Embedded bioprinted multicellular spheroids modeling pancreatic cancer bioarchitecture towards advanced drug therapy
Xiaoyun
Wei†
 *ab,
Yiwen
Wu†
a,
Keke
Chen
*ab,
Yiwen
Wu†
a,
Keke
Chen
 ab,
Ling
Wang
*ab and
Mingen
Xu
*ab
ab,
Ling
Wang
*ab and
Mingen
Xu
*ab
aSchool of Automation, Hangzhou Dianzi University, Hangzhou 310018, China. E-mail: wxyun@hdu.edu.cn; lingw@hdu.edu.cn; xumingen@hdu.edu.cn
bKey Laboratory of Medical Information and 3D Bioprinting of Zhejiang Province, Hangzhou Dianzi University, Hangzhou 310018, China
First published on 10th January 2024
Abstract
The desmoplastic bioarchitecture and microenvironment caused by fibroblasts have been confirmed to be closely related to the drug response behavior of pancreatic ductal adenocarcinoma (PDAC). Despite the extensive progress in developing PDAC models as in vitro drug screening platforms, developing efficient and controllable approaches for the construction of physiologically relevant models remains challenging. In the current study, multicellular spheroid models that emulate pancreatic cancer bioarchitecture and the desmoplastic microenvironment are bioengineered. An extrusion-based embedded dot bioprinting strategy was established to fabricate PDAC spheroids in a one-step process. Cell-laden hydrogel beads were directly deposited into a methacrylated gelatin (GelMA) suspension bath to generate spherical multicellular aggregates (SMAs), which further progressed into dense spheroids through in situ self assembly. By modulating the printing parameters, SMAs, even from multiple cell components, could be manipulated with tunable size and flexible location, achieving tunable spheroid patterns within the hydrogel bath with reproducible morphological features. To demonstrate the feasibility of this printing strategy, we fabricated desmoplastic PDAC spheroids by printing SMAs consisting of tumor cells and fibroblasts within the GelMA matrix bath. The produced hybrid spheroids were further exposed to different concentrations of the drug gemcitabine to verify their potential for use in cell therapy. Beyond providing a robust and facile bioprinting system that enables desmoplastic PDAC bioarchitecture bioengineering, this work introduces an approach for the scalable, flexible and rapid fabrication of cell spheroids or multi-cell-type spheroid patterns as platforms for advanced drug therapy or disease mechanism exploration.
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC), which accounts for 80% of clinical pancreatic cancers, has typically been regarded as the “king of cancer” with a high mortality rate.1,2 Studies have shown that the difficult treatment, low cure rate and poor prognosis of PDAC are closely related to its unique tumor microenvironment (TME), which composed of malignant pancreatic cells, masses of stromal cells, infiltrating immune cells, and the extracellular matrix (ECM).3–5 In particular, stromal cells are abundant and form a unique fibrotic barrier structure that envelops tumor parenchyma, providing a niche for modulating more tumor behaviors, such as tumor growth, invasion and therapeutic drug response.6,7 For this reason, various studies have been devoted to developing in vitro PDAC models that fully replicate the in vivo-like features for the exploration of various diseases and evaluation of therapeutic strategies.8,9Three-dimensional (3D) spheroids, which are formed from aggregates of single cells, represent a step forward in progress in tissue engineering applications for better simulating cellular structures and microenvironments in vivo.10 When compared to two-dimensional (2D) cultured cells, cells in 3D spheroids exhibit improved functions as a result of tight cell–cell and cell–ECM interactions.11 Therefore, 3D tumor spheroids have been pursued and recognized as an optimal model for drug testing.12 Recently, hybrid PDAC spheroids comprising cancer cells and fibroblasts were prepared utilizing ultra-low adhesion plates to reproduce the features of cancer–stroma bioarchitectures.13 Further, in order to include ECM-like features, tumor–stroma hydrogel-based models were developed in a recent approach.14 Researchers utilized superhydrophobic surfaces to produce a core–shell hydrogel bead structure that contained tumor and stroma components, respectively, and demonstrated higher drug resistance compared to monotypic or ECM-free tumor spheroids. Beyond these, 3D bioprinting techniques, such as inkjet bioprinting,15 extrusion bioprinting16 and light-assisted bioprinting,17,18 have emerged as advanced biomanufacturing technologies that allow cell-laden hydrogels to be positioned into specific structures in a programmed manner, and have been widely applied for the construction of 3D tumor models.19–21 A laser-assisted bioprinting process was adopted to generate pancreas spheroid models for studying cancer initiation.22 Additionally, our previous work reported a heterogeneous PDAC microtissue that was fabricated by printing cell-laden hydrogel beads for use as a drug screening platform.23 The drawback of the above-mentioned printed structures is that the cells, which have a scattered distribution within the hydrogel, normally require a long culture period to form cell–cell junctions or eventually spheroidal structures.
In a different approach, embedded bioprinting is an emerging 3D bioprinting strategy that produces complex tissue structures directly inside a reservoir filled with a supporting matrix.24–26 In particular, extrusion-based embedded bioprinting has been utilized to generate tumor spheroid patterns through cell aggregates printed into the support bath. For example, the Kang group presented a dot printing modality to print cell-laden sacrificial biomaterials into a supporting base to fabricate discrete pores after the sacrificial biomaterials were removed; thus, the cells aggregated to form spheroids in situ, which were demonstrated to generate and pattern cell spheroids.27 A more recent work reported that bioinks with high cell density could be directly printed within a supporting hydrogel matrix for mass production of cellular aggregates.28–31 Nevertheless, in these current embedded bioprinting processes, the reported supporting hydrogel baths often depend on soft alginate, collagen or sacrificial pluronic F-127, and limitations such as the lack of tunability of the mechanical properties or undesirable crosslinking rate of the supporting matrix need to be overcome.
In this study, a new embedded dot bioprinting system is developed. The system consists of cell-laden low-concentration type I collagen (5 mg mL−1) with sacrificial gelatin, which is used to prepare a bioink for printing, and a methacrylated gelatin (GelMA) hydrogel, which is used as the support matrix. The developed system was expanded to the application of bioengineering PDAC spheroid patterns. Following this, spherical multicellular aggregates (SMAs) were generated within the GelMA hydrogel bath through a one-step embedded dot printing process, and in situ spheroid formation was easily achieved when the sacrificial gelatin was liquefied, simultaneous accompanied by the promotion of cell assembly process. The excellent properties of GelMA, including biocompatibility, temperature-sensitivity and photo-crosslinkability, make it a robust supporting matrix for embedded bioprinting.32–34 Precise control of the spheroid size and positioning accuracy were evaluated by adjusting the printing parameters. To show the effect of the printing system, a bioink with encapsulated pancreatic cancer cells and fibroblasts was utilized to print SMAs within a GelMA bath. The morphological and phenotypic features of the formed PDAC spheroids were evaluated, as well as the effects of the desmoplastic structure on drug responses using monotypic spheroids and desmoplastic spheroids. Altogether, the new printing process facilitates spheroid patterning, thus providing versatile platforms for diverse biological applications.
2. Materials and methods
2.1. Cell culture
PANC-1, a pancreatic cancer cell line, was purchased from Beijing Beina Chuanglian Biotechnology Institute, China. Normal fibroblast cells from human dermal tissues were kindly provided by Hangzhou Regenovo Biotechnology Co., Ltd. PANC-1 cells were cultured in RPMI-1640 (Wisent, Canada), and fibroblast cells were cultured in high-glucose Dulbecco's modified eagle medium (DMEM, Wisent, Canada). Each basal medium was supplemented with 10% (v/v) fetal bovine serum (FBS, Excell Bio, China) and 1% (v/v) penicillin–streptomycin (Thermo Fisher, USA). Cells were cultured using a T75 cell culture flask and maintained at 37 °C in an incubator with 5% CO2, and split when around 80% confluence was reached.2.2. Preparation of GelMA hydrogel
Different concentrations of GelMA solution, including 5, 8 and 10% (w/v) were prepared by dissolving the corresponding masses of GelMA prepolymer (Regenovo Biotechnology) using the basal medium as the solvent in a 47 °C water bath. For photocuring, corresponding masses of the photoinitiator lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP, Sigma Aldrich, USA) were added and dissolved to reach a concentration of 0.5% (w/v). Afterwards, the prepared GelMA solution was fully sterilized by passing it through a 0.22 μm filter, and then stored at 4 °C for future use.2.3. Rheological evaluation
The rheological properties of GelMA solutions with concentrations of 5, 8 and 10% (w/v) were assessed using a rheometer (Anton Paar, MCR 52) with a 50-mm diameter cone plate geometry as well as a gap size of 104 μm. The shear viscosity at a ramped shear rate from 0.1 to 100 s−1 was measured to evaluate shear-thinning behavior, and the self-recovery property was evaluated through measurement of the storage modulus (G′) and loss modulus (G′′) by alternating the shear strain at 1% and 300% (1 Hz, 100 s) for three cycles.2.4. Preparation of bioink
A mixture of gelatin and collagen solution was prepared and mixed with cells to prepare the bioink for printing. Briefly, an initial concentration of 12% (w/v) gelatin solution was first prepared by dissolving the corresponding mass of gelatin power (Sigma, USA) at 50 °C using basal medium as a solvent, followed by sterilization through a 0.22-μm filter. Afterwards, 600 μL of precursor type I collagen solution (10 mg mL−1, Hangzhou Xiehe Medical Supplies Company), 500 μL of gelatin solution (12% concentration) and 100 μL of HEPES buffer solution (1 M, Thermo Fisher, USA) were quickly mixed to obtain a neutral mixture, in which the final concentrations of type I collagen and gelatin solution were 5 mg mL−1 and 5%, respectively. Finally, the mixed solution was added to the appropriate cell pellets to prepare the bioink for printing.2.5. Embedded dot bioprinting process
A 3D bioprinter (Bio Architect WS) from Regenovo Biotechnology was adopted for bioprinting during the experiment. The bioprinter has performance that allows four kinds of printheads to be simultaneously loaded, as well as an X–Y–Z moving part to ensure flexible mobility. Moreover, a dual-temperature control unit is contained to separately control the temperature of the receiving platform and the printhead. Before printing, the pre-prepared GelMA solution was dispensed according to 300 μL per chamber of the 12-transwell plate, which was subsequently placed on the receiving platform with the temperature set at below 20 °C to convert the GelMA solution into a semi-gelled state to serve as the support bath according to our previous study.35 The bioink was then transferred into a 5-mL syringe and loaded into an extrusion printhead whose temperature was set around 18 °C. The automated printing process began under the pre-designed G-code control, which enables the printhead to work in an intermittent pressure-driven mode, allowing the collagen/gelatin blended hydrogel to be dispensed from a needle with an inner nozzle diameter of 210 μm for a short duration within the GelMA suspension bath. During the printing process, the pneumatic pressure was maintained at 100 kPa, and the speed of the printhead was fixed at 15 mm s−1.2.6. Establishing PDAC spheroids
When the cultured PANC-1 cells reached 80–90% confluency, the cells were dissociated into single cells using 0.05% trypsin-EDTA (Thermo Fisher, USA). The cell suspension was collected and centrifuged, and the cell pellets were encapsulated in 1.2 mL of collagen/gelatin solution to prepare the bioink with a cell density of around 1 × 107 cells per mL for printing. A 4 × 5 array of cell aggregates was printed under the G-code command. Dispensing times of 100, 300 and 500 ms were utilized to obtain SMAs of different sizes. After the printing process finished, the GelMA supporting bath was exposed to 405-nm blue light (200 mW cm−2) for 30 s to achieve photocrosslinking, and then immersed into complete culture medium, with media changes every two days.2.7. Establishing desmoplastic PDAC spheroids
To generate PDAC spheroids with desmoplastic features, PANC-1 cells and fibroblast cells were mixed with the prepared collagen/gelatin solution and prepared into a bioink with cell densities of 1 × 107 cells per mL and 5 × 106 cells per mL, respectively. A dispensing time of 300 ms was adopted to ensure that the printed SMAs were around 500 μm in diameter. Then, the heterogeneous SMAs were subjected to photocuring and put into a coculturing process.A cell plasma membrane staining kit (Beyotime, China) was used to prestain the PANC-1 and fibroblast cells in order to track the cells. Specifically, DiI and DiO were diluted with culture medium at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 to prepare the working solution. After cell passage, the PANC-1 and fibroblast cells were treated with DiI and DiO working solution, respectively, and kept at 37 °C for 20 min. Cells were fully washed with culture medium before being mixed with the printed hydrogel.
1000 to prepare the working solution. After cell passage, the PANC-1 and fibroblast cells were treated with DiI and DiO working solution, respectively, and kept at 37 °C for 20 min. Cells were fully washed with culture medium before being mixed with the printed hydrogel.
2.8. Tracking spheroids proliferation and viability
To characterize the cell proliferation in the spheroids over the five day culture period, F-actin staining was performed on the PDAC spheroids on days 1, 3, and 5 of culturing. Briefly, cell samples were washed using phosphate buffered saline (PBS, Thermo Fisher, USA), and then 4% (v/v) paraformaldehyde (Beyotime, China) was used to fix the cells for 2 h at room temperature. Afterwards, the cells were permeabilized on ice with 0.5% (v/v) Triton X-100 (Beyotime, China) for 30 minutes and blocked with 3% (v/v) bovine serum albumin (BSA, Sigma-Aldrich, USA) for 45 minutes. Alexa Fluor 488 phalloidin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Invitrogen, USA) was then added to incubate the cells for 4 h at room temperature, followed by cell nucleus staining with DAPI solution (10 μg mL−1, Beyotime, China). Cells were observed using a confocal microscope, and the fluorescence area of the spheroids was measured using ImageJ software.
200, Invitrogen, USA) was then added to incubate the cells for 4 h at room temperature, followed by cell nucleus staining with DAPI solution (10 μg mL−1, Beyotime, China). Cells were observed using a confocal microscope, and the fluorescence area of the spheroids was measured using ImageJ software.
Cell viability was assessed using a live/dead viability kit (Thermo Fisher, USA) according to the manufacturer's protocol. Spheroids were stained and analysed on days 1, 3 and 5 of culturing. After 20 minutes of incubation with the live/dead staining solution at 37 °C, the spheroids were washed with PBS and then observed using a confocal microscope (Nikon, A1RHD25, Japan). Live and dead cells were indicated by green and red fluorescence signals, respectively, and the fluorescence images were then processed with ImageJ software and converted to greyscale for area quantification. Cell viability was determined by the ratio of green area to total area.
2.9. Immunofluorescence staining
For the detection of specific proteins, the generated PDAC spheroids containing tumor cells and fibroblasts were fixed with 4% (v/v) paraformaldehyde for 2 h, and then treated with 0.5% (v/v) Triton X-100 for 30 min, followed by blocking with 3% (v/v) BSA for 45 min at room temperature. Then, the spheroids were incubated with the primary antibodies against α-SMA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, Invitrogen, USA) to stain the fibroblasts and cytokeratin 19 (CK19, 1
100, Invitrogen, USA) to stain the fibroblasts and cytokeratin 19 (CK19, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, Invitrogen, USA) to stain tumor cells at 4 °C overnight. The next day, the spheroids were immersed in the secondary antibody solution including Alexa Flour 594 IgG and 488 IgG (1
50, Invitrogen, USA) to stain tumor cells at 4 °C overnight. The next day, the spheroids were immersed in the secondary antibody solution including Alexa Flour 594 IgG and 488 IgG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Invitrogen, USA) for 4 h at room temperature. Finally, DAPI solution (10 μg mL−1) was added for cell nucleus staining and incubated for 10 min. The fluorescence images were captured using a confocal microscope.
200, Invitrogen, USA) for 4 h at room temperature. Finally, DAPI solution (10 μg mL−1) was added for cell nucleus staining and incubated for 10 min. The fluorescence images were captured using a confocal microscope.
2.10 Drug treatment
PDAC mono-spheroids and desmoplastic spheroids were cultured for 5 days before drug treatment. Gemcitabine stock solution (10 mM, MedChemExpress, USA) was diluted in the culture medium and prepared into working fluids at different concentrations of 10, 25, 50, 75 and 100 μM mL−1. Mono-spheroids and desmoplastic spheroids were cultured in the working fluids, as well as in the culture medium containing 0.1% DMSO to serve as control groups. After drug administration and incubation for 48 h, the spheroids were washed with PBS and then subjected to cell viability analysis.2.11. Statistical analysis
Data statistical analysis was conducted using GraphPad Prism 8 software, and reported as mean and standard deviation. Two-way analysis of variance (ANOVA) was generally used to evaluate significant differences between two conditions, and a p value under 0.05 was considered statistically significant.3. Results and discussion
3.1. GelMA suspension bath optimization for embedded dot bioprinting
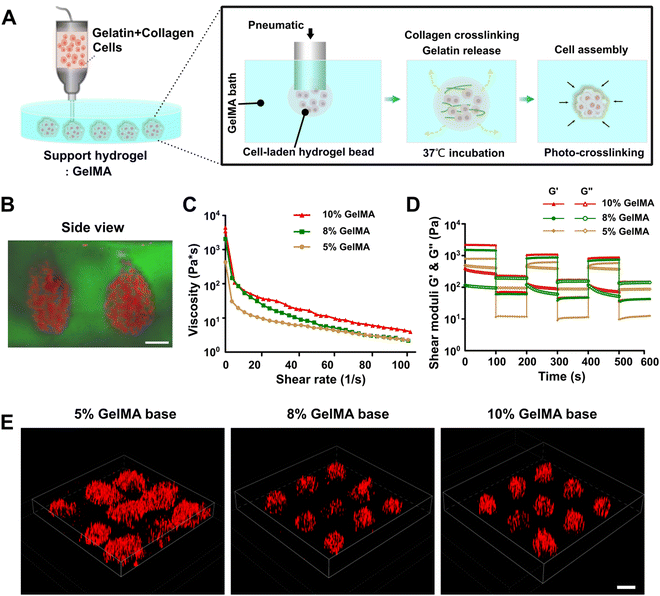
Fig. 1A presents a schematic diagram of printing SMAs utilizing the embedded dot printing system. Soft type I collagen was diluted with thermosensitive gelatin hydrogel and further mixed with cells to prepare the main bioink for printing. Collagen I was utilized to encapsulate cells based on the fact that over 90% of the PDAC ECM is made up of abundant collagen components.36 Additionally, the gelatin hydrogel was introduced here mainly as an auxiliary phase to promote the extrusion printing of low-concentration collagen solutions, because low-concentration collagen solutions are commonly mechanically weak and have low viscosity, which makes the extrusion printing of a single collagen component very challenging.37,38 On the other hand, the thermosensitive gelatin served as a sacrificial phase owing to its ability to liquefy at 37 °C post-printing. In the incubator, the sacrificial gelatin melted while the collagen gradually crosslinked, which synergistically facilitated the process of cell assembly within the GelMA bath. Additionally, GelMA, a modified photocrosslinkable hydrogel derived from gelatin, was used as the suspension bath, both because of its desirable rheological behavior for embedded printing and rapid crosslinking rate to keep the fidelity of the structures and its excellent ECM-mimicking property to support cell growth.26The shear-thinning property and self-healing ability of the suspension hydrogel have been proven to be essential for embedded bioprinting process.39–41 We thus first evaluated the rheological behavior of 5, 8 and 10% GelMA solutions. All the GelMA suspension solutions displayed the requisite shear-thinning in the established shear rate range (Fig. 1C), which facilitated the deposition of bioink during the embedded printing process. The self-healing property of the suspension bath was then tested, as the ability of rapid recovery to the solid state for the bath material is essential to trap the extruded bioink. In our results, the 8 and 10% GelMA solutions exhibited good self-healing ability when the shear strain was increased or decreased; however, although similar recovery phenomena was observed for the 5% GelMA solution, certain points of the storage and loss modulus overlapped, which indicated that it was unsuitable for use as the support bath (Fig. 1D).
On this basis, an embedded dot printing process was applied for positioning the SMAs within the 5, 8 and 10% GelMA suspension baths. For visualization, the cells were pre-stained with DiI (red signal) before printing. The resultant images demonstrated that the printed SMAs were well retained in the GelMA suspension baths with concentrations of 8% or higher, while they were distorted in 5% GelMA (Fig. 1E). The side-view image of the printed SMAs within the GelMA suspension bath visually demonstrated the performance of the embedded dot printing strategy to generate 3D spherical cellular aggregates in an efficient manner (Fig. 1B). Notably, the printed SMAs maintained a good morphology within higher concentrations of GelMA (>5%), which indicated that increasing the GelMA concentration could be beneficial through providing greater mechanical support to the printed SMAs. In addition, during the embedded dot printing process, the uncrosslinked GelMA suspension had insufficient strength to support the deposited bioink. We thus kept the GelMA suspension bath below 20 °C to obtain more viscoplastic conditions before printing, due to its temperature responsiveness and variation in its sol–gel transition properties with temperature, which have been previously reported in many works.42,43
3.2. SMA patterning using embedded dot bioprinting
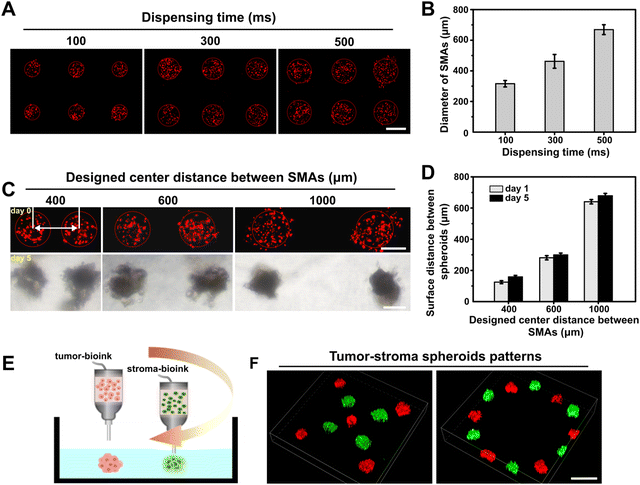
Having demonstrated that we could produce 3D SMAs in the GelMA bath material in one step, we further explored the performance of the proposed embedded dot bioprinting system for precisely controllable patterning of SMAs by specifying the printing parameters. We first attempted to print SMAs with tunable sizes by altering the dispensing time at a fixed pressure (100 kPa), and various dispensing times (from 100 to 500 ms) were adopted. The printed SMAs were imaged and quantified, and diameters ranging from approximately 300 to 700 μm were easily obtained (Fig. 2A and B). In general, the sizes of the SMAs were positively correlated with the dispensing time, because a longer dispensing time allows for a larger amount of bioink to be extruded into the bath material. To further evaluate the patterning ability of the SMAs at different distances, we pre-set the center-to-center distances between each printed dot to 400, 600 or 1000 μm in the G-code. After printing, the patterned SMAs were subsequently cultured for 5 days for the in situ formation of spheroids. The obtained results showed that the location of the cell SMAs was precisely controlled on a micrometer scale and was well maintained at the printed site throughout culturing, with a minimum surface-to-surface distance between spheroids of less than 200 μm. Additionally, after 5 days of culturing, the measured center-to-center distances between spheroids was found to be slightly larger, implying a transformation of the SMAs into a densely packed structure during the process of cultivation (Fig. 2C and D).Embedded dot bioprinting of different types of SMAs was also conducted to determine the ability to spatially arrange multiple types of spheroids within the bath hydrogels. In this case, tumor bioink and stroma bioink were prepared and loaded in two printheads for printing. The tumor-stoma SMAs could be precisely arranged into designed patterns (Fig. 2E). As reported, the reciprocal interactions between cells inside hydrogels mainly depend on direct contact or the paracrine effect, which are both affected by cell–cell distance.44,45 Especially for paracrine signaling, the effective distance for the exchange of cell-secreted signaling molecules has been reported to be approximately 250 μm.46 Thus, our printing system provides an efficient manner for on-demand patterning of multiple types of spheroids at the desired distance, which is expected to promote the exploration of complex cell interactions.
3.3. Bioprinting of PDAC tumor spheroids
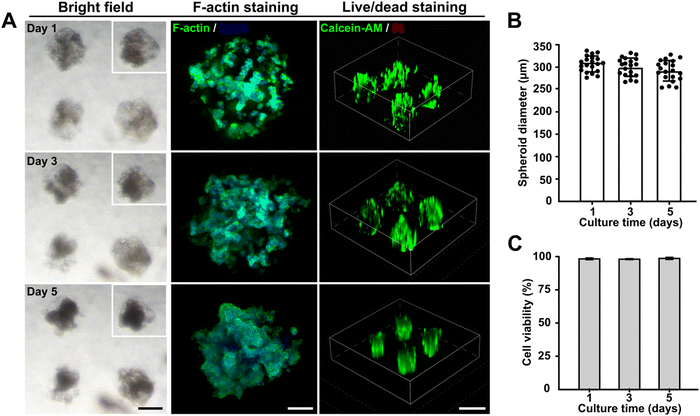
To determine the feasibility of this embedded dot bioprinting system for high-efficiency cell spheroid bioengineering, a collagen/gelatin hydrogel with encapsulated pancreatic cancer cells (1 × 107 cells per mL) was utilized to print SMAs within an 8% GelMA supporting bath. During the printing process, the printing parameters of pneumatic pressure and dispensing time were fixed at 100 kPa and 100 ms, respectively, which allowed the SMAs to be printed at around 300 μm scale. The printed SMAs were cultured for 5 days to evaluate the effects of the embedded bioprinting system on cell behavior. Bright images were observed on days 1, 3, and 5, and F-actin staining was conducted to capture the morphological features of the in situ formed spheroids. Markedly, spheroids were formed after 1 day of culturing and gradually compacted with a slight reduction in their diameter as the number of days of culturing increased. The F-actin and nuclear staining further confirmed that tight cell–cell junctions were achieved with longer culturing (Fig. 3A and B). Cell viability was detected using a standard live/dead kit, and fluorescence images were captured. Calcein-AM and propidium iodide (PI) markers indicated that no necrotic cores were present within spheroids cultured for 5 days, with the cells maintaining a survival rate of more than 95% (Fig. 3C).Thus, the newly developed embedded dot printing system demonstrated the ability to achieve one-step patterning of cell spheroids within a GelMA matrix and provided an highly biocompatible microenvironment to maintain high cell viabilities during the culturing period. Our approach introduced type I collagen and GelMA hydrogel as the final matrices to support cell growth. Collagen has been proven to be the most abundant in vivo ECM protein and is often overexpressed in the majority of solid tumors,47 and GelMA, as a gelatin derivative, possesses inherent biocompatibility.48 Therefore, the embedded dot printing system is broadly applicable for engineering multiple types of tissue models.
3.4. Bioprinting of desmoplastic PDAC spheroids modeling the PDAC bioarchitecture
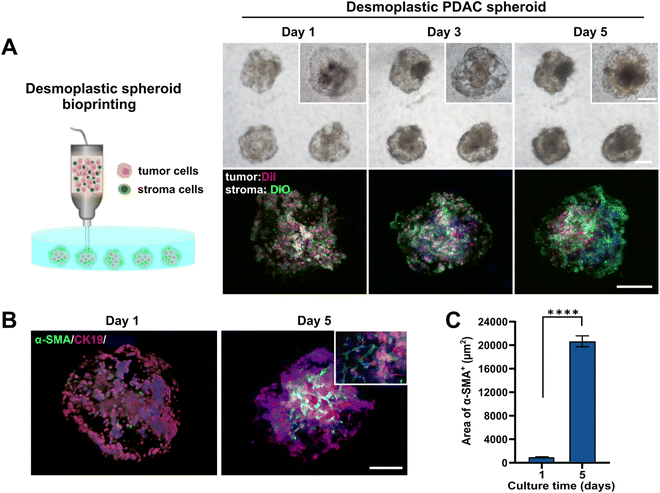
PDAC is usually characterized by a distinctive spatial cellular distribution in which pancreatic cancer cells are encapsulated by a dense fibrotic stroma composed of stromal cells, mainly CAFs, and abundant ECM.49,50 To achieve desmoplastic PDAC spheroid formation using our proposed printing process, here, we mixed 1 × 107 cells per mL of PANC-1 cells and 5 × 106 cells per mL of normal fibroblasts with collagen–gelatin hydrogel to prepare a bioink for patterning SMAs within the GelMA bath (Fig. 4A). To visualize the distribution of tumor and stroma components within the generated spheroids, we pre-labeled PANC-1 cells and fibroblasts using the cell trackers DiI (red signal) and DiO (green signal). During culturing, the morphological changes of the tumor cells and fibroblasts in the spheroids at different time points were observed using bright field and fluorescence microscopy. All cells were homogeneously dispersed throughout the spheroids at day 1. In particular, the fibroblasts initially displayed a round cell morphology within the compact aggregates, but over a period of days, they spread and connected, exhibiting a more fusiform-shaped conformation and eventually forming a dense fibroblastic network that wrapped around the tumor spheroids. The establishment of close cell–cell contacts between fibroblasts was also confirmed through the enhanced green fluorescence signal after 5 days of culturing.We further assessed the α-SMA expression, which is a common biomarker of CAFs,51,52 in the desmoplastic PDAC spheroids to explore whether normal fibroblasts were activated and obtained the CAF phenotype during co-culturing. Fig. 4B shows that on day 1 of culturing, there were fewer α-SMA+ cells in the PDAC spheroids, and the area of α-SMA+ cells significantly increased after 5 days of culturing. Interestingly, α-SMA positive cells were mainly observed in close proximity to the tumor core, beyond which there were relatively few signals of α-SMA+ fibroblasts at the edge of the tumor spheroids. To quantitatively evaluate the expression of α-SMA in the desmoplastic PDAC spheroids, we calculated the area of α-SMA-derived fluorescence signals as shown in Fig. 4C. The area of α-SMA positive cells increased significantly after 5 days of culturing, with an approximately 21.6-fold increase from the beginning (day 1). These observations demonstrated that PDAC spheroids modeling in vivo pancreatic cancer bioarchitecture and the desmoplastic microenvironment could be efficiently produced by the one-step embedded dot printing process.
3.5. Drug screening of bioprinted PDAC spheroids
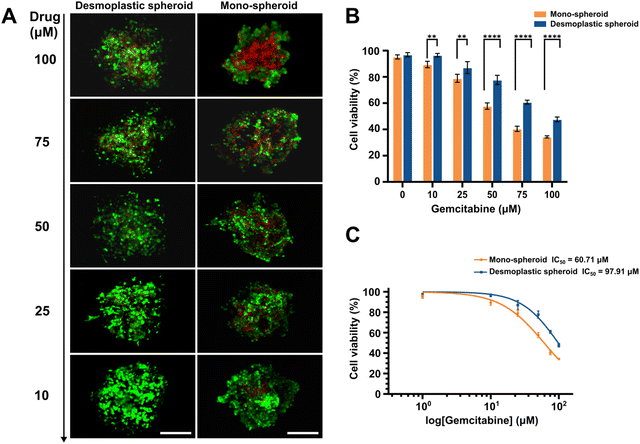
To further verify the applicability of the bioprinted PDAC spheroids as an in-vitro drug screening platform, desmoplastic spheroids and mono-spheroids were established and treated with different concentrations of gemcitabine, a therapeutic drug used in pancreatic cancer treatment,53,54 to verify the influence of the desmoplastic structure and microenvironment provided by activated fibroblasts on pancreatic carcinoma progression and drug resistance. For this, two kinds of PDAC spheroids were printed and cultured for 5 days to obtain a compact spherical morphology. Subsequently, the spheroids were assayed using 10, 25, 50, 75, and 100 μM mL−1 solutions of gemcitabine for 48 h, and a culture medium containing 0.1% DMSO was analyzed as a control. The results demonstrated that heterotypic PDAC spheroids with a desmoplastic component exhibited significant resistance compared to monotypic spheroids at high drug concentrations (over 50 μM). Cell viability was about 47.7% in the desmoplastic spheroids and only 34.3% in mono-spheroids at a high drug concentration of 100 μM (Fig. 5A and B), indicating the strong impact of the fibroblastic bioarchitecture on tumor resistance, as reported previously.55,56 The desmoplastic PDAC spheroids exhibited an IC50 value of around 97.9 μM, which was 1.6-fold higher than that of the monotypic spheroids (60.7 μM).4. Conclusions
In the current study, we presented an embedded dot printing strategy for the on-demand fabrication of multicellular SMAs that could progress into spheroids in a GelMA support bath. Normally, it is difficult to extrude low concentrations of collagen due to their undesirable shear-thinning and poor mechanical properties. Here, a 5-mg mL−1 collagen I solution was successfully printed through blending with a temperature-sensitive gelatin hydrogel, which simultaneously served as a rheology modifier to improve the printability and a sacrificial material to facilitate cell assembly with the suspension matrix. The easy size controllability and precise localization of the printed SMAs for one cell type or even two components were demonstrated, indicating the potential for the generation of spheroid patterns containing different cell types for intercellular crosstalk exploration. As an application example, desmoplastic PDAC spheroids were efficiently fabricated to better mimic the native tumor bioarchitecture and composition. In this process, the tumor cells and fibroblasts closely interacted within collagen and formed tight spheroids with high viabilities in the GelMA support bath. Importantly, the desmoplastic PDAC spheroids were exposed to a range of drug concentrations and exhibited enhanced drug resistance compared to mono-spheroids. Together, the universality and expansibility of the embedded dot printing system may open new paths for the bioengineering of more physiomimetic PDAC models for drug screening with more practical effect.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Funding for this work included the National Natural Science Foundation of China (Grant No. 82303978), the National Key Research and Development Program of China (Grant No. 2022YFA1104600), and the Zhejiang Provincial Natural Science Foundation of China (Grant No. LQ23H160011).References
- J. Kleeff, M. Korc, M. Apte, C. La Vecchia, C. D. Johnson, A. V. Biankin, R. E. Neale, M. Tempero, D. A. Tuveson, R. H. Hruban and J. P. Neoptolemos, Nat. Rev. Dis. Primers, 2016, 2, 16022 CrossRef PubMed.
- M. Orth, P. Metzger, S. Gerum, J. Mayerle, G. Schneider, C. Belka, M. Schnurr and K. Lauber, Radiat. Oncol., 2019, 14, 141 CrossRef PubMed.
- V. P. Balachandran, G. L. Beatty and S. K. Dougan, Gastroenterology, 2019, 156, 2056–2072 CrossRef CAS PubMed.
- M. Ligorio, S. Sil, J. Malagon-Lopez, L. T. Nieman, S. Misale, M. Di Pilato, R. Y. Ebright, M. N. Karabacak, A. S. Kulkarni, A. Liu, N. Vincent Jordan, J. W. Franses, J. Philipp, J. Kreuzer, N. Desai, K. S. Arora, M. Rajurkar, E. Horwitz, A. Neyaz, E. Tai, N. K. C. Magnus, K. D. Vo, C. N. Yashaswini, F. Marangoni, M. Boukhali, J. P. Fatherree, L. J. Damon, K. Xega, R. Desai, M. Choz, F. Bersani, A. Langenbucher, V. Thapar, R. Morris, U. F. Wellner, O. Schilling, M. S. Lawrence, A. S. Liss, M. N. Rivera, V. Deshpande, C. H. Benes, S. Maheswaran, D. A. Haber, C. Fernandez-Del-Castillo, C. R. Ferrone, W. Haas, M. J. Aryee and D. T. Ting, Cell, 2019, 178, 160–175 CrossRef CAS PubMed.
- M. Weniger, K. C. Honselmann and A. S. Liss, Cancers, 2018, 10, 316 CrossRef PubMed.
- W. J. Ho, E. M. Jaffee and L. Zheng, Nat. Rev. Clin. Oncol., 2020, 17, 527–540 CrossRef PubMed.
- A. N. Hosein, R. A. Brekken and A. Maitra, Nat. Rev. Gastroenterol. Hepatol., 2020, 17, 487–505 CrossRef.
- M. A. Heinrich, A. M. R. H. Mostafa, J. P. Morton, L. J. A. C. Hawinkels and J. Prakash, Adv. Drug Delivery Rev., 2021, 174, 265–293 CrossRef CAS PubMed.
- M. V. Monteiro, L. P. Ferreira, M. Rocha, V. M. Gaspar and J. F. Mano, Biomaterials, 2022, 287, 121653 CrossRef CAS PubMed.
- S.-j Kim, E. M. Kim, M. Yamamoto, H. Park and H. Shin, Adv. Healthcare Mater., 2020, 9, 2000608 CrossRef CAS PubMed.
- P. Zhuang, Y.-H. Chiang, M. S. Fernanda and M. He, Int. J. Bioprint., 2021, 7, 444 CrossRef CAS.
- A. Wang, L. A. Madden and V. N. Paunov, J. Mater. Chem. B, 2020, 8, 10487–10501 RSC.
- M. V. Monteiro, V. M. Gaspar, L. Mendes, I. F. Duarte and J. F. Mano, Small Methods, 2021, 5, 2001207 CrossRef CAS PubMed.
- M. V. Monteiro, M. Rocha, V. M. Gaspar and J. F. Mano, Adv. Healthcare Mater., 2022, 11, 2102574 CrossRef CAS PubMed.
- W. L. Ng, X. Huang, V. Shkolnikov, R. Suntornnond and W. Y. Yeong, Bio-Des. Manuf., 2023, 6, 676–690 CrossRef CAS.
- R. Chand, B. S. Muhire and S. Vijayavenkataraman, Int. J. Bioprint., 2022, 8, 545 CrossRef CAS PubMed.
- W. L. Ng, J. M. Lee, M. Zhou, Y.-W. Chen, K.-X. A. Lee, W. Y. Yeong and Y.-F. Shen, Biofabrication, 2020, 12, 022001 CrossRef CAS PubMed.
- W. Guo, B. Li, P. Li, L. Zhao, H. You and Y. Long, J. Mater. Chem. B, 2023, 11, 9572–9596 RSC.
- W. Peng, D. Unutmaz and I. T. Ozbolat, Trends Biotechnol., 2016, 34, 722–732 CrossRef CAS PubMed.
- X. Mi, Z. Su, X. Yue, Y. Ren, X. Yang, L. Qiang, W. Kong, Z. Ma, C. Zhang and J. Wang, Biomater. Sci., 2023, 11, 3813–3827 RSC.
- T. Agarwal, S. Y. Hann, I. Chiesa, H. Cui, N. Celikkin, S. Micalizzi, A. Barbetta, M. Costantini, T. Esworthy, L. G. Zhang, C. De Maria and T. K. Maiti, J. Mater. Chem. B, 2021, 9, 7608–7632 RSC.
- D. Hakobyan, C. Médina, N. Dusserre, M.-L. Stachowicz, C. Handschin, J.-C. Fricain, J. Guillermet-Guibert and H. Oliveira, Biofabrication, 2020, 12, 035001 CrossRef CAS PubMed.
- B. Huang, X. Wei, K. Chen, L. Wang and M. Xu, Int. J. Bioprint., 2023, 9, 676 CrossRef.
- X. Zeng, Z. Meng, J. He, M. Mao, X. Li, P. Chen, J. Fan and D. Li, Acta Biomater., 2022, 140, 1–22 CrossRef CAS.
- H. Budharaju, D. Sundaramurthi and S. Sethuraman, Bioact. Mater., 2024, 32, 356–384 CAS.
- Y. Fang, Y. Guo, B. Wu, Z. Liu, M. Ye, Y. Xu, M. Ji, L. Chen, B. Lu, K. Nie, Z. Wang, J. Luo, T. Zhang, W. Sun and Z. Xiong, Adv. Mater., 2023, 35, 2205082 CrossRef CAS.
- S. Jeon, J.-H. Heo, M. K. Kim, W. Jeong and H.-W. Kang, Adv. Funct. Mater., 2020, 30, 2005324 CrossRef CAS.
- Y. Park, S. T. Ji, U. Yong, S. Das, W. B. Jang, G. Ahn, S.-M. Kwon and J. Jang, Biofabrication, 2021, 13, 045017 CrossRef CAS PubMed.
- J. Han, S. Jeon, M. K. Kim, W. Jeong, J. J. Yoo and H. W. Kang, Biofabrication, 2022, 14, 034102 CrossRef PubMed.
- B. S. Kim, W. W. Cho, G. Gao, M. Ahn, J. Kim and D. W. Cho, Small Methods, 2021, 5, e2100072 CrossRef PubMed.
- J. A. Reid, P. A. Mollica, R. D. Bruno and P. C. Sachs, Breast Cancer Res., 2018, 20, 122 CrossRef PubMed.
- B. J. Klotz, D. Gawlitta, A. Rosenberg, J. Malda and F. P. W. Melchels, Trends Biotechnol., 2016, 34, 394–407 CrossRef CAS PubMed.
- A. G. Kurian, R. K. Singh, K. D. Patel, J. H. Lee and H. W. Kim, Bioact. Mater., 2022, 8, 267–295 CAS.
- K. Yue, G. Trujillo-de Santiago, M. M. Alvarez, A. Tamayol, N. Annabi and A. Khademhosseini, Biomaterials, 2015, 73, 254–271 CrossRef CAS PubMed.
- X. Wei, B. Huang, K. Chen, Z. Fan, L. Wang and M. Xu, Mater. Des., 2022, 223, 111152 CrossRef CAS.
- C. Tian, K. R. Clauser, D. Öhlund, S. Rickelt, Y. Huang, M. Gupta, D. R. Mani, S. A. Carr, D. A. Tuveson and R. O. Hynes, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 19609–19618 CrossRef CAS PubMed.
- D. S. Reynolds, I. de Lázaro, M. L. Blache, Y. Liu, N. C. Jeffreys, R. M. Doolittle, E. Grandidier, J. Olszewski, M. T. Dacus, D. J. Mooney and J. A. Lewis, Adv. Mater., 2023, 35, e2210748 CrossRef PubMed.
- W. Shi, S. Mirza, M. Kuss, B. Liu, A. Hartin, S. Wan, Y. Kong, B. Mohapatra, M. Krishnan, H. Band, V. Band and B. Duan, Adv. Healthcare Mater., 2023, 12, e2300905 CrossRef PubMed.
- K. Zhou, Y. Sun, J. Yang, H. Mao and Z. Gu, J. Mater. Chem. B, 2022, 10, 1897–1907 RSC.
- L. G. Brunel, S. M. Hull and S. C. Heilshorn, Biofabrication, 2022, 14, 032001 CrossRef PubMed.
- Q. Li, L. Ma, Z. Gao, J. Yin, P. Liu, H. Yang, L. Shen and H. Zhou, ACS Appl. Mater. Interfaces, 2022, 14, 41695–41711 CrossRef CAS PubMed.
- Z. Fan, X. Wei, K. Chen, L. Wang and M. Xu, Micromachines, 2023, 14, 878 CrossRef PubMed.
- A. T. Young, O. C. White and M. A. Daniele, Macromol. Biosci., 2020, 20, e2000183 CrossRef PubMed.
- S. H. Kim, J. Turnbull and S. Guimond, J. Endocrinol., 2011, 209, 139–151 CAS.
- P. R. Baraniak and T. C. McDevitt, Regener. Med., 2010, 5, 121–143 CrossRef PubMed.
- S. J. Kim, H. Byun, S. Lee, E. Kim, G. M. Lee, S. J. Huh, J. Joo and H. Shin, Acta Biomater., 2022, 142, 60–72 CrossRef CAS PubMed.
- T. R. Cox, Nat. Rev. Cancer, 2021, 21, 217–238 CrossRef CAS PubMed.
- Z. Chen, H. Xiao, H. Zhang, Q. Xin, H. Zhang, H. Liu, M. Wu, L. Zuo, J. Luo, Q. Guo, C. Ding, H. Tan and J. Li, J. Mater. Chem. B, 2021, 9, 8646–8658 RSC.
- K. P. Pednekar, M. A. Heinrich, J. van Baarlen and J. Prakash, Cancers, 2021, 13, 5006 CrossRef CAS PubMed.
- V. Brancato, V. Comunanza, G. Imparato, D. Corà, F. Urciuolo, A. Noghero, F. Bussolino and P. A. Netti, Acta Biomater., 2017, 49, 152–166 CrossRef CAS PubMed.
- V. Sgarminato, S. L. Marasso, M. Cocuzza, G. Scordo, A. Ballesio, G. Ciardelli and C. Tonda-Turo, Biomater. Sci., 2022, 11, 208–224 RSC.
- H. Takahashi and Y. Kikuchi, Biomater. Sci., 2021, 9, 4448–4458 RSC.
- Y. Binenbaum, S. Na'ara and Z. Gil, Drug Resistance Updates, 2015, 23, 55–68 CrossRef PubMed.
- J. Natu and G. P. Nagaraju, Cancer Lett., 2023, 573, 216382 CrossRef CAS PubMed.
- J. H. Lee, S. K. Kim, I. A. Khawar, S. Y. Jeong, S. Chung and H. J. Kuh, J. Exp. Clin. Cancer Res., 2018, 37, 4 CrossRef PubMed.
- T. Zhang, Y. Ren, P. Yang, J. Wang and H. Zhou, Cell Death Dis., 2022, 13, 897 CrossRef CAS.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |