Open Access Article
Open Access ArticleOn the stability of acid-soluble pea protein-stabilized beverage emulsions against salt addition and heat treatment†
Maryam Nikbakht Nasrabadia,
Michael N. A. Eskinb,
Usha Thiyam-Hollanderb and
Supratim Ghosh *a
*a
aDepartment of Food and Bioproduct Sciences, College of Agriculture and Bioresources, University of Saskatchewan, 51 Campus Drive, Saskatoon, SK S7N 5A8, Canada. E-mail: supratim.ghosh@usask.ca
bDepartment of Food and Human Nutritional Sciences, University of Manitoba, 35 Chancellor's Circle, Winnipeg, MN, Canada
First published on 3rd June 2025
Abstract
This study explores the use of an acidic water-soluble fraction of pea protein as an effective plant-based emulsifier for stabilizing low-pH oil-in-water emulsions. Mildly fractionated soluble pea protein (containing 2.5 wt% protein), recovered by centrifuging pea protein concentrate dispersion at pH 2.0, was directly used to prepare a 5 wt% canola oil-in-water emulsion using a high-pressure homogenizer. Emulsion stability was evaluated at pH 2.0 for 28 days at room temperature as well as against environmental stress conditions, including heat treatment (90 °C, 30 min) in the presence and absence of NaCl (0–1 M). The results showed that the acidic water-soluble pea protein, rich in albumins, exhibited excellent emulsifying properties and improved thermal stability, showing minor changes in droplet size until day 28, even after heat treatment. We proposed that such improved emulsion thermal stability under acidic conditions could be due to the high albumin content, lower surface hydrophobicity and higher β-sheet and random coil secondary structure content of the acidic-soluble fraction compared to the whole pea protein pH 7.0 extracts. However, the emulsions were unstable against high salt concentrations (0.5 and 1 M NaCl), showing extensive aggregation worsened by heating, which was attributed to the charge screening effect of salt and unfavourable conformational change in the albumin-rich proteins. The novel findings of this study can provide essential knowledge and set the stage for the development of pea protein ingredients ideal for utilization in ready-to-drink acidic plant-based beverages.
1. Introduction
A wide variety of food products contain both oil and water, with one phase dispersed within the other. In oil-in-water (O/W) emulsions (e.g., milk, coffee creamer and beverages), oil droplets, stabilized by hydrophilic emulsifiers, are dispersed in an aqueous phase. Synthetic small-molecule emulsifiers such as polysorbates are widely used as water-soluble emulsifying agents. However, due to their health concerns, such as the issues with intestinal barrier permeability during digestion, finding an appropriate alternative for them is highly encouraged.1 Plant-derived materials have recently attracted increasing attention in the food industry owing to their advantages over their animal-based counterparts, including a lower risk of obesity,2,3 and cardiometabolic diseases,4 no limitations in terms of cultural and religious food habits,5 and their economic and environmental advantages.6 Plant-derived proteins are used as functional ingredients with various roles in food formulations, including thickening and gelling agents, stabilizers of emulsions and foams, and binding agents for fat and water.7 Recently, proteins extracted from pulses (such as peas, lentils, and fava beans) have rapidly grown in demand for various food applications. Among the various pulse proteins, pea protein is the most popular and relatively cheaper.8 However, similar to other plant proteins, pea proteins are intractable due to their poor aqueous solubility and sensitivity to pH, salt addition and temperature, which limit their applications.9 Therefore, finding a way to modulate and improve their functionality is highly sought after.Pea seeds are processed into three categories based on protein purity to obtain pea proteins. Pea flour is the milled pea seeds containing about 20–30% proteins.10 Dry fractionation of pea flour produces pea protein concentrate (PPC), where the protein-rich fraction is separated from the starch and fiber components using air classification.11,12 Wet fractionation, the primary process leading to high-purity protein isolates, is based on protein solubilization at high alkaline pH, followed by isoelectric precipitation and centrifugal separation. Recently, a milder route of fractionations compared to conventional wet isolation methods has been proposed, resulting in lower side streams, water footprint and energy consumption, which makes it favourable in terms of cost, environment and sustainability.13 Moreover, it has been reported that the mildly fractionated proteins with lower purities showed similar or even superior functionalities compared to their extremely fractionated and highly purified counterparts due to the preservation of the protein's native properties.14,15 The protein composition can also be tuned by designing and optimizing fractionation routes to achieve specific functionalities.16,17 For instance, in isoelectric precipitation, much of the albumin fraction is lost,15 which can be recovered during a milder fractionation process.18
Most of the previous studies on mild fractionation of pea proteins have evaluated the neutral19 or alkali-soluble fractions.20,21 However, these fractions exhibit limitations in maintaining appropriate solubility and emulsion stability in acidic environments.19 Others have changed the pH of a globulin-rich pea protein isolate (extracted via isoelectric precipitation) to pH 3 before making an emulsion and proposed that a combination of particle-based Pickering and molecular proteins is responsible for the emulsifying property under an acidic environment.22,23 However, there is no knowledge of the emulsification ability and emulsion stability of albumin-rich acidic pea protein fractions under various environmental stresses. Therefore, this study aimed to mildly fractionate pea protein concentrate at an acidic pH 2.0 and utilize the acidic water-soluble pea protein (ASPP) fraction for stabilizing oil-in-water emulsions also in an acidic condition (pH 2.0) appropriate for beverage applications. The stability of these acidic emulsions in the presence of various levels of salt (NaCl) and heat treatment appropriate for beverage processing was also investigated. Moreover, the compositional and structural properties of the ASPP were also evaluated in relation to their functional properties and emulsion stabilization ability under various environmental stresses. The knowledge generated will improve our mechanistic understanding of the functionality of albumin-rich acidic pea proteins and may provide a promising way to manufacture stable acidic beverage emulsions using plant proteins.
2. Materials and methods
2.1 Materials
Pea protein concentrate (PPC) with 51.4 ± 0.5% protein, 34.3 ± 1.0% carbohydrate, 2.1 ± 0.4% lipid, 5.4 ± 0.3% ash, and 6.8 ± 0.7% moisture content24 was kindly donated by AGT Food and Ingredients (Saskatoon, SK, Canada). Canola oil was purchased from a local supermarket in Saskatoon, SK, Canada. Milli-Q™ water (Millipore Corporation, Burlington, MA, USA) was used to prepare reagents for the protein assays and all other experiments. All acids and bases were obtained from Thermo Fisher (Edmonton, AB, Canada). All the other chemicals were purchased from Sigma-Aldrich (Mississauga, ON, Canada).2.2 Extraction and isolation of acidic water-soluble fraction of pea protein
PPC (7 wt% powder-based) was dispersed in deionized water with the pH left unadjusted (∼pH 6.4) and stirred overnight at room temperature for complete hydration. The dispersion was adjusted to pH 2.0 by adding a few drops of 1 M HCl, stirred for another 2 h, followed by centrifugation at 4000 rpm (3220 g) for 20 min (Eppendorf Centrifuge 5810/5810R, Mississauga, ON, Canada). The supernatant was collected as the acid-soluble pea protein (ASPP) fraction, which was directly used for emulsion preparation. A portion of the ASPP solution was freeze-dried using a freeze drier (Labconco, Kansas City, MO), and the powder was kept at 4 °C for further experiments.2.3 ASPP characterization
 | (1) |
2.4 Determination of emulsifying properties
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, followed by high-pressure homogenization (EmulsiFlex-C3, Avestin Inc., Ottawa, ON, Canada) at a pressure of 20
000 rpm, followed by high-pressure homogenization (EmulsiFlex-C3, Avestin Inc., Ottawa, ON, Canada) at a pressure of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 psi (137.9 MPa) for six cycles. About 200 g of emulsions were made for each replicate from a freshly extracted ASPP solution. The emulsions were stored in 50 ml glass vials at room temperature (23 ± 2 °C) for further analysis and visual observation. If required, the emulsion pH was re-adjusted to pH 2.0 with a few drops of 1 M HCl before storage.
000 psi (137.9 MPa) for six cycles. About 200 g of emulsions were made for each replicate from a freshly extracted ASPP solution. The emulsions were stored in 50 ml glass vials at room temperature (23 ± 2 °C) for further analysis and visual observation. If required, the emulsion pH was re-adjusted to pH 2.0 with a few drops of 1 M HCl before storage.2.5 Statistics
ASPP extraction and ASPP-stabilized emulsions were prepared with at least three independent replicates. All measurements were carried out with three replications, and the results are reported as the mean and standard deviation. The experimental data were subjected to a general linear model or one-way analysis of variance (ANOVA) with Tukey's post hoc test using a 95% confidence level where p < 0.05 indicates a significant difference. The statistical analysis was done using Microsoft Excel.3. Results and discussion
3.1 Protein content and protein yield of the mild extraction process
The ASPP was separated from PPC (55.2 ± 0.5% protein content, dry basis) using centrifugation at pH 2.0. The protein content of freeze-dried ASPP was 78.2 ± 1.0% (dry basis). The protein yield (eqn (1)) in ASPP was 66.0 ± 2.6%, which means of the total protein in the PPC, about 66% was successfully extracted into the acidic soluble fraction after dispersing and centrifuging PPC at pH 2.0.3.2 Protein profile of ASPP using SDS PAGE
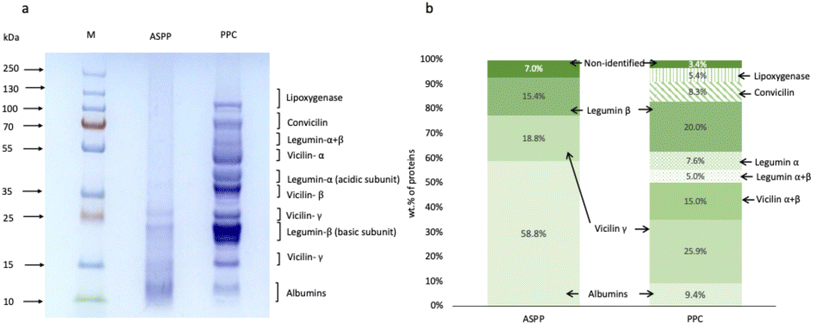
The protein profile of PPC and the extracted ASPP were analyzed by SDS-PAGE (Fig. 1a). The PPC showed all bands expected in pea proteins. Two significant protein fractions in pea protein are albumins and globulins. Albumins are water soluble fractions, relatively smaller in size than the globulins and fall in the molecular weight range 26, 14 and 6 kDa. Globulins are composed of multiple subunits of 11S legumin (42, 23 and 19 kDa), 7S vicilin (50, 36, 28, and 14 kDa), and 7S convicilin (75 kDa).30 As shown in Fig. 1a, the albumin fraction remained in the ASPP after the isolation process at pH 2.0. At lower pH levels, albumins' solubility is relatively higher than other protein fractions, leading to their enrichment in the supernatant during centrifugation.31,32 In contrast, other pea protein fractions, such as lipoxygenase, convicilin, most of the vicilin (vicilin α + β) and the acidic subunit of legumin, disappeared from the ASPP. To better understand the recoverable albumin in ASPP, densitometry analysis was performed on the SDS PAGE profile (Fig. 1b). Albumin content increased from 9.4% in the PPC to 58.8% in the ASPP. The composition of the ASPP was 58.8% albumins, 18.8% vicilin γ, 15.4% legumin β, and the rest 7.0% were non-identified fractions (Fig. 1b). The vicilin α + β and vicilin γ content decreased from 15.0 and 25.9% in PPC to 0% and 18.8% in ASPP, respectively, while the legumin α, legumin α + β and convicilin disappeared in ASPP. Therefore, most of the globulins were separated from PPC during the extraction of ASPP. This could be due to their higher molecular weight, lower solubility, aggregation and precipitation out of the solution as a consequence of the disruption of their native structure and charge distribution under acidic conditions.33,34 Among the disappeared protein fractions is lipoxygenase, an enzyme associated with hexanal production and beany flavour in most of the pulses,35,36 whose removal can be exploited during industrial processing to improve the flavour profile of pulse proteins. The dissociation of legumins at extreme acidic conditions can be another reason for their removal from ASPP.333.3 Structural characterization of ASPP
Fig. 2c also showed the surface hydrophobicity of ASPP (370 ± 100 arbitrary units), which was lower than the PPC at pH 2.0 (907 ± 100 arbitrary units) (p < 0.05). The ASPP was mainly composed of albumins, which typically remain soluble at low pH, which might lead to a conformation that exposes fewer hydrophobic areas. In contrast, globulins, a more significant portion of the whole concentrate, have more hydrophobic amino acids than albumins. Removing most of the globulins during acidic extraction resulted in the soluble fraction having lower surface hydrophobicity. Shen et al.50 reported that pea globulins, especially legumins (11S), showed higher hydrophobicity and fluorescence intensity than albumins (2S), suggesting a scarcity of surface hydrophobic amino acids in the albumin protein structure. Ye et al.44 also reported that chickpea globulins displayed higher levels of aromatic and hydrophobic amino acid residues, in line with their higher surface hydrophobicity, compared to albumins. Also, Ghumman et al.51 and Tang and Ma45 previously reported higher hydrophobic amino acid content in the globulin fraction of lentil and kidney bean protein, respectively, compared to their albumin counterpart.
3.4 Emulsion formation and stabilization ability of ASPP
Many studies have shown a lower emulsifying ability for pea protein at acidic pH than neutral and alkaline pH values.24,56 In another recent study, O/W emulsions were prepared with the soluble fraction of pea protein extracted at pH 7.0, which showed a D4,3 of 0.344 μm at pH 7.0, but upon changing the pH of the emulsion to pH 2.0, D4,3 increased to a much larger size (3.8 μm) due to extensive protein aggregation.19 The higher albumin content of ASPP extracted at low pH could be responsible for its improved emulsion formation ability under acidic pH. Previous studies, such as the one reported by Burgos-Díaz et al.,57 showed that the lupin protein fractions predominantly containing albumins demonstrated notable emulsifying capacity and stability across various pH levels. Kornet et al.58 also noted that the smaller size of the pea albumins enabled them to move rapidly towards the interface and adsorb quicker due to their lower surface adsorption energy, in contrast to the larger globulins. Ye et al.44 also observed smaller droplet sizes of O/W emulsions stabilized by chickpea albumins compared to the globulins at low pH values (pH 3.0 and 5.0). In contrast, at pH 7.0, globulin demonstrated superior emulsion-forming ability compared to albumin.44
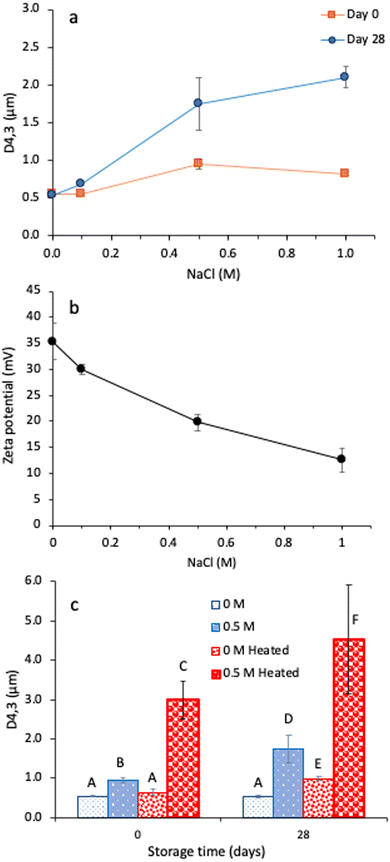
Next, the heat stability of the ASPP-stabilized emulsions was evaluated with and without the presence of 0.5 M salt (Fig. 4c). Without salt, ASPP-stabilized emulsions were highly stable against heat treatment, D4,3 only showed a minor change from 0.551 ± 0.002 to 0.641 ± 0.081 μm (p > 0.05) (Fig. 4c). This behaviour starkly contrasts a previous observation of extensive heat-induced destabilization of emulsions prepared with a soluble fraction of pea proteins recovered at pH 7.19 In the present work, heat treatment in the presence of 0.5 M salt led to a significant increase in D4,3, from 0.950 ± 0.071 μm to 3.00 ± 0.471 μm (p < 0.05), indicating extensive aggregation upon heat treatment in the presence of salt-induced charge screening effect. Compared with the freshly prepared heated emulsion without salt, the extent of droplet flocculation for the heated emulsion with 0.5 M salt addition was much higher, confirmed by comparing their droplet size distribution with and without polysorbate 20 (Fig. 3). Similar destabilization was also observed for the emulsions prepared with pH 7-soluble fraction in the presence of both salt and heat treatment.19 After 28 days, a further increase in D4,3 was observed for both the heat-treated emulsions, with and without salt; however, the effect was much less pronounced without salt.
Transmission profiles give a qualitative expression of emulsion stability under accelerated gravitation. For a quantitative comparison, the area under each transmission curve was calculated using the SEPView software and plotted as integral transmission% as a function of centrifugation time (Fig. 7a). Only the initial 60 min of centrifugation is shown in Fig. 7a as the most of the phase separation happened within the initial phase.61 All emulsions without salt showed a slight change in integral transmission within the first 1 h; the maximum value remained less than 10%, indicating a high stability of emulsions under normal earth gravitation. Heating the emulsion without salt (0 M) led to a slight increase in transmission profiles after 1 h; however, after 28 days of storage, the integral transmission values of the unheated and heated emulsions remained unchanged. In contrast, the integral transmission values sharply increased for all the emulsions with 0.5 M salt, and almost complete phase separation happened after 1 h of centrifugation, indicating unstable emulsions.
Quantitative estimation of the rate of movement of the phase separation during centrifugation can also be obtained from the creaming velocities calculated at the accelerated gravitation (Fig. 7b). The average creaming velocity of the fresh emulsions slightly changed from 48.2 to 56.1 μm min−1 (p > 0.05) with an increase in the salt concentration from 0 to 0.1 M, however, with 0.5 M and 1.0 salt, creaming velocity jumped to 831.9 and 1291.5 μm min−1, respectively. Such a rapid increase in creaming velocity with 0.5 M salt matches the droplet size results (Fig. 3 and 4c) and the visual observation images (Fig. 5). The creaming velocity of the heat-treated emulsion without salt (0 M salt) was 72.75 μm min−1, which did not change significantly from the unheated emulsions (p > 0.05). As a function of time, creaming velocity did not change significantly for the emulsions with 0 M salt (p > 0.05), with and without heat treatment, once again proof of the high stability of ASPP emulsions against heat treatment. For the emulsions with 0.5 M, creaming velocity decreased after 28 days of storage (p < 0.05), possibly due to increased emulsion viscosity due to extensive droplet aggregation. For the emulsions with 1.0 M salt, and heat-treated emulsions with 0.5 M salt, creaming velocity was above 1000 μm min−1, which did not change significantly after 28 days (p > 0.05). However, as indicated in the integral transmission data and the creaming velocity values, all emulsions with 0.5 M salt phase separated rapidly, suggesting they wouldn't be suitable for long-term storage.
3.5 Proposed mechanisms of the effect of various environmental factors on ASPP-emulsion properties
A similar observation of improved thermal stability of heat-modified pea protein-stabilized emulsions was also reported by Devaki and Ghosh,19 where the authors pre-heated the pH 7-extracted pea proteins before making emulsions with the heat-treated proteins at an elevated temperature. The authors proposed that partially denaturing the proteins through pre-heating and preventing aggregation before emulsification enabled the proteins to adsorb at the interface with the exposed hydrophobic patches, prohibiting further denaturation upon post-heating the emulsion. In this study, the intermolecular aggregates between the β-sheet-rich ASPP at the oil droplet surface, formed during heating, could also contribute to the thermal stability of the emulsions. The high surface charge of the droplets stabilized by ASPP at pH 2.0 (+35.3 mV) further inhibited their close approach and provided stability against heat. However, the lower zeta potential of the 0.5 M salt-added ASPP emulsions (+19.8 mV) could not prevent their close approach, resulting in droplet aggregation.
Another reason for the good thermal stability of ASPP-stabilized emulsion at pH 2.0 (without salt) may be attributed to its higher albumin content (Fig. 1). Albumins possess a more flexible conformation with less complex tertiary and quaternary structures compared to globulins, resulting in greater thermal stability68 Globulins are oligomers composed of multiple subunits that can be disrupted by heat, making them prone to thermal denaturation and loss of functionality.30 Based on the DSC results of the ASPP (Fig. 2a), only a minor peak for protein thermal denaturation was observed. Therefore, ASPP did not experience severe unfolding and denaturation at elevated temperatures since it primarily comprises albumin, which enhances the thermal stability of ASPP-stabilized emulsions at pH 2.0, provided they have a sufficiently higher surface charge.
4. Conclusions
This study emphasizes the significant potential of ASPP, the acidic water-soluble fraction of pea protein, in developing plant-based acidic beverage emulsions. The mildly fractionated ASPP could be directly used to develop highly stable 5 wt% canola O/W emulsions pH 2.0. The ASPP-stabilized emulsion was found to be stable during 28 days of storage as well as heat treatment (90 °C for 30 min) without and with 0.1 M salt (NaCl), while higher salt concentrations (0.5 and 1 M) resulted in protein and droplet aggregation. The findings revealed that the acid-soluble fraction was rich in albumins (55.8% albumin compared to 10% in the original pea protein). The acidic condition of the extraction led to a decrease in the intrinsic fluorescence and surface hydrophobicity of the protein due to the removal of most of the globulins, leading to a disappearance of the thermal denaturation peak. The ASPP was also richer in β-sheet and random coil structure than the original pea protein. We proposed that the higher β-sheet and albumin content of ASPP led to improved emulsifying ability and thermal stability under strongly acidic conditions. The observation of extensive droplet aggregation with 0.5 M salt was attributed to the charge screening effect and unfavourable conformational change in the albumin-rich proteins. The development of mildly extracted albumin-rich pulse proteins offers a promising solution to the difficulties previously encountered in formulating acidic emulsions with plant proteins. This endeavour established fundamental knowledge and laid the groundwork for forthcoming research aimed at producing pulse proteins ideal for application in ready-to-drink plant-based low-pH beverages.Data availability
The data supporting the findings of this study are available within the ESI.†Author contributions
Maryam Nikbakht Nasrabadi: conceptualization, methodology, investigation, formal analysis, data curation, visualization, writing – original draft. Michael Eskin: project administration, funding acquisition. Usha Thiyam-Hollander: project administration, funding acquisition. Supratim Ghosh: conceptualization, methodology, visualization, resources, writing – review & editing, supervision, project administration, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by the Agriculture Development Fund grant by the Saskatchewan Ministry of Agriculture, with financial support provided under the Sustainable Canadian Agricultural Partnership, a federal-provincial-territorial initiative. Research instrument support from the Canada Foundation for Innovation (CFI) is also acknowledged.References
- K. F. Csáki, Med. Hypotheses, 2011, 76, 676–681 CrossRef PubMed.
- S.-Y. Chuang, T. H. T. Chiu, C.-Y. Lee, T.-T. Liu, C. K. Tsao, C. A. Hsiung and Y.-F. Chiu, J. Hypertens., 2016, 34, 2164–2171 CrossRef CAS PubMed.
- F. Eichelmann, L. Schwingshackl, V. Fedirko and K. Aleksandrova, Obes. Rev., 2016, 17, 1067–1079 CrossRef CAS PubMed.
- M. Neuenschwander, J. Stadelmaier, J. Eble, K. Grummich, E. Szczerba, E. Kiesswetter, S. Schlesinger and L. Schwingshackl, BMC Med., 2023, 21, 404 CrossRef CAS PubMed.
- U. Fresán, S. Errendal and W. J. Craig, Sustainability, 2020, 12, 9093 CrossRef.
- J. Poore and T. Nemecek, Science, 2018, 360, 987–992 CrossRef CAS PubMed.
- M. Nikbakht Nasrabadi, A. Sedaghat Doost and R. Mezzenga, Food Hydrocolloids, 2021, 118, 106789 CrossRef CAS.
- B. Rajpurohit and Y. Li, J. Future Foods, 2023, 3, 340–356 CrossRef.
- S. N. Warnakulasuriya and M. T. Nickerson, J. Sci. Food Agric., 2018, 98, 5559–5571 CrossRef CAS PubMed.
- C. Tanger, J. Engel and U. Kulozik, Food Hydrocolloids, 2020, 107, 105949 CrossRef CAS.
- P. J. M. Pelgrom, A. M. Vissers, R. M. Boom and M. A. I. Schutyser, Food Res. Int., 2013, 53, 232–239 CrossRef CAS.
- N. D. Asen, R. E. Aluko, A. Martynenko, A. Utioh and P. Bhowmik, Foods, 2023, 12, 3978 CrossRef CAS PubMed.
- A. C. Möller, A. van der Padt and A. J. van der Goot, J. Food Eng., 2021, 291, 110321 CrossRef.
- M. E. Geerts, E. Mienis, C. V. Nikiforidis, A. van der Padt and A. J. van der Goot, Innovative Food Sci. Emerging Technol., 2017, 41, 251–258 Search PubMed.
- C. Kornet, P. Venema, J. Nijsse, E. van der Linden, A. J. van der Goot and M. Meinders, Food Hydrocolloids, 2020, 99, 105332 CrossRef CAS.
- A. C. Möller, A. van der Padt and A. J. van der Goot, Innovative Food Sci. Emerging Technol., 2022, 103144 Search PubMed.
- J. Yang, S. Zamani, L. Liang and L. Chen, Food Hydrocolloids, 2021, 117, 106678 CrossRef CAS.
- J. Yang, R. Kornet, C. F. Diedericks, Q. Yang, C. C. Berton-Carabin, C. V. Nikiforidis, P. Venema, E. van der Linden and L. M. C. Sagis, Food Struct., 2022, 31, 100254 Search PubMed.
- N. D. Devaki and S. Ghosh, J. Am. Oil Chem. Soc., 2024, 101(10), 981–996 CrossRef CAS.
- Z. Gao, P. Shen, Y. Lan, L. Cui, J.-B. Ohm, B. Chen and J. Rao, Food Res. Int., 2020, 131, 109045 CrossRef CAS PubMed.
- F. Schmidt, M. Blankart, J. Wanger, M. Scharfe, T. Scheuerer and J. Hinrichs, J. Food Meas. Char., 2022, 1–10 CAS.
- S. Sridharan, M. B. J. Meinders, J. H. Bitter and C. V. Nikiforidis, Langmuir, 2020, 36, 12221–12229 CrossRef CAS PubMed.
- H.-N. Liang and C.-h. Tang, LWT–Food Sci. Technol., 2014, 58, 463–469 CrossRef CAS.
- F. Keivaninahr, P. Gadkari, K. Z. Benis, M. Tulbek and S. Ghosh, RSC Adv., 2021, 11, 12117–12135 RSC.
- C. D. Doan and S. Ghosh, Nanomaterials, 2019, 9, 949 CrossRef CAS PubMed.
- E. Keuleyan, P. Gélébart, V. Beaumal, A. Kermarrec, L. Ribourg-Birault, S. Le Gall, A. Meynier, A. Riaublanc and C. Berton-Carabin, Food Hydrocolloids, 2023, 141, 108671 CrossRef CAS.
- N. Alizadeh-Pasdar and E. C. Y. Li-Chan, J. Agric. Food Chem., 2000, 48, 328–334 CrossRef CAS PubMed.
- A. Mohanan, Y. R. Tang, M. T. Nickerson and S. Ghosh, RSC Adv., 2020, 10, 14892–14905 RSC.
- M. Primozic, A. Duchek, M. Nickerson and S. Ghosh, Food Chem., 2017, 237, 65–74 CrossRef CAS PubMed.
- M. F. Marcone, Y. Kakuda and R. Y. Yada, Food Chem., 1998, 62, 27–47 CrossRef CAS.
- S. G. Pérez, J. M. Vereijken, G. A. Van Koningsveld, H. Gruppen and A. G. Voragen, J. Food Sci., 2005, 70, C98–C103 CrossRef.
- I. D. Nwachukwu and R. E. Aluko, 2021.
- J. Gueguen, M. Chevalier, J. B. And and F. Schaeffer, J. Sci. Food Agric., 1988, 44, 167–182 CrossRef CAS.
- L. Luo, Z. Wang, Y. Deng, Z. Wei, Y. Zhang, X. Tang, G. Liu, P. Zhou, Z. Zhao and M. Zhang, Food Chem., 2022, 397, 133684 CrossRef CAS PubMed.
- P. J. Shand, H. Ya, Z. Pietrasik and P. K. J. P. D. Wanasundara, Food Chem., 2007, 102, 1119–1130 CrossRef CAS.
- J.-L. Mession, M. L. Chihi, N. Sok and R. Saurel, Food Hydrocolloids, 2015, 46, 233–243 CrossRef CAS.
- X. D. Sun and S. D. Arntfield, Food Res. Int., 2010, 43, 509–515 CrossRef CAS.
- S. D. Arntfield and E. D. Murray, Can. Inst. Food Sci. Technol. J., 1981, 14, 289–294 CrossRef.
- R. Kornet, J. Veenemans, P. Venema, A. J. van der Goot, M. Meinders, L. Sagis and E. van der Linden, Food Hydrocolloids, 2021, 112, 106285 CrossRef CAS.
- N. D. Devaki, M. Sc., University of Saskatchewan, 2022.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer, 2006 Search PubMed.
- K. Gao, F. Zha, Z. Yang, J. Rao and B. Chen, Food Hydrocolloids, 2022, 125, 107409 CrossRef CAS.
- M. Miriani, S. Iametti, F. Bonomi and M. Corredig, Colloids Surf., B, 2012, 93, 41–48 CrossRef PubMed.
- J. Ye, N. Shi, P. Rozi, L. Kong, J. Zhou and H. Yang, Food Bioprocess Technol., 2024, 1–14 Search PubMed.
- C.-H. Tang and C.-Y. Ma, Food Chem., 2009, 115, 859–866 CrossRef CAS.
- R. J. Delahaije, P. A. Wierenga, N. H. van Nieuwenhuijzen, M. L. Giuseppin and H. Gruppen, Langmuir, 2013, 29, 11567–11574 CrossRef CAS PubMed.
- L. G. Phillips, Structure-Function Properties of Food Proteins, Academic Press, 2013 Search PubMed.
- D. E. Igartúa, A. Balcone, F. A. Platania, D. M. Cabezas and G. G. Palazolo, J. Sci. Food Agric., 2024, 104(12), 7291–7300 CrossRef PubMed.
- D. E. Igartúa, M. C. Dichano, M. N. Morales Huanca, G. G. Palazolo and D. M. Cabezas, Food Res. Int., 2024, 188, 114399 CrossRef PubMed.
- Q. Shen, J. Li, X. Shen, X. Zhu, J. Dai, C. Tang, R. Song, B. Li and Y. Chen, Food Hydrocolloids, 2023, 139, 108500 CrossRef CAS.
- A. Ghumman, A. Kaur and N. Singh, Food Hydrocolloids, 2016, 61, 843–850 CrossRef CAS.
- Y. Wang, J. Wang, S. Wang, J. Guo and S. Wang, J. Agric. Food Chem., 2019, 67, 10734–10743 CrossRef CAS PubMed.
- X. Sun, I. C. Ohanenye, T. Ahmed and C. C. Udenigwe, Food Chem., 2020, 329, 127196 CrossRef CAS PubMed.
- A. M. C. Marcelino and L. M. Gierasch, Biopolymers, 2008, 89, 380–391 CrossRef CAS PubMed.
- Z. Wang, Y. Li, L. Jiang, B. Qi and L. Zhou, J. Chem., 2014, 2014, 475389 Search PubMed.
- R. E. Aluko, O. A. Mofolasayo and B. M. Watts, J. Agric. Food Chem., 2009, 57, 9793–9800 CrossRef CAS PubMed.
- C. Burgos-Díaz, J. A. Piornos, T. Wandersleben, T. Ogura, X. Hernández and M. Rubilar, J. Food Sci., 2016, 81, C1699–C1706 CrossRef PubMed.
- R. Kornet, J. Yang, P. Venema, E. van der Linden and L. M. C. Sagis, Food Hydrocolloids, 2022, 126, 107456 CrossRef CAS.
- F. C. L. Almeida, K. Sanches, R. Pinheiro-Aguiar, V. S. Almeida and I. P. Caruso, Front. Mol. Biosci., 2021, 8, 706002 CrossRef CAS PubMed.
- M. Primozic, A. Duchek, M. Nickerson and S. Ghosh, Food Hydrocolloids, 2018, 77, 126–141 CrossRef CAS.
- B. Guldiken, M. Saffon, M. T. Nickerson and S. Ghosh, Food Hydrocolloids, 2023, 145, 109029 CrossRef CAS.
- Y. R. Tang and S. Ghosh, Food Hydrocolloids, 2021, 113, 106399 CrossRef CAS.
- R. E. Aluko and R. Y. Yada, Food Chem., 1995, 53, 259–265 CrossRef CAS.
- M. Carbonaro, P. Maselli and A. Nucara, Amino Acids, 2012, 43, 911–921 CrossRef CAS PubMed.
- K. Shevkani, N. Singh, A. Kaur and J. C. Rana, Food Hydrocolloids, 2015, 43, 679–689 CrossRef CAS.
- S. Damodaran and K. L. Parkin, in Fennema's Food Chemistry, CRC Press, 2017, pp. 235–356 Search PubMed.
- A. A. Schneider, F. Bu and B. P. Ismail, Curr. Res. Food Sci., 2023, 6, 100452 CrossRef CAS PubMed.
- R. I. Monsalve, M. Villalba, M. Rico, P. R. Shewry and R. Rodríguez, Plant Food Allergens, 2003, pp. 42–56 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra06899h |
| This journal is © The Royal Society of Chemistry 2025 |