4D printing for biomedical applications
Arkodip
Mandal†
 and
Kaushik
Chatterjee
and
Kaushik
Chatterjee
 *
*
Department of Materials Engineering, Indian Institute of Science, Bengaluru, Karnataka 560012, India. E-mail: kchatterjee@iisc.ac.in; Tel: +91-80-22933408
First published on 27th February 2024
Abstract
While three-dimensional (3D) printing excels at fabricating static constructs, it fails to emulate the dynamic behavior of native tissues or the temporal programmability desired for medical devices. Four-dimensional (4D) printing is an advanced additive manufacturing technology capable of fabricating constructs that can undergo pre-programmed changes in shape, property, or functionality when exposed to specific stimuli. In this Perspective, we summarize the advances in materials chemistry, 3D printing strategies, and post-printing methodologies that collectively facilitate the realization of temporal dynamics within 4D-printed soft materials (hydrogels, shape-memory polymers, liquid crystalline elastomers), ceramics, and metals. We also discuss and present insights about the diverse biomedical applications of 4D printing, including tissue engineering and regenerative medicine, drug delivery, in vitro models, and medical devices. Finally, we discuss the current challenges and emphasize the importance of an application-driven design approach to enable the clinical translation and widespread adoption of 4D printing.
1 Introduction
Since its inception in the 1980s, advancements in three-dimensional (3D) printing modalities have enabled the rapid and large-scale fabrication of complex, high-resolution architectured matter with desired spatial compositional complexity and tunable properties and functionalities from an expanding arsenal of diverse materials, spanning from soft matter1–3 to ceramics4,5 and metals.6,7 While there are more than fifty different 3D printing technologies according to the American Society for Testing and Materials (ISO/ASTM 52900:2015),8 extrusion9–12 and light-based13,14 printers are the most popular and can process a multitude of raw materials, including photocurable resin/solutions, thixotropic gels, powders, and filaments. Extrusion-based 3D printing systems employ either pneumatic or mechanical dispensing mechanisms, such as pistons or screws, for the precise deposition of materials onto a planar substrate or within a self-healing suspension medium.15,16 Several light-based techniques are available for the 3D printing of photocurable resin/solutions, including stereolithography (SLA), digital light processing (DLP), and volumetric additive manufacturing (VAM), each distinguished by its mode of operation. SLA utilizes a scanning laser beam for point-by-point crosslinking. Conversely, DLP techniques employ photomasks to project 2D patterns for layer-by-layer curing. VAM, a layerless method, achieves individual voxel crosslinking by irradiating dynamically evolving light patterns onto a rotating resin vial, facilitating the creation of 3D structures. In recent years, a slew of scientific breakthroughs exemplified by technologies such as CLIP (Continuous Interface Liquid Production)17 and iCLIP (injection Continuous Interface Liquid Production),18 HARP (High-Area Rapid Printing)19 has transformed 3D printing beyond its traditional role in small-scale prototyping, propelling it into the realm of commercial manufacturing,20 whereas technologies such as CAL (computed axial lithography),21,22 xolography 3D printing,23,24 MM3D (Multimaterial Multinozzle) printing,25 RM-3DP (Rotational Multimaterial 3D Printing)26 have introduced unprecedented capabilities and expanded the horizons of 3D printing.In the biomedical domain, 3D printing has established itself as a versatile technology with diverse applications including, but not limited to, regenerative medicine,27–29 microneedle-based drug/vaccine patches,30–33 flexible bioelectronics,34–36 biohybrid actuators,37–39 customized implants and prosthetics,40–43 and specialized surgical tools.44–46 3D bioprinting has received widespread attention owing to its robust capabilities in recapitulating biomimetic tissue constructs for regenerative medicine,47–51 as well as for developing organotypic models for toxicology, disease modeling, and drug discovery.52–55 Nonetheless, questions persist regarding the requisite threshold of complexity necessary to faithfully emulate the functional characteristics of native tissues within engineered constructs.56 This lingering question arises from the inherent dynamism observed in human tissues, which undergo continuous remodeling and orchestrated morphological adaptations in response to physiological cues. In contrast, engineered tissues produced via 3D printing techniques predominantly exhibit a static nature. They often present lower cellular densities and limited cell–cell interactions, collectively impeding their capacity to mature into fully functional tissue analogs. Consequently, this has motivated investigation into the design of scaffolds endowed with stimuli-responsive attributes aimed at augmenting the physiological relevance and biological functionality of these engineered tissue constructs.57,58 The stimuli-triggered temporal evolution, encompassing time scales ranging from seconds to days, in the structural and functional attributes of 3D-printed constructs has been coined ‘4D printing’ (A broader definition will be presented in the next section). This terminology arises from the incorporation of an additional temporal dimension, complementing the three spatial dimensions, within the fabrication process.
4D printing facilitates controlled morphological transformations in 3D-printed structures, simplifying complex print paths and expediting fabrication. By leveraging 4D printing techniques, it is possible to self-fold 2D planar sheets into anatomically relevant geometries with programmable local curvature for tissue engineering applications in a simple and facile manner, bypassing the need for support structures. Dynamic surface topographical conditioning induced by external stimuli can decide cell fate through mechanotransduction. In hydrogels with embedded cell populations, stimuli-induced changes in the extracellular matrix (ECM) can serve as user-defined cues to guide or direct developmental processes. Microscale structures with shape-changing capabilities hold promise for drug encapsulation and targeted release in response to specific environmental changes. Hydrogel-based drug delivery devices can be designed for controlled degradation under precise conditions, including enzymatic triggers or alterations in temperature and pH, ensuring localized drug release. Furthermore, 4D-printed self-powered biohybrid actuators that can sense and respond to environmental stimuli have garnered significant interest, with applications ranging from artificial muscles to micro-scale robotics.
Given the myriad applications and enormous impact of 4D printing, as evidenced by an exponential increase in publications over the past few years,57 it is imperative to critically assess the current state of 4D printing as it pertains to diverse classes of biomedical materials. In this Perspective, we present a broad definition of 4D printing in the biomedical context, evaluate the current 4D actuation strategies, delve into recent advancements, and offer perspectives into the potential transformative impact of 4D printing across diverse biomedical applications.
2 Defining 4D printing and 4D bioprinting
4D printing encompasses the predictive design and fabrication of constructs ranging in size from the nanoscale to the macroscale via 3D printing techniques, which exhibit the capacity for programmable morphological transformation, modification in physicochemical characteristics, or a shift in functionality within a predefined temporal framework when triggered via exogenous or endogenous stimuli. In the biomedical domain, 4D bioprinting involves the incorporation of single or multiple living cellular components, such as embryoid bodies, spheroids, and organoids, into a bioink matrix and subsequently printed into tissue-mimetic architectures capable of sustaining the long-term viability of encapsulated cells with avenues to induce morphogenic transformations or modulate cellular processes, including proliferation, migration, organization, and differentiation, through the application of biocompatible physiochemical or mechanical cues.3 4D design principles and actuation mechanisms
3.1 Soft matter
Shear-induced alignment during extrusion: Shear-induced alignment of embedded rigid elements (micro/nano-scale fibers/particles) resulting from shear forces at the nozzle tip during the extrusion process (refer Fig. 1(a)-ii) has been exploited for designing anisotropic constructs aimed at orchestrating cellular organization,61 engineering textured composites,62 enhancing electrical conductivity,63 as well as for stimuli-responsive shape-morphing systems.64–68 This approach draws inspiration from the hygroscopic-driven motion in plants, wherein differential shrinkage along distinct axes resulting from the directional orientation of cellulose microfibrils within cell walls of fibrous layers enables seed dispersal at opportune moments.69–71 Biomimetically designed, anisotropic 3D-printed constructs exhibit differences in mechanical properties and swelling behavior between the print path (along the orientation of stiff elements) and perpendicular to the print path. In printed constructs where adjacent layers have different orientations with respect to each other (refer Fig. 1(a)-i), anisotropic swelling induces curvature due to interfacial constraints, which can be theoretically modeled for programmable shape changes as well as for the inverse design of complex, curved architectures64,71–74 (refer Fig. 1(a)-iii).
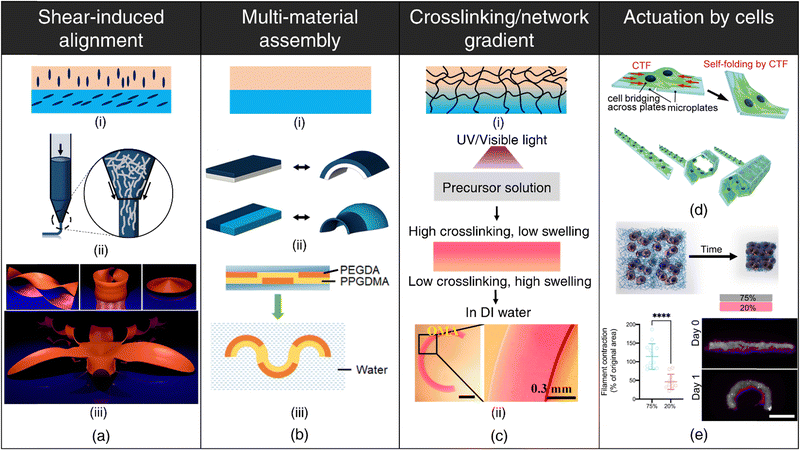 | ||
| Fig. 1 4D printing of hydrogels. (a) Shear-induced alignment. (i) Anisotropic orientation of rigid elements within distinct layers. (ii) Alignment of embedded rigid elements (e.g., fibers) due to shear forces at the nozzle tip. (iii) Numerical results displaying various geometries like helicoid, catenoid, sombrero, and orchid formed by the shape-morphing of flat filament bilayers containing anisotropically oriented fibers. Reproduced with permission from ref. 74. (b) Multi-material assembly. (i) Utilization of hydrogels with varying swelling capacities in different layers. (ii) Configuration variations in hydrogel layers resulting in diverse shape-morphing behaviors. Adapted with permission from ref. 78. (iii) Shape-morphing in water exhibited by a composite beam comprising hydrophobic poly(propylene glycol) dimethacrylate (PPGDMA) and hydrophilic poly(propylene glycol) diacrylate (PEGDA) hydrogels arranged in distinct layers. Reproduced with permission from ref. 99. (c) Crosslinking/network gradient (i) establishment of a gradient in crosslinking or polymer network density throughout the hydrogel thickness. (ii) Development of a photocrosslinking density upon UV or visible light exposure to a precursor solution as a consequence of attenuation of light with increasing depth, resulting in anisotropic water uptake and bending of the hydrogel beam. Reproduced under the terms of CC-BY-NC-ND 4.0 International License from ref. 90. Actuation by cell forces (d), (e) (d) Self-folding of plates facilitated by cell traction forces (CTF). Reproduced under the terms of CC-BY International License from ref. 104. (e) Schematic depicting contraction of fibrous hydrogel assemblies by cells. Differential contraction in fibrous hydrogel composites comprising varying fiber densities, leading to the formation of shape-shifting structures. Reproduced under the terms of CC-BY-NC International License from ref. 105. | ||
The degree of alignment and corresponding swelling anisotropy can be effectively modulated primarily by adjusting the nozzle diameter and print.64,67,75 Smaller nozzle diameters, for instance, lead to more pronounced alignment as shear forces inversely scale with nozzle diameter. However, the adoption of smaller nozzle diameters may be constrained by the rheological properties of the ink. Differences between the print speed (velocity of the nozzle) and the rate of extrusion (feed rate) can lead to die-swelling or thinning of the viscoelastic inks,76 further influencing the extent of alignment.
Shear-induced alignment represents a recurring theme within the domain of 4D printing, with analogous strategies for shape-memory polymers (SMPs), liquid crystal elastomers/polymers, and metals, which will be explored in subsequent sections.
Multi-material assembly: Multi-material assembly is the most straightforward technique to generate complex, programmable bending, buckling, folding, or twisting motions by harnessing the difference in equilibrium water absorption capacities of the constituent hydrogels.77–84 The spatial arrangement of the constituent hydrogels dictates the directions of bending stresses, which, in turn, governs the shape-morphing dynamics (refer Fig. 1(b)). Since the constituent hydrogels typically possess different chemistries, it is important to ensure robust interfacial bonding to prevent delamination.
The inception of multi-material design for inducing stimuli-responsive shape changes can be traced to the mid-1700s, when bimetallic strips were engineered to respond to temperature fluctuations, capitalizing on the disparities in their thermal expansion coefficients. The Timoshenko model, which was originally conceived to elucidate the bending deformation of bimetallic thermostats, has been re-purposed and extended to describe the bending deformation exhibited by bilayer hydrogel-based 4D systems. The modified Timoshenko equation, tailored to describe the curvature of moisture-swelling bilayer models, can be expressed as follows:
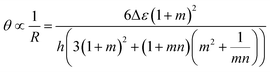 | (1) |
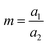 ,
, 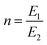 , where a1 and a2 stand for the thickness of the strips while E1 and E2 represent their respective elastic moduli.
, where a1 and a2 stand for the thickness of the strips while E1 and E2 represent their respective elastic moduli.
While the modified Timoshenko equation offers valuable insights into the influence of hydrogel properties and geometry on induced curvature, it is imperative to recognize that the application of the Timoshenko equation comes with several pertinent limitations that necessitate careful consideration.57 (a) The Timoshenko equation operates under the assumption of small, elastic deformations, whereas hydrogels exhibit a viscoelastic nature, often leading to substantial deformations. (b) The Timoshenko equation relies on static values for parameters such as strain mismatch, elastic moduli, and layer thicknesses. However, these attributes in hydrogel bilayers are intrinsically dynamic due to the influence of water swelling. Consequently, the temporal variations in these properties present a significant challenge when employing this equation. (c) Notably, the Timoshenko equation is confined to the analysis of narrow strip-like beams and does not account for the influence of beam length on curvature. Nevertheless, in the case of plate-like assemblies, the aspect ratio of bilayers, defined as the ratio of length to thickness, represents a critical parameter with the potential to induce distinct conformational changes.77,85
Crosslinking/network gradient: For single-component, non-responsive hydrogels, the deliberate induction of structural anisotropy within the hydrogel matrix is a viable strategy to encode shape deformations in 3D-printed constructs86–92 (refer Fig. 1(c)-(i)). This can be accomplished through strategic interventions either during the polymerization process or via subsequent post-processing procedures. This method finds notable applicability within light-based 3D printing techniques such as DLP, where precise modulation of light intensity (via grayscale patterns or photomasks) and exposure time at individual pixel-level dictates monomer conversion and crosslinking and enables fabrication of constructs characterized by a spatial gradient in the degree of crosslinking.86–89,93 Photoabsorber-induced light attenuation is also an up-and-coming technique to generate differential crosslinking along the thickness of the printed construct.90–92 Regions endowed with higher crosslinking density exhibit reduced swelling compared to regions with lower crosslinking, giving rise to anisotropic swelling that facilitates shape-morphing (refer Fig. 1(c)-ii). Photoabsorber-induced light intensity gradient can also lead to constructs that possess internal stresses arising from sequential volume shrinkage during the photopolymerization process. Subsequently, upon mechanical detachment from the substrate, these constructs undergo shrinkage-induced bending deformation autonomously, without the need for external stimulus.94
Post-printing procedures such as dehydration or selective degradation of printed constructs are simple, effective techniques for establishing network and crosslinking gradients, respectively. During the dehydration process, printed constructs undergo sequential, non-uniform moisture loss, with the top region (exposed to air) experiencing a faster loss of water content than the bottom regions (attached to the substrate). Consequently, this temporal variation in water removal results in contractile residual stresses and the concomitant development of a denser polymer network within the upper stratum.95 Upon subsequent rehydration of the dried hydrogels, this gradient in polymer network density drives anisotropic swelling, thereby enabling shape-morphing.95–97 The post-dehydration network gradient can be meticulously engineered by depositing multiple polymer layers in specific regions, yielding higher network densities in targeted areas, and can be tailored to obtain complex, shape deformations.98 Additionally, the application of selective degradation, such as photodegradation induced by UV light exposure in distinct regions, offers a facile avenue to introduce a crosslinking gradient and can be instrumental for developing dynamically tunable, shape-morphing scaffolds.99,100
Integration of different mechanisms: Hydrogels with complex temporal dynamics or superior shape-morphing attributes can be designed through the rational integration of multiple shape-morphing mechanisms within a single construct (multi-material assembly + crosslinking gradient,90 multi-material assembly + dehydration gradient,101 multi-material assembly + degradation gradient,102 multi-material assembly + osmolarity gradient,103 crosslinking gradient + dehydration gradient95,96). For instance, hydrogel constructs featuring a crosslinking gradient can elicit more pronounced bending curvatures due to the synergistic effects of both mechanisms.90 However, it is crucial to ensure that these bending tendencies align harmoniously, preventing counteractive effects that could diminish curvatures. Given the myriad of potential combinations, there are instances where a system may exhibit unexpected behavior, providing fertile grounds for innovative exploration and discovery.
Actuation by living cells: Advances in synthetic biology have enabled the development of genetically engineered cells that can be designed to react to specific biochemical or physical cues and execute predefined functions such as fluoresce, synthesize molecules, proliferate, or contract. The incorporation of engineered living cells as responsive components within polymeric scaffolds has given birth to the field of engineered living materials (ELMs).106–108 ELMs offer a multifaceted platform with compelling potential across applications encompassing biosensing,109–112 drug delivery,113–115 regenerative medicine,116 shape-morphing,115,117 and biohybrid robotics.118
Another area that has received limited attention is harnessing cellular contractile forces to induce macroscale deformations in printed hydrogels (refer Fig. 1(d)). Cells attach to the substrate/ECM via specialized structures known as focal adhesions.119 As these focal adhesions mature, they function as mechanical bridges, linking the intracellular actin cytoskeleton to the ECM.120 This complex network facilitates the transmission of contractile forces generated by the actomyosin machinery, resulting in deformations of the substrate.104 The differential cell-mediated contractility within different ECM networks105 (refer Fig. 1(e)) or spatial cell patterning121 can be utilized to drive programmable deformation in 3D-printed architectures.
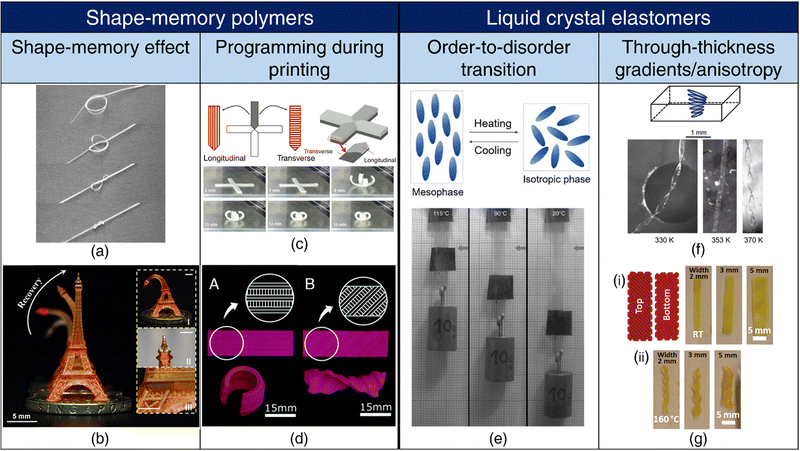 | ||
| Fig. 2 4D printing of SMPs and LCEs. Shape-memory effect (a), (b) (a) A pre-programmed (pre-stretched to 200%) SMP tied into a loose knot tightens above its transition temperature (40 °C), showcasing its potential as a self-tightening suture. Reproduced with permission from ref. 139. (b) Shape-memory effect exhibited by a 3D-printed Eiffel tower. Reproduced under the terms of CC-BY 4.0 International License from ref. 140. Programming during printing (c), (d) (c) Print path designs for a cross shape and a 3D view of anisotropic lamination. (top). Shape-morphing of the cross-shape specimen with an increase in the heating time (bottom). Reproduced under the terms of CC-BY-NC-ND International License from ref. 138. (d) A. Depiction of a monolayer self-rolling element achieved through a parallel arrangement of expanding and shrinking strips. B. Formation of a monolayer self-twisting element via an arrangement of expanding and shrinking strips at a 45° angle with respect to the longitudinal direction. Reproduced under the terms of CC-BY 3.0 International License from ref. 137. (e) Order-to-disorder transition. Representation of chain conformation in a main-chain LCE unit in the mesophase transitioning to a spherical polymer configuration in the isotropic phase upon heating (top). On heating, the LCE undergoes an order-to-disorder transition and contracts, resulting in the lifting of the weight (bottom). Adapted with permission from ref. 141 and 142. Trough thickness gradients/anisotropy (f), (g) (f) Schematics of the director configuration of twisted nematic elastomer (TNE) ribbons (top). Formation of helicoids by narrow TNE films upon heating. Adapted from ref. 143. (g) (i) Schematic of a bilayer rectangular LCE film with the top and bottom layers printed at 90° to each other or ±45° with respect to the long axis of the film. (ii) Transformation of the flat films into helices or helical ribbons upon heating, based on their printed orientations. Reproduced with permission from ref. 144. | ||
Cold drawing or cold programming, on the other hand, allows for deformation into the temporary shape at lower temperatures without requiring external heating. In cold programming, deformation beyond the yield point occurs below Tg, followed by stress relaxation. Upon unloading, some spring-back occurs, but the SMP does not fully revert to its original shape. To regain the permanent shape, heating beyond Tg is necessary.125,131,132
Diverging from conventional dual-shape memory polymers, which involve a permanent shape and a single temporary shape, the development of SMPs exhibiting multiple shape-memory effects is attainable through the design of SMPs with multiple distinct Ttrans or presenting a single broad thermal transition region.133
Programming during printing: Fused depositing modeling (FDM) is one of the prevailing 3D printing techniques employed for thermoplastics. During the printing process, the thermoplastic filament undergoes extrusion at temperatures surpassing its Tg, concurrently experiencing stretching along the print direction. This stretching event functions as the programming step, and as the filament subsequently cools to room temperature, it retains the tensile strain. During the printing of successive layers, reheating of underlying layers occurs upon contact with the molten extrudate, thereby resulting in the release of a portion of the stored pre-strain. As a consequence of the layer-by-layer printing process, a gradient in pre-strain becomes apparent across the thickness of the printed specimen, with the uppermost layer retaining the highest pre-strain. This ensuing anisotropy confers shape-morphing capabilities when subjecting the printed SMP to thermal activation.134–136
Expanding on this concept, the shrinkage associated with the release of pre-strain upon heating beyond Tg can be modulated through adjustments in the printing parameters, such as the nozzle temperature and the layer thickness, as well as the activation temperature. Printed filaments also exhibit different combinations of directional strains depending on the print conditions. Complex shape-morphing behavior can be attained by aligning subsequent layers along different orientations and harnessing the differences in directional strains among various filaments137,138 (refer Fig. 2(c) and (d)).
Through-thickness gradients/anisotropy: The attenuation of UV light as it traverses through the thickness of the LCE leads to differences in light intensity across the thickness of the LCE, which, in turn, induces varying degrees of order-to-disorder transition and mechanical strains within the LCE, resulting in bending of the LCE.152,153 LCEs exhibiting through-thickness variation in mesogen alignment, for instance, in the splay orientation where the nematic director twists from planar alignment on one surface to homeotropic alignment on the other surface, can generate bending moments due to synergistic mechanical responses along the planar and homeotropic axes.154 Similarly, LCEs with twisted nematic orientation, characterized by a 90° rotation in nematic direction across the material thickness, are conducive for twisting and folding deformations143 (refer Fig. 2(f)). Complex Gaussian curvatures, such as cones and anti-cones, can be achieved in LCEs where the nematic director rotates concentrically around a central point.155 The creation of such intricate director profiles involves the use of topological defects and can be realized through photoalignment techniques.156 Furthermore, the application of shear and extensional stresses during the extrusion 3D printing of LCEs enables rheological alignment, resulting in the generation of complex topologies akin to those obtained through photoalignment65,144 (refer Fig. 2(g)). In the case of multimaterial LCEs containing different nematic directors or layers with uniaxial and isotropic alignments, distinct mechanical strains occur along various axes upon stimulation, facilitating the emergence of complex shape-morphing behaviors.157,158
3.2 Ceramics
Shrinkage-mismatch on sintering: Variances in the solid content within ceramic ink, often composed of polymers and nanoparticles, can lead to notable discrepancies in shrinkage behavior on sintering. This differential shrinkage phenomenon can be exploited for the 4D printing of ceramics through the design of bilayer structures characterized by distinct solid content in each layer159 (refer Fig. 3(a)). The magnitude of bending deformation and resultant morphology are influenced by printing parameters such as print speed, as well as design factors including filament diameter, inter-filament spacing, bilayer dimensions, and the angle formed between the printed filaments within each layer.159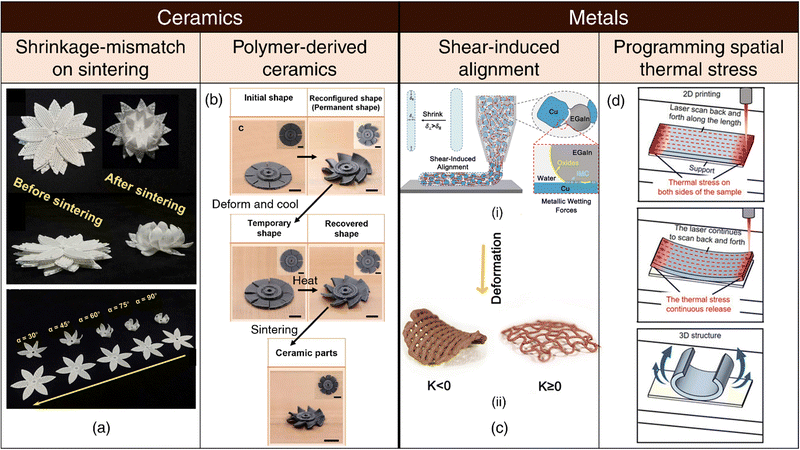 | ||
| Fig. 3 4D printing of ceramics and metals. (a) Shrinkage-mismatch on sintering. Images of top and side views of stacked ceramics pre- and post-sintering, showcasing shrinkage-mismatch phenomena. (top). Diverse shape-morphing behavior in bilayer ceramics resulting from aligning the top and bottom layers at varying angles. Reproduced with permission from ref. 159. (b) Polymer-derived ceramics. Complex 3D architectures including the initial, reconfigured, programmed, recovered, and sintered shapes of 4D-printed blade model. Reproduced with permission from ref. 161. (c) Shear-induced alignment. (i) Illustration of shear-induced alignment of granular metallic inks comprising liquid metal EGaIn. (ii) Shape-morphing upon drying at elevated temperatures. Reproduced with permission from ref. 68. (d) Programming spatial thermal stress. The samples are formed with laser scanning tracks along the length direction (red arrow) resulting in accumulation of thermal stress. Continuous scanning by the laser releases accumulated thermal stress, gradually causing the sample to bend into the expected 3D structure. Adapted under the terms of CC BY 4.0 International license from ref. 163. | ||
Polymer-derived ceramics: Over the past few years, the Lu group has devised multiple methodologies for the 4D printing of polymer-derived ceramics (pyrolyzed from polymeric ceramic precursors).160–162 These innovative techniques rely on the use of programmable shape memory or elastomeric precursors, endowing the printed structures with intrinsic shape-reconfigurable capabilities. Following the successful attainment of their desired configurations, the printed components can be sintered via pyrolysis, yielding ceramic products (refer Fig. 3(b)).
3.3 Metals
Shape-memory alloys: Shape-memory alloys (SMAs) exhibit reversible phase transitions between martensite and austenite phases when subjected to thermal activation and can be classified into three distinct categories: (a) one-way shape memory effect (OWSME), (b) two-way shape memory effect (TWSME), and (c) pseudoelasticity (PE).164 OWSME enables SMAs to return to their original shape upon heating, while TWSME facilitates transitions between two distinct shapes (one in each phase) upon heating and cooling. PE enables SMAs to switch phases in response to mechanical loading or unloading. SMAs exhibit robust mechanical properties and can be thermally actuated in a biocompatible manner with precise control over strength and frequency.165 Additionally, 4D-printed SMAs hold great potential for the design of intricate metallic architectures with shape-recovery and self-healing capabilities.166,167Liquid metals such as eutectic gallium-indium (EGaIn) can be shaped and printed168 and possess unique attributes that can be leveraged for 4D printing, particularly for biomedical applications.169,170 (a) Shear-induced alignment: In a recent report, the Dickey group has explored metallic granular inks comprising metal microparticles interconnected by bridges of liquid metal (in conjunction with water and a polymeric rheological aid) for 3D/4D printing of conducting metallic materials at room temperature.68 Within this framework, the liquid metal component assumes a critical role, since it deforms and elongates under the influence of shear stresses present at the dispensing nozzle and orients itself along the printing direction. Upon drying at elevated temperatures, the printed constructs shrink anisotropically, which drives 4D shape-morphing (refer Fig. 3(c)). (b) Stimuli-responsive shape transformation in micro/nanoparticles: EGaIn nanospheres coated with graphene quantum dots (GQD) exhibit phase separation and undergo a dramatic morphological transformation from nanospheres to hollow rods upon light irradiation by utilizing the photothermal effect of GQDs.171 Magnetic Galinstan liquid metal-iron (GLM-Fe) particles exhibit stochastic fragmentation from larger spheres into smaller spheroids, rods, or star-like geometries upon application of a magnetic field.172 Liquid gallium micro/nanorods, when exposed to acidic conditions, lose their outer oxide layer (critical for maintaining the rod-like morphology) and transition into droplets to minimize surface energy.173 Such micro and nano-scale shape-morphing phenomena hold substantial potential for applications in drug delivery, biosensing, anti-bacterial and anti-cancer therapies and represent an untapped opportunity in the field of 4D printing.
Programming spatial thermal stress: Selective laser melting (SLM) ranks among the foremost techniques for metal 3D printing. One of the major challenges of SLM is the generation of large, anisotropic internal residual stresses that can introduce defects within the final printed part and compromise material properties.174,175 While often perceived as a detriment, laser-induced thermal stresses can be strategically employed to drive self-deformation either during the printing process or post-printing, by releasing accumulated thermal stresses163 (refer Fig. 3(d)). Moreover, manipulating the laser scanning direction enables programmable deformation along specific axes. Importantly, this strategy is not bound by phase transitions, rendering laser-stimulated shape-morphing a versatile and promising approach for advancing 4D printing capabilities in common metallic materials.
Indirect 4D printing of metals via hydrogel infusion: In recent years, the Greer Group has pioneered Hydrogel Infusion Additive Manufacturing (HIAM)176–178 as a transformative platform technology for the 3D printing of architectured metals and alloys. This innovative approach entails the infusion of metallic salt precursors into 3D-printed “blank” hydrogel/organogels matrices fabricated through vat polymerization techniques. Subsequent calcination and reduction processes convert the constructs into metallic structures mirroring the initial hydrogel scaffolds. Notably, material selection occurs post-printing, which allows the utilization of established 4D printing techniques to induce morphological transformations in the hydrogel structures prior to finalizing the choice of the metallic materials. Consequently, this approach opens new horizons for the fabrication of complex, intricate metallic structures that would be unattainable using conventional 3D metal printing.
4 Biomedical applications and scope of innovation
4.1 Tissue engineering and regenerative medicine
In the realm of tissue engineering and regenerative medicine (TERM), 4D printing is a promising biomanufacturing technology with primary applications across three domains:57,179–181 (1) the transformation of initially planar printed constructs into anatomically relevant curved geometries, (2) the emulation of native tissue dynamics and the recapitulation of stimuli-responsive tissue functionality, and (3) the adaptive reconfiguration of printed structures to streamline minimally invasive deployment during surgical procedures.Tubular conduits represent indispensable constituents of numerous organ systems, including the vasculature, pulmonary, nervous, gastrointestinal, and genitourinary systems, spanning in diameter from micrometers to centimeters. Functioning as critical plumbing conduits, these structures facilitate the passive transport of fluids and, under specific circumstances, play a role in modulating fluid composition. Although the intricacies of tube formation during natural tissue development remain partially elucidated, the repertoire of known mechanisms governing tube morphogenesis can provide a foundation for new 4D printing strategies. Tubulogenesis processes encompass diverse mechanisms, such as wrapping, budding, cavitation, cord hollowing, and cell hollowing, each contributing distinctively to the formation of tubular structures through a combination of individual cell behaviors and collective cell rearrangements. For an in-depth understanding of the mechanisms underlying tube morphogenesis, readers are directed to the comprehensive review by Lubarsky and Krasnow.182 Conventional 3D printing techniques encounter inherent complications in stacking annular layers and are further burdened by extended print durations that correlate with the length of the tubular structure. In contrast, 4D printing methodologies enable the rapid self-folding of planar hydrogel sheets into tubes,77,183 even with diameters as small as 20 μm, resembling the tiniest veins.96 This approach has gained considerable traction for the fabrication of vascular grafts,95–97,184 nerve conduits101,185 (refer Fig. 4(b)), skeletal muscle fascicles,186 and tracheal structures187 (refer Fig. 4(a)). Furthermore, homogeneous or patterned cell distribution is more effectively accomplished through the initial seeding of cells on planar surfaces, followed by their transformation into irregular or curved configurations. The initial iterations of 4D-printed self-folding tubular constructs, while conceptually straightforward, faced limitations concerning intricate mesh-like vascular networks and issues with structural integrity, such as loose or leaky connections. However, recent advancements now permit the 4D printing of tight, branched vascular junctions, including T97 and Y95-shaped bifurcations. Continued progress is anticipated to rival or even surpass developments in 3D printing methodologies, particularly regarding entangled, multivascular networks observed in organs like the lungs.188
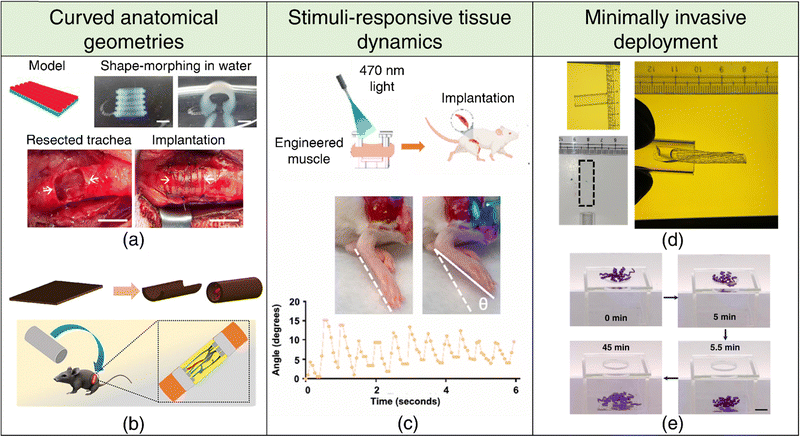 | ||
| Fig. 4 4D printing for tissue engineering and regenerative medicine. Curved anatomical geometries (a), (b) (a) Shape-morphing of DLP-printed constructs in deionized water (top). Implantation of the 4D-printed trachea into damaged trachea rabbit site. Adapted with permission from ref. 187 (b) 4D printing of a flat sheet into a conduit structure (top). Schematic representation of the implantation of a self-folding conduit into a sciatic nerve transected rat model, showcasing sutureless neurorrhaphy possibilities. Adapted with permission from ref. 101. (c) Stimuli-responsive tissue dynamics. Schematic representation of ontogenetically engineered muscle tissue implanted in a volumetric muscle loss mouse model (top). Dorsiflexion in response to 470 nm light, showcasing stimuli-responsive behavior (bottom). Adapted with permission from ref. 116. Minimally invasive deployment (d), (e) (d) Demonstration of the deployability of a rectangular hydrogel through a narrow glass tube of similar dimensions facilitated by 4D shape-morphing. Reproduced with permission from ref. 103. (e) Demonstration of far-from-equilibrium 4D shape-morphing enabling a hydrogel to pass through a small hole, highlighting its potential for minimally invasive applications. Reproduced under the terms of CC-BY 4.0 International License from ref. 198. | ||
Curved tissue interfaces are frequently encountered at the tissue boundaries and the junctions between tissues and bodily lumens, exhibiting diverse scales and degrees of curvature. Understanding the mechanisms underlying curvature emergence in living systems during development can offer clues to better design engineered tissues for transplantation. Apart from genetically programmed growth, physical constraints at tissue interfaces, such as mismatches in mechanical properties or forces, can instigate the development of curved tissue geometries. Bending and buckling-driven morphogenesis constitute the predominant processes that transform a 2D surface into a curved 3D structure in living systems. For instance, epithelial cell sheets exploit differential contractility between cell surfaces to generate bending moments, thereby inducing tissue layer bending or folding. This mechanism has been implicated in various developmental processes across species, including Drosophila mesoderm invagination, vertebrate lens placode invagination, and folding of the intestinal crypt.189 Buckling refers to the abrupt out-of-plane deformation of a beam when subjected to in-plane compressive stress beyond a critical threshold.190 Differences in in-plane mechanical strains experienced by the tissue layer and the substrate it is adhered to can prompt single buckling or the formation of wrinkle-like patterns.191 Notably, bending and buckling can substantially increase the surface-area-to-volume ratios within confined spaces, facilitating nutrient uptake and gas exchange in vital organs like the brain, intestines, and lungs. Hence, local tissue geometry emerges as an important morphological parameter with inherent physiological significance, assuming critical importance in tissue engineering. Traditional 3D printing methods often necessitate the use of support structures to address overhangs when printing curved geometries, a limitation that can be elegantly circumvented through the utilization of 4D printing techniques. This presents a largely untapped potential for the fabrication of biomimetic curved tissue constructs, including but not limited to cardiac patches,192,193 intestinal patches,194 convoluted cortical tissues,195 and cartilaginous and osseous constructs.83,90,91,196 Moreover, the capability of 4D printing capacity to generate complex local curvatures offers an opportunity to investigate the impact of substrate curvature on cellular functionality, which can provide critical insights for advancing tissue engineering strategies.197
Biological tissues possess remarkable adaptability, enabling them to undergo morphological transformations in response to various external and internal cues. Muscle tissues, exemplifying this adaptability, respond to both involuntary and voluntary commands through a finely tuned interplay of chemical and electrical signaling. Engineered, responsive muscle tissue grafts have the potential to restore mobility in individuals affected by volumetric muscle loss stemming from traumatic injuries, tumor resections, or degenerative diseases. It is well-established that mechanical stimulation of engineered tissue accelerates tissue regeneration and rehabilitation.165 In this context, optogenetic engineering allows for spatially precise and non-invasive control over muscle contraction and relaxation.116 By transducing muscle cells to express a light-sensitive cation channel,118 these tissues can be stimulated remotely with light (refer Fig. 4(c)), providing a powerful therapeutic approach for restoring biological function.
The minimally invasive deployment and seamless integration of tissue scaffold into defect cavities is a critical factor for achieving optimal therapeutic outcomes. This necessitates a programmed 4D transformation of tissue scaffolds into temporary, miniaturized configurations to facilitate minimally invasive deployment. Post-implantation, the scaffold can be induced to revert to its original shape or adapt to irregular defect boundaries. SMPs have been widely adopted for this purpose because of their inherent reconfigurability, ability to retain temporary shapes and shape recovery upon stimulation. Nevertheless, there is growing interest in engineering shape memory hydrogels,199,200 reversible shape-morphing hydrogels201,202 (refer Fig. 4(d) and (e)), as well as hydrogels capable of programmed multi-step/far-from-equilibrium shape changes on a single stimulation,103,198 as promising alternatives to SMPs.
4.2 Drug delivery systems
The precise temporal release and spatial delivery of encapsulated therapeutic agents are fundamental design considerations for drug delivery systems (DDSs). Integrating concepts of 4D printing with DDSs represents a transformative approach to address these critical parameters, enabling the targeted delivery of drugs to pathological tissues and regions characterized by distinctive physiological environments (for example, pH).Employing 4D shape-morphing techniques, microgrippers can be engineered to undergo controlled shape transformations in response to specific environmental conditions or subtle biomolecular cues, facilitating the gripping, exposure, and/or subsequent release of encapsulated drugs/cells203–211 (refer Fig. 5(a) and (b)). The triggered degradation/rupture of the outer shell/encasing in the presence of appropriate stimulus is another viable strategy to expose and release core therapeutic payloads.115,212–216 Innovative cage-like microstructures have been engineered for the on-demand capture or release of microparticles, exploiting differences in pore sizes of the expanded and contracted states of the cage, and have exciting applications in drug delivery.217 Nano-scale shape transformations observed in endocytosed liquid metal-based particles featuring a GQD shell, when stimulated by near-infrared (NIR) light, can effectively disrupt cellular structures by breaching the endosomal membrane and result in the accelerated release of drug contents and hold significant promise, particularly in the context of targeted drug delivery to tumor cells.171 Shape-morphing at the microscale is also particularly interesting, as the geometry of the microparticle dictates whether it can be phagocytosed and bears profound implications for biomedicine.
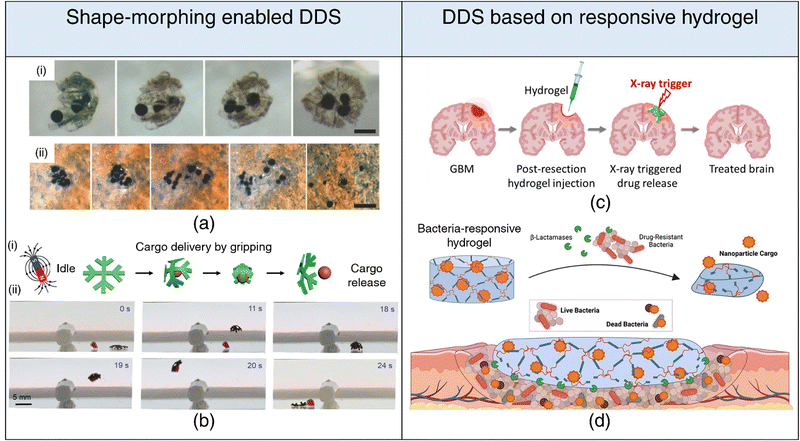 | ||
| Fig. 5 4D printing for drug delivery. Drug delivery systems based on shape-morphing behavior (a), (b) (a) (i) Incorporation of magnetic particles within folding microstructures. (ii) Activation of near-infrared light (NIR) leading to the release of microparticles, showcasing controlled drug delivery capabilities. Adapted with permission from ref. 204. (b) (i) Schematic representation delineating cargo grasping, transport, and release stages. (ii) Demonstration of crossing obstacles while gripping cargo for effective transport and release under magnetic field gradients generated by a permanent magnet. Reproduced under the terms of CC-BY 4.0 International License from ref. 203. Drug delivery systems based on stimuli-responsive hydrogel degradation (c), (d) (c) Schematic illustration of an injectable hydrogel that degrades in response to radiation trigger, facilitating the controlled release of encapsulated drug. Reproduced with permission from ref. 218. (d) Schematic illustration of a β-lactamase-responsive hydrogel designed to degrade in the presence of bacteria, consequently releasing nanoparticle cargo, showcasing targeted and responsive drug delivery systems. Reproduced with permission from ref. 219. | ||
While underexplored, the utility of 4D printing in drug delivery transcends beyond its conventional role in shape-morphing. Stimuli-responsive hydrogels can be engineered to degrade in response to external stimuli (light, electric and magnetic fields, heat, radiation) and release their drug payloads. For instance, a radiation-responsive injectable hydrogel has been proposed to treat glioblastoma, leveraging the synergistic effect of radiation-induced drug release alongside radiotherapy for enhanced therapeutic outcomes218 (refer Fig. 5(c)). By tailoring DDSs to respond to specific biomolecules, initiation of drug release can be triggered exclusively in the presence of these biomolecular cues, enabling highly targeted drug delivery. Enzymatic stimulation has emerged as a potent strategy for achieving precise and targeted release of therapeutic agents at disease sites, effectively minimizing off-target effects. For instance, hydrogels with matrix metalloproteinase (MMP) cleavable crosslinks and sequestered recombinant tissue inhibitor (r-TIMP-3) have been engineered to enable precise and localized delivery of r-TIMP-3 in response to elevated MMP expression, following myocardial infarction.220 Similarly, hydrogels susceptible to degradation by β-lactamase have been synthesized to trigger the release of antimicrobial agents only in the presence of pathogenic bacteria (refer Fig. 5(d)). This innovative approach significantly reduces unwarranted exposure to encapsulated antimicrobials, a key consideration for mitigating the development of antibiotic resistance.219 Furthermore, DDSs can be designed to undergo chemical transformations catalyzed or induced by specific enzymes, leading to the generation of active therapeutic moieties. A prime example is the conversion of prodrugs into their pharmacologically active forms.221,222 This strategy has garnered substantial attention, especially in the domain of cancer therapies, and exhibits considerable promise for clinical translation.223,224
4.3 Biomimicry and dynamic in vitro systems
Mature tissues and organs attain their intricate forms via structural remodeling, reorganization, and morphogenesis through developmental processes. Nevertheless, conventional 3D bioprinting techniques yield static constructs that lack the ability to replicate the dynamic nature of native tissues. Creating 3D structures capable of dynamic shape transformations and emulating native tissues offers a platform for experimentally validating theoretical models of tissue and organ morphogenesis and enhancing our understanding of tissue dynamics. Furthermore, biomimetic 4D printing provides an innovative approach for engineering complex tissues and organs, starting from rudimentary designs and enabling their evolution into final forms through biomimetic processes.Diverse surface patterns ranging from fingerprints225 to buckled tumor spheroids226 and wrinkled white blood cell membranes,227 have their origins in mechanical instabilities. Mechanical instabilities at the interface between epithelial tissues and mesenchyme can lead to the buckling or wrinkling of 1D epithelial tubes and 2D epithelial sheets, resulting in intricate topologies with loops, folds, and undulations observed in physiological contexts like cortical folding in the brain, branching morphogenesis in the lungs (refer Fig. 6(c)), and villus morphogenesis in the intestine.190 When a sheet affixed to an elastic foundation experiences uniaxial compression, wrinkles or folds emerge as a consequence of sheet buckling. These mechanisms underpin 4D printing strategies to generate surface patterns on printed hydrogels.228–230 Recent findings, however, suggest that the fingerprint patterning is better described by Turing reaction-diffusion mechanisms during developmental processes,231 the same mechanism that determines hair follicle spacing,232 lends cheetah its spots,233 or an Ornate Boxfish its hexagon/stripe patterns.234 Patterned surfaces governed by Turing reaction-diffusion mechanisms will represent a new direction in 4D surface morphing.
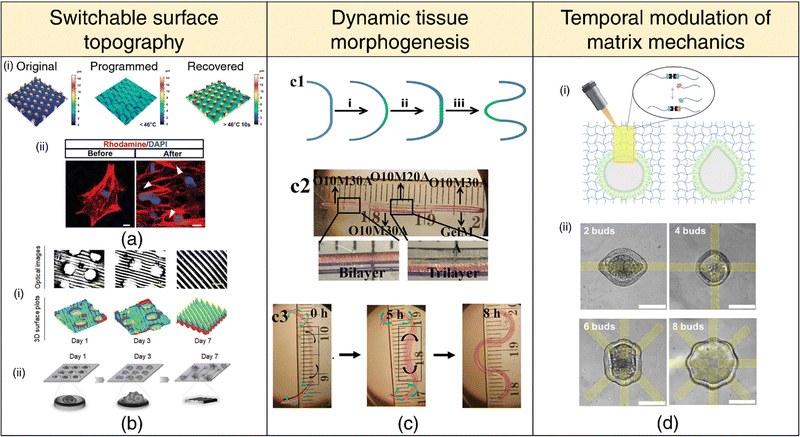 | ||
| Fig. 6 4D printing for dynamic in-vitro systems. Switchable surface topography (a), (b) (a) (i) Laser confocal scanning microscope (LCSM) images depicting the original, programmed, and recovered surface topography. (ii) LCSM images of cells labeled with Rhodamine (red) and DAPI (blue), displaying cytoskeletal distribution above micropillars (white arrows). Adapted with permission from ref. 235. (b) (i) Optical images and 3D surface plots demonstrating 4D shape-morphing polymer (SMP) substrates transitioning from microwell arrays to aligned patterns (microgrooves) over 1, 3, and 7 days. (ii) Schematic illustration of time-dependent cell patterning process during a 7-day culture period. Adapted under the terms of CC-BY 4.0 International License from ref. 236. (c) Mimicking tissue morphogenesis. (1) Schematic representation of lung branching morphogenesis, depicting steps from nascent bud formation to terminal bifurcation. (2) Photomicrograph of a cell-laden trilayer hydrogel bar designed to emulate branching morphogenesis in tissues. (3) Photomicrographs showing the 4D hydrogel system mimicking lung branching morphogenesis during culture at 37 °C. Adapted under the terms of CC-BY 4.0 International License from ref. 102 (d) Temporal modulation of matrix mechanics (i) Utilization of photopatterned light to induce local hydrogel bond rearrangement, causing epithelial deformation within the patterned region. (ii) Selection of pattern spacing to generate up to eight independent deformations within a single organoid. Adapted under the terms of CC-BY 4.0 International License from ref. 237. | ||
Surface topographies of cell culture substrates can govern cell behavior via contact guidance.238 Cells actively respond to their microenvironment, sensing and adapting to topographical cues that influence their migration, alignment, and differentiation. The ability to dynamically manipulate these microenvironmental cues on-demand or through preprogrammed methods offers the potential to create dynamic cell-substrate interfaces that can spatially pattern, orient, or differentiate cells.239–242 Notably, flat topography encourages stem cell adhesion, whereas sub-cellular microstructures promote differentiation. Once a well-adhered and proliferating stem cell population is achieved, the surface topology can be switched on-demand via appropriate stimuli to induce osteogenic differentiation235 (refer Fig. 6(a)). In another context, transitioning from a temporary microwell array to an aligned pattern has proven effective in facilitating the initial aggregation of neural stem cells (NSCs) and subsequent differentiation and the alignment of neurons and glial cells236 (refer Fig. 6(b)). Importantly, the curvature of the substrate's surface plays a pivotal role in determining the spatiotemporal organization of cells,243 laying the groundwork for future investigations into 4D cell-fate switching. Notably, surface topography and dynamic shape changes can also influence bacterial adhesion and prevent biofilm formation via mechano-bactericidal effects and present a promising avenue for engineering 4D-printed antifouling surfaces for medical implants.244–246
The predominant focus within the 4D printing community has been on the advancement of stimuli-responsive mechanisms aimed at facilitating the shape transformation of 3D-printed constructs. We believe that the scope of 4D printing extends beyond mere shape changes to encompass alterations in various material properties, such as stiffness, viscoelasticity, and degradation, among others. Matrix stiffness and viscoelastic characteristics are important parameters for distinguishing various tissue types, including healthy and pathological states. Hence, the spatial and temporal modulation of matrix mechanics247–251 holds immense significance in mimicking dynamic changes in tissue stiffness, as observed during processes such as embryonic development, wound healing, and tumor pathogenesis.252 For instance, the design of hydrogel networks with adhesive moieties and cleavable crosslinks facilitates cellular remodeling and spreading, unlike networks with covalent crosslinks that hinder cell spreading due to their typically sub-cellular mesh sizes. Dynamically transforming the primary hydrogel network into a secondary network through sequential crosslinking mechanisms via external stimuli presents a promising avenue for the advancement of 4D cell-instructive hydrogel printing techniques.253 Controlled patterned light exposure to photodegradable hydrogels has been found to induce local softening or the development of viscoelastic behavior, steering intestinal crypt formation into the degraded region237 (refer Fig. 6(d)). Moreover, by employing stiffening mechanisms, the intestinal crypts can be confined to the desired patterned shape.254
4.4 Soft robotics and medical devices
Soft materials are being increasingly adopted over traditional rigid materials for applications requiring interfacing or adhesion with delicate tissues, owing to their tissue-like mechanics and viscoelastic properties.255–257 This has motivated the development of soft-robotic technologies for applications spanning medical devices (surgical devices, assistive tools), wearable robots and prostheses, actuators, biohybrid devices, and tissue and organ simulators.258 4D printing offers tremendous potential to build self-contained soft-robotic devices that do not need to be tethered to an electrical or pneumatic source but instead harness the material's “intelligence” for actuation on appropriate stimulation.Patients afflicted with neurological disorders that restrict mobility need to perform rehabilitation exercises to restore their range of motion and muscular strength. SMA-based actuators, typically nitinol (NiTi) alloys, have been employed as assistive devices (SHADE and Leia) for repetitive passive mobilization of the ankle joint as an acute post-stroke rehabilitation therapy.259,260 However, SMA actuators require insulation when in contact with biological tissues due to their operational reliance on thermal activation. In recent work, a novel approach of delivering mechanical stimulation directly to tissues through a soft-robotic actuator composed of a NiTi spring embedded within an elastomeric matrix and securely affixed to the tissue via an adhesive hydrogel layer has demonstrated effectiveness in activating mechanosensing pathways and preventing muscular atrophy in rats.165 Additionally, there is also a growing interest in developing soft contractile actuators (based entirely on SMPs, shape-memory hydrogels (SMHs), or LCEs) that can undergo robust, reversible actuation as body-part stimulators.
SMPs have witnessed extensive usage in surgical applications, including intravascular, urogenital, cerebral, ocular, and general surgical procedures.261 Biodegradable SMP-based self-tightening sutures that can respond to physiological heat, enabling them to securely close wounds with a pre-defined pressure, can lead to better wound healing while minimizing scar formation.139 SMP-based stents (cardiaovascular,262,263 neurovascular,264 tracheal,265,266 oesophageal,267,268 biliary,269–271 ureteral272,273) (refer Fig. 7(a)) are amenable to administration via catheters in a miniaturized temporary state.274,275 Subsequently, they can be activated and recovered at body temperature upon reaching the target site, offering vital support for the collapsed vessel.269 In the case of aneurysms, SMP foams present therapeutic advantages over conventional metallic coils. The expansion/recovery of the SMP following implantation in the aneurysm can effectively close the aneurysm, averting potentially lethal rupture.276,277 SMP-based clot removal devices278,279 (refer Fig. 7(d)) and occlusion plugs as contraception devices,280 have also been conceptualized; however, their efficiency and biocompatibility warrant further investigation. SMP-based surgical fasteners,281 microgrippers,282 retrieval devices,283 stomach fillers for obesity control,284 and orthodontic archwires285 are also being explored.
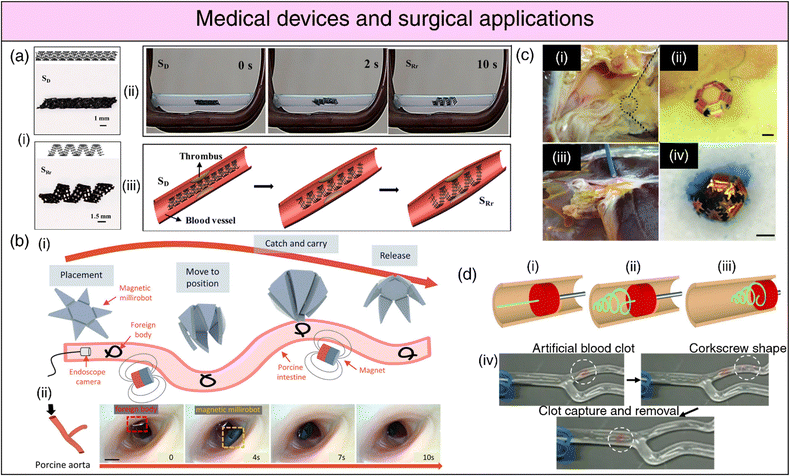 | ||
| Fig. 7 4D printing for medical devices and surgical applications. (a) (i) Deformation and restrictive shape recovery of the printed scaffold. (ii) Demonstration of the restrictive shape recovery process triggered by a 30 kHz alternating magnetic field. (iii) Application of the 4D scaffold as an intravascular stent. Adapted with permission from ref. 292. (b) (i) Schematic representation of foreign body removal experiments. (ii) Ex vivo foreign body removal experiments in porcine aorta. Reproduced under the terms of CC-BY 4.0 International License from ref. 291. (c) (i), (ii) Optical image showing microgrippers distributed at the bile duct opening of a porcine liver. (iii) Optical image of microgripper retrieval using a magnetic catheter. (iv) Image depicting a retrieved μ-gripper with an excised tissue piece after staining with trypan blue. Reproduced with permission from ref. 287. (d) Endovascular thrombectomy using SMP microactuators (i) Deployment of a laser-activated SMP microactuator in its secondary straight rod form through a catheter distal to vascular occlusion. (ii) Transformation of the microactuator into its primary corkscrew form via laser heating. (iii) Retraction of the deployed microactuator to capture the thrombus. (iv) In vitro thrombectomy using the SMP device in a bifurcated vessel model, demonstrating its potential for endovascular interventions. Reproduced under the terms of CC-BY 4.0 International License from ref. 279. | ||
Soft-robotic microgrippers hold substantial promise for microsurgical applications, complementing their already-established role in drug delivery.286 These microgrippers have emerged as a compelling alternative to conventional biopsy procedures, with the pioneering efforts of the Gracias research group prominently advancing this field. Traditional biopsy techniques necessitate substantial forces for tissue excision, often relying on rigid materials. In contrast, untethered microgrippers demonstrate the capability to maneuver through constrained anatomical passages and excise tissue specimens with high-quality DNA and RNA samples suitable for diagnostic analyses287 (refer Fig. 7(c)). Furthermore, shape-morphing hydrogels can function as micro-manipulation systems, streamlining object assembly within microfluidic devices,288 or as precision actuators to impose physiologically relevant compressive stresses on living spheroids for the investigation of their phenotypic response.289 Locomotive milli/micro-bots based on shape deformations can be instrumental in minimally invasive surgical procedures for foreign body removal290,291 (refer Fig. 7(b)) while simultaneously opening avenues for tissue and organ inspection, disease diagnosis, and health monitoring.
5 Outlook
5.1 Current challenges
The current generation of 4D printing techniques suffers from four notable drawbacks that limit their scope of application: (a) simplistic shape transformations, (b) limited materials library, (c) non-biocompatible stimuli, and (d) absence of widely adaptable simulation methods for predictive design. Biological functions governed by stimuli-induced morphological transformations of tissues (e.g., peristaltic contractions) are distinguished by their complexity, which entails a spectrum of actuation forces, temporal dynamics, and the capacity for reversibility. In the context of 4D-printed constructs designed to emulate biological tissues or serve as biomedical devices, the typical prerequisites encompass actuation forces that align with physiological norms and the ability to undergo rapid, repetitive actuation cycles. The attribute of reversibility assumes paramount significance, ensuring that the material swiftly reverts to its initial configuration upon the cessation of the stimulus, thereby facilitating repeated stimulation to invoke varied responses. Realizing the coveted actuation response within physiological environments presents a formidable challenge. Hydrogels, for instance, suffer drastic losses in mechanical properties on swelling. Conversely, shape memory polymers (SMPs) typically lack intrinsic reversible actuation characteristics. Liquid crystal elastomers (LCEs), despite their capability for reversible actuation, have witnessed relatively limited exploration in the context of biomedical applications, primarily due to their reliance on temperatures exceeding the physiological range to achieve meaningful strains. Hence, there is an impetus to design composite architectures to integrate diverse properties and actuation mechanisms, with the overarching objective of paving the way for the next generation of engineered tissues and biomedical devices.The materials library for 4D printing for biomedical applications is rather limited, predominantly owing to concerns regarding biocompatibility, toxicity, and the potential to elicit immune responses. This limitation is further exacerbated when considering materials suitable for facilitating cell encapsulation and the intricacies of bioprinting. While various stimulus modalities, including heat, pH variations, and chemical agents, offer viable means of inducing transformations in 4D-bioprinted constructs, they concurrently introduce concerns over their impact on the viability of encapsulated cells. For realizing in vivo shape transformations, non-invasive or remote stimulation via electric or magnetic fields, NIR, is expected to become the norm. Another alternative is to harness the innate physiological environment surrounding the implanted construct to facilitate the desired shape alterations.
By incorporating theoretical models that capture complex material-stimuli interactions, simulation frameworks offer a means to predict and optimize the behavior of dynamic systems. Hence, a concerted shift toward widely adaptable simulation-guided design methodologies is imperative to enable researchers to efficiently navigate the design space and identify optimal configurations. Central to the success of simulation-guided design approaches is the seamless incorporation of experimental validation to corroborate simulated predictions and refine simulation models. Through systematic experimentation on simplified systems of reduced complexity, researchers can methodically validate the performance of simulation frameworks in capturing the dynamic behavior of 4D printed structures. Furthermore, experimental data serves as invaluable inputs for calibrating material models and fine-tuning simulation parameters, thereby enhancing the predictive accuracy of virtual simulations. However, as 4D printed structures undergo shape transformations, there is a concomitant change in material properties in response to stimuli, necessitating the incorporation of time-dependent properties into simulation models. Failure to accurately capture these dynamic changes can lead to discrepancies between simulated and experimental outcomes, undermining the reliability of predictive simulations.
5.2 Application-driven design
The past decade has witnessed the birth and meteoric rise of 4D printing owing to its broad applicability across a multitude of research domains. This has spurred the development of responsive chemistries, innovative synthesis techniques, and post-processing methods to impart shape-morphing capability, as well as the amalgamation of materials and mechanisms to enable advanced and complex functionalities. In the biomedical domain, a major focus has been dedicated to leveraging these principles utilizing biocompatible materials. However, a dedicated application-driven approach has been largely missing. In many instances, the clinical application is an afterthought, resulting in a design process that lacks clinical input. Consequently, state-of-the-art 4D printing technologies have largely been confined to proof-of-concept stages and have yet to achieve commercial viability. To address this issue, a comprehensive investigation becomes imperative to identify how existing 4D printing techniques can effectively address well-defined clinical challenges. This undertaking should involve close collaboration with clinicians to determine the material choices, specific shape and size transformations, viable actuation methods, actuation forces, and biocompatibility prerequisites for each application. While a diverse array of 4D printing methods and materials is already available, as delineated in this perspective, each problem presents unique complexities necessitating innovative solutions. A combination of experimental data and simulated data (leveraging computational tools such as finite element modeling) can be employed to train machine learning (ML) algorithms to predict forward shape changes as well as navigate the extensive design space to find optimal solutions for inverse shape change problems.293,294 Although this approach may seem tedious, it holds the promise of facilitating the seamless integration of 4D printing technologies into patient care, fostering widespread adoption across domains such as tissue engineering, regenerative medicine, targeted therapeutic delivery, soft robotics, medical devices, and a myriad of other applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Support from the Science and Engineering Research Board (SERB), Government of India (IPA/2020/000025) is gratefully acknowledged.References
- R. L. Truby and J. A. Lewis, Nature, 2016, 540, 371–378 CrossRef CAS.
- J. Li, C. Wu, P. K. Chu and M. Gelinsky, Mater. Sci. Eng., R, 2020, 140, 100543 CrossRef.
- L.-Y. Zhou, J. Fu and Y. He, Adv. Funct. Mater., 2020, 30, 2000187 CrossRef CAS.
- Z. Chen, Z. Li, J. Li, C. Liu, C. Lao, Y. Fu, C. Liu, Y. Li, P. Wang and Y. He, J. Eur. Ceram. Soc., 2019, 39, 661–687 CrossRef CAS.
- Z. Chen, X. Sun, Y. Shang, K. Xiong, Z. Xu, R. Guo, S. Cai and C. Zheng, J. Adv. Ceram., 2021, 10, 195–218 CrossRef CAS.
- J. Ni, H. Ling, S. Zhang, Z. Wang, Z. Peng, C. Benyshek, R. Zan, A. K. Miri, Z. Li, X. Zhang, J. Lee, K.-J. Lee, H.-J. Kim, P. Tebon, T. Hoffman, M. R. Dokmeci, N. Ashammakhi, X. Li and A. Khademhosseini, Mater. Today Bio, 2019, 3, 100024 CrossRef CAS PubMed.
- L. F. Velásquez-García and Y. Kornbluth, Annu. Rev. Biomed. Eng., 2021, 23, 307–338 CrossRef PubMed.
- X. Kuang, D. J. Roach, J. Wu, C. M. Hamel, Z. Ding, T. Wang, M. L. Dunn and H. J. Qi, Adv. Funct. Mater., 2019, 29, 1805290 CrossRef.
- Y. S. Zhang, G. Haghiashtiani, T. Hübscher, D. J. Kelly, J. M. Lee, M. Lutolf, M. C. McAlpine, W. Y. Yeong, M. Zenobi-Wong and J. Malda, Nat. Rev. Methods Primer, 2021, 1, 1–20 CrossRef.
- Z. Jiang, B. Diggle, M. L. Tan, J. Viktorova, C. W. Bennett and L. A. Connal, Adv. Sci., 2020, 7, 2001379 CrossRef CAS.
- J. K. Placone and A. J. Engler, Adv. Healthcare Mater., 2018, 7, 1701161 CrossRef PubMed.
- I. T. Ozbolat and M. Hospodiuk, Biomaterials, 2016, 76, 321–343 CrossRef CAS PubMed.
- R. Levato, O. Dudaryeva, C. E. Garciamendez-Mijares, B. E. Kirkpatrick, R. Rizzo, J. Schimelman, K. S. Anseth, S. Chen, M. Zenobi-Wong and Y. S. Zhang, Nat. Rev. Methods Primer, 2023, 3, 1–19 CrossRef.
- C. Yu, J. Schimelman, P. Wang, K. L. Miller, X. Ma, S. You, J. Guan, B. Sun, W. Zhu and S. Chen, Chem. Rev., 2020, 120, 10695–10743 CrossRef CAS PubMed.
- A. McCormack, C. B. Highley, N. R. Leslie and F. P. W. Melchels, Trends Biotechnol., 2020, 38, 584–593 CrossRef CAS PubMed.
- J. Zhao and N. He, J. Mater. Chem. B, 2020, 8, 10474–10486 RSC.
- J. R. Tumbleston, D. Shirvanyants, N. Ermoshkin, R. Janusziewicz, A. R. Johnson, D. Kelly, K. Chen, R. Pinschmidt, J. P. Rolland, A. Ermoshkin, E. T. Samulski and J. M. DeSimone, Science, 2015, 347, 1349–1352 CrossRef CAS PubMed.
- G. Lipkowitz, T. Samuelsen, K. Hsiao, B. Lee, M. T. Dulay, I. Coates, H. Lin, W. Pan, G. Toth, L. Tate, E. S. G. Shaqfeh and J. M. DeSimone, Sci. Adv., 2022, 8, eabq3917 CrossRef CAS PubMed.
- D. A. Walker, J. L. Hedrick and C. A. Mirkin, Science, 2019, 366, 360–364 CrossRef CAS PubMed.
- M. Zastrow, Nature, 2020, 578, 20–23 CrossRef CAS PubMed.
- B. E. Kelly, I. Bhattacharya, H. Heidari, M. Shusteff, C. M. Spadaccini and H. K. Taylor, Science, 2019, 363, 1075–1079 CrossRef CAS PubMed.
- D. Loterie, P. Delrot and C. Moser, Nat. Commun., 2020, 11, 852 CrossRef CAS.
- M. Regehly, Y. Garmshausen, M. Reuter, N. F. König, E. Israel, D. P. Kelly, C.-Y. Chou, K. Koch, B. Asfari and S. Hecht, Nature, 2020, 588, 620–624 CrossRef CAS.
- L. Stüwe, M. Geiger, F. Röllgen, T. Heinze, M. Reuter, M. Wessling, S. Hecht and J. Linkhorst, Adv. Mater., 2023, 2306716 Search PubMed.
- M. A. Skylar-Scott, J. Mueller, C. W. Visser and J. A. Lewis, Nature, 2019, 575, 330–335 CrossRef CAS.
- N. M. Larson, J. Mueller, A. Chortos, Z. S. Davidson, D. R. Clarke and J. A. Lewis, Nature, 2023, 613, 682–688 CrossRef CAS.
- S. V. Murphy and A. Atala, Nat. Biotechnol., 2014, 32, 773–785 CrossRef CAS.
- S. V. Murphy, P. De Coppi and A. Atala, Nat. Biomed. Eng., 2020, 4, 370–380 CrossRef.
- Y. Wu, M. Qin and X. Yang, J. Mater. Chem. B, 2023, 11(43), 10263–10287 RSC.
- Y. Zhang, J. Xu, Z. Fei, H. Dai, Q. Fan, Q. Yang, Y. Chen, B. Wang and C. Wang, Adv. Mater., 2021, 33, 2106768 CrossRef CAS.
- C. Caudill, J. L. Perry, K. Iliadis, A. T. Tessema, B. J. Lee, B. S. Mecham, S. Tian and J. M. DeSimone, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2102595118 CrossRef CAS.
- A. vander Straeten, M. Sarmadi, J. L. Daristotle, M. Kanelli, L. H. Tostanoski, J. Collins, A. Pardeshi, J. Han, D. Varshney, B. Eshaghi, J. Garcia, T. A. Forster, G. Li, N. Menon, S. L. Pyon, L. Zhang, C. Jacob-Dolan, O. C. Powers, K. Hall, S. K. Alsaiari, M. Wolf, M. W. Tibbitt, R. Farra, D. H. Barouch, R. Langer and A. Jaklenec, Nat. Biotechnol., 2023, 1–8 Search PubMed.
- M. R. Prausnitz, Adv. Drug Delivery Rev., 2004, 56, 581–587 CrossRef CAS.
- T. Kim, Q. Yi, E. Hoang and R. Esfandyarpour, Adv. Mater. Technol., 2021, 6, 2001021 CrossRef CAS.
- K. A. Deo, M. K. Jaiswal, S. Abasi, G. Lokhande, S. Bhunia, T.-U. Nguyen, M. Namkoong, K. Darvesh, A. Guiseppi-Elie, L. Tian and A. K. Gaharwar, ACS Nano, 2022, 16, 8798–8811 CrossRef CAS.
- O. Dadras-Toussi, M. Khorrami, A. S. C. Louis Sam Titus, S. Majd, C. Mohan and M. R. Abidian, Adv. Mater., 2022, 34, 2200512 CrossRef CAS.
- C. Cvetkovic, R. Raman, V. Chan, B. J. Williams, M. Tolish, P. Bajaj, M. S. Sakar, H. H. Asada, M. T. A. Saif and R. Bashir, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 10125–10130 CrossRef CAS.
- L. Ricotti, B. Trimmer, A. W. Feinberg, R. Raman, K. K. Parker, R. Bashir, M. Sitti, S. Martel, P. Dario and A. Menciassi, Sci. Robot., 2017, 2, eaaq0495 CrossRef.
- L. Sun, Y. Yu, Z. Chen, F. Bian, F. Ye, L. Sun and Y. Zhao, Chem. Soc. Rev., 2020, 49, 4043–4069 RSC.
- M. Di Luca, C. Hoskins, F. Corduas, R. Onchuru, A. Oluwasanmi, D. Mariotti, B. Conti and D. A. Lamprou, Int. J. Pharm., 2022, 629, 122363 CrossRef CAS.
- I. Vujaklija and D. Farina, Expert Rev. Med. Devices, 2018, 15, 505–512 CrossRef CAS.
- J. Koprnický, P. Najman and J. Šafka, in 2017 IEEE International Workshop of Electronics, Control, Measurement, Signals and their Application to Mechatronics (ECMSM), 2017, pp. 1–6.
- J. A. Owusu and K. Boahene, Curr. Opin. Otolaryngol. Head Neck Surg., 2015, 23, 261 CrossRef.
- S. V. Krishna, S. K. Gupta, N. Holla and K. Chatterjee, Indian J. Orthop., 2022, 56, 1657–1661 CrossRef.
- P. Honigmann, N. Sharma, B. Okolo, U. Popp, B. Msallem and F. M. Thieringer, Bio Med. Res. Int., 2018, 2018, e4520636 Search PubMed.
- L. Pugliese, S. Marconi, E. Negrello, V. Mauri, A. Peri, V. Gallo, F. Auricchio and A. Pietrabissa, Updat. Surg., 2018, 70, 381–388 CrossRef.
- A. Lee, A. R. Hudson, D. J. Shiwarski, J. W. Tashman, T. J. Hinton, S. Yerneni, J. M. Bliley, P. G. Campbell and A. W. Feinberg, Science, 2019, 365, 482–487 CrossRef CAS.
- M. A. Skylar-Scott, S. G. M. Uzel, L. L. Nam, J. H. Ahrens, R. L. Truby, S. Damaraju and J. A. Lewis, Sci. Adv., 2019, 5, eaaw2459 CrossRef CAS.
- H.-W. Kang, S. J. Lee, I. K. Ko, C. Kengla, J. J. Yoo and A. Atala, Nat. Biotechnol., 2016, 34, 312–319 CrossRef CAS.
- D. B. Kolesky, K. A. Homan, M. A. Skylar-Scott and J. A. Lewis, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 3179–3184 CrossRef CAS.
- T. J. Hinton, Q. Jallerat, R. N. Palchesko, J. H. Park, M. S. Grodzicki, H.-J. Shue, M. H. Ramadan, A. R. Hudson and A. W. Feinberg, Sci. Adv., 2015, 1, e1500758 CrossRef.
- M. V. Monteiro, Y. S. Zhang, V. M. Gaspar and J. F. Mano, Trends Biotechnol., 2022, 40, 432–447 CrossRef CAS.
- S. Correia Carreira, R. Begum and A. W. Perriman, Adv. Healthcare Mater., 2020, 9, 1900554 CrossRef CAS.
- A. C. Juraski, S. Sharma, S. Sparanese, V. A. da Silva, J. Wong, Z. Laksman, R. Flannigan, L. Rohani and S. M. Willerth, Expert Opin. Drug Discovery, 2023, 18, 1043–1059 CrossRef CAS.
- W. Peng, P. Datta, B. Ayan, V. Ozbolat, D. Sosnoski and I. T. Ozbolat, Acta Biomater., 2017, 57, 26–46 CrossRef CAS.
- E. T. Pashuck and M. Stevens, Nat. Biotechnol., 2016, 34, 295–296 CrossRef CAS.
- A. Mandal and K. Chatterjee, Chem. Eng. J., 2023, 455, 140550 CrossRef CAS.
- B. Gao, Q. Yang, X. Zhao, G. Jin, Y. Ma and F. Xu, Trends Biotechnol., 2016, 34, 746–756 CrossRef CAS.
- Y. Qiu and K. Park, Adv. Drug Delivery Rev., 2001, 53, 321–339 CrossRef CAS.
- M. Champeau, D. A. Heinze, T. N. Viana, E. R. de Souza, A. C. Chinellato and S. Titotto, Adv. Funct. Mater., 2020, 30, 1910606 CrossRef CAS.
- S. Choi, K. Y. Lee, S. L. Kim, L. A. MacQueen, H. Chang, J. F. Zimmerman, Q. Jin, M. M. Peters, H. A. M. Ardoña, X. Liu, A.-C. Heiler, R. Gabardi, C. Richardson, W. T. Pu, A. R. Bausch and K. K. Parker, Nat. Mater., 2023, 22, 1039–1046 CrossRef CAS.
- G. Siqueira, D. Kokkinis, R. Libanori, M. K. Hausmann, A. S. Gladman, A. Neels, P. Tingaut, T. Zimmermann, J. A. Lewis and A. R. Studart, Adv. Funct. Mater., 2017, 27, 1604619 CrossRef.
- H. Yuk, B. Lu, S. Lin, K. Qu, J. Xu, J. Luo and X. Zhao, Nat. Commun., 2020, 11, 1604 CrossRef CAS.
- A. Sydney Gladman, E. A. Matsumoto, R. G. Nuzzo, L. Mahadevan and J. A. Lewis, Nat. Mater., 2016, 15, 413–418 CrossRef CAS.
- A. Kotikian, R. L. Truby, J. W. Boley, T. J. White and J. A. Lewis, Adv. Mater., 2018, 30, 1706164 CrossRef.
- J. W. Boley, W. M. van Rees, C. Lissandrello, M. N. Horenstein, R. L. Truby, A. Kotikian, J. A. Lewis and L. Mahadevan, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 20856–20862 CrossRef CAS.
- S. Weng, X. Kuang, Q. Zhang, C. M. Hamel, D. J. Roach, N. Hu and H. Jerry Qi, ACS Appl. Mater. Interfaces, 2021, 13, 12797–12804 CrossRef CAS.
- R. Xing, J. Yang, D. Zhang, W. Gong, T. V. Neumann, M. Wang, R. Huang, J. Kong, W. Qi and M. D. Dickey, Matter, 2023, 6, 2248–2262 CrossRef CAS.
- C. Dawson, J. F. V. Vincent and A.-M. Rocca, Nature, 1997, 390, 668 CrossRef CAS.
- R. Elbaum, L. Zaltzman, I. Burgert and P. Fratzl, Science, 2007, 316, 884–886 CrossRef CAS.
- S. Armon, E. Efrati, R. Kupferman and E. Sharon, Science, 2011, 333, 1726–1730 CrossRef CAS.
- R. M. Erb, J. S. Sander, R. Grisch and A. R. Studart, Nat. Commun., 2013, 4, 1712 CrossRef.
- H. Aharoni, E. Sharon and R. Kupferman, Phys. Rev. Lett., 2014, 113, 257801 CrossRef.
- W. M. van Rees, E. A. Matsumoto, A. S. Gladman, J. A. Lewis and L. Mahadevan, Soft Matter, 2018, 14, 8771–8779 RSC.
- L. Ren, Z. Wang, L. Ren, C. Xu, B. Li, Y. Shi and Q. Liu, Composites, Part B, 2023, 265, 110938 CrossRef.
- H. Yuk and X. Zhao, Adv. Mater., 2018, 30, 1704028 CrossRef.
- G. Stoychev, S. Zakharchenko, S. Turcaud, J. W. C. Dunlop and L. Ionov, ACS Nano, 2012, 6, 3925–3934 CrossRef CAS.
- D. H. Gracias, Curr. Opin. Chem. Eng., 2013, 2, 112–119 CrossRef.
- M. Jamal, S. S. Kadam, R. Xiao, F. Jivan, T.-M. Onn, R. Fernandes, T. D. Nguyen and D. H. Gracias, Adv. Healthcare Mater., 2013, 2, 1142–1150 CrossRef CAS.
- D. Raviv, W. Zhao, C. McKnelly, A. Papadopoulou, A. Kadambi, B. Shi, S. Hirsch, D. Dikovsky, M. Zyracki, C. Olguin, R. Raskar and S. Tibbits, Sci. Rep., 2014, 4, 7422 CrossRef CAS.
- A. Cangialosi, C. Yoon, J. Liu, Q. Huang, J. Guo, T. D. Nguyen, D. H. Gracias and R. Schulman, Science, 2017, 357, 1126–1130 CrossRef CAS.
- Z. Zhao, X. Kuang, C. Yuan, H. J. Qi and D. Fang, ACS Appl. Mater. Interfaces, 2018, 10, 19932–19939 CrossRef CAS.
- Y. B. Lee, O. Jeon, S. J. Lee, A. Ding, D. Wells and E. Alsberg, Adv. Funct. Mater., 2021, 31, 2010104 CrossRef CAS.
- P. J. Díaz-Payno, M. Kalogeropoulou, I. Muntz, E. Kingma, N. Kops, M. D’Este, G. H. Koenderink, L. E. Fratila-Apachitei, G. J. V. M. van Osch and A. A. Zadpoor, Adv. Healthcare Mater., 2023, 12, 2201891 CrossRef.
- J. Kim, C. Kim, Y. Song, S.-G. Jeong, T.-S. Kim and C.-S. Lee, Chem. Eng. J., 2017, 321, 384–393 CrossRef CAS.
- J. Kim, J. A. Hanna, M. Byun, C. D. Santangelo and R. C. Hayward, Science, 2012, 335, 1201–1205 CrossRef CAS.
- L. Huang, R. Jiang, J. Wu, J. Song, H. Bai, B. Li, Q. Zhao and T. Xie, Adv. Mater., 2017, 29, 1605390 CrossRef.
- A. Nojoomi, H. Arslan, K. Lee and K. Yum, Nat. Commun., 2018, 9, 3705 CrossRef.
- Z. Ji, C. Yan, B. Yu, X. Zhang, M. Cai, X. Jia, X. Wang and F. Zhou, Adv. Mater. Technol., 2019, 4, 1800713 CrossRef.
- A. Ding, S. J. Lee, S. Ayyagari, R. Tang, C. T. Huynh and E. Alsberg, Bioact. Mater., 2022, 7, 324–332 CAS.
- A. Ding, O. Jeon, D. Cleveland, K. L. Gasvoda, D. Wells, S. J. Lee and E. Alsberg, Adv. Mater., 2022, 34, 2109394 CrossRef CAS.
- S. B. Gugulothu and K. Chatterjee, ACS Macro Lett., 2023, 12, 494–502 CrossRef CAS.
- Z. Zhao, J. Wu, X. Mu, H. Chen, H. J. Qi and D. Fang, Macromol. Rapid Commun., 2017, 38, 1600625 CrossRef.
- Z. Zhao, J. Wu, X. Mu, H. Chen, H. J. Qi and D. Fang, Sci. Adv., 2017, 3, e1602326 CrossRef.
- L. Zhang, Y. Xiang, H. Zhang, L. Cheng, X. Mao, N. An, L. Zhang, J. Zhou, L. Deng, Y. Zhang, X. Sun, H. A. Santos and W. Cui, Adv. Sci., 2020, 7, 1903553 CrossRef CAS.
- A. Kirillova, R. Maxson, G. Stoychev, C. T. Gomillion and L. Ionov, Adv. Mater., 2017, 29, 1703443 CrossRef.
- W. Kitana, I. Apsite, J. Hazur, A. R. Boccaccini and L. Ionov, Adv. Mater. Technol., 2023, 8, 2200429 CrossRef CAS.
- J. Lai, X. Ye, J. Liu, C. Wang, J. Li, X. Wang, M. Ma and M. Wang, Mater. Des., 2021, 205, 109699 CrossRef CAS.
- E. Käpylä, S. M. Delgado and A. M. Kasko, ACS Appl. Mater. Interfaces, 2016, 8, 17885–17893 CrossRef.
- A. M. Kloxin, A. M. Kasko, C. N. Salinas and K. S. Anseth, Science, 2009, 324, 59–63 CrossRef CAS.
- A. Joshi, S. Choudhury, V. S. Baghel, S. Ghosh, S. Gupta, D. Lahiri, G. K. Ananthasuresh and K. Chatterjee, Adv. Healthcare Mater., 2023, 12, 2300701 CrossRef CAS.
- A. Ding, O. Jeon, R. Tang, Y. B. Lee, S. J. Lee and E. Alsberg, Adv. Sci., 2021, 8, 2004616 CrossRef CAS.
- P. Mondal, A. Mandal and K. Chatterjee, Adv. Mater. Technol., 2023, 8, 2300894 CrossRef CAS.
- K. Kuribayashi-Shigetomi, H. Onoe and S. Takeuchi, PLoS One, 2012, 7, e51085 CrossRef CAS.
- M. D. Davidson, M. E. Prendergast, E. Ban, K. L. Xu, G. Mickel, P. Mensah, A. Dhand, P. A. Janmey, V. B. Shenoy and J. A. Burdick, Sci. Adv., 2021, 7, eabi8157 CrossRef CAS.
- A. Rodrigo-Navarro, S. Sankaran, M. J. Dalby, A. del Campo and M. Salmeron-Sanchez, Nat. Rev. Mater., 2021, 6, 1175–1190 CrossRef.
- L. K. Rivera-Tarazona, Z. T. Campbell and T. H. Ware, Soft Matter, 2021, 17, 785–809 RSC.
- W. V. Srubar, Trends Biotechnol., 2021, 39, 574–583 CrossRef CAS.
- M. Mimee, P. Nadeau, A. Hayward, S. Carim, S. Flanagan, L. Jerger, J. Collins, S. McDonnell, R. Swartwout, R. J. Citorik, V. Bulović, R. Langer, G. Traverso, A. P. Chandrakasan and T. K. Lu, Science, 2018, 360, 915–918 CrossRef CAS.
- X. Liu, T.-C. Tang, E. Tham, H. Yuk, S. Lin, T. K. Lu and X. Zhao, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 2200–2205 CrossRef CAS.
- C. A. Mora, A. F. Herzog, R. A. Raso and W. J. Stark, Biomaterials, 2015, 61, 1–9 CrossRef CAS.
- C. Gilbert, T.-C. Tang, W. Ott, B. A. Dorr, W. M. Shaw, G. L. Sun, T. K. Lu and T. Ellis, Nat. Mater., 2021, 20, 691–700 CrossRef CAS.
- S. Sankaran, J. Becker, C. Wittmann and A. del Campo, Small, 2019, 15, 1804717 CrossRef.
- S. Sankaran and A. del Campo, Adv. Biosyst., 2019, 3, 1800312 CrossRef.
- L. K. Rivera-Tarazona, T. Shukla, K. A. Singh, A. K. Gaharwar, Z. T. Campbell and T. H. Ware, Adv. Funct. Mater., 2022, 32, 2106843 CrossRef CAS.
- E. Rousseau, R. Raman, T. Tamir, A. Bu, S. Srinivasan, N. Lynch, R. Langer, F. M. White and M. J. Cima, Biomaterials, 2023, 302, 122317 CrossRef CAS.
- L. K. Rivera-Tarazona, V. D. Bhat, H. Kim, Z. T. Campbell and T. H. Ware, Sci. Adv., 2020, 6, eaax8582 CrossRef CAS.
- R. Raman, C. Cvetkovic, S. G. M. Uzel, R. J. Platt, P. Sengupta, R. D. Kamm and R. Bashir, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 3497–3502 CrossRef CAS.
- B. Geiger, J. P. Spatz and A. D. Bershadsky, Nat. Rev. Mol. Cell Biol., 2009, 10, 21–33 CrossRef CAS.
- R. Oria, T. Wiegand, J. Escribano, A. Elosegui-Artola, J. J. Uriarte, C. Moreno-Pulido, I. Platzman, P. Delcanale, L. Albertazzi, D. Navajas, X. Trepat, J. M. García-Aznar, E. A. Cavalcanti-Adam and P. Roca-Cusachs, Nature, 2017, 552, 219–224 CrossRef CAS.
- J. M. Viola, C. M. Porter, A. Gupta, M. Alibekova, L. S. Prahl and A. J. Hughes, Adv. Mater., 2020, 32, 2002195 CrossRef CAS.
- A. Lendlein and O. E. C. Gould, Nat. Rev. Mater., 2019, 4, 116–133 CrossRef.
- J. Leng, X. Lan, Y. Liu and S. Du, Prog. Mater. Sci., 2011, 56, 1077–1135 CrossRef CAS.
- M. Behl and A. Lendlein, Mater. Today, 2007, 10, 20–28 CrossRef CAS.
- A. Lendlein and S. Kelch, Angew. Chem., Int. Ed., 2002, 41, 2034–2057 CrossRef CAS.
- C. A. Spiegel, M. Hackner, V. P. Bothe, J. P. Spatz and E. Blasco, Adv. Funct. Mater., 2022, 32, 2110580 CrossRef CAS.
- X. Wan, Y. He, Y. Liu and J. Leng, Addit. Manuf., 2022, 53, 102689 CAS.
- W. Zhang, H. Wang, H. Wang, J. Y. E. Chan, H. Liu, B. Zhang, Y.-F. Zhang, K. Agarwal, X. Yang, A. S. Ranganath, H. Y. Low, Q. Ge and J. K. W. Yang, Nat. Commun., 2021, 12, 112 CrossRef CAS.
- B. Zhang, H. Li, J. Cheng, H. Ye, A. H. Sakhaei, C. Yuan, P. Rao, Y.-F. Zhang, Z. Chen, R. Wang, X. He, J. Liu, R. Xiao, S. Qu and Q. Ge, Adv. Mater., 2021, 33, 2101298 CrossRef CAS.
- 4D Printed Biocompatible Magnetic Composite for Minimally Invasive Deployable Structures|Materials Science|ChemRxiv | Cambridge Open Engage, https://chemrxiv.org/engage/chemrxiv/article-details/63651df4ee31864b167f691c, (accessed October 1, 2023).
- L. Yue, X. Sun, L. Yu, M. Li, S. M. Montgomery, Y. Song, T. Nomura, M. Tanaka and H. J. Qi, Nat. Commun., 2023, 14, 5519 CrossRef CAS.
- G. Li and A. Wang, J. Polym. Sci., Part B: Polym. Phys., 2016, 54, 1319–1339 CrossRef CAS.
- Q. Zhao, M. Behl and A. Lendlein, Soft Matter, 2013, 9, 1744–1755 RSC.
- Y. Wang and X. Li, Mech. Mater., 2020, 151, 103628 CrossRef.
- G. F. Hu, A. R. Damanpack, M. Bodaghi and W. H. Liao, Smart Mater. Struct., 2017, 26, 125023 CrossRef.
- Q. Zhang, K. Zhang and G. Hu, Sci. Rep., 2016, 6, 22431 CrossRef CAS.
- T. van Manen, S. Janbaz and A. A. Zadpoor, Mater. Horiz., 2017, 4, 1064–1069 RSC.
- B. Goo, C.-H. Hong and K. Park, Mater. Des., 2020, 188, 108485 CrossRef CAS.
- A. Lendlein and R. Langer, Science, 2002, 296, 1673–1676 CrossRef.
- Q. Ge, A. H. Sakhaei, H. Lee, C. K. Dunn, N. X. Fang and M. L. Dunn, Sci. Rep., 2016, 6, 31110 CrossRef.
- M. Chen, M. Gao, L. Bai, H. Zheng, H. J. Qi and K. Zhou, Adv. Mater., 2023, 35, 2209566 CrossRef CAS.
- C. Ohm, M. Brehmer and R. Zentel, Adv. Mater., 2010, 22, 3366–3387 CrossRef CAS PubMed.
- Y. Sawa, F. Ye, K. Urayama, T. Takigawa, V. Gimenez-Pinto, R. L. B. Selinger and J. V. Selinger, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 6364–6368 CrossRef CAS.
- C. P. Ambulo, J. J. Burroughs, J. M. Boothby, H. Kim, M. R. Shankar and T. H. Ware, ACS Appl. Mater. Interfaces, 2017, 9, 37332–37339 CrossRef CAS.
- A. Andreu, P.-C. Su, J.-H. Kim, C. S. Ng, S. Kim, I. Kim, J. Lee, J. Noh, A. S. Subramanian and Y.-J. Yoon, Addit. Manuf., 2021, 44, 102024 CAS.
- B. Peng, Y. Yang, K. Gu, E. J. Amis and K. A. Cavicchi, ACS Mater. Lett., 2019, 1, 410–417 CrossRef CAS.
- Q. Mu, L. Wang, C. K. Dunn, X. Kuang, F. Duan, Z. Zhang, H. J. Qi and T. Wang, Addit. Manuf., 2017, 18, 74–83 CAS.
- K. M. Herbert, H. E. Fowler, J. M. McCracken, K. R. Schlafmann, J. A. Koch and T. J. White, Nat. Rev. Mater., 2022, 7, 23–38 CrossRef CAS.
- M. del Pozo, J. A. H. P. Sol, A. P. H. J. Schenning and M. G. Debije, Adv. Mater., 2022, 34, 2104390 CrossRef CAS.
- J. M. McCracken, B. R. Donovan and T. J. White, Adv. Mater., 2020, 32, 1906564 CrossRef CAS.
- D. Corbett and M. Warner, Liq. Cryst., 2009, 36, 1263–1280 CrossRef CAS.
- Y. Yu, M. Nakano and T. Ikeda, Nature, 2003, 425, 145 CrossRef CAS PubMed.
- T. J. White, S. V. Serak, N. V. Tabiryan, R. A. Vaia and T. J. Bunning, J. Mater. Chem., 2009, 19, 1080–1085 RSC.
- A. H. Gelebart, D. Jan Mulder, M. Varga, A. Konya, G. Vantomme, E. W. Meijer, R. L. B. Selinger and D. J. Broer, Nature, 2017, 546, 632–636 CrossRef CAS.
- L. T. de Haan, C. Sánchez-Somolinos, C. M. W. Bastiaansen, A. P. H. J. Schenning and D. J. Broer, Angew. Chem., Int. Ed., 2012, 51, 12469–12472 CrossRef CAS.
- T. H. Ware, M. E. McConney, J. J. Wie, V. P. Tondiglia and T. J. White, Science, 2015, 347, 982–984 CrossRef CAS.
- F. Ge, R. Yang, X. Tong, F. Camerel and Y. Zhao, Angew. Chem., Int. Ed., 2018, 57, 11758–11763 CrossRef CAS.
- X. Lu, H. Zhang, G. Fei, B. Yu, X. Tong, H. Xia and Y. Zhao, Adv. Mater., 2018, 30, 1706597 CrossRef.
- F. Wang, C. Liu, H. Yang, H. Wang, H. Zhang, X. Zeng, C. Wang, W. Zhang, W. Lv, P. Zhu and B. Li, Addit. Manuf., 2023, 63, 103411 CAS.
- G. Liu, Y. Zhao, G. Wu and J. Lu, Sci. Adv., 2018, 4, eaat0641 CrossRef.
- S. Chen, J. Li, H. Shi, X. Chen, G. Liu, S. Meng and J. Lu, Chem. Eng. J., 2023, 455, 140655 CrossRef CAS.
- G. Liu, X. Zhang, X. Lu, Y. Zhao, Z. Zhou, J. Xu, J. Yin, T. Tang, P. Wang, S. Yi, J. Fan, X. Zhuo, Y. H. Chan, W. L. Wong, H. Bian, J. Zuo, Y. Dai, J. Wu and J. Lu, Adv. Mater., 2023, 35, 2302108 CrossRef CAS.
- W. Wu, Y. Zhou, Q. Liu, L. Ren, F. Chen, J. Y. H. Fuh, A. Zheng, X. Li, J. Zhao and G. Li, Adv. Sci., 2023, 10, 2206486 CrossRef CAS.
- J. Mohd Jani, M. Leary, A. Subic and M. A. Gibson, Mater. Des., 2014, 56, 1078–1113 CrossRef CAS.
- S. Nam, B. R. Seo, A. J. Najibi, S. L. McNamara and D. J. Mooney, Nat. Mater., 2023, 22, 249–259 CrossRef CAS.
- D. Kim, I. Ferretto, C. Leinenbach and W. Lee, Adv. Mater. Interfaces, 2022, 9, 2200171 CrossRef CAS.
- S. Saedi, S. E. Saghaian, A. Jahadakbar, N. Shayesteh Moghaddam, M. Taheri Andani, S. M. Saghaian, Y. C. Lu, M. Elahinia and H. E. Karaca, J. Mater. Sci.: Mater. Med., 2018, 29, 40 CrossRef.
- J. Ma, F. Krisnadi, M. H. Vong, M. Kong, O. M. Awartani and M. D. Dickey, Adv. Mater., 2023, 35, 2205196 CrossRef CAS.
- J. Yan, Y. Lu, G. Chen, M. Yang and Z. Gu, Chem. Soc. Rev., 2018, 47, 2518–2533 RSC.
- L. Wang, R. Lai, L. Zhang, M. Zeng and L. Fu, Adv. Mater., 2022, 34, 2201956 CrossRef CAS.
- Y. Lu, Y. Lin, Z. Chen, Q. Hu, Y. Liu, S. Yu, W. Gao, M. D. Dickey and Z. Gu, Nano Lett., 2017, 17, 2138–2145 CrossRef CAS.
- A. Elbourne, S. Cheeseman, P. Atkin, N. P. Truong, N. Syed, A. Zavabeti, M. Mohiuddin, D. Esrafilzadeh, D. Cozzolino, C. F. McConville, M. D. Dickey, R. J. Crawford, K. Kalantar-Zadeh, J. Chapman, T. Daeneke and V. K. Truong, ACS Nano, 2020, 14, 802–817 CrossRef CAS.
- D. Wang, C. Gao, W. Wang, M. Sun, B. Guo, H. Xie and Q. He, ACS Nano, 2018, 12, 10212–10220 CrossRef CAS.
- J. L. Bartlett and X. Li, Addit. Manuf., 2019, 27, 131–149 Search PubMed.
- A. Tabatabaeian, A. R. Ghasemi, M. M. Shokrieh, B. Marzbanrad, M. Baraheni and M. Fotouhi, Adv. Eng. Mater., 2022, 24, 2100786 CrossRef CAS.
- M. A. Saccone, R. A. Gallivan, K. Narita, D. W. Yee and J. R. Greer, Nature, 2022, 612, 685–690 CrossRef CAS.
- D. W. Yee, M. A. Citrin, Z. W. Taylor, M. A. Saccone, V. L. Tovmasyan and J. R. Greer, Adv. Mater. Technol., 2021, 6, 2000791 CrossRef CAS.
- D. W. Yee, M. L. Lifson, B. W. Edwards and J. R. Greer, Adv. Mater., 2019, 31, 1901345 CrossRef.
- Y. Wang, H. Cui, T. Esworthy, D. Mei, Y. Wang and L. G. Zhang, Adv. Mater., 2022, 34, 2109198 CrossRef CAS.
- L. Faber, A. Yau and Y. Chen, Biofabrication, 2023, 16, 012001 CrossRef.
- A. Chen, W. Wang, Z. Mao, Y. He, S. Chen, G. Liu, J. Su, P. Feng, Y. Shi, C. Yan and J. Lu, Adv. Mater., 2023, 2307686 CrossRef.
- B. Lubarsky and M. A. Krasnow, Cell, 2003, 112, 19–28 CrossRef CAS.
- L. Ionov, Macromol. Chem. Phys., 2013, 214, 1178–1183 CrossRef CAS.
- C. Cui, D.-O. Kim, M. Y. Pack, B. Han, L. Han, Y. Sun and L.-H. Han, Biofabrication, 2020, 12, 045018 CrossRef CAS.
- S. Miao, H. Cui, M. Nowicki, L. Xia, X. Zhou, S.-J. Lee, W. Zhu, K. Sarkar, Z. Zhang and L. G. Zhang, Adv. Biosyst., 2018, 2, 1800101 CrossRef.
- G. H. Yang, W. Kim, J. Kim and G. Kim, Theranostics, 2021, 11, 48–63 CrossRef CAS.
- S. H. Kim, Y. B. Seo, Y. K. Yeon, Y. J. Lee, H. S. Park, Md. T. Sultan, J. M. Lee, J. S. Lee, O. J. Lee, H. Hong, H. Lee, O. Ajiteru, Y. J. Suh, S.-H. Song, K.-H. Lee and C. H. Park, Biomaterials, 2020, 260, 120281 CrossRef CAS.
- B. Grigoryan, S. J. Paulsen, D. C. Corbett, D. W. Sazer, C. L. Fortin, A. J. Zaita, P. T. Greenfield, N. J. Calafat, J. P. Gounley, A. H. Ta, F. Johansson, A. Randles, J. E. Rosenkrantz, J. D. Louis-Rosenberg, P. A. Galie, K. R. Stevens and J. S. Miller, Science, 2019, 364, 458–464 CrossRef CAS PubMed.
- B. Schamberger, R. Ziege, K. Anselme, M. Ben Amar, M. Bykowski, A. P. G. Castro, A. Cipitria, R. A. Coles, R. Dimova, M. Eder, S. Ehrig, L. M. Escudero, M. E. Evans, P. R. Fernandes, P. Fratzl, L. Geris, N. Gierlinger, E. Hannezo, A. Iglič, J. J. K. Kirkensgaard, P. Kollmannsberger, Ł. Kowalewska, N. A. Kurniawan, I. Papantoniou, L. Pieuchot, T. H. V. Pires, L. D. Renner, A. O. Sageman-Furnas, G. E. Schröder-Turk, A. Sengupta, V. R. Sharma, A. Tagua, C. Tomba, X. Trepat, S. L. Waters, E. F. Yeo, A. Roschger, C. M. Bidan and J. W. C. Dunlop, Adv. Mater., 2023, 35, 2206110 CrossRef CAS PubMed.
- C. M. Nelson, J. Biomech. Eng., 2016, 138(2), 021005 CrossRef PubMed.
- X. Chen and J. Yin, Soft Matter, 2010, 6, 5667–5680 RSC.
- H. Cui, C. Liu, T. Esworthy, Y. Huang, Z. Yu, X. Zhou, H. San, S. Lee, S. Y. Hann, M. Boehm, M. Mohiuddin, J. P. Fisher and L. G. Zhang, Sci. Adv., 2020, 6, eabb5067 CrossRef CAS PubMed.
- Y. Wang, H. Cui, Y. Wang, C. Xu, T. J. Esworthy, S. Y. Hann, M. Boehm, Y.-L. Shen, D. Mei and L. G. Zhang, ACS Appl. Mater. Interfaces, 2021, 13, 12746–12758 CrossRef CAS PubMed.
- G. Qu, J. Huang, Z. Li, Y. Jiang, Y. Liu, K. Chen, Z. Xu, Y. Zhao, G. Gu, X. Wu and J. Ren, Mater. Today Bio, 2022, 16, 100363 CrossRef CAS PubMed.
- H. Cui, S. Miao, T. Esworthy, S. Lee, X. Zhou, S. Y. Hann, T. J. Webster, B. T. Harris and L. G. Zhang, Nano Res., 2019, 12, 1381–1388 CrossRef.
- A. Ding, S. J. Lee, R. Tang, K. L. Gasvoda, F. He and E. Alsberg, Small, 2022, 18, 2202196 CrossRef CAS.
- D. Baptista, L. Teixeira, C. van Blitterswijk, S. Giselbrecht and R. Truckenmüller, Trends Biotechnol., 2019, 37, 838–854 CrossRef CAS.
- Y. Zhang, K. Liu, T. Liu, C. Ni, D. Chen, J. Guo, C. Liu, J. Zhou, Z. Jia, Q. Zhao, P. Pan and T. Xie, Nat. Commun., 2021, 12, 6155 CrossRef CAS.
- C. Löwenberg, M. Balk, C. Wischke, M. Behl and A. Lendlein, Acc. Chem. Res., 2017, 50, 723–732 CrossRef.
- J. Shang, X. Le, J. Zhang, T. Chen and P. Theato, Polym. Chem., 2019, 10, 1036–1055 RSC.
- Q. Bian, L. Fu and H. Li, Nat. Commun., 2022, 13, 137 CrossRef CAS.
- S. Naficy, R. Gately, R. Gorkin III, H. Xin and G. M. Spinks, Macromol. Mater. Eng., 2017, 302, 1600212 CrossRef.
- S. R. Goudu, I. C. Yasa, X. Hu, H. Ceylan, W. Hu and M. Sitti, Adv. Funct. Mater., 2020, 30, 2004975 CrossRef CAS.
- S. Fusco, M. S. Sakar, S. Kennedy, C. Peters, R. Bottani, F. Starsich, A. Mao, G. A. Sotiriou, S. Pané, S. E. Pratsinis, D. Mooney and B. J. Nelson, Adv. Mater., 2014, 26, 952–957 CrossRef CAS.
- R. Fernandes and D. H. Gracias, Adv. Drug Delivery Rev., 2012, 64, 1579–1589 CrossRef CAS.
- S. Pedron, S. van Lierop, P. Horstman, R. Penterman, D. J. Broer and E. Peeters, Adv. Funct. Mater., 2011, 21, 1624–1630 CrossRef CAS.
- L. J. De Cock, S. De Koker, B. G. De Geest, J. Grooten, C. Vervaet, J. P. Remon, G. B. Sukhorukov and M. N. Antipina, Angew. Chem., Int. Ed., 2010, 49, 6954–6973 CrossRef CAS PubMed.
- Q. Jiang, Y. Shang, Y. Xie and B. Ding, Adv. Mater., 2023, 2301035 CrossRef.
- C. L. Randall, T. G. Leong, N. Bassik and D. H. Gracias, Adv. Drug Delivery Rev., 2007, 59, 1547–1561 CrossRef CAS.
- J. Guan, H. He, L. J. Lee and D. J. Hansford, Small, 2007, 3, 412–418 CrossRef CAS PubMed.
- H. He, J. Guan and J. L. Lee, J. Controlled Release, 2006, 110, 339–346 CrossRef CAS PubMed.
- J. Jiang, J. Xiao, Z. Zhao, M.-S. Yuan and J. Wang, Mater. Chem. Front., 2021, 5, 6027–6040 RSC.
- E. Aznar, L. Mondragón, J. V. Ros-Lis, F. Sancenón, M. D. Marcos, R. Martínez-Máñez, J. Soto, E. Pérez-Payá and P. Amorós, Angew. Chem., Int. Ed., 2011, 50, 11172–11175 CrossRef CAS.
- C. Chen, J. Geng, F. Pu, X. Yang, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2011, 50, 882–886 CrossRef CAS.
- N. Singh, A. Karambelkar, L. Gu, K. Lin, J. S. Miller, C. S. Chen, M. J. Sailor and S. N. Bhatia, J. Am. Chem. Soc., 2011, 133, 19582–19585 CrossRef CAS PubMed.
- R. A. Perez and H.-W. Kim, Acta Biomater., 2015, 21, 2–19 CrossRef CAS.
- Y. Hu, Z. Wang, D. Jin, C. Zhang, R. Sun, Z. Li, K. Hu, J. Ni, Z. Cai, D. Pan, X. Wang, W. Zhu, J. Li, D. Wu, L. Zhang and J. Chu, Adv. Funct. Mater., 2020, 30, 1907377 CrossRef CAS.
- M. Bouché, Y. C. Dong, S. Sheikh, K. Taing, D. Saxena, J. C. Hsu, M. H. Chen, R. D. Salinas, H. Song, J. A. Burdick, J. Dorsey and D. P. Cormode, ACS Biomater. Sci. Eng., 2021, 7, 3209–3220 CrossRef.
- D. Alkekhia, C. LaRose and A. Shukla, ACS Appl. Mater. Interfaces, 2022, 14, 27538–27550 CrossRef CAS.
- B. P. Purcell, D. Lobb, M. B. Charati, S. M. Dorsey, R. J. Wade, K. N. Zellars, H. Doviak, S. Pettaway, C. B. Logdon, J. A. Shuman, P. D. Freels, J. H. Gorman III, R. C. Gorman, F. G. Spinale and J. A. Burdick, Nat. Mater., 2014, 13, 653–661 CrossRef CAS.
- J. Rautio, N. A. Meanwell, L. Di and M. J. Hageman, Nat. Rev. Drug Discovery, 2018, 17, 559–587 CrossRef CAS.
- Y. Yang, H. Aloysius, D. Inoyama, Y. Chen and L. Hu, Acta Pharm. Sin. B, 2011, 1, 143–159 CrossRef CAS.
- A. Xie, S. Hanif, J. Ouyang, Z. Tang, N. Kong, N. Y. Kim, B. Qi, D. Patel, B. Shi and W. Tao, EBioMedicine, 2020, 56, 102821 CrossRef.
- X. Li, F. Huo, Y. Zhang, F. Cheng and C. Yin, J. Mater. Chem. B, 2022, 10, 5504–5519 RSC.
- M. Kücken and A. C. Newell, J. Theor. Biol., 2005, 235, 71–83 CrossRef.
- P. Ciarletta, Phys. Rev. Lett., 2013, 110, 158102 CrossRef CAS.
- L. Wang, C. E. Castro and M. C. Boyce, Soft Matter, 2011, 7, 11319–11324 RSC.
- M. Guvendiren, S. Yang and J. A. Burdick, Adv. Funct. Mater., 2009, 19, 3038–3045 CrossRef CAS.
- C.-Y. Liaw, J. Pereyra, A. Abaci, S. Ji and M. Guvendiren, Adv. Mater. Technol., 2022, 7, 2101118 CrossRef CAS.
- M. Guvendiren and J. A. Burdick, Biomaterials, 2010, 31, 6511–6518 CrossRef CAS.
- J. D. Glover, Z. R. Sudderick, B. B.-J. Shih, C. Batho-Samblas, L. Charlton, A. L. Krause, C. Anderson, J. Riddell, A. Balic, J. Li, V. Klika, T. E. Woolley, E. A. Gaffney, A. Corsinotti, R. A. Anderson, L. J. Johnston, S. J. Brown, S. Wang, Y. Chen, M. L. Crichton and D. J. Headon, Cell, 2023, 186, 940–956 CrossRef CAS.
- S. Sick, S. Reinker, J. Timmer and T. Schlake, Science, 2006, 314, 1447–1450 CrossRef CAS.
- J. B. L. Bard, J. Theor. Biol., 1981, 93, 363–385 CrossRef CAS.
- B. M. Alessio and A. Gupta, Sci. Adv., 2023, 9, eadj2457 CrossRef.
- D. You, G. Chen, C. Liu, X. Ye, S. Wang, M. Dong, M. Sun, J. He, X. Yu, G. Ye, Q. Li, J. Wu, J. Wu, Q. Zhao, T. Xie, M. Yu and H. Wang, Adv. Funct. Mater., 2021, 31, 2103920 CrossRef CAS.
- S. Miao, H. Cui, T. Esworthy, B. Mahadik, S. Lee, X. Zhou, S. Y. Hann, J. P. Fisher and L. G. Zhang, Adv. Sci., 2020, 7, 1902403 CrossRef CAS.
- F. M. Yavitt, B. E. Kirkpatrick, M. R. Blatchley, K. F. Speckl, E. Mohagheghian, R. Moldovan, N. Wang, P. J. Dempsey and K. S. Anseth, Sci. Adv., 2023, 9, eadd5668 CrossRef.
- A. I. Teixeira, G. A. Abrams, P. J. Bertics, C. J. Murphy and P. F. Nealey, J. Cell Sci., 2003, 116, 1881–1892 CrossRef CAS.
- K. A. Davis, K. A. Burke, P. T. Mather and J. H. Henderson, Biomaterials, 2011, 32, 2285–2293 CrossRef CAS.
- M. Ebara, K. Uto, N. Idota, J. M. Hoffman and T. Aoyagi, Adv. Mater., 2012, 24, 273–278 CrossRef CAS.
- S. A. Turner, J. Zhou, S. S. Sheiko and V. S. Ashby, ACS Appl. Mater. Interfaces, 2014, 6, 8017–8021 CrossRef CAS.
- K. Uto, T. Aoyagi, C. A. DeForest, A. S. Hoffman and M. Ebara, Adv. Healthcare Mater., 2017, 6, 1601439 CrossRef.
- S. J. P. Callens, D. Fan, I. A. J. van Hengel, M. Minneboo, P. J. Díaz-Payno, M. M. Stevens, L. E. Fratila-Apachitei and A. A. Zadpoor, Nat. Commun., 2023, 14, 855 CrossRef CAS.
- S. W. Lee, K. S. Phillips, H. Gu, M. Kazemzadeh-Narbat and D. Ren, Biomaterials, 2021, 268, 120595 CrossRef CAS.
- L. C. Hsu, J. Fang, D. A. Borca-Tasciuc, R. W. Worobo and C. I. Moraru, Appl. Environ. Microbiol., 2013, 79, 2703–2712 CrossRef CAS.
- A. Roy and K. Chatterjee, Nanoscale, 2021, 13, 647–658 RSC.
- T. E. Brown and K. S. Anseth, Chem. Soc. Rev., 2017, 46, 6532–6552 RSC.
- M. Guvendiren and J. A. Burdick, Nat. Commun., 2012, 3, 792 CrossRef.
- R. S. Stowers, S. C. Allen and L. J. Suggs, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 1953–1958 CrossRef CAS.
- A. M. Rosales, S. L. Vega, F. W. DelRio, J. A. Burdick and K. S. Anseth, Angew. Chem., Int. Ed., 2017, 56, 12132–12136 CrossRef CAS.
- C. A. DeForest and K. S. Anseth, Nat. Chem., 2011, 3, 925–931 CrossRef CAS.
- J. A. Burdick and W. L. Murphy, Nat. Commun., 2012, 3, 1269 CrossRef.
- S. Khetan, J. S. Katz and J. A. Burdick, Soft Matter, 2009, 5, 1601–1606 RSC.
- F. M. Yavitt, B. E. Kirkpatrick, M. R. Blatchley and K. S. Anseth, ACS Biomater. Sci. Eng., 2022, 8, 4634–4638 CrossRef CAS.
- X. Liu, S. Rao, W. Chen, K. Felix, J. Ni, A. Sahasrabudhe, S. Lin, Q. Wang, Y. Liu, Z. He, J. Xu, S. Huang, E. Hong, T. Yau, P. Anikeeva and X. Zhao, Nat. Methods, 2023, 1–8 Search PubMed.
- N. Li, Y. Li, Z. Cheng, Y. Liu, Y. Dai, S. Kang, S. Li, N. Shan, S. Wai, A. Ziaja, Y. Wang, J. Strzalka, W. Liu, C. Zhang, X. Gu, J. A. Hubbell, B. Tian and S. Wang, Science, 2023, 381, 686–693 CrossRef CAS.
- C. M. Tringides, N. Vachicouras, I. de Lázaro, H. Wang, A. Trouillet, B. R. Seo, A. Elosegui-Artola, F. Fallegger, Y. Shin, C. Casiraghi, K. Kostarelos, S. P. Lacour and D. J. Mooney, Nat. Nanotechnol., 2021, 16, 1019–1029 CrossRef CAS.
- M. Cianchetti, C. Laschi, A. Menciassi and P. Dario, Nat. Rev. Mater., 2018, 3, 143–153 CrossRef.
- S. Pittaccio, S. Viscuso, M. Rossini, L. Magoni, S. Pirovano, E. Villa, S. Besseghini and F. Molteni, J. Mater. Eng. Perform., 2009, 18, 824–830 CrossRef CAS.
- S. Pittaccio and S. Viscuso, J. Mater. Eng. Perform., 2011, 20, 666–670 CrossRef CAS.
- A. Lendlein, M. Behl, B. Hiebl and C. Wischke, Expert Rev. Med. Devices, 2010, 7, 357–379 CrossRef CAS.
- C. M. Yakacki, R. Shandas, C. Lanning, B. Rech, A. Eckstein and K. Gall, Biomaterials, 2007, 28, 2255–2263 CrossRef CAS PubMed.
- H. Holman, M. N. Kavarana and T. K. Rajab, Artif. Organs, 2021, 45, 454–463 CrossRef CAS.
- G. M. Baer, T. S. Wilson, W. Small IV, J. Hartman, W. J. Benett, D. L. Matthews and D. J. Maitland, J. Biomed. Mater. Res., Part B, 2009, 90B, 421–429 CrossRef CAS.
- F. Zhang, N. Wen, L. Wang, Y. Bai and J. Leng, Int. J. Smart Nano Mater., 2021, 12, 375–389 CrossRef.
- W. Zhao, Z. Huang, L. Liu, W. Wang, J. Leng and Y. Liu, Compos. Sci. Technol., 2022, 229, 109671 CrossRef CAS.
- B. V. M. Dasari, D. Neely, A. Kennedy, G. Spence, P. Rice, E. Mackle and E. Epanomeritakis, Ann. Surg., 2014, 259, 852 CrossRef.
- P. Sharma, R. Kozarek and the P. P. C. of the A. C. of Gastroenterology, Off. J. Am. Coll. Gastroenterol. ACG, 2010, 105, 258 CrossRef.
- S. Choudhury, S. Asthana, S. Homer-Vanniasinkam and K. Chatterjee, Biomater. Sci., 2022, 10, 3716–3729 RSC.
- G. Song, H. Q. Zhao, Q. Liu and Z. Fan, Bioact. Mater., 2022, 17, 488–495 Search PubMed.
- A. C. Moss, E. Morris and P. MacMathuna, Cochrane Database Syst. Rev., 2006, 2006(2), CD004200 Search PubMed.
- D. Lange, S. Bidnur, N. Hoag and B. H. Chew, Nat. Rev. Urol., 2015, 12, 17–25 CrossRef.
- A. Mosayyebi, C. Manes, D. Carugo and B. K. Somani, Curr. Urol. Rep., 2018, 19, 35 CrossRef PubMed.
- G. Jordan, US Pat., US8372138B2, 2013.
- C. de Marco, C. C. J. Alcântara, S. Kim, F. Briatico, A. Kadioglu, G. de Bernardis, X. Chen, C. Marano, B. J. Nelson and S. Pané, Adv. Mater. Technol., 2019, 4, 1900332 CrossRef CAS.
- L. De Nardo, R. Alberti, A. Cigada, L. Yahia, M. C. Tanzi and S. Farè, Acta Biomater., 2009, 5, 1508–1518 CrossRef CAS PubMed.
- A. Metcalfe, A.-C. Desfaits, I. Salazkin, L. Yahia, W. M. Sokolowski and J. Raymond, Biomaterials, 2003, 24, 491–497 CrossRef CAS PubMed.
- W. Small, T. S. Wilson, P. R. Buckley, W. J. Benett, J. M. Loge, J. Hartman and D. J. Maitland, IEEE Trans. Biomed. Eng., 2007, 54, 1657–1666 Search PubMed.
- W. S. Iv, T. S. Wilson, W. J. Benett, J. M. Loge and D. J. Maitland, Opt. Express, 2005, 13, 8204–8213 CrossRef PubMed.
- T. Feldman and W. Wang, US Pat., US6550480B2, 2003 Search PubMed.
- M. Bettuchi and R. Heinrich, US Pat., US20090118747A1, 2009.
- D. J. Maitland, A. P. Lee, D. L. Schumann and L. D. Silva, Shape memory polymer (SMP) gripper with a release sensing system, Lawrence Livermore National Laboratory (LLNL), Livermore, CA (United States), 2000 Search PubMed.
- J. A. Teague, US Pat., US20070299456A1, 2007.
- A. Lendlein and R. Langer, US Pat., US20060142794A1, 2006 Search PubMed.
- Y. C. Jung and J. W. Cho, J. Mater. Sci.: Mater. Med., 2010, 21, 2881–2886 CrossRef CAS.
- N. Tanjeem, M. B. Minnis, R. C. Hayward and C. W. Shields IV, Adv. Mater., 2022, 34, 2105758 CrossRef CAS PubMed.
- E. Gultepe, J. S. Randhawa, S. Kadam, S. Yamanaka, F. M. Selaru, E. J. Shin, A. N. Kalloo and D. H. Gracias, Adv. Mater., 2013, 25, 514–519 CrossRef CAS PubMed.
- S. Tasoglu, E. Diller, S. Guven, M. Sitti and U. Demirci, Nat. Commun., 2014, 5, 3124 CrossRef CAS PubMed.
- B. Özkale, R. Parreira, A. Bekdemir, L. Pancaldi, E. Özelçi, C. Amadio, M. Kaynak, F. Stellacci, D. J. Mooney and M. S. Sakar, Lab Chip, 2019, 19, 778–788 RSC.
- M. Sun, C. Tian, L. Mao, X. Meng, X. Shen, B. Hao, X. Wang, H. Xie and L. Zhang, Adv. Funct. Mater., 2022, 32, 2112508 CrossRef CAS.
- B. Sun, R. Jia, H. Yang, X. Chen, K. Tan, Q. Deng and J. Tang, Adv. Intell. Syst., 2022, 4, 2100139 CrossRef.
- H. Wei, Q. Zhang, Y. Yao, L. Liu, Y. Liu and J. Leng, ACS Appl. Mater. Interfaces, 2017, 9, 876–883 CrossRef CAS PubMed.
- X. Sun, L. Yue, L. Yu, H. Shao, X. Peng, K. Zhou, F. Demoly, R. Zhao and H. J. Qi, Adv. Funct. Mater., 2022, 32, 2109805 CrossRef CAS.
- J.-W. Su, D. Li, Y. Xie, T. Zhou, W. Gao, H. Deng, M. Xin and J. Lin, Smart Mater. Struct., 2020, 30, 015028 CrossRef.
Footnote |
| † Current address: Department of Chemical and Biological Engineering, University of Colorado Boulder, Boulder, CO 80303, USA. |
| This journal is © The Royal Society of Chemistry 2024 |
