Dual-mode fluorescence and electrochemiluminescence sensors based on Ru-MOF nanosheets for sensitive detection of apoE genes†
Huiting
Hu‡
a,
Hanfeng
Cui‡
a,
Xia
Yin
b,
Qiqi
Fan
a,
Hai
Shuai
a,
Jing
Zhang
 a,
Fusheng
Liao
a,
Wei
Xiong
a,
Hedong
Jiang
a,
Hao
Fan
a,
Fusheng
Liao
a,
Wei
Xiong
a,
Hedong
Jiang
a,
Hao
Fan
 *a,
Wenming
Liu
*a and
Guobing
Wei
*a,
Wenming
Liu
*a and
Guobing
Wei
 *a
*a
aJiangXi University of Chinese Medicine, Nan Chang, JiangXi 330004, China. E-mail: fanhao11@aliyun.com; abc2481264364@163; weigb@163.com
bJiangXi Province Hospital of Integrated Chinese and Western Medicine, Nan Chang, JiangXi 330004, China
First published on 10th November 2023
Abstract
A fluorescence–electrochemiluminescence (FL–ECL) dual-mode sensor for apoE gene detection has been developed, leveraging the unique properties of ruthenium metal organic framework nanosheets (RuMOFNSs). The system utilizes the quenching effect of the Ru(bpy)32+ ECL signal by ferrocene, leading to the synthesis of a multi-electron electrical signal marker, bisferrocene. By immobilizing the P-DNA on RuMOFNSs, bisferrocene quenches both FL and ECL signals. The addition of T-DNA and the consequent formation of double-stranded DNA enable the ExoIII enzyme to excise the bisferrocene fragment, restoring the signals. The sensor demonstrates wide detection linear ranges (1 fM to 1 nM for FL and 0.01 fM to 10 pM for ECL) and remarkable sensitivity (0.048 fM for FL and 0.016 fM for ECL). The dual-mode design offers enhanced reliability through a self-correction feature, reducing false positives. Compared to single-mode sensors, the dual-mode sensor shows significant advantages. Real-world testing confirms the sensor's capacity for robust detection in actual samples, underscoring its promising application in early disease diagnosis. This innovative approach opens up avenues for multi-signal response sensors, offering significant potential for diagnostic technologies.
1. Introduction
Apolipoprotein E (apoE), an integral lipid-carrying protein in plasma, governs the transformation and metabolism of lipoproteins.1 The apoE ε4 gene, a variant of apoE, has emerged as a pivotal genetic factor for Alzheimer's disease (AD), increasing the risk among carriers.2,3 This link underscores the need for accurate, sensitive, and efficient apoE gene detection methods. While conventional genetic testing techniques such as Southern blotting, polymerase chain reaction (PCR), and high-performance liquid chromatography (HPLC) offer substantial insights, they are hampered by intricate sample preparation, extended procedures, and high costs.4–6In the evolving landscape of material science, optical sensing technologies have significantly advanced. Among these, fluorescence (FL) sensing merits special attention due to its superior sensitivity, which remarkably surpasses colorimetric techniques by an impressive factor of 103.7 Electrochemiluminescence (ECL) further complements the optical sensing domain with its attributes, encompassing facile operation, exceptional sensitivity, minimal background interference, and an expansive detection spectrum.8 Notwithstanding these advancements, it is imperative to underscore that singular signal outputs, typified by FL and ECL, might not consistently match the efficacy of dual signal outputs under certain scenarios. In recent years, dual-mode sensing strategies have risen to prominence,9,10 furnishing multiple signal outputs and melding the complementary strengths of various methods, notably those including electrochemical–colorimetric, electrochemical–electrochemiluminescence, fluorescence–photochemical, and fluorescence–colorimetric, for bolstered analyte detection.11–14 The union of FL and ECL into a unified dual-mode sensing paradigm holds great promise, amplifying the fidelity of apoE gene detection and heralding novel diagnostic avenues.
Although the combined utilization of various materials in sensor architecture has been acknowledged,15 the emphasis on single-material dual-mode sensors has been relatively underexplored, thereby curtailing the expansive potential of FL–ECL dual-mode diagnostics. The luminescent agent tris(2,2′-bipyridyl)ruthenium(II) (Ru(bpy)32+), distinguished by its unparalleled optical properties, has become a staple for both fluorescent and electrochemiluminescence probes.16 However, the luminous efficiency of the traditional Ru(bpy)32+ system is still not high.17,18 Advancements in nanotechnology permit the encapsulation of Ru(bpy)32+ derivatives within nanocarriers.5,19–21 Metal–organic frameworks (MOFs), characterized by their vast surface area, adaptable voids, and robust chemical stability,22 support diverse applications, from luminescence23 and energy storage24 to catalysis.25 Recent focus on molecular-functionalized MOFs26,27 has augmented the loading and luminescence efficiency of luminophores, as showcased by Yan et al.'s innovative coordination of Ru(dcbpy)32+ with Zn2+ in the formulation of two-dimensional MOF nanosheets.28 Ferrocene derivatives, celebrated for their unique reversible redox traits, have found wide acceptance as emission quenchers in FL and ECL sensors.29,30 Recognizing the electron limitation in ferrocene led to the evolution of bisferrocene, a dual ferrocene-unit structure that bolsters quenching efficiency and detection range.31
This research pioneers the utilization of RuMOFNSs' distinct optical attributes and bisferrocene's unparalleled quenching proficiency. Orchestrating a meticulous methodology, we attached bisferrocene groups at the 3′ end and amino groups at the 5′ end of P-DNA, enabling the robust binding of vast P-DNA quantities to RuMOFNSs. This symbiosis causes the suppression of the ECL signal, laying the foundation for a dual-mode apoE gene sensing apparatus. The orchestrated introduction of T-DNA, recognized by ExoIII, and T-DNA's eventual release combine in a sequence of well-coordinated reactions, culminating in the resurgence of the ECL signal. By combining the potent FL and ECL techniques, this avant-garde system forges a robust sensing scaffold. It harmoniously aligns these methods, facilitating simultaneous FL detection due to RuMOFNSs' pronounced emission attributes. This multifaceted strategy promotes cross-verification, assuring a more holistic and accurate biological analysis than its single-mode counterparts. Scheme 1 provides an illustrative overview of RuMOFNSs' formulation and their assembly in sensor design, offering readers insight into the intricate workings of this groundbreaking system. With its potential to reshape the scientific landscape, our study stands as a groundbreaking stride in the precision and reliability of biological detection, encapsulating this research's essence and charting novel pathways.
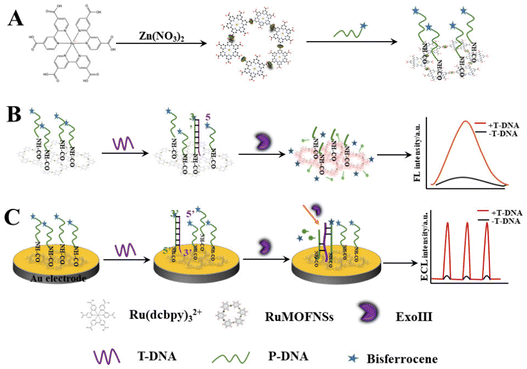 | ||
| Scheme 1 Preparation of RuMOFNSs and their utilization in a bimodal analytical strategy employing both FL and ECL techniques. | ||
2. Experiment section
2.1. Chemicals and materials
Tris(4,4′-dicarboxylidipyridine)ruthenium chloride (Ru(dcbpy)32+), zinc nitrate hexahydrate, and pyrazine were acquired from Xilong Technology Co. Ltd, Guangdong, China. N-Hydroxysuccinimide (NHS), polyvinylpyrrolidone (PVP, Mw = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), 1-ethyl-3-[3-(dimethylamino)propyl]carbodiimide hydrochloride (EDC), and N,N-dimethylformamide (DMF) were obtained from Aladdin Biochemical Technology Co. Ltd, Shanghai. Al2O3 powder was procured from Tianjin Aida Hengsheng Technology Development Co. Ltd. All reagents used were of analytical grade and required no further purification. Ultrapure water (resistivity: 18.2 MΩ cm) was sourced from a Millipore system.
000), 1-ethyl-3-[3-(dimethylamino)propyl]carbodiimide hydrochloride (EDC), and N,N-dimethylformamide (DMF) were obtained from Aladdin Biochemical Technology Co. Ltd, Shanghai. Al2O3 powder was procured from Tianjin Aida Hengsheng Technology Development Co. Ltd. All reagents used were of analytical grade and required no further purification. Ultrapure water (resistivity: 18.2 MΩ cm) was sourced from a Millipore system.
Oligonucleotides and probes, which were modified by the Shanghai Biotechnology Corporation, include:
P-DNA:5′-H2N-TTTCAGGCGGCCGCGCACGTC-bisferrocene-3′
T-DNA: 5′-GACGTGCGCGGCCGCCTG-3′
1MT-DNA: 5′-GACGAGCGCGGCCGCCTG-3′
3MT-DNA: 5′-GACGAGCGCCGCCGTCTG-3′
NCT-DNA: 5′-ACGCACGAATCAGATGTAC-3′
2.2. Apparatus
The FL spectra were recorded using an Edinburgh FLS920 spectrophotometer. The ECL emission measurements were conducted on a model MPI-E electrochemiluminescence analyzer (Xi’An Remax Electronic Science and Technology Co. Ltd, Xi’An, China). All electrochemical measurements were performed on an Autolab electrochemical workstation (Metrohm Instruments Co., Switzerland) at room temperature. Transmission electron microscopy (TEM) images were acquired from a Tecnai G2 F20 S-TWIN electron microscope, operating at 200 kV. Scanning electronmicroscopy (SEM) and energy dispersive X-ray spectroscopy (EDS) were performed using a JEOL-2100F electron microscope. Fourier transform infrared (FT-IR) spectra were obtained using a Bruker VERTEX70 spectrometer. Nuclear magnetic resonance (NMR) spectra were acquired on an Agilent Premium Compact 600 MHz spectrometer. High-resolution mass spectrometry (HRMS) analyses were conducted with an ABSciex instrument, Germany. X-ray diffraction (XRD) measurements were performed using a Rigaku Miniflex-600 system. X-ray photoelectron spectroscopy (XPS) measurements were carried out on a PHI 5000 VersaProbe III.2.3. Synthesis of 3,5-bisferrocene ethoxy benzoic acid
Following the methodologies described in previous research by our group,32 3,5-bisferrocene ethoxybenzoic acid was synthesized. Its characterization was performed using NMR and HRMS. The specific synthesis steps are depicted in Scheme S1 (ESI†). The subsequent ligation of DNA to 3,5-bisferrocene ethoxybenzoic acid was performed by Bioengineering Technologies, located in Shanghai, China.2.4. Preparation of RuMOFNSs
The synthesis of RuMOFNSs followed methodologies outlined in previous studies,28,33,34 as detailed in Scheme 1A. In a 50 mL flask, Ru(dcbpy)32+ (9.0 mg), Zn(NO3)2·6H2O (9.0 mg), PVP (20 mg), and pyridine (1.6 mg) were dissolved in ultrapure water. After 10 minutes of ultrasonication, the solution was deoxygenated with nitrogen and agitated at 60 °C for 2 hours. The resulting crystals were centrifuged, washed, and dried to produce RuMOFNSs.2.5. Fluorescent tests
Scheme 1B illustrates the test procedure of FL sensors. RuMOFNSs (0.025 mg) were dissolved in 10× PBS (pH 4.5), with an EDC to NHS ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and then ultrasonicated for 1 minute until complete dissolution. The solution was incubated at 42 °C for 12 hours, followed by adjusting the pH to 7.5. P-DNA was added until the system concentration reached 10−6 M. The amidation reaction proceeded at 42 °C over a 10 hours period. Subsequently, T-DNA was introduced into the mixture, followed by ExoIII (0.24 U μL−1) after incubation at 37 °C for 2 hours and the FL sensor was assembled. The FL was recorded at an excitation wavelength of 467 nm, the slit width was 5 nm.
1, and then ultrasonicated for 1 minute until complete dissolution. The solution was incubated at 42 °C for 12 hours, followed by adjusting the pH to 7.5. P-DNA was added until the system concentration reached 10−6 M. The amidation reaction proceeded at 42 °C over a 10 hours period. Subsequently, T-DNA was introduced into the mixture, followed by ExoIII (0.24 U μL−1) after incubation at 37 °C for 2 hours and the FL sensor was assembled. The FL was recorded at an excitation wavelength of 467 nm, the slit width was 5 nm.
2.6. Preparation of ECL sensors
Scheme 1C shows the construction of ECL sensors. The gold electrode surface was sequentially polished with 1.0 μm, 0.3 μm, and 0.05 μm Al2O3 powder, and then thoroughly cleaned and dried with ultrapure water. Next, 4 μL of 1% naphthyl phenol and 4 μL of the RuMOFNSs/P-DNA mixture were dried at 37 °C to achieve uniform coverage of RuMOFNSs on the surface. T-DNA was applied to the Au electrode surface and hybridized for 2 hours at 37 °C. Then, it was allowed to digest at 39 °C for 2 hours. Ultimately, the modified gold electrode was submerged in an electrochemical cell to facilitate the necessary measurements. After all steps, the electrodes were rinsed with ultrapure water. See ESI† for details of ECL measurements.3. Results and discussion
3.1. Characterization of RuMOFNSs
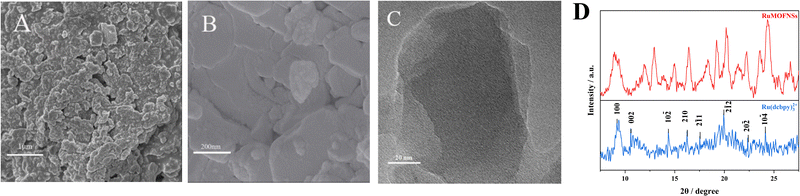
SEM images (Fig. 1A and B) and TEM (Fig. 1C) reveal that RuMOFNSs are stacked in irregular sheets with a higher specific surface area. This structure is conducive to anchoring more P-DNA to the electrode surface. Using XRD analysis (Fig. 1D), it was confirmed that the diffraction peaks of Ru(dcbpy)32+ are in complete agreement with those reported in the literature.35,36 However, certain diffraction peaks observed in the RuMOFNSs were not as sharp and distinct as anticipated.37 This observation is attributable to the flaky structure of the synthesized RuMOFNSs, which somewhat influenced their crystallinity. Nevertheless, the presence of principal characteristic peaks indicates that, despite variations in peak intensities potentially resulting from the synthesis conditions and solvent choices, we have successfully synthesized two-dimensional MOF nanosheets with intact core structural integrity.The synthesized RuMOFNSs were investigated using EDS, XPS, and FT-IR. EDS analysis disclosed the primary elements in RuMOFNSs as C, N, O, Ru, and Zn, consistent with previously reported compositions (Fig. S2A, ESI†). Moreover, Ru is well dispersed in the MOFs, contributing to the material's good optical properties. Further evidence of successful synthesis was provided through XPS analysis, which revealed specific peaks related to C 1s, N 1s, O 1s, Ru 3p, and Zn 2p elements (Fig. S2B, ESI†). In the high-resolution spectrum of Ru 3p, significant peaks were identified at 462.1 eV and 484.2 eV (Fig. S2C, ESI†), while the corresponding Zn 2p3/2 and Zn 2p1/2 peaks appeared at 1021.1 eV and 1044.2 eV (Fig. S2D, ESI†). These observations further confirm the successful creation of RuMOFNSs.
FT-IR analysis of the synthesized RuMOFNSs (Fig. S2E, ESI†) identified distinct peaks at 1716 cm−1 and 1552 cm−1, associated with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of the –COOH groups on the pyridine ring. The simultaneous absence of the C
O stretching vibration of the –COOH groups on the pyridine ring. The simultaneous absence of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration, paired with asymmetric (COO–) and symmetric (COO–) stretching vibrations at 1628 cm−1 and 1356 cm−1, confirms the full deprotonation of the carboxyl groups in Ru(dcbpy)32+. This deprotonation leads to their coordination with metal ions, further affirming the successful synthesis. Distinct peaks at 1650 cm−1 and 1750 cm−1 were observed as shown in Fig. S2F (ESI†), indicating the formation of amide bonds between P-DNA and RuMOFNSs. These peaks provide evidence of the successful immobilization of P-DNA on the RuMOFNS surface.
O stretching vibration, paired with asymmetric (COO–) and symmetric (COO–) stretching vibrations at 1628 cm−1 and 1356 cm−1, confirms the full deprotonation of the carboxyl groups in Ru(dcbpy)32+. This deprotonation leads to their coordination with metal ions, further affirming the successful synthesis. Distinct peaks at 1650 cm−1 and 1750 cm−1 were observed as shown in Fig. S2F (ESI†), indicating the formation of amide bonds between P-DNA and RuMOFNSs. These peaks provide evidence of the successful immobilization of P-DNA on the RuMOFNS surface.
3.2. RuMOFNS FL and ECL performance
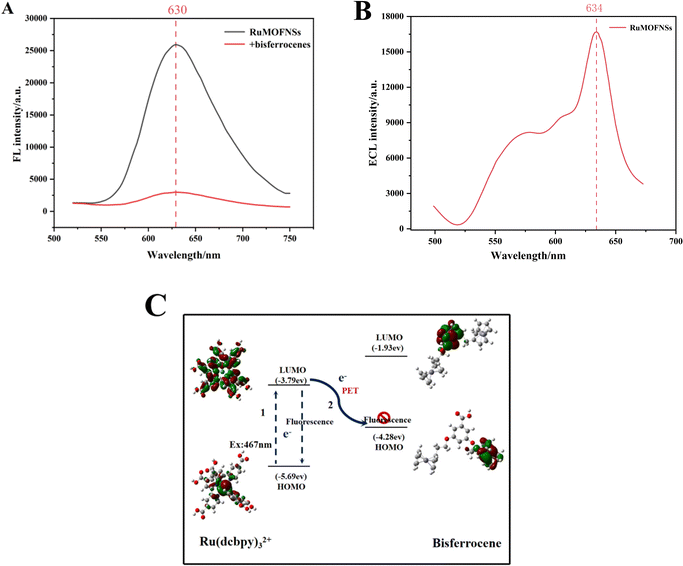
The sensing potential of RuMOFNSs was evaluated through analysis of their FL and ECL capabilities. As shown in Fig. 2A, excitation at 467 nm results in an emission peak at 630 nm in the spectra. The introduction of bisferrocene-modified P-DNA to the system led to a noticeable decline in the FL signal at 630 nm. To understand the underlying cause of the FL quenching by bisferrocene in RuMOFNSs, the principles of frontier orbital theory38,39 were applied, uncovering a photoinduced electron transfer (PET) mechanism at play. Further validation of this quenching mechanism was sought through density functional theory (DFT) calculations using the Gaussian 09 code. Optimization of the geometry for main-group elements was carried out employing the M06L/6-31G(d,p) basis set, while the SDD basis set was utilized specifically for Fe and Ru atoms. These results are visualized in Fig. 2C. The results indicate that the highest occupied molecular orbital (HOMO) of bisferrocene lies between the HOMO and LUMO of Ru(dcbpy)32+. This confirms the hypothesis that excited electrons from Ru(dcbpy)32+ can easily transition to the bisferrocene's HOMO, resulting in the luminescence quenching of Ru(dcbpy)32+.In Fig. 2B, we present the ECL spectrum obtained using optical filter technology. The emission spectrum of RuMOFNSs in the 634 nm region closely resembles the residual fluorescence emission, indicating a significant ECL signal exhibited by RuMOFNSs. However, RuMOFNSs exhibit a pronounced ECL signal, which is dampened with the introduction of bisferrocene-modified P-DNA. The ECL quenching mechanism involving bisferrocene may be inferred from previous studies on the attenuation of the Ru(bpy)32+ ECL signal by ferrocene. This is generally attributed to a charge-transfer mechanism.40 Within the ECL system, the Fc+ interferes with the excited state of Ru(bpy)32+, resulting in a decrease in the ECL signal intensity of Ru(bpy)32+ as follows:
| Fc − e− → Fc+ | (1) |
| Fc+ + Ru(bpy)32+* → Fc+ + Ru(bpy)33+ | (2) |
The quenching mechanism can vary depending on the ECL potential. At a lower ECL potential, Ru(bpy)32+* is derived from co-reactants like TPA. Here, Fc+ competes with co-reactants. However, the energy difference (ΔE) between Fc+ and Ru(bpy)3+ is not sufficient to produce the excited state Ru(bpy)32+*. At higher ECL potentials, the oxidized form, Fc+, competes with Ru(bpy)33+ and Ru(bpy)32+ to interact with co-reactants.
Furthermore, comparative analyses were conducted on the FL and ECL signals of RuMOFNSs and Ru(bpy)32+. The efficiency of bisferrocene's quenching was compared with that of ferrocene (details in the ESI†). The gathered data underscore that the synthesized RuMOFNSs possess a robust FL signal, as well as a consistent, high-intensity ECL signal.
3.3. Characterization of biosensors
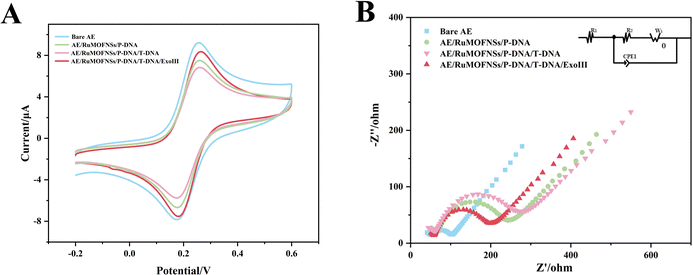
The step-by-step preparation of the biosensor is monitored by cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) measurements.As shown in Fig. 3A, the [Fe(CN)6]3−/4− redox peak exhibits the largest redox current value on the bare Au electrode (AE). After anchoring P-DNA and RuMOFNSs, followed by complementary pairing of P-DNA and T-DNA, the CV redox current gradually decreased. This reduction is due to P-DNA and T-DNA occupying and obstructing the electron transport channel, thus reducing the electron transfer rate. However, when ExoIII is added, the single nucleotide and bisferrocene fragment at the blunt 3′ end of P-DNA are gradually removed, and T-DNA is released, leading to an increase in the redox current.
EIS further confirms this process. The semicircles in the Nyquist plots (Fig. 3B) correspond to the charge transfer resistance (Ret) and reflect the electron transfer kinetics of the redox probe on the electrode surface. The Ret of the naked Au electrode is smaller, demonstrating excellent conductivity. However, when P-DNA is fixed on the electrode surface, the Ret value gradually increases, echoing the CV results. When ExoIII is added, the Ret value decreases significantly, reinforcing the observations obtained from CV.
3.4. Optimization of experimental conditions
The connection between RuMOFNSs and P-DNA is an important factor affecting the performance of the dual-mode sensor. Several reaction conditions are investigated to optimize the system, including enzyme digestion time, digestion temperature, pH of PBS (phosphate-buffered saline), and the proportion of activators (EDC:NHS). These experiments aim to identify the optimal conditions for maximizing sensor performance. When the optimal conditions were determined (time 120 min, temperature 39 °C, pH 7.5, and an EDC:NHS ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), subsequent experiments were performed under these conditions. See ESI† for details.
1), subsequent experiments were performed under these conditions. See ESI† for details.
3.5. Analytical performance of dual-mode sensors
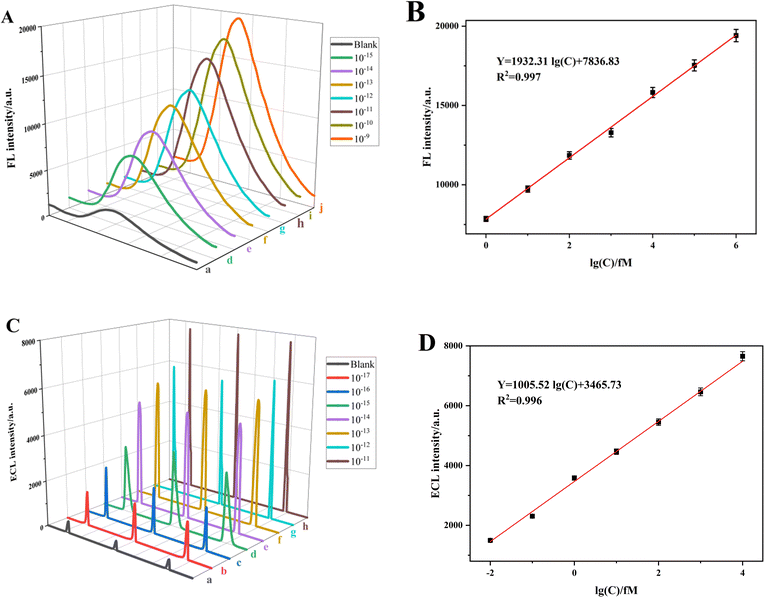
The FL and ECL signal intensities exhibited a positive correlation with the concentrations of the apoE gene in the range of 1 fM to 1 nM and 0.01 fM to 10 pM, respectively, as depicted in Fig. 4A and C. The dual-signal sensor demonstrated a strong linear relationship within these concentration ranges. The linear regression fitting was described by the following equations: Y = 1932.31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log(C) + 7836.83, R2 = 0.997 and Y = 1005.52
log(C) + 7836.83, R2 = 0.997 and Y = 1005.52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log(C) + 3465.73, R2 = 0.996, as depicted in Fig. 4B and D, respectively. These equations correspond to detection limits of 0.048 fM and 0.016 fM. The formula used to calculate the detection limit is detailed below:
log(C) + 3465.73, R2 = 0.996, as depicted in Fig. 4B and D, respectively. These equations correspond to detection limits of 0.048 fM and 0.016 fM. The formula used to calculate the detection limit is detailed below:| L = 3Sb/k0 | (3) |
| Method | Luminescent molecules | Quenching molecules | Linear range | Detection limit | Ref. |
|---|---|---|---|---|---|
| Electrochemiluminescence | Met-AuNCs | Ferrocene | 1 pM–10 nM | 0.48 pM | 41 |
| Electrochemiluminescence | Ru(bpy)32+ | AuNPs | 0.5–50 nM | 12 fM | 42 |
| Fluorescence | FAM | PGO | 0–0.25 μM | 3.39 nM | 43 |
| Fluorescence | FAM | DABCYL | 0.5–10 nM | 0.38 nM | 44 |
| FL–ECL | RuMOFNSs | Bisferrocene | 0.01 fM–10 pM | 0.048 fM | This work |
| 0.016 fM |
3.6. Selectivity, stability, and repeatability of dual-mode sensors
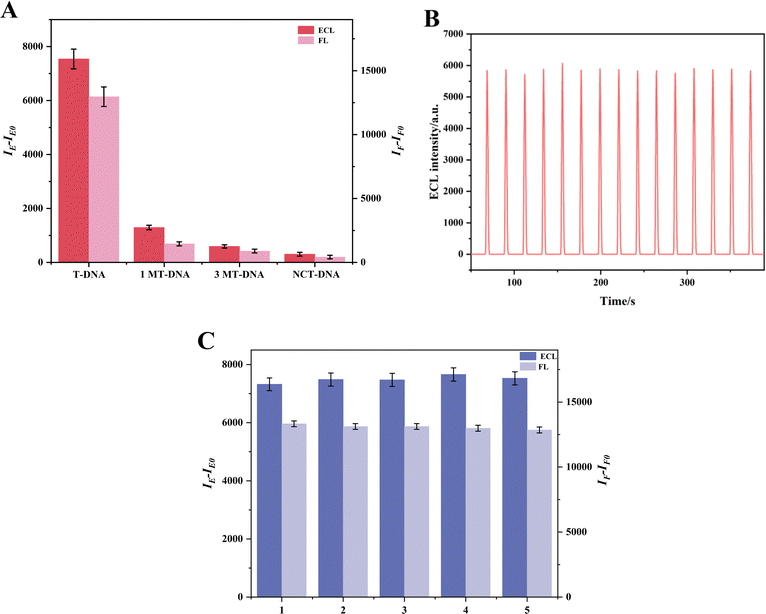
To evaluate the selectivity, stability and repeatability of the dual-mode sensor, various experiments were carried out.For selectivity, several DNA base sequences were examined, such as T-DNA, which is fully complementary to P-DNA, as well as single-base mismatch DNA (1MT-DNA), three-base mismatch DNA (3MT-DNA), and a non-complementary random base sequence DNA (NCT-DNA). The FL and ECL detection results are presented in Fig. 5A. Notably, the changes in FL and ECL signals (expressed as IF–IF0 for FL and IE–IE0 for ECL, where IF and IE represent the FL and ECL intensities, and IF0 and IE0 represent blank values) are significant only when T-DNA is present, underscoring the sensor's selectivity. Moreover, the stability of the apoE gene was assessed through 15 consecutive electrode scans at a concentration of 10−12 M for the apoE genes' ECL signal. The results revealed that the ECL signal remained nearly constant, with a relative standard deviation (RSD) of just 1.30%, thereby indicating good stability. Furthermore, Fig. 5C depicts the repeatability of the dual-mode sensors. The FL was tested five times for repeatability, achieving an RSD of 1.3%. Concurrently, ECL intensities of five distinct electrodes exhibited an RSD of 1.6%, underscoring the exceptional reproducibility of the dual-mode sensors.
3.7. Real sample detection
Real sample analysis is a critical metric for evaluating the practical application potential of the designed sensing system. The suitability of the sensor for real sample analysis was confirmed by calculating the recovery of apoE genes at different concentrations (5, 10, and 20 fM) in healthy human serum samples, diluted 10-fold, using the standard addition method (with three parallel measurements per sample). Table S1 (ESI†) reveals that the recovery rates for both FL and ECL tests range between 98.2% to 102.2% and 97.8% to 102.1%, respectively, displaying relative standard deviations (RSD) of 1.85% to 2.40% for FL and 0.88% to 2.19% for ECL. These findings indicate that the dual-mode sensor proficiently reduced background disturbances and matrix effects present in the biological samples. Furthermore, to confirm the utility of the dual-mode sensor in a PCR setting, it was utilized to amplify the gene of Corydalis yanhusuo through the PCR process. Table S2 (ESI†) lists the sequences of oligonucleotides, indicating that the constructed sensor can successfully recognize the Corydalis yanhusuo gene in the PCR environment, displaying a sensitivity of 24.85 fM for FL and 1.46 fM for ECL.4. Conclusion
A dual-mode sensor employing both FL and ECL has been successfully developed using RuMOFNSs, showcasing robust FL and ECL functionalities. Utilizing bisferrocene as an efficient signal-quenching material, the system is able to proficiently quench both FL and ECL signals emitted by RuMOFNSs. Under meticulously optimized conditions, this innovative dual-mode sensing platform has demonstrated the ability to detect the apoE gene within the concentration range of 1 fM to 1 nM (for FL) and 0.01 fM to 10 pM (for ECL), maintaining a linear response. The respective detection limits for FL and ECL are set at 0.048 fM and 0.016 fM (S/N = 3). The integration of FL and ECL testing offers an enhanced self-correction capability, effectively mitigating the risk of false-positive results. Furthermore, the sensor's adept performance in real-sample detection presents a promising avenue for multi-signal response sensors, laying the groundwork for potential applications in early-stage disease diagnosis.Author contributions
Huiting Hu: writing – original draft; Hanfeng Cui: supervision; Xia Yin: project administration; Qiqi Fan: data curation and methodology; Hai Shuai: software; Jing Zhang: investigation; Fusheng Liao: investigation; Wei Xiong: software; Hedong Jiang: methodology and investigation; Wenming Liu: supervision; Guobing Wei: funding acquisition; and Hao Fan: supervision and funding acquisition.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was financially supported by the National Science Foundation of China (grant no. 82160729), the Natural Science Foundation of JiangXi Province (20212ACB206005, 20192BAB205103, and 20202BABL206152), the Science and Technology Project of Education Department of JiangXi Province (GJJ2200974 and GJJ2200906), the Doctoral Research Foundation project of Jiangxi University of Traditional Chinese Medicine (2022WBZR005), the Innovation and Entrepreneurship Training Program for College Students of Jiangxi University of Chinese Medicine (S202210412040, 202210412076, and 202110412164) and the Jiangxi University of Chinese Medicine School-level Science and Technology Innovation Team Development Program (CXTD22005), and the NATCM's Project of High-level Construction of Key TCM Disciplines (jzzdxk-2023019).References
- F. Chen, Q. Ke, W. Wei, L. Cui and Y. Wang, Mol. Ther. – Nucleic Acids, 2023, 33, 529–542 CrossRef CAS PubMed.
- K. R. Bales and S. M. Paul, Neurosci. Lett., 2019, 709, 134366 CrossRef CAS PubMed.
- K. L. Rasmussen, J. Luo, B. G. Nordestgaard, A. Tybjærg-Hansen and R. Frikke-Schmidt, Atherosclerosis, 2023, 117218 CrossRef CAS PubMed.
- A. Mouchard, M. C. Boutonnet, C. Mazzocco, N. Biendon and N. Macrez, Sci. Rep., 2019, 9, 3989 CrossRef PubMed.
- D. A. Al-Quwaie, A. Allohibi, M. Aljadani, A. M. Alghamdi, A. A. Alharbi, R. S. Baty, S. H. Qahl, O. Saleh, A. O. Shakak, F. S. Alqahtani, O. S. F. Khalil, M. T. El-Saadony and A. M. Saad, Molecules, 2023, 28, 5859 CrossRef CAS PubMed.
- B. Weis, J. Schmidt, F. Lyko and H. G. Linhart, BMC Biotechnol., 2010, 10, 75 CrossRef PubMed.
- G. Capilli, S. Cavalera, L. Anfossi, C. Giovannoli, M. Minella, C. Baggiani and C. Minero, Nano Res., 2019, 12, 1862–1870 CrossRef CAS.
- X. Z. Wu, Y. J. Sun, T. He, Y. L. Zhang, G. J. Zhang, Q. M. Liu and S. Chen, Langmuir, 2021, 37, 11309–11315 CrossRef CAS PubMed.
- L. Zheng, Q. Guo, C. Yang, J. Wang, X. Xu and G. Nie, Sens. Actuators, B, 2023, 379, 133269 CrossRef CAS.
- J. Xu, Y. Zhang, L. Li, Q. Kong, L. Zhang, S. Ge and J. Yu, ACS Appl. Mater. Interfaces, 2018, 10, 3431–3440 CrossRef CAS PubMed.
- Y. Hu, L. Zhu, X. C. Mei, J. S. Liu, Z. P. Yao and Y. C. Li, Anal. Chem., 2021, 93, 12367–12373 CrossRef CAS PubMed.
- X. B. Feng, N. Gan, H. R. Zhang, T. H. Li, Y. T. Cao, F. T. Hu and Q. L. Jiang, Biosens. Bioelectron., 2016, 75, 308–314 CrossRef CAS PubMed.
- L. Zhang, F. Z. Chen, H. D. Sun, R. Z. Meng, Q. S. Zeng, X. X. Wang and H. Zhou, ACS Appl. Mater. Interfaces, 2022, 14, 46103–46111 CrossRef CAS PubMed.
- K. Y. He, L. P. Sun, L. Wang, W. Li, G. X. Hu, X. F. Ji, Y. M. Zhang and X. H. Xu, J. Hazard. Mater., 2022, 423, 126962 CrossRef CAS PubMed.
- S. S. Cong, Z. Y. Jiang, R. Z. Zhang, H. P. Lv, J. N. Guo, L. B. Zhang and X. Q. Lu, Anal. Chem., 2022, 94, 6695–6702 CrossRef CAS PubMed.
- D. D. Hu, T. Y. Zhan, Z. Y. Guo, S. Wang and Y. F. Hu, Sens. Actuators, B, 2021, 327, 128896 CrossRef CAS.
- F. K. Du, H. Zhang, X. C. Tan, J. Yan, M. Liu, X. Chen, Y. Y. Wu, D. F. Feng, Q. Y. Chen, J. M. Cen, S. G. Liu, Y. Q. Qiu and H. Y. Han, Biosens. Bioelectron., 2018, 106, 50–56 CrossRef CAS PubMed.
- H. J. Wang, Y. L. Yuan, Y. Zhuo, Y. Q. Chai and R. Yuan, Anal. Chem., 2016, 88, 2258–2265 CrossRef CAS PubMed.
- G. Valenti, E. Rampazzo, S. Bonacchi, L. Petrizza, M. Marcaccio, M. Montalti, L. Prodi and F. Paolucci, J. Am. Chem. Soc., 2016, 138, 15935–15942 CrossRef CAS PubMed.
- X. J. Li, S. Q. Yu, T. Yan, Y. Zhang, B. Du, D. Wu and Q. Wei, Biosens. Bioelectron., 2017, 89, 1020–1025 CrossRef CAS PubMed.
- G. H. Zhao, Y. G. Wang, X. J. Li, X. Dong, H. Wang, B. Du, W. Cao and Q. Wei, ACS Appl. Mater. Interfaces, 2018, 10, 22932–22938 CrossRef CAS PubMed.
- L. Bianchi, A. Amorati, R. Basosi, C. Zambonin and F. Armaroli, Chem. Commun., 2021, 57, 1583–1586 Search PubMed.
- B. B. Guo, J. C. Yin, N. Li, Z. X. Fu, X. Han, J. Xu and X. H. Bu, Adv. Opt. Mater., 2021, 9, 2100283 CrossRef CAS.
- F. X. Wang, X. W. Wu, X. H. Yuan, Z. C. Liu, Y. Zhang, L. J. Fu, Y. S. Zhu, Q. M. Zhou, Y. P. Wu and W. Huang, Chem. Soc. Rev., 2017, 46, 6816–6854 RSC.
- A. H. Chughtai, N. Ahmad, H. A. Younus, A. Laypkov and F. Verpoort, Chem. Soc. Rev., 2015, 44, 6804–6849 RSC.
- H. Dong, S. H. Liu, Q. Liu, Y. Y. Li, Z. Xu, Y. Y. Li and Q. Wei, Anal. Chem., 2022, 94, 12852–12859 CrossRef CAS PubMed.
- S. Li, L. Yu, L. Xiong and Y. X. Xiao, Sens. Actuators, B, 2022, 355, 131282 CrossRef CAS.
- M. X. Yan, J. Ye, Q. J. Zhu, L. P. Zhu, J. S. Huang and X. R. Yang, Anal. Chem., 2019, 91, 10156–10163 CrossRef CAS PubMed.
- Y. Li, H. L. Qi, Y. G. Peng, Q. Gao and C. X. Zhang, Electrochem. Commun., 2008, 10, 1322–1325 CrossRef CAS.
- R. Q. Wang, T. Zhou, A. Li, J. Qu, X. Zhang, X. F. Zhu and S. Jing, Dalton Trans., 2022, 51, 15330–15338 RSC.
- H. H. Xu, H. F. Cui, Z. J. Yin, G. B. Wei, F. S. Liao, Q. X. Shu, G. Q. Ma, L. Cheng, N. Hong, J. Xiong and H. Fan, Bioelectrochemistry, 2020, 134, 107522 CrossRef CAS PubMed.
- Y. Fan, F. S. Liao, W. T. Wu, N. Hong, L. Cheng, Y. Q. Zhong, H. F. Cui, G. Q. Ma, J. Xiong and H. Fan, Sens. Actuators, B, 2020, 318, 128045 CrossRef CAS.
- L. Zheng, Q. F. Guo, C. N. Yang, J. J. Wang, X. J. Xu and G. M. Nie, Sens. Actuators, B, 2023, 379, 133269 CrossRef CAS.
- E. Villani, K. Sakanoue, Y. Einaga, S. Inagi and A. Fiorani, J. Electroanal. Chem., 2022, 921, 116677 CrossRef CAS.
- Q. Li, J. Y. Zheng, Y. L. Yan, Y. S. Zhao and J. N. Yao, Adv. Mater., 2012, 24, 4745–4749 CrossRef CAS PubMed.
- H. Q. Yin, J. C. Yang and X. B. Yin, Anal. Chem., 2017, 89, 13434–13440 CrossRef CAS PubMed.
- C. A. Kent, B. P. Mehl, L. Q. Ma, J. M. Papanikolas, T. J. Meyer and W. B. Lin, J. Am. Chem. Soc., 2010, 132, 12767–12769 CrossRef CAS PubMed.
- X. M. Liu, Y. T. Qi, S. H. Pu, Y. Wang and Z. Q. Gao, RSC Adv., 2021, 11, 22214–22220 RSC.
- W. Sun, M. Li, J. L. Fan and X. J. Peng, Acc. Chem. Res., 2019, 52, 2818–2831 CrossRef CAS PubMed.
- W. D. Cao, J. P. Ferrance, J. Demas and J. P. Landers, J. Am. Chem. Soc., 2006, 128, 7572–7578 CrossRef CAS PubMed.
- P. F. Liu, K. R. Zhao, Z. J. Liu, L. Wang, S. Y. Ye and G. X. Liang, Biosens. Bioelectron., 2021, 176, 112954 CrossRef CAS PubMed.
- M. Zhang, H. Hai, F. Y. Zhou, J. C. Zhong and J. P. Li, Chin. J. Inorg. Anal. Chem., 2018, 46, 203–209 CrossRef CAS.
- T. Sun, X. Li, X. C. Jin, Z. Y. Wu, X. C. Chen and J. Q. Qiu, Int. J. Mol. Sci., 2022, 23, 6326 CrossRef CAS PubMed.
- S. Y. He, P. Li, L. Tang, M. J. Chen, Y. X. Yang, Z. Y. Zeng, W. Xiong, X. Wu and J. H. Huang, Microchem. J., 2022, 172, 106946 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb01934a |
| ‡ Huiting Hu and Hanfeng Cui contributed equally to this paper. |
| This journal is © The Royal Society of Chemistry 2024 |