Carbon-based nanozymes: catalytic mechanisms, performance tuning, and environmental and biomedical applications
Hongying
Ye
,
Yongquan
Lai
,
Zhaodi
Wu
,
Guizhen
Li
,
Qingqing
Hua
and
Wenyuan
Zhu
 *
*
Guangxi Key Laboratory of Electrochemical and Magneto-Chemical Functional Materials, College of Chemistry and Bioengineering, Guilin University of Technology, Guilin 541006, P. R. China. E-mail: wyzhu@glut.edu.cn
First published on 14th July 2025
Abstract
Carbon-based nanozymes—a significant subclass of nanomaterials mimicking natural enzyme catalytic functions—offer superior thermal/chemical stability and enhanced biocompatibility compared to natural enzymes and other nanozyme types, owing to their unique carbon matrix. This review comprehensively examines research progress in carbon-based nanozymes. It details their classification into three categories: (1) carbon nanozymes (e.g., fullerenes, graphene, carbon dots, and nanotubes), (2) heteroatom-doped variants (N, P, S, and Se), and (3) metal/metal oxide-supported types, including single-atom nanozymes with M–Nx–C sites. The primary catalytic mechanisms, focusing on peroxidase (POD)-like and superoxide dismutase (SOD)-like activities, alongside oxidase (OXD)-like and catalase (CAT)-like mechanisms, are discussed. Strategies for regulating nanozyme performance—such as size/morphology control, composition/structure tuning (doping and defect engineering), surface modification, biomolecular interactions, and external environment manipulation (pH and temperature)—are highlighted. The review emphasizes the broad applications of carbon-based nanozymes in environmental engineering (pollutant detection/degradation), biosensing (H2O2, biomolecules, antioxidants, and tumor markers), and biomedicine (antioxidant/anti-inflammatory therapy, tumor treatment, antibacterial/antiviral applications, and bioimaging). Finally, it addresses existing challenges, including relatively lower activity/specificity compared to natural enzymes, limited enzyme-mimicking types, unclear atomic-scale mechanisms, synthesis control difficulties, biocompatibility/safety concerns, and the need for standardized research frameworks. This overview underscores the immense potential and future research directions for carbon-based nanozymes.
1. Introduction
Natural enzymes, which are proteins or RNA with high specificity and catalytic efficiency produced by living cells,1 are widely used in industrial and biomedical fields.2–5 Although natural enzymes play irreplaceable roles in various fields owing to their biological catalytic functions, they exhibit inevitable limitations.6,7 For instance, they are easily inactivated under non-physiological conditions,6 thus irreversibly losing their catalytic activity. In addition, the extraction and purification processes of natural enzymes are complex, time-consuming and costly.6 These inherent drawbacks significantly hinder the practical applications of natural enzymes. In order to avoid the above-mentioned limitations and broaden the application fields, researchers have successively proposed substitutes with catalytic activity comparable to that of natural enzymes—nanozymes.Nanozymes are a novel class of nanomaterials with catalytic functions8 combining the unique properties of nanomaterials with the catalytic characteristics of natural enzymes.9 They are now widely used in biosensing,9,10 disease treatment,11,12 and environmental engineering.8 Since the discovery of Fe3O4 nanoparticles exhibiting peroxidase (POD)-like activity in 2007,13 various nanomaterials with enzyme-mimicking properties, including metals, metal oxides, carbon-based nanomaterials, and metal–organic frameworks (MOFs),14 have been developed. These materials achieve efficient catalysis through surface effects and size effects, among other mechanisms. Compared to natural enzymes, nanozymes exhibit superior stability and are less affected by harsh conditions such as high temperatures and extreme pH.9 Furthermore, their preparation is typically simple and cost-effective,15 facilitating large-scale production.
Carbon-based nanozymes are a new class of synthetic nanomaterials16 and a significant category within nanomaterial research. They hold unique advantages in the field, being primarily composed of carbon-based materials, including fullerenes, graphene,17 carbon nanotubes, carbon quantum dots, and graphene quantum dots.14,18 Through specific designs and techniques, these materials can imitate the catalytic functions of natural enzymes.
Generally speaking, carbon-based nanozymes exhibit the following characteristics:18 first, they provide abundant active sites,7 thereby enabling efficient catalysis of various reactions to meet diverse catalytic needs. Furthermore, compared with natural enzymes, they exhibit better thermal and chemical stability and can maintain stable catalytic activity even in harsh environments.7 Additionally, carbon-based nanozymes demonstrate good biocompatibility19 and biosafety,12 implying low risks and side effects in biomedical applications. Notably, their catalytic activity can be regulated by methods, such as chemical modification and doping,20 optimizing the performance and specificity to meet different application requirements. Moreover, their manufacturing costs are relatively low, aligning with sustainability principles. Specifically, carbon materials are abundant, environmentally friendly, and low-cost, fitting well with sustainable development principles. Last but not least, carbon-based nanozymes often possess catalytic properties along with additional functionalities such as conductivity and adsorption.
This article provides a comprehensive overview of carbon-based nanozymes, including their classification, diverse catalytic mechanisms, and performance regulation strategies. Furthermore, research on their applications in multiple domains will be discussed in detail, and the existing challenges will be summarized.
2. Categorization of carbon-based nanozymes
Carbon, a fundamental component of all living things on Earth, can form stable covalent bonds with various atoms to construct complex structures. So far, a variety of carbon-based nanozymes have been developed. Based on composition and structural characteristics, they are primarily classified into three categories: carbon nanozymes, heteroatom-doped carbon-based nanozymes, and metal/metal oxide-supported carbon-based nanozymes.21 The synthesis of these carbon-based nanozymes not only reflects the structural diversity of carbon-based materials but also fosters in-depth research into the catalytic mechanisms of carbon-based materials and their application potential in various fields.2.1. Carbon-based nanozymes
Carbon nanozymes can be traced back to 1996 when Dugan et al.22 discovered that fullerene derivatives possess superoxide dismutase-like activity and can scavenge superoxide anion radicals. Since then, research has been conducted on carbon-based materials, including fullerenes, graphene,17 carbon dots, carbon nanotubes, carbon nanospheres and carbon nanofibers.23 Their enzyme-like activities have increasingly been studied,14 often focusing on the construction of their derivatives.Among these carbon nanozymes:
• Fullerene nanozymes often exhibit nuclease-like activity and possess rich chemical composition, small size, and uniform dispersibility. However, their closed structure limits the overall number of active sites, resulting in relatively low catalytic activity.
• Graphene nanozymes exhibit excellent peroxidase (POD)-like activity. For instance, Huang et al.17 prepared POD-active graphene-based materials using hydrothermal reduction with Ganoderma lucidum polysaccharides. Carboxylated graphene oxide (GO–COOH) exhibits stronger affinity for organic TMB due to the π–π conjugation effect.24
• Carbon nanotube nanozymes (Fig. 1c) like their carbon dot counterparts exhibit enhanced peroxidase (POD)-like activity when modified with functional groups. For example, Wang et al.25 synthesized oxidized carbon nanotubes (o-CNTs) using green methods (Fig. 2),25 confirming their POD-like activity with abundant surface oxygen-containing functional groups identified as the catalytic active sites.
 | ||
| Fig. 1 Classification of carbon-based nanozymes: (a) fullerene nanozymes, (b) graphene nanozymes, (c) carbon nanotube nanozymes, and (d) carbon dot nanozymes. | ||
• Carbon dot nanozymes commonly bear abundant hydrophilic functional groups (e.g., carbonyl, carboxyl, and hydroxyl25) on their surfaces. These groups provide numerous active sites for interacting with substrates, thereby enhancing the catalytic efficiency. Additionally, they confer high water solubility and ease of functionalization. Leveraging these characteristics, carbon dot nanozymes and related carbon nanomaterials show broad application prospects in biomedicine, clinical diagnosis, and biocatalysis.
• Carbon nanosphere nanozymes can demonstrate various enzyme-like activities through elemental doping, while their closed structure helps minimize interference from oxygen defects.
• Carbon nanofiber nanozymes are a class of artificial enzyme-mimicking materials based on carbon nanofibers. Their catalytic activity and selectivity can be modulated through strategies such as doping (e.g., with nitrogen or boron) or surface functionalization (e.g., by introducing metal nanoparticles). Research has shown that defective sites and edge carbon atoms on the surface of carbon nanofibers can catalyze the decomposition of hydrogen peroxide to generate reactive oxygen species (ROS). This property enables their application in areas like tumor therapy and pollutant degradation.26
Collectively, these studies provide valuable insights for the rational design of carbon-based nanozymes and their derivatives. However, for pure carbon materials, their inherent rigid structure may result in relatively low activity or insufficient substrate recognition. In addition, these carbon-based nanozymes incorporate responsive carbon-based materials, which can undergo controllable structural or property changes in response to external stimuli such as light, heat, pH, electric fields, or magnetic fields. Crucially, the electron distribution within these carbon materials can be modulated through heteroatom doping (e.g., N and S). This modulation is particularly effective in enhancing their responsiveness to specific stimuli like light and electric fields. Therefore, constructing enzyme-like active sites through element doping presents a promising strategy to enhance their activity, specificity and application fields.
2.2. Hetero-atom-doped carbon-based nanozymes
Carbon-based nanozymes regulate their specificity and catalytic activity by introducing heteroatoms into carbon materials. These heteroatoms, such as nitrogen (N), phosphorus (P), sulfur (S), and selenium (Se), alter the electronic structure and surface properties of materials like carbon dots, graphene, and carbon nanotubes. Single heteroatom doping is the most fundamental strategy to modulate the electronic structure of carbon-based nanozymes. By precisely controlling the type, number and position of the dopant atoms, the electronic distribution and energy band structure of carbon materials can be targeted to modulate their enzyme-like catalytic activity. Among the many dopant elements, nitrogen (N) is the most widely studied, primarily due to its similar atomic radius to carbon and unique electron-donating properties. The nitrogen atom (five valence electrons) replaces a carbon atom (four valence electrons) in the lattice, contributing an additional electron to the π-conjugated system.27 This shifts the Fermi energy level upward and increases the electron density,28 endowing nitrogen-doped carbon materials with enhanced electron-donating capacity29 that facilitates electron transfer in catalysis.Beyond nitrogen, other non-metallic heteroatoms exhibit distinct modulation effects. For example, boron (B) doping introduces electron holes due to its three valence electrons30—one less than carbon. Consequently, this electron-deficient state enables p-type semiconductor properties, allowing attraction of electrons from oxygen or H2O2 to facilitate their decomposition. Sulfur (S) and phosphorus (P) doping introduce more complex electronic effects.31,32 Their larger atomic radii (vs. carbon) induce localized strains and lattice defects that alter the charge distribution. Sulfur-doped materials specifically show excellent redox activity,32 attributed to sulfur-induced spin density redistribution and charge polarization.
In conclusion, heteroatom-doped carbon-based nanozymes demonstrate superior selectivity in mimicking natural enzymes compared to undoped analogues, showing significant research and application potential.
The synthesis of these nanozymes employs diverse methods, including hydrothermal treatment, chemical vapor deposition (CVD), and solution-based approaches. For example, nitrogen-doped carbon dots (N-CDs), nitrogen/sulfur-doped carbon dots (N,S-CDs), and sulfur-doped reduced graphene oxide (S-rGO)20 can be synthesized hydrothermally. Wu et al.20 prepared sulfur-doped graphene oxide (S-GO) via a simple hydrothermal reaction, eliminating the need for templates and surfactants. Compared to its undoped precursor, this material showed significantly enhanced peroxidase (POD)-like activity, indicating that sulfur-containing groups serve as major active sites. The chemical vapor deposition (CVD) method produces large-area, high-quality graphene and carbon nanotubes. Solution-based methods are straightforward and scalable. Lou et al.33 synthesized a highly N-doped carbon nanozyme (Fig. 3a), with XPS analysis revealing four nitrogen configurations: pyridinic N, graphitic N, pyrrolic N, and oxidized N. Yan et al.34 employed secondary N-doping to fabricate porous carbon nanozymes with a tunable pyridinic-N content, exhibiting peroxidase-like activity (Fig. 3b). Their study demonstrated a positive correlation between the pyridinic-N content and peroxidase (POD)-like activity (Fig. 3e–g). In contrast, Gu et al.35 reported distinct catalytic roles for nitrogen species in iron-coordinated systems: pyrrolic-N coordinates Fe(III) as the dominant active site, while pyridinic-N coordinates Fe(II) with minimal activity contribution. The low D-band center position at pyrrolic-N–Fe sites facilitates Fe–oxo intermediate formation, enhancing the catalytic performance.
Beyond nitrogen doping, non-metallic sites (e.g., Se) in graphene confer enzyme-mimicking activities. For example, GO–Se nanozymes (Fig. 3c) exhibit glutathione peroxidase (GPx)-like activity.36 Co-doping synergistically enhances activity versus single-heteroatom doping. Chen et al.37 achieved N,S-co-doped porous carbon nanozymes showing 3.9-fold higher specific activity and 11.8-fold greater H2O2 affinity than N-doped analogs, enabling TAC detection in beverages via POD-like activity. Kim et al.38 synthesized N,B-co-doped rGO (NB-rGO), exhibiting 40-fold and 35-fold higher catalytic activity toward TMB and H2O2, respectively, versus undoped rGO. DFT calculations revealed N,B co-doping activates pyridinic-N, with boron serving as a co-active site. Tripathi et al.39 developed N,P,S-co-doped CQDs (Fig. 3d), where multi-heteroatom synergy enhanced the catalytic activity and broadened the operational pH range.
2.3. Metal- or metal oxide-supported carbon-based nanozymes
Metal/metal oxide doping into carbon-based nanozymes addresses the easy aggregation and high cost of pure metal nanozymes. Additionally, metal-carbon synergies further enhance the catalytic activity and stability. Gao et al.40 immobilized Fe3O4 nanoparticles on aminopyridine-functionalized multi-walled carbon nanotubes (MWCNT-AP), obtaining well-dispersed superparamagnetic Fe3O4/MWCNT-AP nanocomposites. The enhanced POD-like activity enables phenolic compound degradation in environmental waters. Similarly, in situ anchoring of copper nanoparticles onto aminated graphene quantum dot frameworks modulate enzyme-like activity and enhances H2O2 affinity. At 7.66% loading, Cu single-atom nanozymes (SAN) exhibited 3.4-fold higher POD activity and 8.88-fold greater SOD activity than low-loading counterparts,41 demonstrating loading-dependent selectivity. Xie et al.42 prepared ultrathin Cu/CN nanosheets (Fig. 4a) via supramolecular pre-organization/calcination, achieving 14.3 wt% Cu loading. High-density atomic Cu sites confer exceptional peroxidase (POD)-like activity. | ||
| Fig. 4 Synthesis of (a) Cu/CN, (b) Fe–N–C single-atom catalysts (SACs), (c) Fe single-atom nanozymes (Fe-SANs), and (d) Mo single atoms in N-doped carbon (MoSA–Nx–C). | ||
Researchers have discovered single-atom nanozymes (SAzymes) with M–Nx–C sites resembling natural metalloenzyme centers, where M is a transition metal (Fe, Co, Pt, Mn, and Cu). These SAzymes mimic the homogeneous active centers of natural enzymes, exhibiting ultrahigh atomic utilization efficiency and tunable coordination environments.43 Under optimized conditions, they demonstrate catalytic activities and reaction kinetics comparable to natural enzymes.44 During high-temperature pyrolysis, metal precursors enable direct synthesis of SAzymes via strong interactions with pyrolytic intermediates. Consequently, numerous studies employed pyrolytic synthesis (Fig. 4a–d). Wu et al.45 used ZIF-8 to disperse atomic Fe uniformly in N-doped carbon matrices. Subsequent carbonization/activation (Fig. 4b) yielded 2D ultrathin Fe–N–C SAzymes with high surface area and hierarchical porosity. Jiao et al.46 developed atomic-scale SAzymes using salt templating (Fig. 4c), achieving 13.5 wt% Fe loading in N-doped carbon nanosheets. Zn (Co)–N–C SAzymes prepared similarly showed lower POD-like activity than Fe–N–C. The superior activity of FeN4 sites originates from their strong H2O2 adsorption, lowering the energy barrier for ˙OH formation.
Biomimetic construction of enzyme-like active sites enhances nanozyme activity and specificity (Fig. 5a–c). Song et al.24 synthesized Cu(II)-modified graphitic carbon nitride (g-C3N5–Cu2+), demonstrating a rational design strategy for high-efficiency biomimetic nanozymes. Huang et al.47 prepared cytochrome P450-mimetic FeN5 SA/CNF (Fig. 5a), exhibiting stronger electron-withdrawing effects than FeN4 that facilitate O–O bond cleavage and enhance the enzymatic activity. Its catalytic rate constant exceeded commercial Pt/C by 70-fold, conferring antibacterial properties. Remarkably, their encapsulation-pyrolysis strategy extends to Co, Ni, Cu, and Mn atoms, generating thermodynamically stable M-N5/C structures. Wang et al.48 constructed MoSA–Nx–C (x = 2, 3, 4) (Fig. 4d), demonstrating that atomic configuration tuning via coordination number or metal center modification modulates SAzyme activity. Heteroatom substitution (P, S, and Bi) for nitrogen forms Fe/CoN3P49,50 (Fig. 5b and c), FeN3S,51 FeNBC,52 and FeN3Bi53 compounds. This narrows interatomic distances and enhances substrate interactions. In FeNPC systems, P-doping induces long-range charge transfer that activates metal centers and reduces the energy barrier for ROS formation.52 Bi-doping creates high-density dual catalytic sites for enhanced activity modulation. In summary, carbon-based nanozymes are classified based on composition and structure, with distinct types offering unique catalytic advantages and broad applicability.
3. Catalytic mechanisms of carbon-based nanozymes
Natural enzyme catalysis, underpinned by transition state theory, involves stabilizing high-energy transition states to reduce activation energy. Analogously, nanozyme catalysis proceeds through three fundamental steps: substrate adsorption, reaction, and product desorption. Based on catalytic function, nanozymes are classified into four types (Fig. 6a): peroxidase (POD), superoxide dismutase (SOD), oxidase (OXD), and catalase (CAT). Carbon-based nanozymes predominantly exhibit POD-like and SOD-like activities. The POD-like mechanism involves hydrogen peroxide (H2O2) as an electron acceptor generating hydroxyl radicals (˙OH). Conversely, some nanozymes function as electron carriers. For SOD-like activity, superoxide anions (˙O2−) coordinate with active sites to form transient complexes that disproportionate to O2 and H2O2via coupled electron/proton transfer.3.1. Catalytic mechanism of peroxidase (POD)
Peroxidase (POD)-like nanozyme catalysis primarily involves interactions between nanomaterial surface active sites and reactants through three steps: (1) substrate adsorption: active sites (e.g., metal ions and defects) bind to substrates (H2O2 and TMB). (2) Electron transfer: mediated by catalytic centers (metal ions/organic cofactors). (3) Product release and cycle regeneration: active sites undergo redox reactions to release products, sustaining catalytic cycles.Notably, researchers widely recognize hydroxyl radicals (˙OH) as active intermediates in carbon nanozyme catalytic reactions.54 However, Yan et al.34 investigated the generation and capture of ˙OH during the catalytic process of N-CMNs by using electron spin resonance (ESR) spectroscopy and fluorescence spectroscopy, and the results were not consistent with expectations. Therefore, Yan et al.34 speculated that the catalytic mechanism of N-CMNs is similar to that of natural HRP. The high oxidation state intermediate dominates its POD-like catalytic activity, and the catalytic cycle reaction pathway is shown in Fig. 6d. Specifically, steps 1 to 3 represent the adsorption and decomposition processes of H2O2, steps 4 to 5 are the transfer processes of proton and electron pairs, and step 6 is the desorption process of H2O2. In addition, the catalytic mechanisms of different types of nanozymes are slightly different. The following are some common peroxidase (POD)-like nanozyme materials and their catalytic mechanisms, for instance, carbon quantum dots (CQDs) simulate peroxidase (POD)-like activity through surface functional groups and electron transfer processes.14 Graphene nanozymes follow a non-free radical mechanism. They adsorb and catalyze the decomposition of H2O2 through surface defect sites and π–π interactions. GO acts as an electron carrier, and when it combines with the amino groups in TMB, it will increase its electron density, which is conducive to electron migration.24 Metal-supported carbon-based nanozymes, such as Fe–Nx–C, catalyze the decomposition of H2O2 to produce hydroxyl radicals (˙OH) through Fenton reactions or Fenton-like reactions55 and then oxidize reducing substrates such as TMB, ABTS, and OPD.46,56
3.2. Catalytic mechanism of superoxide dismutase (SOD)
Natural superoxide dismutase (SOD) catalyzes the disproportionation of superoxide radicals (˙O2−) to O2 and H2O2. Unlike natural SOD, artificial SOD-mimicking nanozymes typically exhibit multimodal antioxidant activities that more efficiently eliminate reactive oxygen species (ROS).57 Zhang et al.58 demonstrated that Fe–Cu–N6 single-atom nanozymes significantly surpass natural enzymes in catalytic performance, with exceptional SOD-like selectivity. For C-dot SOD-mimics, Gao et al.59 proposed a catalytic cycle (Fig. 6e) involving seven distinct states of π-conjugated carbonyl groups, including two transition states (TS1/TS2). Hydroxyl-functionalized C-dots exhibit stronger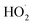 radical capture capability due to substantially lower binding energies.
radical capture capability due to substantially lower binding energies.
3.3. Catalytic mechanism of oxidase (OXD)
Oxidases (OXD) catalyze hydrogen donor oxidation using O2 as the electron acceptor, generating H2O2 and H2O in the absence of exogenous H2O2.60 Chen et al.53 verified the role of O2 in TMB oxidation by purging N2/O2 into FeBiNC–TMB systems. Under N2-saturated conditions, FeBiNC activity increased by 160% (versus air) but decreased by 93% under O2 saturation, confirming the critical role of O2 in oxidase (OXD)-like catalysis. Xu et al.43 simulated the four-electron reduction mechanism (Fig. 6f). Electron spin resonance revealed ultrahigh O2 adsorption energy on FeNSC sites, inducing competitive adsorption that facilitates catalytic cycle completion.3.4. Catalytic mechanism of catalase (CAT)
Catalase decomposes H2O2 into H2O and O2, protecting cells from ROS-mediated oxidative damage. Thus, catalase (CAT)-like nanozymes alleviate tumor hypoxia as radiosensitizers in cancer therapy.5 Chen et al.50 proposed a CAT-like mechanism where H2O2 adsorption at neutral pH leads to heterolytic cleavage forming ˙OH or homolytic cleavage yielding HOO˙ intermediates (reaction pathway 1: H2O2 → ˙OH + OH−; pathway 2: H2O2 → HOO˙ + H+). These intermediates react with water or O˙ species, ultimately regenerating O2 and H2O via H abstraction from additional H2O2. In addition, catalase (CAT)-like enzymes exhibit higher activity under neutral or alkaline conditions. By comparing the free energy of the catalase-like catalytic mechanism of four catalyst models, namely Co-N3PS, Co-N3P, CoN4 and Co3O4 (Fig. 6g), the results show that the doped S atom has a stronger affinity for the H atom, while the P atom has a stronger affinity for the hydroxyl radical (˙OH). Moreover, the Co–S coordination bond is of great significance to the progress of H2O2 dissociation in Co-N3PS.3.5. Challenges in the research of catalytic mechanisms
The following are the primary challenges in nanozyme mechanistic studies.4. Fundamental basis for nanozyme catalysis
While diverse nanozymes may exhibit identical catalytic activities, their underlying mechanisms often differ fundamentally. For carbon-based nanozymes, catalytic properties are primarily derived from the following three key attributes: high surface area enabling dense active site exposure, versatile adsorption/loading capacity for catalytic modifiers and superior electron-transfer kinetics that facilitate redox processes.4.1. Active sites
Carbon-based nanozymes host abundant active sites primarily comprising oxygen-containing functional groups: hydroxyl (–C–OH), carboxyl (–COOH), and carbonyl (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O).59 These surface/defect-bound groups interact with substrates via hydrogen bonding or serve as catalytic centers, forming stable active sites. For instance, carbon nanotubes and graphene quantum dots leverage high surface areas for substrate adsorption and catalysis. Gao et al.59 introduced hydroxyl groups (–C–OH), carboxyl groups (HO–C
O).59 These surface/defect-bound groups interact with substrates via hydrogen bonding or serve as catalytic centers, forming stable active sites. For instance, carbon nanotubes and graphene quantum dots leverage high surface areas for substrate adsorption and catalysis. Gao et al.59 introduced hydroxyl groups (–C–OH), carboxyl groups (HO–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), and carbonyl groups (–C
O), and carbonyl groups (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) on the surface of carbon dots (CDs), endowing them with superoxide dismutase (SOD)-like activity through the synergistic effect of multiple groups. Specifically, the hydroxyl and carboxyl groups combine with the superoxide anion through hydrogen bonds, while the carbonyl group takes electrons from the superoxide anion, thereby generating oxygen and a reduced carbon dot. Subsequently, the reduced carbon dot will then be oxidized by another superoxide anion to produce hydrogen peroxide. The research of Sun et al.61 confirmed that the carboxyl group (HO–C
O) on the surface of carbon dots (CDs), endowing them with superoxide dismutase (SOD)-like activity through the synergistic effect of multiple groups. Specifically, the hydroxyl and carboxyl groups combine with the superoxide anion through hydrogen bonds, while the carbonyl group takes electrons from the superoxide anion, thereby generating oxygen and a reduced carbon dot. Subsequently, the reduced carbon dot will then be oxidized by another superoxide anion to produce hydrogen peroxide. The research of Sun et al.61 confirmed that the carboxyl group (HO–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and carbonyl group (–C
O) and carbonyl group (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) on the outer surface of GQDs served as the substrate binding site and catalytic active site, respectively, while the hydroxyl group (–C–OH) inhibited its peroxidase (POD)-like activity.
O) on the outer surface of GQDs served as the substrate binding site and catalytic active site, respectively, while the hydroxyl group (–C–OH) inhibited its peroxidase (POD)-like activity.
4.2. Electron transfer
Catalytic processes typically involve electron transfer and redox reactions. Consequently, nanozyme activity correlates directly with charge transfer efficiency. Owing to superior conductivity, graphene and carbon nanotubes enable H2O2 decomposition by serving as electron carriers, generating ˙OH radicals. For example, graphene oxide (GO) exhibits oxidase-like activity25 through electron transfer from its valence band to the LUMO (lowest unoccupied molecular orbital) of H2O2.4.3. Synergistic effect
Carbon-based nanozyme activity can be enhanced through heteroatom doping (N, P, and S) or multi-metal loading, which provide additional active sites and improve the catalytic efficiency. For instance, Li et al.44 incorporated sulfur into Fe–N4–C systems, introducing sulfoxide functional groups (–S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) onto the carbon plane. This transforms the traditional planar Fe–N4–C centers into three-dimensional architectures. Studies confirm that these oxidized sulfur groups assist in substrate binding orientation and promote product desorption, yielding specific activity that is 6.8-fold higher than that of Fe–N–C SAzymes.
O) onto the carbon plane. This transforms the traditional planar Fe–N4–C centers into three-dimensional architectures. Studies confirm that these oxidized sulfur groups assist in substrate binding orientation and promote product desorption, yielding specific activity that is 6.8-fold higher than that of Fe–N–C SAzymes.
5. Regulating carbon-based nanozyme performance
Similar to natural enzymes, nanozyme activity depends on the pH and temperature. Nevertheless, their catalytic performance can be modulated by strategies such as altering their size, morphology, composition, coordination structure, surface modification, interaction with biomolecules, or changing reaction conditions. These performance regulation strategies (size/morphology control, composition tuning, etc.) are rigorously implemented by leveraging the fundamental elements—active sites, electron transfer, and synergistic effects—established in Section 4.5.1. Tailoring the size and morphology
Nanomaterial size and morphology critically influence the catalytic activity. Tuning these parameters optimizes the surface area distribution and active site exposure, thereby enhancing the performance. Typically, smaller nanozymes with higher specific surface areas exhibit greater substrate binding probability and superior catalytic activity. Porous carbon-based nanozymes are commonly synthesized using metal–organic frameworks (MOFs) as sacrificial templates.62 Controlled carbonization of MOFs enables precise regulation of pore architecture and morphology.5.2. Engineering the chemical composition
Nanomaterial catalytic activity can be modulated by tailoring the composition and structure. Heteroatom doping (N, P, and S) or phase engineering alters the charge density and induces structural distortion, thereby tuning catalytic performance. Density functional theory (DFT) calculations confirm that defect engineering diversifies nanozyme activities.63 Introducing P/S heteroatoms to replace coordinating nitrogen forming Fe/CoN3P49,50 and FeN3S51 enables precise geometric configuration control.Atomically dispersing active centers maximizes the catalytic efficiency. In carbon-supported single-atom catalysts (SACs), M–Nx–C configurations form stable M–N bonds that mimic natural heme enzymes (e.g., Fe–N4 of HRP and cytochrome P450). This structural biomimicry achieves near-native enzyme simulation. For instance, inspired by heme cofactors, Li et al.44 incorporated sulfur into Fe–N4–C, grafting sulfoxide (–S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) groups onto the carbon plane. This transforms 2D Fe–N4 sites into 3D architectures, optimizing substrate interaction geometries.
O) groups onto the carbon plane. This transforms 2D Fe–N4 sites into 3D architectures, optimizing substrate interaction geometries.
5.3. Implementing surface modifications
Catalytic reactions occur primarily on nanozyme surfaces. Modifying surfaces with coordinating components, functional groups, or polymers enhances substrate binding, significantly altering the activity and selectivity. For example, Chen et al.64 immobilized Pt clusters on layered Fe–N–C carriers, synthesizing FeSA–PtC conjugates with 4.5-fold higher peroxidase-like activity than Fe–N–C and 7-fold higher than Pt clusters. DFT calculations attributed this to electron transfer from Pt clusters to Fe single-atom sites (not vice versa), enhancing O–O bond cleavage. Liu et al.65 confirmed that carboxyl, hydroxyl, and amino groups on carbon dots govern SOD-like activity through radical quenching experiments. Huo et al.66 synthesized PSAF NCs via ZIF-8 pyrolysis and PEG modification. These selectively trigger Fenton reactions in acidic tumor microenvironments (TME), generating cytotoxic ˙OH for tumor therapy.5.4. Tuning interactions with biomolecules
Biomolecular interactions modulate nanozyme catalytic activity. Specifically, conjugating proteins or DNA regulates nanozyme activity and stability. Li et al.67 demonstrated this by anchoring ferroheme (hemin) onto Zn–N–C single-atom nanozymes to construct Zn–N–C@heme SAzymes, which detect propyl gallate (PG) and formaldehyde (HCHO) via peroxidase (POD)-like activity. Mechanistically, in the presence of H2O2, the SAzyme catalyzes the oxidation of colorless PG to a yellow quinone product and HCHO competitively inhibits this reaction by binding to the active site. This principle establishes a dual-analyte detection platform for PG and HCHO.5.5. Optimizing reaction conditions
Nanozyme catalytic activity is modulated by external environmental factors, including the pH, temperature, and ionic strength. Optimizing these parameters enhances the catalytic efficiency. Among these, pH exerts the most significant influence, thus shifting pH values fundamentally alters activity profiles and enzyme-mimicking types. Experimental evidence confirms acidic conditions favor peroxidase (POD)-like activity, while neutral/alkaline conditions enhance superoxide dismutase (SOD)-like and catalase (CAT)-like activities.685.6. Current challenges in regulating nanozyme performance
However, these regulatory strategies face the following key challenges: (1) size-dependent activity enhancement involves inherent trade-offs, with nanoscale dimensions boosting the activity via the increased surface area but reducing structural stability. (2) Electron transfer mechanisms, governed by composition/surface modifications, remain elusive due to hybrid system complexity. Precisely tracking electron paths and quantifying synergy are challenging. (3) Biomolecular/environmental regulation exhibits strong pH/temperature dependence. Consequently, most nanozymes function optimally only under specific conditions, which constrains their physiological adaptability and application scope.6. Applications of carbon-based nanozymes
Carbon-based nanozymes exhibit broad applications across environmental engineering, biosensing, and biomedicine. Specifically, carbon-based nanozymes excel at biosensing and are capable of detecting small molecules, proteins, viruses, and cells. In the environmental field, carbon-based nanozymes are mainly used for the detection and degradation of various pollutants, such as organic dyes, heavy metal ions, antibiotics, etc. In the field of biomedicine, carbon-based nanozymes can catalyze reactions to produce reactive oxygen species (ROS), thereby achieving the purpose of antibacterial and killing tumor cells. Thus, they hold significant potential for integrated sensing, diagnostics, and therapeutics.6.1. Field of environmental engineering
Carbon-based nanozymes enable sensitive detection and efficient degradation of environmental pollutants,2,7 including heavy metal ions, like Pb(II), Hg(II), and Cr(VI), and organic pollutants, such as phenolic compounds, antibiotics (e.g., tetracycline), and pesticides (e.g., glyphosate). These compounds constitute major sources of soil and water contamination. Conventional remediation technologies (physical adsorption, chemical oxidation, and bioremediation) face persistent limitations: oxide byproduct generation, high operational costs, poor reaction selectivity and limited recyclability. Nanozymes overcome these constraints owing to their exceptional physicochemical properties—including high specific surface area, enhanced adsorption capacity, abundant catalytic active sites, robust stability, and strong toxicant resistance—enabling efficient ROS-mediated degradation of textile dyes, antibiotics, and pesticide residues.| Nanozyme (g L−1) | Pollutant (mg L−1) | Activation (g L−1 M−1) | k (min−1) | k nor (min−1 g−1 L) | Ref. |
|---|---|---|---|---|---|
| FeCo-NC-1,0.1 | BPA,20 | PMS,0.2 | 1.179 | 11.79 | 72 |
| FeCo-NC-2,0.1 | BPA,20 | PMS,0.2 | 1.252 | 12.52 | |
| FeCo-NC-3,0.1 | BPA,20 | PMS,0.2 | 0.465 | 4.65 | |
| Bi-NC,0.03 | RhB,10 | PMS,15 | 0.467 | 15.57 | 73 |
| Fe-NC,0.03 | RhB,10 | PMS,15 | 0.505 | 16.83 | |
| FeBi-NC,0.03 | RhB,10 | PMS,15 | 0.733 | 24.43 | |
| Fe/Cu-NC,2 | MG,10 | PMS,30 | 0.8218 | 0.411 | 74 |
| Fe/Cu-NC,2 | MB,10 | PMS,30 | 0.5702 | 0.285 | |
| Fe/Cu-NC,2 | RhB,10 | PMS,30 | 0.5943 | 0.297 | |
| γ-Fe2O3/Mn3O4,0.05 | RhB | PMS,50 | — | — | 75 |
| Mn-SAzyme,0.025 | MB,10 | H2O2,0.1 | ∼0.0096 | ∼0.384 | 76 |
| Mn-SAzyme,0.025 | RhB,10 | H2O2,0.1 | <0.0096 | <0.384 | |
| MOF-545(Fe),10 | MO,10 | H2O2,0.1 | — | — | 77 |
| MOF-545(Fe),10 | MB,10 | H2O2,0.1 | — | — |
Carbon-based nanozymes mimic peroxidase activity by catalyzing H2O2 decomposition to generate hydroxyl radicals (˙OH), oxidizing organic dyes.78,79 Chen et al.80 synthesized Fe/Cu–N–C single-atom nanozymes via folic acid-templated pyrolysis. This layered material degrades Rhodamine B (RhB) in environmental matrices (Fig. 7c), demonstrating wastewater treatment potential. Chen et al.53 doped Fe into Bi-MOF and then carried out high-temperature pyrolysis to form the FeBi-NC SAzyme. The dual-site single-atom nanozyme, featuring Fe–N4 and Bi–N4, is loaded on a carbon matrix. This nanozyme can be used in multi-step catalytic reactions. In other words, redox reactions and activation of persulfate to efficiently degrade the organic dye Rhodamine B. This cascaded catalytic characteristic enables it to play an important role in environmental remediation and wastewater treatment. Feng et al.81 synthesized a Mn-SAzyme with metallic manganese as the active center. In this study, based on the Fenton-like reaction, methylene blue (MB) was used as the organic dye model (Fig. 7d), and the Mn-SAzyme was used as the catalyst. The degradation rate of methylene blue could reach 90.1% within 300 minutes under the given reaction conditions. Nanozymes exhibit higher catalytic activity and stability than natural enzymes in the degradation process of organic dyes while maintaining efficient catalytic performance under complex environmental conditions. In addition, nanozymes can exert excellent catalytic activity under mild conditions, including normal temperature, pressure and near-neutral pH conditions, thereby reducing the negative impact on the environment. Furthermore, nanozymes usually have good recyclability and reusability, reducing degradation costs and enhancing feasibility in practical applications. In conclusion, nanozymes have demonstrated high efficiency, stability, and environmental friendliness in the degradation of organic dye, providing a promising strategy for solving the problem of organic dye pollution.
6.2. Field of biosensing
Biosensors based on carbon-based enzyme-like materials have been successfully applied to detect H2O2 and small biomolecules that can release H2O2, such as glucose24 and choline.52 They have also been used to detect other reducing small biomolecules like ascorbic acid, cysteine, biothiols, and tumor markers.6.3. Field of biomedicine
Carbon-based nanozymes exhibit promising biomedical applications, including cancer therapy, anti-inflammatory/antibacterial treatment, wound disinfection, cellular protection, and bioimaging.Leveraging their tunable enzyme-like activity, high stability, and multifunctional synergy, carbon-based nanozymes implement HDAC modulation through direct inhibition mechanisms. Studies confirm that nitrogen-containing functional groups (e.g., pyridinic-N and graphitic-N) on N-doped nanozymes bind Zn(II) in HDAC catalytic centers, blocking enzymatic function.103,104 This inhibition exhibits subtype selectivity; for instance, S-doped nanozymes show enhanced HDAC6 affinity,105 attributed to specific interactions between sulfur atoms and HDAC6's zinc-finger ubiquitin-binding domain (ZnF-UBP) within its dual-catalytic domains (CD1/CD2).
6.4. Adaptation challenges in practical Applications
Nanozymes face three key implementation barriers:• Complex matrix interference: in biosensing applications (e.g., blood analysis), endogenous components like proteins and ions inhibit nanozyme catalytic activity, compromising detection accuracy. Similarly, complex environmental matrices impair pollutant degradation efficiency in aqueous remediation systems. Critically, comprehensive datasets on nanozyme environmental persistence and ecotoxicological risks remain scarce.
• Biocompatibility–activity trade-off: the optimal equilibrium between biocompatibility and catalytic efficacy remains elusive. Key unresolved challenges include: undefined in vivo metabolic pathways, precious metal-induced cytotoxicity (e.g., mitochondrial apoptosis), and scarce long-term biosafety data (>6 months exposure models). This fundamental conflict severely restrains clinical translation.
• Lack of standardization: a unified evaluation framework is absent, leading to inconsistent testing protocols across studies. Critical metrics—including Michaelis constants (Km) and turnover frequencies (kcat)—lack standardized assay methodologies, severely compromising cross-study comparability.
7. Conclusion
While carbon-based nanozymes demonstrate marked advantages over natural enzymes in terms of cost efficiency109 (>100-fold reduction), scalable production,110 extended storage stability109 (≥5 months110,111), and robustness under harsh conditions,112 three fundamental limitations persist: inferior activity and specificity compared to natural enzymes (kcat/Km values <15–20%113 of native analogs), limited enzyme-mimic diversity (with >90% studies focused on oxidoreductases versus <5% on transferases, lyases, ligases, and isomerases114), and unresolved recyclability115 (showing >90% activity loss after 5 reuse cycles111,116).The heterogeneity of material structures leads to divergent catalytic mechanisms, while atomic-scale reaction pathways with definitive structure–activity relationships require further elucidation (see Section 3). Although Section 5 outlines performance modulation strategies, current synthesis methods fail to precisely control active-site density (<±10% variance) and coordination environments (e.g., M–N4versus M–N2O2 configurations), necessitating universal synthetic protocols, and sensing applications dominate research (>70% of publications117), yet large-scale in vivo behavior studies remain scarce (<5% of studies report pharmacokinetics118,119). Critically, the mismatch between optimized catalytic conditions (e.g., pH 4.5) and physiological environments (pH 7.4) exacerbates unresolved biocompatibility challenges (Section 6), demanding interdisciplinary convergence across computational modeling, surface science, and nanotoxicology.
Nanozyme research and application require establishing standards such as ISO-compliant testing protocols with defined reaction conditions and substrate ranges. Concurrently, advanced fundamental research infrastructure (e.g., in situ characterization platforms) and scaling instrumentation are paramount to build a vertically integrated nanozyme value chain spanning innovation discovery, industrial production, and clinical translation.
In summary, this work highlights carbon-based nanozymes as transformative catalytic platforms, demonstrating quantifiable advancements in sustainability (reducing enzyme production costs by >90%34) and multimodal therapeutic efficacy (e.g., ROS generation rates >50 μM min−1). These innovations establish foundational frameworks for next-generation nanobiocatalysts, with proven versatility in environmental remediation, precision diagnostics, and targeted therapeutics. Their continued evolution will catalyze cross-disciplinary convergence across materials science, synthetic biology, and nanomedicine, positioning this field in the vanguard of sustainable nanotechnology innovation.
Data availability
All relevant data are within the manuscript and its additional files.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the study reported in this document.Acknowledgements
We are thankful for financial support from Guangxi Key Laboratory of Electrochemical and Magnetochemical Functional Materials, National Natural Science Foundation of China (No. 21565011), the Guangxi Key Laboratory of Environmental Pollution Control Theory and Technology, and Guangxi Colleges and Universities Key Laboratory of Food Safety and Detection.References
- W. L. Li and T. Head-Gordon, ACS Cent. Sci., 2021, 7, 72–80 CrossRef CAS PubMed.
- R. Singh, A. Umapathi, G. Patel, C. Patra, U. Malik, S. K. Bhargava and H. K. Daima, Sci. Total Environ., 2023, 854, 158771 CrossRef CAS PubMed.
- L. H. Shen, D. X. Ye, H. B. Zhao and J. J. Zhang, Anal. Chem., 2021, 93, 1221–1231 CrossRef CAS PubMed.
- J. Shen, J. Chen, Y. P. Qian, X. Q. Wang, D. S. Wang, H. G. Pan and Y. G. Wang, Adv. Mater., 2024, 36, 2313406 CrossRef CAS PubMed.
- G. J. Hou, Y. K. Xu, C. H. Wang, C. Lu, A. Thakur, K. Zhang, W. Li and Z. J. Xu, Aging Dis., 2025 DOI:10.14336/AD.2025.0011.
- X. Ma, G. Sun, A. Leng, X. Wei, J. Xu, W. Ding, H. Liu and H. Fang, Talanta, 2025, 296, 128398 CrossRef PubMed.
- A. Das, R. V. S. Uppaluri and S. Mitra, Chem. Eng. J., 2025, 507, 160762 CrossRef CAS.
- E. Z. Gomaa, J. Cluster Sci., 2022, 33, 1275–1297 CrossRef CAS.
- A. M. Ashrafi, Z. Bytesnikova, J. Barek, L. Richtera and V. Adam, Biosens. Bioelectron., 2021, 192, 113494 CrossRef CAS PubMed.
- L. Feng, M. C. Zhang and Z. Y. Fan, Anal. Methods, 2024, 16, 6771–6792 RSC.
- X. D. Zhang, X. K. Chen and Y. L. Zhao, Nano-Micro Lett., 2022, 14, 165–191 CrossRef PubMed.
- M. Cordani, J. Fernández-Lucas, A. Khosravi, E. N. Zare, P. Makvandi, A. Zarrabi and S. Iravani, Int. J. Biol. Macromol., 2025, 297, 139704 CrossRef CAS PubMed.
- L. Z. Gao, J. Zhuang, L. Nie, J. B. Zhang, Y. Zhang, N. Gu, T. H. Wang, J. Feng, D. L. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- J. He, Y. Hou, Z. Zhang, J. Zhang, X. Yan, K. Fan and M. Liang, Nano Biomed. Eng., 2024, 16, 28–47 CrossRef CAS.
- W. P. Yang, X. Yang, L. J. Zhu, H. S. Chu, X. Y. Li and W. T. Xu, Coord. Chem. Rev., 2021, 448, 214170 CrossRef CAS.
- Z. Q. Zhuang, Y. N. Yu, S. P. Dong, X. L. Sun and L. Mao, Anal. Bioanal. Chem., 2024, 416, 5949–5964 CrossRef CAS PubMed.
- C. Liu, Y. M. Zhao, D. Xu, X. X. Zheng and Q. Huang, Anal. Bioanal. Chem., 2021, 413, 4013–4022 CrossRef CAS PubMed.
- I. Batool, A. Anwar, M. Imran and Z. I. Alvi, Top. Catal., 2025, 68, 823–855 CrossRef CAS.
- M. A. Ansari, S. Shoaib, W. Chauhan, R. M. Gahtani, U. Hani, M. N. Alomary, G. Alasiri, N. Ahmed, R. Jahan, N. Yusuf and N. Islam, Environ. Res., 2024, 241, 117522 CrossRef CAS PubMed.
- K. Y. Wu, Y. Feng, Y. S. Li, L. Li, R. Liu and L. D. Zhu, Anal. Bioanal. Chem., 2020, 412, 5477–5487 CrossRef CAS PubMed.
- Y. F. Chen, Y. Zhang, L. Jiao, H. Y. Yan, W. L. Gu and C. Z. Zhu, Chin. J. Anal. Chem., 2021, 49, 907–921 CAS.
- L. L. Dugan, D. M. Turetsky, C. Du, D. Lobner, M. Wheeler, C. R. Almli, C. K. F. Shen, T. Y. Luh, D. W. Choi and T. S. Lin, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 9434–9439 CrossRef CAS PubMed.
- C. Zhao, J. Kang, Y. W. Li, Y. Wang, X. Y. Tang and Z. Q. Jiang, Cyborg Bionic Syst., 2023, 4, 0022 CrossRef CAS PubMed.
- Y. J. Song, K. G. Qu, C. Zhao, J. S. Ren and X. G. Qu, Adv. Mater., 2010, 22, 2206–2210 CrossRef CAS PubMed.
- H. Wang, P. H. Li, D. Q. Yu, Y. Zhang, Z. Z. Wang, C. Q. Liu, H. Qiu, Z. Liu, J. S. Ren and X. G. Qu, Nano Lett., 2018, 18, 3344–3351 CrossRef CAS PubMed.
- Z. Zhuang, Y. Yu, S. Dong, X. Sun and L. Mao, Anal. Bioanal. Chem., 2024, 416, 5949–5964 CrossRef CAS PubMed.
- X. Wan, Y. Ge, J. Zhang, W. Pan, N. Li and B. Tang, ACS Appl. Mater. Interfaces, 2023, 15, 44763–44772 CrossRef CAS PubMed.
- L. Feng, L. Zhang, S. Zhang, X. Chen, P. Li, Y. Gao, S. Xie, A. Zhang and H. Wang, ACS Appl. Mater. Interfaces, 2020, 12, 17547–17556 CrossRef CAS PubMed.
- J. Shen, J. Chen, Y. P. Qian, X. Q. Wang, D. S. Wang, H. G. Pan and Y. G. Wang, Adv. Mater., 2024, 36, 2313406 CrossRef CAS PubMed.
- E. Perez Mayoral, M. G. Ojer, M. Ventura and I. Matos, Nanomaterials, 2023, 13, 2013 CrossRef PubMed.
- X. M. Dang, H. G. Zhang, X. M. Chen and H. M. Zhao, Sep. Purif. Technol., 2024, 330, 125312 CrossRef CAS.
- Y. Qu, W. Zhang, B. Guang, Y. Xiao, X. Liu, C. Zhai and Y. Liu, J. Alloys Compd., 2022, 912, 165261 CrossRef CAS.
- Z. P. Lou, S. Zhao, Q. Wang and H. Wei, Anal. Chem., 2019, 91, 15267–15274 CrossRef CAS PubMed.
- H. Y. Yan, L. Z. Wang, Y. F. Chen, L. Jiao, Y. Wu, W. Q. Xu, W. L. Gu, W. Y. Song, D. Du and C. Z. Zhu, Research, 2020, 2020, 8202584 CAS.
- Y. Xiong, H. C. Li, C. W. Liu, L. R. Zheng, C. Liu, J. O. Wang, S. J. Liu, Y. H. Han, L. Gu, J. S. Qian and D. S. Wang, Adv. Mater., 2022, 34, 2110653 CrossRef CAS PubMed.
- Y. Y. Huang, C. Q. Liu, F. Pu, Z. Liu, J. S. Ren and X. G. Qu, Chem. Commun., 2017, 53, 3082–3085 RSC.
- Y. F. Chen, L. Jiao, H. Y. Yan, W. Q. Xu, Y. Wu, H. J. Wang, W. L. Gu and C. Z. Zhu, Anal. Chem., 2020, 92, 13518–13524 CrossRef CAS PubMed.
- M. S. Kim, S. Cho, S. H. Joo, J. Lee, S. K. Kwak, M. I. Kim and J. Lee, ACS Nano, 2019, 13, 4312–4321 CrossRef CAS PubMed.
- K. M. Tripathi, H. T. Ahn, M. Chung, X. A. Le, D. Saini, A. Bhati, S. K. Sonkar, M. I. Kim and T. Kim, ACS Biomater. Sci. Eng., 2020, 6, 5527–5537 CrossRef CAS PubMed.
- Y. Y. Gao, H. X. Li, Z. Z. Ou, P. Hao, Y. Li and G. Q. Yang, Acta Phys.-Chim. Sin., 2011, 27, 2469–2477 CAS.
- X. Z. Chen, X. Y. Liu, S. F. Zhang, X. Y. Mu and X. D. Zhang, Prog. Biochem. Biophys., 2023, 50, 2684–2696 Search PubMed.
- X. Y. Xie, X. F. Chen, Y. H. Wang, M. S. Zhang, Y. X. Fan and X. P. Yang, Talanta, 2023, 257, 124387 CrossRef CAS PubMed.
- Y. Xu, Y. J. Ma, X. H. Chen, K. Q. Wu, K. Y. Wang, Y. F. Shen, S. Q. Liu, X. J. J. Gao and Y. J. Zhang, Angew. Chem., Int. Ed., 2024, 136, 124387 Search PubMed.
- G. M. Li, H. Liu, T. D. Hu, F. Pu, J. S. Ren and X. G. Qu, J. Am. Chem. Soc., 2023, 145, 16835–16842 CrossRef CAS PubMed.
- W. W. Wu, L. Huang, X. Y. Zhu, J. X. Chen, D. Y. Chao, M. H. Li, S. L. Wu and S. J. Dong, Chem. Sci., 2022, 13, 4566–4572 RSC.
- L. Jiao, J. B. Wu, H. Zhong, Y. Zhang, W. Q. Xu, Y. Wu, Y. F. Chen, H. Y. Yan, Q. H. Zhang, W. L. Gu, L. Gu, S. P. Beckman, L. Huang and C. Z. Zhu, ACS Catal., 2020, 10, 6422–6429 CrossRef CAS.
- L. Huang, J. X. Chen, L. F. Gan, J. Wang and S. J. Dong, Sci. Adv., 2019, 5, 5490 CrossRef PubMed.
- Y. Wang, G. R. Jia, X. Q. Cui, X. Zhao, Q. H. Zhang, L. Gu, L. R. Zheng, L. H. Li, Q. Wu, D. J. Singh, D. Matsumura, T. Tsuji, Y. T. Cui, J. Zhao and W. T. Zheng, Chem, 2021, 7, 436–449 CAS.
- S. F. Ji, B. Jiang, H. G. Hao, Y. J. Chen, J. C. Dong, Y. Mao, Z. D. Zhang, R. Gao, W. X. Chen, R. F. Zhang, Q. Liang, H. J. Li, S. H. Liu, Y. Wang, Q. H. Zhang, L. Gu, D. M. Duan, M. M. Liang, D. S. Wang, X. Y. Yan and Y. D. Li, Nat. Catal., 2021, 4, 407–417 CrossRef CAS.
- Y. J. Chen, B. Jiang, H. G. Hao, H. J. Li, C. Y. Qiu, X. Liang, Q. Y. Qu, Z. D. Zhang, R. Gao, D. M. Duan, S. F. Ji, D. S. Wang and M. M. Liang, Angew. Chem., Int. Ed., 2023, 62, e202301879 CrossRef CAS PubMed.
- L. Jiao, Y. K. Kang, Y. F. Chen, N. N. Wu, Y. Wu, W. Q. Xu, X. Q. Wei, H. J. Wang, W. L. Gu, L. R. Zheng, W. Y. Song and C. Z. Zhu, Nano Today, 2021, 40, 101261 CrossRef CAS.
- L. Jiao, W. Q. Xu, Y. Zhang, Y. Wu, W. L. Gu, X. X. Ge, B. B. Chen, C. Z. Zhu and S. J. Guo, Nano Today, 2020, 35, 100971 CrossRef CAS.
- Q. M. Chen, Y. Liu, Y. W. Lu, Y. J. Hou, X. D. Zhang, W. B. Shi and Y. M. Huang, J. Hazard. Mater., 2022, 422, 126929 CrossRef CAS PubMed.
- H. J. Sun, Y. Zhou, J. S. Ren and X. G. Qu, Angew. Chem., Int. Ed., 2018, 57, 9224–9237 CrossRef CAS PubMed.
- N. Cheng, J. C. Li, D. Liu, Y. H. Lin and D. Du, Small, 2019, 15, 1901485 CrossRef CAS PubMed.
- Y. Li, R. Javed, R. Li, Y. Y. Zhang, Z. Y. Lang, H. B. Zhao, X. Liu, H. M. Cao and D. X. Ye, Food Chem., 2023, 406, 135017 CrossRef CAS PubMed.
- Q. D. Zeng, H. H. Zhong, J. H. Liao, Q. Huo, B. P. Miao, L. Zeng, B. Zhang and G. H. Nie, Biomater. Sci., 2024, 12, 5150–5163 RSC.
- S. F. Zhang, Y. H. Li, S. Sun, L. Liu, X. Y. Mu, S. H. Liu, M. L. Jiao, X. Z. Chen, K. Chen, H. Z. Ma, T. Li, X. Y. Liu, H. Wang, J. N. Zhang, J. Yang and X. D. Zhang, Nat. Commun., 2022, 13, 4744 CrossRef CAS PubMed.
- W. H. Gao, J. Y. He, L. Chen, X. Q. Meng, Y. N. Ma, L. L. Cheng, K. S. Tu, X. F. Gao, C. Liu, M. Z. Zhang, K. L. Fan, D. W. Pang and X. Y. Yan, Nat. Commun., 2023, 14, 160 CrossRef CAS PubMed.
- Y. Chong, Q. Liu and C. C. Ge, Nano Today, 2021, 37, 101076 CrossRef CAS.
- H. J. Sun, A. D. Zhao, N. Gao, K. Li, J. S. Ren and X. G. Qu, Angew. Chem., Int. Ed., 2015, 54, 7176–7180 CrossRef CAS PubMed.
- X. H. Niu, Q. R. Shi, W. L. Zhu, D. Liu, H. Y. Tian, S. F. Fu, N. Cheng, S. Q. Li, J. N. Smith, D. Du and Y. H. Lin, Biosens. Bioelectron., 2019, 142, 111495 CrossRef CAS PubMed.
- M. C. Hu, Z. H. Yao and X. Q. Wang, Abstr. Pap. Am. Chem. Soc., 2017, 254, 3477–3502 Search PubMed.
- Y. F. Chen, L. Jiao, H. Y. Yan, W. Q. Xu, Y. Wu, L. R. Zheng, W. L. Gu and C. Z. Zhu, Anal. Chem., 2021, 93, 12353–12359 CrossRef CAS PubMed.
- C. Liu, W. B. Fan, W. X. Cheng, Y. P. Gu, Y. M. Chen, W. H. Zhou, X. F. Yu, M. H. Chen, M. R. Zhu, K. L. Fan and Q. Y. Luo, Adv. Funct. Mater., 2023, 33, 2213856 CrossRef CAS.
- M. F. Huo, L. Y. Wang, Y. W. Wang, Y. Chen and J. L. Shi, ACS Nano, 2019, 13, 2643–2653 CAS.
- H. Li, Q. Li, Q. Shi, Y. Wang, X. Liu, H. Tian, X. Wang, D. Yang and Y. Yang, Food Chem., 2022, 389, 132985 CrossRef CAS PubMed.
- J. J. X. Wu and H. Wei, Prog. Chem., 2021, 33, 42–51 CAS.
- Y. Y. Qi, B. J. Li, D. D. Song, F. R. Xiu and X. Gao, Spectrochim. Acta, Part A, 2023, 297, 122722 CrossRef CAS PubMed.
- Y. W. Wang, Q. Liu, L. X. Wang, S. R. Tang, H. H. Yang and H. B. Song, Microchim. Acta, 2019, 186, 7 CrossRef PubMed.
- P. K. Boruah and M. R. Das, J. Hazard. Mater., 2020, 385, 121516 CrossRef CAS PubMed.
- W. X. Zhang, M. Yang, H. Zhang, X. J. Yu, W. Zhang, A. T. S. Wee, X. Yan, J. W. Qi and J. S. Li, Chem. Eng. J., 2022, 428, 131080 CrossRef CAS.
- Q. M. Chen, Y. Liu, Y. W. Lu, Y. J. Hou, X. D. Zhang, W. B. Shi and Y. M. Huang, J. Hazard. Mater., 2022, 422, 126929 CrossRef CAS PubMed.
- X. F. Chen, Y. Wang, M. Feng, D. Deng, X. Y. Xie, C. X. Deng, K. N. Khattak and X. P. Yang, Chin. Chem. Lett., 2023, 34, 107969 CrossRef CAS.
- Q. L. Ma, X. Y. Zhang, R. N. Guo, H. X. Zhang, Q. F. Cheng, M. Z. Xie and X. W. Cheng, Sep. Purif. Technol., 2019, 210, 335–342 CrossRef CAS.
- Q. H. Feng, G. Wang, L. H. Xue, Y. S. Wang, M. L. Liu, J. Liu, S. T. Zhang and W. P. Hu, ACS Appl. Nano Mater., 2023, 6, 4844–4853 CrossRef CAS.
- C. Zhang, H. C. Li, C. Li and Z. Q. Li, Molecules, 2020, 25, 168 CrossRef CAS PubMed.
- N. Y. Donkadokula, A. K. Kola, I. Naz and D. Saroj, Rev. Environ. Sci. Bio/Technol., 2020, 19, 543–560 CrossRef CAS.
- S. Varjani, P. Rakholiya, H. Y. Ng, S. M. You and J. A. Teixeira, Bioresour. Technol., 2020, 314, 123728 CrossRef CAS PubMed.
- X. F. Chen, Y. Wang, M. Feng, D. Deng, X. Y. Xie, C. X. Deng, K. N. Khattak and X. P. Yang, Chin. Chem. Lett., 2023, 34, 107969 CrossRef CAS.
- Q. H. Feng, G. Wang, L. H. Xue, Y. S. Wang, M. L. Liu, J. Liu, S. T. Zhang and W. P. Hu, ACS Appl. Nano Mater., 2023, 6, 4844–4853 CrossRef CAS.
- X. Y. Xie, Y. Zhao, Y. X. Fan, L. Jiang, W. Liu and X. P. Yang, Langmuir, 2024, 40, 12671–12680 CrossRef CAS PubMed.
- J. Wang, J. J. Zhang, K. Y. Guo, Q. Y. Yue, K. X. Yin, X. Xu, Y. W. Li, Y. Gao and B. Y. Gao, Adv. Funct. Mater., 2024, 34, 2406790 CrossRef CAS.
- L. Jiao, W. Q. Xu, H. Y. Yan, Y. Wu, C. R. Liu, D. Du, Y. H. Lin and C. Z. Zhu, Anal. Chem., 2019, 91, 11994–11999 CrossRef CAS PubMed.
- Y. Wu, J. B. Wu, L. Jiao, W. Q. Xu, H. J. Wang, X. Q. Wei, W. L. Gu, G. X. Ren, N. A. Zhang, Q. H. Zhang, L. Huang, L. Gu and C. Z. Zhu, Anal. Chem., 2020, 92, 3373–3379 CrossRef CAS PubMed.
- X. H. Gao, J. J. Tang, H. R. Liu, L. B. Liu and Y. Z. Liu, Drug Dev. Res., 2019, 80, 438–445 CrossRef CAS PubMed.
- Y. W. Mao, J. Zhang, R. Zhang, J. Q. Li, A. J. Wang, X. C. Zhou and J. J. Feng, Anal. Chem., 2023, 95, 8640–8648 CrossRef CAS PubMed.
- W. Q. Xu, W. Y. Song, Y. K. Kang, L. Jiao, Y. Wu, Y. F. Chen, X. L. Cai, L. R. Zheng, W. L. Gu and C. Z. Zhu, Anal. Chem., 2021, 93, 12758–12766 CrossRef CAS PubMed.
- Y. J. Song, Y. Chen, L. Y. Feng, J. S. Ren and X. G. Qu, Chem. Commun., 2011, 47, 4436–4438 RSC.
- W. J. Ma, J. J. Mao, X. T. Yang, C. Pan, W. X. Chen, M. Wang, P. Yu, L. Q. Mao and Y. D. Li, Chem. Commun., 2019, 55, 159–162 RSC.
- F. F. Cao, L. Zhang, Y. W. You, L. R. Zheng, J. S. Ren and X. G. Qu, Angew. Chem., Int. Ed., 2020, 59, 5108–5115 CrossRef CAS PubMed.
- Y. J. Zhang, W. H. Gao, Y. A. Ma, L. L. Cheng, L. Zhang, Q. G. Liu, J. Y. Chen, Y. R. Zhao, K. S. Tu, M. Z. Zhang and C. Liu, Nano Today, 2023, 49, 101768 CrossRef CAS.
- H. Zhou, J. Lu and X. L. Yin, Biomaterials, 2025, 123465 Search PubMed.
- Y. T. Su, F. Wu, Q. X. Song, M. J. Wu, M. Mohammadniaei, T. W. Zhang, B. L. Liu, S. S. Wu, M. Zhang, A. Li and J. Shen, Biomaterials, 2022, 281, 121325 CrossRef CAS PubMed.
- P. Malla, Y. M. Wang and C. H. Su, J. Nanobiotechnol., 2025, 23, 130 CrossRef PubMed.
- M. Sha, W. Q. Xu, Z. C. Wu, W. L. Gu and C. Z. Zhu, Chem. J. Chin. Univ.s, 2022, 43, 1751–1756 Search PubMed.
- S. F. Cai, J. M. Liu, J. W. Ding, Z. Fu, H. L. Li, Y. L. Xiong, Z. Lian, R. Yang and C. Y. Chen, Angew. Chem., Int. Ed., 2022, 61, e202204502 CrossRef CAS PubMed.
- Y. Wang, Y. Y. Xu, J. Y. Song, X. T. Liu, S. J. Liu, N. Yang, L. Wang, Y. J. Liu, Y. W. Zhao, W. H. Zhou and Y. Y. Zhang, Int. J. Nanomed., 2024, 19, 5837–5858 CrossRef PubMed.
- H. S. Han and K. Y. Choi, Biomedicines, 2021, 9, 305 CrossRef CAS PubMed.
- M. F. Huo, L. Y. Wang, H. X. Zhang, L. L. Zhang, Y. Chen and J. L. Shi, Small, 2019, 15, 1901834 CrossRef PubMed.
- J. R. Zhu, Q. L. Li, X. Li, X. M. Wu, T. Yuan and Y. L. Yang, Langmuir, 2022, 38, 6860–6870 CrossRef CAS PubMed.
- D. F. Cao, X. Y. Zhou, Q. Guo, M. Y. Xiang, M. H. Bao, B. S. He and X. Y. Mao, Biomark. Res., 2024, 12, 142 CrossRef PubMed.
- L. Asaad, B. Pepperrell, E. McErlean and F. Furlong, Int. J. Mol. Sci., 2025, 26, 1274 CrossRef CAS PubMed.
- B. Riddhidev, K. Endri, L. Sabitri, N. K. Lauren, K. Nishanth, I. Dragan, H. P. M. Kay, S. James, T. William and T. L. M. Viranga, Eur. J. Med. Chem., 2022, 244, 114807 CrossRef CAS PubMed.
- Y. Sun, B. Xu, X. Pan, H. Wang, Q. Wu, S. Li, B. Jiang and H. Liu, Coord. Chem. Rev., 2023, 475, 214896 CrossRef CAS.
- Y. A. Ma, J. J. Zhao, L. L. Cheng, C. Li, X. J. Yan, Z. C. Deng, Y. J. Zhang, J. Liang, C. Liu and M. Z. Zhang, Carbon, 2023, 204, 526–537 CrossRef CAS.
- J. Y. Lu, Y. L. Mao, S. P. Feng, X. Li, Y. K. Gao, Q. F. Zhao and S. L. Wang, Acta Biomater., 2022, 148, 310–322 CrossRef CAS PubMed.
- J. Shin, N. Y. Kang, B. Kim, H. Y. S. Hong, L. Yu, J. Kim, H. M. Kang and J. S. Kim, Chem. Soc. Rev., 2023, 52, 4488–4514 RSC.
- P. Malla, Y. M. Wang and C. H. Su, J. Nanobiotechnol., 2025, 23, 7786–7824 CrossRef PubMed.
- Y. Cai, Z. Xiao, T. Cheng, B. Yuan, Y. Cui, J. L. Chen, Y. Zhao, P.-T. Chou and Y.-K. Peng, ACS Appl. Mater. Interfaces, 2025, 17, 37577–37585 CrossRef PubMed.
- D. Q. Zhu, M. L. Zhang, C. Wang, P. P. Gai and F. Li, Chem. Mater., 2022, 34, 11072–11080 CrossRef CAS.
- S. H. Chen, S. W. Guan, J. Tan, W. Lu, H. F. Zhang, J. J. Qiu, H. Q. Zhu and X. Y. Liu, Adv. Funct. Mater., 2024, 34, 2411202 CrossRef CAS.
- Y. J. Ji, Y. Wang, H. Wang, J. S. Bai, J. S. Ren and X. G. Qu, Angew. Chem., Int. Ed., 2025, e202505742 Search PubMed.
- H. Wei and E. K. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- Y. H. Hu, X. J. J. Gao, Y. Y. Zhu, F. Muhammad, S. H. Tan, W. Cao, S. C. Lin, Z. Jin, X. F. Gao and H. Wei, Chem. Mater., 2018, 30, 6431–6439 CrossRef CAS.
- M. Q. Xu, W. W. Wang, H. Han, Y. P. Wei, J. Q. Sha and G. D. Liu, Chem. Eng. J., 2023, 475, 146324 CrossRef CAS.
- M. W. Liu, W. Q. Xu, Y. J. Tang, Y. Wu, W. L. Gu, D. Du, Y. H. Lin and C. Z. Zhu, Angew. Chem., Int. Ed., 2025, 64, e202424070 CrossRef CAS PubMed.
- R. Zhang, B. Jiang, K. Fan, L. Gao and X. Yan, Nat. Rev. Bioeng., 2024, 2, 849–868 CrossRef CAS.
- S. Khan, M. Sharifi, S. H. Bloukh, Z. Edis, R. Siddique and M. Falahati, Talanta, 2021, 224, 121805 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |







