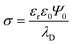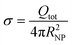Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Modelling bionano interactions and potential health risks for environmental nanoplastics: the case of functionalized polystyrene
Julia Subbotina *,
Oran McElligott and
Vladimir Lobaskin
*,
Oran McElligott and
Vladimir Lobaskin
School of Physics, College of Science, University College Dublin, Dublin 4, Ireland. E-mail: yulia.subbotina1@ucd.ie
First published on 28th January 2026
Abstract
Micro- and nanoplastic pollution has been raising increasing concern due to their adverse environmental and potential human health effects. The impact of plastic particulates, especially in their nanoforms, on the health of living organisms is not fully understood. Based on substantial evidence, it can be assumed that the key processes underlying the bioaccumulation and toxicity of nano-sized materials are controlled by bionano interactions, particularly through the formation of protein coronas. Understanding the composition of such biocoronas and the factors governing their formation can aid in material risk assessment and the development of safety measures. In this study, we report on novel parametrization of the UA/CoronaKMC coarse-grained multiscale approach for predicting protein corona composition that can be formed on pristine (PS) and modified forms (PS-NH2 and PS-COOH) of polystyrene nanoplastics in blood plasma. The reported methodology extends the use of the UA/CoronaKMC method for further implementation into digital machine-learning SSbD frameworks for pre-assessments of the nanotoxicity of novel polymers.
Environmental significanceMicro- and nanoplastics are significant environmental pollutants with increasing human exposure. While chemically inert, these particles pose potential health risks through environmental modifications and as carriers of toxic substances. Understanding nanoplastic surface properties' influence on bionano interactions and particle circulation is crucial. Limited information exists on plastic–biomolecule interactions due to polymer variability and diverse physicochemical characteristics of degraded particles, while experimental research costs create barriers. To advance nanosafety assessment of polymeric nanomaterial–protein interactions, we extended the previously presented multiscale simulation approach for modeling polymeric materials. This extension focuses on physics-based parametrization of protein–nanoparticle interfaces for noncrystalline polymers, particularly polystyrenes. The reported computational approach predicts protein corona compositions for the evaluation of polystyrene nanotoxicity and protein orientations on nanoparticles, modeling biomolecular adsorption based on bionano interface parameters. It can be used as a “Safe and Sustainable by Design” (SSbD) nanoinformatics toolkit for designing safer polymers. |
1 Introduction
To date, numerous polymeric materials have been introduced to the global market, and polystyrene (PS) consistently represents about 5% of the total annual global polymer production by volume.1 They offer significant advantages by providing cost-effective solutions for the production of various consumable goods. However, the increase in polymeric consumables combined with the inadequate waste management (note that global recycling rates are low 9%2 leads to the accumulation of enormous volumes of polymer waste in landfills and aquatic environments at the end of their life cycle.2–5 Without proper recycling practices, plastic waste degrades under exposure to environmental conditions initially to microparticles (microplastic) and subsequently to nanoparticles (nanoplastic).6 This process can be relatively fast, e.g., it might take less than two months to reach the nanoparticle (NP) state with sizes smaller than 250 nm for disposable PS-made coffee lids.7 The environmental spread of nanoplastics is undeniable. Their presence was recorded in aquatic ecosystems,8,9 in the atmosphere,10 and in the soil.11,12 It was even observed in the ice samples obtained from the remote Arctic and Antarctic regions.13,14 As a result of this widespread pollution, nanoplastics penetrate into the human body through various pathways: food (oral ingestion), water (absorption by the skin or oral ingestion), or the air (inhalation).15–17 Plastics, detected in human faeces,18 in human placenta (referred to as “plasticenta”),19 or in the brains of humans and other mammals20,21 point to the alarming scale of this exposure.A concerning consequence of nanoplastic pollution is caused by environmental degradation of polymers that produces not only different particle sizes but also numerous variations of surface chemistry, as a result of physical processes and reactions with chemicals co-located with polymeric waste.22–30 Surface modifications enhance the colloidal stability of the nanoplastics and can transform a relatively inert polymer bulk material into a distribution of “activated” NPs. Surface-modified NPs can be internalized by living organisms and pass through the blood–brain barrier (BBB) in a manner similar to more reactive inorganic NPs.31–37
The primary factor facilitating this process is the adsorption of proteins onto the hydrophobic surface of the NP as it enters the bloodstream. Exogenous agents are typically encountered by opsonins, such as antibodies or complement proteins, which bind to these agents, marking them for destruction by phagocytes and facilitating their clearance. However, during this process, opsonins compete with other blood proteins, and the ultimate composition of the protein layer on the NP's surface, known as the protein “corona”, will determine the fate of the NPs.38–41 This protein makeup, especially if enriched with dysopsonins, gives the NP a “new identity” that can help avoid the recognition process by the mononuclear phagocyte system (MPS).42,43 As a result, this competition may help the NP remain undetected by the MPS for longer, increasing its blood circulation times and its persistence.44
The impact of nanoplastic pollution on various living organisms has been extensively examined in the recent literature.45–51 Specifically, with regard to PS, research has demonstrated that its nanotoxicity can differ between organisms.52 In humans,53 particular emphasis was placed on the health of women and fetal development during pregnancy.54–57 Moreover, research has consistently demonstrated a robust correlation between metabolic disorders and exposure to PS NPs in both animal models and cellular systems.58–62
PS NPs have also been increasingly noted for their neurotoxic effects. There is increasing evidence from animal studies on the negative role of PS NP exposure in anxiety, depression, cognitive impairment, and neurodevelopmental disorders both in adults and after developmental or maternal exposure.63–67 The exposure time of PS NPs was shown to be irrelevant for health outcomes, as short-term exposure was associated with long-term cognitive decline, neuronal damage, and inflammation.68 PS NPs, specifically anionic ones, may exacerbate Parkinson's disease pathology by disrupting the gut–brain axis, facilitating α-synuclein aggregation, and inducing neuroinflammation and mitochondrial dysfunction.69–74
Understanding the factors governing nanotoxicity is crucial for predictive environmental toxicology as well as for developing safe biomedical applications based on PS NPs. The protein corona composition was shown to be a crucial determinant of nanotoxicity.75–77 It shapes how NPs interact with cells and organs, and ultimately modulates the NP's toxicity and immune responses. At the same time, protein corona composition is an imprint of the NP characterised by a set of intrinsic (material type, surface chemistry, size) and extrinsic (zeta potential, colloidal stability, and solubility) properties that are affected by specific environments (chemical compositions, pH, ionic strength).78–80 Manipulation of the intrinsic and extrinsic properties of NPs experimentally can help identify safer materials that degrade to NPs with the preferred corona composition. However, experimental assessments of many combinations of protein corona–NP systems might be a tedious task. Recent advances in modelling the protein corona facilitate the rational design of safer nanomaterials. These methodologies, ranging from multiscale physics-based modelling81–85 to machine-learning approaches,39,86–88 are revolutionising the material design and safety assessment.
In our previous work, we introduced and applied a multiscale United Atom (UA) method combined with a kinetic Monte Carlo approach to model protein corona compositions (CoronaKMC) and predict the nanotoxicity of crystalline materials with varying complexities as a function of the composition of the protein corona.89–93 In this study, we extend this methodology to include a polymeric material, PS. Our multiscale simulations are designed to model key molecular events of individual and collective protein–NP interactions by examining protein adsorption and biocorona formation on functionalized PS NPs with different charges. The adsorption of the five most prevalent proteins in blood plasma, namely serum albumin (HSA), fibrinogen (FG), immunoglobulin IgG (IGH), complement component 3 (C3), and apolipoprotein A1 (APO-AI), onto small, 10-nm PS NPs is modelled. New UA parameters for coarse-grain (CG) modelling of protein-PS NPs are developed. The results of the CG simulations are compared with existing experimental data and higher-resolution computational simulations. The novel parametrization expands the capacity of UA/CoronaKMC for modelling common industrial materials. The results obtained are then used to assess the potential biomedical application of PS NPs.
2 Methods
We first introduce the protocol for building a CG model of PS surfaces. Modified PS NPs will be designed as decorated raspberry core–shell CG structures originally described in ref. 92 to mimic the roughness of weathered materials. The CG mapping for proteins is set at one-bead-per-amino-acid (AA) resolution (see subsection 2.1), while each PS bead represents a styrene tetramer. The parametrization protocol for obtaining CG interaction potentials for the novel UA beads used in constructing the raspberry models is described in section 2.4.3. Protein corona formation (subsection 2.2) is modelled using a combination of the UA method with kinetic Monte Carlo (KMC) simulations.90,93,942.1 General concepts of the multiscale model for predicting individual protein adsorption affinities
In this work, we use the UA model to evaluate the average protein adsorption energy, as described in our previous publications.89,91–98 In this model, the complete protein–NP interaction potential Up-NP is approximated by a sum of non-bonded pairwise-additive interaction terms for each AA in the protein and the NP modelled as a set of CG beads with assigned UA material-specific parameters:
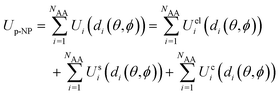 | (1) |
Within the UA paradigm, both materials constituting the bionano interface – the protein and the NP – are modelled using a rigid-body approximation, i.e., we neglect any possible change of the protein conformation (no side-chain flexibility or backbone motion is allowed) or PS chains at the NP surface. Interfacial water at the bionano interface is included explicitly at short distances via the short-range surface potential and implicitly beyond the cut-off distance of the atomistic model and defined by the force field selected for MD simulation (see section 2.4.3 for more information).
The potential Up-NP depends on the position of the protein with respect to the NP surface and is determined by di, the distance between the NP surface and the protein surface (surface–surface distance or SSD) and by the orientation of the protein characterized by two rotational angles, θ and ϕ, relative to the initial orientation of the protein and the normal of the surface (z-axis). The angles θ and ϕ correspond to an initial vector in the reference frame of the protein, which is rotated so that this vector faces the surface of the NP when the centre of mass of the protein is in a position (0, 0, z) relative to the surface of the NP. The rotational transformation is sequential with the first rotation of the protein by − ϕ around the z-axis, followed by 180° – θ rotation around the y-axis. In a reference frame (at [ϕ, θ] = [0°, 0°]), the protein is initially aligned to the principal axis with the longest axis along z and the second longest along the y-axis. The plot of adsorption energy values Eads(ϕ, θ), obtained by integrating the interaction potential along the z-axis in a given protein orientation in the θ, ϕ coordinate space,
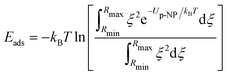 | (2) |
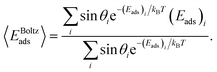 | (3) |
2.2 CoronaKMC model of protein corona formation (competitive adsorption)
The competitive binding of blood plasma proteins during corona formation is a multifaceted, dynamic process that takes place in two regions of the bionano interface.101 It is accepted that the corona hierarchy is multilayered, yet the majority of published studies focus on monolayered coronas.102 Proteins binding to the surface may do so irreversibly, resulting in the formation of a “hard” corona. In contrast, proteins that are weakly bound undergo reversible adsorption, remaining in a “soft” corona region until they are supplanted by incoming proteins with higher adsorption affinity. In addition, proteins from solution may bind to the adsorbed proteins of the “hard” corona, thus forming further adsorption layers.103 Proteins in both categories compete for available binding sites on the NP surface, a phenomenon known as the Vroman effect. This competitive process persists until the system reaches equilibrium, at which point proteins with the highest individual binding energy saturate the adsorption sites. Protein concentration also influences this process: proteins present in higher concentrations are more likely to occupy the surface, whereas those arriving later and in lower concentrations can only fill the remaining gaps left by earlier arrivals. In case of relatively weak interactions between PS and proteins, the timescales of corona equilibration can reach hours and days due to the higher energy barriers related to the change of protein conformations that adopt their shape to have a stronger interaction with the surface as compared to the energy barrier for reversible desorption.104,105 As a result, modelling the complete corona formation process with standard MD methods is practically unfeasible, although it has been attempted.74,106 For example, in ref. 74 it took 75 ns of production run for human α-synuclein to make the initial contact with the PS surface, while the first signs of protein structural changes occurred after 400 ns of stimulation. To overcome the limits of corona dimensions and extreme timescales, we employ a KMC method,81,90,107–109 which proved to be more efficient in modelling adsorption/desorption events through the set of reaction rate equations for diffusion-driven collision events. However, that approach would miss the information on protein unfolding and the formation of further layers, which would require more detailed models of the proteins and their interactions.103Recently, we applied the hard-sphere (HS) KMC methodology90,93 for predicting the formation of milk protein films on iron and aluminium surfaces.94,110 The HS KMC method considers protein adsorption events as physical interactions between a patch of the protein and the NP surface. It also assumes that this process can be reversible. The attachment and detachment rates are calculated for different protein orientations chosen at random from the energy heatmaps, so proteins can bind to the NP surface on different sides; the method calculates the occupied NP surface fraction by calculating the projections of proteins as hard spheres with radius RP corresponding to the radius of gyration of the molecule; the availability of binding sites on the NP surface for incoming protein is scaled according to the surface area occupied by other proteins.
A single event of protein adsorption is modelled as adsorption in random orientations. The probability of such a binding event is predicted from AES UA heatmaps. The latter are pre-calculated for individual proteins separately. In the steady state and with the assumption that desorption is a first-order reaction and the adsorption is a pseudo-first-order, the rate constants for adsorption (ka)/desorption (kd) can be evaluated from the following equation:
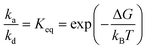 | (4) |
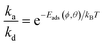 | (5) |
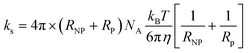 | (6) |
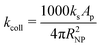 | (7) |
| kd = eEads/kBTkcoll | (8) |
2.3 CoronaKMC simulations setup and parameters
In our HS KMC simulations, we employed rate constants and adsorption areas for each biomolecule orientation, which were obtained from the adsorbate parameters. This was accomplished by automatically converting .uam output and .pdb structures using the BuildCoronaParams.py script. This script takes a list of biomolecules and their concentrations in mol L−1 as input, locates the corresponding structures and .uam binding energy tables, and computes rate constants and adsorption areas for each biomolecule orientation. During the time-resolved KMC simulation, events are generated that correspond to the adsorption or desorption of adsorbates. Adsorption events involve choosing a potential adsorbate based on a probability proportional to its collision rate with the NP and generating a random position on the NP surface. Proteins were allowed to adsorb and be replaced during the evolution of the protein corona. As a result, only the final adsorbed protein fractions obtained after the system reaches equilibrium were considered to predict the corona composition. Time-dependent saturation of protein binding sites by all and individual proteins was used to monitor corona equilibration convergence. The KMC simulation time was set to 3 min for all three types of 10 nm PS NPs. The selected timescale was deemed sufficient to obtain converged protein corona compositions and was consistent with the experimental timescales reported for corona formation in PS NPs105,112,113 when the initial protein corona composition remained mostly unchanged from the moment of initial protein adsorption within the 0.5 min frame.To predict the corona composition, we applied realistic concentrations of the selected most abundant blood plasma proteins that were reported in the literature (see Table 2 for more information). A detailed description of the CoronaKMC model can be found in the original work.90,93
2.4 Predicting individual protein adsorption affinities with the UA model
To construct adsorption energy heatmaps for the KMC model and to estimate individual protein affinity to PS NPs, several steps must be executed. First, a raspberry model of unmodified or altered PS NPs with the Cartesian coordinates of NP raspberry beads should be developed. Second, UA parameters should be assigned to each NP raspberry bead. These parameters include the ζ potential, the Hamaker constant, charge, and radius. Third, Cartesian coordinates for proteins should be established. These coordinates can be obtained through experimental or computational methods. To ensure accurate AA content for UA calculations, it is necessary to evaluate the protonation state of the protein at the selected pH. Lastly, non-bonded pairwise Usi potentials should be provided in tabular format for every combination of the NP raspberry bead and AA in the protein (potential mean force profiles or PMFs), including ionized AA states when necessary. If the UA distribution lacks potentials for specific material/AA combinations, they can be pre-calculated using all-atom MD simulations. The subsequent sections will provide comprehensive explanations on the implementation of these steps for novel parametrization of the UA model to characterize bionano interactions at PS interfaces.Taking into account all these factors, we model a PS NP as a single smooth uniform-density sphere of the corresponding radius, decorated with smaller neutral PS and charged beads placed around the smooth sphere mimicking a low-roughness surface with chemical modifications. For comparison purposes, we used similar parameters of the previously proposed MD model of PS NPs for protein adsorption.106 The total number (198) of charged surface beads was placed in the outer layer to model charged PS NPs. The ζ-potentials shown in Table 1 were evaluated via the Grahame equation129 from the corresponding surface charge densities σ at the slipping plane where surface potential Ψ0 ≈ ζ:
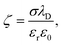 | (9) |
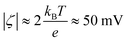 , suggesting that the surface charge density σ = 0.63 e/nm2 corresponds to moderate to strongly charged surfaces.130 Furthermore, considering that the carboxylic group has a pKa of about 2–5, it is expected that at pH 7.4 it should be fully ionized. In this case, the PMFs for the COO− fragment (see Fig. S1c in SI) were assigned to the decorating beads. At the same time, the NH2 group is basic (pKa ≈ 10.0) and thus it is also expected to be fully ionized at a neutral pH. The PMFs for NH3+ groups shown in Fig. S1d in SI were assigned to decorating beads in PS-NH2 NP models.
, suggesting that the surface charge density σ = 0.63 e/nm2 corresponds to moderate to strongly charged surfaces.130 Furthermore, considering that the carboxylic group has a pKa of about 2–5, it is expected that at pH 7.4 it should be fully ionized. In this case, the PMFs for the COO− fragment (see Fig. S1c in SI) were assigned to the decorating beads. At the same time, the NH2 group is basic (pKa ≈ 10.0) and thus it is also expected to be fully ionized at a neutral pH. The PMFs for NH3+ groups shown in Fig. S1d in SI were assigned to decorating beads in PS-NH2 NP models.
The radii of the beads in the outer raspberry shell were set to 0.36 nm, as estimated from the radius of gyration (rg) of the chosen PS oligomer fragments used for parametrization of PS-AA side chain (SCA) PMFs (see section 2.4.3). The resulting CG models of PS NPs are shown in Fig. S1 in the SI. UA simulations require a set of parameters describing NP-SCA interaction (the short-range NP-SCA potentials, Hamaker constant, and specific volume values) that are assigned for each CG bead comprising the NP, including decorating beads.131 The procedure for obtaining short-range potentials for SCA–PS, SCA–COOH, and SCA–NH2 interactions is described in the following section. The Hamaker constants and specific volume values required for pairwise interactions were evaluated using the protocol described in detail in ref. 93. To improve the sampling of protein adsorption energies for varying roughness values of the PS surface, 10 random rotational configurations of NP coordinates were used to obtain average adsorption affinities by the MultiSurfaceAverage.py tool in the UA package as previously described in ref. 92.
| Proteins | Name | PDB ID | UniProt ID | MW, kDaa | QP, eb | CPc, g L−1 | PDB resolution, Å | Chainsd |
|---|---|---|---|---|---|---|---|---|
| a Molecular weight, calculated from PDB structure.b Total charge, calculated at pH 7.4.c Protein concentration in blood plasma.d Chains included in PDB of biological assembly. | ||||||||
| HSA | Human serum albumin | 1AO6 | P02768 | 131 | −15.0 | 35–50 (ref. 139) | 2.50 | A |
| C3 | Complement component C3 | 2A73 | P01024 | 183 | −24.0 | 1.2 (ref. 140) | 3.30 | A, B |
| FG | Fibrinogen | 3GHG | P02671 | 228 | −14.0 | 2–4 (ref. 141) | 2.90 | A, D, G, J, B, E, H, K |
| APO-AI | Apo-lipoprotein | 3K2S | P02647 | 56 | −8.0 | 1.58 (ref. 142 and 143) | N/A | A, B |
| IGH | Immunoglobulin | 1HZH | P01857 | 150 | +22.0 | 5.6–18 (ref. 144) | 2.70 | A–D |
w(r) = −kBT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln[g(r)] ln[g(r)]
| (10) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of interacting beads was considered in this study. To model the PS-SCA interaction, a single CG bead of PS chain was represented by the (C8H8)n=4 fragment. The simulation systems were composed of one biomolecular fragment and one PS fragment solvated by TIP3P water and neutralized by 0.15 M KCl. The temperature was set at 298.15 K in the NPT and NVT ensembles, and the pressure was at 1 bar in the NPT ensemble. The Nose–Hoover thermostat was employed for NVT simulations, while Berendsen's weak coupling thermostat and barostat were applied for NPT runs. The system was modeled using periodic boundary conditions with an approximate primary cell size of 8.0 nm × 8.0 nm × 8.0 nm. These boxes were pre-equilibrated to obtain a proper density/pressure and were subject to 200 ns production runs to collect RDFs. All-atom molecular dynamics simulations in this study were performed with the GROMACS package146 and the CHARMM36m147 force field parameters with a cutoff distance 1.2 nm. RDFs for each set of SCA-material pair were calculated by the gmx rdf tool in GROMACS, ensuring that the distance between the two centers of mass (COM) was selected. To reduce the noise in RDF curves, a 1D Gaussian filter with a standard deviation for the Gaussian kernel equal to 2–4 is applied (scipy.ndimage.gaussian_filter1d Python library). RDFs were inverted as per eqn (10) to obtain CG PMFs (in kBT). The CG potentials used in the UA model are referenced with respect to the surface–surface separation distances, which are obtained from the potentials constructed along the distance between the planar surface of the material and the COM of SCA. To harmonize the potentials obtained from the inverted RDFs, we shifted them by the distances corresponding to the radius of gyration of the material fragment, replicating a single CG bead. The radii of gyration were obtained with the gmx gyrate GROMACS tool for analysis of MD trajectories. Short-range UA potentials for the decorating beads were obtained in a similar way. To cover various pH regimes in the UA CG models, ionised and nonionised analogues of COOH and NH2 CG beads were parametrized. Final PMF sets used in UA simulations are shown in Fig. S1.
1 ratio of interacting beads was considered in this study. To model the PS-SCA interaction, a single CG bead of PS chain was represented by the (C8H8)n=4 fragment. The simulation systems were composed of one biomolecular fragment and one PS fragment solvated by TIP3P water and neutralized by 0.15 M KCl. The temperature was set at 298.15 K in the NPT and NVT ensembles, and the pressure was at 1 bar in the NPT ensemble. The Nose–Hoover thermostat was employed for NVT simulations, while Berendsen's weak coupling thermostat and barostat were applied for NPT runs. The system was modeled using periodic boundary conditions with an approximate primary cell size of 8.0 nm × 8.0 nm × 8.0 nm. These boxes were pre-equilibrated to obtain a proper density/pressure and were subject to 200 ns production runs to collect RDFs. All-atom molecular dynamics simulations in this study were performed with the GROMACS package146 and the CHARMM36m147 force field parameters with a cutoff distance 1.2 nm. RDFs for each set of SCA-material pair were calculated by the gmx rdf tool in GROMACS, ensuring that the distance between the two centers of mass (COM) was selected. To reduce the noise in RDF curves, a 1D Gaussian filter with a standard deviation for the Gaussian kernel equal to 2–4 is applied (scipy.ndimage.gaussian_filter1d Python library). RDFs were inverted as per eqn (10) to obtain CG PMFs (in kBT). The CG potentials used in the UA model are referenced with respect to the surface–surface separation distances, which are obtained from the potentials constructed along the distance between the planar surface of the material and the COM of SCA. To harmonize the potentials obtained from the inverted RDFs, we shifted them by the distances corresponding to the radius of gyration of the material fragment, replicating a single CG bead. The radii of gyration were obtained with the gmx gyrate GROMACS tool for analysis of MD trajectories. Short-range UA potentials for the decorating beads were obtained in a similar way. To cover various pH regimes in the UA CG models, ionised and nonionised analogues of COOH and NH2 CG beads were parametrized. Final PMF sets used in UA simulations are shown in Fig. S1.3 Results
3.1 Individual protein adsorption onto pristine and functionalized PS NPs
 | ||
| Fig. 1 Adsorption energy surfaces (heatmaps) for one configuration of selected functionalised NPs and five proteins: serum albumin (HSA), fibrinogen (FG), immunoglobulin (IGH), complement component 3 (C3), and apolipoprotein A1 (APO-AI). This set of multiple heatmaps was used to obtain the Boltzmann-average energy values shown in Fig. 3. The positions of the minima are marked with the “star” signs. Contour maps were generated using identical energy ranges to emphasize the comparative intensity of interactions between specific proteins and NPs. The AESs have some common features, however, it depends on the functionalization and the charge of the NPs. | ||
3.1.1.1 Adsorption of HSA. A multi-modal mechanism of HSA adsorption on 99-nm pristine, carboxylate- and amine-modified PS NPs was noted in ref. 149. The authors suggested that although HSA has an overall negative charge, its surface contains a mix of positively and negatively charged AAs, creating localised binding patches accessible for oppositely charged particles. They suggested that in solution with 150 mM monovalent ions, the Debye length (κ−1) is about 0.8 nm, thus, proteins and NPs should be in proximity of each other to experience strong electrostatic forces. However, at these distances, van der Waals attraction is expected to become the primary driving mechanism for HSA adsorption, facilitated by the heterogeneous charge distribution on the protein's surface. In the present study, the HSA's AAs nearest to the NP surface were Glu16, Glu17, Hsd128, Glu131, Lys162, Thr166, and Gln170 for PS NP (Fig. 2a–d) and Glu17, Lys162, Gln170, and Pro282 for PS-NH2 (Fig. 2c–f), while for PS-COOH the closest side chains were located at the loops of the α-helixes near Glu501, Lys538, Gln580, and Ala581 (Fig. 2b–e). The NP-binding site near the residues 540–580 of the homologous BSA protein was previously reported by MD simulations of BSA interacting with pristine and modified PS NPs.150 In the case of human serum albumin (HSA), the corresponding lowest energy binding configuration was located at the coordinates (ϕ = 125°; θ = 55°) for negatively charged PS-COOH nanoparticles (see Fig. 1a). The authors of the MD study also noted an overlap of NP-binding sites in BSA for aminated and pristine PS plastic models vs. the carboxylated surface model of PS. Previously, it was also shown that BSA would adopt a side-on orientation while binding to pristine PS NPs with radii of 30, 100, and 110 nm.113 A side-on orientation was also predicted by UA simulations for HSA binding onto all three models of modified PS NPs used in the current work (Fig. 1a–c). Experimental studies151 have shown that the fluorescence quenching of HSA-PS complexes, reflecting the interaction strength between the fluorophore and the quencher, decreases in the order PS-NH2 > PS-COOH > PS. In Förster resonance energy transfer (FRET), the efficiency of energy transfer is highly dependent on the distance (rFRET) between the donor and acceptor molecules.152 The efficiency is inversely proportional to the sixth power of the distance (1/rFRET6) between NP and tryptophan (Trp) and tyrosine (Tyr) residues in the protein. For HSA the closest to the PS surfaces was Tyr138 (6.7 nm for pristine and 6.8 nm for PS-NH2 NP) and Tyr497 (5.8 nm for PS-COOH). Furthermore, the reported stronger quenching effect for amine-functionalized PS was assigned to enhanced local hydrophobicity near Tyr138 and Tyr150 AAs that can be induced by additional rearrangement of the interacting nearby protein's region. These aromatic AAs were calculated to be close to the binding site near Lys162 for HSA@PS-NH2 UA docking pose with (ϕ = 305°; θ = 95°) rotational coordinates as shown at Fig. 2i (Tyr138 and Tyr150 are highlighted in cadmium yellow colors). At least two well-defined HSA drug binding sites have been described in the literature.153 Sudlow drug sites I and II are located near Trp214 and Arg410 AAs (highlighted at Fig. 2b in forest green and red colors, respectively). It is predicted that these residues do not directly interact with the NP surface in either PS modification, suggesting that their interaction with the NP may be irrelevant for preserving these sites. However, in the case of PS-COOH NP, disruption of this pocket may occur due to protein adsorption onto the NP via adjacent Glu501 and Lys538, potentially affecting drug–HSA interactions. This hypothesis, however, necessitates further investigation, including unfolding processes as part of bionano interfacial processes, although such simulations exceed the capabilities of the UA framework.
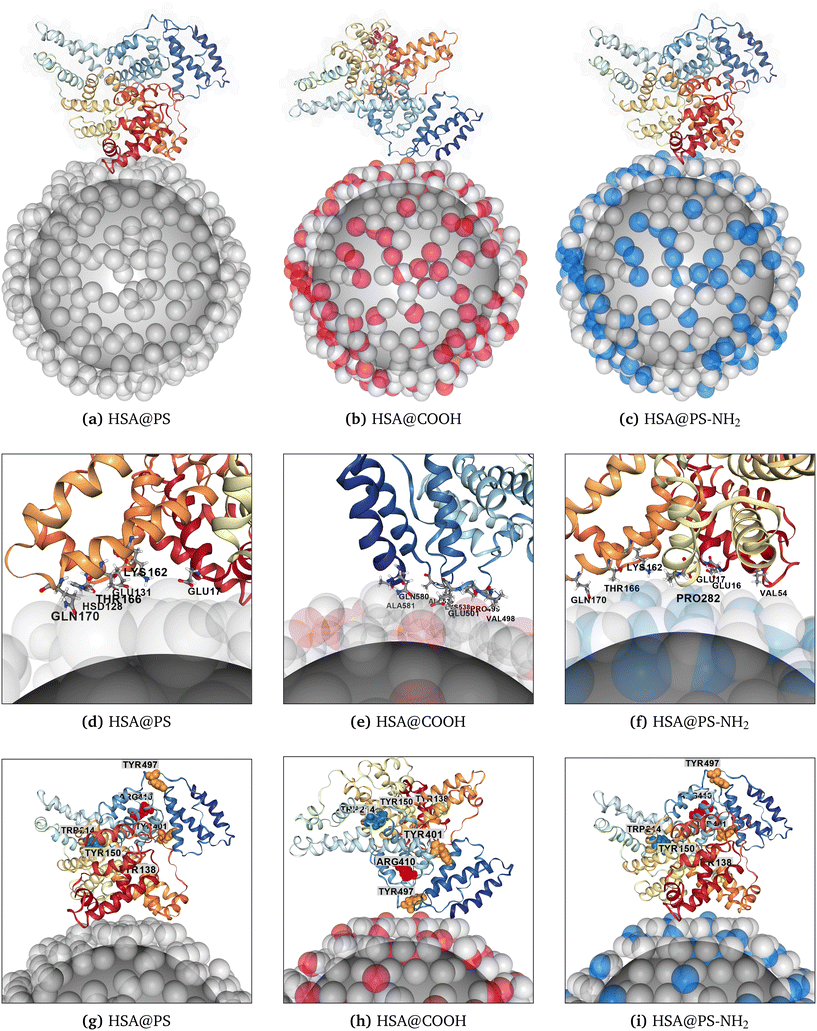 | ||
| Fig. 2 The lowest energy configurations of HSA protein bound to various PS NPs as predicted by the UA modeling. Here, the coordinates of the minima (marked with the “star” symbol) were taken from adsorption heatmaps shown in Fig. 1. AAs closest to the nanosurface (within 5 Å cut-off) are labeled (d–f). Aromatic residues relevant for fluorescence quenching of HSA-PS complexes discussed in text, Tyr138/Tyr150 and Tyr401/Tyr497, are highlighted in cadmium yellow and ochre orange colors (g–i). The Sudlow drug sites I and II near Trp214 and Arg410 AAs are also shown in forest green and red colors, respectively. The residue identifiers are the same as in the original protein PDB files. | ||
3.1.1.2 Adsorption of C3. The human C3 protein comprises several critical functional structural elements, including the β (residues 1–645) and α (residues 650–1641) chains.154,155 C3 convertases cleave C3 at Ser726–Arg727, initiating the conversion of the anaphylatoxin domain or ANA (residues 650–726) into anaphylatoxin C3a and activating the C3b fragment with the thioester-containing domain (TED, residues 963–1268). The covalent attachment of C3b to antigen surfaces via a thioester bond in TED activates the complement component C3 defense mechanism against pathogens. The thioester bond (Cys988–Gln991) is shielded from reactions with small amino or hydroxyl nucleophiles (e.g., water, ammonia, methyl amines) by a hydrophobic/aromatic pocket composed of Met1378, Tyr1425, Tyr1460, and Phe1047 in native C3, becoming exposed only after proteolytic cleavage. Upon activation, C3b binds to the sugar-rich cell surfaces of viruses and pathogens, although this opsonization does not discriminate against host surface receptors. The surface-bound C3b subsequently induces phagocytosis. Residues 1496–1641 constitute a carboxy-terminal C345C or NRT domain with a netrin-like fold. The NRT domain is a target for complement inhibitors under development as therapeutics for various diseases. Inhibitor binding to this domain can prevent the formation or stability of the convertase enzymes. The domain undergoes significant structural changes during complement activation and binding events.156,157 The C345C domain mediates key complement interactions, and dysregulation of C3 activation involving this region has been implicated in Alzheimer's disease-related neuroinflammation and synaptic loss.158 Complement activation by polymer-coated surfaces was previously reported.159 The activation can occur as a result of C3 direct binding to the surface (alternative pathway). For C3 protein bound to PS NPs, three favourable docking poses were observed (see Fig. S4 in SI). For pristine PS the protein was bound via Leu292, Lys283 and the α-helix between Lys700 and Glu711 on the ANA fragment. A similar orientation with the α-helix between Lys700 and Arg718 on the ANA fragment was predicted for PS-COOH NP. And in the case of PS-NH2 NP the interaction occurred via the α-helix between Gln1616 and Phe1637 at the NRT domain. Predicted poses resonate with experimentally observed behavior of C3 protein. For example, it was shown that binding of the NRT domain to PS-NH2 NP could work as a nanobody inhibiting amplification of complement system activation at the C3 level.160 Furthermore, for C3 to effectively target pathogens or damaged cells, its ANA domain must be appropriately exposed. Thus, if ANA binds to PS and PS-COOH NPs as predicted by our simulations, this interaction can potentially disrupt ANA's movement and prevent complement activation in response to pathogens. None of the PS NPs targeted the compstatin binding site near the Ile389–Ser394, Arg456–Met457, and Gly489–Asp491 pocket located on the β-chain161–163 and thus shall not alter its ability to inhibit C3 activation.
3.1.1.3 Adsorption of IGH. Activation of complement can also proceed via a classical activation pathway: initial binding of immunoglobulins to biomaterials, followed by binding of complement component C1 to the FC fragment of immunoglobulins.159,164,165 Thus, complement activation of the classic pathway in response to contact with artificial materials suggests the possibility of Fab-on surface orientation of immunoglobulins. It was also shown that pre-adsorption of antibodies before blood plasma/serum incubation enhances the uptake of functionalized PS NPs of 230 nm and 150 nm by target cells of moDCs with the CD3 receptor.166 This uptake enhancement suggests the optimal orientation of immunoglobulin bound to PS NPs to target the CD3 receptor. Superior targeting efficacy results from substantial exposure to functional receptor binding sites on Fab1 and Fab2 in outer space (Fc-on surface bound orientation). In simulations of all-atoms,167 the three possible orientations for the adsorption of IGH onto a flat PS surface were predicted: Fab-Fab-on, Fc-on, and Fab-Fc-on orientations. In our calculations, we observed only the Fab-on or Fc-on orientation. The simultaneous interaction between two subunits and the PS surface was not predicted due to a mismatch in protein and NP size, resulting in an insufficient surface area available to accommodate both subunits. Our calculations indicate that the preferred orientation of IGH on pristine and functionalized PS NPs differs (see Fig. S5 and Tables S1–S3 in SI). However, a very shallow minimum (Eads(ϕ, θ) = −2.6kBT) was predicted in the case of PS-NH2 NP where the IGH was oriented with both Fab fragments facing outward and Ile266, Gln330–Lys336 residues on the Fc facing the NP surface (ϕ = 345°; θ = 110°). While this orientation was also anticipated for pristine PS NPs (Eads(ϕ, θ) = −13.7kBT), a stronger adsorption minimum (Eads(ϕ, θ) = −24.6kBT) was expected for a different orientation (ϕ = 330°; θ = 35°). In this orientation Arg18, Arg24, Arg29, Ser67, and Asp70 in the Fab1 fragment were the closest to the surface. A similar orientation was predicted for PS-COOH NP.
3.1.1.4 Adsorption of FG. The adsorption of FG at the solid–liquid interfaces has been widely studied because of its crucial role in blood coagulation. According to SFG spectroscopy, FG forms a complete layer on pristine PS surfaces, with absorption reaching equilibrium at approximately 20
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 seconds.168,169 Upon adsorption, FG adopts a bent conformation, where one E-D fragment lies flat and the other E-D segment stretches outward. This bending, which deviates by 30° from its linear form, occurs near the center of the FG molecule. Once bound to PS, the orientation and shape of fibrinogen do not change over time. The change of SFG intensity of N–H stretching signals (3270 cm−1) from FG was also observed as a result of adsorption onto PS. At the same time, for a blank polyester surface, FG was predicted to bind to the PET surface through the D-terminus170 similar to FG adsorption at the silicon oil–water interface. All-atom MD simulation studies of FG adsorption to mica and graphite surfaces have shown that the attachment to the surface occurred through the D the E regions.171 In ref. 172, the exploration of micro-electrophoretic behavior and concentration depletion revealed two main mechanisms for FG adsorption. The first mechanism, which is more prevalent at lower ionic strengths, involves random orientation adsorption that allows FG to penetrate deeply into the fuzzy polymeric layer on the surface of NP. The second mechanism, referred to as a side-on adsorption, becomes more prevalent at pH > 5.8 and an ionic strength of 0.15 M. The results of our calculations show (see Fig. S6) that for pristine and positively charged PS, the lowest energy of adsorption was predicted for orientations where FG approaches the PS surface at the coiled coil region between E and D domains and near the residues Phe74, Lys78, Lys81, Asp82, or Ser85 or Phe35, Ser37, Asp38, Glu39, Asp40 (see Tables S1–S3 in SI). This orientation can be considered as a side-on orientation predicted for experimental conditions close to our theoretical ones (pH = 7.0, I = 0.15 M). Similar residues were predicted to mediate the attachment of the E-domain to the surface of the EMT zeolite.173 The adsorption onto negatively charged PS-COOH takes place near the Lys173, Asn175, and Pro260, Ala271, Asp272 residues within one of the D domains, and this can be described as an orientation that involves penetration into the polymer's surface layer.
000 seconds.168,169 Upon adsorption, FG adopts a bent conformation, where one E-D fragment lies flat and the other E-D segment stretches outward. This bending, which deviates by 30° from its linear form, occurs near the center of the FG molecule. Once bound to PS, the orientation and shape of fibrinogen do not change over time. The change of SFG intensity of N–H stretching signals (3270 cm−1) from FG was also observed as a result of adsorption onto PS. At the same time, for a blank polyester surface, FG was predicted to bind to the PET surface through the D-terminus170 similar to FG adsorption at the silicon oil–water interface. All-atom MD simulation studies of FG adsorption to mica and graphite surfaces have shown that the attachment to the surface occurred through the D the E regions.171 In ref. 172, the exploration of micro-electrophoretic behavior and concentration depletion revealed two main mechanisms for FG adsorption. The first mechanism, which is more prevalent at lower ionic strengths, involves random orientation adsorption that allows FG to penetrate deeply into the fuzzy polymeric layer on the surface of NP. The second mechanism, referred to as a side-on adsorption, becomes more prevalent at pH > 5.8 and an ionic strength of 0.15 M. The results of our calculations show (see Fig. S6) that for pristine and positively charged PS, the lowest energy of adsorption was predicted for orientations where FG approaches the PS surface at the coiled coil region between E and D domains and near the residues Phe74, Lys78, Lys81, Asp82, or Ser85 or Phe35, Ser37, Asp38, Glu39, Asp40 (see Tables S1–S3 in SI). This orientation can be considered as a side-on orientation predicted for experimental conditions close to our theoretical ones (pH = 7.0, I = 0.15 M). Similar residues were predicted to mediate the attachment of the E-domain to the surface of the EMT zeolite.173 The adsorption onto negatively charged PS-COOH takes place near the Lys173, Asn175, and Pro260, Ala271, Asp272 residues within one of the D domains, and this can be described as an orientation that involves penetration into the polymer's surface layer.
3.1.1.5 Adsorption of APO-AI. According to our UA model simulations, fragments between Gln41–Lys45–Leu46–Leu47 were found near pristine and COOH-modified PS NP's surface (see Fig. S7). In the case of amino-modified PS, multiple orientations with similar Eads(ϕ, θ) = −22 ± 3kBT were located on the AES (see Tables S1–S3 in SI). The alternative conformation was located at ϕ = 310°; θ = 55° and had the Lys226–Glu234 patch near the surface. Unfortunately, no information on the orientation of APO-AI at the NP's surface was found in the literature to validate these docking poses.
In a study177 involving 23 nm PS, 24 nm PS-COOH, and 57 nm PS-NH2 NPs, it was observed that APO-AI has a greater affinity for PS and PS-COOH NPs than HSA or reconstituted HDL proteins. In line with this observation, our calculations have also shown that APO-AI has a higher individual binding energy than HSA for pristine and COOH-modified PS (see Fig. 3). For apolipoprotein APO-AI, the UA-calculated binding affinity values were exothermic, ranging from −60.5kBT (PS) to −48.4kBT (PS-COOH) to −24.9kBT (PS-NH2).
It was shown experimentally178 that the fluorescence quenching efficiency for interaction between FG and 80 nm PS NP decreased by 45.5%, 49.2%, and 55.9% for PS-COOH, PS, and PS-NH2 NPs, suggesting that positively charged PS NPs are more likely to combine with FG and change its structure. Thermodynamic measurements indicated that the interaction between FG and PS occurs spontaneously (ΔG < 0) and is mainly driven by hydrophobic interactions (ΔH > 0 and ΔS > 0). According to UA-simulations, FG also exhibits the strongest interactions with positively charged PS-NH2, followed by PS-COOH and then pristine PS NPs (see Fig. 3).
3.2 Formation of protein corona on PS NPs
Previous studies have indicated that incubation of 80 nm PS NPs without any functionalization with human plasma results in a protein corona that significantly enhances placental transfer.179 This effect was more pronounced compared to the PS-corona formed with BSA and dextran, which was used as a control in the experiments. After incubation of PS particles with plasma, the most abundant proteins were albumin, immunoglobulins, and apolipoproteins. At the same time, a high mean corona intensity was also observed corresponding to fibrinogen and C3 (see Table S5 in ref. 179). The results of our CoronaKMC simulations of competitive adsorption of selected proteins onto smaller 10 nm PS NP echo similar results. Calculations have predicted that the NP's coverage with proteins occurs from the initial seconds (Fig. 4) and that the maximum load capacity is reached within 60 s of simulation time or less. Taking into account the geometric constraints, the maximum protein load capacity for a 10 nm PS NP is expected to range from 9 to 16 proteins, depending on their dimensions (see the maxLoadGeom.ipynb notebook in the GitHub archive in SI).As shown in Fig. S8 in the supporting materials, the first 9 HSA entities are adsorbed at t = 0.01 s (Fig. S8a) and an APO-AI binds at t = 0.03 s (Fig. S8j) for pristine PS. In the case of PS-NH2 NP, 12 HSA molecules are adsorbed at t = 0.01 s (Fig. S8c) and later the number of HSA molecules drops to 0 at t = 14.3 s, while 1 C3 molecule binds to the NP at t = 0.03 s and reaches its maximum load (17 molecules) at t = 60.4 s (Fig. S8f). Meanwhile, 1–2 molecules of FG absorb/desorb at the surface of PS-NH2 NP. For PS-COOH NP, the corona composition was more diverse – only APO-AI was excluded. At first, 1 APO-AI and 5 IGH molecules bind to the surface at t = 0.01 s (see Fig. S8k and n). IGH reaches its maximum load at t = 0.02 s by replacing APO-AI and remains unchanged throughout the trajectory. Reversible binding of single C3, FG, and HSA is also observed (see Fig. S8b, e, h, k in SI).
The calculated total protein load number (nmax) on the surface of NP ranging from 8 to 17 was in line with the estimated maximum protein load number. The composition of the protein corona stabilized within a 60–100 second simulation frame before reaching an adsorption plateau. The sole exception was negatively charged PS-COOH, where the maximum load of the immunoglobulin heavy chain (IGH) remained nearly constant from the initial frames of the simulations, and proteins with low maximum load numbers underwent continuous exchange.
The highest total protein coverage Ωt of PSNPs was approximately 72–81%, and this loading capacity was achieved within the first 30 seconds of KMC simulation (see Fig. 5). The results obtained echo the study in ref. 112 where the rapid formation of a protein corona comprising the most abundant plasma proteins was experimentally observed at the earliest exposure time point (0.5 min) for PS NPs charged positively (115 nm diameter) and negatively (112 nm). The authors further noted that the composition of the protein corona was established at the earliest exposure time point and did not undergo significant qualitative changes even with extended plasma exposure (480 minutes), although quantitative alterations were observed. Notably, the rankings of individual binding affinities align with the composition of the protein corona. The concentration of blood plasma proteins emerged as the second most significant factor. For example, in the case of PS-COOH, APO-AI exhibited a slightly stronger binding to the surface compared to IGH and C3. However, the immunoglobulin IGH (CP = 5.6–18 g L−1) outperformed APO-AI (CP = 1.58 g L−1) and C3 (CP = 1.2 g L−1). Furthermore, albumin HSA (CP = 35–50 g L−1), despite having the lowest binding energy, demonstrated a presence similar to that of lipoprotein APO-AI. The interplay of additional factors, such as the shape and size of the protein, in conjunction with the associated surface charge distribution that complements the shape, charge, and surface charge distribution of NPs, is anticipated to collectively determine the final “new identity” of PS NPs.
 | ||
| Fig. 5 The 3 min of KMC simulation evolution of total protein load onto 10 nm PS (a), PS-COOH (b), and PS-NH2 (c) NPs. | ||
4 Discussion
4.1 Takeaways from CG modelling of individual protein adsorption: trends and methodology
In our previous reports, we have examined the potential limitations of the UA's predictive capabilities, which arise from the inherent constraints of the “rigid body” description of proteins and other approximations. This methodology does not account for alterations in protein conformation during the adsorption process (refer to section “Increasing the Resolution of the UANanoDock Predictions” in ref. 148 for further discussion). Moreover, within the additivity approximation, the interaction of each AA with the NP surface is optimised separately for each individual AAs, while in reality the optimal conformation may be impossible due to steric constraints within the protein globule. The model also neglects the entropic contributions associated with protein adsorption, such as protein conformational entropy loss, solvent rearrangement, and counterion release. The inconsistency in predicting binding energies, as compared to reported values, can be attributed to these simplifications. The 〈EBoltzads〉 represents an average of potential energies over sampled orientations, providing only enthalpic estimates. Consequently, UA-calculated adsorption affinities 〈EBoltzads〉 cannot be directly compared to experimental binding free energies. The exclusion of entropic effects typically results in weaker binding predictions; thus, the UA model is anticipated to overestimate the absolute strength of NP–protein interactions. Variations in experimental conditions, such as larger NPs, pH, and ionic strength of the buffers used, may further exacerbate these discrepancies. Experimental investigations into protein adsorption onto PS NPs indicate the potential for protein unfolding; however, the secondary structure elements can be either preserved or even stabilised in the presence of NPs. Consequently, the errors associated with “rigid” proteins can vary in absolute value, contingent upon the preserved stability of the secondary structure. The study by Alemie et al.180 demonstrated that the adsorption of immunoglobulin IGH onto PS NPs with various coatings containing amino and carboxylic groups, such as allylamine, 2-methyl-2-oxazoline, or acrylic acid, resulted in denaturation. The circular dichroism (CD) spectra indicated that IGH's secondary structure exhibited a substantial presence of β-sheets and turns on NH2-coatings, whereas some α-helices were stabilized on coatings with COOH groups. In a study,177 it was shown that when APO-AI binds to PS or PS-COOH particles, there is an increase in the α-helical signal in CD spectra, suggesting that this part of the structure is either stabilized or enhanced. In contrast, the formation of the APO-AI-PS-NH2 complex reduces the CD signal for APO-AI, with this effect becoming more significant at higher particle concentrations. This indicates that the binding with positively charged particles may disrupt the secondary structure of APO-AI. On the other hand, protein flexibility can impact the stoichiometry of adsorption: for example it was shown that FG due to its high flexibility at the coiled-coil region171,181 can bind to two separate NPs, potentially changing the particle aggregation state.182 Hinge bending in FG may also play an important role in fibrin polymerisation, as it can provide the necessary flexibility to accommodate new molecules in the growing fibre and thus can impact the regulation of blood clotting. Thus, it is advisable to include multiple protein conformations to evaluate protein adsorption with the “rigid body” approach implemented in UA or introduce other treatments of protein flexibility. Regarding the potential rearrangements of polymeric NPs upon interaction with proteins, the application of the “rigid body” approximation to the NP may introduce additional errors in estimations of adsorption energies. This is due to the significant energy changes associated with such interactions, which cannot be directly captured in the multiscale UA model. Technically, these degrees of freedom can be accounted for through extended simulations with enhanced sampling within an all-atom MD framework. However, in practice, such simulations require enormous computing resources and can only be applied to study a limited number of proteins. A further NP-linked source of discrepancy between the results obtained from UA and those reported in all-atom MD simulations, as documented in the benchmarking report,106 is the relatively high sphericity of the UA CG raspberry models of NPs compared to the shape of MD-simulated polymeric nanoclusters. It is important to note, however, that the UA values account for the contributions of proteins interacting with various segments of the NP surface (10 NP orientations were included), potentially offering a more accurate representation of experimental conditions. Facilitating such comprehensive scanning of the entire NP surface in MD simulations is challenging due to the substantial system sizes involved.The UA adsorption affinities partially correlated to the protein charge (see Fig. 6). The stronger adsorption for positively-charged PS-NH2 NPs (as compared to pristine PS NPs) was predicted for negatively charged proteins, for example C3, HSA, and FG proteins with a total protein charge QP = −24.0e, −15.0e, and −14.0e, respectively. Similarly, the most positively charged protein IGH (QP = +22.0e) had the strong preference for negatively-charged PS-COOH NPs and the least affinity for positively-charged PS-NH2 NPs. Thus, the electrostatic attraction and repulsion are expected to play a substantial role in protein adsorption onto modified PS NPs. However this trend is not linear. For example, the adsorption of C3 protein (QP = −24.0e) was also improved when the PS NPs become functionalised with negatively charged carboxylate groups. At the same time, the APO-AI protein, regardless of its moderate negative charge (QP = −8.0e), has shown the least preference for positively-charged PS-NH2 NPs as compared to pristine. These trends can indicate that protein adsorption onto PS NPs is a complex process that is regulated by various contributions and not exclusively by electrostatic forces. Furthermore, the sensitivity of the CoronaKMC model to initial parameters indicates that varying blood plasma compositions, represented by different initial protein concentrations, will result in different coronas. Moreover, as previously noted, PS nanotoxicity demonstrates size-dependent characteristics. To accurately predict NP toxicity, it is essential to consider a realistic distribution of NP sizes present in the environment, rather than assuming a uniform 10 nm NP size. Such variations in NP size may impact not only the electrostatic interactions between proteins and NPs but also potentially lead to NP agglomeration under real-world conditions. This agglomeration could alter the size of PS clusters and further change the content of the corona.183,184
4.2 Corona of PS NPs and possible implications for environmental nanotoxicology and medicine
The APO-AI apolipoprotein, immunoglobulin IGH, complement component C3 and serum albumin HSA were predicted as the most abundant in the protein corona of pristine and modified 10 nm PS NP in our calculations. These abundances can result in various outcomes. For example, the presence of IGH in the protein corona can enhance the opsonisation of NPs, facilitating the immune system's identification and removal of PS NPs, and potentially increasing the NPs' immunotoxicity by triggering an undesirable immune response,185 a complication undesirable for drug nanocarriers. A minimal number of surface-bound immunoglobulin molecules were sufficient to trigger complement activation in response to superparamagnetic iron oxide (SPIO) nanoworms,186 indicating that natural antibodies play a crucial role in C3 opsonization. Analogous immune complement activation was observed for PS-coated QCM-D sensor surfaces.187 Experimental observations of the activation of C-related pathways in pig and human blood serum in response to exposure to PS NPs were also reported.188 Activation of the complement system via the C3-associated pathway is commonly associated with the immune response to pathogens and with pro-inflammatory effects.189,190 The prolonged activation of this mechanism can lead to inflammation and tissue damage.191 The same pathway was also linked to neurological disorders.192,193 Interaction with the C3 proteins was reported for various nanomaterials, and it was shown that C3 protein bound directly to the foreign surface can be replaced.194 In our KMC simulations, the C3 adsorption dominated in the case of negatively charged PS-NH2 NPs and was less dominant for negatively charged PS-COOH NPs. As a consequence of C-activation triggered by interaction between complement component C3 and the NP, the use of PS NPs as intravenous materials for biomedical NP-based applications requires caution due to potential immunological risks. Yet, these NPs might be useful, as it was shown that the C3-activation process can be harnessed to regulate tumour growth.195 Apolipoproteins, as well as lipoprotein particles, were found to be the main adsorbates on polymeric biomaterial surfaces.196–198 The adsorption of the APO-AI protein from the bloodstream onto silica NPs (SiNPs) has demonstrated an initial reduction in the cytotoxic and pro-inflammatory effects of SiNPs. The subsequent rapid clearance of NPs coated with the protein corona resulted in a depletion of APO-AI, leading to cardiovascular damage and atherosclerosis.199 Moreover, the interaction of APO-AI with NPs can be influenced by the mode of exposure. The respiratory exposure of mice to SiNPs led to the pre-adsorption of pulmonary surfactants, which ultimately resulted in stronger adsorption of APO-AI from the bloodstream. Due to the presence of APO-AI in the corona of pristine hydrophobic PS NPs (Fig. 4a), effects similar to SiNPs may be expected to occur from exposure to engineered or freshly synthesized PS NPs. The expected primary adsorption of HSA can be viewed as a beneficial factor if PS NPs are intended to act as drug carriers, as HSA adsorption is known to promote NP uptake.200 Moreover, albumin plays a role in transporting both exogenous and endogenous ligands in the bloodstream, such as fatty acids, hormones, and AAs, which typically bind to its two sites,201 effectively functioning as a “nano-drug” itself. The combination of HSA with the NP can be used to develop a versatile drug delivery system, offering advantages such as reduced toxicity, improved drug distribution, and the potential to overcome multidrug resistance.202,203 Nevertheless, there are drawbacks to employing PS NPs as drug nanocarriers, as albumin's interaction with the NP can negatively impact the pharmacodynamics and pharmacokinetics of drugs.204,205 For example, the presence of NPs led to a reduction in the stoichiometry coefficient of HSA-paracetamol from 2 to 0.3 due to the loss of paracetamol binding sites in HSA.206 Docking studies207 have identified two binding sites in HSA for paracetamol–Sudlow sites I and II (see Fig. 2). Although there might not be a direct interaction with the sites, the NP binding in proximity to the paracetamol site could cause structural changes in Sudlow's sites, thereby affecting the efficiency of paracetamol binding. The presence of FG in the corona was predicted only for PS-COOH NP (see Fig. 4b). As a result of this interaction, the NP can weakly deplete FG from the blood. This can modulate thrombosis and inflammation without compromising hemostasis, demonstrating the potential to treat fibrinogen-driven pathologies.208 At the same time, triggering of inflammatory cascades can be expected. It was demonstrated for negatively charged poly(acrylic acid)-conjugated gold NPs, that protein unfolding can improve FG interactions with the Mac-1 integrin receptor.182,209 In conclusion, the processes previously discussed may have detrimental effects on human health as a result of long-term exposure to polymeric waste in the environment. As the accumulation of polymeric materials in natural settings increases, so does the bioaccumulation of these substances, potentially leading to the processes mentioned earlier. The extensive distribution of micro and nanoplastic particles, along with other pollutants,151,210 raises the probability of cell uptake of PS NPs with co-adsorbed environmental contaminants, which can intensify the nanotoxicity of PS particles. Moreover, studies have indicated that the adsorption of plasma blood proteins onto PS NPs slows their aggregation and significantly enhances their colloidal stability.211–215 This situation could pose a potential threat to human health as it may result in prolonged environmental exposure, necessitating further research.Conclusions
The key novelty of the reported work is an application of a multiscale UA technique to predict interactions between PS nanoplastics, a prevalent industrial and environmental polymeric pollutant, and the five most abundant human blood plasma proteins (albumin, fibrinogen, immunoglobulin, apolipoprotein, and complement component C3 proteins). As a part of our ongoing efforts in developing UA, we have introduced a new set of coarse-grained parameters of PS-AA interactions, required to evaluate the binding affinity for other proteins with functionalized PS NPs. Using novel parameters, we predicted the protein coronas of charged and neutral PS NPs through a competitive adsorption model (UA/NPCoronaPredict) incorporating the most abundant human blood plasma proteins. Our analysis has shown that apolipoprotein APO-AI, immunoglobulin IGH, and complement component C3 are the predominant proteins in the coronas of pristine, negatively, and positively charged PS NPs, respectively. Consequently, PS NPs are anticipated to elicit an immune response. Consistent with previous predictions, our findings suggest that charged PS NPs are likely to exhibit stronger interactions with plasma proteins. Therefore, natural surface wear processes can chemically and physically alter the outer layer of NPs, potentially leading to an increase in the nanotoxicity of PS NPs following their inhalation or ingestion. The presented parametrization extends the capacity of the UA/CoronaKMC method for modelling micro- and nanoplastics and provides data for machine learning and “safe and sustainable by design” pre-assessment of novel polymers.Author contributions
Conceptualization, J. S.; methodology, J. S.; testing, O. M.; supervision, V. L., J. S.; writing – original draft preparation, J. S.; writing – review and editing, J. S., V. L.; funding acquisition, V. L.Conflicts of interest
There are no conflicts to declare.Data availability
Additional data is available from supporting information (SI) online: LINK. The PDF file contains a visualisation of the PS NP multiscale models used in the work, a visualization of the lowest energy complexes corresponding to minima found at the adsorption heatmaps; tables for multiple adsorption poses of protein–NP complexes and corresponding AAs, nearest to the NP's surface. The PDB structures of predicted complexes (lowest energies) and the Jupiter Notebook for their visualization, PS potentials, and UA configurational files are available via GitHub link: https://github.com/juliasubbotina/uaCoronaPS. The latest official release of UA/NPCoronaPredict package (including material parameters, examples of required files, and post-processing tools) can be found here: https://github.com/ucd-softmatterlab/NPCoronaPredict. Supplementary information (SI) is available. See DOI: https://doi.org/10.1039/d5en00784d.Acknowledgements
The authors acknowledge the Irish Centre for High-End Computing (ICHEC) and the ResearchIT Sonic cluster, which was funded by UCD IT Services and the Research Office, for the provision of computational facilities and support. This work was funded by Science Foundation of Ireland grant, 16/IA/4506 “Quantitative Modeling of Bionano Interface”, EU Horizon 2020 grant NanoSolveIT, grant agreement 814572, EU Horizon Europe grant nanoPASS, grant agreement 101092741, and EU Marie Sklodowska-Curie Action CompSafeNano (grant agreement 101008099).References
- B. J. Seewoo, E. V. Wong, Y. R. Mulders, L. M. Goodes, E. Eroglu, M. Brunner, A. Gozt, P. Toshniwal, C. Symeonides and S. A. Dunlop, Heliyon, 2024, 10, e32912 CrossRef CAS PubMed.
- OECD, Global Plastics Outlook, 2022, p. 201 Search PubMed.
- H. S. Charlton-Howard, A. L. Bond, J. Rivers-Auty and J. L. Lavers, J. Hazard. Mater., 2023, 450, 131090 CrossRef CAS PubMed.
- L. Lebreton, B. Slat, F. Ferrari, B. Sainte-Rose, J. Aitken, R. Marthouse, S. Hajbane, S. Cunsolo, A. Schwarz, A. Levivier, K. Noble, P. Debeljak, H. Maral, R. Schoeneich-Argent, R. Brambini and J. Reisser, Sci. Rep., 2018, 8, 4666 CrossRef CAS PubMed.
- T. K. Dey, M. Rasel, T. Roy, M. E. Uddin, B. K. Pramanik and M. Jamal, Sci. Total Environ., 2023, 867, 161390 CrossRef CAS PubMed.
- C. C. Gaylarde, J. A. Baptista Neto and E. M. da Fonseca, Environ. Pollut., 2021, 272, 115950 Search PubMed.
- S. Lambert and M. Wagner, Chemosphere, 2016, 145, 265 Search PubMed.
- M. Shen, Y. Zhang, Y. Zhu, B. Song, G. Zeng, D. Hu, X. Wen and X. Ren, Environ. Pollut., 2019, 252, 511–521 CrossRef CAS PubMed.
- K. Mattsson, S. Jocic, I. Doverbratt and L.-A. Hansson, Microplastic Contamination in Aquatic Environments, 2018, pp. 379–399 Search PubMed.
- M. A. Bhat, K. Gedik and E. O. Gaga, Air Qual., Atmos. Health, 2022, 16(2), 233–262 CrossRef PubMed.
- A. Wahl, C. Le Juge, M. Davranche, H. El Hadri, B. Grassl, S. Reynaud and J. Gigault, Chemosphere, 2021, 262, 127784 Search PubMed.
- Y. Liu, H. Shao, J. Liu, R. Cao, E. Shang, S. Liu and Y. Li, Soil Use Manage., 2021, 37, 224–242 CrossRef.
- D. Materić, H. A. Kjar, P. Vallelonga, J.-L. Tison, T. Rockmann and R. Holzinger, Environ. Res., 2022, 208, 112741 CrossRef PubMed.
- M. Bergmann, F. Collard, J. Fabres, G. W. Gabrielsen, J. F. Provencher, C. M. Rochman, E. van Sebille and M. B. Tekman, Nat. Rev. Earth Environ., 2022, 3, 323–337 CrossRef CAS.
- Q. Liu, Z. Chen, Y. Chen, F. Yang, W. Yao and Y. Xie, J. Agric. Food Chem., 2021, 69, 10450–10468 CrossRef CAS PubMed.
- H. Lai, X. Liu and M. Qu, Nanomaterials, 2022, 12, 1298 CrossRef CAS PubMed.
- S. Kihara, I. Koper, J. P. Mata and D. J. McGillivray, Adv. Colloid Interface Sci., 2021, 288, 102337 CrossRef CAS PubMed.
- N. Zhang, Y. B. Li, H. R. He, J. F. Zhang and G. S. Ma, Sci. Total Environ., 2021, 767, 144345 CrossRef CAS PubMed.
- A. Ragusa, A. Svelato, C. Santacroce, P. Catalano, V. Notarstefano, O. Carnevali, F. Papa, M. C. A. Rongioletti, F. Baiocco and S. Draghi, et al., Environ. Int., 2021, 146, 106274 CrossRef CAS PubMed.
- A. Nihart, M. Garcia and E. El Hayek, et al., Nat. Med., 2025, 31, 1114–1119 CrossRef CAS PubMed.
- Q. Shi, J. Tang, R. Liu and L. Wang, Crit. Rev. Environ. Sci. Technol., 2022, 52, 3863–3895 CrossRef CAS.
- A. Katsumiti, M. P. Losada-Carrillo, M. Barros and M. P. Cajaraville, Sci. Rep., 2021, 11, 22396 CrossRef CAS PubMed.
- N. Meides, T. Menzel, B. Poetzschner, M. G. Loder, U. Mansfeld, P. Strohriegl, V. Altstaedt and J. Senker, Environ. Sci. Technol., 2021, 55, 7930–7938 CrossRef CAS PubMed.
- L. Ding, Z. Ouyang, P. Liu, T. Wang, H. Jia and X. Guo, Sci. Total Environ., 2022, 802, 149840 CrossRef CAS PubMed.
- A. Delacuvellerie, S. Benali, V. Cyriaque, S. Moins, J. Raquez, S. Gobert and R. Wattiez, J. Hazard. Mater., 2021, 419, 126526 CrossRef CAS PubMed.
- T. Morohoshi, T. Oi, H. Aiso, T. Suzuki, T. Okura and S. Sato, Microbes Environ., 2018, 33, 332–335 CrossRef PubMed.
- C. Odobel, C. Dussud, L. Philip, G. Derippe, M. Lauters, B. Eyheraguibel, G. Burgaud, A. ter Halle, A. Meistertzheim, S. Bruzaud, V. Barbe and J. Ghiglione, Front. Microbiol., 2021, 12, 734782 CrossRef PubMed.
- V. Viljakainen and L. Hug, Comput. Struct. Biotechnol. J., 2021, 19, 6191–6200 CrossRef CAS PubMed.
- C. Rex M, A. Debroy, M. Nirmala and A. Mukherjee, RSC Adv., 2023, 13, 22905–22917 RSC.
- P. A. Athulya and N. Chandrasekaran, J. Mol. Liq., 2023, 369, 120950 CrossRef CAS.
- H.-J. Zhang, S. Li, X.-L. Wang, K.-D. Zhang, H.-T. Fang, X. Wu, Z. Huang, W. Jiang, L. Yang and Q.-G. Tan, et al., Nat. Commun., 2025, 17(1), 1116 CrossRef PubMed.
- S. Shan, Y. Zhang, H. W. Zhao, T. Zeng and X. Zhao, Chemosphere, 2022, 134261 CrossRef CAS PubMed.
- M. Ayer, M. Schuster, I. Gruber, C. Blatti, E. Kaba, G. U. Enzmann, O. Burri, R. Guiet, A. Seitz, B. Engelhardt and H. Klok, Adv. Healthcare Mater., 2021, 10, 2001375 CrossRef CAS PubMed.
- S. Shan, Y. Zhang, H. Zhao, T. Zeng and X. Zhao, Chemosphere, 2022, 298, 134261 CrossRef CAS PubMed.
- M. Prust, J. Meijer and R. H. S. Westerink, Part. Fibre Toxicol., 2020, 17, 24 CrossRef PubMed.
- V. Kopatz, K. Wen, T. Kovacs, A. S. Keimowitz, V. Pichler, J. Widder, A. D. Vethaak, O. Holloczki and L. Kenner, Nanomaterials, 2023, 13, 1404 CrossRef CAS PubMed.
- X. Liu, Y. Zhao, J. Dou, Q. Hou, J. Cheng and X. Jiang, Nano Lett., 2022, 22, 1091–1099 Search PubMed.
- M. P. Monopoli, C. Aberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed.
- C. D. Walkey, J. B. Olsen, F. Song, R. Liu, H. Guo, D. W. H. Olsen, Y. Cohen, A. Emili and W. C. W. Chan, ACS Nano, 2014, 8, 2439–2455 Search PubMed.
- Y. K. Lee, E.-J. Choi, T. J. Webster, S.-H. Kim and D. Khang, Int. J. Nanomed., 2015, 10, 97–113 Search PubMed.
- D. Westmeier, R. H. Stauber and D. Docter, Toxicol. Appl. Pharmacol., 2016, 299, 53–57 CrossRef CAS PubMed.
- A. Lesniak, F. Fenaroli, M. P. Monopoli, C. Aberg, K. A. Dawson and A. Salvati, ACS Nano, 2012, 6, 5845–5857 CrossRef CAS PubMed.
- V. Francia, K. Yang, S. Deville, C. Reker-Smit, I. Nelissen and A. Salvati, ACS Nano, 2019, 13, 11107–11121 CrossRef CAS PubMed.
- N. Bertrand, P. Grenier, M. Mahmoudi, E. Lima, E. A. Appel, F. Dormont, J.-M. Lim, R. Karnik, R. Langer and O. Farokhzad, Nat. Commun., 2017, 8, 777 CrossRef PubMed.
- H. Leslie and M. Depledge, Environ. Int., 2020, 142, 105807 CrossRef CAS PubMed.
- K. Kadac-Czapska, E. Knez, M. Gierszewska, E. Olewnik-Kruszkowska and M. Grembecka, Materials, 2023, 16, 674 CrossRef CAS PubMed.
- T. Kogel, O. Bjoroy, B. Toto, A. M. Bienfait and M. Sanden, Sci. Total Environ., 2020, 709, 136050 CrossRef CAS PubMed.
- T. Roy, T. K. Dey and M. Jamal, Environ. Monit. Assess., 2022, 195, 27 CrossRef PubMed.
- A. Banerjee and W. L. Shelver, Sci. Total Environ., 2021, 755, 142518 CrossRef CAS PubMed.
- D. M. Mitrano, P. Wick and B. Nowack, Nat. Nanotechnol., 2021, 1–10 Search PubMed.
- EU Commission Report, Nanoplastics: state of knowledge and environmental and human health impacts, 2023.
- L. M. A. Martin, N. Gan, E. Wang, M. Merrill and W. Xu, Environ. Pollut., 2022, 292, 118442 CrossRef CAS PubMed.
- W. Bu, Y. Cui, Y. Jin, X. Wang, M. Jiang, R. Huang, J. Egbobe, X. Zhao and J. Tang, Toxics, 2024, 12(12), 908 CrossRef CAS PubMed.
- C. M. Cary, G. M. DeLoid, Z. Yang, D. Bitounis, M. Polunas, M. J. Goedken, B. Buckley, B. Cheatham, P. A. Stapleton and P. Demokritou, Nanomaterials, 2023, 13(4), 720 CrossRef CAS PubMed.
- S. B. Fournier, J. N. D'Errico, D. S. Adler, S. Kollontzi, M. J. Goedken, L. Fabris, E. J. Yurkow and P. A. Stapleton, Part. Fibre Toxicol., 2020, 17, 55 CrossRef CAS PubMed.
- B. Jeong, J. Y. Baek, J. Koo, S. Park, Y.-K. Ryu, K.-S. Kim, S. Zhang, C. Chung, R. Dogan, H.-S. Choi, D. Um, T.-K. Kim and S.-L. Wang, et al., J. Hazard. Mater., 2022, 426, 127815 CrossRef CAS PubMed.
- L. Zeng, C. Zhou, W. Xu, Y. Huang, W. Wang, Z. Ma, J. Huang, J. Li, L. Hu, Y. Xue, T. Luo and L. Zheng, Ecotoxicol. Environ. Saf., 2023, 257, 114941 CrossRef CAS PubMed.
- Y. Wang, K. Xu, X. Gao, Z. Wei, Q. Han, S. Wang, W. Du and M. Chen, Part. Fibre Toxicol., 2024, 21, 21 CrossRef CAS PubMed.
- J. Zhao, N. V. Adiele, D. Gomes, M. V. Malovichko, D. J. Conklin, A. Ekuban, J. Luo, T. C. Gripshover, W. H. Watson, M. Banerjee, M. L. Smith, E. Rouchka, R. Xu, X. Zhang, D. D. Gondim, M. C. Cave and T. E. O’Toole, Toxicol. Sci., 2024, 198(2), 210–220 CrossRef CAS PubMed.
- X. Fan, X. Wei, H. Hu, B. Zhang, D. Yang, H. Du, R. Zhu, X. Sun, Y. Oh and N. Gu, Chemosphere, 2022, 288, 132607 CrossRef CAS PubMed.
- B. Meesaragandla, D. O. Blessing, S. Karanth, A. Rong, N. Geist and M. Delcea, Macromol. Biosci., 2023, e2200464 CrossRef PubMed.
- L. Rubio, I. Barguilla, J. Domenech, R. Marcos and A. Hernandez, J. Hazard. Mater., 2020, 398, 122900 Search PubMed.
- Y. Kitagishi, M. Kobayashi, K. Kikuta and S. Matsuda, Depress Res. Treat., 2012, 2012, 752563 Search PubMed.
- C. Chu, Y. Zhang, Q. Liu, Y. Pang, Y. Niu and R. Zhang, Ecotoxicol. Environ. Saf., 2022, 241, 113785 CrossRef CAS PubMed.
- J. Chen, L. Yan, Y. Zhang, X. Liu, Y. Wei, Y. Zhao, K. Li, Y. Shi, H. Liu, W. Lai, L. Tian and B. Lin, J. Nanobiotechnol., 2024, 22(1), 651 CrossRef CAS PubMed.
- L. Zou, X. Xu, Y. Wang, F. Lin, C. Zhang, R. Liu, X. Hou, J. Wang, X. Jiang, Q. Zhang and L. Li, Environ. Sci. Technol., 2024, 58(27), 11945–11957 Search PubMed.
- Y. Chen, Y. Nan, L. Xu, A. Dai, R. M. M. Orteg, M. Ma, Y. Zeng and J. Li, Part. Fibre Toxicol., 2025, 22, 13 CrossRef CAS PubMed.
- S. Shan, D. Cheng, H. Li, W. Yao, R. Kou, J. Ji, N. Liu, T. Zeng and X. Zhao, J. Hazard. Mater., 2025, 489, 137615 CrossRef CAS PubMed.
- X. Liang, G. Huang, Y. Wang, N. Andrikopoulos, H. Tang, F. Ding, Y. Li and P. C. Ke, ACS Nano, 2025, 19, 5475–5492 CrossRef CAS PubMed.
- A. Jeong, S. J. Park, E. J. Lee and K. W. Kim, J. Hazard. Mater., 2024, 465, 133289 CrossRef CAS PubMed.
- X. Liang, N. Andrikopoulos, H. Tang, Y. Wang, F. Ding and P. C. Ke, Small, 2024, 20, 2308753 CrossRef CAS PubMed.
- B. Liang, Y. Huang, Y. Zhong, Z. Li, R. Ye, B. Wang, B. Zhang, H. Meng, X. Lin and J. Du, et al., J. Hazard. Mater., 2022, 430, 128459 CrossRef CAS PubMed.
- Z. Liu, A. Sokratian, A. M. Duda, E. Xu, C. Stanhope, A. Fu, S. Strader, H. Li, Y. Yuan, B. G. Bobay, J. Sipe, K. Bai, I. Lundgaard, N. Liu, B. Hernandez, C. Bowes Rickman, S. E. Miller and A. B. West, Sci. Adv., 2023, 9(46), eadi8716 CrossRef CAS PubMed.
- N. Tripathi, F. Saudrais, M. Rysak, L. Pieri, S. Pin, G. Roma, J.-P. Renault and Y. Boulard, Biomacromolecules, 2025, 26(3), 1476–1497 CrossRef CAS PubMed.
- S. J. Park, Int. J. Nanomed., 2020, 15, 5783–5802 Search PubMed.
- N. Liu, M. Tang and J. Ding, Chemosphere, 2019, 245, 125624 CrossRef PubMed.
- C. Corbo, R. Molinaro, A. Parodi, N. T. T. Furman, F. Salvatore and E. Tasciotti, Nanomedicine, 2016, 11(1), 81–100 CrossRef CAS PubMed.
- E. Wyrzykowska, A. Mikolajczyk, I. Lynch, N. Jeliazkova, N. Kochev, H. Sarimveis, P. Doganis, P. Karatzas, A. Afantitis, G. Melagraki, A. Serra, D. Greco, J. Subbotina, V. Lobaskin, M. A. Banares, E. Valsami-Jones, K. Jagiello and T. Puzyn, Nat. Nanotechnol., 2022, 17, 924–932 CrossRef CAS PubMed.
- G. Mancardi, A. Mikolajczyk, V. K. Annapoorani, A. Bahl, K. Blekos, J. Burk, Y. A. Cetin, K. Chairetakis, S. Dutta and L. Escorihuela, et al., Mater. Today, 2023, 67, 344–370 CrossRef.
- D. Power, S. Poggio, H. Lopez and V. Lobaskin, Bionano Interactions: A Key to Mechanistic Understanding of Nanoparticle Toxicity, Computational Nanotoxicology: Challenges and Perspectives, ed. T. Puzyn and A. Gajewicz, Jenny Stanford Publishing, 2019, pp. 189–215 Search PubMed.
- O. Vilanova, J. J. Mittag, P. M. Kelly, S. Milani, K. A. Dawson, J. O. Radler and G. Franzese, ACS Nano, 2016, 10, 10842–10850 CrossRef CAS PubMed.
- I. Hasenkopf, R. Mills-Goodlet, L. Johnson, I. Rouse, M. Geppert, A. Duschl, D. Maier, V. Lobaskin, I. Lynch and M. Himly, Nano Today, 2022, 46, 101561 CrossRef CAS.
- P. Sarker, M. S. J. Sajib, X. Tao and T. Wei, J. Phys. Chem. B, 2022, 126(3), 601–608 CrossRef CAS PubMed.
- H. Lee, Pharmaceutics, 2024, 16(11), 1419 CrossRef CAS PubMed.
- O. Vilanova, A. Martinez-Serra, M. Monopoli and G. Franzese, Front. Nanotechnol., 2024, 6, 1531039 CrossRef.
- Z. Ban, P. Yuan, F. Yu, T. Peng, Q. Zhou and X. Hu, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 10492–10499 CrossRef CAS PubMed.
- J. Huzar, R. Coreas, M. Landry and G. Tikhomirov, ACS Nano, 2025, 19, 4333–4345 CrossRef CAS PubMed.
- X. Fu, C. Yang, Y. Su, C. Liu, H. Qiu, Y. Yu, G. Su, Q. Zhang, L. Wei, F. Cui, Q. Zou and Z. Zhang, Research, 2024, 7, 0487 CrossRef CAS PubMed.
- D. Power, I. Rouse, S. Poggio, E. Brandt, H. Lopez, A. Lyubartsev and V. Lobaskin, Modell. Simul. Mater. Sci. Eng., 2019, 27, 084003 CrossRef CAS.
- I. Rouse and V. Lobaskin, Biophys. J., 2021, 120, 4457–4471 CrossRef PubMed.
- J. Subbotina and V. Lobaskin, J. Phys. Chem. B, 2022, 126, 1301–1314 CrossRef CAS PubMed.
- J. Subbotina, I. Rouse and V. Lobaskin, Nanoscale, 2023, 15, 13371–13383 RSC.
- I. Rouse, D. Power, J. Subbotina and V. Lobaskin, J. Chem. Inf. Model., 2024, 64, 7525–7543 CrossRef CAS PubMed.
- P. Mosaddeghi Amini, J. Subbotina and V. Lobaskin, Nanomaterials, 2023, 13, 1857 CrossRef PubMed.
- H. Lopez and V. Lobaskin, J. Chem. Phys., 2015, 143, 243138 CrossRef PubMed.
- H. Lopez, E. G. Brandt, A. Mirzoev, D. Zhurkin, A. Lyubartsev and V. Lobaskin, Advances in Experimental Medicine and Biology, Springer International Publishing, 2017, pp. 173–206 Search PubMed.
- S. A. Alsharif, D. Power, I. Rouse and V. Lobaskin, Nanomaterials, 2020, 10, 1967 CrossRef CAS PubMed.
- I. Rouse and V. Lobaskin, Faraday Discuss., 2023, 244, 306–335 RSC.
- J. N. Israelachvili, The Handbook of Surface Imaging and Visualization, CRC Press, 2022, pp. 793–816 Search PubMed.
- I. Rouse, D. Power, E. G. Brandt, M. Schneemilch, K. Kotsis, N. Quirke, A. P. Lyubartsev and V. Lobaskin, Phys. Chem. Chem. Phys., 2021, 23, 13473–13482 RSC.
- B. Kharazian, N. Hadipour and M. Ejtehadi, Int. J. Biochem. Cell Biol., 2016, 75, 162–174 CrossRef CAS PubMed.
- K. Nienhaus and G. U. Nienhaus, Small, 2023, 19, 2301663 CrossRef CAS PubMed.
- V. Karunakaran Annapoorani, S. Dutta, O. Ajia, O. A. Petrisor, V. Lobaskin and N.-V. Buchete, J. Phys. Chem. Lett., 2025, 16, 11356–11364 CrossRef CAS PubMed.
- H. Pan, M. Qin, W. Meng, Y. Cao and W. Wang, Langmuir, 2012, 28, 12779–12787 CrossRef CAS PubMed.
- A. F. Lima and A. A. Sousa, Physchem, 2023, 3, 385–410 Search PubMed.
- H. Lee, Small, 2020, 16, 1906598 CrossRef CAS PubMed.
- J. Talbot, X. Jin and N.-H. L. Wang, Langmuir, 1994, 10, 1663–1666 CrossRef CAS.
- J. Talbot, G. Tarjus, P. Van Tassel and P. Viot, Colloids Surf., A, 2000, 165, 287–324 CrossRef CAS.
- V. P. Zhdanov and N.-J. Cho, Math. Biosci., 2016, 282, 82–90 CrossRef CAS PubMed.
- P. Mosaddeghi Amini, I. Rouse, J. Subbotina and V. Lobaskin, Beilstein J. Nanotechnol., 2024, 15, 215–229 Search PubMed.
- M. v. Smoluchowski, Z. fur Phys. Chem., 1918, 92, 129–168 Search PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi, C. Reinhardt, K. Landfester, H. Schild, M. Maskos, S. Knauer and R. Stauber, Nat. Nanotechnol., 2013, 8(10), 772–781 Search PubMed.
- P.-L. Latreille, J.-M. Rabanel, M. Le Goas, S. Salimi, J. Arlt, S. A. Patten, C. Ramassamy, P. Hildgen, V. A. Martinez and X. Banquy, Adv. Mater., 2022, 34, 2203354 CrossRef CAS PubMed.
- S. M. Bhattacharjee, A. Giacometti and A. Maritan, J. Phys.: Condens. Matter, 2013, 25, 503101 CrossRef PubMed.
- P. J. Flory, Principles of Polymer Chemistry, Cornell University Press, Ithaca, NY, 1953 Search PubMed.
- L. Ramirez, S. Ramseier Gentile, S. Zimmermann and S. Stoll, Water, 2019, 11, 721 CrossRef CAS.
- A. Lappala, S. Mendiratta and E. M. Terentjev, Macromolecules, 2015, 48, 1894–1900 CrossRef CAS.
- T. I. Morozova and A. Nikoubashman, J. Phys. Chem. B, 2018, 122, 2130–2137 CrossRef CAS PubMed.
- W.-L. Chen, R. Cordero, H. Tran and C. K. Ober, Macromolecules, 2017, 50, 4089–4113 CrossRef CAS.
- T. Trumpour and J. A. Forrest, Eur. Phys. J. E, 2022, 45, 85 Search PubMed.
- N. Hueppe, F. R. Wurm and K. Landfester, Macromol. Rapid Commun., 2022, 2200611 Search PubMed.
- G.-B. Im, Y. G. Kim, I.-S. Jo, T. Y. Yoo, S.-W. Kim, H. S. Park, T. Hyeon, G.-R. Yi and S. H. Bhang, J. Hazard. Mater., 2022, 430, 128411 Search PubMed.
- T. Du, X. Yu, S. Shao, T. Li, S. Xu and L. Wu, Environ. Sci. Technol., 2023, 57, 3206–3217 CrossRef CAS PubMed.
- S. Ducoli, S. Federici, R. Nicsanu, A. Zendrini, C. Marchesi, L. Paolini, A. Radeghieri, P. Bergese and L. E. Depero, Environ. Sci.: Nano, 2022, 9, 1414–1426 RSC.
- J. Gigault, H. El Hadri, B. Nguyen, B. Grassl, L. Rowenczyk, N. Tufenkji, S. Feng and M. Wiesner, Nat. Nanotechnol., 2021, 1–7 Search PubMed.
- D. Choi, J. Hwang, J. Bang, S. Han, T. Kim, Y. Oh, Y. Hwang, J. Choi and J. Hong, Sci. Total Environ., 2021, 752, 142242 CrossRef CAS PubMed.
- A. Schrade, V. Mailander, S. Ritz, K. Landfester and U. Ziener, Macromol. Biosci., 2012, 12, 1459–1471 Search PubMed.
- S. Lerch, M. Dass, A. Musyanovych, K. Landfester and V. Mailander, Eur. J. Pharm. Biopharm., 2013, 84, 265–274 Search PubMed.
- H.-J. Butt, K. Graf and M. Kappl, Physics and chemistry of interfaces, John Wiley & Sons, 2023 Search PubMed.
- S. Mahanta, P. Vallejo-Ramirez, N. Karedla, P. Puczkarski and M. Krishnan, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2209955119 CrossRef CAS PubMed.
- United Atom/NPCoronaPredict software at GitHub, 2024, https://github.com/ucd-softmatterlab/NPCoronaPredict.
- S. Sugio, A. Kashima, S. Mochizuki, M. Noda and K. Kobayashi, Protein Eng., Des. Sel., 1999, 12, 439–446 CrossRef CAS PubMed.
- J. M. Kollman, L. Pandi, M. R. Sawaya, M. Riley and R. F. Doolittle, Biochemistry, 2009, 48, 3877–3886 CrossRef CAS PubMed.
- E. O. Saphire, P. W. H. I. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton and I. A. Wilson, Science, 2001, 293, 1155–1159 CrossRef CAS PubMed.
- B. J. C. Janssen, E. G. Huizinga, H. C. A. Raaijmakers, A. Roos, M. R. Daha, K. Nilsson-Ekdahl, B. Nilsson and P. Gros, Nature, 2005, 437(7058), 505–511 CrossRef CAS PubMed.
- Z. Wu, V. Gogonea, X. Lee, M. A. Wagner, X.-M. Li, Y. Huang, A. Undurti, R. P. May, M. Haertlein, M. Moulin, I. Gutsche, G. Zaccai, J. A. DiDonato and S. L. Hazen, J. Biol. Chem., 2009, 284, 36605–36619 CrossRef CAS PubMed.
- S. Jo, X. Cheng, J. Lee, S. Kim, S.-J. Park, D. S. Patel, A. H. Beaven, K. I. Lee, H. Rui and S. Park, et al., J. Comput. Chem., 2017, 38, 1114–1124 CrossRef CAS PubMed.
- M. H. Olsson, C. R. Sondergaard, M. Rostkowski and J. H. Jensen, J. Chem. Theory Comput., 2011, 7, 525–537 CrossRef CAS PubMed.
- V. Mishra and R. J. Heath, Int. J. Mol. Sci., 2021, 22, 8411 CrossRef CAS PubMed.
- A. Sahu and J. D. Lambris, Immunol. Rev., 2001, 180, 35–48 Search PubMed.
- X. Yu, F. Hu, Q. Yao, C. Li, H. Zhang and Y. Xue, BMC Cancer, 2016, 16, 1–12 Search PubMed.
- J. B. Karlin, D. J. Juhn, J. Starr, A. M. Scanu and A. H. Rubenstein, J. Lipid Res., 1976, 17, 30–37 CrossRef CAS.
- A. Leino, O. Impivaara, M. Kaitsaari and J. Jarvisalo, Clin. Chem., 1995, 41, 1633–1636 Search PubMed.
- M. H. Jazayeri, A. A. Pourfathollah, M. J. Rasaee, Z. Porpak and M. E. Jafari, Biomed. Aging Pathol., 2013, 3, 241–245 CrossRef CAS.
- M. J. Sippl, M. Ortner, M. Jaritz, P. Lackner and H. Flockner, Folding Des., 1996, 1, 289–298 Search PubMed.
- S. Pronk, S. Pall, R. Schulz, P. Larsson, P. Bjelkmar, R. Apostolov, M. R. Shirts, J. C. Smith, P. M. Kasson, D. van der Spoel, B. Hess and E. Lindahl, Bioinformatics, 2013, 29, 845–854 Search PubMed.
- J. Huang, S. Rauscher, G. Nawrocki, T. Ran, M. Feig, B. L. de Groot, H. Grubmuller and A. D. MacKerell, Nat. Methods, 2016, 14, 71–73 CrossRef PubMed.
- J. Subbotina, P. D. Kolokathis, A. Tsoumanis, N. K. Sidiropoulos, I. Rouse, I. Lynch, V. Lobaskin and A. Afantitis, J. Chem. Inf. Model., 2025, 65, 3142–3153 Search PubMed.
- N. Ganji and G. D. Bothun, Environ. Sci.: Nano, 2021, 8, 160–173 Search PubMed.
- U. Janke, N. Geist, E. Weilbeer, W. Levin and M. Delcea, ChemBioChem, 2024, e202400188 Search PubMed.
- D. Rajendran and N. Chandrasekaran, ACS Omega, 2023, 8, 43719–43731 Search PubMed.
- J. Shi, F. Tian, J. Lyu and M. Yang, J. Mater. Chem. B, 2015, 3, 6989–7005 RSC.
- J. Ghuman, P. Zunszain, I. Petitpas, A. A. Bhattacharya, M. Otagiri and S. Curry, J. Mol. Biol., 2005, 353(1), 38–52 CrossRef CAS PubMed.
- B. J. Janssen, E. G. Huizinga, H. C. Raaijmakers, A. Roos, M. R. Daha, K. Nilsson-Ekdahl, B. Nilsson and P. Gros, Nature, 2005, 437, 505–511 CrossRef CAS PubMed.
- P. Gros, F. J. Milder and B. J. Janssen, Nat. Rev. Immunol., 2008, 8, 48–58 CrossRef CAS PubMed.
- Z. Chen, M. Wang, W. Duan, Y. Xia, H. Liu and F. Qian, J. Biol. Chem., 2025, 301, 108250 Search PubMed.
- B. V. Geisbrecht, J. D. Lambris and P. Gros, Semin. Immunol., 2022, 101627 CrossRef CAS PubMed.
- B. Wang, X. Zhong, L. Fields, H. Lu, Z. Zhu and L. Li, J. Am. Soc. Mass Spectrom., 2023, 34, 459–471 CrossRef CAS PubMed.
- Y. Arima, M. Toda and H. Iwata, Adv. Drug Delivery Rev., 2011, 63, 988–999 Search PubMed.
- H. Pedersen, R. K. Jensen, J. M. B. Jensen, R. Fox, D. V. Pedersen, H. G. Olesen, A. G. Hansen, D. Christiansen, S. M. Mazarakis and N. Lojek, et al., J. Immunol., 2020, 205, 2287–2300 Search PubMed.
- B. J. Janssen, E. F. Halff, J. D. Lambris and P. Gros, J. Biol. Chem., 2007, 282, 29241–29247 Search PubMed.
- C. Lamers, D. C. Mastellos, D. Ricklin and J. D. Lambris, Trends Pharmacol. Sci., 2022, 43, 629–640 Search PubMed.
- T. E. Mollnes, B. S. Storm, O. L. Brekke, P. H. Nilsson and J. D. Lambris, Semin. Immunol., 2022, 101604 CrossRef CAS PubMed.
- S. M. Moghimi, A. J. Andersen, D. Ahmadvand, P. P. Wibroe, T. L. Andresen and A. C. Hunter, Adv. Drug Delivery Rev., 2011, 63, 1000–1007 Search PubMed.
- A. J. Pinto, A. Chouquet, I. Bally, V. Rossi, N. M. Thielens, C. Dumestre-Pérard, R. Kunert, C. Gaboriaud, W. L. Ling and J.-B. Reiser, FEBS J., 2025 DOI:10.1111/febs.70309.
- M. Tonigold, J. Simon, D. Estupinan, M. Kokkinopoulou, J. Reinholz, U. Kintzel, A. Kaltbeitzel, P. Renz, M. P. Domogalla and K. Steinbrink, et al., Nat. Nanotechnol., 2018, 13, 862–869 Search PubMed.
- N. Javkhlantugs, H. Bayar, C. Ganzorig and K. Ueda, Int. J. Nanomed., 2013, 2487–2496 Search PubMed.
- J. Wang, X. Chen, M. L. Clarke and Z. Chen, J. Phys. Chem. B, 2006, 110, 5017–5024 Search PubMed.
- T. Lu and Z. Chen, J. Phys. Chem. Lett., 2023, 3139–3145 Search PubMed.
- J. Gao, P. Stengel, T. Lu, Y. Wu, D. D. Hawker, K. E. Gutowski, J. M. Hankett, M. Kellermeier and Z. Chen, Langmuir, 2023, 39, 12270–12282 CrossRef CAS PubMed.
- S. Kohler, F. Schmid and G. Settanni, Langmuir, 2015, 31, 13180–13190 CrossRef PubMed.
- A. Bratek-Skicki, P. Zeliszewska and Z. Adamczyk, Colloids Surf., B, 2013, 103, 482–488 CrossRef CAS PubMed.
- H. Derakhshankhah, A. Hosseini, F. Taghavi, S. Jafari, A. Lotfabadi, M. R. Ejtehadi, S. Shahbazi, A. Fattahi, A. Ghasemi and E. Barzegari, et al., Sci. Rep., 2019, 9, 1558 Search PubMed.
- G. Dežecalić, N. Deželić and Ž. Telišman, Eur. J. Biochem., 1971, 23, 575–581 Search PubMed.
- S. McColman, R. Li, S. Osman, A. Bishop, K. P. Wilkie and D. T. Cramb, Nanoscale, 2021, 13, 20550–20563 RSC.
- D. Prozeller, C. Rosenauer, S. Morsbach and K. Landfester, Biointerphases, 2020, 15(3), 031009 CrossRef CAS PubMed.
- R. Cukalevski, M. Lundqvist, C. Oslakovic, B. Dahlback, S. Linse and T. Cedervall, Langmuir, 2011, 27(23), 14360–14369 CrossRef CAS PubMed.
- X. Wang, J. Zhao, S. Ding and H. Zhang, Int. J. Biol. Macromol., 2023, 238, 124049 CrossRef CAS PubMed.
- M. M. Gruber, B. Hirschmugl, N. Berger, M. Holter, S. Radulović, G. Leitinger, L. Liesinger, A. Berghold, E. Roblegg and R. Birner-Gruenberger, et al., J. Nanobiotechnology, 2020, 18, 1–14 CrossRef PubMed.
- M. N. Alemie, R. Bright, N. H. Nguyen, V. K. Truong, D. Palms, J. D. Hayball and K. Vasilev, ACS Appl. Mater. Interfaces, 2024, 16(38), 50507–50523 Search PubMed.
- S. Kohler, F. Schmid and G. Settanni, PLoS Comput. Biol., 2015, 11, e1004346 CrossRef PubMed.
- Z. J. Deng, M. Liang, I. Toth, M. J. Monteiro and R. F. Minchin, ACS Nano, 2012, 6, 8962–8969 CrossRef CAS PubMed.
- S. Yu, M. Shen, S. Li, Y. Fu, D. Zhang, H. Liu and J. Liu, Environ. Pollut., 2019, 255, 113302 CrossRef CAS PubMed.
- M. Auguste, T. Balbi, A. Miglioli, S. Alberti, S. Prandi, R. Narizzano, A. Salis, G. Damonte and L. Canesi, Nanomaterials, 2021, 11, 3291 CrossRef CAS PubMed.
- S. Panico, S. Capolla, S. Bozzer, G. Toffoli, M. Dal Bo and P. Macor, Pharmaceutics, 2022, 14(12), 2605 CrossRef CAS PubMed.
- V. Vu, G. B. Gifford, F. Chen, H. Benasutti, G. Wang, E. Groman, R. Scheinman, L. Saba, S. Moghimi and D. Simberg, Nat. Nanotechnol., 2018, 14, 260–268 Search PubMed.
- A. Sellborn, M. Andersson, J. Hedlund, J. Andersson, M. Berglin and H. Elwing, Mol. Immunol., 2005, 425, 569–574 CrossRef PubMed.
- T. Meszaros, G. T. Kozma, T. Shimizu, K. Miyahara, K. Turjeman, T. Ishida, Y. Barenholz, R. Urbanics and J. Szebeni, Int. J. Nanomed., 2018, 13, 6345–6357 Search PubMed.
- M. Noris and G. Remuzzi, Semin. Nephrol., 2013, 33, 479–492 Search PubMed.
- N. S. Merle, S. E. Church, V. Fremeaux-Bacchi and L. T. Roumenina, Front. Immunol., 2015, 6, 262 Search PubMed.
- R. Franzin, A. Stasi, M. Fiorentino, G. Stallone, V. Cantaluppi, L. Gesualdo and G. Castellano, Front. Immunol., 2020, 11, 734 CrossRef CAS PubMed.
- D. Perez Sierra, A. Tripathi and A. Pillai, Biomark. Neuropsychiatry, 2022, 7, 100056 CrossRef PubMed.
- O. Fatoba, T. Itokazu and T. Yamashita, FEBS J., 2022, 289, 2085–2109 CrossRef CAS PubMed.
- F. Chen, G. Wang, J. I. Griffin, B. Brenneman, N. K. Banda, V. M. Holers, D. S. Backos, L. Wu, S. M. Moghimi and D. Simberg, Nat. Nanotechnol., 2016, 12(4), 387–393 CrossRef PubMed.
- S. Moghimi and T. Andresen, Mol. Immunol., 2009, 46, 1571–1572 Search PubMed.
- R. Cornelius, J. Archambault and J. Brash, Biomaterials, 2002, 23(17), 3583–3587 Search PubMed.
- R. Cornelius, J. Macri and J. Brash, J. Biomed. Mater. Res., Part A, 2011, 99(1), 109–115 CrossRef PubMed.
- R. Cornelius, J. Macri, K. Cornelius and J. Brash, Langmuir, 2015, 31(44), 12087–12095 CrossRef CAS PubMed.
- X. Liu, W. Wei, Z. Liu, E. Song, J. Lou, L. Feng, R. Huang, C. Chen, P. C. Ke and Y. Song, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2108131118 CrossRef CAS PubMed.
- L. Treuel, S. Brandholt, P. Maffre, S. Wiegele, L. Shang and G. Nienhaus, ACS Nano, 2014, 8(1), 503–513 CrossRef CAS PubMed.
- K. Yamasaki, V. T. G. Chuang, T. Maruyama and M. Otagiri, Biochim. Biophys. Acta, Gen. Subj., 2013, 1830, 5435–5443 CrossRef CAS PubMed.
- N. H. Moghadam, S. Salehzadeh, J. Rakhtshah, A. H. Moghadam, H. Tanzadehpanah and M. Saidijam, Int. J. Biol. Macromol., 2019, 125, 931–940 Search PubMed.
- X. Zhang, B. Niebuur, P. Chytil, T. Etrych, S. K. Filippov, A. Kikhney, D. C. F. Wieland, D. Svergun and C. Papadakis, Biomacromolecules, 2018, 19(2), 470–480 CrossRef CAS PubMed.
- F. Yang, Y. Zhang and H. Liang, Int. J. Mol. Sci., 2014, 15, 3580–3595 CrossRef CAS PubMed.
- Z. Zhivkova, Curr. Pharm. Des., 2015, 21(14), 1817–1830 CrossRef CAS PubMed.
- Z. Sunil, J. Thomas, M. Shivashankar and N. Chandrasekaran, Environ. Sci.: Nano, 2024, 11, 1012–1022 RSC.
- P. Daneshgar, A. A. Moosavi-Movahedi, P. Norouzi, M. R. Ganjali, A. Madadkar-Sobhani and A. A. Saboury, Int. J. Biol. Macromol., 2009, 45, 129–134 CrossRef CAS PubMed.
- L. J. Juang, W. S. Hur, L. M. Silva, A. W. Strilchuk, B. Francisco, J. Leung, M. K. Robertson, D. J. Groeneveld, B. La Prairie, E. M. Chun, A. P. Cap, J. P. Luyendyk, J. S. Palumbo, P. R. Cullis, T. H. Bugge, M. J. Flick and C. J. Kastrup, Blood, 2022, 139(9), 1302–1311 CrossRef CAS PubMed.
- Z. J. Deng, M. Liang, M. Monteiro, I. Toth and R. F. Minchin, Nat. Nanotechnol., 2011, 6, 39–44 CrossRef CAS PubMed.
- J. Liu, Y. Ma, D. Zhu, T. Xia, Y. Qi, Y. Yao, X. Guo, R. Ji and W. Chen, Environ. Sci. Technol., 2018, 52(5), 2677–2685 CrossRef CAS PubMed.
- X. Li, E. He, K. Jiang, W. Peijnenburg and H. Qiu, Water Res., 2020, 190, 116742 CrossRef PubMed.
- Z. Dong, Y. Hou, W. Han, M. Liu, J. Wang and Y. Qiu, Water Res., 2020, 182, 115978 CrossRef CAS PubMed.
- S. Kihara, N. J. van der Heijden, C. Seal, J. Mata, A. Whitten, I. Koper and D. McGillivray, Bioconjugate Chem., 2019, 304, 1067–1076 CrossRef PubMed.
- R. Cukalevski, S. A. Ferreira, C. Dunning, T. Berggard and T. Cedervall, Nano Res., 2015, 8, 2733–2743 CrossRef CAS.
- M. Kendall, P. Ding and K. Kendall, Nanotoxicology, 2011, 5, 55–65 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |