Simple and fast electrochemical detection of sequence-specific DNA via click chemistry-mediated labeling of hairpin DNA probes with ethynylferrocene
Abstract
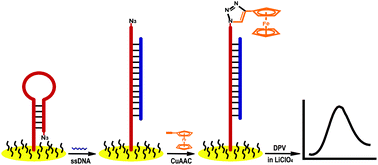
A universal and straightforward electrochemical biosensing strategy for the detection and identification of sequence-specific DNA via click chemistry-mediated labeling of hairpin DNA probes (hairpins) with ethynylferrocene was reported. In the target-unbound form, the immobilized hairpins were kept in the folded stem–loop configuration with their azido terminals held in close proximity of the electrode surface, making them difficult to be labeled with ethynylferrocene due to the remarkable steric hindrance of the densely packed hairpins. Upon hybridization, they were unfolded and underwent a large conformational change, thus enabling the azido terminals to become available for its subsequent conjugation with ethynylferrocene via the Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC). After that, the quantitatively labeled ethynylferrocene could be exploited as the electroactive probes to monitor the DNA hybridization. As the unfolded hairpins were labeled in a stoichiometric ratio of 1 : 1, the electrochemical measurement based on differential pulse voltammetry enabled a reliable quantification of sequence-specific DNA. Under optimal conditions, the strategy could detect target single-stranded DNA (ssDNA) down to 0.296 pM with a good linear response over the range from 1 pM to 1 nM, and had excellent specificity in the genotyping of single-nucleotide polymorphisms. Furthermore, it also exhibited good detection reliability in serum samples and required no complicated protocols. More importantly, the simplicity of this strategy together with its compatibility with microfluidic chips makes it show great potential in clinical applications, where simple procedures are generally preferred.

 Please wait while we load your content...
Please wait while we load your content...