Specific removal of protein using protein imprinted polydopamine shells on modified amino-functionalized magnetic nanoparticles†
Ruixia Gao*a,
Lili Zhangb,
Yi Haob,
Xihui Cuib and
Yuhai Tang*ab
aInstitute of Analytical Science, School of Science, Xi’an Jiaotong University, Xi’an 710049, P. R. China. E-mail: ruixiagao@mail.xjtu.edu.cn; tyh57@mail.xjtu.edu.cn; Fax: +86 029 8265 5399; Tel: +86 029 8265 5399
bCollege of Pharmacy, Xi’an Jiaotong University, Xi’an 710061, P. R. China
First published on 7th November 2014
Abstract
Thin imprinted shells over functionalized magnetic nanoparticles is an effective solution to weaken mass transfer resistance, achieve high binding capacity, and attain rapid separation. In this work, a simple, green, and effective approach was developed to imprint bovine serum albumin (BSA) on the surface of amino-modified Fe3O4 nanoparticles (Fe3O4@NH2) using dopamine as monomer through a two-step immobilized template strategy. The results of X-ray diffraction and vibrating sample magnetometry indicated that the as-synthesized nanomaterials exhibited high crystallinity and satisfactory superparamagnetic properties. Transmission electron microscopy and Fourier transform infrared spectroscopy of the products showed that polydopamine shells successfully attached onto Fe3O4@NH2. The polydopamine shells with a thickness of about 10 nm enable the template recognition sites to be accessed easily, and exhibit fast kinetics and high adsorption capacity in aqueous solution. Meanwhile, an excellent selectivity towards BSA has been presented when bovine hemoglobin (BHb), transferrin, and immunoglobulin G (IgG) were employed as competitive proteins. Good recovery after a reasonably mild elution and successful capture of the target protein from a real sample of bovine blood suggests its potential value in practical applications. In addition, the resultant polymers were stable and had no obvious deterioration after six adsorption–regeneration cycles. The versatility of the proposed method has also been verified by choosing four other proteins with different isoelectric points as the templates.
1. Introduction
Affinity separation and removal of high-abundant proteins from blood plasma is often used as a sample pretreatment technique before clinical and pharmaceutical proteomic studies. Several feasible methods, such as immobilized metal affinity chromatography (IMAC),1,2 dielectrophoresis,3,4 and immunoaffinity chromatography5,6 have been applied to remove high-abundant proteins from biological samples. However, there are some drawbacks, including cost, time and labor consumption, poor stability, limitations of the targets, and non-reusability that prohibit the widespread applicability of these approaches. Therefore, to develop a more effective and general application method for the specific isolation and depletion of high-abundant proteins is of vital significance.Molecularly imprinted polymers (MIPs), tailor-made receptors with specific three-dimensional recognition sites complementary in shape, size, and functionality with the templates,7,8 are considered one of the most promising affinity matrices. MIPs have also been regarded as a satisfactory adsorbent for the separation and removal of target proteins,9,10 as a result of good physical, chemical, and thermal stability, simple and economical production, durability, and reusability, as well as high specificity. With respect to protein imprinting, a big problem is the large molecular size which restricts the mass transfer of proteins between the cross-linked polymer network and the solution. Another problem is aqueous media where proteins prefer to exist, but the noncovalent interactions of templates and monomers can be reduced remarkably. Other problems include flexible conformation, complexity, and numbers of functional groups, all of which are unfavorable elements for imprinting proteins.
Some imprinting approaches have been developed to solve the mass transfer difficulty of proteins, for example, surface imprinting,11,12 epitope imprinting,13,14 and metal-coordination polymerization.15,16 Among these, surface imprinting with the recognition cavities located at the surface of the MIPs, is regarded as the most promising method for imprinting proteins, as it is expected to weaken mass transfer resistance and make the removal of the template protein easy and complete.17,18 Nanomaterials are ideal supports for surface imprinting, especially magnetic Fe3O4 nanoparticles (NPs), due to their superparamagnetism and simple preparation.19,20 These excellent properties make imprinted Fe3O4 NPs useful for improving the accessibility of the recognition cavities generated and the separation efficiency of polymers from solution. Several protein molecularly imprinted works based on Fe3O4 NPs have been reported, in which silane or other active groups (such as –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, –COOH, –CHO, and –NH2) were introduced, often involving harsh conditions and/or multiple functionalization steps.21–23 Furthermore, the synthesis process was usually performed in organic solvents, which is a minus point for protein imprinting. Therefore, more effective and simpler strategies for protein imprinting on the surface of Fe3O4 NPs are still required. Our previous work24 developed a much easier synthetic process for BHb imprinting employing amino group directly-functionalized Fe3O4 NPs as carriers. Though this work displayed some advantages in aspects of preparation technology, the hydrophilicity of the obtained products was unsatisfactory in virtue of adopting hydrophobic monomers.
C, –COOH, –CHO, and –NH2) were introduced, often involving harsh conditions and/or multiple functionalization steps.21–23 Furthermore, the synthesis process was usually performed in organic solvents, which is a minus point for protein imprinting. Therefore, more effective and simpler strategies for protein imprinting on the surface of Fe3O4 NPs are still required. Our previous work24 developed a much easier synthetic process for BHb imprinting employing amino group directly-functionalized Fe3O4 NPs as carriers. Though this work displayed some advantages in aspects of preparation technology, the hydrophilicity of the obtained products was unsatisfactory in virtue of adopting hydrophobic monomers.
Considering the solubility of protein, monomers with multifunctional groups and properties of hydrophilicity and biocompatibility are appropriate for imprinting proteins. Dopamine (DA), containing catechol and amine groups, is accepted as a popular adhesive for proteins.25,26 The catechol groups are liable to be oxidized to generate o-quinone functionalities which make DA form thin polydopamine (PDA) films onto different kinds of materials in weak alkaline media at room temperature. To date, a certain number of successful examples using DA as the monomer and cross-linker for protein imprinting have been reported.27–29 However, to the best of our knowledge, adopting DA as a monomer for the imprinting of proteins on the surface of amino group directly-functionalized Fe3O4 NPs has not been reported yet.
Taking into account the advantages of surface imprinting technology, magnetic separation, and PDA, the key idea of this work is to develop a facile approach to imprint protein on the surface of amino group directly-functionalized Fe3O4 NPs using dopamine as monomer through a two-step immobilized template process. The procedure of this preparation method was rather green and simple. The obtained nanomaterials could be easily separated from the solution under an external magnet within three seconds. In order to obtain the best recognition performance, the polymerization and adsorption conditions were investigated in detail. In addition, the practicability of the as-prepared imprinted polymers for biological application was further investigated by the separation and removal of a target protein from a real bovine blood sample. Furthermore, four other proteins with different isoelectric points were chosen as the templates to evaluate the versatility of the approach developed in this work.
2. Experimental
2.1. Materials
Dopamine (DA) was purchased from Alfa Aesar Chemical Company. 1,6-Diaminohexane (DAH), anhydrous sodium acetate (NaOAc), ferric chloride hexahydrate (FeCl3·6H2O), ethylene glycol (EG), diethylene glycol (DEG), ethanol, trihydroxymethyl aminomethane (Tris), sodium dodecyl sulfonate (SDS), and acetic acid (HAc) were provided by Xi’an Chemicals Ltd. Bovine serum albumin (BSA; pI = 4.9, Mw = 66.0 kDa), bovine hemoglobin (BHb; pI = 6.9, Mw = 64.0 kDa), bovine pancreas ribonuclease A (RNase A; pI = 9.4, Mw = 13.7 kDa), hemoglobin (Hb; pI = 6.8, Mw = 65.0 kDa), transferrin (transferrin; pI = 5.5, Mw = 77.0 kDa), immunoglobulin G (IgG; pI = 8.0, Mw = 150 kDa), and lysozyme (Lyz; pI = 11.2, Mw = 13.4 kDa) were obtained from Sigma. The highly purified water (18.0 MΩ cm−1) was prepared with a WaterPro water system (Axlwater Corporation, TY10AXLC1805-2, China) and used throughout the experiments. The bovine blood sample was kindly gifted from the local market. All reagents were of at least analytical grade and used without further treatment.2.2. Preparation of Fe3O4@BSA–MIPs and Fe3O4@NIPs
The amino-modified Fe3O4 nanoparticles (denoted as Fe3O4@NH2) were synthesized according to a previous method30 with some modifications. FeCl3·6H2O (1.0 g), NaOAc (3.5 g), and DAH (6.0 g) were dissolved in a mixture of EG (10 mL) and DEG (10 mL) in a Teflon-lined stainless-steel autoclave, which was sealed and heated at 200 °C. After reaction for 8 h, the autoclave was cooled to room temperature. The resultant black products were washed with highly purified water and ethanol to remove the solvent and the unreacted DAH, and then dried in a vacuum for further use.The optimal experimental conditions for the polymerization of the BSA surface-imprinted magnetic nanoparticles were as follows: (1) the mass of immobilized template BSA (10 mg, 20 mg, 30 mg, 40 mg, and 50 mg, respectively) was dissolved in 10 mM Tris–HCl buffer (pH = 8.5, 30 mL) in a three-neck round-bottom flask, and mixed with Fe3O4@NH2 (200 mg). The mixture was stirred for 30 min to obtain the Fe3O4@NH2–BSA complex. The complex was collected by an external magnetic field and left in the bottom of the flask, and the concentration of BSA in the supernatant was detected by a UV-vis spectrophotometer at about 280 nm. (2) The amount of the functional monomer DA (50 mg, 75 mg, 100 mg, 125 mg, and 150 mg, respectively) and 30 mL of Tris–HCl buffer (pH = 8.5, 10 mM) were added to the above flask, and stirred for 5 h at room temperature. The obtained products were rinsed with highly purified water until the supernatant was clear, and then washed with SDS–HAc (2%) to remove the template protein until no adsorption was detected by the UV-vis spectrophotometer. The resulting imprinted polymers (denoted as Fe3O4@BSA–MIPs) were collected by an external magnetic field, washed with highly purified water, and then dried under vacuum. Non-imprinted polymers (denoted as Fe3O4@NIPs) were prepared following the same procedure in the absence of the template protein BSA. Furthermore, the immobilization time of the BSA and the polymerization time of DA were also investigated.
Moreover, to testify the efficiency of Fe3O4@NH2 as the support, bare magnetic molecularly imprinted and non-imprinted polymers (denoted as B-Fe3O4@BSA–MIPs and B-Fe3O4@NIPs) were also prepared by the same way as that of Fe3O4@BSA–MIPs and Fe3O4@NIPs except for employing bare magnetic nanoparticles as the support.
2.3. Binding experiments
To investigate the adsorption kinetics of Fe3O4@BSA–MIPs and Fe3O4@NIPs, 10 mg of adsorbent was suspended in 10 mL of BSA Tris–HCl buffer (pH = 7.0, 10 mM) solution at a concentration of 0.30 mg mL−1. The mixture was incubated at regular time intervals from 5 min to 60 min at room temperature, and the polymers were magnetically separated from the solution. Then, the concentration of BSA in the supernatant was measured by UV-vis spectrophotometry.The adsorption capacity (Q) of the template protein or the competitive protein bound to the imprinted polymers is calculated as follows:
 | (1) |
The isothermal adsorption experiment was operated through changing the concentration of BSA from 0.050 to 0.50 mg mL−1 in Tris–HCl buffer (pH = 7.0, 10 mM) while employing 10 mg of Fe3O4@BSA–MIPs or Fe3O4@NIPs and shaking for 20 min at room temperature. Then, the adsorbent was isolated by a magnet and the residue BSA in the supernatant was determined by UV-vis spectrophotometry.
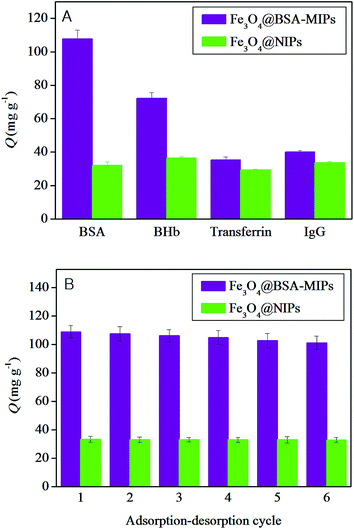
To evaluate the specific adsorption capability of the as-prepared polymers, 10 mg of Fe3O4@BSA–MIPs or Fe3O4@NIPs was incubated in 3 mL of Tris–HCl (10 mM, pH = 7.0) solution of BSA, BHb, transferrin, and IgG at a concentration of 0.30 mg mL−1 respectively at room temperature for 20 min, then the separation and determination procedures were conducted as described earlier in the adsorption kinetics experiments.
The imprinting factor (IF) and selectivity coefficient (SC) are usually used to determine the selectivity properties of the imprinted polymers towards the template protein and the competitive protein. The IF and SC are calculated from the following equations:
 | (2) |
 | (3) |
To evaluate if the amino groups on Fe3O4@NH2 could enhance the adsorption performance of the imprinted nanoparticles, we compared the binding capabilities of Fe3O4@BSA–MIPs, Fe3O4@NIPs, B-Fe3O4@BSA–MIPs, and B-Fe3O4@NIPs. In detail, 10 mg of the above four polymers was incubated with 10 mL of the BSA solution at a concentration of 0.30 mg mL−1 (pH = 7.0, 10 mM Tris–HCl) at room temperature for 20 min. Then, the extraction and detection procedures were conducted as described earlier in the adsorption experiments.
2.4. Reusability of the resultant imprinted nanomaterials
To estimate the reusability of Fe3O4@BSA–MIPs and Fe3O4@NIPs, the BSA adsorption–regeneration procedure was repeated 6 times using the same polymers. Briefly, 10 mg of the polymer was added to 10 mL of the BSA solution at a concentration of 0.30 mg mL−1 (pH = 7.0, 10 mM Tris–HCl) and incubated at room temperature for 20 min. Then, Fe3O4@BSA–MIPs or Fe3O4@NIPs were removed by a magnet and the bound amount of BSA was quantified by UV-vis spectrophotometry. The reused polymers were eluted with SDS–HAc (2%) for 3 h to ensure complete removal of the adsorbed BSA. The recovered products were then reused for the adsorption of BSA for another 5 cycles, and for every cycle the supernatant was collected and determined by UV-vis spectrophotometry at about 280 nm.2.5. Real sample analysis
10 mg of Fe3O4@BSA–MIPs was merged with 10 mL of the standard protein mixture (containing 0.30 mg mL−1 BSA and 0.30 mg mL−1 BHb) and the bovine blood sample diluted 150-fold with Tris–HCl buffer solution (pH = 7.0, 10 mM), respectively. After incubation for 20 min under gentle shaking, SDS–HAc (2%) was employed to elute the adsorbed protein for 3 h. The diluted, adsorbed, and eluted samples were used for SDS-PAGE analysis.2.6. Versatility investigation
The versatility of this approach was evaluated by using another three magnetic imprinted polymers generated in the same way as that of Fe3O4@BSA–MIPs except for adopting BHb, Hb, RNase A, and Lyz as the template protein, which were denoted as Fe3O4@BHb–MIPs, Fe3O4@Hb–MIPs, Fe3O4@RNase A–MIPs, and Fe3O4@Lyz–MIPs, respectively. The full cross-reaction characterizations of the four protein-imprinted nanomaterials along with the non-imprinted Fe3O4@NIPs were investigated by adsorbing the corresponding template protein and the three other proteins, namely, 10 mg of the polymer was incubated with 10 mL of the Tris–HCl buffer (pH = 7.0, 10 mM) solution of BSA, BHb, Hb, RNase A, and Lyz at a concentration of 0.30 mg mL−1 at room temperature for 20 min. Then, the extraction and detection procedures were conducted as described earlier in the binding experiments.2.7. Characterization
The morphology of the obtained nanomaterials was studied using a Tecnai G2 T2 S-TWIN transmission electron microscope. Fourier transform infrared (FT-IR) spectra were obtained via a Nicolet AVATAR 330 FT-IR spectrophotometer. The identification of the crystalline phase was carried out by a Rigaku D/max/2500v/pc (Japan) X-ray diffractometer with Cu Kα radiation. The magnetic properties were analyzed with a vibrating sample magnetometer (VSM) (LDJ 9600-1, USA). The adsorption data were measured using a UV-2450 spectrophotometer (Shimadzu, Japan). Electrophoretic analysis of the protein samples was performed using regular SDS-PAGE (Bio-Rad, Hercules, CA) with 10% running and 5% stacking gels. The proteins were stained with Coomassie Brilliant Blue R-250.3. Results and discussion
3.1. Preparation of Fe3O4@BSA–MIPs
The procedure for the preparation of Fe3O4@BSA–MIPs, combining the merits of surface imprinting technology, a two-step immobilized template strategy, functionalized magnetic nanomaterials, and the hydrophilicity of DA, is displayed in Fig. 1. First, Fe3O4@NH2, the surface of which was full of numerous amino groups, was synthesized through a modified one-step solvothermal method, using FeCl3·6H2O as the single iron source and DAH as the ligand. Next, the template protein BSA was immobilized on the surface of Fe3O4@NH2 through multi-hydrogen bonding interactions between the amino groups of Fe3O4@NH2 and the amino acids on BSA to form the Fe3O4@NH2–template complex. Then, a thin adherent polydopamine (PDA) shell was deposited on the surface of Fe3O4@NH2 to further immobilize the template protein by using DA as the monomer and cross-linker. Notably, the dopamine units possess amino, hydroxyl, and phenyl groups which can interact with BSA through multi-hydrogen bonds and hydrophobic effects. The two immobilized template processes can orientate template proteins orderly and keep the imprinted sites uniform for improving the imprinting effect.31 Finally, after washing with the 2% SDS–HAc solution, Fe3O4@BSA–MIPs with imprinted recognition sites complementary to BSA in shape, size, and functional group orientation were obtained.3.2. Optimization of the polymerization conditions
When we carried out the optimal experimental conditions, the effect of different mass of immobilization of BSA onto Fe3O4@NH2 in the range of 10–50 mg was investigated. As shown in Table S1,† the QIM increased with increasing QAD from 10 mg to 30 mg, indicating that a higher QAD might make Fe3O4@NH2 interact adequately with BSA through forming multi-hydrogen bonds. However, the QIM remained almost unchanged with increasing QAD from 30 mg to 50 mg. A possible reason was that the amino groups on Fe3O4@NH2 had been saturated with BSA. Excessive QAD could not increase the immobilization. Thus, the QAD of 30 mg was chosen in this work. Furthermore, we also evaluated the immobilization time of BSA by varying from 10 min to 50 min. The results are presented in Table S2,† from which we found that the best immobilization time was 30 min.To optimize the recognition performance of Fe3O4@BSA–MIPs, the thickness of the PDA shell, which is controlled by the amount of DA, was adjusted from 50 mg to 150 mg. The results of the adsorption capacity (Q) and imprinting factor (IF) are presented in Fig. 2A. Both Q and IF increased with an increasing amount of DA from 50 mg to 100 mg, indicating that enough functional monomers could react adequately with template proteins and create enough recognition cavities in the network of polymers to enhance the imprinting ability of Fe3O4@BSA–MIPs towards BSA. However, a decrease of Q and IF was observed when the amount of DA was further increased. Too thick PDA imprinted shells may make it hard for the protein to access the recognition cavities. Therefore, 100 mg of DA was chosen for the polymerization of Fe3O4@BSA–MIPs in accordance with the experimental results.
 | ||
| Fig. 2 Effect of the amount (A) and polymerization time (B) of DA on the imprinting performance of Fe3O4@BSA–MIPs and Fe3O4@NIPs. | ||
The polymerization time of DA was varied from 3 h to 7 h to seek the shortest experimental period and the best binding performance. The results are displayed in Fig. 2B. It is indicated that the optimal recognition sites and the most suitable shell thickness were formed when the polymerization time was 5 h. The polymerization degree was incomplete when the polymerization time was less than 5 h, because the thin PDA shell could not immobilize much template and created less imprinted cavities. Conversely, too many monomers self-polymerized and blocked the recognition sites, impacting the imprinting effect of the polymers when the polymerization time was over 5 h. Therefore, 5 h was chosen as the polymerization time in this work.
3.3. Characterization of the obtained nanomaterials
The morphological structure and size of Fe3O4@NH2 and Fe3O4@BSA–MIPs were characterized by TEM. As can be seen from Fig. 3, the obtained nanomaterials possessed a spherical structure with a narrow particle size distribution. Through statistical analysis of the particles, the average diameter of Fe3O4@NH2 (Fig. 3A) was about 60 nm. It was found that Fe3O4@BSA–MIPs with a distinguishable core–shell structure were successfully prepared after a two-step template immobilization process, and whose diameter increased to approximately 80 nm (Fig. 3B), indicating that the thickness of the PDA imprinting shell was just about 10 nm. The thickness was suitable for the three-dimensional structure of the BSA,32 making the mass transport between the solution and the surface of Fe3O4@BSA–MIPs easier.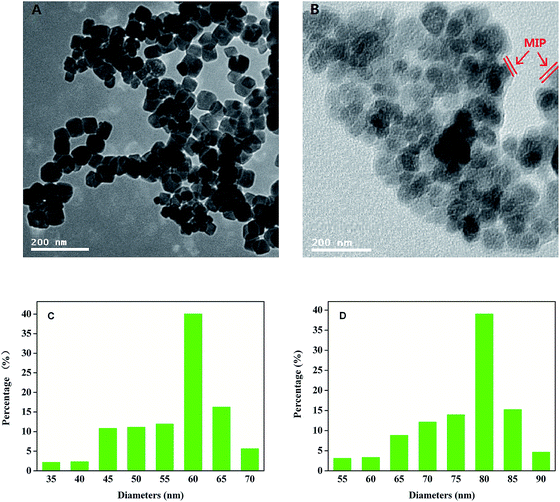 | ||
| Fig. 3 TEM images of Fe3O4@NH2 (A) and Fe3O4@BSA–MIPs (B); the size distribution histograms of Fe3O4@NH2 (C) and Fe3O4@BSA–MIPs (D). | ||
The chemical groups of the pristine Fe3O4@NH2 and Fe3O4@BSA–MIPs (Fig. 4) were characterized by FT-IR spectroscopy. The strong peak at 575 cm−1 observed in both samples (curves a and b) was attributed to the stretch of the Fe–O vibration. The peaks at around 1080 cm−1 and 1640 cm−1, corresponding to the C–N stretch and the bending vibration of N–H24 respectively, indicate that the amino functionalization was on the surface of Fe3O4@NH2 (curve a). When compared with Fe3O4@NH2, the new appearance of typical peaks in the spectra of Fe3O4@BSA–MIPs (curve b), such as phenylic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching at 1500 cm−1, the enhanced adsorption peak intensity of C–N bond at 1640 cm−1, and the lessened characteristic peak of the Fe–O bond at 575 cm−1 should be attributed to the coating of the PDA layer on the magnetic nanoparticles.
C stretching at 1500 cm−1, the enhanced adsorption peak intensity of C–N bond at 1640 cm−1, and the lessened characteristic peak of the Fe–O bond at 575 cm−1 should be attributed to the coating of the PDA layer on the magnetic nanoparticles.
The crystalline structures of Fe3O4@NH2 and Fe3O4@BSA–MIPs were characterized by XRD in the 2θ range of 20–70° (Fig. 5). Six characteristic peaks of Fe3O4 (2θ = 30.38°, 35.58°, 43.14°, 53.48°, 57.08°, and 62.66°) were observed in both samples. The peak positions at the corresponding 2θ values were indexed as (220), (311), (400), (422), (511), and (440), respectively, which matched well with the database of magnetite in the JCPDS-International Center for Diffraction Data (JCPDS card: 19-629). The distinctive diffraction peaks of Fe3O4@BSA–MIPs were in agreement with those of Fe3O4, illustrating that the obtained imprinted polymers containing Fe3O4 with a spinel structure and the phase of Fe3O4 did not change during the synthetic process.33
The magnetic properties of Fe3O4@NH2 and core–shell Fe3O4@BSA–MIPs were studied using a vibrating sample magnetometer at room temperature. Fig. 6 illustrates the plots of magnetization versus magnetic field (M–H loop) of Fe3O4@NH2 (curve a) and Fe3O4@BSA–MIPs (curve b) from which we found there was no remanence and coercivity, demonstrating that both polymers were superparamagnetic. It is noteworthy that the saturation magnetization of Fe3O4@BSA–MIPs (40.7 emu g−1) was lower than that of Fe3O4@NH2 (53.5 emu g−1), which was ascribed to the shielding effect of the PDA shell on the surface of Fe3O4@NH2. The experimental results show that the obtained imprinted polymers possess sufficient magnetic force to be quickly separated from the solution under an external magnet within three seconds.
3.4. Adsorption performance
To further investigate the kinetic mechanisms of adsorption, the obtained data were calculated by the second-order rate equation that can be expressed as follows:
 | (4) |
As shown in Table 1, the correlation coefficients (r) of Fe3O4@BSA–MIPs and Fe3O4@NIPs are 0.9998 and 0.9964, indicating that the second-order model fitted the binding data well. The values of v0 implied that the initial adsorption rate of Fe3O4@BSA–MIPs was much faster than that of Fe3O4@NIPs, which was in keeping with Fig. 7A. The results also illustrated that the adsorption capacity was proportional to the number of active recognition sites on the surface of the imprinted nanomaterials, and the chemical adsorption was the rate-limiting step in the recognition process of the resulting polymers.36
| Model | Equations and parameters | Fe3O4@BSA–MIPs | Fe3O4@NIPs |
|---|---|---|---|
| Second-order kinetics | Equation | t/Qt = 0.0082 + 0.0091t | t/Qt = 0.0989 + 0.0292t |
| Qe (mg g−1) | 109.9 | 34.13 | |
| K (g mg−1 min−1) | 0.0101 | 0.0087 | |
| v0 (mg g−1 min−1) | 122.0 | 10.11 | |
| r | 0.9998 | 0.9964 | |
| Langmuir isotherm | Equation | Ce/Q = 2.1162 × 10−4 + 0.0085Ce | Ce/Q = 0.0019 + 0.0254Ce |
| Qmax (mg g−1) | 117.6 | 39.37 | |
| KL (mL mg−1) | 40.18 | 13.37 | |
| r | 0.9969 | 0.9954 | |
| Freundlich isotherm | Equation | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Q = 2.2525 + 0.3381 Q = 2.2525 + 0.3381![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ce Ce |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Q = 1.7022 + 0.3843 Q = 1.7022 + 0.3843![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ce Ce |
| KF (mg g−1) | 178.9 | 50.37 | |
| n | 0.3381 | 0.3843 | |
| r | 0.9811 | 0.9339 |
The saturation binding data was further processed by the Freundlich and Langmuir isothermal models to estimate the adsorption properties of the resultant nanomaterials. The two models are expressed as follows:
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Q = log Q = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KF + n KF + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ce Ce
| (5) |
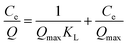 | (6) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Q versus log
Q versus log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ce, respectively.
Ce, respectively.
The Langmuir isotherm model assumes that the adsorption takes place at specific homogeneous sites as well as monolayer sorption, and each site can bind only one molecule.37 While the Freundlich isotherm model is applied to multi-layer adsorption and unfavorable adsorption on heterogeneous surfaces.38 Through comparison of the r values displayed in Table 1, we can conclude that the experimental data are better fitted with the Langmuir isotherm model (r > 0.99) than the Freundlich model (r < 0.94). The maximum adsorption capacities obtained from the experiment are also close to the apparent maximum adsorption capacities (117.6 mg g−1 for Fe3O4@BSA–MIPs and 39.37 mg g−1 for Fe3O4@NIPs) calculated using the Langmuir isotherm model. In light of the results, the binding of BSA onto the resultant nanomaterials may be all monolayer adsorption, and no further binding takes place at the binding site once a template molecule occupies this site.
| Polymers | Q (mg g−1) | IF |
|---|---|---|
| a In this experiment, 10 mg of Fe3O4@BSA–MIPs, B-Fe3O4@BSA–MIPs, Fe3O4@NIPs, and B-Fe3O4@NIPs was incubated in 10 mL of a Tris–HCl (10 mM, pH = 7.0) solution of BSA at a concentration of 0.30 mg mL−1 at room temperature for 20 min. (n = 5). | ||
| Fe3O4@BSA–MIPs | 107.8 | — |
| Fe3O4@NIPs | 32.15 | 3.35 |
| B-Fe3O4@BSA–MIPs | 53.19 | — |
| B-Fe3O4@NIPs | 31.84 | 1.67 |
3.5. Reusability
The property of reusability is significant for the actual application of polymers. To test the stability of the prepared magnetic nanomaterials, the adsorption–regeneration cycle was repeated six times using the same Fe3O4@BSA–MIPs and Fe3O4@NIPs. As shown in Fig. 8B, the adsorption capacity of Fe3O4@BSA–MIPs was reduced by only 7.1% after six cycles, which might be because some recognition cavities of the imprinted polymers were clogged after regeneration or destroyed after washing, and thus, they were no longer fit for the template molecules. Whereas, the affinity of Fe3O4@NIPs remained almost unchanged, because their recognition was nonspecific and the effect of washing could be ignored. After six cycles of adsorption–regeneration, the adsorption performance of the imprinted and non-imprinted magnetic nanomaterials prepared in this work were stable, which is an outstanding advantage for practical application.3.6. Application
Serum albumin accounts for approximately 50% of total blood plasma proteins,39 which is considered to act as a shield in the analysis of biomarkers that are commonly at lower concentration levels in plasma samples. Therefore, we chose fresh bovine blood diluted 150-fold with Tris–HCl (pH = 7.0, 10 mM) as the real sample to further evaluate the practicability of Fe3O4@BSA–MIPs. The SDS-PAGE analysis is illustrated in Fig. 9. There were two bands in lane 1, indicating the mixture of BSA and BHb. It was found from lane 2 that the band of BSA faded after treatment with Fe3O4@BSA–MIPs, while the band of BHb had little change. The BSA band reappeared in lane 3 after elution with 2% SDS–HAc solution, revealing that the BSA was selectively captured by Fe3O4@BSA–MIPs. Lane 5 displayed the supernatant of the fresh bovine blood diluted 150-fold (lane 4) after treatment with Fe3O4@BSA–MIPs, in which BSA almost disappeared while the other bands were retained, suggesting that Fe3O4@BSA–MIPs had specific recognition for BSA in the bovine blood sample and less co-adsorption with others. After elution with 2% SDS–HAc solution, only the band for BSA was observed (lane 6), indicating that the BSA was selectively isolated and recovered from the real sample after elution. The above results confirmed the specificity and practicability of Fe3O4@BSA–MIPs for the separation of BSA.3.7. Method validation
The generality of the proposed imprinting approach was further investigated through the full cross-selectivity test of four protein-imprinted nanomaterials along with non-imprinted Fe3O4@NIPs, and the results are summarized in Table 3. It was clearly observed that the adsorption capacity, imprinting factor, and selectivity coefficient of different imprinted polymers to corresponding template proteins were all higher than those of the other three proteins, demonstrating that the developed method was valid to imprint different kinds of proteins with different isoelectric points. It is worth mentioning that the BHb-imprinted nanomaterials used in this work have a larger adsorption capacity (181.1 mg g−1) towards BHb and a higher imprinting factor (4.96) than those of our previous work (37.58 mg g−1, 3.60).24 The greatest difference lies in that the latter adopted two types of silane coupling agent as monomers while the former employed DA as monomer. It is also worth noting that the Hb-imprinted nanoparticles used in this work have a considerably larger adsorption capacity (187.9 mg g−1) towards Hb and a relatively higher imprinting factor (5.48) than those of Zhou’s work (17.50 mg g−1, 5.01).20 The main contrast was that the latter employed bare Fe3O4 nanoparticles as the support while the former used Fe3O4@NH2 instead. Therefore, NH2-modification on the surface of Fe3O4 nanoparticles enhances the performance of the imprinted nanomaterials.| Polymers | Proteins | Q (mg g−1) | IF | SC |
|---|---|---|---|---|
| a In these experiments, 10 mg of Fe3O4@BSA–MIPs, Fe3O4@BHb–MIPs, Fe3O4@Hb–MIPs, Fe3O4@RNase A–MIPs, Fe3O4@Lyz–MIPs, and Fe3O4@NIPs was incubated in 10 mL of a Tris–HCl (10 mM, pH = 7.0) solution of BSA, BHb, Hb, RNase A, and Lyz at a concentration of 0.30 mg mL−1 respectively at room temperature for 20 min (n = 5). | ||||
| Fe3O4@BSA–MIPs | BSA | 107.8 ± 2.7 | 3.35 ± 0.02 | — |
| BHb | 72.13 ± 1.8 | 1.97 ± 0.08 | 1.70 ± 0.07 | |
| Hb | 70.15 ± 1.9 | 2.05 ± 0.03 | 1.63 ± 0.04 | |
| RNase A | 82.61 ± 2.2 | 2.01 ± 0.07 | 1.67 ± 0.11 | |
| Lyz | 99.1 ± 3.4 | 2.10 ± 0.04 | 1.59 ± 0.09 | |
| Fe3O4@BHb–MIPs | BSA | 73.89 ± 1.7 | 2.30 ± 0.09 | 2.16 ± 0.08 |
| BHb | 181.1 ± 3.6 | 4.96 ± 0.23 | — | |
| Hb | 146.3 ± 3.8 | 4.27 ± 0.26 | 1.16 ± 0.02 | |
| RNase A | 93.82 ± 2.9 | 2.28 ± 0.07 | 2.18 ± 0.25 | |
| Lyz | 105.7 ± 3.1 | 2.24 ± 0.05 | 2.21 ± 0.32 | |
| Fe3O4@Hb–MIPs | BSA | 71.41 ± 2.0 | 2.22 ± 0.04 | 2.47 ± 0.16 |
| BHb | 159.2 ± 3.7 | 4.36 ± 0.06 | 1.26 ± 0.35 | |
| Hb | 187.9 ± 3.5 | 5.48 ± 0.03 | — | |
| RNase A | 91.85 ± 2.4 | 2.24 ± 0.02 | 2.45 ± 0.26 | |
| Lyz | 109.3 ± 2.9 | 2.31 ± 0.09 | 2.37 ± 0.12 | |
| Fe3O4@RNase A–MIPs | BSA | 47.61 ± 2.2 | 1.48 ± 0.06 | 4.06 ± 0.12 |
| BHb | 66.22 ± 3.6 | 1.81 ± 0.03 | 3.32 ± 0.29 | |
| Hb | 63.01 ± 3.9 | 1.84 ± 0.05 | 3.27 ± 0.37 | |
| RNase A | 246.8 ± 8.3 | 6.01 ± 0.26 | — | |
| Lyz | 136.5 ± 2.7 | 2.89 ± 0.04 | 2.08 ± 0.08 | |
| Fe3O4@Lyz–MIPs | BSA | 43.94 ± 1.4 | 1.37 ± 0.08 | 4.50 ± 0.13 |
| BHb | 62.73 ± 2.5 | 1.72 ± 0.03 | 3.58 ± 0.46 | |
| Hb | 59.42 ± 1.9 | 1.73 ± 0.02 | 3.56 ± 0.39 | |
| RNase A | 119.6 ± 3.6 | 2.91 ± 0.05 | 2.12 ± 0.25 | |
| Lyz | 291.1 ± 11.2 | 6.16 ± 0.31 | — | |
| Fe3O4@NIP | BSA | 32.15 ± 0.94 | — | — |
| BHb | 36.54 ± 1.2 | — | — | |
| Hb | 34.26 ± 1.4 | — | — | |
| RNase A | 41.06 ± 2.9 | — | — | |
| Lyz | 47.26 ± 1.3 | — | — | |
4. Conclusion
In this work, a facile, general, and economical approach was successfully developed to imprint proteins on the surface of Fe3O4@NH2 using dopamine as monomer. Because of combining the advantages of surface imprinting technology, immobilized template strategy, magnetic separation, and PDA, these resulting imprinted nanomaterials exhibited fast kinetics, a large adsorption capacity, high selectivity, and satisfactory reusability. Furthermore, the favorable versatility for proteins with different isoelectric points, and the successful application in the specific capture of the target protein from a real sample of bovine blood and good recovery after a reasonably mild elution, indicate that the proposed method could be an alternative solution for the selective removal of high-abundant proteins from complex biological samples.Acknowledgements
The authors are grateful for financial support from the National Natural Science Foundation of China (no. 21305107), the Fundamental Research Funds for the Central Universities (no. 08143081, 08142034), and China Postdoctoral Science Foundation (no. 2014M562388).References
- F. R. Wang, C. Chmil, F. Pierce, K. Ganapathy, B. B. Gump, J. A. MacKenzie, Y. Mechref and K. Bendinskas, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2013, 934, 26–33 CrossRef CAS PubMed.
- J. L. Cao, X. H. Zhang, X. W. He, L. X. Chen and Y. K. Zhang, J. Mater. Chem. B, 2013, 1, 3625–3632 RSC.
- M. Javanmard, S. Emaminejad, C. Gupta, J. Provine, R. W. Davis and R. T. Howe, Sens. Actuators, B, 2014, 193, 918–924 CrossRef CAS PubMed.
- M. Javanmard, S. Emaminejad, R. W. Dutton and R. W. Davis, Anal. Chem., 2012, 84, 1432–1438 CrossRef CAS PubMed.
- D. N. Gunasena and Z. El Rassi, J. Sep. Sci., 2011, 34, 2097–2105 CAS.
- R. L. Gundry, M. Y. White, J. Nogee, I. Tchernyshyov and J. E. Van Eyk, Proteomics, 2009, 9, 2021–2028 CrossRef CAS PubMed.
- B. Li, J. J. Xu, A. J. Hall, K. Haupt and B. Tse Sum Bui, J. Mol. Recognit., 2014, 27, 559–565 CrossRef CAS PubMed.
- A. Mirmohseni, M. Shojaei and R. Pourata, RSC Adv., 2014, 4, 20177–20184 RSC.
- E. Yildirim, E. Turan and T. Caykara, J. Mater. Chem., 2012, 22, 636–642 RSC.
- K. El Kirat, M. Bartkowski and K. Haupt, Biosens. Bioelectron., 2009, 24, 2618–2624 CrossRef CAS PubMed.
- J. Bognár, J. Szücs, Z. Dorkó, V. Horváth and R. E. Gyurcsányi, Adv. Funct. Mater., 2013, 23, 4703–4709 CrossRef.
- T. Wangchareansak, C. Sangma, K. Choowongkomon, F. Dickert and P. Lieberzeit, Anal. Bioanal. Chem., 2011, 400, 2499–2506 CrossRef CAS PubMed.
- M. Bosserdt, N. Gajovic-Eichelman and F. W. Scheller, Anal. Bioanal. Chem., 2013, 405, 6437–6444 CrossRef CAS PubMed.
- A. M. Bossi, P. S. Sharma, L. Montana, G. Zoccatelli, O. Laub and R. Levi, Anal. Chem., 2012, 84, 4036–4041 CrossRef CAS PubMed.
- V. Saumya, K. P. Prathish and T. P. Rao, Talanta, 2011, 85, 1056–1062 CrossRef CAS PubMed.
- L. Uzun, R. Uzek, S. Şenel, R. Say and A. Denizli, Mater. Sci. Eng., C, 2013, 33, 3432–3439 CrossRef CAS PubMed.
- F. Kartal and A. Denizli, J. Sep. Sci., 2014, 37, 2077–2086 CrossRef CAS PubMed.
- E. Moczko, A. Guerreiro, E. Piletska and S. Piletsky, Langmuir, 2013, 29, 9891–9896 CrossRef CAS PubMed.
- R. X. Gao, X. R. Mu, J. J. Zhang and Y. H. Tang, J. Mater. Chem. B, 2014, 2, 783–792 RSC.
- W. H. Zhou, C. H. Lu, X. C. Guo, F. R. Chen, H. H. Yang and X. R. Wang, J. Mater. Chem., 2010, 20, 880–883 RSC.
- W. W. Huang, X. Yang, S. Zhao, M. Zhang, X. L. Hu, J. Wang and H. T. Zhao, Analyst, 2013, 138, 6653–6661 RSC.
- X. J. Li, B. L. Zhang, L. Tian, W. Li, T.J. Xin, H. P. Zhang and Q. Y. Zhang, Sens. Actuators, B, 2014, 196, 265–271 CrossRef CAS PubMed.
- T. Jing, H. R. Du, Q. Dai, H. Xia, J. W. Niu, Q. L. Hao, S. R. Mei and Y. K. Zhou, Biosens. Bioelectron., 2010, 26, 301–306 CrossRef CAS PubMed.
- R. X. Gao, X. R. Mu, Y. Hao, L. L. Zhang, J. J. Zhang and Y. H. Tang, J. Mater. Chem. B, 2014, 2, 1733–1741 RSC.
- Y. Zhang, K. Panneerselvam, R. Ogaki, L. Hosta-Rigau, R. V. D. Westen, B. E. B. Jensen, B. M. Teo, M. F. Zhu and B. Städler, Langmuir, 2013, 29, 10213–10222 CrossRef CAS PubMed.
- A. Nematollahzadeh, A. Shojaei, M. J. Abdekhodaie and B. Sellergren, J. Colloid Interface Sci., 2013, 404, 117–126 CrossRef CAS PubMed.
- A. Tretjakov, V. Syritski, J. Reut, R. Boroznjak, O. Volobujeva and A. Öpik, Microchim. Acta, 2013, 180, 1433–1442 CrossRef CAS.
- Z. W. Xia, Z. A. Lin, Y. Xiao, L. Wang, J. N. Zheng, H. H. Yang and G. N. Chen, Biosens. Bioelectron., 2013, 47, 120–126 CrossRef CAS PubMed.
- R. Liu, M. Sha, S. S. Jiang, J. Luo and X. Y. Liu, Talanta, 2014, 120, 76–83 CrossRef CAS PubMed.
- L. Y. Wang, J. Bao, L. Wang, F. Zhang and Y. D. Li, Chem.–Eur. J., 2006, 12, 6341–6347 CrossRef CAS PubMed.
- L. J. Liu, J. J. Zheng, G. J. Fang and W. H. Xie, Anal. Chim. Acta, 2012, 726, 85–92 CrossRef CAS PubMed.
- X. J. Li, B. L. Zhang, W. Li, X. F. Lei, X. L. Fan, L. Tian, H. P. Zhang and Q. Y. Zhang, Biosens. Bioelectron., 2014, 51, 261–267 CrossRef CAS PubMed.
- F. X. Chen, S. L. Xie, J. H. Zhang and R. Liu, Mater. Lett., 2013, 112, 177–179 CrossRef CAS PubMed.
- D. R. Kryscio and N. A. Peppas, Anal. Chim. Acta, 2012, 718, 109–115 CrossRef CAS PubMed.
- M. S. Zhang, J. R. Huang, P. Yu and X. Chen, Talanta, 2010, 81, 162–166 CrossRef CAS PubMed.
- I. Koç, G. Baydemir, E. Bayram, H. Yavuz and A. Denizli, J. Hazard. Mater., 2011, 192, 1819–1826 CrossRef PubMed.
- S. Y. Tao, C. Wang, W. Ma, S. Wu and C. G. Meng, Microporous Mesoporous Mater., 2012, 147, 295–301 CrossRef PubMed.
- R. Dan, Y. Z. Wang, L. Du, S. H. Du, M. D. Huang, S. Yang and M. Zhang, Analyst, 2013, 138, 3433–3443 RSC.
- M. A. O. da Silva and M. A. Z. Arruda, Talanta, 2009, 77, 985–990 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra07965e |
| This journal is © The Royal Society of Chemistry 2014 |