Fluorescent graphene quantum dots for biosensing and bioimaging
Zetan Fan
,
Shuhua Li
,
Fanglong Yuan
and
Louzhen Fan
*
Department of Chemistry, Beijing Normal University, Beijing, 100875, China. E-mail: lzfan@bnu.edu.cn
First published on 6th February 2015
Abstract
Fluorescent carbon-based nanomaterials, including carbon dots (C-Dots) and the emerging graphene quantum dots (GQDs) are attracting increasing interest in the field of biosensing and bioimaging for their high aqueous solubility, stable photoluminescence (PL), low cytotoxicity, good biocompatibility as well as superior resistance to photobleaching. Compared with C-Dots, the quasi zero-dimensional (0D) GQDs with a few atomic layers can exhibit strong PL without passivation. With these extraordinary properties, GQDs are suitable alternatives as both a versatile platform for biosensing and a promising class of bioimaging probes. In this review, we introduce the synthesis, structure, PL properties and currently available applications of GQDs in biosensing and bioimaging field. We hope this review will provide critical insights to inspire more exciting researches on GQDs for biological applications to better realize the potential of the intriguing GQDs in the near future.
1. Introduction
Graphene, a novel single-atom-thick and two-dimensional (2D) nano-graphitic carbon sheet, exhibits characteristics including large surface area, high carrier transport mobility, superior mechanical flexibility and excellent thermal and chemical stability, which are suitable for various applications in optoelectronic devices, energy-storage media and drug-delivery vehicles.1–4 Since graphene is a zero-bandgap semiconductor, no optical photoluminescence (PL) is observed in pristine form, which limits its application in optoelectronics. However, both theoretical and experimental studies show that the band gap of graphene can be tuned by converting 2D graphene sheets into zero-dimensional (0D) graphene quantum dots (GQDs).5–8 Due to quantum confinement and edge effects, GQDs, as a new kind of quantum dots (QDs), have attracted tremendous research interest due to their high aqueous solubility, stable PL, low cytotoxicity, good biocompatibility as well as superior resistance to photobleaching compared with traditional semiconductor QDs and organic dyes.9,10Another example of 0D fluorescent carbon-based nanomaterials, carbon dots (C-Dots) have also attracted much attention in recent years.11–13 Though GQDs are even considered as a kind of C-Dots, some differences should be pointed out. C-Dots are either amorphous or crystalline,14 while GQDs clearly possess graphene lattices inside the dots produced from graphene-based starting materials or the rigid synthetic chemistry of graphene-like smaller polycyclic aromatic hydrocarbon molecules.15,16 Unlike the quasi-spherical C-Dots, GQDs with a few atomic layer graphene sheets can exhibit strong PL without passaivation.17 Moreover, the unique PL properties of GQDs can be effectively tuned by doping heteroatoms to the π-conjugated system.18–20
With these extraordinary properties, GQDs are suitable alternatives as both a versatile platform for biosensing and a promising class of bioimaging probes. To date, various GQDs-based probes for the detection of ions, biomolecules and small organic molecules with aromatic structures have been designed,21 and biomarkers based on blue and green fluorescent GQDs have been widely used.22 However, much work is still needed to explore the full potential of these nanomaterials for developing advanced smart sensors and biomarkers.
In this review, we introduce the synthesis, structure, PL properties and currently available applications of GQDs in biosensing and bioimaging field. We hope this review will provide critical insights to inspire more exciting researches on GQDs for biological applications to better realize the potential of the intriguing GQDs in the near future.
2. Synthesis, structure and PL properties of GQDs
2.1 Synthesis and structure
Up to now, the synthesis of GQDs with tunable size can be generally divided into two strategies: top-down and bottom-up methods. The typical methods are listed in Table 1. The top-down routes are implemented via either physical or chemical techniques to cut down large graphene or graphene oxide (GO) sheets, carbon nanotubes (CNTs), carbon fibers or graphite into small pieces of graphene sheets while in the bottom-up methods, GQDs are controllably prepared by stepwise reactions of small molecules. For top-down approaches, acidic oxidation,23–25 hydrothermal17,26 or solvothermal,27,28 microwave-assisted,29,30 sonication-assisted,31 electrochemical,12,19,20,32–35 chemical exfoliation,36,37 photo-fenton reaction15 methods have been used to synthesize GQDs. Conversely, the bottom-up methods offer us exciting opportunities to control the GQDs with well-defined molecular size, shape and properties. Stepwise solution chemistry methods using benzene derivatives,38–41 precursor pyrolysis,42–44 microwave-assisted hydrothermal treatment of carbohydrates,45 ruthenium-catalyzed cage-opening of fullerenes46 and carbonization of hexaperihexabenzocoronene (HBC)16 have been utilized to prepare GQDs successfully.| Methods | Subclassification | Raw materials | Size (nm) | Color | Quantum yield (QY, %) | References |
|---|---|---|---|---|---|---|
| Top-down | Acid oxidation | Carbon black | 15–18 | Green, yellow | 4.04, 2.09 | 23 |
| Carbon fibers | 1–4 | Blue, green, yellow | 24 | |||
| Coal | 2.96 ± 0.96 | Green | 25 | |||
| Hydrothermal | GO | 1.5–5 | Green | 7.5 | 26 | |
| GO | 5–13 | Blue | 6.9 | 17 | ||
| Solvothermal | GO | ∼3 | Green | 31 | 27 | |
| GO | 3–5 | Blue, green | 29–19 | 28 | ||
| Microwave | GO | 2–7 | Blue, green | 29 | ||
| GO | 3 | Blue | 22.9 | 30 | ||
| Sonication | Graphene | 3–5 | Blue | 3.4 | 10 | |
| Electrochemistry | Graphite rod | 5–10 | Yellow | 14 | 33 | |
| Graphite rod | 3–7 | Green, yellow | 13 | 19 | ||
| Graphite rod | 3 | Green | 10.6 | 20 | ||
| Graphene | ∼3.0 | Blue | 10 | 35 | ||
| Chemical exfoliation | Multi-wall carbon nanotubes (MWCNTs) | 20 | Blue | 6.53 | 36 | |
| Graphite nanoparticles | 4 | Blue | 37 | |||
| Bottom-up | Hydrothermal | Pynene | 3.6 ± 1.2 | Green | 23 | 40 |
| Polythiophene derivatives | 2–6 | Red | 5.4 | 41 | ||
| Microwave-assist pyrolysis | Glycerol | 1–5 | Green | 45 | ||
| Catalyzed cage-opening | C60 | 2.7–10 | 46 | |||
| Precursor pyrolysis | L-Glutamic acid | 4.66 ± 1.24 | Blue | 54.5 | 44 | |
| Citric acid | 15 | Blue | 9.0 | 42 | ||
| Glucose | 1.6–21 | Blue, UV | 7–11 | 43 |
In general, the average sizes of GQDs are mostly below 10 nm, and the largest diameter of GQDs reported so far is 60 nm.16 Similar to the size, the height of GQDs depends on the preparation methods, mostly ranging from 0.5 to 5 nm. However, the heights of GQDs prepared through different methods don't monotonously relate to their sizes. Most of GQDs consist of no more than 5 layers, and a great deal of monolayer GQDs have been prepared successfully.23,26,33 GQDs prepared by our group via electrochemical exfoliation of graphite followed by room temperature reduction with hydrazine (N2H4) were uniform and monodisperse (Fig. 1a and b), with an average diameter of 5 to 10 nm (Fig. 1c) and a topographic height below 0.5 nm (Fig. 1d and e), mostly consisting of a single graphene layer.33
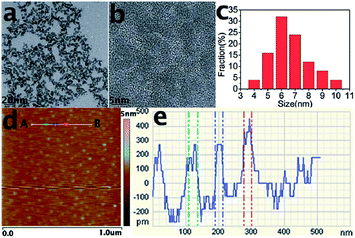 | ||
| Fig. 1 TEM (a) and HRTEM (b) images and size distribution (c) of GQDs. (d) AFM image of the GQDs deposited on freshly cleaved mica substrates. (e) Height profile along the line from A to B in (d). Reprinted with permission from: ref. 33, Copyright 2012 Royal Society of Chemistry. | ||
The high resolution transmission electron microscopy (HRTEM) images of GQDs feature two kinds of lattice fringes, namely (002) interlayer spacing and (1120) in-plane lattice spacing. The former centered at about 0.34 nm has been observed from GQDs prepared by acidic oxidation of carbon black23 and electrochemical cutting method,32 the latter mostly centered at about 0.24 nm has been observed from GQDs synthesized via acidic oxidation from carbon fiber,24 hydrothermal treatment29 and photo-fenton reaction,15 excepting for 0.21 nm (Fig. 2a) via hydrothermal cutting strategy.26 Interestingly, the two kinds of lattice fringes (Fig. 2b) have been simultaneously been observed in the case of electrochemical method from MWCNTs.34
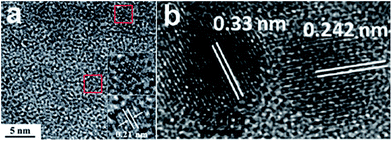 | ||
| Fig. 2 (a) TEM image of GQDs. (b) Fringe patterns of GQDs. Reprinted with permission from: (a) ref. 26, Copyright 2012 Royal Society of Chemistry, (b) ref. 34, Copyright 2012 Wiley-VCH. | ||
GQDs are generally composed of a series of surface groups, such as carboxyl, hydroxyl and epoxy groups.9,10 Fourier transform infrared spectroscopy (FTIR) and X-ray photoelectron spectroscopy (XPS) are commonly adopted to analyze their components. For sulfur-doped GQDs (S-GQDs) prepared by electrolysis of graphite in sodium p-toluenesulfonate aqueous solution,20 the XPS survey spectrum (Fig. 3a) showed dominant graphitic C1s peak at 284 eV, S2p peak at 167 eV and O1s peak at 531 eV. The presence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–O, C
C, C–O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–S bonds indicated that GQDs were functionalized with hydroxyl, carbonyl, carboxyl and epoxy groups, and the high-resolution S2p spectrum confirmed the successful incorporation of S atoms into GQDs (Fig. 3b and c). The results were further confirmed by the corresponding FTIR spectrum (Fig. 3d).
O, C–S bonds indicated that GQDs were functionalized with hydroxyl, carbonyl, carboxyl and epoxy groups, and the high-resolution S2p spectrum confirmed the successful incorporation of S atoms into GQDs (Fig. 3b and c). The results were further confirmed by the corresponding FTIR spectrum (Fig. 3d).
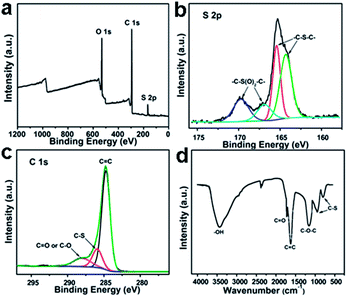 | ||
| Fig. 3 XPS (a) S2p (b) C1s (c) and FTIR (d) spectra of S-GQDs. Reprinted with permission from: ref. 20, Copyright 2014 American Chemical Society. | ||
Raman spectroscopy is also a powerful and non-destructive tool for the characterization of GQDs. The G band is assigned to the E2g vibration modes of the aromatic domains, whereas the D band arises from the breathing modes of graphitic domains. Traditionally, the intensity ratio of disorder D to crystalline G (ID/IG) is used to compare the structural order between crystalline and amorphous graphitic systems. The ID/IG values of GQDs vary significantly depending on the preparation methods. The electrochemically prepared boron-doped GQDs (B-GQDs) from high purity graphite rods have a high ID/IG value of 1.23,19 indicating numerous defects on the B-GQDs.
2.2 Absorbance and PL properties
For the UV-vis absorption spectrum of GQDs, two typical peaks can be observed: a peak at around 230 nm assigned to π–π* transition of aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds and a shoulder at about 300 nm due to n–π* transition of C
C bonds and a shoulder at about 300 nm due to n–π* transition of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds.23,33,37 The most fascinating feature of GQDs is their PL. Variously sized GQDs with deep ultraviolet,38 blue,15,17,24,28,31,34,36 green,12,23,26–28 yellow,33 and red41 colors have been prepared via different synthetic approaches. The PL mechanism may derive from intrinsic state emission and defect state emission,47 which arise from quantum confinement effect, emissive traps, excitons of carbon, aromatic structures, oxygen-containing groups, free zigzag sites and edge defects.48–50
O bonds.23,33,37 The most fascinating feature of GQDs is their PL. Variously sized GQDs with deep ultraviolet,38 blue,15,17,24,28,31,34,36 green,12,23,26–28 yellow,33 and red41 colors have been prepared via different synthetic approaches. The PL mechanism may derive from intrinsic state emission and defect state emission,47 which arise from quantum confinement effect, emissive traps, excitons of carbon, aromatic structures, oxygen-containing groups, free zigzag sites and edge defects.48–50
The PL of GQDs are influenced by many factors, such as size,16,23,24,51 excitation wavelength,52,53 pH,17,24,52,53 solvent polarity,52,54 surface oxidation degree,30,53 surface functionalizations28,51 and heteroatom doping.19,20 Three kinds of GQDs with different sizes of 1–4, 4–8, and 7–11 nm via varying the reaction temperatures, emitted different PL changing from blue and green to yellow (Fig. 4a and b).24 GQDs prepared via acidic oxidation from carbon black exhibited PL transition from green to yellow as their sizes increased from 15 to 18 nm.23 The size-dependent PL of GQDs mainly attributed to the quantum confinement effect. However, it is worth noting that the size-independent PL of the GQDs (Fig. 4c) has also been observed from glucose carbonization,43 due to the surface state emission. The largest GQDs prepared from unsubstituted hexa-peri-hexabenzocoronene (HBC) also showed blue PL (Fig. 4d),16 which could be ascribed to the zigzag edge sites and defect effect.17
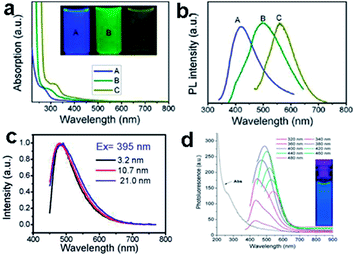 | ||
| Fig. 4 (a) UV-vis spectra of GQDs A, B, and C, corresponds to synthesized reaction temperature at 120, 100, and 80 °C, respectively. Inset of panel is a photograph of the corresponding GQDs under UV light with 365 nm excitation. (b) PL spectra of GQDs with different emission color excited at 318, 331, and 429 nm, respectively. (c) The normalized PL spectra of the GQDs with various sizes. (d) UV-vis absorption and PL emission spectra of GQDs. Reprinted with permission from: (a) and (b) ref. 24, Copyright 2012 American Chemical Society, (c): ref. 43, Copyright 2012 American Chemical Society, (d) ref. 16, Copyright 2011 American Chemical Society. | ||
GQDs prepared via hydrothermal method exhibited excitation-dependent PL properties17 (Fig. 5a) which might result from different size or emissive sites.54,55 Meanwhile, the excitation-independent PL behavior has also been reported (Fig. 5b).31,33,42 This could be explained by the high uniformity emissive sites contained in GQDs. Interestingly, the PL of GQDs has also been found to be sensitive to solvent and pH. The emission peak of GQDs synthesized via solvothermal method shifted from 475 to 515 nm in THF, acetone, DMF, and water, respectively (Fig. 5c).54 Under alkaline conditions, the hydrothermally synthesized GQDs emitted strong blue PL while under acidic conditions it was nearly completely quenched (Fig. 5d). When pH was switched repeatedly between 13 and 1, the PL intensity varied reversibly.17 In addition, the pH had nearly no influence on the PL wavelength, but just affected the PL intensities of GQDs, which was also observed in GQDs with emissive zigzag sites.24,26
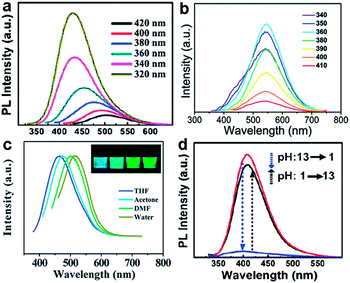 | ||
| Fig. 5 (a) PL spectra of the GQDs at different excitation wavelengths. (b) PL spectra of the GQDs at different excitation wavelengths. (c) Effect of solvents on the fluorescence of GQDs. (d) pH-dependent PL spectra when pH is switched between 13 and 1. Reprinted with permission from: (a) and (d) ref. 17, Copyright 2010 Wiley-VCH, (b) ref. 33, Copyright 2012 Royal Society of Chemistry, (c) ref. 54, Copyright 2011 Royal Society of Chemistry. | ||
Doping GQDs with heteroatoms could effectively modulate their band gap and electronic density, enhancing chemical activity and QY of GQDs for practical applications, which were proved through theoretical calculations and detailed experiments.17,19,20,38 B-GQDs gave rise to rich PL owing to their peculiar interaction with the surrounding media of borax (Fig. 6a and b)19 and S-GQDs exhibited bright green PL (Fig. 6c).20 Upconversion PL, another exciting feature of GQDs, was reported by many groups,47,49,56 but some results were artificial. Gan et al. revealed that the upconversion PL in GQDs under excitation from a xenon lamp was excited by the second-order diffraction light of wavelength λ/2 coexisting in the excitation light of wavelength λ. The real upconversion from GQDs was obtained under excitation with a femtosecond pulsed laser (Fig. 6d).57
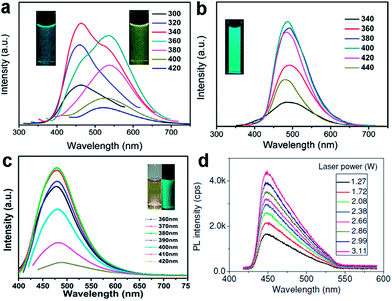 | ||
| Fig. 6 PL spectra of the B-GQDs aqueous solution in the absence (a) and presence (b) of borax. (c) PL spectra of S-GQD. (d) Upconversion photoluminescence spectra obtained from GQDs under excitation of a femtosecond pulsed laser at 800 nm. Reprinted with permission from: (a) and (b) ref. 19, Copyright 2014 Elsevier, (c) ref. 20, Copyright 2014 American Chemical Society, (d) ref. 57, Copyright 2013 Wiley-VCH. | ||
The QY of GQDs varies with the fabrication method and the surface chemistry involved. For the unpassivated GQDs, their QYs are always low, due to the carboxylic and epoxide groups contained in GQDs acting as the non-radiative electron–hole recombination centers.58 Therefore, the removal of these oxygen-containing groups may improve the QY, either by reduction or surface passivation.59 Li et al. improved the QY of GQDs from 11.7 to 22.9% by gentle reduction with NaBH4.30 Our group obtained yellow fluorescent GQDs with a relative high QY about 14% by reduction with N2H4.33 Tetsuka et al. prepared amino-functionalized GQDs with QY ranging from 29 to 19%, which decreased with quantity of amine functionalization.28
3. Biosensing
In view of the high aqueous solubility, stable PL, low cytotoxicity, good biocompatibility, superior resistance to photobleaching as well as unique electrochemical and electrochemiluminescence (ECL) emission properties of GQDs, PL, electrochemical and ECL biosensors have been explored to detect ions, biomaterials, and small organic molecules with aromatic structures.3.1 PL biosensors
Various GQDs-based PL sensors have been fabricated with either signal-off or signal-on process. Recently, our group presented a novel PL sensor using S-GQDs for the detection of Fe3+ in human serum samples (Fig. 7a).20 Doping GQDs with S atom could effectively modulate the band gap and electronic density of GQDs, which promoted the coordination interaction between Fe3+ and phenolic hydroxyl groups of S-GQDs, leading to significant PL quenching. Such PL probe exhibited highly sensitive and selective detection of Fe3+ in the concentration range of 0.01–0.70 μM, and the limit of detection (LOD) was as low as 4.2 nM. Taking advantage of the PL quenching of GQDs, a turn-off sensor for Ag+ and biothiol detection was developed by Ran et al.60 The formation of Ag nanoparticles (AgNPs) on GQDs was able to quench the PL of GQDs and the addition of biothiols could cause a further turn-off phenomenon due to their strong interaction through the formation of Ag–S bonds. The LOD was 3.5 nM for Ag+, 6.2 nM for cysteine (Cys), 4.5 nM for homocysteine (Hcy) and 4.1 nM for glutathione (GSH) in the range 0–100 nM. He et al. reported a glucose biosensor via the PL quenching of hemin-functionalized GQDs.61 According to the surface quenching states (SQS) induced mechanism, 3-aminobenzeneboronic acid functionalized GQDs (APBA-GQDs) were also used for glucose detection by Qu et al.62 In addition, Wang et al. developed a simple PL off assay for the activity of protein kinase CK2 based on the selective aggregation of phosphorylated peptide-GQD conjugates triggered by Zr4+ coordination.63 When the substrate peptide was phosphorylated by CK2, the introduced Zr4+ could serve as a linkage between the phosphorylated sites of phosphopeptides via the multi-coordinative interactions between Zr4+ and phosphate groups, resulting in extensive aggregation of the GQDs and effective PL quenching (Fig. 7b), thus the activity of protein kinase could be facilely monitored with an LOD of 0.03 unit mL−1.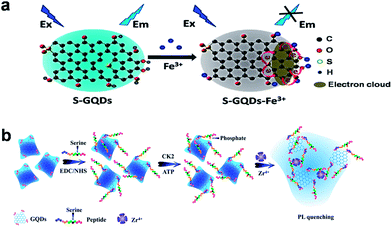 | ||
| Fig. 7 (a) Schematic illustration of the PL quenching mechanism of the S-GQDs in the presence of Fe3+. (b) Schematic representation of the CK2 kinase assay based on the aggregation and PL quenching of phosphorylated peptide-GQD conjugates via Zr4+ linkages. Reprinted with permission from: (a) ref. 20, Copyright 2014 American Chemical Society, (b) ref. 63, Copyright 2013 American Chemical Society. | ||
Apart from the signal-off PL sensors described above, signal-on PL sensors have been designed as well.19,64–69 An aggregation-induced PL increasing sensor for label-free detection of glucose was proposed by Zhang et al.64 The reaction of the two cis-diol units in glucose with the two boronic acid groups on the boron-doped GQDs (BGQDs) surfaces created structurally rigid BGQDs-glucose aggregates, restricting the intramolecular rotations and thus resulting in a great boost in the PL intensity (Fig. 8a). In another study, Li et al. designed a glucose sensing system using anionic GQDs and a cationic boronic acid-substituted bipyridinium salt (BBV).65 The pristine GQDs acted as luminescent reporting units, while BBV served as a PL quencher and a glucose receptor simultaneously. It was postulated that the electrostatic attraction between GQDs and BBV resulted in the formation of a ground-state complex which facilitated the excited-state electron transfer from GQDs to bipyridinium and quenched the PL intensity of the GQDs. When glucose was added to the system, the boronic acids were converted to tetrahedral anionic glucoboronate esters, which effectively neutralized the net charge of the cationic bipyridinium, greatly diminishing its quenching efficiency, and the PL intensity of GQDs recovered. Li et al. demonstrated a label-free PL signal-on assay for the detection of trypsin with an LOD of 33 ng mL−1 based on cytochrome c (Cyt c)-induced self-assembled GQDs.66 Cyt c which is rich in Fe3+ could quench the fluorescence of GQDs. When trypsin was present, Cyt c was cleaved into smaller fragments on the C-terminal side of arginine and lysine residues, and Fe3+ in Cyt c was reduced to Fe2+ with the aid of the digestive enzyme. Thus the cleaved Cyt c was unable to quench the PL of GQDs, which resulted in the restoration of PL. Moreover, trypsin cleaved the peptide bonds of Cyt c to gave lysine and arginine residues, both of which chemically reduced GQDs into reduced-GQDs (r-GQDs), inducing a subsequent increase in PL intensity. Liu et al. proposed a PL “off-to-on” mechanism of glutathione-functionalized GQDs (GQDs@GSH) for the assay of adenosine triphosphate (ATP).67 Fe3+ quenched the fluorescence of GQDs@GSH by electron transfer and then were disassociated from GQDs@GSH by the phosphate ions through the strong interactions, which resulted in the restoration of PL. Since ATP was the major phosphate-containing metabolite in cell lysates and blood serum, the proposed sensing assay was successfully applied to estimate ATP level in cell lysates and blood serum. Wu et al. designed a PL turn-on sensor for biothiols detection and the LOD for GSH, Cys and Hcy were 5 nM, 2.5 nM and 5 nM, respectively.68 Hg2+ could efficiently quench the blue PL of GQDs and biothiols selectively bonded to Hg2+ through Hg–S bonding interactions to recover the PL of GQDs. Our group demonstrated B-GQDs used as PL turn-on sensor for detecting Al3+ (Fig. 8b).19 With increasing Al3+ concentration from 0 to 100 μM, the PL intensity of B-GQDs was enhanced linearly. The LOD of B-GQDs towards Al3+ was estimated as 3.64 μM, which met the limit for drinking water according to the World Health Organization standard (7.41 μM).
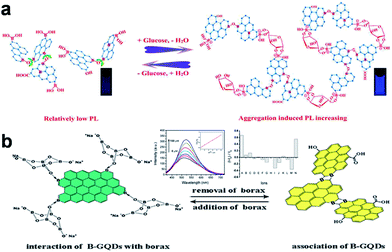 | ||
| Fig. 8 (a) Proposed “Aggregation-Induced PL Increasing” Mechanism for the glucose-specific sensing by BGQDs. (b) The structure illustration of B-GQDs in the presence (left) and absence (right) of borax, respectively. Inset: PL emission spectra of B-GQDs aqueous solution upon adding different concentrations of Al3+ ions at 405 nm excitation and maximum luminescent response of B-GQDs upon addition of different metal ions (100 μM) listed from left to right: (A) Al3+, (B) Ag+, (C) Na+, (D) K+, (E) Mg2+, (F) Ca2+, (G) Ba2+, (H) Cd2+, (I) Pb2+, (J) Fe2+, (K) Zn2+, (L) Fe3+, (M) Cr3+ and (N) Al3+ + Na+ + K+ + Mg2+ + Ca2+ + Ba2+ + Fe3+. Reprinted with permission from: (a) ref. 64, Copyright 2014 American Chemical Society, (b) ref. 19, Copyright 2014 Elsevier. | ||
Since π–π stacking interaction between graphene and GQDs could bring them into luminescence resonance-energy transfer (LRET) proximity and lead to PL quenching of GQDs, a series of LRET sensors were developed.70–72 Zhao et al. presented a LRET immunosensor using graphene as acceptor and mouse anti-human immunoglobulin G (mIgG, antibody) conjugated-GQDs as donor for the sensitive detection of human immunoglobulin G (IgG, antigen) with an LOD of 10 ng mL−1.70 When adding graphene into the mIgG-GQDs solution, both the π–π stacking interaction between graphene and GQDs and the nonspecific binding interaction between mIgG and the graphene surface brought graphene and GQDs into LRET proximity to facilitate the PL quenching of GQDs. The addition of human IgG would bind the mIgG due to the specific antibody–antigen interaction, which effectively increased the distance between mIgG-GQDs and graphene surface, thus hindering the LRET process and then producing a restoration of PL. Qian et al. established a PL sensing platform for DNA detection by regulating the interaction between the reduced GQDs (rGQD) labeled DNA probe and GO (Fig. 9).71 When ssDNA-rGQDs probe was absorbed on the surface of GO, both electrostatic attractions and π–π stacking interactions between ssDNA-rGQDs and the GO surface resulted in substantial PL quenching of ssDNA-rGQDs. Upon the addition of target DNA (tDNA), the tDNA could capture the ssDNA-rGQDs in the assembled ssDNA-rGQDs/GO to produce dsDNA-rGQDs through specific base pairing, leading to the liberation and detachment of the dsDNA-rGQDs from the GO and PL recovery from the free dsDNA-rGQDs. The established method for DNA detection had a broad linear range of 6.7–46.0 nM with an LOD of 75.0 pM. Furthermore, Qian et al. constructed a dual-functional biosensor for simultaneous detection of DNA and bovine α-thrombin by taking advantage of the intense dual-color PL of GQDs, efficient quenching effect of GO, specific recognition between probes and targets, and unique interaction between GQDs and GO.72 Like graphene and GO, CNTs were also used as acceptor in the LRET process for DNA detection by Qian et al.73 The specific PL “on-off-on” process originated from PL quenching owing to LRET between GQDs-labeled probe and CNTs and subsequent PL recovery due to liberation of free double-stranded DNA probe was employed to achieve quantitative analysis of target DNA.
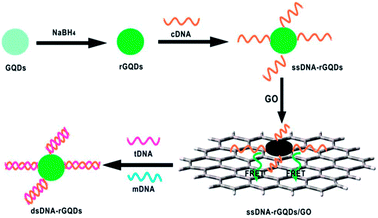 | ||
| Fig. 9 Schematic illustration of a universal PL sensing platform for the detection of DNA based on LRET between GQDs and GO. Reprinted with permission from ref. 71, Copyright 2014 Royal Society of Chemistry. | ||
3.2 Electrochemical biosensors
GQDs, which can act as multivalent redox species using cyclic voltammetry (CV), chronoamperometry (CA) and differential pulse voltammetry (DPV) measurements, present exciting opportunities for building electrochemical sensors.74–91 The first electrochemical sensor was designed for the detection of ssDNA by Zhao et al.74 The probe ssDNA (ssDNA-1) immobilized on GQDs inhibited the electron transfer effect between the electro-active species [Fe(CN)6]3−/4− and the electrode via electrostatic repulsion, resulting in a drastic decrease of the electrochemical signal. However, when the target ssDNA (ssDNA-2) or protein was present, the ssDNA-1 would bind the target, allowing the electron transfer and leading to the increase of peak currents. Muthurasu et al. demonstrated a simple strategy to detect and quantify H2O2 using electrochemical methods by employing horseradish peroxidase (HRP) functionalized GQDs.75 CV and CA electrochemical techniques were employed to investigate redox property, electron transfer behaviour and H2O2 sensing experiments. This particular electrochemical biosensor showed the sensitivity values of 0.905 and 7.057 μA mM−1 and LOD of ∼530 and 2.16 μM along with a fast response time of ∼2–3 s. Jiang et al. fabricated a nonenzymatic H2O2 sensor based on AgNPs deposited porous g-C3N4 (Ag/p-g-C3N4) using nitrogen-doped GQDs (N-GQDs) as effective photocatalyst (Fig. 10).76 The CV and amperometry results showed that the Ag/p-g-C3N4 composite responded to H2O2 with wide linear range and low LOD. Since GQDs exhibited similar peroxidase-like activity, another non-enzymatic H2O2 detection in living cells was also realized on a covalently assembled GQDs/Au electrode.77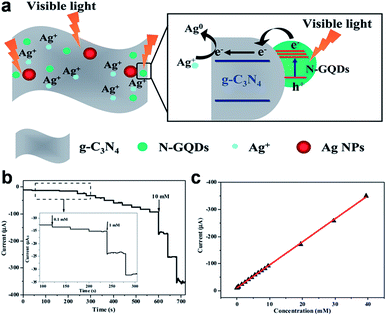 | ||
| Fig. 10 (a) A schematic diagram illustrating the proposed mechanism for the photochemical synthesis of Ag/p-g-C3N4 nanocomposites. (b) Typical steady-state response of the Ag/p-g-C3N4/GCE to successive injection of H2O2 into the stirred N2-saturated 0.2 M PBS at pH 7.4 (applied potential: −0.30 V), and (c) the corresponding calibration curve. Reprinted with permission from ref. 76, Copyright 2014 Royal Society of Chemistry. | ||
Razmi et al. presented a new, simple and low cost glucose biosensor using glucose oxidase (GOx) and GQDs.78 They immobilized GOx on the GQDs modified carbon ceramic electrode (CCE) and direct electrochemistry of GOx was realized. The developed biosensor (Gox-GQD/CCE) responded efficiently to glucose presence over the concentration range 5–1270 mM with the LOD of 1.73 mM. The high performance of the biosensor was attributed to the large surface-to-volume ratio, excellent biocompatibility of GQDs, porosity of GQDs/CCE, and the abundance of hydrophilic edges as well as hydrophobic plane in GQDs which enhanced the enzyme absorption on the electrode surface.
Wang et al. designed a novel sandwich-type electrochemical immunosensor using Fe3O4@GQDs hybrid and apoferritin-encapsulated Cu (Cu-apoferritin) nanoparticles as labels for highly selective and sensitive detection of avian leukosis virus subgroup J (ALVs-J) (Fig. 11).79 GQDs were used both for the conjugation of primary ALVs-Jantibodies (Ab1) and immobilization of secondary ALVs-Jantibodies (Ab2) after compounded with Fe3O4. Cu-apoferritin nanoparticles were first selected to immobilize onto Fe3O4@GQDs hybrid as electroactive probes. After the sandwich-type assembly, Cu was released from the apoferritin cavity, and then was detected by DPV. The huge surface area of GQDs increased the loading of antibodies and Cu-apoferritin nanoparticles, and a large amount of electroactive probes could be capsulated into the cavity of apoferritin, which increased the electrochemical signal significantly. Moreover, the proposed immunosensor displayed excellent analytical performance for the detection of ALVs-J ranging from 102.08 to 104.50 TCID50 mL−1 with an LOD of 115 TCID50 mL−1.
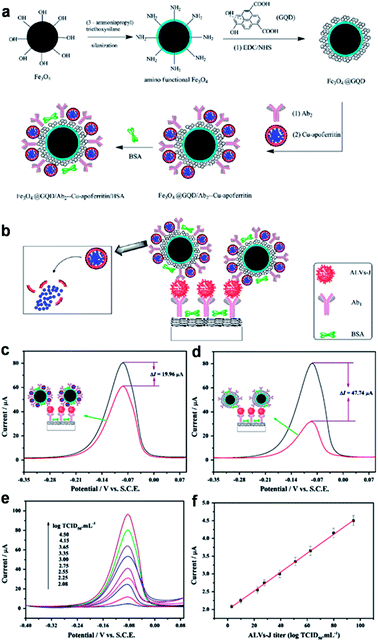 | ||
| Fig. 11 Schematic representation of the preparation of Fe3O4@GQDs/Ab2-Cu-apoferritin/BSA (a) and immunosensor (b). The DPV comparisons for the proposed immunosensors with (upper curve) and without (lower curve) GQDs (c) and apoferritin (d). (e) DPV responses of the immunosensor with increasing ALVs-J concentrations under the optimized conditions (102.08, 102.25, 102.55, 102.75, 103.00, 103.35, 103.65, 104.15, 104.50 TCID50 mL−1). (f) Calibration curve. Reprinted with permission from ref. 79, Copyright 2013 Elsevier. | ||
3.3 ECL biosensors
ECL, a smart combination of chemiluminescence (CL) and electrochemistry, is a valuable detection method and becoming increasingly recognized in analytical chemistry due to its high sensitivity, simplified set-up, label-free nature and low background signal. A series of semiconductor nanocrystals (such as semiconductor QDs, C-Dots and silicon QDs), have been reported to exhibit ECL emission, which show promise for the construction of ECL biosensors.80,81 ECL emission was observed from GQDs as well,30,82–85 and the considerable ECL mechanism of the GQDs was believed to form the excited state GQDs* with the help of the coreactant (such as S2O82−, H2O2 or TPA).30,82–84Yang et al. designed a novel sandwich-type ECL immunosensor by using gold–silver nanocomposite-functionalized graphene (GN-Ag–Au) as sensor platform and GQDs functionalized porous PtPd nanochains (pPtPd) as signal amplifiers for the detection of a tumor marker (Fig. 12).82 The possible response mechanism for the anodic ECL was proposed in the presence of coreactant TPA. The proposed ECL immunosensor exhibited good precision, acceptable stability and reproducibility. Under optimal conditions, the ECL immunosensors exhibited a wide detection range (0.002–70 U mL−1) and a low LOD (0.96 mU mL−1) for carbohydrate antigen 199 (CA199) in real serum samples.
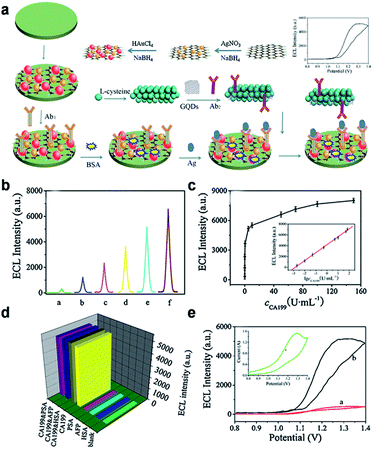 | ||
| Fig. 12 (a) Schematic illustration of the fabrication procedure of the immunosensor. (b) ECL of the immunosensor at various concentrations of CA199 (a: 0.002, b: 0.01, c: 0.05, d: 0.5, e: 5, f: 50 U mL−1, respectively). (c) Relationship between ECL and the CA199 concentration, each point was the average of ten measurements. (d) The selectivity of the ECL immunosensor. (e) ECL-potential curves of pure GQDs (red curve), pPtPd@GQD (black curve) and cyclic voltammogram (inset) of pPtPd@GQD on the electrode in pH 7.4 PBS containing TPA. Reprinted with permission from ref. 82, Copyright 2013 Elsevier. | ||
Using K2S2O8 as coreactant, Lu et al. constructed an ECL sensor via an electrochemiluminescence resonance energy transfer (ERET) between GQDs and gold nanoparticles (AuNPs) for DNA damage detection.83 The bright blue luminescent GQDs prepared by thermal reduction coupled with UV-light irradiation displayed strong and stable ECL behavior. Au-NPs were linked with a probe of ssDNA (cp53 ssDNA) to form AuNPs-ssDNA. The ECL signal of the GQDs could be quenched by non-covalent binding of the AuNPs-ssDNA to the GQDs, due to the occurrence of an ERET between GQDs and AuNPs. When AuNPs-ssDNA was then hybridized with target p53 DNA to form AuNPs-dsDNA, the non-covalent interaction between GQDs and ds-DNA weakened and the ECL of the GQDs recovered. Therefore, the difference between the quenching efficiencies of AuNPs-ssDNA and AuNPs-dsDNA to GQDs engendered an ECL sensor for detection of target p53 ssDNA with an LOD of 13 nM. In addition, the resultant ECL sensor could also be used for DNA damage detection based on the different bonding abilities of damaged target p53 ssDNA and cp53 ssDNA linked AuNPs.
By combining intense ECL of GQDs and aptamer technique, Lu et al. proposed a novel ECL aptamer sensor for measuring ATP.84 A newly anodic ECL was observed from the water-soluble GQDs with H2O2 as coreactant, and the ECL induced a strong light emission at a low potential. As an amplified element, GQDs doped SiO2 nanospheres (SiO2/GQDs) were used to produce an amplified ECL signal and improved the sensitivity of the sensor. The proposed ECL aptamer sensor exhibited excellent analytical performance for ATP determination, ranging from 5.0 × 10−12 to 5.0 × 10−9 mol L−1 with the LOD of 1.5 × 10−12 mol L−1.
In addition, Dong et al. presented an ECL sensor for the detection of H2O2 using hydrazide-modified single-layer GQDs (HM-SGQDs) (Fig. 13).85 Due to the introduction of hydrazine groups and the formation of abundant luminol-like units, HM-SGQDs exhibited unique PL, CL and anodic ECL properties. The hydrazide groups of HM-SGQDs could be electrochemically oxidized in alkaline solutions, producing a strong anodic ECL signal. The ECL intensity could be enhanced sensitively by H2O2, and the enhanced ECL intensity was proportional to the concentration of H2O2 in a wide range of 3 to 500 mM with an LOD of 0.7 mM.
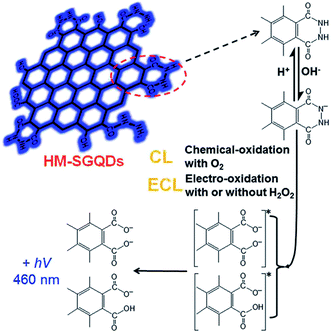 | ||
| Fig. 13 CL and ECL mechanisms of HM-SGQDs. Reprinted with permission from ref. 85, Copyright 2014 Royal Society of Chemistry. | ||
4. Bioimaging
Fluorescent organic dyes and semiconductor QDs have been widely used for bioimaging or visualization of living cells.86–89 However, photobleaching of organic dyes and toxicity of heavy metal contained in semiconductor materials prompt us to find better substitutes. GQDs, which exhibit stable PL, low toxicity and good biocompatibility, provide a much better choice for both in vitro and in vivo bioimaging.4.1 In vitro imaging
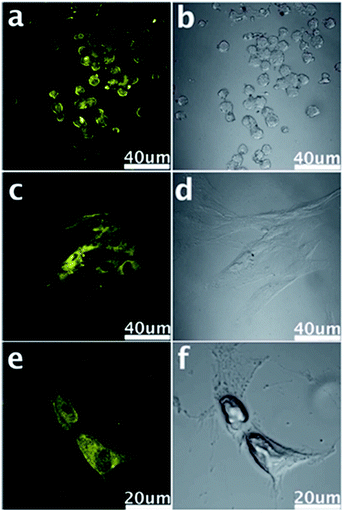 | ||
| Fig. 14 Confocal fluorescence microscopy images of stem cells of NSCs (a), PPCs (c) and CPCs (e) with the fluorescent GQDs incorporated at the excitation wavelength of 405 nm and corresponding images under bright field (b, d and f). Reprinted with permission from ref. 33, Copyright 2012 Royal Society of Chemistry. | ||
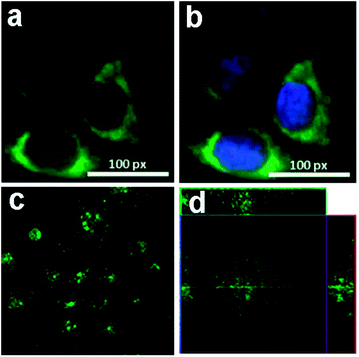
We further investigated the uptake mechanism and biocompatibility of such GQDs with human neural stem cells (hNSCs).95 TEM images (Fig. 15) confirmed the GQDs were indeed internalized by hNSCs via the endocytosis mechanism and were located in the cytoplasm. We found that GQDs did not affect the self-renewal capacity of hNSCs. Single cell dissociated from GQDs-labeled hNSCs finally formed large neurospheres, and the cells expressing the cell type-specific marker for hNSCs, nestin, were identified. Furthermore, no significant change was found in the viability, proliferation, metabolic activity of NSCs after treatment with GQDs, and GQDs did not affect the differentiation potential of hNSCs. After 14 days of differentiation, no difference was detected in the hNSC growth between the control group and the 25 μg mL−1 GQDs treatment group (Fig. 16a and b). Cells from both groups showed elongated cell shapes with neurite outgrowths, resulting in the formation of an interconnected neuronal network. Then glial fibrillary acidic protein (GFAP) and neuron-specific class III beta-tubulin (tubulin βIII) were examined using immunocytochemistry, and hNSCs could spontaneously differentiate into neurons and glial cells (Fig. 16c–f). The above results open a promising avenue for labeling stem cells with GQDs and also offer a potential opportunity to develop GQDs for biomedical applications.
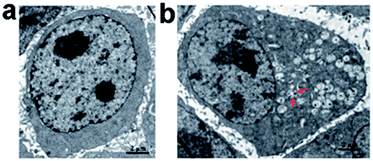 | ||
| Fig. 15 TEM images of hNSCs after 24 h incubation with 25 μg mL−1 of GQDs. (a) Control and (b) GQDs. Red arrows in (b) indicate intracellular vesicles engulfing GQDs. Reprinted with permission from ref. 95, Copyright 2014 Royal Society of Chemistry. | ||
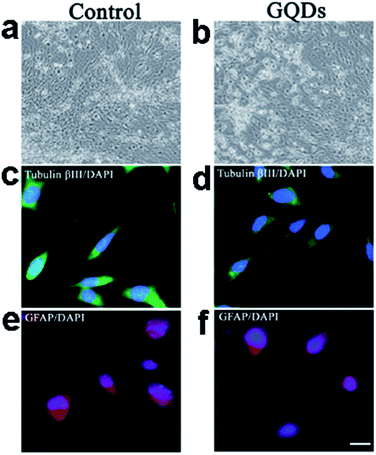 | ||
| Fig. 16 Bright-field images of the hNSCs differentiated for 14 days (a: control; b: 25 μg mL−1 GQDs). Representative microphotographs demonstrated tubulin βIII+ (green, c and d) and GFAP+ (red, e and f) cells. The nuclei were counterstained with DAPI (blue). Scale bar: 20 μm. Reprinted with permission from ref. 95, Copyright 2014 Royal Society of Chemistry. | ||
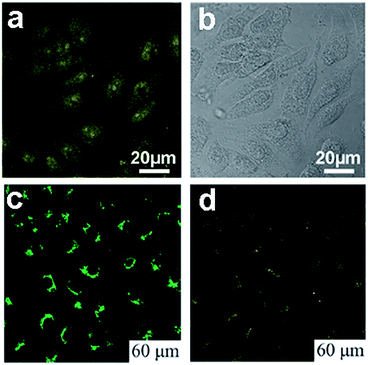 | ||
| Fig. 17 (a) Confocal fluorescence microscopy images of Hela cells with B-GQDs coated with liposomes at the excitation wavelength of 405 nm and corresponding image under bright field (b). (c) and (d) are washed MG-63 cells imaged under bright field, 405 nm, 488 nm excitations, respectively. Reprinted with permission from: (a) and (b) ref. 19, Copyright 2014 Elsevier, (c) and (d) ref. 54, Copyright 2011 Royal Society of Chemistry. | ||
Peng et al. chose the green luminescent GQDs to incubate with human breast cancer cell lines T47D with the nucleus stained with DAPI,24 The obtained images (Fig. 18a and b) clearly visualized the phase contrast image of T47D cells, nucleus stained blue with DAPI, agglomerated high contrast fluorescent image of green GQDs around each nucleus and overlay image of cell with phase contrast, DAPI and green GQDs. Dong et al. incubated human breast cancer MCF-7 cells with GQDs prepared by acidic oxidation,23 and bright green color could be observed when imaged on the confocal laser scanning microscope with excitation at 488 nm (Fig. 18c). The section analysis of one MCF-7 cell indicated that GQDs were able to label not only the cell membrane and the cytoplasm, but also the nucleus (Fig. 18d). That was the first time for luminescent carbon nanomaterials to label the cell nucleus. In addition, Hela cells,96–100 A-549 cells,101,102 macrophages and hepatocellular cells103,104 were also adopted to incubate with fluorescent GQDs by several groups. All these studies suggested the low cytotoxicity and excellent biocompatibility of GQDs by MTT assay, thus they could be used as an eco-friendly material in biolabeling.
 | ||
| Fig. 18 Fluorescent images of human breast cancer cell T47D after incubation with green GQDs for 4 h (a) agglomerated green GQDs surrounding each nucleus. (b) The overlay high contrast image of nucleolus stained with blue DAPI and GQDs (green) staining. (c) Images of human breast cancer MCF-7 cells labeled with GQDs taken by a confocal laser scanning microscope (CLSM) (c) fluorescent image; (d) section analysis. Reprinted with permission from (a) and (b) ref. 24, Copyright 2012 American Chemical Society, (c) and (d) ref. 23, Copyright 2012 Royal Society of Chemistry. | ||
In an early study, Sun et al. realized selective targeted cellular imaging by conjugating a B-cell specific antibody Rituxan (anti-CD20) to the PEGylated nanosized graphene oxide (NGO-PEG). After incubating B-cells and T-cells with NGO-PEG-Rituxan, the intrinsic near-infrared (NIR) PL of NGO-PEG was detected selectively on positive Raji B-cell surfaces and not on negative CEM T-cells (Fig. 19a and b). Then a widely used cancer drug doxorubicin (DOX) was loaded onto NGO-PEG-Rituxan for selective killing of cancer cells in vitro.107
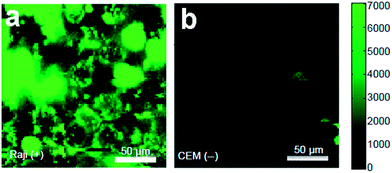 | ||
| Fig. 19 NIR fluorescence images of CD20 positive Raji B-cells (a) and CD20 negative CEM B-cells (b) treated with NGO-PEG-Rituxan conjugate. The scale bar shows the intensity of total NIR emission (in the range 1100–2200 nm). Reprinted with permission from ref. 107, Copyright 2008 Springer. | ||
Nigam et al. reported GQDs conjugated with hyaluronic acid functionalized human serum albumin nanoparticles (HA-HSA-NPs) for pancreatic cancer specific bioimaging.108 Panc-1 cell lines were rich in CD-44, which was a receptor for hyaluronic acid and responsible for chemotherapy-resistant pancreatic cancer cells. After incubated with GQDs tagged HA-HSA-NPs, strong green fluorescence was observed inside the pancreatic cells resulting from the efficient uptake of HA-HSA-NPs while unfunctionalized HSA-NPs exhibited weak fluorescence signals (Fig. 20), indicating that HA-HSA-NPs could efficiently target CD-44 by expressing pancreatic cells via HA-mediated endocytosis pathways.
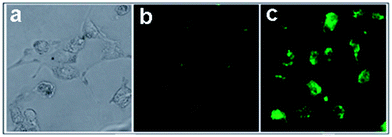 | ||
| Fig. 20 Fluorescence imaging of Panc-1 cells after 24 h incubation in the presence of GQD-conjugated HSA nanoparticles in Dulbecco's modified Eagle's (DMEM) media with 10% fetal bovine serum in CO2 incubator at 37 °C; (a) cells without staining (control); (b) HSA nanoparticles with GQDs only; (c) HSA NPs functionalized with HA and GQDs. Reprinted with permission from ref. 108, Copyright 2014 Royal Society of Chemistry. | ||
In a more recent study, fluorescent GQDs were prepared and conjugated with folic acid (FA) to demonstrate highly selective and specific tumour cell imaging by Wang et al.109 The fluorescence of the GQDs in FA overexpressed HeLa cells was considerably stronger than that in the A549 and HEK293A cells (Fig. 21), which expressed FA receptor (FR) at a low level, indicating that GQD-FA was internalized via FR-induced endocytosis. The FA-conjugated GQDs were then employed as a carrier of the antitumor drug DOX for targeted cell delivery.
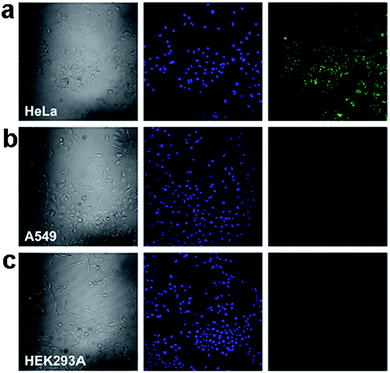 | ||
| Fig. 21 Confocal laser scanning microscopy of (a) HeLa human cervical carcinoma cells, (b) A549 adenocarcinomic human alveolar basal epithelial cells, and (c) HEK293A normal human embryonic kidney cells incubated with GQD-FA (20.0 μg mL−1) at 37 °C for 10 min. Left column: bright field images. Middle column: fluorescence image with Hoechst nuclear stained (blue). Right column: GQD fluorescence image (green). Reprinted with permission from ref. 109, Copyright 2014 Elsevier. | ||
Insulin-conjugated GQDs were synthesized and utilized for specific labeling and dynamic tracking of insulin receptors in 3T3-L1 adipocytes by Zheng et al.110 They investigated the dynamic trafficking of insulin receptors using total internal reflection fluorescence microscopy (TIRFM), which evanescently and selectively illuminated the thin plasmalemmal region. After preincubating adipocytes with insulin-GQDs to allow endocytosis and exocytosis of insulin receptors, time-lapse images were taken under total TIRFM (Fig. 22). It demonstrated that GQDs could be readily conjugated with a wide range of biomolecules without interfering with their activities and then used to specifically label and track molecular targets involved in dynamic cellular process in live cells.
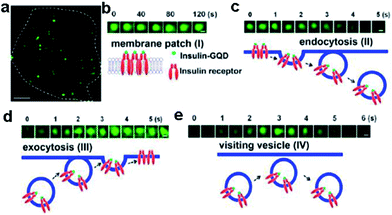 | ||
| Fig. 22 Tracking the dynamics of insulin receptors in living adipocytes using TIRFM. (a) Typical TIRFM image of a 3T3-L1 adipocyte after 1 h incubation of insulin-GQDs. Scale bar = 5 μm. (b) Membrane patch consisting of insulin-GQD/insulin receptor clusters (type I). (c) Endocytosis of fluorescent membrane patches into a vesicle (type II). (d) Exocytosis of a vesicle containing insulin-GQD/insulin receptor complexes (type III). (e) Transient approaching and retrieval of insulin-GQD/insulin receptor containing vesicle (type IV). Scale bars = 0.2 μm. Reprinted with permission from ref. 110, Copyright 2013 American Chemical Society. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Göppert-Mayer units, which far surpassed that of the organic dyes and was comparable to that of the high performance semiconductor QDs.
000 Göppert-Mayer units, which far surpassed that of the organic dyes and was comparable to that of the high performance semiconductor QDs.
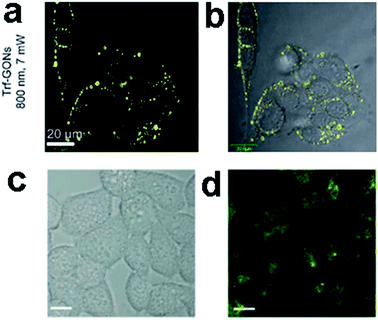 | ||
| Fig. 23 In vitro two-photon luminescence of GONs (a and b). Two-photo cell imaging under (c) bright field and (d) 800 nm excitation. (All scale bars: 10 μm.) Reprinted with permission from: (a) and (b) ref. 111, Copyright 2012 Wiley-VCH, (c) and (d) ref. 18, Copyright 2013 American Chemical Society. | ||
4.2 In vivo imaging
Since Yang et al. firstly explored the feasibility of C-Dots as a fluorescence contrast agent in mice,112 a variety of C-Dots have been synthesized and used for in vivo imaging.113–115 GQDs, similar to C-Dots, can also be applied for in vivo imaging. PEGylated nanographene sheet (NGS-PEG) with an average size of 30 nm prepared by Yang et al. was administered into the mice after labeling with a commonly used NIR fluorescent dye Cy7. In vivo fluorescence imaging revealed high tumor uptake of NGS in several xenograft tumor mouse models (Fig. 24). Then the strong optical absorbance of NGS in the NIR region was utilized for in vivo photothermal therapy.116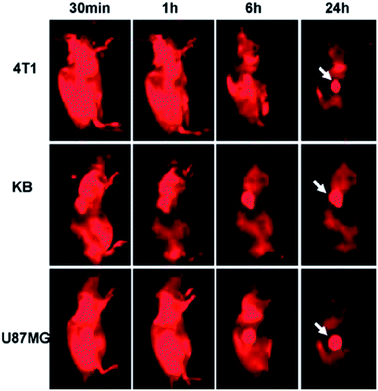 | ||
| Fig. 24 Spectrally unmixed in vivo fluorescence images of 4T1 tumor bearing Balb/c mice, KB, and U87MG tumor bearing nude mice at different time points post injection of NGS-PEG-Cy7. Reprinted with permission from: ref. 116, Copyright 2010 American Chemical Society. | ||
The feasibility of GQDs prepared by a one-step pyrolysis of L-glutamic acid for in vivo bioimaging was investigated in mice by Wu et al.44 After injecting GQDs subcutaneously into the back of nude mice and intramuscularly into the right back leg, the fluorescence images of the mice under different excitation and emission filters were obtained (Fig. 25). The detectable fluorescence region extended with the longer excitation and emission wavelengths from the intramuscular injection spot of the right back leg, demonstrating that longer wavelengths for in vivo fluorescence imaging had better penetration ability than the short ones.
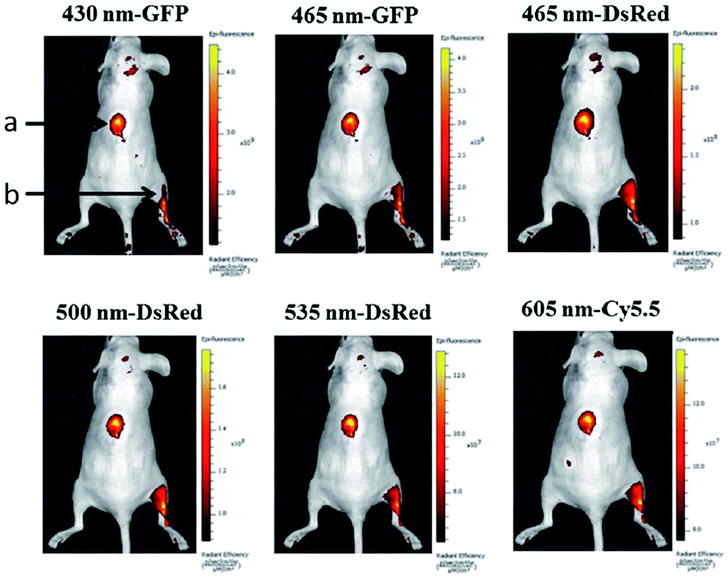 | ||
| Fig. 25 In vivo fluorescence imaging of mice injected GQDs subcutaneously (spot a) and intramuscularly (spot b). The images were taken at various excitation wavelengths and emission wavelengths indicated at the top of each image. A 100 mL aliquot of 25 mg mL−1 GQDs was injected into each spot. The following emission bandpass filters were used: GFP: 515–575 nm; DsRed: 575–650 nm; Cy5.5: 695–770 nm. Reprinted with permission from ref. 44, Copyright 2013 Royal Society of Chemistry. | ||
Ge et al. investigated the in vivo fluorescence imaging capability of red fluorescent GQDs prepared via a hydrothermal method with polythiophene derivatives.41 After the injection of GQDs aqueous solution into the back of a nude mouse, the injection sites showed much higher fluorescence intensity than the background signal produced by the mouse skin as depicted in Fig. 26a and b. Furthermore, no apparent fluorescence intensity decay was observed at the injection sites, and the injected GQDs did not show evidence of obvious diffusion even 1 week after injection. More importantly, GQDs could be applied as a photodynamic therapy (PDT) agent for the simultaneous imaging and highly efficient treatment of cancer. Using female BALB/nu mice with subcutaneous breast cancer xenografts as an animal model, three groups, the PDT group, the control groups included mice that received a GQDs injection at the same dose but were not irradiated (the C1 group) and mice that did not received a GQDs injection but were irradiated (the C2 group) were chosen to evaluate the performance of the GQDs for in vivo PDT. In the PDT group, the tumours first turned dark and festered, which increased the tumour size slightly (Fig. 26c). The tumours began to decompose after 9 days and were destroyed after 17 days, leaving black scars at the original sites, which fell off ∼1 week later. No tumour regrowth was observed in the PDT group over the course of 50 days. In contrast, the tumours in the C1 and C2 groups grew significantly during the study period (Fig. 26d), indicating that neither light irradiation nor GQDs injection alone inhibited the tumor growth.
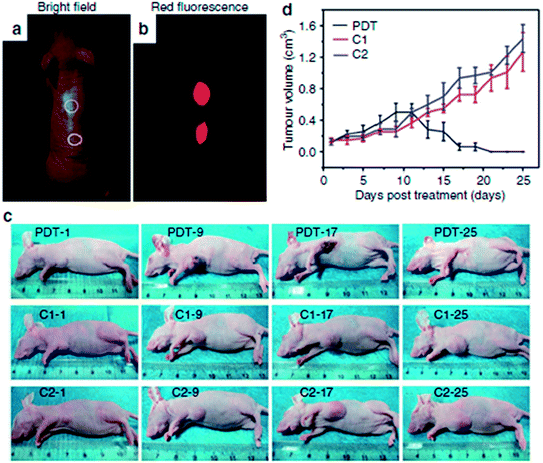 | ||
| Fig. 26 In vivo imaging and PDT. (a) Bright-field image and (b) red-fluorescence image after subcutaneous injection of GQDs in different areas. The excitation wavelength was 502–540 nm, and the collected fluorescence channel was 695–775 nm. (c) Photographs of mice after various treatmentsion the 1st, 9th, 17th and 25th day. (PDT: GQDs + light irradiation; C1: GQDs only; C2: light irradiation only) (d) Time-dependent tumour growth curves (n = 5) after different treatments. P < 0.05 for each group. Reprinted with permission from ref. 41, Copyright 2014 Nature Publishing Group. | ||
5. Conclusions and future perspectives
In this review, GQDs have been comprehensively introduced from the aspects of synthetic approaches, structure, PL properties and applications for biosensing and bioimaging. We have discussed GQDs-based PL, electrochemical and ECL biosensors with high sensitivity and selectivity for the detection of ions, biomolecules and small organic molecules with aromatic structures. In addition, GQDs are suitable materials for both in vitro and in vivo bioimaging. The use of GQDs for biosensing and bioimaging has achieved great success within a short time.117,118 However, GQDs have yet to be exploited because of a number of current problems such as short wavelength emission, low yield and QY.119 Meanwhile, strong two/three-photon fluorescent GQDs for live deep-tissue imaging and special biomolecule modified GQDs for targeted cancer imaging, diagnosis and therapy are particularly desired. Herein, we speculate the future perspectives on GQDs-based biosensors and biomarkers as follows.5.1 QY improvement
The highest QY of GQDs reported so far is 54.5% with blue fluorescence emission,44 but the QY of GQDs with other color fluorescence emission is less than 20%, much lower than traditional semiconductor QDs and organic dyes. Therefore the QY improvement in longer wavelength emission of GQDs is challenging. With the achievement of uniform size, shape and edge, the multicolor PL and high QY of GQDs will be more promising for biosensing and bioimaging.5.2 Red/near-IR and the second near-IR (NIR-II) PL emission GQDs for bioimaging
Due to the auto-fluorescence and light scattering background of biological specimens in the short-wavelength region, fluorescent materials emitting in the long-wavelength region are expected to provide a deeper light penetration into the specimens with a higher imaging contrast. Biological imaging with fluorescent agents in the long NIR region (1000–1700 nm, terrned the second near-IR region), which has received growing interests in recent years, can afford even deeper penetration than the traditional NIR (750–900 nm) imaging techniques.120–124 GQDs with bright fluorescence emissions emphasized in the red/near-IR as well as the NIR-II spectral regions are desired for effective tissue penetration and in vivo imaging.5.3 Biosensing and bioimaging based on the upconversion PL of GQDs
Upconversion PL of GQDs provides the advantage in exciting near-IR light using multi-photon excitation. It has received a great deal of attention for its promising applications in both basic biological researches and clinical diagnostics with the avoidance of harmful UV or blue excitations and the reduction of photo damage in biotissues. Therefore, upconversion PL will expand the applications of GQDs for biosensing and bioimaging.5.4 GQDs functionalization for specific labeling
Researches on potential applications of GQDs as a cell imaging agent mainly concentrate on the uptake of bare GQDs by cells without considering the selectivity and function of the agent. Functional groups on the GQDs can be modified or conjugated with a wide range of biomolecules, thus contributing to specifically label and track molecular targets. It is of great significance for the exploitation of surface functionalization in GQDs for controlled coupling with bioactive species to enable specific targeting in cellular, in vivo imaging, noninvasively detecting disease of living biosystems and related biomedical applications.Acknowledgements
This work is supported by NSFC (21073018), the Major Research Plan of NSFC (21233003), the Fundamental Research Funds for the Central Universities, Key Laboratory of Theoretical and Computational Photochemistry.Notes and references
- C. Chung, Y. K. Kim, D. Shin, S. R. Ryoo, B. H. Hong and D. H. Min, Acc. Chem. Res., 2013, 46, 2211–2224 CrossRef CAS PubMed.
- V. Georgakilas, M. Otyepka, A. B. Bourlinos, V. Chandra, N. Kim, K. C. Kemp, P. Hobza, R. Zboril and K. S. Kim, Chem. Rev., 2012, 112, 6156–6214 CrossRef CAS PubMed.
- K. S. Novoselov, A. K. Geim, S. V. Morozov, D. Jiang, Y. Zhang, S. V. Dubonos, I. V. Grigorieva and A. A. Firsov, Science, 2004, 306, 666–669 CrossRef CAS PubMed.
- K. S. Novoselov, Z. Jiang, Y. Zhang, S. V. Morozov, H. L. Stormer, U. Zeitler, J. C. Maan, G. S. Boebinger, P. Kim and A. K. Geim, Science, 2007, 315, 1379–1379 CrossRef CAS PubMed.
- S. Fujii and T. Enoki, J. Am. Chem. Soc., 2010, 132, 10034–10041 CrossRef CAS PubMed.
- K. A. Ritter and J. W. Lyding, Nat. Mater., 2009, 8, 235–242 CrossRef CAS PubMed.
- X. Li, X. Wang, L. Zhang, S. Lee and H. Dai, Science, 2008, 319, 1229–1232 CrossRef CAS PubMed.
- L. A. Ponomarenko, F. Schedin, M. I. Katsnelson, R. Yang, E. W. Hill, K. S. Novoselov and A. K. Geim, Science, 2008, 320, 356–358 CrossRef CAS PubMed.
- J. H. Shen, Y. H. Zhu, X. L. Yang and C. Z. Li, Chem. Commun., 2012, 48, 3686–3699 RSC.
- S. J. Zhu, S. J. Tang, J. H. Zhang and B. Yang, Chem. Commun., 2012, 48, 4527–4539 RSC.
- Y.-P. Sun, B. Zhou, Y. Lin, W. Wang, K. A. S. Fermando, P. Pathak, M. J. Meziani, B. A. Harruff, X. Wang, H. Wang, P. G. Luo, H. Yang, M. E. Kose, B. Chen, L. M. Veca and S.-Y. Xie, J. Am. Chem. Soc., 2006, 128, 7756–7757 CrossRef CAS PubMed.
- Y. Li, Y. Hu, Y. Zhao, G. Shi, L. Deng, Y. Hou and L. Qu, Adv. Mater., 2011, 23, 776–780 CrossRef CAS PubMed.
- S. Zhu, Q. Meng, L. Wang, J. Zhang, Y. Song, H. Jin, K. Zhang, H. Sun, H. Wang and B. Yang, Angew. Chem., Int. Ed., 2013, 52, 3953–3957 CrossRef CAS PubMed.
- S. N. Baker and G. A. Baker, Angew. Chem., Int. Ed., 2010, 49, 6726–6744 CrossRef CAS PubMed.
- X. Zhou, Y. Zhang, C. Wang, X. Wu, Y. Yang, B. Zheng, H. Wu, S. Guo and J. Zhang, ACS Nano, 2012, 6, 6592–6599 CrossRef CAS PubMed.
- R. Liu, D. Wu, X. Feng and K. Muellen, J. Am. Chem. Soc., 2011, 133, 15221–15223 CrossRef CAS PubMed.
- D. Y. Pan, J. C. Zhang, Z. Li and M. H. Wu, Adv. Mater., 2010, 22, 734–738 CrossRef CAS PubMed.
- Q. Liu, B. D. Guo, Z. Y. Rao, B. H. Zhang and J. R. Gong, Nano Lett., 2013, 13, 2436–2441 CrossRef CAS PubMed.
- Z. T. Fan, Y. C. Li, X. H. Li, L. Z. Fan, S. X. Zhou, D. C. Fang and S. H. Yang, Carbon, 2014, 70, 149–156 CrossRef CAS PubMed.
- S. H. Li, Y. C. Li, J. Cao, J. Zhu, L. Z. Fan and X. H. Li, Anal. Chem., 2014, 86, 10201–10207 CrossRef CAS PubMed.
- H. Sun, L. Wu, W. Wei and X. Qu, Mater. Today, 2013, 16, 433–442 CrossRef CAS PubMed.
- Y. B. Song, S. J. Zhu and B. Yang, RSC Adv., 2014, 4, 27184–27200 RSC.
- Y. Dong, C. Chen, X. Zheng, L. Gao, Z. Cui, H. Yang, C. Guo, Y. Chi and C. M. Li, J. Mater. Chem., 2012, 22, 8764–8766 RSC.
- J. Peng, W. Gao, B. K. Gupta, Z. Liu, R. Romero-Aburto, L. Ge, L. Song, L. B. Alemany, X. Zhan, G. Gao, S. A. Vithayathil, B. A. Kaipparettu, A. A. Marti, T. Hayashi, J. J. Zhu and P. M. Ajayan, Nano Lett., 2012, 12, 844–849 CrossRef CAS PubMed.
- R. Ye, C. Xiang, J. Lin, Z. Peng, K. Huang, Z. Yan, N. P. Cook, E. L. G. Samuel, C. C. Hwang, G. Ruan, G. Ceriotti, A.-R. O. Raji, A. A. Marti and J. M. Tour, Nat. Commun., 2013, 4, 2943–2948 Search PubMed.
- D. Pan, L. Guo, J. Zhang, C. Xi, Q. Xue, H. Huang, J. Li, Z. Zhang, W. Yu, Z. Chen, Z. Li and M. Wu, J. Mater. Chem., 2012, 22, 3314–3318 RSC.
- Q. Liu, B. D. Guo, Z. Y. Rao, B. H. Zhang and J. R. Gong, Nano Lett., 2013, 13, 2436–2441 CrossRef CAS PubMed.
- H. Tetsuka, R. Asahi, A. Nagoya, K. Okamoto, I. Tajima, R. Ohta and A. Okamoto, Adv. Mater., 2012, 24, 5333–5338 CrossRef CAS PubMed.
- S. Chen, J. W. Liu, M. L. Chen, X. W. Chen and J. H. Wang, Chem. Commun., 2012, 48, 7637–7639 RSC.
- L. L. Li, J. Ji, R. Fei, C. Z. Wang, Q. Lu, J. R. Zhang, L. P. Jiang and J. J. Zhu, Adv. Funct. Mater., 2012, 22, 2971–2979 CrossRef CAS.
- S. Zhuo, M. Shao and S. T. Lee, ACS Nano, 2012, 6, 1059–1064 CrossRef CAS PubMed.
- Y. Li, Y. Zhao, H. H. Cheng, Y. Hu, G. Q. Shi, L. M. Dai and L. T. Qu, J. Am. Chem. Soc., 2012, 134, 15–18 CrossRef CAS PubMed.
- M. Zhang, L. Bai, W. Shang, W. Xie, H. Ma, Y. Fu, D. Fang, H. Sun, L. Fan, M. Han, C. Liu and S. Yang, J. Mater. Chem., 2012, 22, 7461–7467 RSC.
- D. B. Shinde and V. K. Pillai, Chem.–Eur. J., 2012, 18, 12522–12528 CrossRef CAS PubMed.
- A. Ananthanarayanan, X. Wang, P. Routh, B. Sana, S. Lim, D. H. Kim, K. H. Lim, J. Li and P. Chen, Adv. Funct. Mater., 2014, 24, 3021–3026 CrossRef CAS.
- F. Liu, M. H. Jang, H. D. Ha, J. H. Kim, Y.-H. Cho and T. S. Seo, Adv. Mater., 2013, 25, 3657–3662 CrossRef CAS PubMed.
- L. Lin and S. Zhang, Chem. Commun., 2012, 48, 10177–10179 RSC.
- Q. Li, S. Zhang, L. Dai and L. Li, J. Am. Chem. Soc., 2012, 134, 18932–18935 CrossRef CAS PubMed.
- X. Yan, X. Cui and L. Li, J. Am. Chem. Soc., 2010, 132, 5944–5945 CrossRef CAS PubMed.
- L. Wang, Y. Wang, T. Xu, H. Liao, C. Yao, Y. Liu, Z. Li, Z. Chen, D. Pan, L. Sun and M. Wu, Nat. Commun., 2014, 5, 5357–5365 CrossRef CAS PubMed.
- J. Ge, M. Lan, B. Zhou, W. Liu, L. Guo, H. Wang, Q. Jia, G. Niu, X. Huang, H. Zhou, X. Meng, P. Wang, C. S. Lee, W. Zhang and X. Han, Nat. Commun., 2014, 5, 4596–4603 CAS.
- Y. Q. Dong, J. W. Shao, C. Q. Chen, H. Li, R. X. Wang, Y. W. Chi, X. M. Lin and G. N. Chen, Carbon, 2012, 50, 4738–4743 CrossRef CAS PubMed.
- L. Tang, R. Ji, X. Cao, J. Lin, H. Jiang, X. Li, K. S. Teng, C. M. Luk, S. Zeng, J. Hao and S. P. Lau, ACS Nano, 2012, 6, 5102–5110 CrossRef CAS PubMed.
- X. Wu, F. Tian, W. Wang, J. Chen, M. Wu and J. X. Zhao, J. Mater. Chem. C, 2013, 1, 4676–4684 RSC.
- Z. Huang, Y. Shen, Y. Li, W. Zheng, Y. Xue, C. Qin, B. Zhang, J. Hao and W. Feng, Nanoscale, 2014, 6, 13043–13052 RSC.
- J. Lu, P. S. E. Yeo, C. K. Gan, P. Wu and K. P. Loh, Nat. Nanotechnol., 2011, 6, 247–252 CrossRef CAS PubMed.
- S. J. Zhu, J. H. Zhang, S. J. Tang, C. Y. Qiao, L. Wang, H. Y. Wang, X. Liu, B. Li, Y. F. Li, W. L. Yu, X. F. Wang, H. C. Sun and B. Yang, Adv. Funct. Mater., 2012, 22, 4732–4740 CrossRef CAS.
- L. S. Fan, Y. W. Hu, X. Wang, L. Zhang, F. Li, D. Han, Z. Li, Q. Zhang, Z. Wang and L. Niu, Talanta, 2012, 101, 192–197 CrossRef CAS PubMed.
- J. Shen, Y. Zhu, C. Chen, X. Yang and C. Li, Chem. Commun., 2011, 47, 2580–2582 RSC.
- S. J. Chen, Y. N. Hong, Y. Liu, J. Z. Liu, C. W. T. Leung, M. Li, R. T. K. Kwok, E. G. Zhao, J. W. Y. Lam, Y. Yu and B. Z. Tang, J. Am. Chem. Soc., 2013, 135, 4926–4929 CrossRef CAS PubMed.
- F. Yang, M. Zhao, B. Zheng, D. Xiao, L. Wu and Y. Guo, J. Mater. Chem., 2012, 22, 25471–25479 RSC.
- D. Y. Pan, J. C. Zhang, Z. Li, C. Wu, X. M. Yan and M. H. Wu, Chem. Commun., 2010, 46, 3681–3683 RSC.
- S. J. Zhu, J. H. Zhang, X. Liu, B. Li, X. F. Wang, S. J. Tang, Q. N. Meng, Y. F. Li, C. Shi, R. Hu and B. Yang, RSC Adv., 2012, 2, 2717–2720 RSC.
- S. Zhu, J. Zhang, C. Qiao, S. Tang, Y. Li, W. Yuan, B. Li, L. Tian, F. Liu, R. Hu, H. Gao, H. Wei, H. Zhang, H. Sun and B. Yang, Chem. Commun., 2011, 47, 6858–6860 RSC.
- L. Bao, Z. L. Zhang, Z. Q. Tian, L. Zhang, C. Liu, Y. Lin, B. P. Qi and D. W. Pang, Adv. Mater., 2011, 23, 5801–5806 CrossRef CAS PubMed.
- J. Zong, Y. Zhu, X. Yang, J. Shen and C. Li, Chem. Commun., 2011, 47, 764–766 RSC.
- Z. Gan, X. Wu, G. Zhou, J. Shen and P. K. Chu, Adv. Opt. Mater., 2013, 1, 554–558 CrossRef.
- K. P. Loh, Q. L. Bao, G. Eda and M. Chhowalla, Nat. Chem., 2010, 2, 1015–1024 CrossRef CAS PubMed.
- J. Shen, Y. Zhu, X. Yang, J. Zong, J. Zhang and C. Li, New J. Chem., 2012, 36, 97–101 RSC.
- X. Ran, H. J. Sun, F. Pu, J. S. Ren and X. G. Qu, Chem. Commun., 2013, 49, 1079–1081 RSC.
- Y. Z. He, X. X. Wang, J. Sun, S. F. Jiao, H. Q. Chen, F. Gao and L. Wang, Anal. Chim. Acta, 2014, 810, 71–78 CrossRef CAS PubMed.
- Z. B. Qu, X. G. Zhou, L. Gu, R. M. Lan, D. D. Sun, D. J. Yu and G. Y. Shi, Chem. Commun., 2013, 49, 9830–9832 RSC.
- Y. Wang, L. Zhang, R. P. Liang, J. M. Bai and J. D. Qiu, Anal. Chem., 2013, 85, 9148–9155 CrossRef CAS PubMed.
- L. Zhang, Z. Y. Zhang, R. P. Liang, Y. H. Li and J. D. Qiu, Anal. Chem., 2014, 86, 4423–4430 CrossRef CAS PubMed.
- Y. H. Li, L. Zhang, J. Huang, R. P. Liang and J. D. Qiu, Chem. Commun., 2013, 49, 5180–5182 RSC.
- X. Li, S. J. Zhu, B. Xu, K. Ma, J. H. Zhang, B. Yang and W. J. Tian, Nanoscale, 2013, 5, 7776–7779 RSC.
- J. J. Liu, X. L. Zhang, Z. X. Cong, Z. T. Chen, H. H. Yang and G. N. Chen, Nanoscale, 2013, 5, 1810–1815 RSC.
- Z. Z. Wu, W. Y. Li, J. Chen and C. Yu, Talanta, 2014, 119, 538–543 CrossRef CAS PubMed.
- J. Zong, X. Yang, A. Trinchi, S. Hardin, I. Cole, Y. Zhu, C. Li, T. Muster and G. Wei, Biosens. Bioelectron., 2014, 51, 330–335 CrossRef CAS PubMed.
- H. M. Zhao, Y. Y. Chang, M. Liu, S. Gao, H. T. Yu and X. Quan, Chem. Commun., 2013, 49, 234–236 RSC.
- Z. S. Qian, X. Y. Shan, L. J. Chai, J. J. Ma, J. R. Chen and H. Feng, Nanoscale, 2014, 6, 5671–5674 RSC.
- Z. S. Qian, X. Y. Shan, L. J. Chai, J. R. Chen and H. Feng, Nanotechnology, 2014, 25, 415–501 Search PubMed.
- Z. S. Qian, X. Y. Shan, L. J. Chai, J. J. Ma, J. R. Chen and H. Feng, Biosens. Bioelectron., 2014, 60, 64–70 CrossRef CAS PubMed.
- J. Zhao, G. F. Chen, L. Zhu and G. X. Li, Electrochem. Commun., 2011, 13, 31–33 CrossRef CAS PubMed.
- A. Muthurasu and V. Ganesh, Appl. Biochem. Biotechnol., 2014, 174, 945–959 CrossRef CAS PubMed.
- D. L. Jiang, Y. Zhang, H. Y. Chu, J. Liu, J. Wan and M. Chen, RSC Adv., 2014, 4, 16163–16171 RSC.
- Y. Zhang, C. Y. Wu, X. J. Zhou, X. C. Wu, Y. Q. Yang, H. X. Wu, S. W. Guo and J. Y. Zhang, Nanoscale, 2013, 5, 1816–1819 RSC.
- H. Razmi and R. Mohammad-Rezaei, Biosens. Bioelectron., 2013, 41, 498–504 CrossRef CAS PubMed.
- X. D. Wang, L. J. Chen, X. R. Su and S. Y. Ai, Biosens. Bioelectron., 2013, 47, 171–177 CrossRef CAS PubMed.
- L. Wu, J. S. Wang, J. S. Ren, W. Li and X. G. Qu, Chem. Commun., 2013, 49, 5675–5677 RSC.
- C. Y. Zhang, L. Wang, A. M. Wang, S. Y. Zhang, C. J. Mao, J. M. Song, H. L. Niu, B. K. Jin and Y. P. Tian, Appl. Surf. Sci., 2014, 315, 22–27 CrossRef CAS PubMed.
- H. M. Yang, W. Y. Liu, C. Ma, Y. Zhang, X. Wang, J. H. Yua and X. R. Song, Electrochim. Acta, 2014, 123, 470–476 CrossRef CAS PubMed.
- Q. Lu, W. Wei, Z. X. Zhou, Z. X. Zhou, Y. J. Zhang and S. Q. Liu, Analyst, 2014, 139, 2404–2410 RSC.
- J. J. Lu, M. Yan, L. Ge, S. G. Ge, S. W. Wang, J. X. Yan and J. H. Yu, Biosens. Bioelectron., 2013, 47, 271–277 CrossRef CAS PubMed.
- Y. Q. Dong, R. P. Dai, T. Q. Dong, Y. W. Chi and G. N. Chen, Nanoscale, 2014, 6, 11240–11245 RSC.
- N. Chen, Y. He, Y. Su, X. Li, Q. Huang, H. Wang, X. Zhang, R. Tai and C. Fan, Biomaterials, 2012, 33, 1238–1244 CrossRef CAS PubMed.
- M. C. Mancini, B. A. Kairdolf, A. M. Smith and S. Nie, J. Am. Chem. Soc., 2010, 130, 10836–10837 CrossRef PubMed.
- X. H. Gao, L. L. Yang, J. A. Petros, F. F. Marshal, J. W. Simons and S. M. Nie, Curr. Opin. Biotechnol., 2005, 16, 63–72 CrossRef CAS PubMed.
- R. Hardman, Environ. Health Perspect., 2006, 114, 165–172 CrossRef.
- P. J. Fairchild, K. F. Nopan, S. Cartland and H. Waldmann, Int. Immunopharmacol., 2005, 5, 13–21 CrossRef CAS PubMed.
- D. J. Prockop, Science, 2001, 293, 211–212 CrossRef CAS.
- G. C. Parker, M. Nastassova-Kristeva, L. M. Eisenberg, M. S. Rao, M. A. Williams, P. R. Sanberg and D. English, Stem Cells Dev., 2005, 14, 463–469 CrossRef CAS PubMed.
- A. Solanki, J. D. Kim and K. B. Lee, Nanomedicine, 2008, 3, 567–578 CrossRef CAS PubMed.
- Y. E, L. Bai, L. Fan, M. Han, X. Zhang and S. Yang, J. Mater. Chem., 2011, 21, 819–823 RSC.
- W. Shang, X. Zhang, M. Zhang, Z. Fan, Y. Sun, M. Han and L. Fan, Nanoscale, 2014, 6, 5799–5806 RSC.
- Z. Qian, J. Ma, X. Shan, L. Shao, J. Zhou, J. Chen and H. Feng, RSC Adv., 2013, 3, 14571–14579 RSC.
- X. Zhu, X. Xiao, X. Zuo, Y. Liang and J. Nan, Part. Part. Syst. Charact., 2014, 31, 801–809 CrossRef CAS.
- C. Hu, Y. Liu, Y. Yang, J. Cui, Z. Huang, Y. Wang, L. Yang, H. Wang, Y. Xiao and J. Rong, J. Mater. Chem. B, 2013, 1, 39–42 RSC.
- S. Chen, X. Hai, C. Xia, X.-W. Chen and J.-H. Wang, Chem.–Eur. J., 2013, 19, 15918–15923 CrossRef CAS PubMed.
- N. Li, X. Liang, L. Wang, Z. Li, P. Li, Y. Zhu and J. Song, J. Nanopart. Res., 2012, 14, 1177–1185 CrossRef.
- W. J. Xie, Y. Y. Fu, H. Ma, M. Zhang and L. Z. Fan, Acta Chim. Sin., 2012, 70, 2169–2172 CrossRef CAS.
- H. Sun, L. Wu, N. Gao, J. Ren and X. Qu, ACS Appl. Mater. Interfaces, 2013, 5, 1174–1179 CAS.
- V. Kumar, V. Singh, S. Umrao, V. Parashar, S. Abraham, A. K. Singh, G. Nath, P. S. Saxena and A. Srivastava, RSC Adv., 2014, 4, 21101–21107 RSC.
- L. Zhang, Y. Xing, N. He, Y. Zhang, Z. Lu, J. Zhang and Z. Zhang, J. Nanosci. Nanotechnol., 2012, 12, 2924–2928 CrossRef CAS PubMed.
- J. Nicolas, S. Mura, D. Brambilla, N. Mackiewicz and P. Couvreur, Chem. Soc. Rev., 2013, 42, 1147–1235 RSC.
- M. Shi, J. Lu and M. S. Shoichet, J. Mater. Chem., 2009, 19, 5485–5498 RSC.
- X. Sun, Z. Liu, K. Welsher, J. Robinson, A. Goodwin, S. Zaric and H. Dai, Nano Res., 2008, 1, 203–212 CrossRef CAS PubMed.
- P. Nigam, S. Waghmode, M. Louis, S. Wangnoo, P. Chavan and D. Sarkar, J. Mater. Chem. B, 2014, 2, 3190–3195 RSC.
- X. Wang, X. Sun, J. Lao, H. He, T. Cheng, M. Wang, S. Wang and F. Huang, Colloids Surf., B, 2014, 122, 638–644 CrossRef CAS PubMed.
- X. T. Zheng, A. Than, A. Ananthanaraya, D.-H. Kim and P. Chen, ACS Nano, 2013, 7, 6278–6286 CrossRef CAS PubMed.
- J.-L. Li, H.-C. Bao, X.-L. Hou, L. Sun, X.-G. Wang and M. Gu, Angew. Chem., Int. Ed., 2012, 51, 1830–1834 CrossRef CAS PubMed.
- S. T. Yang, L. Cao, P. G. Luo, F. Lu, X. Wang, H. Wang, M. J. Meziani, Y. Liu, G. Qi and Y. P. Sun, J. Am. Chem. Soc., 2009, 131, 11308–11309 CrossRef CAS PubMed.
- H. Tao, K. Yang, Z. Ma, J. Wan, Y. Zhang, Z. Kang and Z. Liu, Small, 2012, 8, 281–290 CrossRef CAS PubMed.
- P. G. Luo, S. Sahu, S.-T. Yang, S. K. Sonkar, J. Wang, H. Wang, G. E. LeCroy, L. Cao and Y.-P. Sun, J. Mater. Chem. B, 2013, 1, 2116–2127 RSC.
- L. Cao, S. T. Yang, X. Wang, P. G. Luo, H.-H. Liu, S. Sahu, Y. Liu and Y. P. Sun, Theranostics, 2012, 2, 295–301 CrossRef CAS PubMed.
- K. Yang, S. Zhang, G. Zhang, X. Sun, S.-T. Lee and Z. Liu, Nano Lett., 2010, 10, 3318–3323 CrossRef CAS PubMed.
- Y. Jing, Y. Zhu, X. Yang, J. Shen and C. Li, Langmuir, 2010, 27, 1175–1180 CrossRef PubMed.
- A. Zhu, C. Ding and Y. Tian, Sci. Rep., 2013, 3, 2933–2939 Search PubMed.
- J. Zong, X. Yang, A. Trinchi, S. Hardin, I. Cole, Y. Zhu, C. Li, T. Muster and G. Wei, Nanoscale, 2013, 5, 11200–11206 RSC.
- G. Hong, J. C. Lee, J. T. Robinson, U. Raaz, L. Xie, N. F. Huang, J. P. Cooke and H. Dai, Nat. Med., 2012, 18, 1841–1846 CrossRef CAS PubMed.
- N. Vogt, Nat. Methods, 2014, 11, 988–989 CrossRef CAS.
- A. M. Smith, M. C. Mancini and S. Nie, Nat. Nanotechnol., 2009, 4, 710–711 CrossRef CAS PubMed.
- Nature Publishing Group, Nature, 2014, 512, 117–117 Search PubMed.
- G. Hong, S. Diao, J. Chang, A. L. Antaris, C. Chen, B. Zhang, S. Zhao, D. N. Atochin, P. L. Huang, K. I. Andreasson, C. J. Kuo and H. Dai, Nat. Photonics, 2014, 8, 723–730 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2015 |




