Intracellular biosensors by functional nanomaterial-integrated CRISPR technologies for real-time molecular sensing
Min Yu
Choi
 a,
Chenzhong
Li
b,
Jin-Ha
Choi†
a,
Chenzhong
Li
b,
Jin-Ha
Choi†
 *a and
Jeong-Woo
Choi†
*a and
Jeong-Woo
Choi†
 *c
*c
aSchool of Chemical Engineering, Clean Energy Research Center, Jeonbuk National University, Jeonju 54896, Republic of Korea. E-mail: jhchoi@jbnu.ac.kr
bBiomedical Engineering, School of Medicine, The Chinese University of Hong Kong, Shenzhen 518172, China
cDepartment of Chemical and Biomolecular Engineering, Sogang University, Seoul 04107, Republic of Korea. E-mail: jwchoi@sogang.ac.kr
First published on 19th November 2025
Abstract
CRISPR technology, originally developed as a genome-editing tool, has recently emerged as a powerful platform for intracellular biosensing. By harnessing the programmability and target specificity of CRISPR-associated proteins, such as Cas9, Cas12, and Cas13, researchers have engineered biosensors capable of detecting a wide array of intracellular signals, including nucleic acids, non-coding RNAs, and small-molecule metabolites. This review discusses the recent advancements in CRISPR-powered biosensors for real-time, dynamic monitoring of cellular processes and molecular events. Particular focus is given to the integration of nanotechnology, which plays a crucial role in enhancing the delivery efficiency, signal amplification, and sensor stability. Nanomaterials such as gold nanoparticles, quantum dots, DNA nanostructures, and upconversion nanoparticles have been strategically employed to improve the intracellular transport of CRISPR components, facilitate signal readouts, and enable multimodal sensing in complex cellular environments. Additionally, we delve into how CRISPR-nanotechnology hybrids can be adapted for multiplex analysis and single-cell resolution. This review also addresses the current challenges in intracellular biosensing, including precise delivery, biocompatibility, and long-term monitoring, and outlines future directions for the application of these systems in precision medicine, synthetic biology, and advanced therapeutic monitoring. Through the convergence of gene-editing systems and nanotechnology, CRISPR-based intracellular biosensors are anticipated to revolutionize next-generation diagnostic and therapeutic strategies.
Introduction
Detection and imaging of biomolecules within living cells play a central role in elucidating the complex spatiotemporal distribution and dynamic changes in cellular components.1 Such analyses extend beyond simple identification of molecular presence and contribute to the understanding of the functional mechanisms of intracellular molecular networks, including protein–nucleic acid interactions, signaling pathways, and metabolic fluxes. Direct molecular imaging of living cells minimizes artifacts that may arise from fixation or destructive treatments, thereby providing reliable data that more accurately reflect physiological conditions. Moreover, sensitive and precise monitoring of intracellular molecules enables real-time tracking of the expression patterns and dynamics of disease-associated biomarkers.2,3 This capability can be applied to the early diagnosis and staging of diseases such as cancer, neurodegenerative disorders, and viral infections and provides critical evidence for evaluating drug responsiveness and optimizing therapeutic strategies.4 High-resolution and ultrasensitive biosensing technologies are gaining increasing importance as they are directly linked not only to clinical diagnostics but also to the advancement in personalized medicine.Consequently, there has been a growing interest in systems capable of sensing and monitoring nucleic acids, proteins, and ions within the cellular environment, with particular attention given to clustered regularly interspaced short palindromic repeat (CRISPR)-based platforms that generate highly specific signals in response to even single-nucleotide variations. CRISPR-based genome-editing technologies have attracted significant attention for potential therapeutic applications, including the treatment of genetic disorders and cancer.5 Moreover, certain types of CRISPR exhibit collateral cleavage activity upon target sequence recognition, which has demonstrated significant potential in biosensing applications where the specific recognition of a target must generate a measurable signal.6–8 In this context, CRISPR/Cas12a and Cas13a have been extensively used and have shown high detection accuracies for DNA and RNA targets, respectively.9,10
Another notable advantage is their functionality at mild temperatures, suggesting the feasibility of their operation in living cells. These features highlight the potential of CRISPR technology as an alternative to PCR, which is regarded as the gold standard for nucleic acid detection. Although PCR offers high accuracy and is used in clinical diagnostics, it is limited by its complexity, lengthy processing time, and requirement for trained personnel. Moreover, PCR cannot capture dynamic changes in living systems and is prone to environmental interference during sample preparation.11 Conversely, CRISPR systems can be flexibly integrated into intracellular biosensing for real-time and dynamic monitoring.11,12
However, there are still limitations in delivery efficiency and stability in vivo, and it remains challenging to obtain sufficient signals in the complex biological environment.13 In addition, the intrinsic off-target effects of the CRISPR system, which refer to unintended recognition and cleavage of non-target sequences, restrict its performance in living cells.14,15 Thus, various nanomaterial integration methods have been proposed to overcome these limitations. Various types of nanoparticles have been developed to enable the stable intracellular delivery of the CRISPR system, which is relatively large in size and susceptible to degradation.16–18 Depending on the design, these nanoparticles may serve a single function or perform multiple roles, such as enhancing signal intensity through optical phenomena. Such improvements significantly increase the detection sensitivity of intracellular analytes.19–22
This review first examines the mechanisms of CRISPR systems that have been actively investigated for biosensing, along with the unique characteristics associated with each CRISPR type. It then discusses the classes of biomolecules that are commonly targeted at the intracellular level and highlights the CRISPR-based applications developed for their detection. Particular attention is given to strategies addressing the delivery efficiency and stability of CRISPR systems, as well as the integration of nanomaterials designed to further enhance sensitivity. Finally, it outlines the current challenges and future perspectives of intracellular biosensing and monitoring using CRISPR technology.
CRISPR systems as intracellular biosensors
Mechanism and programmability of CRISPR-Cas systems
The CRISPR-Cas system originates from the bacterial immune response against invading nucleic acids and functions through a mechanism that recognizes and cleaves foreign sequences.23,24 The activation and cleavage activity of CRISPR has been widely exploited as a powerful gene-editing technology in the life sciences.25 CRISPR-based genome-editing technologies have been applied for transcriptional regulation, epigenome modification, genome-wide screening, and even chromosome imaging26. Furthermore, certain CRISPR types possess trans-cleavage activity, in which nucleic acid recognition triggers the nonspecific cleavage of nearby single-stranded sequences. This property has enabled integration into various readout systems for biomarker sensing. Furthermore, CRISPR systems provide the programmability of the platform enables facile retargeting of different biomarkers simply by altering the crRNA sequence.3CRISPR systems are highly diverse and are generally classified into two major classes: class 1 and class 2. Class 1 includes types I, III, and IV, whereas class 2 comprises types II, V, and VI.27,28 Among these, Cas9, Cas12, and Cas13, which belong to class 2, have been extensively investigated as molecular diagnostic tools owing to their unique cleavage activities.29–31
However, RNA probes required for Cas13a-based sensing are more susceptible to degradation than DNA probes, which increases the risk of false-positive results.36
Numerous studies have been conducted to expand the target range, enhance the cleavage efficiency, and reduce the off-target effects of Cas12a. First, it is well established that single-stranded DNA (ssDNA) can also activate Cas12a.38 This property has allowed its application in the detection of various biomarkers without the need for a Protospacer Adjacent Motif (PAM) sequence.39 Furthermore, the discovery that Cas12a can be activated by extending the 3′ terminus of short sequences with random sequences has increased the flexibility in designing activator strands. A method was also introduced to detect RNA targets directly without prior RNA amplification by utilizing the relatively stable Cas12a. This approach involves supplying short ssDNA or PAM-containing double-stranded DNA (dsDNA) to the seed region of the crRNA to enable RNA target detection at the crRNA terminus, although this still requires the addition of exogenous DNA.40
Owing to their high target specificity and ability to generate measurable signals through collateral cleavage, CRISPR-Cas systems have been integrated with a broad range of reporter strategies and extensively studied in biosensing applications (Fig. 1).41
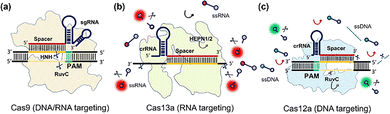 | ||
| Fig. 1 Mechanisms of CRISPR-Cas systems. (a) Cas9 and sgRNA bind with target dsDNA, and the nuclease domains cleave the target strands. (b) Cas13a activated by target RNA and enables trans-cleavage to adjacent ssRNAs. (c) Cas12a and crRNA recognized target DNA and induced trans-cleavage to near ssDNAs. Reproduced from ref. 33 with permission from the Royal Society of Chemical, copyright 2023. | ||
Types of intracellular targets
The intracellular environment contains a variety of biomolecules, some of which serve as biomarkers reflecting the state of the cell or disease. To discuss intracellular biosensing using CRISPR, it is important to understand which types of markers can be targeted in order to emphasize the necessity of intracellular monitoring.DNA methylation is an epigenetic modification involved in various cellular functions and is regarded as a potential biomarker for disease diagnosis and monitoring.45 This modification primarily occurs at cytosine bases within CpG islands, where it prevents the binding of transcription factors and RNA polymerase to promoter regions, ultimately leading to transcriptional repression.46 Aberrant DNA methylation and the resulting dysregulation of gene expression have been reported to be associated with various diseases, including cancer.47 Monitoring DNA methylation at the single-cell level provides valuable insight into dynamic cellular states and enables the characterization of cell heterogeneity, which is often obscured in bulk analyses.48
The localization and translation of mRNA impact cellular physiology at various levels. At the single-cell level, mRNA trafficking enables rapid and localized responses to both intracellular and extracellular signals, thereby affecting cell motility and differentiation. Therefore, it is crucial to analyze mRNA imaging from the intracellular to multicellular level, extending beyond cytoplasmic localization.50
Among noncoding RNAs, microRNAs (miRNAs) are short RNAs that exert significant regulatory functions in numerous biological pathways. They are frequently upregulated in cancer and are implicated in tumorigenesis, rendering intracellular sensing indispensable.51 For instance, the aberrant expression of the well-known oncogenic biomarker miR-21 is commonly associated with tumor initiation and metastasis. Similarly, the overexpression of miRNA-196 and miRNA-106 has been reported as an indicator of cancer metastasis.19 However, miRNA biomarkers that are overexpressed in tumor cells often remain present at non-negligible levels in normal cells, rendering them susceptible to interference from extracellular signals. Thus, advanced imaging strategies with improved spatial resolution are required to enable the accurate detection of miRNAs, specifically within tumor cells.52 Circular RNAs (circRNAs), a class of noncoding RNAs, have been implicated in cancer initiation and progression through multiple regulatory mechanisms. Owing to their broad presence in human cells and their tissue specificity, circRNAs have emerged as valuable biomarkers for cancer diagnosis.52 Specifically, circRNAs are widely distributed in human cells and have been reported to participate in numerous malignancy-related processes, including cell cycle regulation, tumorigenesis, invasion, metastasis, apoptosis, and angiogenesis.53 Long noncoding RNAs (lncRNAs), typically defined as transcripts longer than 200 nucleotides, regulate gene expression either through direct interactions with mRNAs in the cytoplasm or by modulating mRNA translation.54 They have been shown to influence cell proliferation, invasion, and apoptosis, while imbalances in lncRNAs are associated with autoimmune diseases, such as systemic lupus erythematosus, rheumatoid arthritis, and desiccation syndrome. Furthermore, lncRNAs are increasingly recognized as promising biomarkers for the early diagnosis of cancers.55
Nanotechnology-enabled CRISPR biosensing
The stable delivery of CRISPR into cells is a prerequisite for monitoring of various biomolecules in the intracellular environment. However, the delivery of CRISPR systems is generally hindered by the large size of Cas proteins and the inefficiency caused by endosomal and lysosomal degradation pathways.63 Once delivered, CRISPR systems also face the risk of enzymatic or RNA degradation and interference owing to their highly complex intracellular environment.19 This limitation critically affects sensor performance, motivating ongoing research on nanomaterials that exhibit high biocompatibility, efficient loading capacity, stability, and specific catalytic activity.64Achieving stable delivery of the CRISPR system using nanoparticles also contributes to mitigating off-target effects. CRISPR inherently exhibits off-target activity, in which non-target sequences are mistakenly recognized, cleaved, and activated. To reduce such effects, extensive efforts have been made to engineer Cas proteins and guide RNAs (gRNAs).65 Additionally, delivering CRISPR in the form of ribonucleoproteins (RNPs) or as a combination of mRNA and sgRNA can further improve specificity.66 However, these approaches are often limited by challenges in delivery due to the large size and negative charge of the components. Therefore, technologies that protect CRISPR systems and enable their efficient delivery to the desired intracellular locations using nanoparticles are essential for successful in vivo biosensing applications.67
Furthermore, when the CRISPR system recognizes targets and generates signals inside cells, the signal-to-noise ratio (SNR) is often compromised because of the complex intracellular environment. To address this issue, it is necessary to integrate various nanomaterials capable of enhancing optical readout signals. This chapter examines the commonly used nanocarriers for intracellular delivery and evaluates their potential for CRISPR/Cas system delivery. We also explored different types of nanomaterials designed to amplify the signal intensity within cells and reviewed how these materials have been integrated with CRISPR systems in ex vivo models.
Delivery enhancement of CRISPR components
For intracellular gene editing or target recognition, specific components should be delivered across the cell membrane and, in the case of gene editing, be taken up by the nucleus. The materials commonly employed for this purpose include metallic nanomaterials, DNA, and lipid-based biomolecules. These carriers should exhibit low cytotoxicity, allow the efficient loading of various probes for intracellular sensing, and maintain stability within the cellular environment. Optimizing nanocarriers for the delivery of the CRISPR system ensures its efficient activation, which has a significant impact on the specificity and sensitivity of biosensing.Thus, the potential of gold nanomaterials as delivery vectors for CRISPR systems has been demonstrated. Yuan et al. proposed a delivery strategy in which both the CRISPR system and the signal probe were immobilized onto AuNPs, thereby ensuring intracellular transport and effective activation of the sensing mechanism.73 Mout et al. employed arginine-functionalized AuNPs to co-deliver CRISPR-Cas9 and sgRNA, thereby reducing the risk of immunogenic responses typically associated with plasmid-based delivery. This strategy enabled efficient gene editing within HeLa cells, in which the nanocomplexes persisted for up to 30 h without compromising cell viability (Fig. 2).74
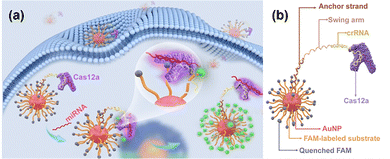 | ||
| Fig. 2 Imaging of specific miRNA using CRISPR nanorobot. (a) Schematic of the detection system operated by CRISPR immobilized onto AuNPs. (b) Structure of a nanorobot for localization and enhancing detection efficiency. Reproduced from ref. 73 with permission from the American Chemical Society, copyright 2024. | ||
Accordingly, DNA nanostructures have also been explored for CRISPR system delivery. Tang et al. engineered a DNA origami template with PAM-rich regions to promote higher Cas9 loading and designed the origami structure with lockable features, enabling encapsulation of sgRNA/Cas9 complexes during intracellular delivery for more efficient transport.80 Similarly, Xu et al. developed a DNA origami nanocage (DONC) capable of confining Cas9 RNPs within its internal space. This structure incorporates two chain locks that are selectively released by ATP and miR-21, allowing the complexes to be protected during delivery and subsequently released upon target recognition.81 Such DNA nanostructure-based delivery systems can be designed in diverse forms, and additional sequence designs can introduce multiple functionalities to ensure CRISPR activation at desired intracellular locations.
Nevertheless, there are limitations in achieving sufficient sensitivity for the detection of targets present at extremely low concentrations in living cells.55
However, when using the aforementioned ionizable cationic lipids, cargoes may not be effectively encapsulated at neutral pH due to the lack of charge. Wei et al. addressed this issue by incorporating additional cationic lipids that remain positively charged at neutral pH, preserving the tertiary structure and stability of Cas9/sgRNA RNPs during delivery. Using this LNP-based delivery system, they achieved selective gene editing in liver and lung tissues in mice.86 Chen et al. employed a thermally stable, highly negatively charged iGeoCas9 mutant and optimized the LNP formulation to achieve tissue-specific therapeutic gene editing. These LNPs contain biodegradable and acid-degradable lipids, which help ensure the stable delivery of the CRISPR system in vivo.87
Signal amplification and readout platforms
Optical readout methods convert biological interactions into optical signals94 and are widely employed for rapid and straightforward sensing and monitoring owing to non-ionizing radiation and high resolution.95,96 Various nanomaterials with unique optical surface properties can further enhance signals and reduce background noise, thereby enabling sensitive detection. Representative optical detection methods used in intracellular sensing include fluorescence and Raman spectroscopy.QDs are nanocrystalline semiconductors that exhibit quantum confinement effects and provide unique optoelectronic properties.98 They typically exhibit high quantum efficiency, strong photostability, broad excitation ranges, and narrow emission peaks.99,100 A key advantage of QDs is that their emission wavelengths can be finely tuned by controlling the particle size and composition, covering the visible to NIR spectrum.101 This NIR window reduces the light scattering in biological tissues, thereby improving the resolution.96 Another NIR-compatible fluorescent material, upconversion nanoparticles, sequentially absorb NIR photons and emit a single photon of higher energy, typically in the visible region.102 UCNPs offer adjustable size, low fluorescence background, deep tissue penetration, low cytotoxicity, and strong photochemical stability.103,104 By adjusting the type and concentration of dopants, photophysical properties such as the emission wavelength, intensity, and lifetime can be tailored, enabling the design of detection systems for specific biological targets.105–108 UCNPs absorbing 980 nm NIR energy can be further modified with Nd3+-doped shells to respond to a more tissue-compatible 808 nm laser. Gong et al. employed UCNPs as carriers and activators of DNA to analyze miR-21 in living cells and designed peanut-shaped UCNPs for efficient tumor cell delivery. In this study, the fabricated PUCNPs-NH2/PEG-ZL-DNA successfully detected targets in cells under 808 nm light.11
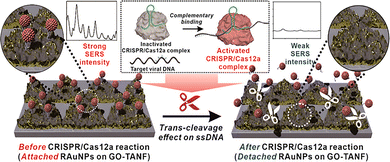 | ||
| Fig. 3 Schematic of RAuNPs immobilized GO/TANF array. Reproduced from ref. 113 with permission of the American Chemistry Society, copyright 2021. | ||
Notably, several studies have demonstrated the use of MEF for intracellular biomarker detection. Choi et al. sensitively detected caspase-3, a biomarker for various diseases, inside cells by dual-conjugating AuNPs with fluorophore via a peptide and single-stranded DNA (ssDNA). Verification of this system in MCF-7 cells showed that it did not affect cell viability and produced strong fluorescence emission even at low intracellular target concentrations.123 Gold nanorods conjugated with fluorescently labeled nucleic acid probes can be delivered into cells in a quenched state; upon target binding, probe unfolding induces fluorescence amplification through MEF, enabling intracellular target detection.71 Similarly, molecular beacons conjugated to gold nanorods have been delivered into stem cells to monitor miR-124 in real time, allowing for the observation of neuronal differentiation in human iPSC-derived neural stem cells.72 Silver nanomaterials provide higher plasmonic frequencies and stronger MEF effects than gold nanomaterials, making them suitable for ultrasensitive single-molecule detection. Their performance can be further enhanced through the structural engineering of nanocubes, nanowires, or nanoplatelets, although surface stabilization often requires protective coatings.124–126
GO, a carbon-based nanomaterial, possesses intrinsic properties such as flexibility and biocompatibility, which have made it widely utilized in the development of biosensors.127–129 The oxygen-containing functional groups of GO, including epoxides, hydroxyls, and carboxyls, confer hydrophilicity, negative surface charge, and chemical versatility, enabling interactions with biomolecules via electrostatic forces, hydrogen bonding, and π–π stacking.130–132 Moreover, the amphiphilic, nanoscopic, and high surface area properties of GO promote cell adhesion, enhancing cellular attachment in substrate-based differentiation assays and contributing to improved sensor stability.133 These properties also allow efficient immobilization of multiple biorecognition elements on a single sheet, facilitating multiplexed detection and preconcentration of low-abundance targets from complex samples.134,135 Surface passivation strategies further reduce nonspecific adsorption, enhancing selectivity. These properties make GO an effective platform for simultaneous detection of multiple biomarkers, which is particularly valuable in analyzing complex disease states.135,136 Moreover, GO can quench fluorescence through FRET, thereby enabling highly sensitive detection in fluorescence-based sensing applications.
These materials can be applied in combinations of two or more components to enhance sensitivity while maintaining stability or to generate hotspots that maximize the signal output. Mahani et al. developed a quenching-based fluorescent probe with high selectivity, sensitivity, and low toxicity by combining AuNPs with sulfhydryl-functionalized Cu-doped carbon quantum dots (S-Cu@CQD) and successfully imaged GSH in breast cancer cells.61 Plasmonic nanoassemblies utilizing hotspots induced by the simultaneous use of AuNPs and AuNRs enabled the sensitive detection of telomerase (TE) and miR-21 within cells, facilitating early cancer detection. In addition to combining two distinct materials, hotspot generation has also been achieved through core–shell particle designs. Xu et al. employed Au@4MBN@AuNPs, in which the Raman reporter molecule 4MBN was inserted between the metal core and the shell while using ROX particles for signal generation, significantly enhancing the ROX Raman signals and improving the accuracy and reliability of quantification (Fig. 4).4
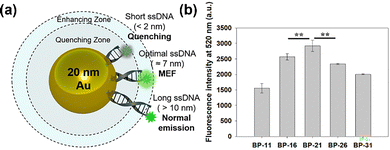 | ||
| Fig. 4 Optimization of the distance between AuNPs and fluorescence. (a) Schematic of the MEF phenomenon depending on the distance from AuNP surface. (b) Fluorescence intensity data for MEF verification based on the length of sequences. Reproduced from ref. 121 with permission of the American Chemical Society, copyright 2021. | ||
Applications in live-cell and single-cell biosensing
Multiplexed analysis of intracellular targets
Assessing multiple variables simultaneously within a single cell is considered the key to understanding complex cellular functions.137 Wang et al. developed a CRISPR/Cas9 system-based fluorescent probe for chromatin imaging of living cells. By labeling sgRNA with a fluorophore, they demonstrated the ability to image multiple targets simultaneously using different colors.138 Extensive efforts have been devoted to imaging RNA within living cells because this approach enables a deeper understanding of cellular functions and provides new insights into disease therapy. Conventional RNA imaging strategies often require multiple fluorescent tags that may perturb normal cellular physiology. Jia et al. developed an RNA-imaging platform based on the dCas12a orthogonal system by combining a crRNA-based conformational switch and a controllable CRISPR activation (CRISPRa) system. This method enables simultaneous detection of cancer-related HER2 and survivin mRNAs in living cells.49 Ke et al. developed a hybrid-engineered nuclease, deactivated AsCas12a (dAsCas12a), and hairpin-structured crRNA (h-CAP) to enhance the specificity and efficiency of Cas12a, enabling multiplex gene expression regulation in human cells.139 Shen et al. established a CRISPR-Cas13a platform combined with T7 transcriptional amplification for simultaneous detection of FEN1 and APE1 using an AND logic gate. FEN1 is the central enzyme in the base excision repair (BER) pathway, whereas APE1 initiates downstream repair processes. Concurrent detection of both enzymes reduces false-positive errors in tumor cell identification.59Disease biomarker sensing in living cells
Recently, CRISPR has been frequently applied for the sensitive detection and imaging of cancer biomarkers within the intracellular environment, often for visualizing the tumor microenvironment (TME). Intracellular RNA targeting may cause non-specific cleavage events that can induce cytotoxicity. Chen et al. addressed this issue by engineering a modified CRISPR-Cas12a system and introducing artificial bubble structures into the seed region to eliminate the PAM sequence dependency on target recognition and cis-cleavage activity. This optimized system was validated for telomerase, ATP, and miR-21 detection. Sufficient trans-cleavage activity in living cells was confirmed using confocal laser scanning microscopy combined with a fluorescence-quencher reporter. Delivery was achieved using a commercial transfection reagent without any assistance.51 Considering the complexity of immune responses and the heterogeneity of the tumor microenvironment, Liu et al. designed a CRISPR/Cas12a-based system for sensitive detection of IFN-γ. Conventional antibody-based probes are often limited by background interference and poor tumor penetration. To overcome these limitations, the system incorporates an NIR photoactivatable aptamer, a CRISPR module, and UCNPs functionalized with cationic polymers for electrostatic assembly and intracellular delivery. The CRISPR system followed an AND logic design, being activated only in the presence of both light and IFN-γ, which enabled quantitative in vivo imaging of exogenous IFN-γ in a PBMC-engrafted mouse model.57 Zhang et al. reported a fluorescent CRISPR (fCRISPR) system that enables multicolor genome imaging. This system employs fluorogenic proteins intrinsically unstable owing to a peptide-derived degron domain (tDeg), leading to rapid degradation. Binding to a specific RNA hairpin stabilizes proteins, allowing fluorescence emission and dynamic RNA visualization.44Moreover, for the efficient operation of CRISPR systems within living cells, sufficient metal ions, such as Mg2+, Mn2+, and Co2+, are essential to support trans-cleavage activity. Two-dimensional nanosheet-based nanomaterials have also been used for CRISPR-Cas12a delivery. Liu et al. used cobalt oxyhydroxide (CoOOH) nanosheets, which are positively-charged layered materials, to detect intracellular miR-21. In the absence of ascorbic acid (AA), steric hindrance prevents Cas12a activation; however, intracellular AA reduces CoOOH to Co2+, relieving the steric blockade. Co2+ ions further act as cofactors to enhance Cas12a catalytic activity, thereby improving the detection efficiency.19 Similarly, Wang et al. synthesized ultrathin MnO2 nanosheets capable of binding to CRISPR-Cas12a components through π–π interactions. With their large surface area, excellent biocompatibility, and low cytotoxicity, MnO2 nanosheets are suitable for biosensing applications. Inside cells, the nanosheets are reductively degraded by endogenous biomolecules, releasing the CRISPR system for target detection.140 MnO2 nanosheets are reduced by GSH to produce Mn2+, which in turn enhances the trans-cleavage activity of CRISPR/Cas12a, thereby improving sensitivity (Fig. 5).141
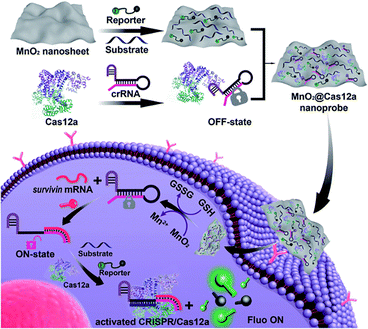 | ||
| Fig. 5 Schematic of live-cell biosensing analysis of mRNA using MnO2 as the carrier and accelerator of CRISPR/Cas12a. This image reproduced from Wang et al.140 and licensed under a Creative Commons Attribution 3.0 Unported Licence. Copyright 2022, Royal Society of Chemistry. | ||
Single-cell CRISPR biosensing
Real-time high-resolution imaging is essential for understanding the diverse dynamic behaviors of individual RNA molecules in a single cell. However, single-molecule live-cell imaging technologies capable of tracking RNA and DNA at a single-molecule resolution enable the investigation of biological phenomena that are difficult to capture through bulk analysis. To address this, research has been conducted using single-molecule live-cell fluorescence in situ hybridization (smLiveFISH), which combines the CRISPR-Csm complex with multiplexed gRNAs. When applied to RNA labeling in cells, this approach provides sufficient signal intensity while reducing the high background noise that commonly occurs in conventional Cas system-based methods. This system successfully tracks individual native NOTCH2 and MAP1B transcripts in living cells.142Another study developed a CRISPR-mediated fluorescence in situ hybridization amplification (CRISPR FISHer) system to image specific DNA sequences in live cells. This method was used to record the movements of extrachromosomal circular DNA (eccDNA) and invading DNA, enabling the analysis of dynamic behavioral differences between the chromosomal and extrachromosomal loci.43 This study enabled real-time imaging based on Cas9, allowing a deeper understanding of biological events within living cells. Furthermore, by facilitating the investigation of invading DNA replication and innate immune responses at the single-cell level, it holds potential to aid biomedical diagnostics and clinical therapeutics.
scCLEAN was developed to enhance the biological resolution of low-abundance transcripts. This system increases the complexity of lowly expressed molecules, thereby improving the sensitivity. Using this approach, scCLEAN enables the discrimination of subtle transitional states within a homogeneous cell population, highlighting its potential to advance single-cell methodologies.143 This study achieved enhanced resolution of biological signals and provides a promising approach for advancing single-cell methodologies.
Perturb-FISH combines CRISPR perturbation with in situ gRNA detection, providing simultaneous spatial transcriptomic information at the single-cell level and revealing the molecular changes associated with specific genetic perturbations.144
CRISPR-based single-cell detection and imaging approaches have made it possible to monitor the dynamic behavior of RNAs, DNAs, and other biomolecules sensitively and precisely, thus serving as powerful tools for advancing our understanding of complex biological processes within individual cells.
Challenges and future perspectives
The integration of CRISPR/Cas systems with nanomaterial-based biosensors has ushered in a new era of intracellular diagnostics and therapeutic monitoring. However, several challenges remain to be addressed to fully realize their potential.Achieving precise delivery of CRISPR components to target cells remains a critical hurdle. Recent advancements in nanomaterial engineering have shown promise in enhancing delivery efficiency and specificity. These innovations aim to minimize off-target effects and improve therapeutic outcomes.
Biocompatibility of nanomaterials is important for their clinical applications. Ongoing research focuses on optimizing surface modifications to reduce immunogenic responses and enhance biocompatibility. The dynamic nature of the cellular environment poses challenges for the stability of biosensors. Strategies to improve sensor longevity include the development of robust nanomaterial coatings and the incorporation of protective biomolecular layers to shield the sensors from enzymatic degradation and environmental fluctuations. Enhancing the SNR is crucial to biosensor sensitivity. The path to clinical applications involves navigating complex regulatory landscapes. Standardization of fabrication processes, rigorous safety evaluations, and alignment with regulatory frameworks are essential steps. Additionally, the scalability of production methods is necessary to meet clinical demands.
Strategies to address these limitations are currently being actively explored. For instance, recent studies have employed nucleus-specific photoactivatable CRISPR system to monitor metal ions in vivo, and further incorporated an AND logic gate strategy to achieve more specific imaging.145 Enabling such precise spatiotemporal control contributes to accurate monitoring of the target. Moreover, the integration of CRISPR/Cas-based biosensors into wearable and implantable devices is a promising strategy for continuous health monitoring. Recent developments in microfluidic systems and flexible electronics have facilitated the creation of noninvasive real-time biosensing platforms for personalized healthcare. CRISPR/Cas-based biosensors have the potential to revolutionize personalized medicine by enabling the real-time monitoring of therapeutic targets and disease biomarkers. This capability allows for tailored treatment strategies and timely adjustments to therapeutic regimens. The application of artificial intelligence and machine learning algorithms to biosensor data can enhance the data interpretation and decision-making processes. These technologies enable the analysis of complex datasets, leading to improved diagnostic accuracy and predictive modeling in clinical settings.
Conclusions
Recent developments in CRISPR-based intracellular biosensing, combined with advanced nanomaterials, have significantly improved the sensitivity, specificity, and versatility of molecular detection. Gold and lipid nanoparticles, DNA nanostructures, and smart plasmonic assemblies have enabled the efficient delivery of CRISPR components into live cells, while minimizing cytotoxicity and off-target effects. Concurrently, the integration of optical readouts such as fluorescence and Raman spectroscopy with plasmonic enhancement strategies has enhanced SNR, allowing real-time monitoring of diverse cellular analytes. These advancements collectively establish a robust foundation for next-generation diagnostic and therapeutic applications, enabling personalized and responsive healthcare solutions that leverage CRISPR-nanotechnology biosensing.Author contributions
Conceptualization, M. C., J-H. C., and J-W. C.; investigation, M. C. and C. L.; writing – original draft, M. C. and C. L.; writing – review and editing, M. C. and J-H. C.; supervision, J-H. C. and J-W. C.; project administration, J-H. C. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT)(RS-2024-00344633), and by GRDC Cooperative Hub through the National Research Foundation of Korea funded by the Ministry of Science and ICT (Grant number RS-2023-00259341), and by National R&D Program through the National Research Foundation of Korea (NRF) funded by Ministry of Science and ICT (NRF-2022M3H4A1A01005271).References
- L.-Z. Yang, Y. Wang, S.-Q. Li, R.-W. Yao, P.-F. Luan, H. Wu, G. G. Carmichael and L.-L. Chen, Mol. Cell, 2019, 76, 981–997.e7 CrossRef CAS PubMed
.
- Y. He, Q. Wang, C. Hong, R. Li, J. Shang, S. Yu, X. Liu and F. Wang, Angew. Chem., Int. Ed., 2023, 62, e202307418 CrossRef CAS
.
- Y. Yin, Z. Li, C. Liu, X.-B. Zhang and Q. Ou, ACS Sens., 2025, 10, 5456–5472 CrossRef CAS
.
- C. Liu, J. Huang, R. Liu, Y. Zhu and T. Xu, Sens. Actuators, B, 2025, 437, 137759 CrossRef CAS
.
- Q.-N. Li, Y.-X. Cui, Z.-Q. Dai, Z.-L. Yao, M.-Y. Li, Q.-L. Cai and D.-M. Kong, Anal. Chem., 2025, 97, 4194–4201 CrossRef CAS
.
- G. Cao, J. Dong, X. Chen, P. Lu, Y. Xiong, L. Peng, J. Li, D. Huo and C. Hou, Chem. Commun., 2022, 58, 6328–6331 RSC
.
-
S. Liu, X. Rao, R. Zhao and W. Han, Front. Genome Ed., 2022, 4, 929929 Search PubMed
.
- Y. Wang, L. Fu, D. Tao, X. Han, B. Xu, M. Deng, S. Li, C. Zhao, X. Li, S. Zhao, P. Gong, Y. Yang, E. M. Khazalwa, Y. Ma, J. Ruan, C. Li and S. Xie, ACS Synth. Biol., 2023, 12, 2051–2060 CrossRef CAS
.
- H. Cao, Y. Wang, N. Zhang, S. Xia, P. Tian, L. Lu, J. Du and Y. Du, Front. Cell Dev. Biol., 2022, 10, 866820 CrossRef
.
- C. Wang, X. Xu, W. Yao, L. Wang, X. Pang, S. Xu and X. Luo, Nano Lett., 2025, 25, 1666–1672 CrossRef CAS
.
- Y. Gong, H. Yang and C. Ding, Biosens. Bioelectron., 2025, 267, 116755 CrossRef CAS
.
- J. Yoon, B. M. Conley, M. Shin, J.-H. Choi, C. K. Bektas, J.-W. Choi and K.-B. Lee, ACS Nano, 2022, 16, 5764–5777 CrossRef CAS PubMed
.
- Z. Dong, R. Su, Y. Fu, Y. Wang and L. Chang, ACS Biomater. Sci. Eng., 2024, 10, 5595–5608 CrossRef CAS
.
- M. Naeem, S. Majeed, M. Z. Hoque and I. Ahmad, Cells, 2020, 9, 1608 CrossRef CAS PubMed
.
- W. Zhang, R. Shi, K. Dong, H. Hu, W. Shu, Y. Mu, B. Yan, L. Li, X. Xiao and H. Wang, Anal. Chem., 2022, 94, 8596–8604 CrossRef CAS PubMed
.
- S. T. Kim, A. Chompoosor, Y. Yeh, S. S. Agasti, D. J. Solfiell and V. M. Rotello, Small, 2012, 8, 3253–3256 CrossRef CAS PubMed
.
- E. M. Clarissa, M. Karmacharya, H. Choi, S. Kumar and Y. Cho, Small, 2025, 2409353 CrossRef
.
- C. Wiraja, Y. Zhu, D. C. S. Lio, D. C. Yeo, M. Xie, W. Fang, Q. Li, M. Zheng, M. Van Steensel, L. Wang, C. Fan and C. Xu, Nat. Commun., 2019, 10, 1147 CrossRef PubMed
.
- Z. Liu, S. Zhou, D. Zhang and M. Yang, Sens. Actuators, B, 2025, 424, 136897 CrossRef CAS
.
- Q. Su, J. Li, M. Fu, R. Sun, J. Chen, F. Xing and L. Sun, Anal. Chem., 2025, 97, 5575–5584 CrossRef CAS
.
- Q. Zhang, J. Li, Y. Li, G. Tan, M. Sun, Y. Shan, Y. Zhang, X. Wang, K. Song, R. Shi, L. Huang, F. Liu, Y. Yi and X. Wu, Biosens. Bioelectron., 2022, 202, 113978 CrossRef CAS PubMed
.
- L. Yang, J.-H. Lee, C. Rathnam, Y. Hou, J.-W. Choi and K.-B. Lee, Nano Lett., 2019, 19, 8138–8148 CrossRef CAS
.
- J. E. van Dongen, J. T. W. Berendsen, R. D. M. Steenbergen, R. M. F. Wolthuis, J. C. T. Eijkel and L. I. Segerink, Biosens. Bioelectron., 2020, 166, 112445 CrossRef CAS
.
- H. Zhao, Y. Sheng, T. Zhang, S. Zhou, Y. Zhu, F. Qian, M. Liu, W. Xu, D. Zhang and J. Hu, Nat. Commun., 2024, 15, 2901 CrossRef CAS
.
- R. Fu and Y. Xianyu, Small, 2023, 19, 2300057 CrossRef CAS PubMed
.
- J.-H. Choi, J. Yoon, M. Chen, M. Shin, L. L. Goldston, K.-B. Lee and J.-W. Choi, Biochip J, 2025, 19, 167–181 CrossRef CAS
.
- L. Wei, Z. Wang, Y. She and H. Fu, Anal. Chem., 2025, 97, 11943–11958 CrossRef PubMed
.
- K. S. Makarova, Y. I. Wolf, J. Iranzo, S. A. Shmakov, O. S. Alkhnbashi, S. J. J. Brouns, E. Charpentier, D. Cheng, D. H. Haft, P. Horvath, S. Moineau, F. J. M. Mojica, D. Scott, S. A. Shah, V. Siksnys, M. P. Terns, Č. Venclovas, M. F. White, A. F. Yakunin, W. Yan, F. Zhang, R. A. Garrett, R. Backofen, J. van der Oost, R. Barrangou and E. V. Koonin, Nat. Rev. Microbiol., 2020, 18, 67–83 CrossRef CAS PubMed
.
- M. Huang, X. Zhou, H. Wang and D. Xing, Anal. Chem., 2018, 90, 2193–2200 CrossRef CAS PubMed
.
- J. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins and F. Zhang, Science, 2018, 360, 439–444 CrossRef CAS PubMed
.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky and J. A. Doudna, Science, 2018, 360, 436–439 CrossRef CAS PubMed
.
- E. Deltcheva, K. Chylinski, C. M. Sharma, K. Gonzales, Y. Chao, Z. A. Pirzada, M. R. Eckert, J. Vogel and E. Charpentier, Nature, 2011, 471, 602–607 CrossRef CAS PubMed
.
- F. X. Liu, J. Q. Cui, Z. Wu and S. Yao, Lab Chip, 2023, 23, 1467–1492 RSC
.
- Q. Zheng, W. Li, L. Mao and M. Wang, Biomater. Sci., 2021, 9, 7024–7033 RSC
.
- J. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy, J. Joung, V. Verdine, N. Donghia, N. M. Daringer, C. A. Freije, C. Myhrvold, R. P. Bhattacharyya, J. Livny, A. Regev, E. V. Koonin, D. T. Hung, P. C. Sabeti, J. J. Collins and F. Zhang, Science, 2017, 356, 438–442 CrossRef CAS PubMed
.
- Y. Chen, X. Wang, J. Zhang, Q. Jiang, B. Qiao, B. He, W. Yin, J. Qiao and Y. Liu, Nat. Commun., 2024, 15, 8342 CrossRef
.
- T. Kasputis, Y. He, Q. Ci and J. Chen, Anal. Chem., 2024, 96, 2676–2683 CrossRef CAS
.
- M. Y. Choi, I. Haizan and J. H. Choi, Sens. Actuators, B, 2025, 439, 137830 CrossRef CAS
.
- J.-Y. Ma, S.-Y. Wang, Y.-C. Du, D.-X. Wang, A.-N. Tang, J. Wang and D.-M. Kong, Anal. Chem., 2022, 94, 8050–8057 CrossRef CAS PubMed
.
- S. R. Rananaware, E. K. Vesco, G. M. Shoemaker, S. S. Anekar, L. S. W. Sandoval, K. S. Meister, N. C. Macaluso, L. T. Nguyen and P. K. Jain, Nat. Commun., 2023, 14, 5409 CrossRef CAS
.
- H. Wu, R. Zhang, H. Lai, L. Lai, Y. Liu, Y. Fan, Y. Zhang, J. Shi and M. Yang, Sens. Actuators, B, 2025, 427, 137120 CrossRef CAS
.
- Q. Ain, C. Schmeer, D. Wengerodt, O. W. Witte and A. Kretz, Int. J. Mol. Sci., 2020, 21, 2477 CrossRef CAS
.
- X.-Y. Lyu, Y. Deng, X.-Y. Huang, Z.-Z. Li, G.-Q. Fang, D. Yang, F.-L. Wang, W. Kang, E.-Z. Shen and C.-Q. Song, Cell Res., 2022, 32, 969–981 CrossRef CAS PubMed
.
- Z. Zhang, X. Rong, T. Xie, Z. Li, H. Song, S. Zhen, H. Wang, J. Wu, S. R. Jaffrey and X. Li, Nat. Commun., 2024, 15, 934 CrossRef CAS
.
- Z. Wei Han, F. Ma and C. Yang Zhang, Anal. Chim. Acta, 2023, 1251, 340996 CrossRef PubMed
.
- M. Dadmehr, M. Hosseini, S. Hosseinkhani, M. R. Ganjali, M. Khoobi, H. Behzadi, M. Hamedani and R. Sheikhnejad, Biosens. Bioelectron., 2014, 60, 35–44 CrossRef CAS PubMed
.
- F. Ma, W. Liu, B. Tang and C. Zhang, Chem. Commun., 2017, 53, 6868–6871 RSC
.
- N. J. Spix, W. A. Habib, Z. Zhang, E. Eugster, H. Milliron, D. Sokol, K. Lee, P. A. Nolte, J. L. Endicott, K. F. Krzyzanowski, T. Hinoue, J. Morrison, B. K. Johnson, W. Zhou, H. Shen and P. W. Laird, Nat. Commun., 2025, 16, 6273 CrossRef CAS
.
- H.-Y. Jia, S.-Y. Yao, Y.-F. Li, B.-C. Ye and B.-C. Yin, Biosens. Bioelectron., 2025, 273, 117185 CrossRef CAS
.
- S. Das, M. Vera, V. Gandin, R. H. Singer and E. Tutucci, Nat. Rev. Mol. Cell Biol., 2021, 22, 483–504 CrossRef CAS PubMed
.
- S. Chen, R. Wang, S. Peng, S. Xie, C. Lei, Y. Huang and Z. Nie, Chem. Sci., 2022, 13, 2011–2020 RSC
.
- L. Li, S. Xu, M. Chen, P. Lu, M. Zhao, J. Jiao and J. Jiao, Chem. Eng. J., 2025, 505, 159310 CrossRef CAS
.
- M. Yang, H. Li, X. Li, K. Huang, W. Xu and L. Zhu, Biosens. Bioelectron., 2022, 209, 114258 CrossRef CAS PubMed
.
- X. You, R. He, S.-Y. Liu and Z. Dai, Biomed. Anal., 2024, 1, 1–27 CrossRef CAS
.
- J.-Y. Tang, M.-L. Zhao, X.-M. Zhou, Y.-Q. Chai, R. Yuan, Y.-M. Lei and Y. Zhuo, Anal. Chem., 2025, 97, 3378–3386 CrossRef CAS
.
- Y. Pan, X. Luan, F. Zeng, Q. Xu, Z. Li, Y. Gao, X. Liu, X. Li, X. Han, J. Shen and Y. Song, Biosens. Bioelectron., 2022, 209, 114239 CrossRef CAS PubMed
.
- Z. Liu, X. Duan, Y. Yun, S. Li, Z. Feng, J. Zhan, R. Liu, Y. Li and J. Zhang, ACS Nano, 2024, 18, 3826–3838 CrossRef CAS
.
- L. Li, L. Huang, M. Li, S. He, H. Xu, C. Lu and C. Xing, Biosens. Bioelectron., 2025, 282, 117501 CrossRef CAS
.
- D. Shen, H. Guo, F. Zhang, X. Chen, X. Tong, H. Li, W. Wu and S. Mei, Biosens. Bioelectron., 2025, 280, 117406 CrossRef CAS PubMed
.
- S. Zhang, W. Xiong, S. Xu and R. Qian, Chemosensors, 2025, 13, 17 CrossRef CAS
.
- M. Mahani and L. Montazer, Luminescence, 2025, 40, e70225 CrossRef CAS
.
- J. Yoon, M. Shin, D. Kim, J. Lim, H.-W. Kim, T. Kang and J.-W. Choi, Biosens. Bioelectron., 2022, 196, 113725 CrossRef CAS PubMed
.
- B.-S. Jeong, H. C. Kim, C. M. Sniezek, S. Berger, J. M. Kollman, D. Baker, J. C. Vaughan and X. Gao, J. Controlled Release, 2025, 381, 113651 CrossRef CAS
.
- S. Xie, B. Xu, R. Tang, S. Chen, C. Lei and Z. Nie, Anal. Chem., 2022, 94, 10159–10167 CrossRef CAS
.
- C. Guo, X. Ma, F. Gao and Y. Guo, Front. Bioeng. Biotechnol., 2023, 11, 1143157 CrossRef
.
- H. Manghwar, B. Li, X. Ding, A. Hussain, K. Lindsey, X. Zhang and S. Jin, Adv. Sci., 2020, 7, 1902312 CrossRef CAS PubMed
.
- J. Padayachee and M. Singh, Nanobiomedicine, 2020, 7, 1849543520983196 CrossRef PubMed
.
- J. Lim, J. Yoon, M. Shin, K. Lee and J. Choi, Small Methods, 2022, 6, 2100912 CrossRef CAS
.
- H. Shaat, A. Mostafa, M. Moustafa, A. Gamal-Eldeen, A. Emam, E. El-Hussieny and M. Elhefnawi, Int. J. Pharm., 2016, 504, 125–133 CrossRef CAS
.
- B. D. Chithrani, A. A. Ghazani and W. C. W. Chan, Nano Lett., 2006, 6, 662–668 CrossRef CAS
.
- Y. Song, J. Soto, X. Lin, T. Hoffman, E. Hu, N. Zhu, J. Zarubova, Y. Wu, J. Tian, P. K. Wong and S. Li, Adv. Funct. Mater., 2025, 35, 2410910 CrossRef CAS
.
- Y. Hou, M. Chen, L. Yang and K.-B. Lee, Nano Lett., 2025, 25, 10402–10411 CrossRef CAS
.
- A. Yuan, R. Sha, W. Xie, G. Qu, H. Zhang, H. Wang, X. C. Le, G. Jiang and H. Peng, J. Am. Chem. Soc., 2024, 146, 26657–26666 CrossRef CAS
.
- R. Mout, M. Ray, G. Yesilbag Tonga, Y.-W. Lee, T. Tay, K. Sasaki and V. M. Rotello, ACS Nano, 2017, 11, 2452–2458 CrossRef CAS
.
- L.-Y. Duan, J.-W. Liu, R.-Q. Yu and J.-H. Jiang, Biosens. Bioelectron., 2021, 177, 112976 CrossRef CAS PubMed
.
- K.-R. Kim, S. J. Kang, A.-Y. Lee, D. Hwang, M. Park, H. Park, S. Kim, K. Hur, H. S. Chung, C. Mao and D.-R. Ahn, Biomaterials, 2019, 195, 1–12 CrossRef CAS
.
- X. Fu, G. Ke, F. Peng, X. Hu, J. Li, Y. Shi, G. Kong, X.-B. Zhang and W. Tan, Nat. Commun., 2020, 11, 1518 CrossRef CAS
.
- Y. Du, P. Peng and T. Li, ACS Nano, 2019, 13, 5778–5784 CrossRef CAS
.
- H. Tan, J. Zhao, S. Liu, R. Yuan and S. Chen, Sens. Actuators, B, 2025, 442, 138067 CrossRef CAS
.
- W. Tang, T. Tong, H. Wang, X. Lu, C. Yang, Y. Wu, Y. Wang, J. Liu and B. Ding, Angew. Chem., Int. Ed., 2023, 62, e202315093 CrossRef CAS PubMed
.
- Z. Xu, Y. Dong, N. Ma, X. Zhu, X. Zhang, H. Yin, S. Chen, J.-J. Zhu, Y. Tian and Q. Min, J. Am. Chem. Soc., 2023, 145, 26557–26568 CrossRef CAS
.
- P. Kazemian, S.-Y. Yu, S. B. Thomson, A. Birkenshaw, B. R. Leavitt and C. J. D. Ross, Mol. Pharm., 2022, 19, 1669–1686 CrossRef CAS
.
- M. Schlich, R. Palomba, G. Costabile, S. Mizrahy, M. Pannuzzo, D. Peer and P. Decuzzi, Bioeng. Transl. Med., 2021, 6, e10213 CrossRef CAS
.
- L. Xu, X. Wang, Y. Liu, G. Yang, R. J. Falconer and C.-X. Zhao, Adv. Nanobiomed. Res., 2022, 2, 2100109 CrossRef CAS
.
- J. M. Johansson, H. Du Rietz, H. Hedlund, H. C. Eriksson, E. Oude Blenke, A. Pote, S. Harun, P. Nordenfelt, L. Lindfors and A. Wittrup, Nat. Commun., 2025, 16, 5354 CrossRef PubMed
.
- T. Wei, Q. Cheng, Y.-L. Min, E. N. Olson and D. J. Siegwart, Nat. Commun., 2020, 11, 3232 CrossRef CAS
.
- K. Chen, H. Han, S. Zhao, B. Xu, B. Yin, A. Lawanprasert, M. Trinidad, B. W. Burgstone, N. Murthy and J. A. Doudna, Nat. Biotechnol., 2025, 43, 1445–1457 CrossRef CAS
.
- S. Chen, M. Triki, S. Pinto Carneiro and O. M. Merkel, Nanoscale, 2025, 17, 6604–6619 RSC
.
- S. Deng, X. Li, S. Liu, J. Chen, M. Li, S. Y. Chew, K. W. Leong and D. Cheng, Sci. Adv., 2020, 6, eabb4005 CrossRef CAS
.
- S. Wang, W. Song, S. Wei, S. Zeng, S. Yang, C. Lei, Y. Huang, Z. Nie and S. Yao, Anal. Chem., 2019, 91, 8622–8629 CrossRef CAS PubMed
.
- G. Frigerio, S. Motta, P. Siani, E. Donadoni and C. Di Valentin, J. Controlled Release, 2025, 379, 344–362 CrossRef CAS PubMed
.
- Y. Tian, Q. Lu, X. Guo, S. Wang, Y. Gao and L. Wang, Nanoscale, 2020, 12, 7776–7781 RSC
.
- F. Li, Z. Zhang, X. Wang, X. Yin, M. Fu, T. Qin, X. Ji, G. Yang and S. Sun, Int. J. Biol. Macromol., 2025, 289, 138875 CrossRef CAS PubMed
.
- K. Duy Mac and J. Su, npj Biosensing, 2025, 2, 20 CrossRef PubMed
.
- B. Kaur, S. Kumar and B. K. Kaushik, Biosens. Bioelectron., 2022, 197, 113805 CrossRef CAS PubMed
.
- S. Liu, W. Dong, H. Gao, Z. Song and Z. Cheng, Acc. Chem. Res., 2025, 58, 543–554 CrossRef CAS PubMed
.
- E. K. Herkert and M. F. Garcia-Parajo, ACS Photon., 2025, 12, 1259–1275 CrossRef CAS PubMed
.
- K. Agarwal, H. Rai and S. Mondal, Mater. Res. Express, 2023, 10, 062001 CrossRef CAS
.
- C. S. M. Martins, A. P. LaGrow and J. A. V. Prior, ACS Sens., 2022, 7, 1269–1299 CrossRef CAS PubMed
.
- K. D. Wegner and N. Hildebrandt, TrAC, Trends Anal. Chem., 2024, 180, 117922 CrossRef CAS
.
- W. Shen, Y. Wang and L. Liao, Laser Photon. Rev., 2025, 19, 2401947 CrossRef
.
- J. Chen and J. X. Zhao, Sensors, 2012, 12, 2414–2435 CrossRef CAS PubMed
.
- J. Máčala, E. Makhneva, A. Hlaváček, M. Kopecký, H. H. Gorris, P. Skládal and Z. Farka, Anal. Chem., 2024, 96, 10237–10245 CrossRef
.
- S. Cheng, B. Huang, K. Lu, J. Li, H. Wen and J. Zhang, Anal. Methods, 2025, 17, 5563–5578 RSC
.
- L. Yang, C. Che, M. Guo, C. Fan, L. Sun and S. Chen, ACS Omega, 2024, 9, 47156–47166 CrossRef CAS PubMed
.
- C. Würth, B. Grauel, M. Pons, F. Frenzel, P. Rissiek, K. Rücker, M. Haase and U. Resch-Genger, Nano Res., 2022, 15, 9639–9646 CrossRef
.
- W. Jiang, J. Yi, X. Li, F. He, N. Niu and L. Chen, Biosensors, 2022, 12, 1036 CrossRef CAS PubMed
.
- D. Huang, F. Li, H. Ågren and G. Chen, Nat. Commun., 2025, 16, 4218 CrossRef CAS
.
- W.-H. Chang, S.-Q. Zhang, Z.-Y. Yang and C.-H. Lin, Nanomaterials, 2025, 15, 64 CrossRef CAS
.
- E. Lenzi, D. Jimenez de Aberasturi and L. M. Liz-Marzán, ACS Sens., 2019, 4, 1126–1137 CrossRef CAS PubMed
.
- C. Zhang, J. Tan, B. Du, C. Ji, Z. Pei, M. Shao, S. Jiang, X. Zhao, J. Yu, B. Man, Z. Li and K. Xu, ACS Appl. Mater. Interfaces, 2024, 16, 12085–12094 CrossRef CAS PubMed
.
- A. Li, X. Mo, Y. Lu, G. Zhu, C. Liu, X. Yang, Y. Huang, J. Sheng, H. Zhang, D. Meng and X. Zhao, Biosens. Bioelectron., 2025, 270, 116973 CrossRef CAS PubMed
.
- J.-H. Choi, M. Shin, L. Yang, B. Conley, J. Yoon, S.-N. Lee, K.-B. Lee and J.-W. Choi, ACS Nano, 2021, 15, 13475–13485 CrossRef CAS PubMed
.
- M. Li, S. K. Cushing and N. Wu, Analyst, 2015, 140, 386–406 RSC
.
- M. P. Singh and G. F. Strouse, J. Am. Chem. Soc., 2010, 132, 9383–9391 CrossRef CAS
.
- H. Yu, Y. Peng, Y. Yang and Z.-Y. Li, NPJ Comput. Mater., 2019, 5, 45 CrossRef
.
- V. Pellas, D. Hu, Y. Mazouzi, Y. Mimoun, J. Blanchard, C. Guibert, M. Salmain and S. Boujday, Biosensors, 2020, 10, 146 CrossRef CAS PubMed
.
- Z. Mei and L. Tang, Anal. Chem., 2017, 89, 633–639 CrossRef CAS
.
- E. Tóth, D. Ungor, T. Novák, G. Ferenc, B. Bánhelyi, E. Csapó, M. Erdélyi and M. Csete, Nanomaterials, 2020, 10, 1048 CrossRef PubMed
.
- D. Chakraborty, A. Mukherjee and K. R. Ethiraj, Anal. Methods, 2022, 14, 3614–3622 RSC
.
- J.-H. Choi, J. Lim, M. Shin, S.-H. Paek and J.-W. Choi, Nano Lett., 2021, 21, 693–699 CrossRef CAS PubMed
.
- J.-H. Lee, J.-H. Choi, S.-T. D. Chueng, T. Pongkulapa, L. Yang, H.-Y. Cho, J.-W. Choi and K.-B. Lee, ACS Nano, 2019, 13, 8793–8803 CrossRef CAS PubMed
.
- J.-H. Choi and J.-W. Choi, Nano Lett., 2020, 20, 7100–7107 CrossRef CAS PubMed
.
- Y. Saad, M. H. Gazzah, K. Mougin, M. Selmi and H. Belmabrouk, Plasmonics, 2022, 17, 1489–1500 CrossRef CAS PubMed
.
- B. J. Yun, J. E. Kwon, K. Lee and W. G. Koh, Sens. Actuators, B, 2019, 284, 140–147 CrossRef CAS
.
- S. K. Gahlaut, A. Pathak and B. D. Gupta, Biosensors, 2022, 12, 713 CrossRef CAS PubMed
.
- J. Yoon, T. Lee, G. B. Bapurao, J. Jo, B.-K. Oh and J.-W. Choi, Biosens. Bioelectron., 2017, 93, 14–20 CrossRef CAS PubMed
.
- T. Ha, S. Park, M. Shin, J.-Y. Lee, J.-H. Choi and J.-W. Choi, Chem. Eng. J., 2023, 463, 142284 CrossRef CAS
.
- J. Yoon, S. N. Lee, M. K. Shin, H.-W. Kim, H. K. Choi, T. Lee and J.-W. Choi, Biosens. Bioelectron., 2019, 140, 111343 CrossRef CAS PubMed
.
- W. Yu, L. Sisi, Y. Haiyan and L. Jie, RSC Adv., 2020, 10, 15328–15345 RSC
.
- A. Jiříčková, O. Jankovský, Z. Sofer and D. Sedmidubský, Materials, 2022, 15, 920 CrossRef
.
- W. Gul and H. Alrobei, Polymers, 2021, 13, 1818 CrossRef CAS PubMed
.
- J. Lee, H. K. Choi, L. Yang, S. D. Chueng, J. Choi and K. Lee, Adv. Mater., 2018, 30, 1802762 CrossRef
.
- A. Baruah, R. Newar, S. Das, N. Kalita, M. Nath, P. Ghosh, S. Chinnam, H. Sarma and M. Narayan, Discover. Nano, 2024, 19, 103 CrossRef CAS PubMed
.
- Y. Zhang, X. Chen, S. Yuan, L. Wang and X. Guan, Anal. Chem., 2020, 92, 15042–15049 CrossRef CAS
.
- Y. Wu, C. Wang, C. Wang, P. Wang, X. Chang, L. Han and Y. Zhang, Micromachines, 2022, 13, 2046 CrossRef PubMed
.
- C. Chen, Z. Zhao, N. Qian, S. Wei, F. Hu and W. Min, Nat. Commun., 2021, 12, 1–13 CrossRef
.
- S. Wang, Y. Hao, L. Zhang, F. Wang, J. Li, L. Wang and C. Fan, CCS Chem., 2019, 1, 278–285 CrossRef CAS
.
- Y. Ke, S. Zhang, Y. Gao, X. Chen, J. He, Y. Chen, Q. Zhang and X. Ding, Anal. Chem., 2024, 96, 2637–2642 CrossRef CAS PubMed
.
- D.-X. Wang, Y.-X. Wang, J. Wang, J.-Y. Ma, B. Liu, A.-N. Tang and D.-M. Kong, Chem. Sci., 2022, 13, 4364–4371 RSC
.
- J. X. Liu, X. M. Sun, D. Liu, Y. H. Liu and C. Y. Li, Biosens. Bioelectron., 2022, 216, 114646 CrossRef CAS PubMed
.
- C. Xia, D. Colognori, X. S. Jiang, K. Xu and J. A. Doudna, Nat. Biotechnol., 2025, 1–8 Search PubMed
.
- A. C. Pandey, J. Bezney, D. DeAscanis, E. B. Kirsch, F. Ahmed, A. Crinklaw, K. S. Choudhary, T. Mandala, J. Deason, J. S. Hamidi, A. Siddique, S. Ranganathan, K. Brown, J. Armstrong, S. Head, P. Ordoukhanian, L. M. Steinmetz and E. J. Topol, Nat. Commun., 2025, 16, 4664 CrossRef CAS PubMed
.
- L. Binan, A. Jiang, S. A. Danquah, V. Valakh, B. Simonton, J. Bezney, R. T. Manguso, K. B. Yates, R. Nehme, B. Cleary and S. L. Farhi, Cell, 2025, 188, 2141–2158.e18 CrossRef CAS PubMed
.
- R. Liu, D. Jiang, Y. Yun, Z. Feng, F. Zheng, Y. Xiang, H. Fan and J. Zhang, Angew. Chem., Int. Ed., 2024, 63, e202315536 CrossRef CAS PubMed
.
Footnote |
| † The authors equally contributed to this work. |
| This journal is © The Royal Society of Chemistry 2026 |




