Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Isoindoline-based fluorogenic probes bearing a self-immolative linker for the sensitive and selective detection of O-GlcNAcase activity†
Yuan-Hao Wu‡
a,
Guan-Jun Wang‡a,
Chen Guo‡a,
Pei-Pei Wang ce,
Jun-Yi Wangl,
Xi-Le Hu
ce,
Jun-Yi Wangl,
Xi-Le Hu *a,
Yi Zang
*a,
Yi Zang *dfg,
Tony D. James
*dfg,
Tony D. James *jk,
Jia Li
*jk,
Jia Li *cefghi and
Xiao-Peng He
*cefghi and
Xiao-Peng He *ab
*ab
aKey Laboratory for Advanced Materials and Joint International Research Laboratory of Precision Chemistry and Molecular Engineering, Feringa Nobel Prize Scientist Joint Research Center, School of Chemistry and Molecular Engineering, East China University of Science and Technology, 130 Meilong Rd, Shanghai 200237, China. E-mail: xphe@ecust.edu.cn; xlhu@ecust.edu.cn
bThe International Cooperation Laboratory on Signal Transduction, Eastern Hepatobiliary Surgery Hospital, National Center for Liver Cancer, Shanghai 200438, China
cNational Center for Drug Screening, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China. E-mail: jli@simm.ac.cn
dLingang Laboratory, Shanghai 201203, China. E-mail: yzang@lglab.ac.cn
eSchool of Chinese Materia Medica, Nanjing University of Chinese Medicine, Nanjing 210023, China
fSchool of Pharmaceutical Science and Technology, Hangzhou Institute for Advanced Study, University of Chinese Academy of Sciences, Hangzhou, 310024, China
gState Key Laboratory of Chemical Biology, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, 201203, China
hUniversity of Chinese Academy of Sciences, Beijing, 100049, China
iZhongshan Institute for Drug Discovery, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Guangdong 528400, China
jDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk
kSchool of Chemistry and Chemical Engineering, Henan Normal University, Xinxiang 453007, China
lShanghai Starriver Bilingual School, No. 2588 Jindu Road, Minhang District, Shanghai, China
First published on 15th July 2024
Abstract
O-GlcNAcase (OGA) is implicated in several important biological and disease-relevant processes. Here, we synthesized fluorogenic probes for OGA by grafting GlcNAc directly or using a self-immolative linker to the hydroxyl position of 4-hydroxylisoindoline (BHID), a typical excited-state intramolecular proton transfer (ESIPT) probe. The probe was used for a fluorogenic assay to determine the half maximal inhibitory concentration of a known OGA inhibitor and differentiate between OGA and hexosaminidase when GlcNAc is replaced by GlcNPr, where a propionyl group is used instead of an acetyl group.
O-GlcNAcylation is a typical post-translational modification implicated in a diverse range of biological processes by introducing N-acetylglucosamine (GlcNAc) to the serine or threonine residue of proteins. Two key enzymes are involved in this modification process O-GlcNAc transferase (OGT) that transfers GlcNAc to biomolecules, and O-GlcNAcase (OGA) that deglycosylates GlcNAc reversibly. Both enzymes have been shown to be associated with a variety of biological processes, and abnormal OGA expression was found to be linked with a number of diseases including diabetes, cancer, Alzheimer's disease and cardiovascular disease.1,2 This highlights the importance of developing effective sensing tools for OGA.
In addition to traditional biochemical techniques such as quantitative real-time polymerase chain reaction and western blot, activity-based assays are commonly used for the analysis of enzymatic activities. While colorimetric assays are generally used for enzymatic assays, fluorogenic probes have been extensively developed to visualize the activity of an enzyme of interest in live cells.3,4 A general strategy to design such probes relies on the introduction of the substrate molecule of a specific enzyme to a fluorescent dye, inhibiting the electron or charge transfer process of the dye system, thus quenching the fluorescence.5 Then after selective removal of the substrate by the enzyme, the fluorescence is restored. With this rationale, fluorogenic probes for many diverse glycosidases have been developed for the intracellular and in vivo imaging of their activities.6,7 Glycosidase-activatable prodrugs have also been developed for therapy of cancer and senescence.8,9
Several fluorogenic probes based on the sensing mechanisms described above have been reported for OGA detection.10–15 However, it is still necessary to expand the range of designs that can be incorporated into molecular probes for OGA. Excited-state intramolecular proton transfer (ESIPT) is a photochemical process that describes the switch of a molecule from the enol to keto form upon light excitation, causing its fluorescence emission features to change. Fluorescent probes based on the ESIPT mechanism are advantageous in terms of large Stokes shift and flexibility in fine-tuning the ESIPT states by simply blocking the proton donor site.16 A survey of the literature indicates the importance and popularity of using fluorescent ESIPT dyes for biosensing and bioimaging.17
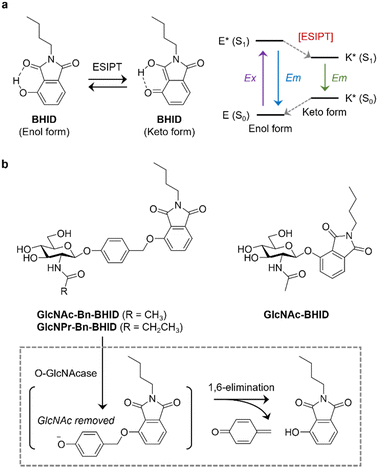
Here, we develop ESIPT-based OGA probes, and demonstrate the importance of incorporating a self-immolative linker to improve sensitivity (Fig. 1). We chose 2-butyl-4-hydroxyisoindoline-1,3-dione (BHID), which has been used to develop fluorogenic probes for detecting hydrazine and peroxynitrite,18,19 as the fluorophore because of its accessibility and simplicity for structural modification. As such, to the hydroxyl position of BHID GlcNAc was added with or without a “self-immolative” spacer.20 It has been reported that the presence of the spacer enhances the sensitivity of fluorogenic probes for glycosidases probably because of a reduced steric hindrance.21 GlcNAc was introduced to the hydroxyl group of p-hydroxybenzaldehyde by stirring in acetonitrile with silver oxide and potassium iodide, followed by reduction and bromination to produce intermediate GlcNAc-Bn-Br. Then, this intermediate underwent Williamson ether reaction with BHID, followed by deacetylation to afford GlcNAc-Bn-BHID in moderate yield (Scheme S1, ESI†). A control probe GlcNAc-BHID without the spacer was also prepared by directly introducing GlcNAc to the phenolic OH of BHID followed by deprotection (Scheme S1, ESI†).
It is known that in addition to OGA, other endogenous enzymes such as N-acetyl-β-D-hexosaminidases A (HexA) can also unselectively remove GlcNAc from proteins. A previous report indicated that the replacement of the N-acetyl group with an N-propionyl group in the GlcNAc structure achieved selective binding to OGA over HexA due to better ligand-binding.22–24 Therefore, a GlcNPr (in which a propionyl was used to modify the 2-amino group of the sugar instead of acetyl group) donor was used to glycosylate the spacer, followed by coupling to BHID by the same protocol as described above. The resulting GlcNPr-Bn-BHID probe was used to achieve selectivity sensing of OGA over a commercial Hex.
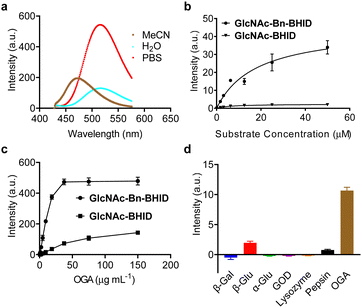
With the probes in hand, we determined their sensing properties. The ESIPT properties of BHID were determined in different solvents by fluorescence spectroscopy (Fig. 2a). We found that it exhibited two emission peaks with different Stokes shift. In aprotic polar solvent (acetonitrile) a short emission was observed, while a longer emission was seen in protic solvent (deionized water and PBS). This dual-emission reflected the enol and keto form in the ESIPT process, confirming the ESIPT mechanism of our fluorophore.
Then, we expressed OGA by BL21(DE3) Competent Escherichia coli for enzymatic analysis. To establish the activity-based assay, a fluorescence microplate reader was used. GlcNAc-BHID and GlcNAc-Bn-BHID were incubated with OGA in an optimized buffer system of NaCl–NaH2PO4-BSA (bovine serum albumin) at pH 7.0.22,25 We first evaluated the fluorescence response of the two probes with increasing substrate concentrations at an OGA concertation of 1 μg mL−1 (Fig. 2b). A substrate concentration of 50 μM was found to be required for the fluorescence to reach equilibrium. Over the entire substrate concentration range, the fluorescence of GlcNAc-Bn-BHID was constantly more intense than that of GlcNAc-BHID, suggesting the former being more sensitive. Indeed, a ∼3-fold larger Km (Michalies constant) was determined for GlcNAc-Bn-BHID than GlcNAc-BHID by using a nonlinear curve fitting method in GraphPad Prism 9.5.0 (Table. S1, ESI†), indicating a larger catalytic efficiency of GlcNAc-Bn-BHID. Then, with a fixed substrate concentration (50 μM), increasing concentrations of OGA was added to the system. We determined that equilibrium was reached for GlcNAc-Bn-BHID with 37.5 μg mL−1 OGA, and a larger OGA concentration was required to fully activate the fluorescence of GlcNAc-BHID (Fig. 2c). These results corroborate the importance of the spacer for improving OGA sensitivity. Measurement using fluorescence spectroscopy also confirmed the fluorescence “turn-on” response of GlcNPr-Bn-BHID in the presence of OGA (Fig. S1, ESI†).
Next, the selectivity of GlcNAc-Bn-BHID for OGA was determined. A range of other glycosidases including β-galactosidase (β-Gal), α-glucosidase (α-Glu) and β-glucosidase (β-Glu), and other physiological enzymes including lysozyme, pepsin and glucose oxidase (GOD) were used. All enzymes were separately incubated with the probe in microplate wells for 90 min, followed by fluorescence detection in the microplate reader. As shown in Fig. 2d, the results indicated that the unselective enzymes hardly enhanced the fluorescence of GlcNAc-Bn-BHID. A slightly enhanced fluorescence was observed for β-glucosidase, which might be the result of structural similarity between glucose and GlcNAc.
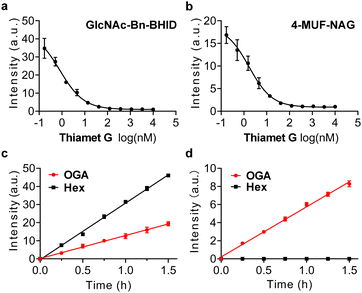
To prove its real-world applicability, we sought to exploit the probe for evaluating the inhibitory activity of OGA inhibitors. A commercial OGA probe, 4-MUF-NAG, was used as reference. In addition, a known OGA inhibitor, Thiamet-G, was used for the experiments.26 Lineweaver–Burk plots were obtained for GlcNAc-Bn-BHID (Fig. 3a) and 4-MUF-NAG (Fig. 3b) with different inhibitor concentrations (0.17 nM to 10 μM) in the presence of OGA (Table. S2, ESI†). The fluorescence of both probes decreased gradually when the concentration of inhibitor increased. The half maximal inhibitory concentration (IC50) of Thiamet-G was determined to be 0.8 nM and 1.7 nM for the GlcNAc-Bn-BHID and 4-MUF-NAG-based assay, respectively. The IC50 determined from the GlcNAc-Bn-BHID-based assay agrees with that reported previously,26 suggesting the reliability of our ESIPT-based probe for screening OGA inhibitors.
Finally, the probes were used to differentiate OGA and HexA. It is important to achieve selective OGA detection over other enzymes to better elaborate the biological functions. In particular, HexA unselectively hydrolyzes GlcNAc and N-acetylgalactosamine (GalNAc) from biomacromolecules in the lysosome.27 A previous report indicated that N-propionyl substitution significantly enhances OGA binding of N-glucosamines over HexA. Thus, we determined whether our GlcNAc-Bn-BHID and GlcNPr-Bn-BHID could be used to differentiate the enzymes in a fluorogenic assay.
We found that in the presence of both OGA and a commercial hexosaminidase (from Canavalia ensiformis) used as a homogenous surrogate for human HexA,28 the fluorescence of GlcNAc-Bn-BHID enhanced in a concentration-dependent manner (Fig. 3c). However, when GlcNPr-Bn-BHID was used as the substrate, a time-dependent fluorescence enhancement was only observed for OGA, and the presence of commercial hexosaminidase did not cause the probe to fluoresce with time (Fig. 3d). The better sensitivity of GlcNAc-Bn-BHID than GlcNPr-Bn-BHID was probably a result of the steric effects of the propionyl group that partially compromises glucosamine binding. However, the observation that GlcNPr-Bn-BHID does not respond to commercial hexosaminidase suggests its potential to be used for OGA-selective biosensing and bioimaging in complicated biological samples where both enzymes co-exist. As such, our ongoing research will focus on developing fluorogenic OGA probes suitable for cell imaging.4,29–31
In summary, we have developed the first ESIPT-based fluorogenic probe for OGA, which is an important glycosidase implicated in a variety of disease-relevant biological processes. The presence of a self-immolative linker was shown to significantly improve the sensitivity of the probe. We also established an enzymatic assay that exhibited time- and concentration-dependent fluorescence recovery in the presence of recombinant OGA. We also determined the IC50 of a known OGA inhibitor and achieved the differentiation between OGA and commercial hexosaminidase. We believe that the chemical probe developed as part of this research can provide useful strategies for the study of OGA-based biological events and OGA-based inhibitor screening.
The authors thank the Natural National Science Foundation of China (NSFC) (No. 92253306, 82130099 and 22108077), the Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX03), the International Cooperation Program of Shanghai Science and Technology (No. 23490711600), the Fundamental Research Funds for the Central Universities (222201717003), the Programme of Introducing Talents of Discipline to Universities (B16017), the Open Funding Project of the State Key Laboratory of Bioreactor Engineering, State Key Laboratory of Drug Research (SKLDR-2023-KF-10) and Ministry of Education Key Laboratory on Signalling Regulation and Targeting Therapy of Liver Cancer (Naval Medical University) (Grant. 2023-MEKLLC-MS/ZD-00*) for financial support. TDJ wishes to thank the University of Bath and the Open Research Fund of the School of Chemistry and Chemical Engineering, Henan Normal University (2020ZD01) for support. The Research Center of Analysis and Test of East China University of Science and Technology was gratefully acknowledged for assistance in analytical experiments.
Data availability
All data supporting this research are included in the main article and/or ESI.†Conflicts of interest
The authors declare no conflict of interest.Notes and references
- Y. Li, M. Xie, L. Men and J. Du, Int. J. Mol. Med., 2019, 44, 363–374 CAS.
- J. Ma, C. Hou and C. Wu, Chem. Rev., 2022, 122, 15822–15864 CrossRef CAS PubMed.
- J. J. Zhang, X. Z. Chai, X. P. He, H. J. Kim, J. Y. Yoon and H. Tian, Chem. Soc. Rev., 2019, 48, 683–722 RSC.
- W. T. Dou, H. H. Han, A. C. Sedgwick, G. B. Zhu, Y. Zang, X. R. Yang, J. Yoon, T. D. James, J. Li and X. P. He, Sci. Bull., 2022, 67, 853–878 CrossRef CAS PubMed.
- Y. Kim, H. Li, J. Choi, J. Boo, H. Jo, J. Y. Hyun and I. Shin, Chem. Soc. Rev., 2023, 52, 7036–7070 RSC.
- X. Chai, H. H. Han, A. C. Sedgwick, N. Li, Y. Zang, T. D. James, J. Zhang, X. L. Hu, Y. Yu, Y. Li, Y. Wang, J. Li, X. P. He and H. Tian, J. Am. Chem. Soc., 2020, 142, 18005–18013 CrossRef CAS PubMed.
- H. Tian, W. Lin, X. L. Hu, J. B. Wang, M. Y. Zhang, Y. Zang, X. Y. Wu, J. Li, T. D. James and X. P. He, Org. Chem. Front., 2023, 10, 2913–2917 RSC.
- D. L. Shi, W. W. Liu, Y. Gao, X. M. Li, Y. Y. Huang, X. K. Li, T. D. James, Y. Guo and J. Li, Nat. Aging., 2023, 3, 297–312 CrossRef CAS PubMed.
- M. Chang, Y. Dong, H. Xu, A. B. Cruickshank-Taylor, J. S. Kozora, B. Behpour and W. Wang, Angew. Chem., Int. Ed., 2024, 63, e202315425 CrossRef CAS PubMed.
- J. Y. Hyun, S. H. Park, C. W. Park, H. B. Kim, J. W. Cho and I. Shin, Org. Lett., 2019, 21, 4439–4442 CrossRef CAS PubMed.
- H. Jung, S. H. Park, W. H. Yang, J. W. Cho and I. Shin, Sens. Actuators, B, 2022, 367, 132093 CrossRef CAS.
- J. Boo, J. Lee, Y. H. Kim, C. H. Lee, B. Ku and I. Shin, Front. Chem., 2023, 11, 1133018 CrossRef CAS PubMed.
- S. Cecioni and D. J. Vocadlo, J. Am. Chem. Soc., 2017, 139, 8392–8395 CrossRef CAS PubMed.
- E. J. Kim, D. O. Kang, D. C. Love and J. A. Hanover, Carbohydr. Res., 2006, 341, 971–982 CrossRef CAS PubMed.
- V. S. Borodkin, K. Rafie, N. Selvan, T. Aristotelous, I. Navratilova, A. T. Ferenbach and D. M. F. Van Aalten, ACS Chem. Biol., 2018, 13, 1353–1360 CrossRef CAS PubMed.
- A. C. Sedgwick, L. Wu, H. H. Han, S. D. Bull, X. P. He, T. D. James, J. L. Sessler, B. Z. Tang, H. Tian and J. Yoon, Chem. Soc. Rev., 2018, 47, 8842–8880 RSC.
- H. Gu, W. J. Wang, W. Y. Wu, M. L. Wang, Y. R. Liu, Y. J. Jiao, F. Wang, F. Wang and X. Q. Chen, Chem. Commun., 2023, 59, 2056–2071 RSC.
- S. Hu, J. Wang, M. Luo, Z. Wu, Y. Hou and X. Chen, Anal. Bioanal. Chem., 2021, 413, 5463–5468 CrossRef CAS PubMed.
- L. Wu, X. Tian, D. J. Lee, J. Yoon, C. S. Lim, H. M. Kim and T. D. James, Chem. Commun., 2021, 57, 11084–11087 RSC.
- S. Gnaim and D. Shabat, Acc. Chem. Res., 2014, 47, 2970–2984 CrossRef CAS PubMed.
- Q. Wu, Q. H. Zhou, W. Li, T. B. Ren, X. B. Zhang and L. Yuan, ACS Sens., 2022, 7, 3829–3837 CrossRef CAS PubMed.
- M. S. Macauley, G. E. Whitworth, A. W. Debowski, D. Chin and D. J. Vocadlo, J. Biol. Chem., 2005, 280, 25313–25322 CrossRef CAS PubMed.
- M. González-Cuesta, P. Sidhu, R. A. Ashmus, A. Males, C. Proceviat, Z. Madden, J. C. Rogalski, J. A. Busmann, L. J. Foster, J. M. García Fernández, G. J. Davies, C. Ortiz Mellet and D. J. Vocadlo, J. Am. Chem. Soc., 2022, 144, 832–844 CrossRef PubMed.
- B. Li, H. Li, C. W. Hu and J. Jiang, Nat. Commun., 2017, 8, 666 CrossRef PubMed.
- Y. Gao, L. Wells, F. I. Comer, G. J. Parker and G. W. Hart, J. Biol. Chem., 2001, 276, 9838–9845 CrossRef CAS PubMed.
- S. A. Yuzwa, M. S. Macauley, J. E. Heinonen, X. Shan, R. J. Dennis, Y. He, G. E. Whitworth, K. A. Stubbs, E. J. McEachern, G. J. Davies and D. J. Vocadlo, Nat. Chem. Biol., 2008, 4, 483–490 CrossRef CAS PubMed.
- J. J. Morsby and B. D. Smith, Bioconjugate Chem., 2022, 33, 544–554 CrossRef CAS PubMed.
- Q. Chen, P. Guo, L. Liu, T. Liu, X. H. Qian and Q. Yang, Biochimie, 2014, 97, 152–162 CrossRef CAS PubMed.
- X.-P. He and H. Tian, Chem, 2018, 4, 246–268 CAS.
- L. Dong, M.-Y. Zhang, H.-H. Han, Y. Zang, G.-R. Chen, J. Li, X.-P. He and S. Vidal, Chem. Sci., 2022, 13, 247–256 RSC.
- J.-B. Jiao, G.-Z. Wang, X.-L. Hu, Y. Zang, S. Maisonneuve, A. C. Sedgwick, J. L. Sessler, J. Xie, J. Li, X.-P. He and H. Tian, J. Am. Chem. Soc., 2020, 142, 1925–1932 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc02845g |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |