Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Heteromolecular compounds in binary systems of amino acids with opposite and same chiralities†
Anton I.
Isakov
a,
Heike
Lorenz
 *b,
Andrey A.
Zolotarev
Jr
*b,
Andrey A.
Zolotarev
Jr
 a and
Elena N.
Kotelnikova
a and
Elena N.
Kotelnikova
 a
a
aDepartment of Crystallography, Saint Petersburg State University, Universitetskaya emb. 7/9, 199034 Saint Petersburg, Russia
bMax Planck Institute for Dynamics of Complex Technical Systems, Sandtorstrasse 1, 39106 Magdeburg, Germany. E-mail: lorenz@mpi-magdeburg-mpg.de; Tel: +49 391 6110 293
First published on 14th January 2020
Abstract
A classification of discrete compounds in binary chiral systems of single and different substances (homo- and heteromolecular compounds) is presented. It considers both chemical and crystallographic characteristics of such compounds. Using amino acids as chiral model systems, features of crystal structures of equimolar and non-equimolar heteromolecular compounds are reviewed. In this connection, the concept of homo- and heteromolecular dimers in compounds formed by amino acids is introduced and analyzed. As a result, a correlation between the molecular dimer type and the side chain structure (linear or branched) and the conformation (extended or folded) of the relevant compounds' molecules is derived. Two non-equimolar discrete heterocompounds discovered in the chiral systems L-valine–L-isoleucine and L-valine–L-leucine are discussed using the concepts proposed. The previously studied first system features a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Val
1 (Val![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ile) compound. A newly investigated second system is found to form a 3
Ile) compound. A newly investigated second system is found to form a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Val
1 (Val![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Leu) compound, and its crystal structure is described and evaluated.
Leu) compound, and its crystal structure is described and evaluated.
1. Introduction
Many organic substances are chiral molecules, which can exist as levo and dextrorotatory optical antipodes. These molecular entities being mirror images of each other and non-superposable are called enantiomers.1 In their molecular structures, enantiomers possess a chiral element, which in most cases is a chiral center and less frequently, a chiral plane or a chiral axis. Usually, the chiral center is a so-called asymmetric carbon atom, i.e. a carbon atom interlinked by “tetrahedral” bonds to four different substituents (ligands). Chiral molecules may have various numbers of chiral centers (n), with n defining the number of possible configurations of the molecule (2n). In a symmetric environment, all the physical and chemical properties of enantiomers are identical, except for the direction of plane-polarized light rotation.Size and shape of molecules and geometry of intermolecular hydrogen bonds are general factors influencing the molecular packing in crystal structures of organic substances.2,3 In the case of chiral substances, the molecule configuration becomes the main additional factor for molecular packing.
Enantiomers are widely used in pharmaceuticals, food industries and electronics. For example, in 2004, nine out of the ten most sold drugs contained chiral active ingredients.4 Furthermore, in the same year, among the 16 newly approved synthetic drugs, 13 were chiral with all of them being single enantiomers.5 However, products of non-stereoselective industrial synthesis are mixtures of both enantiomers. Therefore, the problem of applicability of chiral substances (for example, for API's production) is closely connected to the problem of resolution of enantiomeric mixtures. The most profitable resolution methods are crystallization methods that require the understanding of phase equilibria in a system and plotting its phase diagram.6
The largest part of published information on chiral systems refers to enantiomers of a single substance, while reports on enantiomeric systems of different substances are rather rare. Amino acids are suitable model compounds to investigate binary chiral systems of the latter type. The great diversity of amino acids and relatively simple structures of their molecules made them already model compounds, for example, for the determination of hydrogen bond lengths in proteins and other biopolymers.7,8
The vast variety of proteins is a result of the combination of twenty proteinogenic α-amino acids, with nineteen being chiral compounds. Among them, seventeen molecules contain only one asymmetric carbon atom and, consequently, exist as two enantiomers. Molecules of the residual amino acids, threonine and isoleucine, contain two asymmetric carbon atoms and thus, exist as four stereoisomers forming two enantiomer and four diastereomer pairs, respectively. Eight proteinogenic α-amino acids are essential amino acids, which cannot be produced by the human body and, therefore, must be consumed. The subjects of our study, the aliphatic amino acids valine (C5H11NO2), leucine (C6H13NO2), and isoleucine (C6H13NO2), belong to this group.
In this present work, we (1) introduce a classification of discrete compounds formed in chiral systems (section 2), (2) discuss the chiral molecular packing in crystal structures of amino acids and their equimolar heteromolecular compounds (sections 3 and 4), and (3) present detailed studies of mainly two levorotatory amino acid systems, L-valine–L-leucine and L-valine–L-isoleucine, as examples of binary chiral systems forming non-equimolar heteromolecular compounds (section 5).
2. Classification of discrete compounds in binary systems of chiral molecules
A first classification attempt for such compounds was proposed by the present authors earlier.9 We considered the classification necessary, since in the literature, there is plenty of information available on various types of chiral compounds, but, at the same time, the compounds are described using “spontaneously arisen” terms which are not self-sufficient, i.e. they do not reflect the proper place of a particular compound in the general hierarchy of chiral substances.The revised classification is shown as a flow diagram in Fig. 1. We implemented significant changes, namely, the proposed concepts for homo- and heteromolecular dimers (see sections 4.1 and 4.2), and considered diastereomers to be different substances, differing on molecular structures and physical characteristics. However, the key change is an improved order of basic chemical and crystallographic classifying features that define the hierarchy of chiral compounds.
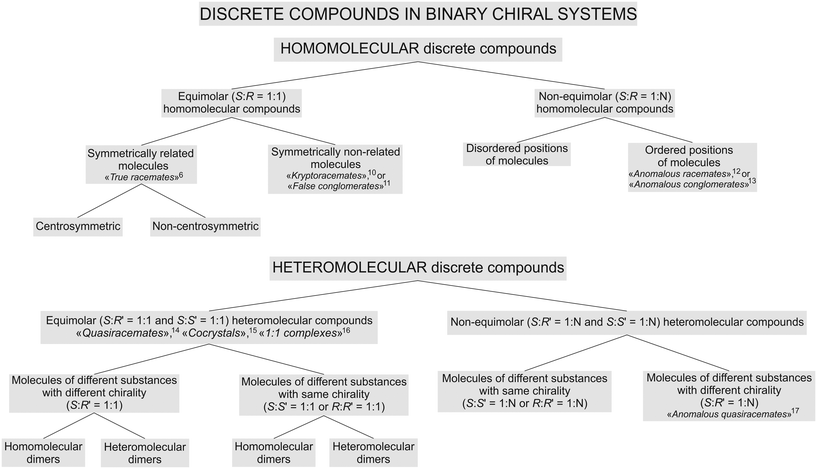 | ||
| Fig. 1 Classification of discrete compounds in binary systems of chiral molecules. Corresponding related and historical terms6,10–17 are given with references. | ||
The chemical composition of a particular compound became the first classifying feature, and the molecular ratio of the components constituting a compound as the second classifying feature. Consequently, in Fig. 1, compounds formed in binary chiral systems were first divided into two main types: homomolecular and heteromolecular discrete compounds. Afterwards, they were split into equimolar and non-equimolar compounds, respectively.
In the case of homomolecular compounds (homocompounds), the third classifying feature is based on the molecular packings' character in the crystal structure. Accordingly, equimolar homocompounds are divided into compounds with symmetry related molecular components (centrosymmetric and non-centrosymmetric ones) and compounds with symmetrically independent molecular components. Exemplary centrosymmetric and non-centrosymmetric equimolar homocompounds are two polymorphs of malic acids' true racemate, RSI and RSII, respectively.9 An exemplary homocompound containing symmetrically independent molecules is DL-allylglycine.18
Non-equimolar homocompounds, in turn, can be divided into compounds having ordered and disordered molecular positions. An exemplary compound with ordered molecular positions is the non-equimolar discrete compound S3R of malic acid.9,19 To the present authors' knowledge, there is no available information in the literature about any non-equimolar homocompounds with disordered molecular positions.
In the case of heteromolecular compounds (heterocompounds), the third classifying feature is the chirality of the molecular components. Accordingly, equimolar heterocompounds are divided into compounds composed of molecular components with different chiralities and ones with the same chirality. The crystal structure of heterocompounds of either type can consist of homomolecular or heteromolecular dimers.
In other words, systems with components having different chiralities can produce compounds containing homomolecular dimers (e.g., L-valine–D-norvaline, see section 4.1) and compounds containing heteromolecular dimers (e.g., D-valine–L-leucine, see section 4.2). Heterocompounds with homomolecular dimers are found not only among amino acids but also among other chiral organic compounds.20–25 As well as systems of components having the same chirality can form compounds containing heteromolecular dimers (e.g., L-malic acid–L-tartaric acid26) and, possibly, those consisting of homomolecular dimers (no examples have been found yet in the published literature).
Similarly, non-equimolar heterocompounds are divided into compounds composed of molecular components with the same or different chiralities. The latter case is found in heterocompounds formed in the (+)-2,4-dimethylglutaric acid–(−)-dilactic acid and (−)-2,4-dimethylglutaric acid–(+)-2-methylglutaric acid systems.14 In contrast, the molecular components of non-equimolar heterocompounds found by the present authors in the systems of L-valine–L-isoleucine9,27 and L-valine–L-leucine (see section 5) have the same chirality.
3. Crystal structure of amino acids
The publications7–9,16,27–35 that appeared in the last decades made a considerable contribution to the concept of molecular structures of amino acids.For the description of the amino acids' crystal structure, the terms “dimer” and “molecular layer”9,27,36–38 will be used. The enantiomer L-valine is applied here as a representative example of a typical aliphatic amino acid. Fig. 2a shows the projection of the L-valine crystal structure onto the ac plane. In the crystal structure, it is possible to distinguish dimers consisting of two levorotatory valine molecules interlinked with hydrogen bonds (Fig. 2b). Pairing of molecules with formation of a dimer is typical for hydrophobic amino acids.7 Each dimer is bonded to neighbor dimers in the ab plane via hydrogen bonds. Thus, the thickness of the formed molecular layer in the direction of the c axis is equal to one dimer molecule (two valine molecules)‡ (Fig. 2a). The layers are interlinked via the van der Waals bonds.
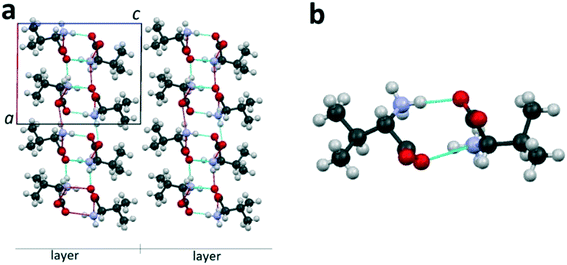 | ||
| Fig. 2 Projection of the L-valine crystal structure on the ac plane of the monoclinic cell (a) and its dimer molecule (b). Hereinafter, dotted lines are the hydrogen bonds. Images are constructed in the program Mercury40 using the structural data CSD LVALIN01.33 | ||
4. Crystal structure of equimolar heterocompounds of amino acids
In the literature, there are no data found on equimolar heterocompounds of amino acids having the same chirality. Therefore, this present article concentrates on compounds with molecular components that possess different chiralities, i.e. different signs of optical activity S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R′ = 1
R′ = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (see Fig. 1). A considerable contribution to the investigation of such heterocompounds was made by C. H. Görbitz, B. Dalhus and co-workers.7,16,28–32,41 They systematically studied equimolar compounds composed of molecules of different amino acids using SCXRD techniques. These compounds have been mentioned under various names including “1:1 complexes”,16,29,31,32 “diastereomeric complexes”,30 “pseudoracemic complexes”,41 and “quasiracemates”.7,28 To describe the interactions of different molecules in crystal structures of these compounds, C. H. Görbitz7 used designations LD–LD and L1–D1 in accordance with the geometry of the hydrogen bonds formed. In the following, the terms homo- and heteromolecular dimers are used to describe the crystal structures of amino acid heterocompounds.
1 (see Fig. 1). A considerable contribution to the investigation of such heterocompounds was made by C. H. Görbitz, B. Dalhus and co-workers.7,16,28–32,41 They systematically studied equimolar compounds composed of molecules of different amino acids using SCXRD techniques. These compounds have been mentioned under various names including “1:1 complexes”,16,29,31,32 “diastereomeric complexes”,30 “pseudoracemic complexes”,41 and “quasiracemates”.7,28 To describe the interactions of different molecules in crystal structures of these compounds, C. H. Görbitz7 used designations LD–LD and L1–D1 in accordance with the geometry of the hydrogen bonds formed. In the following, the terms homo- and heteromolecular dimers are used to describe the crystal structures of amino acid heterocompounds.
4.1. Equimolar heterocompounds of amino acids with homomolecular dimers
Heterocompounds of this type will be examined on the example of an equimolar discrete compound formed in the amino acids' system L-valine–D-norvaline.31Fig. 3a and b show the homomolecular dimer molecules of L-valine and D-norvaline, respectively. Fig. 3c presents the projection onto the ac plane of the crystal structure (S. G. C2) of the equimolar discrete compound of L-valine and D-norvaline molecules. In its crystal structure, it is possible to distinguish molecular layers having a thickness of one dimer molecule. The homomolecular dimers of valine and norvaline are alternating within each layer. The molecular layers are practically perpendicular to the c axis of the monoclinic cell. Neighbor layers are shifted along the a axis with respect to each other and, therefore, the translation along the c axis includes two molecular layers. Consequently, the monoclinic cell of the heterocompound doubled in size in comparison to that of L-valine (see Fig. 2).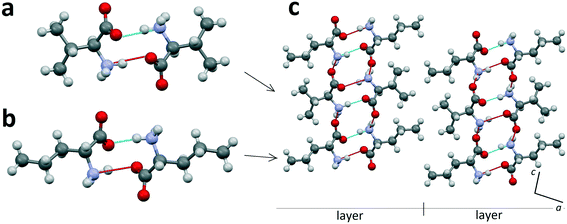 | ||
| Fig. 3 Homomolecular dimers of L-valine (a) and D-norvaline (b) and projection on the ac plane of the crystal structure of the equimolar discrete compound in the L-valine–D-norvaline system (c). Images are constructed in the program Mercury40 using the structural data CSD BERQEU.31 | ||
4.2. Equimolar heterocompounds of amino acids with heteromolecular dimers
Heterocompounds of this type will be examined on the example of an equimolar discrete compound formed in the amino acids' system D-valine–L-leucine.31Fig. 4a shows a heteromolecular dimer molecule composed of D-valine and L-leucine molecules. In Fig. 4b, the projection onto the bc plane of the crystal structure (S. G. P21) of the equimolar compound of D-valine and L-leucine molecules is presented. In this crystal structure, it is also possible to distinguish the molecular layers having a thickness of one dimer molecule. It is very important to note that in this case, all the dimeric molecules are identical. It can be seen that the neighbor layers are rotated by 180° in the ab plane with respect to each other (Fig. 4b). Consequently, in this case, the translation along the c axis also includes two molecular layers.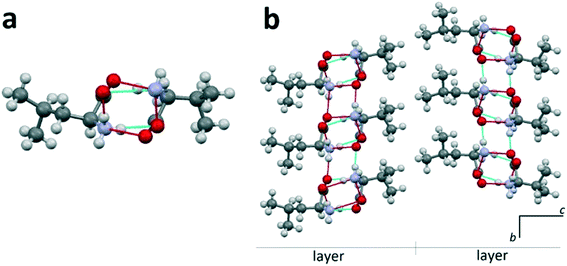 | ||
| Fig. 4 L-Leucine–D-valine dimer molecule (a) and projection on the bc plane of the crystal structure of the equimolar heterocompound in the D-valine–L-leucine system (b). Images are constructed in the program Mercury40 using the structural data CSD BERPET.31 | ||
4.3. Discussion
The concept of homo- and heteromolecular dimers is considered in the classification of discrete compounds in binary chiral systems (Fig. 1). This concept is also reflected in Tables 1 and 2, created using the results obtained by C. H. Görbitz, B. Dalhus and co-workers.7,16,28–32 It should be noted that only the equimolar heterocompounds of amino acids having components with different chiralities were studied. Among them, we consider here only the compounds whose crystal structures have been deciphered.| 1b. Different types of side chains in molecular components | |||
|---|---|---|---|
| No. | Branched molecule (component 1) | Linear molecule (component 2) | Ref. |
| 1 | L-Valine | D-α-Aminobutyric acid | 31 |
| 2 | L-Valine | D-Norvaline | 31 |
| 3 | L-Valine | D-Methionine | 31 |
| 4 | L-Valine | D-Norleucine | 16 |
| 5 | L-Isoleucine | D-Alanine | 29 |
| 6 | L-Isoleucine | D-Norleucine | 29 |
| 7 | L-Isoleucine | D-Norvaline | 29 |
| 8 | L-Isoleucine | D-Methionine | 29 |
| 9 | L-Isoleucine | D-α-Aminobutyric acid | 29 |
| 10 | L-allo-Isoleucine | D-Norleucine | 16 |
| 2. Equimolar heterocompounds of amino acids with heteromolecular dimers | |||
|---|---|---|---|
| 2a. Same type of side chains in molecular components | |||
| No. | Branched molecule (component 1) | Branched molecule (component 2) | Ref. |
| 1 | L-Leucine | D-Valine | 31 |
| 2 | L-Leucine | D-allo-Isoleucine | 7 |
| 3 | L-Isoleucine | D-allo-Isoleucine | 30 |
| 4 | L-Isoleucine | D-Valine | 29 |
| 5 | L-Isoleucine | D-Leucine | 29 |
| Amino acid | Conformation of molecule | ||
|---|---|---|---|
| Enantiomer L | True racemate LD | Heterocompound LD′ | |
| Valine | Extended and folded | Folded | Extended |
| Isoleucine | Extended and folded | Folded | Extended |
| allo-Isoleucine | Extended and folded | No data | Extended |
In Table 1, the L and D components of a heterocompound are arranged in separate columns. The equimolar heterocompounds of aliphatic α-amino acids with homo- and heteromolecular dimers are listed in parts 1 and 2 of Table 1, respectively. Each part comprises two categories, distinguished in accordance with the structure of radical R or the structure of the side chain of the corresponding α-amino acid (see Fig. 5 and 6). The side chain can be branched or linear. For example, molecules of valine (C5H11NO2), leucine (C6H13NO2), and isoleucine (C6H13NO2) (Fig. 6a) have a branched side chain, while molecules of norvaline (C5H11NO2), norleucine (C6H13NO2), and methionine (C5H11NO2S) (Fig. 6b) possess a linear one.
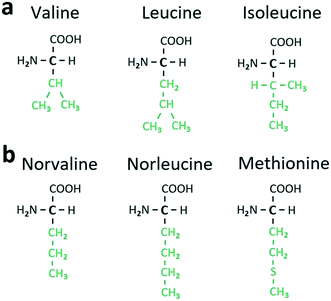 | ||
| Fig. 6 Structural formulae of certain aliphatic α-amino acids with branched (a) and linear (b) side chains (colored in green). | ||
In the heterocompounds with homomolecular dimers (Table 1, part 1), category 1a includes two compounds, wherein both molecular components are linear. Category 1b comprises ten compounds with the L component being branched and the D component linear. In the heterocompounds with heteromolecular dimers (Table 1, part 2), category 2a includes five compounds, wherein both constituents are branched. Category 2b comprises four compounds wherein the L component is branched, while the D component is linear.
This method of examining the equimolar heterocompounds of amino acids reveals a connection between the type of molecular dimers (homo- or heteromolecular) and the side chain structure of the molecular components (branched or linear). As seen in Table 1, all the heterocompounds composed of molecular components with linear side chains belong to group 1, i.e. have homomolecular dimers. In turn, all the heterocompounds consisting of components with branched side chains belong to group 2, i.e. feature heteromolecular dimers.
More complex is the case when one of the heterocompound components is characterized by a branched molecule and the other by a linear molecule. Compounds of such a type are present in both parts in Table 1. Affinity of a particular compound toward part 1 or 2 likely depends on the nature of the branched molecular component. It turned out that if the branched component is valine, isoleucine, or allo-isoleucine (part 1b, Table 1), then the heterocompound has homomolecular dimers. In turn, if the branched component is leucine (part 2b, Table 1), then the resulting heterocompound has heteromolecular dimers.
A probable explanation is based on the following. There are two conformations of molecules in the crystal structures of valine, isoleucine, and allo-isoleucine (Table 2). Relative to each other, one of the conformations can be considered as “extended” (Fig. 7a) and the other one as “folded” (Fig. 7b). The differences between the extended and folded conformations are clearly seen from the comparison of the corresponding torsion angles in Fig. 7.
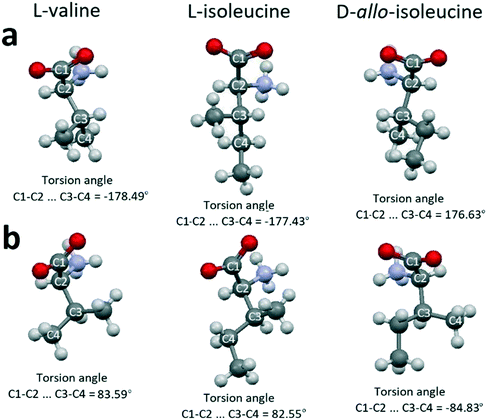 | ||
| Fig. 7 Extended (a) and folded (b) conformations of valine, isoleucine and allo-isoleucine molecules. Images are constructed in the program Mercury40 using the structural data CSD LVALIN01, LISLEU02 and DAILEU01, respectively. | ||
In the case of the equimolar homocompounds (true racemates LD, Fig. 1), molecules of valine, isoleucine, and allo-isoleucine adopt the folded conformation. In contrast, in the case of the equimolar heterocompounds (“quasiracemates” LD′, Fig. 1), they have the extended conformation. This is confirmed by the examples of four heterocompounds containing valine, five heterocompounds containing isoleucine, and one heterocompound comprising allo-isoleucine (see Tables 1 and 2). Another situation occurs in the case of leucine. All the molecules in the crystal structure of L-leucine are identical and, consequently, there is no need to attribute a conformation. It was found that all the heterocompounds containing leucine and linear molecules of other amino acids form heteromolecular dimers (see 2b in Table 1).
Table 3 summarizes the equimolar heterocompounds with the L component being the aromatic α-amino acid phenylalanine (C9H11NO2) and the D component as an aliphatic amino acid. Similar to Table 1, Table 3 consists of two parts comprising heterocompounds with homo- and heteromolecular dimers, respectively. The phenylalanine molecule (component 1) is considered as branched, since it contains a phenyl group. The molecules of the aliphatic amino acids (component 2) are both branched and linear molecules.
Heterocompounds with homomolecular dimers (Table 3, part 1) are represented exclusively by compounds whose both the components have branched molecules and one of the components is aromatic, while the other one is aliphatic. It should be noted that when both branched components are aliphatic molecules (see 2a in Table 1), the resulting heterocompound, in contrast, is composed of heteromolecular dimers.
Heterocompounds with heteromolecular dimers (Table 3, part 2) are represented solely by compounds whose one of the components has a branched molecule (aromatic component), while the other one has a linear molecule (aliphatic component). Thus, as in the case of heterocompounds composed of two aliphatic amino acids, the configuration of the molecular side chain greatly affects the structure of the equimolar heterocompounds containing an aromatic phenylalanine molecule.
5. Crystal structure of non-equimolar heterocompounds of amino acids
In the literature, we did not find examples of non-equimolar heterocompounds of amino acids with different chiralities where their crystal structure is known (see also Fig. 1). Therefore, in the following we focus on compounds composed of amino acids of the same chirality. One has already been reported by the authors, while the other one is new and its crystal structure will be presented and discussed below.5.1. Non-equimolar heterocompounds with molecules of the same chirality
Table 4 comprises the two amino acid systems, namely L-valine–L-isoleucine and L-valine–L-leucine, together with the associated non-equimolar compounds found. In the first one, a molecular component ratio of Val![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ile = 2
Ile = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1§ was determined and the compound was designated as V2I.27 The related results, including the crystal structure of compound V2I, limits of solid solutions in the system, and thermal deformations of the system components and compound V2I, are discussed in detail in the previous studies.9,27,37 The newly discovered compound in the L-valine–L-leucine system has a molecular ratio of Val
1§ was determined and the compound was designated as V2I.27 The related results, including the crystal structure of compound V2I, limits of solid solutions in the system, and thermal deformations of the system components and compound V2I, are discussed in detail in the previous studies.9,27,37 The newly discovered compound in the L-valine–L-leucine system has a molecular ratio of Val![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Leu = 3
Leu = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and is denoted V3L.
1 and is denoted V3L.
5.2. Crystal structure of the non-equimolar heterocompound V3L
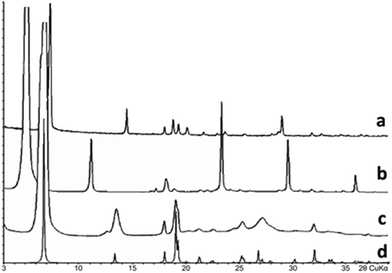
Samples of mixtures containing various contents of valine and leucine were prepared by isothermal crystallization. The reactants used were L(+)-valine and L(+)-leucine of 99% purity, purchased from Alfa Aesar, Massachusetts, USA. Pre-weighted mixtures of the reactants were put into a weighing bottle and dissolved in distilled water at 25 °C under continuous stirring with a glass rod. Water was added until complete dissolution of the substance. Then, the bottle was loosely covered with a cap and kept undisturbed for a period of at least three weeks at 25 °C until the solvent was completely evaporated. Precipitates were then analyzed by means of powder X-ray diffraction (PXRD) techniques. Fifteen samples of the prepared valine/leucine mixtures having a varying composition were studied using a Rigaku MiniFlex II diffractometer with the following settings: CuKα radiation, a scan speed of 2° min−1, a step of 0.02°, and 2θ range = 3–40°. The obtained PXRD data suggested the formation of the new compound V3L having a molar ratio of Val![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Leu = 3
Leu = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. As seen in Fig. 8, the diffraction pattern of this compound differs from those of the system components Val and Leu and is not their combination. Accordingly, two two-phase regions were observed in-between monophasic areas corresponding to compounds V3L, Val and Leu. The PXRD data showed that the major volume of the 75 mol% Val/25 mol% Leu sample belongs to phase V3L, and the diffraction peaks of other phases are practically absent. The most well-shaped crystal found in the sample was taken for structural analysis.
1. As seen in Fig. 8, the diffraction pattern of this compound differs from those of the system components Val and Leu and is not their combination. Accordingly, two two-phase regions were observed in-between monophasic areas corresponding to compounds V3L, Val and Leu. The PXRD data showed that the major volume of the 75 mol% Val/25 mol% Leu sample belongs to phase V3L, and the diffraction peaks of other phases are practically absent. The most well-shaped crystal found in the sample was taken for structural analysis.
Structural analysis of a V3L single crystal was performed using a diffractometer Agilent Technologies SuperNova with CuKα radiation and at a temperature of 100 K. The structure has been solved by direct methods and refined by means of the SHELX program incorporated in the OLEX2 program package. The carbon- and nitrogen-bound H atoms were placed in calculated positions and included in the refinement in the ‘riding’ model approximation. The experimental conditions applied and crystal structure parameters of the non-equimolar heterocompound V3L are summarized in Table 5. Fig. 9 shows the projection of the V3I compounds' crystal structure onto the ac plane of the monoclinic cell (Fig. 9a) and the V3L dimer molecule (Fig. 9b).
| Empirical formula | C21H46N4O8 |
|---|---|
| Formula weight | 482.62 |
| Temperature/K | 100 |
| Crystal system | Monoclinic |
| Space group | P21 |
| a/Å | 9.6267(7) |
| b/Å | 5.2704(2) |
| c/Å | 13.8290(17) |
| α/° | 90 |
| β/° | 109.943(11) |
| γ/° | 90 |
| Volume/Å3 | 659.56(11) |
| Z | 4 |
| ρ calc/g cm−3 | 1.215 |
| μ/mm−1 | 0.764 |
| F(000) | 264.0 |
| Radiation | CuKα (λ = 1.54184) |
| 2θ range for data collection/° | 6.8–139.962 |
| Index ranges | −11 ≤ h ≤ 11, −6 ≤ k ≤ 6, −16 ≤ l ≤ 16 |
| Reflections collected | 5431 |
| Independent reflections | 2475 [Rint = 0.0515, Rsigma = 0.0500] |
| Data/restraints/parameters | 2475/1/189 |
| Goodness-of-fit on F2 | 1.025 |
| Final R indices [I ≥ 2σ(I)] | R 1 = 0.0687, wR2 = 0.1864 |
| Final R indices [all data] | R 1 = 0.0829, wR2 = 0.2039 |
| Largest diff. peak/hole / e Å−3 | 0.35/−0.26 |
| Flack parameter | 0.0(2) |
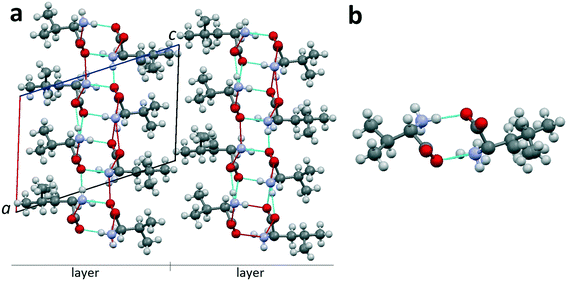 | ||
| Fig. 9 (a) Projection of the crystal structure of non-equimolar discrete heterocompound V3L on the ac plane of the monoclinic cell.42 (b) Dimer molecule of heterocompound V3L; occupation degree of the left position is 100% Val; occupancy of the right position is mixed: 50% Val and 50% Leu. Images are constructed in the program Mercury40 using the structural data CCDC 1903257. | ||
Heterocompound V3L contains the molecules of valine and leucine in a ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Two out of four molecular positions in the unit cell (Z = 4) are independent. One of the independent positions is occupied by a valine molecule (1 Val) (Fig. 9b, left part). The other independent position is disordered, i.e. it is characterized by mixed occupation. It can be occupied with equal probability by either a valine or leucine molecule (1/2 Val + 1/2 Leu) (Fig. 9b, right part). Therefore, the total number of valine molecules in the compounds' unit cell is (1 + 1/2) × 2 = 3, while the total number of leucine molecules is 1/2 × 2 = 1. Consequently, the heterocompound has a general formula of V3L. Since one of the independent positions has a mixed occupation in the crystal structure, two kinds of dimers exist: homomolecular Val–Val and heteromolecular Val–Leu ones. The dimers are connected to each other via hydrogen bonds in the bc plane and, therefore, form a layer with a thickness of one dimer molecule. The layers, in turn, are interlinked via van der Waals bonds (Fig. 9a).
1. Two out of four molecular positions in the unit cell (Z = 4) are independent. One of the independent positions is occupied by a valine molecule (1 Val) (Fig. 9b, left part). The other independent position is disordered, i.e. it is characterized by mixed occupation. It can be occupied with equal probability by either a valine or leucine molecule (1/2 Val + 1/2 Leu) (Fig. 9b, right part). Therefore, the total number of valine molecules in the compounds' unit cell is (1 + 1/2) × 2 = 3, while the total number of leucine molecules is 1/2 × 2 = 1. Consequently, the heterocompound has a general formula of V3L. Since one of the independent positions has a mixed occupation in the crystal structure, two kinds of dimers exist: homomolecular Val–Val and heteromolecular Val–Leu ones. The dimers are connected to each other via hydrogen bonds in the bc plane and, therefore, form a layer with a thickness of one dimer molecule. The layers, in turn, are interlinked via van der Waals bonds (Fig. 9a).
5.3. Discussion
In Fig. 10, the projections of the crystal structures of Leu, Val and Ile are opposed to those of the respective heterocompounds V3L and V2I. In addition, Table 6 contains the corresponding monoclinic cell parameters for the mentioned compounds.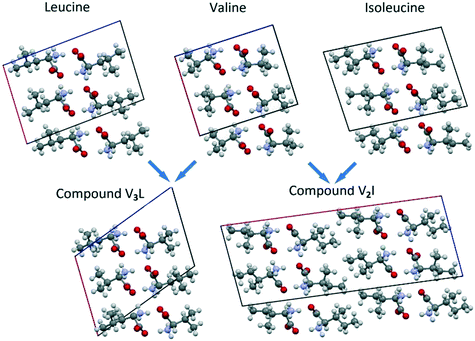 | ||
| Fig. 10 Projections of Leu, Val, Ile, V3L and V2I crystal structures on the corresponding planes of their monoclinic cells. | ||
| Compound | S. G. | a, Å | b, Å | c, Å | β, deg. | V, Å3 | Ref. |
|---|---|---|---|---|---|---|---|
| L-Ile | P21 | 9.75(2) | 5.32(2) | 14.12(2) | 95.8(2) | 723 | 43 |
| L-Val | P21 | 9.71(1) | 5.27(2) | 12.06(2) | 90.8(2) | 617.07 | 33 |
| L-Leu | P21 | 9.562(2) | 5.301(1) | 14.519(3) | 94.20(2) | 733.965 | 35 |
| V3L | P21 | 9.6267(7) | 5.2704(2) | 13.829(2) | 109.94(1) | 659.6 | This work |
| V2I | C2 | 25.7697(14) | 5.2445(2) | 9.6681(6) | 97.215(5) | 1296.29 | 27 |
As shown, the crystal structure of compound V3L closely resembles the structures of Leu, Val, and Ile enantiomers (Fig. 10). In the four compounds, the monoclinic cell comprises four molecules, and its asymmetric unit includes two molecules. Linear parameters a, b, and c and volume V of the monoclinic cells have close values as well. The angular parameter β of heterocompound V3L is notably larger than those of the Leu, Val, and Ile enantiomers (Fig. 10 and Table 6).
The molecular components of both non-equimolar heterocompounds V3L and V2I have the same (L) chirality and both have disordered molecular positions. Nevertheless, the comprehensive analysis of the crystal structures of V3L and V2I revealed substantial differences. First, in the case of compound V3L, only a half of the molecular positions show mixed occupation (Fig. 11a), while in compound V2I, all the molecular positions exhibit mixed occupation (Fig. 11b). Second, the monoclinic cell of V3L comprises four molecules, while that of V2I includes eight molecules, i.e. it is doubled in the direction of the longest axis of the cell.27
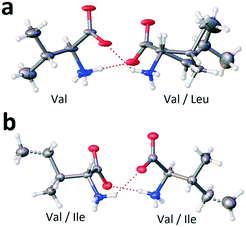 | ||
| Fig. 11 Occupancy of molecular positions in the dimers of non-equimolar heterocompounds V3L (a) and V2I (b). Ellipsoids reflect the 20% and 50% probabilities in the case of V3L and V2I, correspondingly. Images are constructed in the service OLEX2.44 | ||
The molecular components' conformations of non-equimolar heterocompounds V3L and V2I are discussed below. For convenience, the extended conformation of molecules is marked with superscript e (Vale and Ilee) and the folded conformation with symbol f (Valf and Ilef). As it was already mentioned, there are no conformations attributed to Leu molecules.
As given above, heterocompound V3L is characterized by one ordered and one disordered positions in the asymmetric unit. A 50/50 statistically mixed population of the disordered position implies that in every monoclinic cell, one dimer is composed solely of Val molecules (homomolecular dimer) and the other dimer is composed of both Val and Leu molecules (heteromolecular dimer) (Fig. 11a). One of the valine molecules in the homomolecular dimer has extended conformation Vale, while the other molecule has folded conformation Valf (dimer Vale–Valf). The valine molecule in the heteromolecular dimer shows extended conformation Vale (dimer Vale–Leu). It means that the ordered position is occupied by valine molecules having the extended conformation only, while the disordered position is populated with valine molecules having the folded conformation and leucine molecules (Table 7).
| System | Heterocompound | Dimer types | Number of independent molecular positions | Number of ordered molecular positions | Character of molecule in ordered position | Number of disordered molecular positions | Character of molecules in disordered position |
|---|---|---|---|---|---|---|---|
| Val–Leu | V3L | Vale–Valf | 2 | 1 | Vale | 1 | Valf, Leu |
| Vale–Leu | |||||||
| Val–Ile | V2I | Vale–Valf | 2 | 0 | — | 2 | Ilee, Ilef, Vale, Valf |
| Ilee–Ilef | |||||||
| Vale–Ilef | |||||||
| Valf–Ilee |
The monoclinic cell of heterocompound V2I has two independent molecular positions and both are disordered, i.e. characterized by mixed occupation. Molecules occupying one of these independent positions have extended conformation, while molecules present in the other independent position have folded conformation. Consequently, the crystal structure contains dimers of four types, namely, homomolecular dimers Vale–Valf and Ilee–Ilef and heteromolecular dimers Vale–Ilef and Valf–Ilee (Table 7).
Therefore, it can be concluded that the currently known non-equimolar heterocompounds of amino acids can be divided into two groups. V3L as the compound of the first group contains two independent molecular positions and only one of them is disordered, while V2I as the compound belonging to the second group has all its molecular positions disordered. It should be noted that in the L-isoleucine–L-leucine system, the recently found compound I3L belongs, apparently, to the first group. The results of its investigation were presented at a conference45 and will be published later.
6. Summary
Peculiarities of the crystal chemistry of heterocompounds formed by α-amino acids, i.e. compounds consisting of enantiomers of two different α-amino acids, have been reviewed and studied. The consideration is complicated by the lack of a common approach for terminology and allocation of compounds with chiral molecules in the literature. This complication for multi-component crystals, in general, was mentioned also, for example, by G. R. Desiraju et al.46 To improve this situation, a classification of discrete compounds formed in binary chiral systems either by a single substance (homomolecular compounds) or by different substances (heteromolecular compounds) is proposed. The classification is based on both chemical and crystallographic characteristics of the discrete compounds. The concept of homo- and heteromolecular dimers is introduced based on the analysis of published results for amino acid equimolar heterocompounds. Heterocompounds composed of homomolecular dimers are characterized by alternating the dimer molecules of each substance, while those consisting of heteromolecular dimers are characterized by the dimer molecules of different substances. The correlation found between the dimer type and the side chain structure (linear or branched) and the conformation (extended or folded) of the relevant heterocompounds' molecules is discussed.Equimolar heterocompounds of amino acids having different chiralities are known from the published literature. Their crystallochemical characteristics were analyzed based on the data available from the CSD and other sources. The proposed concept of homo- and heteromolecular dimers allowed the division of the known equimolar heterocompounds into two groups: those with homo- and those with heteromolecular dimers. It was found that the molecules of the same substance have different conformations in the homo- and heteromolecular dimers. In the case of the equimolar homocompounds (true racemates), the branched side chain molecules have a folded conformation, while in the case of the equimolar heterocompounds, they are characterized by an extended conformation. Equimolar heterocompounds of amino acids having the same chirality have not yet been found in the published literature.
Non-equimolar heterocompounds are very rare. Only three examples of the compounds of chiral substances have been reported in the literature. Two compounds consist of molecules having different chiralities and their crystal structures are unknown.14 The third compound was described by the present authors27 and is an example of a non-equimolar heterocompound composed of the same chirality molecules. The crystal structure of this compound V2I, formed in the system L-Val–L-Ile, is characterized by two disordered molecular positions in the asymmetric unit of its monoclinic cell (S. G. C2). A recently revealed further example is compound V3L in the system L-Val–L-Leu. Its crystal structure exhibits one ordered and one disordered molecular positions in the asymmetric unit of its monoclinic cell (S. G. P21), thus differing from the crystal structure of heterocompound V2I. The crystal structures of V2I and V3L were analyzed using the concepts of homo- and heteromolecular dimers, side chain types (linear or branched) and molecular conformations (extended or folded). Of course, further assessment of the presented structural trends requires more data. Thus, in view of the poor quantity of known crystal structures of non-equimolar compounds, there is an obvious need for continuation of their investigations.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The investigations were performed using equipment of the Resource Centre “Centre for X-ray Diffraction Studies” of St. Petersburg State University. The authors appreciate the financial support provided by the Russian Foundation for Basic Research (Project 18-35-00183 mol_a). Open Access funding provided by the Max Planck Society.Notes and references
- A. D. McNaught and A. Wilkinson, Compendium of Chemical Terminology, Blackwell Scientific Publications, Oxford, 2nd edn, 1997, p. 464 Search PubMed.
- A. I. Kitaigorodsky, Molecular crystals, Nauka, Moscow, 1971, p. 424(Rus) Search PubMed.
- G. R. Desiraju, Crystal Engineering: From Molecule to Crystal, J. Am. Chem. Soc., 2013, 135(27), 9952–9967 CrossRef CAS PubMed.
- A. M. Rouhi, Chiral Chemistry, Chem. Eng. News, 2004, 82(24), 47–62 CrossRef.
- H. Murakami, From Racemates to Single Enantiomers – Chiral Synthetic Drugs over the last 20 Years, Top. Curr. Chem., 2007, 269, 273–299 CrossRef CAS PubMed.
- J. Jacques, A. Collet and S. H. Wilen, Enantiomers, Racemates and Resolutions, J. Wiley & Sons, New York, 1981, p. 233 Search PubMed.
- C. H. Görbitz, Crystal structures of amino acids: from bond lengths in glycine to metal complexes and high pressure polymorphs, Crystallogr. Rev., 2015, 21(3), 160–212 CrossRef.
- E. V. Boldyreva, Combined X-ray diffraction and Raman spectroscopy studies of phase transitions in crystalline amino acids at low temperatures and high pressures: selected examples, Phase Transitions, 2009, 82(4), 303–321 CrossRef CAS.
- E. N. Kotelnikova, A. I. Isakov and H. Lorenz, Non-equimolar discrete compounds in binary chiral systems of organic substances, CrystEngComm, 2017, 19, 1851–1869 RSC.
- G. A. Morales and F. R. Fronczek, A Kryptoracemic Hydroperoxide, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1996, 52(5), 1266–1268 CrossRef.
- R. Bishop and M. L. Scudder, Multiple Molecules in the Asymmetric Unit (Z′ > 1) and the Formation of False Conglomerate Crystal Structures, Cryst. Growth Des., 2009, 9(6), 2890–2894 CrossRef CAS.
- M. Bergmann and M. Lissitzin, Die überzähligen Stereoisomeren der γ-Amino-β-oxy-buttersäure, Ber. Dtsch. Chem. Ges., 1930, 63(2), 310–313 CrossRef.
- A. A. Bredikhin, Z. A. Bredikhina, D. V. Zakharychev, A. I. Samigullina and A. T. Gubaidullin, 4-Benzoylamino-3-hydroxybutyric Acid, Historically First “Anomalous Racemate”: Reinvestigation, Cryst. Growth Des., 2015, 15(3), 1362–1373 CrossRef CAS.
- A. Fredga, Steric correlations by the quasi-racemate method, Tetrahedron, 1960, 8, 126–144 CrossRef CAS.
- M. D. Eddleston, M. Arhangelskis, T. Fričšić and W. Jones, Solid state grinding as a tool to aid enantiomeric resolution by cocrystallization, Chem. Commun., 2012, 48, 11340–11342 RSC.
- B. Dalhus and C. H. Görbitz, Molecular aggregation in selected crystalline 1:1 complexes of hydrophobic D- and L-amino acids. II. The D-norleucine series, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1999, 55(7), 1105–1112 CrossRef.
- M. Andersson, A. Fredga and B. Jerslev, Anomalous Racemates of Malic Acid, Acta Chem. Scand., 1966, 20, 1060–1063 CrossRef CAS.
- B. Dalhus and C. H. Görbitz, Non-centrosymmetric racemates: space-group frequencies and conformational similarities between crystallographically independent molecules, Acta Crystallogr., Sect. B: Struct. Sci., 2000, 56(4), 715–719 CrossRef CAS PubMed.
- A. I. Isakov, E. N. Kotelnikova and H. Lorenz, Non-Equimolar Discrete Phases Formed in the System of Malic Acid Enantiomers, Chem. Eng. Technol., 2015, 38(6), 1047–1052 CrossRef CAS.
- M. E. Breen, S. L. Tameze, W. G. Dougherty, W. S. Kassel and K. A. Wheeler, Structural Studies of Enantiomers, Racemates, and Quasiracemates. 2-(3-Bromophenoxy)propionic Acid and 2-(3-Methoxyphenoxy)propionic Acid, Cryst. Growth Des., 2008, 8(10), 3863–3870 CrossRef CAS.
- M. S. Hendi, R. E. Davis, V. M. Lynch and K. A. Wheeler, Structural studies of enantiomers, racemates, and quasiracemates. 2-(2,4-dichlorophenyl)propanoic acid and 2-(2-chloro-4-nitrophenyl)propanoic acid, Cryst. Eng., 2001, 4, 11–24 CrossRef CAS.
- M. S. Hendi, P. Hooter, R. E. Davis, V. M. Lynch and K. A. Wheeler, Structural Studies of Enantiomers, Racemates, and Quasiracemates: N-(4-Methylbenzoyl)methylbenzylamine and N-(4-Nitrobenzoyl)methylbenzylamine, Cryst. Growth Des., 2004, 4(1), 95–101 CrossRef CAS.
- S. L. Fomulu, M. S. Hendi, R. E. Davis and K. A. Wheeler, Structural Studies of Enantiomers, Racemates, and Quasiracemates. 2-(2,4,5-Trichloroanilino)propanoic Acid and 2-(2,4,5-Trichlorophenoxy)propanoic Acid, Cryst. Growth Des., 2002, 2(6), 637–644 CrossRef CAS.
- S. L. Fomulu, M. S. Hendi, R. E. Davis and K. A. Wheeler, Structural Studies of Enantiomers, Racemates, and Quasiracemates. N-(2-Chlorobenzoyl)methylbenzylamine and N-(2-Bromobenzoyl)methylbenzylamine, Cryst. Growth Des., 2002, 2(6), 645–651 CrossRef CAS.
- J. T. Cross, N. A. Rossi, M. Serafin and K. A. Wheeler, Tröger's base quasiracemates and crystal packing tendencies, CrystEngComm, 2014, 16, 7251–7258 RSC.
- C. B. Aakeröy, T. I. Cooke and M. Nieuwenhuyzen, The crystal structure of the molecular cocrystal L-malic acid L-tartaric acid (1/1), Supramol. Chem., 1996, 7(2), 153–156 CrossRef.
- A. I. Isakov, E. N. Kotelnikova, S. Muenzberg, S. N. Bocharov and H. Lorenz, Solid Phases in the System L-Valine – L-Isoleucine, Cryst. Growth Des., 2016, 16, 2653–2661 CrossRef CAS.
- C. H. Görbitz and P. Karen, Twin Displacive Transitions in Amino Acid Quasiracemates, J. Phys. Chem. B, 2015, 119(15), 4975–4984 CrossRef PubMed.
- B. Dalhus and C. H. Görbitz, Molecular aggregation in crystalline 1:1 complexes of hydrophobic D- and L-amino acids. I. The L-isoleucine series, Acta Crystallogr., Sect. B: Struct. Sci., 1999, 55(3), 424–431 CrossRef PubMed.
- B. Dalhus and C. H. Görbitz, Structural relationships in crystals accommodating different stereoisomers of 2-amino-3-methylpentanoic acid, Acta Crystallogr., Sect. B: Struct. Sci., 2000, 56(4), 720–727 CrossRef CAS PubMed.
- B. Dalhus and C. H. Görbitz, Molecular aggregation in selected crystalline 1:1 complexes of hydrophobic D- and L-amino acids. III. The L-leucine and L-valine series, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1999, 55(9), 1547–1555 CrossRef.
- C. H. Görbitz, K. Rissanen, A. Valkonen and Å. Husabø, Molecular aggregation in selected crystalline 1:1 complexes of hydrophobic D- and L-amino acids. IV. The L-phenylalanine series, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2009, 65(6), o267–o272 CrossRef PubMed.
- B. Dalhus and C. H. Görbitz, Crystal Structures of Hydrophobic Amino Acids. I. Redeterminations of L-Methionine and L-valine at 120 K, Acta Chem. Scand., 1996, 50, 544–548 CrossRef CAS.
- B. Dalhus and C. H. Görbitz, Triclinic Form of DL-Valine, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1996, 52(7), 1759–1761 CrossRef.
- C. H. Görbitz and B. Dalhus, Redetermination of L-Leucine at 120K, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1996, 52(7), 1754–1756 CrossRef.
- E. N. Kotelnikova, A. I. Isakov, L. Yu. Kryuchkova and H. Lorenz, Acids with Chiral Molecules as Essential Organic Compounds of Biogenic–Abiogenic Systems, in Processes and Phenomena on the Boundary between Biogenic and Abiogenic Nature, ed. O. V. Frank-Kamenetskaya, D. Y. Vlasov, E. G. Panova and S. N. Lessovaia, Springer, 2019, Lecture Notes in Earth System Sciences Search PubMed.
- A. I. Isakov, E. N. Kotelnikova, S. N. Bocharov, A. A. Zolotarev Jr and H. Lorenz, Thermal deformations of the crystal structures of L-valine, L-isoleucine and discrete compound V2I, in BIWIC 2013 – 23rd International Workshop on Industrial Crystallization, ed. H. Lorenz and H. Buchholz, Cuvillier Verlag, Göttingen, 2016, pp. 7–12 Search PubMed.
- A. I. Isakov, E. N. Kotelnikova, S. N. Bocharov, A. A. Zolotarev Jr and H. Lorenz, Thermal deformations of the crystal structures of L-valine, L-isoleucine and discrete compound V2I, in BIWIC 2013 – 23rd International Workshop on Industrial Crystallization, ed. H. Lorenz and H. Buchholz, Cuvillier Verlag, Göttingen, 2016, pp. 7–12 Search PubMed.
- C. H. Görbitz, P. Karen, M. Dušek and V. Petříček, An exceptional series of phase transitions in hydrophobic amino acids with linear side chains, IUCrJ, 2016, 3(5), 341–353 CrossRef PubMed.
- C. F. Macrae, I. J. Bruno, J. A. Chisholm, P. R. Edgington, P. McCabe, E. Pidcock, L. Rodriguez-Monge, R. Taylor, J. van de Streek and P. A. Wood, Mercury CSD 2.0 – new features for the visualization and investigation of crystal structures, J. Appl. Crystallogr., 2008, 41(2), 466–470 CrossRef CAS.
- C. H. Görbitz, B. Dalhus and G. M. Day, Pseudoracemic amino acid complexes: blind predictions for flexible two-component crystals, Phys. Chem. Chem. Phys., 2010, 12, 8466–8477 RSC.
- CCDC 1903257.
- C. H. Görbitz and B. Dalhus, L-isoleucine, Redetermination at 120K, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1996, 52(6), 1464–1466 CrossRef.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, OLEX2: a complete structure solution and analysis program, J. Appl. Crystallogr., 2009, 42(2), 339–341 CrossRef CAS.
- L. Yu. Kryuchkova, E. N. Kotelnikova, A. A. Zolotarev Jr and H. Lorenz, Limits of solid solutions in the L-leucine—L-isoleucine system according to PXRD and SCXRD data, in BIWIC 2018 – 25th International Workshop on Industrial Crystallization, ed. Y. Cartigny and N. Couvrat, Rouen, 2018, p. 225 Search PubMed.
- G. R. Desiraju, J. J. Vittal and A. Ramanan, Crystal Engineering: A Textbook, World Scientific Publishing Co. Pte. Ltd., Singapore, 2011, p. 131 Search PubMed.
Footnotes |
| † CCDC 1903257. For crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ce01333d |
| ‡ C. H. Görbitz7,39 uses the term “bilayer” to define such layers. |
| § Hereinafter, international abbreviations of the amino acid names are used: Val for valine, Ile for isoleucine, and Leu for leucine. |
| This journal is © The Royal Society of Chemistry 2020 |