 2007, 286(4), 365–376 CrossRef.
- T. Akagi, P. Piyapakorn and M. Akashi, Formation of unimer nanoparticles by controlling the self-association of hydrophobically modified poly(amino acid)s, Langmuir, 2012, 28(11), 5249–5256 CrossRef CAS PubMed.
- P. Piyapakorn, T. Akagi, M. Hachisuka, T. Onishi, H. Matsuoka and M. Akashi, Structural Analysis of Unimer Nanoparticles Composed of Hydrophobized Poly(amino acid)s, Macromolecules, 2013, 46(15), 6187–6194 CrossRef CAS.
- S. K. Hamilton and E. Harth, Molecular Dendritic Transporter Nanoparticle Vectors Provide Efficient Intracellular Delivery of Peptides, ACS Nano, 2009, 3(2), 402–410 CrossRef CAS PubMed.
- Y. Koda, T. Terashima, M. Sawamoto and H. D. Maynard, Amphiphilic/fluorous random copolymers as a new class of non-cytotoxic polymeric materials for protein conjugation, Polym. Chem., 2015, 6(2), 240–247 RSC.
- R. Gracia, M. Marradi, G. Salerno, R. Pérez-Nicado, A. Pérez-San Vicente, D. Dupin, J. Rodriguez, I. Loinaz, F. Chiodo and C. Nativi, Biocompatible single-chain polymer nanoparticles loaded with an antigen mimetic as potential anticancer vaccine, ACS Macro Lett., 2018, 7(2), 196–200 CrossRef CAS.
- L. M. Campos, K. L. Killops, R. Sakai, J. M. J. Paulusse, D. Damiron, E. Drockenmuller, B. W. Messmore and C. J. Hawker, Development of Thermal and Photochemical Strategies for Thiol−Ene Click Polymer Functionalization, Macromolecules, 2008, 41(19), 7063–7070 CrossRef CAS.
- A. Gruber, L. Navarro and D. Klinger, Reactive Precursor Particles as Synthetic Platform for the Generation of Functional Nanoparticles, Nanogels, and Microgels, Adv. Mater. Interfaces, 2020, 7(5), 1901676 CrossRef CAS.
- Y. Lee, J. Pyun, J. Lim and K. Char, Modular synthesis of functional polymer nanoparticles from a versatile platform based on poly(pentafluorophenylmethacrylate), J. Polym. Sci., Part A: Polym. Chem., 2016, 54(13), 1895–1901 CrossRef CAS.
- A. Gruber, D. Işık, B. B. Fontanezi, C. Böttcher, M. Schäfer-Korting and D. Klinger, A versatile synthetic platform for amphiphilic nanogels with tunable hydrophobicity, Polym. Chem., 2018, 9(47), 5572–5584 RSC.
- K. Han, R. Tiwari, T. Heuser and A. Walther, Simple Platform Method for the Synthesis of Densely Functionalized Microgels by Modification of Active Ester Latex Particles, Macromol. Rapid Commun., 2016, 37(16), 1323–1330 CrossRef CAS PubMed.
- R. Kakuchi and P. Theato, Post-Polymerization Modifications via Active Esters, Funct. Polym. Post-Polym. Modif., 2013, 45–64 CAS.
- A. Das and P. Theato, Activated Ester Containing Polymers: Opportunities and Challenges for the Design of Functional Macromolecules, Chem. Rev., 2016, 116(3), 1434–1495 CrossRef CAS PubMed.
- M. Eberhardt, R. Mruk, R. Zentel and P. Théato, Synthesis of pentafluorophenyl(meth)acrylate polymers: New precursor polymers for the synthesis of multifunctional materials, Eur. Polym. J., 2005, 41(7), 1569–1575 CrossRef CAS.
- H. Son, J. Ku, Y. Kim, S. Li and K. Char, Amine-Reactive Poly(pentafluorophenyl acrylate) Brush Platforms for Cleaner Protein Purification, Biomacromolecules, 2018, 19(3), 951–961 CrossRef CAS PubMed.
- M. Zamfir, P. Theato and J.-F. Lutz, Controlled folding of polystyrene single chains: design of asymmetric covalent bridges, Polym. Chem., 2012, 3(7), 1796–1802 RSC.
- Y. Liu, T. Pauloehrl, S. I. Presolski, L. Albertazzi, A. R. Palmans and E. W. Meijer, Modular Synthetic Platform for the Construction of Functional Single-Chain Polymeric Nanoparticles: From Aqueous Catalysis to Photosensitization, J. Am. Chem. Soc., 2015, 137(40), 13096–13105 CrossRef CAS PubMed.
- J. Chen, K. Li, J. S. L. Shon and S. C. Zimmerman, Single-Chain Nanoparticle Delivers a Partner Enzyme for Concurrent and Tandem Catalysis in Cells, J. Am. Chem. Soc., 2020, 142(10), 4565–4569 CrossRef CAS PubMed.
- J. Chen, J. Wang, K. Li, Y. Wang, M. Gruebele, A. L. Ferguson and S. C. Zimmerman, Polymeric “Clickase” Accelerates the Copper Click Reaction of Small Molecules, Proteins, and Cells, J. Am. Chem. Soc., 2019, 141(24), 9693–9700 CrossRef CAS PubMed.
- S. H. Thang, Y. K. Chong, R. T. A. Mayadunne, G. Moad and E. Rizzardo, A novel synthesis of functional dithioesters, dithiocarbamates, xanthates and trithiocarbonates, Tetrahedron Lett., 1999, 40(12), 2435–2438 CrossRef CAS.
- A. P. P. Kröger, R. J. E. A. Boonen and J. M. J. Paulusse, Well-defined single-chain polymer nanoparticles via thiol-Michael addition, Polymer, 2017, 120, 119–128 CrossRef.
- R. Nicolaÿ, Synthesis of Well-Defined Polythiol Copolymers by RAFT Polymerization, Macromolecules, 2011, 45(2), 821–827 CrossRef.
- J. T. Gerig, Fluorine NMR, in Biophys. Textb. Online, 2001, pp. 1–35 Search PubMed.
- J. Seuring and S. Agarwal, First Example of a Universal and Cost-Effective Approach: Polymers with Tunable Upper Critical Solution Temperature in Water and Electrolyte Solution, Macromolecules, 2012, 45(9), 3910–3918 CrossRef CAS.
- A. Gandhi, A. Paul, S. O. Sen and K. K. Sen, Studies on thermoresponsive polymers: Phase behaviour, drug delivery and biomedical applications, Asian J. Pharm. Sci., 2015, 10(2), 99–107 CrossRef.
- L. S. Campbell-Verduyn, L. Mirfeizi, A. K. Schoonen, R. A. Dierckx, P. H. Elsinga and B. L. Feringa, Strain-promoted copper-free “click” chemistry for 18F radiolabeling of bombesin, Angew. Chem., Int. Ed., 2011, 50(47), 11117–11120 CrossRef CAS PubMed.
- J. A. Bornhorst and J. J. Falke, Purification of Proteins Using Polyhistidine Affinity Tags, Methods Enzymol., 2000, 326, 245–254 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0py00922a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2020 |