The role of protein folding in prenucleation clusters on the activity of enzyme@metal–organic frameworks†
Brooke P.
Carpenter‡
a,
Ben
Rose‡
 a,
Elisa M.
Olivas
a,
Mariana X.
Navarro
a,
A. Rain
Talosig
a,
Elisa M.
Olivas
a,
Mariana X.
Navarro
a,
A. Rain
Talosig
 a,
Paul J.
Hurst
a,
Giuseppe
Di Palma
a,
Li
Xing
e,
Rweetuparna
Guha
a,
Paul J.
Hurst
a,
Giuseppe
Di Palma
a,
Li
Xing
e,
Rweetuparna
Guha
 b,
Stacy M.
Copp
bcd and
Joseph P.
Patterson
b,
Stacy M.
Copp
bcd and
Joseph P.
Patterson
 *a
*a
aDepartment of Chemistry, University of California, Irvine, Irvine, CA 92697-2025, USA. E-mail: patters3@uci.edu
bDepartment of Materials Science and Engineering, University of California, Irvine, Irvine, CA 92697, USA
cDepartment of Physics and Astronomy, University of California, Irvine, Irvine, CA 92697, USA
dDepartment of Chemical and Biomolecular, Engineering University of California, Irvine, Irvine, CA 92697, USA
eIrvine Materials Research Institute, University of California, Irvine, Irvine, CA 92697, USA
First published on 11th December 2023
Abstract
Enzymes present a promising green chemistry alternative for accelerating chemical reactions in industry. However, many industrial applications require harsh synthetic conditions (i.e. high temperature and/or organic solvents), causing enzymes to denature. Metal–organic frameworks (MOFs) offer a promising method to overcome the structural limitations of potentially revolutionary enzymes through enzymatic immobilization. However, for high activity of immobilized enzymes to be achieved, a comprehensive understanding of the factors influencing enzyme performance is critical. While extensive efforts have demonstrated that final MOF properties such as crystallinity, porosity, and network defects significantly influence enzyme activity, here we propose a separate, but equally impactful, factor in determining enzyme@MOF activity: enzyme folding in prenucleation clusters. To investigate both MOF structural property and enzyme folding factors, two model enzymes, glucose oxidase and catalase, were encapsulated into ZIF-8. The biocomposite formation and prenucleation clusters were examined through electron microscopy, advanced spectroscopy, and scattering techniques, respectively. Results from these studies indicate enzyme folding with MOF precursors is a crucial aspect to consider when pairing enzymes with selected MOF precursors. To assist in this consideration, we provide alternative strategies for retaining enzyme activity in denaturing conditions. Finally, a roadmap is provided for tuning activity of immobilized enzymes, paving the way for industrial-scale production of enzyme@MOFs.
Introduction
Enzymes are highly efficient and selective biological catalysts that hold immense potential in various industries, including drug delivery,1,2 bioremediation,3 and CO2 reduction.4 Enzyme-catalyzed industrial reactions hold many benefits from a sustainability perspective, as enzymes are biosynthesized and biodegradable, breaking down into amino acids and metal ions which are generally not cytotoxic. However, the widespread application of enzymes in industrial catalysis is limited due to the propensity of biomolecules to denature or unfold in harsh industrial conditions. Metal–organic frameworks (MOFs), porous coordination polymers consisting of metal nodes and organic linkers, offer a promising solution by protecting and immobilizing enzymes in their active conformations.1–3 Unlike other protein immobilization methods, MOFs provide a wide range of metal and ligand building units, allowing customization of crystal size, structure, and porosity along with optimization for different enzymes and applications.4 Some MOFs enable one-pot integration of enzymes in aqueous synthetic conditions that favor protein stability and retain activity in the composite.2,3 The properties of the encapsulating MOF, such as network topology,5 morphology, defects,6 and size7 play an important role in catalytic performance of the encapsulated enzyme.8,9 While little is known about a given enzyme environment and its exact conformation during MOF formation, enzymatic tertiary structure, location, and orientation within the crystal are known to play key roles in the activity of the immobilized enzyme.10–12Zeolitic imidazolate framework-8 (ZIF-8) has emerged as a widely studied MOF for enzyme immobilization due to its precursor availability, ease of synthesis, and environmentally friendly synthetic conditions.13,14 ZIF-8 forms a sodalite (sod) topology consisting of zinc metal nodes connected by 2-methylimidazole (HmIM) linkers. The crystal formation and final crystal properties in ZIF-8 can be tuned through variations in the ligand-to-metal ratio (HmIM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn) with the aim of achieving high enzyme activity and encapsulation efficiency.6 At low ratios of HmIM
Zn) with the aim of achieving high enzyme activity and encapsulation efficiency.6 At low ratios of HmIM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn, enzymes with low isoelectric points (<7) are thought to drive nucleation through attraction of the positively charged metal ions to the negatively charged enzyme surface.3,15,16 This approach, coined biomimetic mineralization, has yielded various ZIF-8 morphologies and network topologies, such as the most observed polymorph, sodalite (sod), along with ZIF-C and ZIF-L.5 Through the biomimetic mineralization process, mixtures of different ZIF-8 polymorphs are often obtained, with the exact polymorphs varying from enzyme to enzyme, indicating that biomolecular differences play significant roles in MOF polymorph determination and enzyme activity.
Zn, enzymes with low isoelectric points (<7) are thought to drive nucleation through attraction of the positively charged metal ions to the negatively charged enzyme surface.3,15,16 This approach, coined biomimetic mineralization, has yielded various ZIF-8 morphologies and network topologies, such as the most observed polymorph, sodalite (sod), along with ZIF-C and ZIF-L.5 Through the biomimetic mineralization process, mixtures of different ZIF-8 polymorphs are often obtained, with the exact polymorphs varying from enzyme to enzyme, indicating that biomolecular differences play significant roles in MOF polymorph determination and enzyme activity.
While final MOF polymorph and crystallinity have been extensively examined as driving factors behind activity of MOF-encapsulated enzymes, a biomolecule-focused approach examining enzyme folding in prenucleation clusters has not previously been offered. This understanding is critical because enzymatic catalysis is dependent on proper enzyme folding. In previous research, we have shown that biomolecule folding affects the crystallization mechanism, kinetics, and structural properties of MOFs synthesized in situ.17 This study also found that the MOF precursors, specifically zinc acetate, can influence the initial protein folding. Other studies have shown that hydrophilic ligands can stabilize enzymes in their active conformation, retaining activity of the enzyme in the final enzyme@MOF,18 whereas more hydrophobic ligands lead to a loss of functionality.19 Thus, a more comprehensive understanding of biomolecular folding during enzyme@MOF synthesis is needed. Here, we reemphasize the role of MOF crystallinity in enzyme@ZIF-8 activity, while introducing a novel factor relevant to activity: enzymatic interactions with MOF building blocks. More specifically, the enzyme interactions with MOF precursors, particularly upon formation of prenucleation clusters consisting of enzymes and MOF precursors, are shown to influence the folding, encapsulation efficiency, and activity of the enzymes glucose oxidase (GOx) and catalase (CAT) within the ZIF-8 system. To exhibit enzyme activity, we show that building units must form with minimal enzyme denaturation. If this requirement is met, the physical properties of the crystal can then be optimized for high activity, as crystallinity and structure influence the diffusion and accessibility of the substrate to the immobilized enzyme. Time-resolved cryogenic-transmission electron microscopy (cryoTEM) is utilized to elucidate two distinct mechanisms resulting in either highly or poorly active enzyme@ZIF-8 biocomposites.
Results
Enzyme@MOF synthesis and structural characterization
GOx and CAT were each encapsulated into ZIF-8 by varying the HmIM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn ratio to achieve two different mechanistic pathways: biomimetic mineralization and solid-state transformation.15 To synthesize the biocomposites, separate stock solutions of the enzymes (5 mg mL−1, 0.75 mL), 2-methylimidazole (HmIM) (320 mM and 2800 mM, 0.75 mL), and zinc acetate (40 mM, 1.50 mL) were prepared in nanopure water. The different concentrations of HmIM were combined with each of the enzymes and zinc acetate solutions to produce systems with HmIM
Zn ratio to achieve two different mechanistic pathways: biomimetic mineralization and solid-state transformation.15 To synthesize the biocomposites, separate stock solutions of the enzymes (5 mg mL−1, 0.75 mL), 2-methylimidazole (HmIM) (320 mM and 2800 mM, 0.75 mL), and zinc acetate (40 mM, 1.50 mL) were prepared in nanopure water. The different concentrations of HmIM were combined with each of the enzymes and zinc acetate solutions to produce systems with HmIM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn ratios of 4
Zn ratios of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (biomineralization approach) and 35
1 (biomineralization approach) and 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (solid-state transformation approach) with final enzyme concentrations of 1.25 mg mL−1. After solutions were combined and aged for 24 hours without mixing, the samples were centrifuged and washed 3× with either water or methanol. Fourier transform-infrared (FTIR) spectroscopy confirmed the encapsulation of each enzyme for all CAT and GOx biocomposites through the presence of the amide carbonyl (amide I) stretch at 1660 cm−1, which was not present in ZIF-8 synthesized without protein (ESI Fig. S1†).3
1 (solid-state transformation approach) with final enzyme concentrations of 1.25 mg mL−1. After solutions were combined and aged for 24 hours without mixing, the samples were centrifuged and washed 3× with either water or methanol. Fourier transform-infrared (FTIR) spectroscopy confirmed the encapsulation of each enzyme for all CAT and GOx biocomposites through the presence of the amide carbonyl (amide I) stretch at 1660 cm−1, which was not present in ZIF-8 synthesized without protein (ESI Fig. S1†).3
At 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
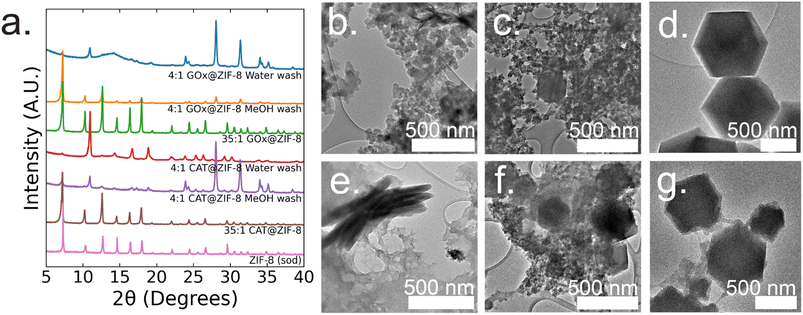
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 synthesis conditions with water washes, crystals have an encapsulation efficiency (EE%) of ∼100% (ESI Fig. S2†). Powder X-ray diffraction (P-XRD) verifies the product to be a heterogeneous mixture of both an amorphous phase, which can be observed in the broad peak around 14°, and crystalline ZIF-CO3-1 (ZIF-C), a ZIF-8 polymorph which incorporates CO2 into its crystal structure as carbonate ion.5,20 Dry-state TEM shows small, ∼20 nm, spherical particles along with sheets for 4
1 GOx@ZIF-8 synthesis conditions with water washes, crystals have an encapsulation efficiency (EE%) of ∼100% (ESI Fig. S2†). Powder X-ray diffraction (P-XRD) verifies the product to be a heterogeneous mixture of both an amorphous phase, which can be observed in the broad peak around 14°, and crystalline ZIF-CO3-1 (ZIF-C), a ZIF-8 polymorph which incorporates CO2 into its crystal structure as carbonate ion.5,20 Dry-state TEM shows small, ∼20 nm, spherical particles along with sheets for 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 (Fig. 1b). At 4
1 GOx@ZIF-8 (Fig. 1b). At 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 synthesis conditions with methanol washes, P-XRD verifies phase transformation of the ZIF-C crystals to ZIF-8 (sod) with no obvious amorphous phase present in the pattern. Dry-state TEM images indicate multiple phases in the final product; specifically, particles of ZIF-8 sod with rhombic dodecahedron morphologies and amorphous spherical particles are present (Fig. 1b). We believe that this phase transformation occurs as a result of methanol being a stronger coordinating solvent and displacing the carbonate ligands in ZIF-C.5,20 As the carbonate ligands are displaced, the coordination environment around the metal nodes changes, causing the structural rearrangement to sodalite topology with minimal coordination defects. At 35
1 GOx@ZIF-8 synthesis conditions with methanol washes, P-XRD verifies phase transformation of the ZIF-C crystals to ZIF-8 (sod) with no obvious amorphous phase present in the pattern. Dry-state TEM images indicate multiple phases in the final product; specifically, particles of ZIF-8 sod with rhombic dodecahedron morphologies and amorphous spherical particles are present (Fig. 1b). We believe that this phase transformation occurs as a result of methanol being a stronger coordinating solvent and displacing the carbonate ligands in ZIF-C.5,20 As the carbonate ligands are displaced, the coordination environment around the metal nodes changes, causing the structural rearrangement to sodalite topology with minimal coordination defects. At 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 synthesis conditions, crystals have a low EE% of 30% (ESI Fig. S2†). P-XRD verifies the absence of an amorphous phase and the presence of only sodalite (sod) with coordination defects; this is seen in the lower relative intensity of the first peak (visible around 7°) compared to the higher angle peaks. This is in contrast to pure sod, where the first peak is significantly more intense than all subsequent peaks (Fig. 1a). Dry-state TEM images show biocomposites with rhombic dodecahedron morphology, which is characteristic of sod, while also indicating the presence of both defects and polycrystallinity (Fig. 1d and ESI S8†).
1 GOx@ZIF-8 synthesis conditions, crystals have a low EE% of 30% (ESI Fig. S2†). P-XRD verifies the absence of an amorphous phase and the presence of only sodalite (sod) with coordination defects; this is seen in the lower relative intensity of the first peak (visible around 7°) compared to the higher angle peaks. This is in contrast to pure sod, where the first peak is significantly more intense than all subsequent peaks (Fig. 1a). Dry-state TEM images show biocomposites with rhombic dodecahedron morphology, which is characteristic of sod, while also indicating the presence of both defects and polycrystallinity (Fig. 1d and ESI S8†).
At 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CAT@ZIF-8 conditions with water washes, crystals have an EE% of 100% (ESI Fig. S2†). P-XRD verifies the product to be ZIF-C with a slight amorphous peak present. Dry-state TEM images show a continuous film for CAT@ZIF-8 along with stacked sheets found throughout the film (Fig. 1e). At 4
1 CAT@ZIF-8 conditions with water washes, crystals have an EE% of 100% (ESI Fig. S2†). P-XRD verifies the product to be ZIF-C with a slight amorphous peak present. Dry-state TEM images show a continuous film for CAT@ZIF-8 along with stacked sheets found throughout the film (Fig. 1e). At 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CAT@ZIF-8 with methanol washes, P-XRD verifies phase transformation of ZIF-C crystals to ZIF-8 (sod) with no obvious amorphous phase present in the pattern. Dry-state TEM shows multiple phases with concentrated regions of amorphous particles surrounding crystals with rhombic dodecahedron morphology (Fig. 1f). At 35
1 CAT@ZIF-8 with methanol washes, P-XRD verifies phase transformation of ZIF-C crystals to ZIF-8 (sod) with no obvious amorphous phase present in the pattern. Dry-state TEM shows multiple phases with concentrated regions of amorphous particles surrounding crystals with rhombic dodecahedron morphology (Fig. 1f). At 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CAT@ZIF-8, crystals have an EE% of 100%. P-XRD confirms no amorphous phase present and only sodalite (sod). Dry-state TEM images show biocomposites with rhombic dodecahedron morphology, and confirm the presence of coordination defects (Fig. 1g). SEM of both 4
1 CAT@ZIF-8, crystals have an EE% of 100%. P-XRD confirms no amorphous phase present and only sodalite (sod). Dry-state TEM images show biocomposites with rhombic dodecahedron morphology, and confirm the presence of coordination defects (Fig. 1g). SEM of both 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 35
1 and 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CAT@ZIF-8 confirm these results (ESI Fig. S3†).
1 CAT@ZIF-8 confirm these results (ESI Fig. S3†).
In summary, both CAT and GOx drive similar final crystal structures when varying the HmIM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn ratios. Both enzymes have low isoelectric points (<7); thus, the ability for each enzyme to drive the formation of a kinetic product when below the normal supersaturation conditions for ZIF-8 is expected. However, each enzyme drives the formation of kinetic products with different morphologies at 4
Zn ratios. Both enzymes have low isoelectric points (<7); thus, the ability for each enzyme to drive the formation of a kinetic product when below the normal supersaturation conditions for ZIF-8 is expected. However, each enzyme drives the formation of kinetic products with different morphologies at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 conditions with water washes. Additionally, CAT consistently becomes encapsulated into ZIF-8 at any HmIM
1 conditions with water washes. Additionally, CAT consistently becomes encapsulated into ZIF-8 at any HmIM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn ratio, whereas the EE% for GOx decreases with increasing HmIM
Zn ratio, whereas the EE% for GOx decreases with increasing HmIM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn.
Zn.
Enzyme activity in MOFs
Enzymatic activity assays were performed on GOx@ZIF-8 systems. The 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 was observed to have a slightly lower activity compared to the free enzyme (Fig. 2a), and the activity of GOx@ZIF-8 decreased even further when synthesized at 35
1 GOx@ZIF-8 was observed to have a slightly lower activity compared to the free enzyme (Fig. 2a), and the activity of GOx@ZIF-8 decreased even further when synthesized at 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 synthetic conditions. These results are supported by literature, which have found amorphous MOFs and MOFs with large defects to have greater activity than highly crystalline sod crystals.6,8 To investigate how phase transformation affects activity, activity measurements were then performed on the 4
1 synthetic conditions. These results are supported by literature, which have found amorphous MOFs and MOFs with large defects to have greater activity than highly crystalline sod crystals.6,8 To investigate how phase transformation affects activity, activity measurements were then performed on the 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 with methanol washes, and no activity was observed. Enzymatic activity assays were then performed on CAT@ZIF-8 systems. No enzymatic activity could be observed in any of the experimented synthetic conditions for CAT@ZIF-8 (Fig. 2b and ESI Fig. S4†).
1 GOx@ZIF-8 with methanol washes, and no activity was observed. Enzymatic activity assays were then performed on CAT@ZIF-8 systems. No enzymatic activity could be observed in any of the experimented synthetic conditions for CAT@ZIF-8 (Fig. 2b and ESI Fig. S4†).
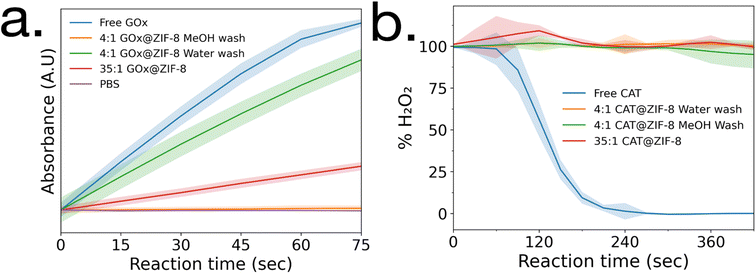 | ||
| Fig. 2 Enzyme activity assays of (a) GOx@ZIF-8 and (b) CAT@ZIF-8 at a variety of synthetic conditions. Standard deviations are shown as highlighted sections. | ||
Enzyme folding in prenucleation clusters and activity relationship
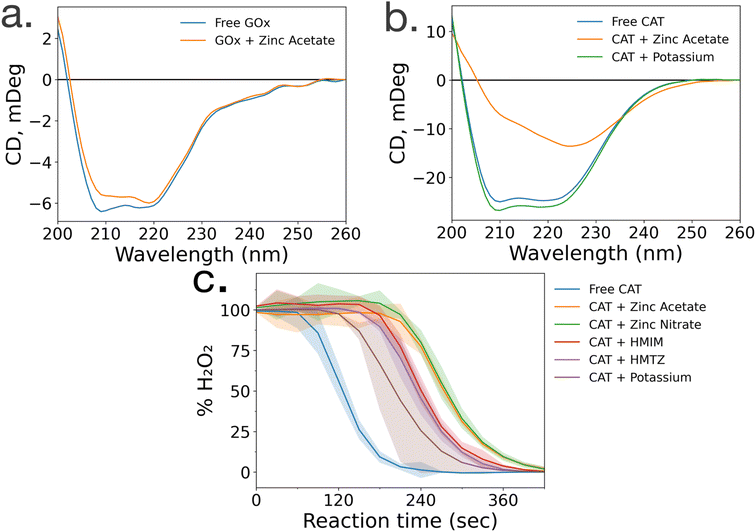
To gain insight into the lack of enzymatic activity in CAT@ZIF-8 systems, protein folding studies were undertaken. Characterization of the protein folding within MOF prenucleation clusters is challenging, as the species are transient and advanced spectroscopic techniques often require long scan times to achieve sufficient resolution, surpassing the lifespan of the transient species. Additionally, imidazole rings absorb light at 280 nm, the same wavelength as proteins, making it impossible to utilize light absorption techniques like UV-visible and circular dichroism (CD) spectroscopy to probe protein folding in the presence of HmIM.21,22 Instead, we used CD spectroscopy to examine enzyme folding in the presence of divalent zinc (Zn), as previous studies have found that proteins can form stable complexes with each of the precursors.17 CD spectroscopy shows that GOx/Zn solutions exhibit a slight reduction in alpha helical character as compared to GOx without Zn (Fig. 3a). In contrast, CD spectra of CAT/Zn solutions show a dramatic decrease in alpha helical character and increase in beta sheet character, indicating general denaturation of the biomolecule (Fig. 3b).As a control, we compared the CD spectra of free CAT and CAT/Zn mixture to CAT in the presence of potassium acetate, to determine that stability of CAT was dependent on the specific metal cation and not the presence of any counterion, potassium is a common biological cation found in high concentrations in most living systems, unlike zinc which is usually found in low concentrations; as such, potassium is expected to lead to very little denaturation.23 Furthermore, CAT activity has been reported in a potassium-based MOF system in a recent publication by Di Palma et al.24 The folding of CAT incubated with zinc acetate and potassium acetate was examined via CD spectroscopy, which showed that potassium acetate altered the secondary structure of the protein minimally compared to zinc acetate (Fig. 3b). Based on these results, we propose that the Zn2+ ions cause unfolding in prenucleation clusters, not the counterions.
While CD studies are useful when determining unfolding in specific enzymatic secondary structural elements, these structural changes do not necessarily correspond to changes in activity. To better understand the effects of MOF precursors on CAT activity, enzymatic assays were performed with CAT incubated with HmIM or zinc acetate, with potassium acetate as a control. CAT incubated with potassium acetate had the best activity, followed by CAT incubated with HmIM, then finally zinc acetate (Fig. 3c). While we note that these studies only deal with prenucleation clusters and not the whole MOF, our findings support previous studies which showed that CAT exhibits excellent activity when confined in a potassium-based MOF.24 Furthermore, the activity and folding of CAT with MAF-7 – which shares the zinc metal ion and sodalite topology of ZIF-8 but uses a more hydrophilic linker, 3-methyl-1H-1,2,4 triazole (HmTz)18 – precursors were also investigated. MAF-7 was investigated specifically because it shares many features with ZIF-8, yet encapsulated CAT shows activity (ESI Fig. S9†). While MAF-7 and ZIF-8 share zinc metal ions, the metal salts used are different, with ZIF-8 being commonly made with zinc acetate and MAF-7 being commonly made with zinc nitrate. Activity assays confirmed CAT incubated with either salt to have similar activity (Fig. 3c). CAT was then incubated with either ligand, HmIM or HmTz, and enzymatic activity was also similar with each ligand.
Mechanistic and structural relationships to enzyme activity
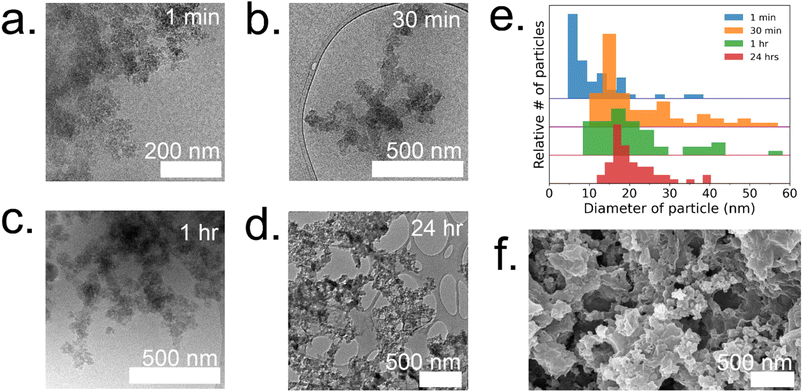
To gain a deeper understanding of the evolution of each enzyme@metal–organic framework (MOF) and to eventually customize their properties, time-resolved cryoTEM analysis was conducted on the GOx@ZIF-8 system during the initial hour of crystallization. The choice of GOx@ZIF-8 systems was motivated by the fact that each synthetic condition yields a distinct enzymatic outcome, unlike CAT@ZIF-8, which exhibits zero activity at any synthetic condition. In situ time-resolved cryoTEM was utilized to image the crystal formation reaction and found small irregular amorphous aggregate networks at 1 minute (Fig. 4a). The aggregates then evolved into larger particulates by 30 minutes (Fig. 4b). The particles appear to be stable at 1 h (Fig. 4c) and remain in the sample after 24 h (Fig. 4d and g). Through particle size analysis, the particles at 1 min averaged ∼10 ± 7 nm while the particles at 30 min, 1 h, and 24 h averaged ∼20 ± 10 nm (Fig. 4d and ESI Fig. S6†). While these particles are hypothesized to be amorphous based on the large amorphous backbone in the P-XRD, the ZIF-C region is believed to be the dense sheet-like region surrounded by the amorphous particles (Fig. 4d and f).5 This is consistent with previous findings on the published morphology and crystal structure of ZIF-C, which – much like the sheet-like polymorph dia – consists of Zn-HmIM sheets, but instead of being bridged by HmIM like in dia, ZIF-C's sheets are bridged by carbonate ligands.5,25To understand how the mechanistic and structural properties of the 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 differentiated from the previous system, we performed similar in situ time-resolved cryoTEM studies. CryoTEM images reveal that at 1 minute, aggregates similar to the structure found in the 4
1 GOx@ZIF-8 differentiated from the previous system, we performed similar in situ time-resolved cryoTEM studies. CryoTEM images reveal that at 1 minute, aggregates similar to the structure found in the 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
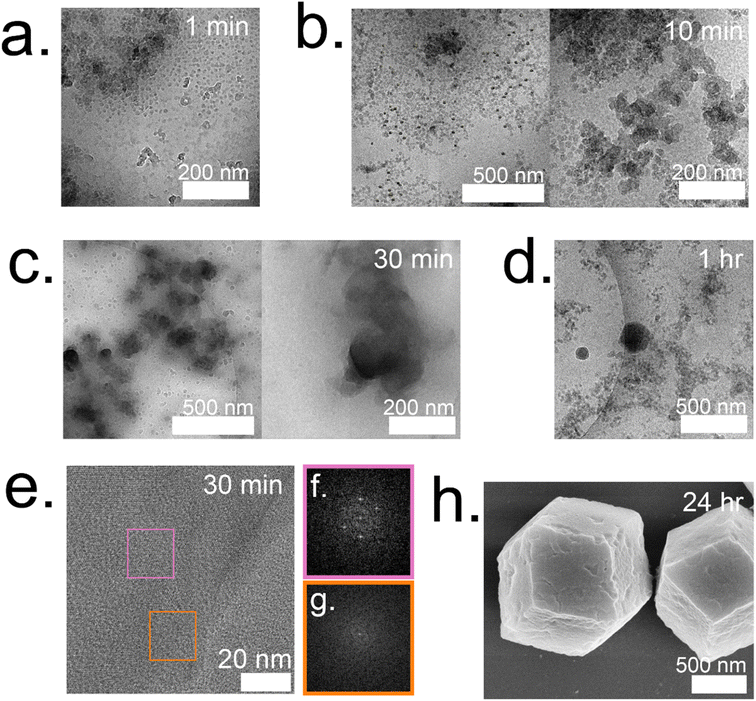
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 system form and are surrounded by individual particles that are 12 ± 2 nm (Fig. 5a). These particles continue to grow and double in size (21 ± 4 nm) by 10 min (Fig. 5b). At 10 minutes, these particles are localized and surround a highly concentrated region. The amorphous phases continue to aggregate and condense to form dense phases surrounded by a cloud, which is hypothesized to serve as a rich reservoir of MOF precursors for the growing dense phase (Fig. 5c). Outlines of irregular crystals can be observed within these clouds and closer inspection into the regions with high-resolution TEM reveals crystallinity as indicated by the lattice spacing (Fig. 5e) and fast Fourier transform (FFT) (Fig. 5f).13 The crystal appears poorly crystalline at 30 min with lattices appearing in multiple directions along with gaps without lattices (Fig. 5g). These gaps in the lattice are believed to be coordination defects containing enzymes. By 1 hour, well-defined rhombic dodecahedron crystals that are believed to be monocrystalline can be observed along with branched networks of aggregates dispersed throughout the sample (Fig. 5d and ESI S7†). As commonly observed with this system, amorphous phases contributing to the crystal growth undergo solid-state transformation to achieve a monocrystalline structure. While topological and structural characteristics of ZIF-8 (sod) are observed after 24 h, indentions and pores can also be observed on the surface of the crystals (Fig. 5h). Highly porous, irregular crystals can also be observed after 24 h in addition to the lesser porous, regular crystals (ESI Fig. S8†).
1 system form and are surrounded by individual particles that are 12 ± 2 nm (Fig. 5a). These particles continue to grow and double in size (21 ± 4 nm) by 10 min (Fig. 5b). At 10 minutes, these particles are localized and surround a highly concentrated region. The amorphous phases continue to aggregate and condense to form dense phases surrounded by a cloud, which is hypothesized to serve as a rich reservoir of MOF precursors for the growing dense phase (Fig. 5c). Outlines of irregular crystals can be observed within these clouds and closer inspection into the regions with high-resolution TEM reveals crystallinity as indicated by the lattice spacing (Fig. 5e) and fast Fourier transform (FFT) (Fig. 5f).13 The crystal appears poorly crystalline at 30 min with lattices appearing in multiple directions along with gaps without lattices (Fig. 5g). These gaps in the lattice are believed to be coordination defects containing enzymes. By 1 hour, well-defined rhombic dodecahedron crystals that are believed to be monocrystalline can be observed along with branched networks of aggregates dispersed throughout the sample (Fig. 5d and ESI S7†). As commonly observed with this system, amorphous phases contributing to the crystal growth undergo solid-state transformation to achieve a monocrystalline structure. While topological and structural characteristics of ZIF-8 (sod) are observed after 24 h, indentions and pores can also be observed on the surface of the crystals (Fig. 5h). Highly porous, irregular crystals can also be observed after 24 h in addition to the lesser porous, regular crystals (ESI Fig. S8†).
Discussion
Enzyme folding and activity studies emphasize the importance of the biological relationship between structure and function when designing a successful biocomposite. As GOx is resistant to unfolding by zinc, it is able to remain in an active conformation and exhibit activity in ZIF-8 (Fig. 3a). CAT, however, is not resistant to unfolding by zinc, and therefore, no active CAT@ZIF-8 complex can be formed. Considering these results, we hypothesize that enzymes must be resistant to unfolding in prenucleation clusters to be active in MOF biocomposites.One way to prevent metal-induced unfolding from occurring is by using a hydrophilic ligand. Previous studies and our own work have shown CAT to remain active in the zinc-based MAF-7 (ESI Fig. S9†),18 which we believe is attributed to the ability of the more hydrophilic ligand to stabilize an active protein conformation. While CAT exhibited activity when incubated with both HmTz or HmIM, we hypothesize that HmTz, with an additional nitrogen group (ESI Fig. S10†), will exhibit enhanced binding strength to the protein complex, while the two nitrogen groups in HmIM are more likely to bind to zinc. Supporting this hypothesis, the solution of HmTz and CAT becomes turbid rapidly, indicating large aggregates, whereas the solution of HmIM and CAT remains clear (ESI Fig. S5†). Importantly, these CAT/HmTz complexes still retain somewhat high activity, meaning that significant protein denaturation is not occurring (Fig. 2c). We suggest that these strong protein/HmTz complexes stabilize CAT in a more active conformation and protect it against denaturation by zinc. Additionally, previous studies have demonstrated that the addition of hydrophilic groups to organic ligands can influence the restoration of the active protein conformation in final MOF crystals. These studies have shown that hydrophilic groups facilitate hydration effects within MOF pores, which reduce chelation interactions between the metal and the protein, thereby preventing unfolding.26
The understanding of how each individual MOF precursor and their combination affects enzyme folding and orientation in initial complexes is indeed crucial when initially designing a biocomposite. This knowledge helps in determining the appropriate enzyme/MOF precursor pairing for a specific application. By studying the interactions between the enzyme and different MOF precursors, researchers can identify compatible combinations that promote favorable enzyme folding and orientation within the MOF framework. However, in some cases, a desired enzyme/MOF precursor pairing may be inherently incompatible. In such situations, extra steps can be employed to induce an active enzyme conformation, like the addition of a hydrophilic ligand,18,26 or protecting polymer,27 along with subjecting the protein to ultrasound waves.28 Whatever the case, all techniques and enzyme@MOF synthesis processes must take into account the effect of MOF precursors on the stability of the enzyme in prenucleation clusters. However, this is only the first step in ensuring enzyme@MOF activity.
Recently, Wu et al. have found that GOx packaged in amorphous MOFs (aMOFs) exhibits high enzymatic activity due to large defects and interconnected mesopores, allowing access to substrate by enzymes inside of the composite.8 While defects are present in our aMOFs, the particles are 5× smaller (∼20 nm) than the aMOFs in the previously mentioned study (100 nm). As GOx has an average length of ∼6 nm and the aMOFs are only ∼20 nm, a large surface of the enzyme is likely to be exposed to the particle surface and accessible to substrate.29 These findings support that the activity of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 is partially due to the lower diffusion barrier that the substrate must overcome to enter the biocomposite and reach the enzyme. While GOx may be present on the surface of 4
1 GOx@ZIF-8 is partially due to the lower diffusion barrier that the substrate must overcome to enter the biocomposite and reach the enzyme. While GOx may be present on the surface of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8, it is also immobilized inside the MOF, as confirmed through FTIR analysis of biocomposites washed with sodium dodecyl sulfate, a detergent known to remove surface-bound protein (ESI Fig. S11†). Furthermore, along with EE% measurements that assess the overall enzyme content in an enzyme@MOF, loading capacity (LC%) studies were performed to quantify the concentration of encapsulated enzymes within the MOF interior. LC% studies determined that 4
1 GOx@ZIF-8, it is also immobilized inside the MOF, as confirmed through FTIR analysis of biocomposites washed with sodium dodecyl sulfate, a detergent known to remove surface-bound protein (ESI Fig. S11†). Furthermore, along with EE% measurements that assess the overall enzyme content in an enzyme@MOF, loading capacity (LC%) studies were performed to quantify the concentration of encapsulated enzymes within the MOF interior. LC% studies determined that 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 contains significant GOx content within the amorphous structure, while 35
1 GOx@ZIF-8 contains significant GOx content within the amorphous structure, while 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 contains almost no GOx within the crystalline structure. Thus, for 35
1 GOx@ZIF-8 contains almost no GOx within the crystalline structure. Thus, for 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8, most of the enzyme concentration determined by EE% is immobilized either on the surface or just under the surface of the MOF (ESI Table 1†).7
1 GOx@ZIF-8, most of the enzyme concentration determined by EE% is immobilized either on the surface or just under the surface of the MOF (ESI Table 1†).7
While the precise mechanism of ZIF-C formation remains somewhat elusive throughout the in situ cryoTEM studies, it is hypothesized to occur through the initial aggregation followed by the subsequent rearrangement of amorphous particles. While the inclusion of ZIF-C, a polymorph that has been reported in solvothermal synthesis,20 in biomimetically mineralized CAT and GOx ZIF-8 composites is a novel result, we hypothesize that the ZIF-C composites are not active, and activity comes from aMOFs present in the sample. Previous studies have shown that ZIF-C is not porous to N2, making it unlikely that the composite is porous to glucose, the substrate of GOx, which is larger and more polar.5 Thus, we believe the smaller size of the aMOFs, coupled with the disordered aMOF, to be the major factors in driving the high activity for this system, and the ZIF-C samples to be much less relevant in conferring activity. This is confirmed by comparing enzymatic activity assays of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 aged at 1 h (before ZIF-C has formed) and 24 h (after some ZIF-C has formed). The 24 h aged 4
1 GOx@ZIF-8 aged at 1 h (before ZIF-C has formed) and 24 h (after some ZIF-C has formed). The 24 h aged 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 exhibited lower activity than the 1 h aged 4
1 GOx@ZIF-8 exhibited lower activity than the 1 h aged 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 (ESI Fig. S12†), which supports a direct correlation between amorphous character and enzyme activity. We believe such amorphous characteristics also influence the activity of the 35
1 GOx@ZIF-8 (ESI Fig. S12†), which supports a direct correlation between amorphous character and enzyme activity. We believe such amorphous characteristics also influence the activity of the 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 biocomposite. Irregular crystals observed through TEM are hypothesized to have remained polycrystalline with coordination defects within them. As the size of glucose (∼10 Å) is much larger than the 3.4 Å window typically observed in ZIF-8 (sod),6,13,30 such defects are required for accessibility of enzymes to substrate. By increasing the enzyme loading concentration, we can expect to have an increase in amorphous character and defects in the crystal resulting in greater activity.6,15 Furthermore, such a system is expected to have some enzymatic activity as a result of the defects, but not as high as the free enzyme or at 4
1 GOx@ZIF-8 biocomposite. Irregular crystals observed through TEM are hypothesized to have remained polycrystalline with coordination defects within them. As the size of glucose (∼10 Å) is much larger than the 3.4 Å window typically observed in ZIF-8 (sod),6,13,30 such defects are required for accessibility of enzymes to substrate. By increasing the enzyme loading concentration, we can expect to have an increase in amorphous character and defects in the crystal resulting in greater activity.6,15 Furthermore, such a system is expected to have some enzymatic activity as a result of the defects, but not as high as the free enzyme or at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 biocomposites. This claim is supported by our enzymatic assay results, which show low but present activity for the 35
1 GOx@ZIF-8 biocomposites. This claim is supported by our enzymatic assay results, which show low but present activity for the 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 sample (Fig. 2a). However, when enzymatic activity assays of 35
1 GOx@ZIF-8 sample (Fig. 2a). However, when enzymatic activity assays of 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 were replicated in Tris–HCl buffer, the biocomposites showed a marked decrease in activity, as opposed to 4
1 GOx@ZIF-8 were replicated in Tris–HCl buffer, the biocomposites showed a marked decrease in activity, as opposed to 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 which retained high activity (ESI Fig. S13†). This observation can be supported by previous studies which have reported the phosphate anions in PBS to degrade crystalline ZIF-8.31 Additionally, these enzyme activities studies, along with the aforementioned loading capacity studies (ESI Table 1†), indicate that 35
1 GOx@ZIF-8 which retained high activity (ESI Fig. S13†). This observation can be supported by previous studies which have reported the phosphate anions in PBS to degrade crystalline ZIF-8.31 Additionally, these enzyme activities studies, along with the aforementioned loading capacity studies (ESI Table 1†), indicate that 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 requires defects to showcase activity of enzymes that are immobilized just under the MOF surface.15 On the other hand, this data suggests 4
1 GOx@ZIF-8 requires defects to showcase activity of enzymes that are immobilized just under the MOF surface.15 On the other hand, this data suggests 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 does not require PBS degradation to showcase activity as the biocomposite contains pores large enough for glucose to reach the enzyme.8
1 GOx@ZIF-8 does not require PBS degradation to showcase activity as the biocomposite contains pores large enough for glucose to reach the enzyme.8
While outside of the scope of this study, another factor that could influence the activity of 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 is the orientation of the enzymes located within or near the defects. A study by Pan et al. used electron paramagnetic resonance spectroscopy to investigate the orientation of metalloenzymes within MOF surfaces.10 Although these enzymes were not immobilized onto the MOF in a similar mechanism, the idea of being able to control the orientation and exposure of enzymes to substrates would allow enhanced selectivity of substrates of varying sizes.
1 GOx@ZIF-8 is the orientation of the enzymes located within or near the defects. A study by Pan et al. used electron paramagnetic resonance spectroscopy to investigate the orientation of metalloenzymes within MOF surfaces.10 Although these enzymes were not immobilized onto the MOF in a similar mechanism, the idea of being able to control the orientation and exposure of enzymes to substrates would allow enhanced selectivity of substrates of varying sizes.
This work focuses on the effect that MOF precursors have on enzymatic folding and activity. Moreover, biomolecular unfolding from precursor interactions can additionally affect the formation mechanism and final MOF structure, which has been demonstrated in an FITC-BSA@ZIF-8 model.17,32 Furthermore, while the amorphous structure that we report in 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 aligns with some reports that have also observed amorphous ZIF-8 at this condition,8 other studies have also reported crystalline sod products at the same L
1 GOx@ZIF-8 aligns with some reports that have also observed amorphous ZIF-8 at this condition,8 other studies have also reported crystalline sod products at the same L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M ratio.6 We suggest that these differences may be due to the differing stabilities, structures, and purities of GOx obtained by different laboratories. Inherent to any biomolecular study is the understanding that many biomolecules – even those purchased from the same retailer and produced by the same organism line – are not structurally and compositionally identical, and studies on protein folding offer an opportunity to bridge this gap. The study of enzyme@MOFs is an inherently interdisciplinary field, and while our study only focuses on two enzymes, we believe that we have established a new protocol to take into consideration when designing and producing an enzyme@MOF.
M ratio.6 We suggest that these differences may be due to the differing stabilities, structures, and purities of GOx obtained by different laboratories. Inherent to any biomolecular study is the understanding that many biomolecules – even those purchased from the same retailer and produced by the same organism line – are not structurally and compositionally identical, and studies on protein folding offer an opportunity to bridge this gap. The study of enzyme@MOFs is an inherently interdisciplinary field, and while our study only focuses on two enzymes, we believe that we have established a new protocol to take into consideration when designing and producing an enzyme@MOF.
Conclusions
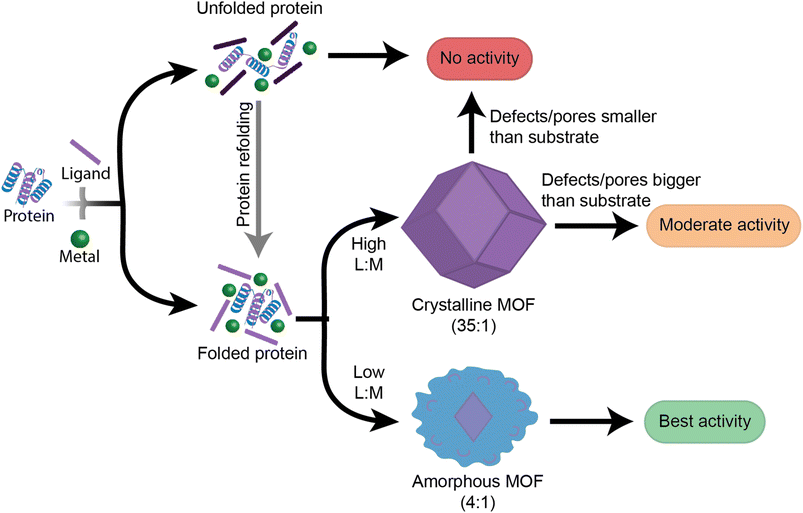
This work provides insight into new, as well as previously established, factors that should be followed when designing high performance enzyme@MOF biocomposites (Fig. 6). While no single MOF is suitable for immobilizing every enzyme, we have demonstrated that studying the folding and activity of the enzymes within stable clusters using each MOF precursor is a key initial screening of compatible precursors. Additionally, stabilizing interactions between precursors and enzymes are key to understanding the final activity in a MOF. Once precursors pass the initial screening with MOF precursors, the MOF design can then be redirected towards manipulating the crystal formation mechanism so that the final physical properties of the crystal align with the desired applications. We aim that providing this step-by-step flowchart emphasizes the role of enzyme folding in prenucleation clusters and will lead to exponential growth in the design and use of high performance protein-MOFs composites. Future studies should take a more enzyme-oriented approach to enzyme@MOF synthesis, focusing on the real-time folding of biomolecules in prenucleation clusters and the enzymatic structural changes that take place once an enzyme is encapsulated into a MOF.Experimental
Materials
2-Methylimidazole, zinc acetate dihydrate, tetrasodium ethylenediaminetetraacetic acid, and bovine liver catalase were purchased from Sigma Aldrich. Glucose oxidase from A. niger, concentrated sulfuric acid, and potassium acetate were purchased from VWR international. Horseradish peroxidase, methanol, dimethyl sulfoxide, and 30% hydrogen peroxide were obtained from ThermoFisher. 3-Methyl-1H-1,2,4-triazole and 3,3′,5,5′-tetramethylbenzidine were purchased from TCI chemicals. Zinc nitrate dihydrate, xylenol orange, sodium phosphate dibasic anhydrous, phosphate buffered saline 10×, and ferrous ammonium sulfate were purchased from Strem Chemicals, Abcam, Chem-Impex International, Boston Bioproducts, and EMD Serono respectively.GOx@ZIF-8 and CAT@ZIF-8 synthesis
Individual solutions of 2-methylimidazole (HmIM) (2800 mM and 320 mM, 0.750 mL), zinc acetate (Zn) (40 mM), and enzyme (5 mg mL−1, variable volume) were prepared in Milli-Q water (18 MΩ). For the high HmIM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn conditions (35
Zn conditions (35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), the more concentrated solution of HmIM was used to achieve a final HmIM
1), the more concentrated solution of HmIM was used to achieve a final HmIM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn concentration ratio of 700
Zn concentration ratio of 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 mM. For low HmIM
20 mM. For low HmIM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn conditions (4
Zn conditions (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), the less concentrated solution of HmIM was used to achieve a final HmIM
1), the less concentrated solution of HmIM was used to achieve a final HmIM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn concentration ratio of 80
Zn concentration ratio of 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 mM. The final enzyme concentration in each solution was 1.25 mg mL−1. To synthesize the enzyme@MOF composite, 750 μL of HmIM solution was added to 750 μL of enzyme solution, after which 1.5 mL of zinc solution was added in a glass vial, and the reaction was aged for 24 h without stirring. After 24 h, the precipitate was retrieved by centrifuging the reaction at 10
20 mM. The final enzyme concentration in each solution was 1.25 mg mL−1. To synthesize the enzyme@MOF composite, 750 μL of HmIM solution was added to 750 μL of enzyme solution, after which 1.5 mL of zinc solution was added in a glass vial, and the reaction was aged for 24 h without stirring. After 24 h, the precipitate was retrieved by centrifuging the reaction at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. While the supernatant was separated and stored for EE% measurements, the precipitates were washed 3× with either water or methanol prior to further characterization. For the non-crystalline 4
000 rpm. While the supernatant was separated and stored for EE% measurements, the precipitates were washed 3× with either water or methanol prior to further characterization. For the non-crystalline 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GOx@ZIF-8 sample, the reaction was aged for 1 h, but all other steps remained consistent.
1 GOx@ZIF-8 sample, the reaction was aged for 1 h, but all other steps remained consistent.
Enzymatic activity of catalase
The ferrous oxidation-xylenol orange (FOX) assay was used to measure the enzymatic activity of both catalase and CAT@ZIF-8 systems. The FOX reagent was made using a solution of methanol (180 mL), Milli-Q water (20 mL), and sulfuric acid 98% (278 μL). After stirring to ensure the solution was homogeneous, ferrous ammonium sulfate (19.6 mg) and then xylenol orange (15.2 mg) were added in their solid forms to the solution and allowed to stir for 1 h to ensure complete dissolution. Solutions of catalase and CAT@ZIF-8 systems were diluted to 0.01 mg mL−1 in 1× phosphate buffered saline (PBS), to a final volume of 2 mL. To initiate the reaction, 10 μL of 30% hydrogen peroxide was added to 2 mL of CAT or CAT@ZIF-8 system while stirring (320 rpm). Aliquots of the reaction mixture (50 μL) were taken, first at 15 seconds after the beginning of the reaction, then every 30 seconds, and added to 950 μL of FOX reagent. The absorbance of the samples were then measured at 560 nm using a Nanodrop absorption spectrophotometer, and plotted using the matplotlib, pandas, scipy, and numpy python libraries in an author-written python script.Enzymatic activity of glucose oxidase
An enzymatic assay based on the oxidation of tetramethylbenzidine (TMB) by horseradish peroxidase (HRP) was employed to measure the activity of glucose oxidase (GOx). First, a phosphate-citrate buffer was prepared by dissolving sodium phosphate dibasic anhydrous (8.15 mg mL−1) and citric acid monohydrate (9.60 mg mL−1) into nanopure water under vigorous stirring to ensure complete dissolution. Then, a solution of TMB (0.1 mg mL−1) was prepared in this buffer by first dissolving TMB (10 mg) into dimethyl sulfoxide (10 mL), stirring to fully dissolve the indicator. This solution was then diluted 10× in the previously synthesized phosphate-citrate buffer. A solution of HRP (5 mg mL−1) was then prepared in this TMB buffer, and diluted 100× with more TMB buffer to a final concentration of 0.05 mg mL−1. EE% measurements were used to dilute each bio-MOF composite to 2 mL of 0.015 mg mL−1 GOx in PBS, based on the assumption that all GOx not present in the supernatant was present in the final bio-MOF composite. 200 μL of this bio-MOF PBS solution were added to 400 μL of the HRP/TMB solution in a plastic cuvette, and then 200 μL of a 10 mM aqueous glucose solution were added to start the reaction. The reaction was measured at 650 nm every 15 seconds over 300 seconds in a Nanodrop UV-visible spectrophotometer using the “kinetics” mode. Results were plotted using the pandas and pyplot python libraries in an author-written python script.Circular dichroism
Solutions of enzymes and enzymes with MOF precursors were made at a concentration of 0.25 mg mL−1, and placed into a quartz cuvette with 1 cm pathlength. Measurements for catalase were taken in triplicate in the 200 to 260 nm wavelength range with 1 nm bandwidth and 1 nm scanning intervals with integration time of 0.75 s on a Chirascan V100 circular dichroism spectrometer from applied photophysics at 20 °C. The circular dichroism data for GOx was collected using a Jasco J-810 circular dichroism spectrometer. Results were smoothed and plotted using the pandas, numpy, scipy and matplotlib python modules respectively in an author-written python script. Plots were adjusted via array addition and subtraction so absorbance values at 260 nm read zero, and smoothed using a 1d Gaussian filter with a sigma value of 0.8.TEM
Dry-state TEM samples were prepared by diluting each sample 10× in nanopure water. Samples were then pipetted onto either 400 mesh carbon grids from TedPella or Quantifoil R2/2 Holey Carbon from electron microscopy sciences. A JEOL-2100 TEM equipped with a Schottky type field emission gun was used to image samples. SerialEM software was used to obtain samples using a Gatan Oneview camera.CryoTEM
CryoTEM samples were pipetted onto 400 mesh carbon grids from TedPella or Quantifoil R2/2 holey carbon from electron microscopy sciences. Prior to sample application, the grids underwent glow discharge for 70 seconds to enhance grid hydrophilicity and spreading of sample. Time-resolved samples were taken at 1 minute, 30 minutes, and 1 h were samples and centrifuged for 5 seconds where the supernatants were then vitrified using an Automatic Plunge Freezer EMGP2 (Leica Microsystems) at each time point. The plunger was set to 95% humidity in the sample chamber and blotted for 2.5 s prior to auto-plunging into liquid propane. A JEOL-2100 TEM equipped with a Schottky type field emission gun was used to image samples. SerialEM software in low dose imaging mode was used to obtain samples using a Gatan K3 and a Gatan Oneview camera.SEM
Samples (10 μL) were pipetted onto 1 mm glass slides. Prior imaging, samples were coated with 5–10 nm iridium (Quorum Q150T) to combat charging effects. A Magellan 400 XRH system was used to obtain secondary electrons images while operating at an accelerating voltage of 5 kV.Intrinsic tryptophan fluorescence
Intrinsic tryptophan fluorescence was performed on the MOF washing supernatant in a manner similar to earlier published works on bio-MOFs to determine encapsulation efficiency (EE%)10,11 first, supernatants were diluted 10× in EDTA-PBS, which was prepared by making a 16.88 mg mL−1 solution of ethylenediaminetetraacetate tetrasodium salt (EDTA) in phosphate buffered saline (PBS). This diluted supernatant was measured using a Cary-60 UV-visible fluorimeter, exciting at 280 nm and measuring the fluorescence at 340 nm. The EE% was determined by referencing measured values to the slope of a previously prepared calibration curve, which was validated with a Bradford assay for GOx (ESI Fig. S14†).Powder X-ray diffraction
Samples were first dried in a vacuum oven with heating turned off to ensure proper dehydration of the bio-MOF composite. Depending on the sample being run, 3.9 mg of a zirconium(iv) oxide standard was added to each sample before grinding. These samples were crushed into a fine powder using a mortar and pestle, then placed on a zero-background crystal attachment and diffracted using a Rigaku Ultima III X-ray diffractometer in Bragg–Brentano mode, measuring from 5–50 degrees at variable scan speeds. The data was plotted using the matplotlib, numpy, and pandas python libraries in an author-written python script.Author contributions
J. P. P. conceived and supervised the study. B. P. C. and B. R. equally contributed to the manuscript regarding ideas, investigation, data analysis, and writing. E. M. O aided in SEM imaging and imaging analysis as well as in enzyme activity measurements. M. X. N. and G. P. aided in establishing the methodology of enzyme activity assays. A. R. T. aided in SEM imaging. P. J. H. aided in performing cryoTEM imaging. L. X. aided in high-resolution cryoTEM imaging and image analysis. R. G. aided in circular dichroism and UV-visible measurements. S. M. C. provided resources for circular dichroism and UV-visible measurements and aided in data processing. All authors contributed toward writing and editing the manuscript, and all approved of the final version.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was primarily funded by the National Science Foundation CBET Award #2102033. This research was partially funded by the University of California Irvine Undergraduate Research Opportunities Program (UROP) and Beckman Scholars Program (BSP). The authors acknowledge the use of a circular dichroism spectrometer acquired with support from AFOSR DURIP FA9550-22-1-0206. Furthermore, the authors acknowledge the use of facilities and instrumentation at the UC Irvine Materials Research Institute (IMRI), which is supported in part by the National Science Foundation through the UC Irvine Materials Research Science and Engineering Center (DMR-2011967). The authors further acknowledge Professor Jennifer A. Prescher and Professor Kenneth J. Shea for the use of their laboratories for enzymatic assays. The authors also acknowledge Professor Dmitry Fishman for the use of his facility for fluorescence spectroscopy and FTIR spectroscopy experiments. The authors further acknowledge the teams behind the matplotlib, pandas, numpy, and scipy python libraries for the use of these libraries for figure creation and data analysis.References
- H. D. Lawson, S. P. Walton and C. Chan, ACS Appl. Mater. Interfaces, 2021, 13, 7004–7020 CrossRef CAS.
- H. Zheng, Y. Zhang, L. Liu, W. Wan, P. Guo, A. M. Nyström and X. Zou, J. Am. Chem. Soc., 2016, 138, 962–968 CrossRef CAS PubMed.
- K. Liang, R. Ricco, C. M. Doherty, M. J. Styles, S. Bell, N. Kirby, S. Mudie, D. Haylock, A. J. Hill, C. J. Doonan and P. Falcaro, Nat. Commun., 2015, 6, 7240 CrossRef CAS.
- H. Furukawa, K. E. Cordova, M. O'Keeffe and O. M. Yaghi, Science, 2013, 341, 1230444 CrossRef.
- F. Carraro, M. de J. Velásquez-Hernández, E. Astria, W. Liang, L. Twight, C. Parise, M. Ge, Z. Huang, R. Ricco, X. Zou, L. Villanova, C. O. Kappe, C. Doonan and P. Falcaro, Chem. Sci., 2020, 11, 3397–3404 RSC.
- L. Tong, S. Huang, Y. Shen, S. Liu, X. Ma, F. Zhu, G. Chen and G. Ouyang, Nat. Commun., 2022, 13, 951 CrossRef CAS PubMed.
- P. Li, S.-Y. Moon, M. A. Guelta, L. Lin, D. A. Gómez-Gualdrón, R. Q. Snurr, S. P. Harvey, J. T. Hupp and O. K. Farha, ACS Nano, 2016, 10, 9174–9182 CrossRef CAS.
- X. Wu, H. Yue, Y. Zhang, X. Gao, X. Li, L. Wang, Y. Cao, M. Hou, H. An, L. Zhang, S. Li, J. Ma, H. Lin, Y. Fu, H. Gu, W. Lou, W. Wei, R. N. Zare and J. Ge, Nat. Commun., 2019, 10, 5165 CrossRef PubMed.
- S. Pyreddy, A. Poddar, F. Carraro, S. A. Polash, C. Dekiwadia, B. Murdoch, Z. Nasa, T. S. Reddy, P. Falcaro and R. Shukla, Biomater. Adv., 2023, 149, 213420 CrossRef CAS.
- Y. Pan, H. Li, J. Farmakes, F. Xiao, B. Chen, S. Ma and Z. Yang, J. Am. Chem. Soc., 2018, 140, 16032–16036 CrossRef CAS PubMed.
- X. Wang, P. C. Lan and S. Ma, ACS Cent. Sci., 2020, 6, 1497–1506 CrossRef CAS.
- D. Fernando, M. Mathesh, J. Cai and W. Yang, Langmuir, 2023, 39, 7979–7985 CrossRef CAS PubMed.
- K. S. Park, Z. Ni, A. P. Côté, J. Y. Choi, R. Huang, F. J. Uribe-Romo, H. K. Chae, M. O'Keeffe and O. M. Yaghi, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10186–10191 CrossRef CAS.
- M. Jian, B. Liu, R. Liu, J. Qu, H. Wang and X. Zhang, RSC Adv., 2015, 5, 48433–48441 RSC.
- A. F. Ogata, A. M. Rakowski, B. P. Carpenter, D. A. Fishman, J. G. Merham, P. J. Hurst and J. P. Patterson, J. Am. Chem. Soc., 2020, 142, 1433–1442 CrossRef CAS PubMed.
- N. K. Maddigan, A. Tarzia, D. M. Huang, C. J. Sumby, S. G. Bell, P. Falcaro and C. J. Doonan, Chem. Sci., 2018, 9, 4217–4223 RSC.
- B. P. Carpenter, A. R. Talosig, J. T. Mulvey, J. G. Merham, J. Esquivel, B. Rose, A. F. Ogata, D. A. Fishman and J. P. Patterson, Chem. Mater., 2022, 34, 8336–8344 CrossRef CAS PubMed.
- W. Liang, H. Xu, F. Carraro, N. K. Maddigan, Q. Li, S. G. Bell, D. M. Huang, A. Tarzia, M. B. Solomon, H. Amenitsch, L. Vaccari, C. J. Sumby, P. Falcaro and C. J. Doonan, J. Am. Chem. Soc., 2019, 141, 2348–2355 CrossRef CAS PubMed.
- N. Német, Y. Miele, G. Shuszter, E. L. Tóth, J. E. Maróti, P. J. Szabó, F. Rossi and I. Lagzi, React. Kinet., Mech. Catal., 2022, 135, 15–28 CrossRef.
- S. A. Basnayake, J. Su, X. Zou and K. J. Jr Balkus, Inorg. Chem., 2015, 54, 1816–1821 CrossRef CAS PubMed.
- M. F. Pignataro, M. G. Herrera and V. I. Dodero, Molecules, 2020, 25, 4854 CrossRef CAS.
- S. M. Kelly, T. J. Jess and N. C. Price, Biochim. Biophys. Acta, 2005, 1751, 119–139 CrossRef CAS.
- M. Stillman, Angew. Chem., Int. Ed., 2007, 46, 8741–8742 CrossRef CAS.
- G. Di Palma, S. Geels, B. P. Carpenter, R. A. Talosig, C. Chen, F. Marangoni and J. P. Patterson, Biomater. Sci., 2022, 10, 6749–6754 RSC.
- Z. Huang, M. Ge, F. Carraro, C. Doonan, P. Falcaro and X. Zou, Faraday Discuss., 2021, 225, 118–132 RSC.
- H. Zhang, Y. Lv, T. Tan and D. van der Spoel, J. Phys. Chem. B, 2016, 120, 477–484 CrossRef CAS PubMed.
- F. Lyu, Y. Zhang, R. N. Zare, J. Ge and Z. Liu, Nano Lett., 2014, 14, 5761–5765 CrossRef CAS PubMed.
- J. Liang, M. Y. Bin Zulkifli, J. Yong, Z. Du, Z. Ao, A. Rawal, J. A. Scott, J. R. Harmer, J. Wang and K. Liang, J. Am. Chem. Soc., 2022, 144, 17865–17875 CrossRef CAS PubMed.
- S. Libertino, V. Aiello, A. Scandurra, M. Renis and F. Sinatra, Sensors, 2008, 8, 5637–5648 CrossRef CAS.
- T. M. Schwarz, C. A. Dietrich, J. Ott, E. M. Weikum, R. Lawitzki, H. Solodenko, E. Hadjixenophontos, B. Gault, J. Kästner, G. Schmitz and P. Stender, Sci. Rep., 2021, 11, 11607 CrossRef CAS.
- N. K. Maddigan, O. M. Linder-Patton, P. Falcaro, C. J. Sumby, S. G. Bell and C. J. Doonan, ACS Appl. Mater. Interfaces, 2021, 13, 51867–51875 CrossRef CAS.
- B. P. Carpenter, A. R. Talosig, B. Rose, G. D. Palma and J. P. Patterson, Chem. Soc. Rev., 2023, 52, 6918–6937 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ta05397k |
| ‡ Indicates equal contributions to this manuscript. |
| This journal is © The Royal Society of Chemistry 2024 |