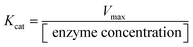Construction of a highly efficient DNA nanotube sensor with peroxide-like activity†
Ying
Zhang
ab,
Lingqi
Wu
ab,
Xin
Su
*b and
Hao
Liang
 *ab
*ab
aState Key Laboratory of Chemical Resource Engineering, Beijing University of Chemical Technology, Beijing 100029, P. R. China. E-mail: lianghao@mail.buct.edu.cn
bCollege of Life Science and Technology, Beijing University of Chemical Technology, Beijing 100029, P. R. China
First published on 12th December 2023
Abstract
The G-quadruplex/heme complexes are special DNA-based artificial metalloenzymes with peroxidase-like activity and are widely used in biosensing and biocatalysis. However, their peroxidase-like activity is not satisfactory. Due to the high programmability and good stability of DNA, DNA as a scaffold material is promising for enhancing the activity of artificial metalloenzymes. In this work, an effective DNA nanotube-based peroxidase was constructed using a self-assembly strategy. To improve the activity of G-quadruplex/heme complexes, a new method for the construction of G-quadruplex/heme complex arrays was proposed in a simple and inexpensive way. By designing the toes of DNA nanotubes as G-quadruplexes, G-quadruplex arrays could be formed on pure DNA nanotubes, and then the G-quadruplex arrays bind to heme to form a nanotube-supported DNAzyme termed as DNTzyme. Agarose gel electrophoresis, circular dichroism, and fluorescence microscopy were used to characterize DNTzyme. What is more, because the loading of DNAzyme on DNA nanotubes can increase their biological stability, a hydrogen peroxide detection sensor was constructed using the enhanced enzymatic activity and excellent stability of DNTzyme. The sensor could accurately and efficiently detect peroxide and show enhanced fluorescence with a detection limit of 49 nM for H2O2 and 1.4 μM for TBHP, and a color development time of about 5 min. This sensor is expected to have applications in bio-detection, biocatalysis, and drug delivery.
1. Introduction
Conventional catalyst synthesis methods require large amounts of hazardous and toxic reagents, as well as dangerous reaction conditions such as high temperatures and pressures. These conditions result in low reaction efficiency, exorbitant cost, and severe pollution problems. The idea of biomimetic catalysis has been proposed to solve these issues. Biomimetic catalysis, which synthesizes artificial catalysts to achieve environmentally friendly, efficient, cost-effective, and highly selective reactions, mimics biological catalysis based on a profound comprehension of catalytic reactions in living organisms.1–3 Among them, artificial metalloenzymes are a significant subject of investigation in the biomimetic catalysis field. They have the catalytic efficiency and specificity of natural enzymes, as well as greater flexibility and controllability.4,5 In comparison to traditional catalysts, artificial metalloenzymes have the capability to catalyze reactions under mild conditions and offer superior application prospects.6 Nonetheless, artificial metalloenzymes present certain problems, including low activity, poor biocompatibility, etc. Therefore, building artificial metalloenzymes with biomolecules can solve these problems efficiently.6,7 At present, there are few reports on artificial metalloenzymes with nucleic acid as the backbone.In recent years, the study of DNA-based biomimetic catalysts has received much attention because they can mimic the catalytic function of natural enzymes. These catalysts offer several advantages, including high efficiency, high selectivity, easy synthesis, and customizable structure.8 At present, scientists have proficiently employed DNA molecules to fabricate diverse biomimetic catalysts, including DNAzyme,9 DNA metal–organic frameworks,10 and DNA-based artificial metalloenzymes.8 DNA-based artificial metalloenzymes are promising artificial catalysts that combine the advantages of DNA and metal complex catalysts, and they have a wide range of applications in biomedical and chemical synthesis, etc. The DNA backbone has the advantages of high programmability, stability, and structural diversity.8 Moreover, the DNA groove is a unique structure that can elicit a distinctive influence on chemical reactions.11–13 Nevertheless, the accurate controlling of the binding sites of metal complexes to DNA is typically challenging, thereby restricting further research and development in this field.
DNA nanotubes are nanoscale tubes composed of DNA molecules with excellent mechanical properties, chemical stability, and biocompatibility. DNA nanotube preparation methods currently include rolled ring amplification13 and self-assembly.14 Of both these methods, the most prevalent method is self-assembly. DNA nanotubes with different morphologies and functions can be constructed by designing DNA sequences and assembly conditions using the self-assembly between DNA molecules.15,16 However, there are few reports on DNA nanotubes being used to construct artificial metalloenzymes for biomimetic catalysis. We speculate whether DNA nanotubes can be used to mimic enzymatic properties by non-covalent binding to metal complexes. The peroxidase activity of G-quadruplex/iron porphyrin complexes has received much attention.17 Although the G-quadruplex/iron porphyrin complex is expected to serve as an alternative to horseradish peroxidase (HRP) in many fields, its low catalytic activity greatly limits its further application. Achieving a more efficient catalytic system based on G-quadruplex/iron porphyrin complexes remains an important and valuable challenge.18 The G-quadruplex/heme complex is a specialized DNA-based artificial metalloenzyme in which heme binds to the G-quadruplex precisely and non-covalently.19 The G-quadruplex/heme complex is a promising model DNA-based mimetic catalyst, owing to the specific binding of heme to the G-quadruplex. The long-term goal of researchers is to figure out how to improve the peroxidase-like activity of the G-quadruplex/heme complex. However, no reports exist on constructing arrays of G-quadruplex/heme complexes on pure DNA nanotubes for enhancing their activities.
In this paper, an effective and highly active DNA nanotube-like peroxidase was constructed by using a self-assembly strategy. To enhance the activity of G-quadruplex/heme complexes, a novel approach was presented for constructing G-quadruplex/heme complex arrays in a convenient and inexpensive way. G-quadruplex arrays could be created on pure DNA nanotubes by designing the toes of DNA nanotubes as G-quadruplexes, and then the G-quadruplex arrays bind to heme to form G-quadruplex/heme complex arrays. DNTzyme was characterized using agarose gel electrophoresis, circular dichroism, and fluorescence microscopy. And a variety of factors affecting the peroxidase activity were explored to guide the design and optimization of the DNTzyme. Then, the stability of the DNTzyme was compared to that of HRP. Finally, the loading of DNAzyme on DNA nanotubes can increase their biological stability by increasing their resistance to biological enzymes,20,21 and we tried to apply it to the detection of peroxide and to achieve enhanced fluorescence. This study was anticipated to have implications for bioassays, biocatalyses, and drug delivery.
2. Experiments
2.1. Preparation of materials
a. Preparation of blank DNA nanotubes. Single-stranded DNA (ssDNA) used in the experiments was pre-quantified using a UV spectrophotometer at 260 nm. The total volume was set to 1 mL. Five ssDNAs from S1–S5 (1 or 5 μM each), MgCl2 (12.5 mM), and KCl (100 mM) were added to 1× TAE buffer. The system was heated to 90 °C in a PCR instrument and kept for 5 min, and then slowly cooled down to 4 °C at a rate of −0.1 °C min−1 to produce DNA nanotube samples. The DNA nanotubes were stored at 4 °C.
b. Preparation of DNA nanotubes loaded with G-quadruplex arrays. Five ssDNAs S1–S5 (1 or 5 μM each), where the S2 was replaced with G4-S2, MgCl2 (12.5 mM), and KCl (100 mM) were added to 1× TAE buffer. The procedure discussed in subsection ‘a’ was repeated.
c. Preparation of fluorescently labelled DNA nanotubes. Five ssDNAs S1–S5, where the S3 was replaced with fluorescently labelled FAM-S3, MgCl2 (12.5 mM), and KCl (100 mM) were added to 1× TAE buffer. The procedure discussed in subsection ‘a’ was repeated. The nanotubes were characterized by agarose gel electrophoresis.
2.2. Activity determination
The activity of DNTzyme was determined by measuring the efficiency of oxidation of ABTS by DNTzyme in the presence of H2O2. The total volume of the reaction system was set to 1 mL. The DNTzyme (0.5 μM), H2O2 (1 mM), and ABTS (1 mM) were added to buffer A (see Table S1 for details, ESI†). The solution was left at room temperature for 5 min. The absorbance change values were determined at 420 nm using a UV spectrophotometer after reacting the system for 5 min. The initial velocity of the reaction V0 was calculated based on the extinction coefficient of 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1. Three parallel sets of experiments were performed. Kinetic parameters were determined by varying the concentration of the H2O2 system (0–3 mM), and Km and Vmax were obtained by fitting the kinetic curves. Kcat was obtained using the formula:
000 M−1 cm−1. Three parallel sets of experiments were performed. Kinetic parameters were determined by varying the concentration of the H2O2 system (0–3 mM), and Km and Vmax were obtained by fitting the kinetic curves. Kcat was obtained using the formula:2.3. Study on activity factors of DNTzyme
2.4. Stability experiment
2.5. Application of the DNTzyme sensor
DNTzyme-like peroxidase activity was measured by oxidizing ADHP in the presence of H2O2 to produce Resorufin that emitted bright red fluorescence. The total volume of the assay system was set at 1 mL. The fluorescence intensity of Resorufin was evaluated by incubating ADHP (8 μM), DNTzyme (1 μM), and various concentrations of H2O2 in buffer A for 5 min.3. Results and discussion
3.1. Preparation and characterization of DNTzyme arrays
DNA nanotubes loaded with the G-quadruplex were assembled from five ssDNAs, and then combined with hemin to form a complex DNTzyme. The synthesis process is shown in Scheme 1A and B.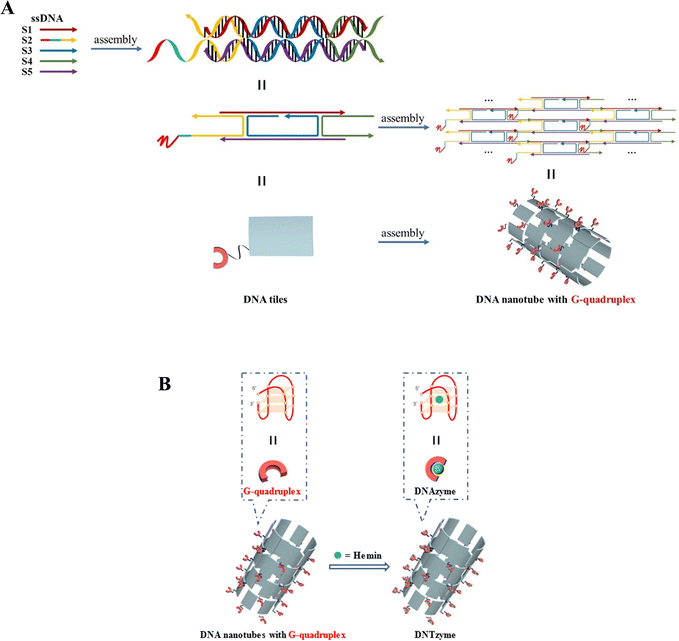 | ||
| Scheme 1 (A) Schematic diagram of the synthesis of DNA nanotubes loaded with G-quadruplexes; (B) G-quadruplex array combined with hemin to construct DNTzyme. | ||
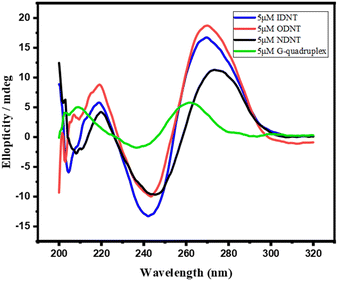
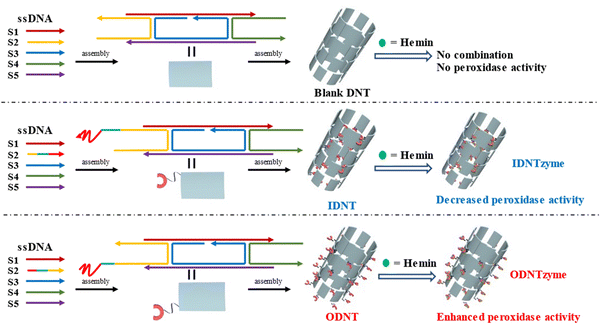
DNA nanotubes were assembled from nanoscale DNA tiles formed by annealing five ssDNAs. The ssDNAs involved in the production of DNA nanotubes were extended in order to establish accurate G-quadruplexes on the DNA nanotubes. After these lengthened ssDNAs were assembled into the DNA nanotubes, the lengthened portions manifested themselves on the DNA nanotubes as regularly aligned branched strands. It was feasible to generate G-quadruplexes with diverse topologies on DNA nanotubes by tailoring the sequences of these branching strands into different G-quadruplex sequences (Scheme 1A). It has been reported that lengthening the 3′-end of ssDNA S2 created a branched strand on the inner surface of DNA nanotubes, whereas lengthening its 5′-end created a branched strand on the outer surface of DNA nanotubes.22 Arrays of G-quadruplexes could thus be formed on the inner or outer surface of DNA nanotubes, termed IDNT and ODNT, respectively. Because the assembly of nanotubes relies on hydrogen bonding between bases, the addition of branched strands could be detrimental to the assembly of nanotubes compared to the absence of branched strands. Therefore, after synthesizing DNA nanotubes with G-quadruplex arrays, the products were analyzed by agarose gel electrophoresis, which confirmed that the addition of branched strands had no substantial negative influence on nanotube synthesis. As demonstrated in Fig. 1A, the mobility of the ODNT and IDNT bands was much lower than that of the ssDNA bands, implying the presence of assembly. Fluorescence labeling was used to visualize the DNA nanotubes. Green lines several tens of microns long could be seen in Fig. 1C and D, indicating the successful synthesis of DNA nanotubes. The TEM image of DNA nanotubes is shown in Fig. S1 (ESI†). To further demonstrate the successful modification of the G-quadruplexes on DNA nanotubes, the CD plots of DNA nanotube samples loaded with the G-quadruplex on the inner or outer surface (IDNT, ODNT) and samples without G-quadruplex (NDNT) were compared. The peaks of the DNA nanotubes loaded with G-quadruplexes became higher at 260–280 nm, as shown in Fig. 2, and this wavelength range corresponded to the peak location of the G-quadruplex standard samples. All these results further verified that the G-quadruplex could be successfully arranged on the DNA nanotubes.
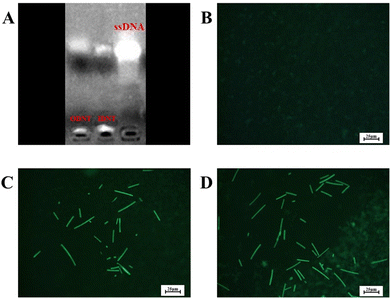 | ||
| Fig. 1 Electrophoretic analysis of DNTs (A) and fluorescence microscopy images of the blank control (B), ODNT (C) and IDNT (D). | ||
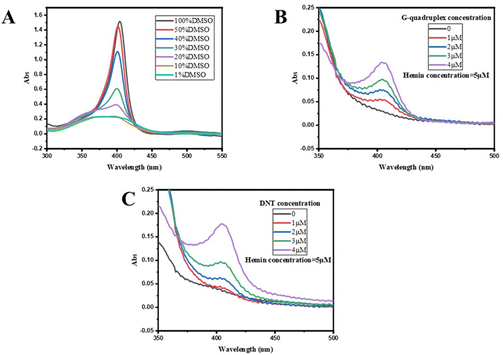
Subsequently, the binding of DNA nanotubes to hemin was confirmed by UV absorption spectroscopy. The UV absorption spectra of hemin mixtures with various concentrations of the G-quadruplex (ssDNA G4-S2 1–4 μM) or DNAzyme (DNT) are presented in Fig. 3B and C. As the concentration of the G-quadruplex or DNTs increased, the UV absorption of the sample was enhanced, i.e. a colour-enhancing effect occurred. As illustrated in Fig. 3A, the UV absorption of hemin and DNA nanotubes in 1% DMSO solution was weak near 410 nm. However, a strong absorption peak was observed for the mixture of hemin and DNA nanotubes in 1% DMSO solution, indicating successful binding of hemin and DNTs. The binding ratio of hemin to DNA nanotubes was then optimized (Fig. S2, ESI†). When the concentration of hemin was low, the activity of DNTzyme was continuously enhanced by the addition of hemin. When the hemin-to-DNT concentration ratio reached 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the addition of hemin no longer significantly increased DNTzyme activity. This result indicated that the binding of hemin to DNTs reached saturation when their concentration ratio reached 3
1, the addition of hemin no longer significantly increased DNTzyme activity. This result indicated that the binding of hemin to DNTs reached saturation when their concentration ratio reached 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Therefore, the hemin-to-DNT concentration ratio was set at 3
1. Therefore, the hemin-to-DNT concentration ratio was set at 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for the synthesis of DNTzyme.
1 for the synthesis of DNTzyme.
3.2. Factors affecting the catalytic activity of DNTzyme
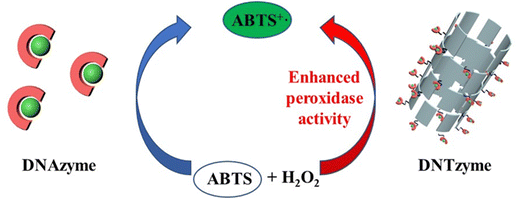
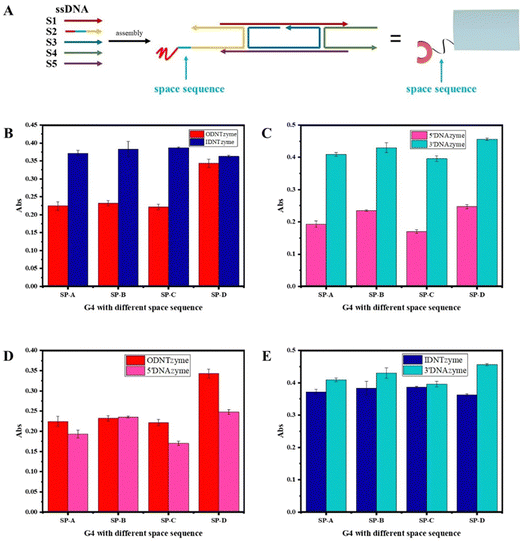
We investigated the parameters influencing the activity of DNTzyme-like peroxidase after demonstrating the successful synthesis of DNTzyme. The activity was determined by measuring the efficiency of DNTzyme in catalyzing the oxidation of ABTS by H2O2, as illustrated in Scheme 2. We investigated five key factors affecting DNTzyme activity: array loading position (inner or outer surface), spatial spacer sequence, proportion of single-strand concentration, G-quadruplex sequence, and array density.The peroxidase activity of IDNTzyme and ODNTzyme was compared with that of the free enzymes 3′DNAzyme and 5′DNAzyme, respectively.
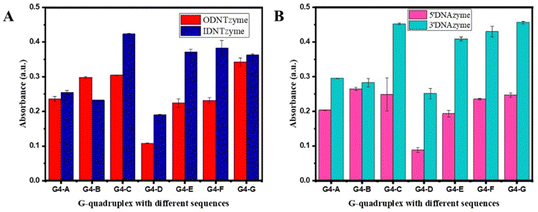 | ||
| Fig. 5 (A) Comparison of peroxidase-like activities of ODNTzyme and IDNTzyme; (B) comparison of peroxidase-like activities of 5′DNAzyme and 3′DNAzyme. | ||
DNTzyme and the equivalent free DNAzyme activity were then compared. As shown in Fig. 6, when the G-quadruplexes were loaded on the outer surface of the DNA nanotubes, the activities of DNTzyme were all higher than those of DNAzyme. However, when G-quadruplexes were loaded on the inner surface of the DNA nanotubes, the activities of DNTzyme were all lower than those of DNAzyme. Loading the G-quadruplexes on the inner or outer surface of the DNA nanotubes resulted in different activity changes. This might be because when the G-quadruplexes were loaded on the outer surface of the DNA nanotubes, the array increased the local concentration of DNAzyme, which in turn increased its activity. When the G-quadruplexes were loaded on the inner surface of the DNA nanotubes, the narrow space restricted G-quadruplex folding and binding to hemin in the DNA nanotubes, thus inhibiting their activity.
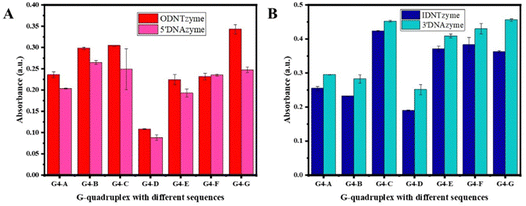 | ||
| Fig. 6 (A) Comparison of peroxidase-like activities of ODNTzyme and 5′DNAzyme; (B) comparison of peroxidase-like activities of IDNTzyme and 3′DNAzyme. | ||
The kinetic parameters of DNTzyme were determined to investigate the mechanism of the changes in the apparent activity of DNTzyme. Table 1 and Fig. S3 (ESI†) reveal that the ODNTzyme exhibited a higher affinity for the substrate and a higher catalytic efficiency than the free DNAzyme. The increased catalytic efficiency was attributed to the high spatiotemporal density on the outside of G-quadruplex/hemin complexes, which increased the affinity of the substrate. In contrast, the catalytic efficiency of IDNTzyme was lower than that of free DNAzyme, although the formation of DNAzyme arrays should have increased its catalytic effectiveness. The lower catalytic efficiency of IDNTzyme compared to free DNAzyme suggested that the limited space prevented contact with the substrates.
| K m (mM) | V max (nM s−1) | K cat (s−1) | |
|---|---|---|---|
| ODNTzyme | 0.672 | 1746 | 1.746 |
| 5′DNAzyme | 1.021 | 1271 | 1.271 |
| IDNTzyme | 1.297 | 1474 | 1.474 |
| 3′DNAzyme | 1.247 | 1784 | 1.784 |
3.3. Stability of DNTzyme
The stability of the catalyst was an essential measure of its excellence. We compared DNTzyme's temperature, metal ions, and organic reagent stability to those of HRP.The activities of DNTzyme, DNAzyme, and HRP were assessed after heat treatment at 20–70 °C for 1 h. As shown in Fig. S7A (ESI†), the activities of DNTzyme and DNAzyme were almost unchanged after heat treatment at 60 °C or less for 1 h, whereas the activity of HRP decreased significantly after heat treatment at 50 °C for 1 h. The activities of DNTzyme, DNAzyme, and HRP were also determined for heat treatment at 60 °C for 0–4 h. As shown in Fig. S7B (ESI†), the activities of DNTzyme and DNAzyme were almost unchanged, while the activity of HRP was almost completely lost with increasing heat treatment time. These results suggested that DNTzyme had much better thermal stability than HRP.
The activities of DNTzyme, DNAzyme, and HRP were determined after 1 h treatment with various metal ions and organic reagents. Fig. S7C (ESI†) shows that the activities of DNTzyme, DNAzyme, and HRP were all lost to variable degrees after metal ion treatment, because the different salt ions affected the structure of DNA and proteins. The activity of DNTzyme, DNAzyme, and HRP was lost to variable degrees after treatment with various organic reagents (Fig. S7D, ESI†), and the activities of DNTzyme and DNAzyme were lost more. This could be because organic reagents were more destructive to the DNA structure.
3.4. Application of DNTzyme in detecting peroxide explosives
DNAzyme is widely used in biosensors. Recognition elements can be constructed for sensitive detection of target substances using G-quadruplex/hemin complexes and catalysis by H2O2. Based on this, DNTzyme was used to create a simple, quick, and cost-effective sensor for the detection of peroxides and achieving enhanced fluorescence. The principle of peroxide detection was that peroxides oxidize ADHP under the action of DNTzyme to produce resorufin, which could produce a strong red fluorescence (Fig. 9A). The effectiveness of DNTzyme and free DNAzyme in detecting explosives was compared. As shown in Fig. 9B, both DNTzyme and free DNAzyme had good linearity for H2O2 detection in the range of 0–1000 nM. The detection limit of H2O2 for DNTzyme was 49 nM, whereas the detection limit of H2O2 for DNAzyme was 77 nM, which meant that DNTzyme had a better detection effect. The same method was used to detect TBHP. As shown in Fig. 9C, its linearity was still good, and the detection limits of TBHP for DNTzyme and DNAzyme were 1.4 μM and 2.4 μM, respectively. The color development reaction lasted 5 min, which was quite efficient. These results indicated that the constructed peroxide sensor was sensitive, accurate, and efficient.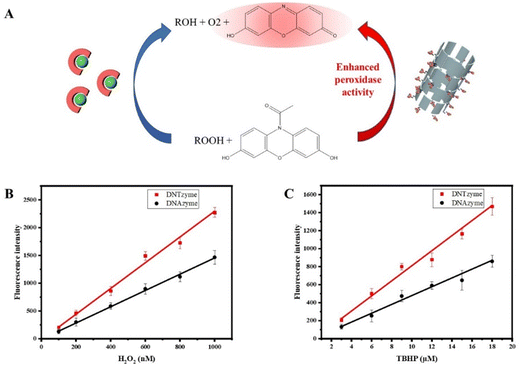 | ||
| Fig. 9 (A) Schematic diagram of DNTzyme-catalyzed oxidation of ADHP by H2O2. The sensors constructed by DNTzyme and DNAzyme were used to detect H2O2 (B) and TBHP (C). | ||
4. Conclusion
We successfully constructed G-quadruplex arrays on DNA nanotubes and confirmed by agarose gel electrophoresis, fluorescence microscopy, and circular dichroism. The binding of G-quadruplex arrays to hemin was confirmed by UV absorption spectroscopy, and the binding ratio of hemin to G-quadruplex arrays was optimized. Some key factors affecting the catalytic activity of DNTzyme were then investigated, including the array loading position, spacer sequence, single-strand concentration ratio, G-quadruplex sequence, and array density. Finally, a peroxide detection sensor was constructed based on the enhanced enzymatic activity and excellent temperature stability of DNTzyme. The sensor can reliably and efficiently detect peroxide and fluorescence enhancement levels with good linearity. The detection limit for H2O2 was 49 nM, the detection limit for TBHP was 1.4 μM, and the color development time was about 5 min. Using the G-quadruplex/hemin complex as an example, this research highlighted the value and potential of DNTs as backbone materials by constructing arrays to effectively enhance the activity of DNAzyme. Because of the functional diversity of G-quadruplexes, G-quadruplex arrays on DNTs were projected to have a higher application value in bio-detection, biocatalysis, and drug delivery.Author contributions
Ying Zhang: experimental design and planning, performing the experiment, writing the initial draft and revising the manuscript; Lingqi Wu: performing the experiments and drawing of the diagram; Xin Su: ideas, oversight and leadership responsibility for the research activity planning and execution and revised the initial draft; Hao Liang: ideas, oversight and leadership responsibility for the research activity planning and execution, revised the initial draft and funding acquisition.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This study was funded by the National Natural Science Foundation of China (22078014).References
- R. J. Hooley, Nat. Chem., 2016, 8, 202–204 CrossRef CAS PubMed.
- W. Xu, L. Jiao, Y. Wu, L. Hu, W. Gu and C. Zhu, Adv. Mater., 2021, 33, 2005172 CrossRef CAS PubMed.
- H. Zhang, S. Li, A. Qu, C. Hao, M. Sun, L. Xu, C. Xu and H. Kuang, Chem. Sci., 2020, 11, 12937–12954 RSC.
- B. Saif and P. Yang, ACS Appl. Bio. Mater., 2021, 4, 1156–1177 CrossRef CAS PubMed.
- T. Sun, S. He, Z. Xu, J. Zuo, Y. Yu and W. Yang, Org. Biomol. Chem., 2021, 19, 3589–3594 RSC.
- B. J. Bloomer, D. S. Clark and J. F. Hartwig, Biochemistry, 2023, 62, 221–228 CrossRef CAS PubMed.
- F. Schwizer, Y. Okamoto, T. Heinisch, Y. Gu, M. M. Pellizzoni, V. Lebrun, R. Reuter, V. Köhler, J. C. Lewis and T. R. Ward, Chem. Rev., 2018, 118, 142–231 CrossRef CAS PubMed.
- J. H. Yum, H. Sugiyama and S. Park, Chem. Rec., 2022, 22, e202100333 CrossRef CAS PubMed.
- J. Dong, X. Qiu, M. Huang, X. Chen and Y. Li, Talanta, 2023, 257, 124384 CrossRef CAS PubMed.
- L. Gao, Y. Deng, H. Liu, K. Solomon, B. Zhang and H. Cai, Biosensors, 2022, 12, 745 CrossRef CAS PubMed.
- S. K. Silverman, Angew. Chem., Int. Ed., 2010, 49, 7180–7201 CrossRef CAS PubMed.
- S. Park and H. Sugiyama, Molecules, 2012, 17, 12792–12803 CrossRef CAS PubMed.
- S. Park and H. Sugiyama, Angew. Chem., Int. Ed., 2010, 49, 3870–3878 CrossRef CAS PubMed.
- O. I. Wilner, R. Orbach, A. Henning, C. Teller, O. Yehezkeli, M. Mertig, D. Harries and I. Willner, Nat. Commun., 2011, 2, 540 CrossRef PubMed.
- P. W. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- A. Rangnekar, K. V. Gothelf and T. H. LaBean, Nanotechnology, 2011, 22, 235601 CrossRef PubMed.
- J. H. Yum, S. Park and H. Sugiyama, Org. Biomol. Chem., 2019, 17, 9547–9561 RSC.
- Y. Chen, D. Qiu, X. Zhang, Y. Liu, M. Cheng, J. Lei, J. L. Mergny, H. Ju and J. Zhou, Anal. Chem., 2022, 94, 2212–2219 CrossRef CAS PubMed.
- S. Haldar, Y. Zhang, Y. Xia, B. Islam, S. Liu, F. L. Gervasio, A. J. Mulholland, Z. A. E. Waller, D. Wei and S. Haider, J. Am. Chem. Soc., 2022, 144, 935–950 CrossRef CAS PubMed.
- J. Wang, D.-X. Wang, B. Liu, X. Jing, D.-Y. Chen, A.-N. Tang, Y.-X. Cui and D.-M. Kong, Adv. Mater., 2022, 17, e202101315 CAS.
- D.-X. Wang, J. Wang, Y.-X. Wang, Y.-C. Du, Y. Huang, A.-N. Tang, Y.-X. Cui and D.-M. Kong, Chem. Sci., 2021, 12, 7602–7622 RSC.
- T. Chang, H. Gong, P. Ding, X. Liu, W. Li, T. Bing, Z. Cao and D. Shangguan, Chemistry, 2016, 22, 4015–4021 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb01984e |
| This journal is © The Royal Society of Chemistry 2024 |