Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Boosting sugarcane trash decomposition: synergistic action and proximity effect of xylanase and feruloyl esterase co-displayed on the cell surface of Pichia pastoris (Komagataella phaffi)
Apisan
Phienluphon
 ab,
Keiko
Kondo
abc,
Hiroyuki
Okano
de,
Takashi
Watanabe
de,
Takashi
Nagata
ab,
Keiko
Kondo
abc,
Hiroyuki
Okano
de,
Takashi
Watanabe
de,
Takashi
Nagata
 *abcd and
Masato
Katahira
*abcd
*abcd and
Masato
Katahira
*abcd
aInstitute of Advanced Energy, Kyoto University, Gokasho, Uji, Kyoto 611-0011, Japan. E-mail: katahira.masato.6u@kyoto-u.ac.jp; nagata.takashi.6w@kyoto-u.ac.jp
bGraduate School of Energy Science, Kyoto University, Gokasho, Uji, Kyoto 611-0011, Japan
cIntegrated Research Center for Carbon Negative Science, Kyoto University, Gokasho, Uji, Kyoto, 611-0011, Japan
dBiomass Product Tree Industry-Academia Collaborative Research Laboratory, Kyoto University, Gokasho, Uji, Kyoto 611-0011, Japan
eResearch Institute for Sustainable Humanosphere, Kyoto University, Gokasho, Uji, Kyoto 611-0011, Japan
First published on 19th January 2024
Abstract
Yeast surface display technology offers a promising avenue for enhancing lignocellulosic biomass degradation and bioconversion processes. In this study, we investigated the synergistic action of cell-surface-displayed xylanase (XYN) and feruloyl esterase (FAE) using Pichia pastoris (Komagataella phaffi) and also explored the impact of the proximity effect when both enzymes are co-displayed on the same cell surface. We engineered three P. pastoris strains: X-Pichia displaying XYN, F-Pichia displaying FAE, and X/F-Pichia co-displaying both enzymes. Immunofluorescence confirmed successful cell-surface display. In hydrolysis experiments using acid-pretreated sugarcane trash, a clear synergy emerged. The mixing of X-Pichia and F-Pichia (“X-Pichia + F-Pichia”) exhibited an approximately 1.2 times higher reducing sugar yield compared to X-Pichia alone. Moreover, X/F-Pichia surpassed this mixture due to the proximity effect, yielding roughly 1.5 times more reducing sugar than X-Pichia alone. Additionally, X/F-Pichia produced about 1.1 times more ferulic acid than F-Pichia alone or “X-Pichia + F-Pichia.” These results underscore the potential of co-displaying multiple enzymes on the P. pastoris cell surface to significantly enhance biomass degradation. The enzyme co-display system with synergy and proximity effects holds promise for efficient lignocellulosic biomass utilization and sustainable bioprocessing. This work contributes to the development of more efficient and sustainable bioconversion processes for lignocellulosic biomass.
1. Introduction
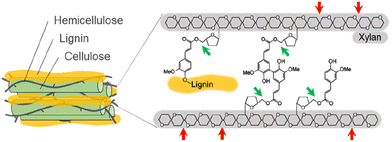
Lignocellulosic biomass, derived from plant materials such as agricultural waste, wood, and grasses, represents a vast and renewable resource with significant potential for producing biofuels and valuable chemicals.1 However, its intricate structure, which stabilizes the plant cell wall, presents challenges for effective breakdown into constituent sugars and aromatic compounds.2 Hemicellulose plays a crucial role in bridging cellulose fibers/microfibrils through hydrogen bonding, while lignin fills the spaces between cellulose and hemicellulose in the cell wall.3 Hemicellulose is a branched heteropolysaccharide made up from monomers (glucose, mannose, galactose, xylose, and arabinose) and various modification groups (ferulic, p-coumaric, and acetic acids).3 The complex composition and cross-linked nature of hemicellulose pose challenges for its enzymatic degradation.Xylanase (XYN) and feruloyl esterase (FAE) are essential enzymes with distinct catalytic functions that play a critical role in efficient hemicellulose degradation.4,5 XYN is an enzyme that specifically hydrolyzes the β-1,4-glycosidic linkages within the xylan backbone, yielding shorter xylooligosaccharides and xylose molecules (Fig. 1).4 This enzymatic activity significantly enhances the accessibility of hemicellulose for further degradation and conversion. In parallel, FAE specializes in cleaving the ester linkages between hemicellulose and phenolic moieties, liberating ferulic acid (FA) and disrupting the lignin-carbohydrate complex (Fig. 1).5 By hydrolyzing the ester bonds within the lignocellulosic matrix, FAE not only improves the accessibility of other enzymes to the substrate for biomass degradation6 but also generates a valuable phenolic compound, FA, which can be used in the food, pharmaceutical, and nutraceutical industries.7–9
Previous studies have shown that XYN and FAE work synergistically in biomass degradation, underscoring their potential to enhance the efficiency of lignocellulosic biomass conversion for sustainable bioprocessing.6,10–12 For instance, the addition of a crude extract from Aspergillus oryzae, containing FAE, to Celluclast 1.5L, which includes xylanase, cellulase, and pectinase, resulted in observable synergistic effects.10 Additionally, synergistic effects were demonstrated through the incorporation of FAEs from termite metagenomes with XYN11 from Thermomyces lanuginosus.11 Likewise, the combined utilization of FAE-1 and various XYNs also showed synergistic effects.12
Yeast surface display (YSD) is a technique employed for the presentation of enzymes on the yeast cell surface through fusion with an anchoring domain.13 This method has been employed to create whole-cell biocatalysts capable of producing valuable chemicals and biofuels from lignocellulosic biomass.14 The potential of YSD has been further augmented through genetic engineering strategies, including the optimization of promoters,15 signal peptides,16 anchoring proteins,17,18 yeast cell wall modification,18,19 and modification of the secretory pathway.20,21
YSD, while demonstrating notable advantages, does face certain limitations that warrant consideration. The finite surface space on a yeast cell poses challenges in achieving optimal enzyme display levels for biomass hydrolysis. Furthermore, excess enzyme production to the yeast cell surface can lead to cell metabolism imbalances and eventually decrease the functionality of enzymes.22,23 Balancing enzyme display for optimal hydrolysis may lack the quantitative precision which is seen in systems relying on secreted enzymes.
However, it is crucial to note that in the context of a cell factory system, YSD offers unique practical benefits such as reusability and streamlined downstream purification, underscoring its potential in scaled bioprocessing and compound production.24 Thus, YSD offers several potential advantages over enzyme secretion for applications in lignocellulosic biomass degradation.24 Immobilization on the cell surface can enhance enzyme stability and prevent degradation compared to free enzymes in solution.25,26 Concentration of enzymes on the cell surface can result in higher local enzyme concentrations, thereby increasing catalytic efficiency.27,28 Yeast cells with surface-displayed enzymes can be easily separated from the reaction mixture and reused, simplifying enzyme recycling.29–31
Furthermore, YSD facilitates the co-display of multiple different enzymes on the same cell surface, introducing the concept of a proximity effect among these enzymes.32,33 This concept has gained prominence due to its ability to accelerate the reaction rates of enzyme cascades.30,33–36 Notably, the close proximity of three different kinds of cellulase co-displayed on the yeast cell surface has been reported to increase ethanol yield through the simultaneous saccharification and fermentation of phosphoric acid-swollen cellulose,30,32,37 microcrystalline cellulose38 and alkali-pretreated sugarcane bagasse.21 Additionally, scaling up of the simultaneous saccharification and fermentation of pretreated bagasse in a 1 L bioreactor using cellulase-displaying yeast together with commercial cellulase achieved fermentability of up to 86.5% of the theoretical yield.21 It is worth noting that the depolymerization of lignocellulosic biomass can be enhanced by sequential actions using peroxidases and cellulases.39
Previously, YSD of XYNs was shown to be effective in biomass degradation. Xylanase Orf6-unm, when displayed on the yeast cell surface, exhibited enhanced xylanolytic activity compared to the enzyme purified from Escherichia coli, leading to improved corn stover digestion by rumen cultures.40 In a separate study, the surface display of a XYN from Lentinula edodes on yeast cells demonstrated its capacity to efficiently hydrolyze wheat residue.41
However, it is worth noting that, to date, there have been no reported instances of YSD for FAEs. YSD of both XYN and FAE on the same yeast surface has not been previously investigated, either. Since the coexistence of XYN and FAE leads to their synergistic action, we hypothesized that employing YSD to immobilize XYN and FAE on the same yeast surface could introduce not only synergistic effects but also proximity effects.
Recent studies have highlighted the utilization of the engineered Pichia pastoris (Komagataella phaffi) as a versatile chassis biocatalyst to produce biofuels, including ethanol,42 isobutanol,43 and biodiesel.44
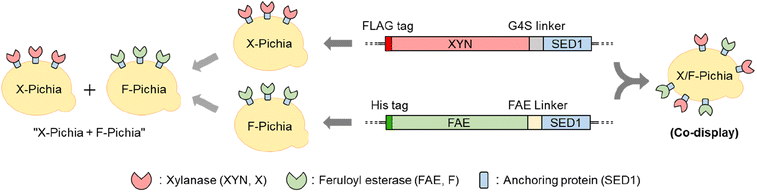
In this study, we individually fused XYN from Thermomyces lanuginosus and FAE from Acremonium alcalophilum with the Saccharomyces cerevisiae SED1 anchoring domain.45 This anchoring domain is known for its ability to effectively localize fusion proteins on the P. pastoris cell surface.31,41,46–48 After successfully generating strains that display XYN alone (X-Pichia), FAE alone (F-Pichia), and both enzymes together (X/F-Pichia) (Fig. 2), we examined the localization, stability, and relative activity of the displayed enzymes under various conditions, including temperature, pH level, and the presence of different additives.
This study represents the first instance of displaying FAE on the yeast cell surface and exploring the synergistic and proximity effects between XYN and FAE in biomass degradation. Specifically, sugarcane trash, defined as the upper shoots and leaf sheath with the leaves remaining on the ground and constituting approximately 15% of the total aboveground biomass,49 was used as the substrate. When we used a mixture of X-Pichia and F-Pichia (“X-Pichia + F-Pichia”), we observed synergistic enhancement of the hydrolysis of acid-pretreated sugarcane trash leading to increased production of reducing sugars compared to X-Pichia alone. Moreover, X/F-Pichia, benefiting from the proximity effect between XYN and FAE, further amplified the synergistic effect observed in “X-Pichia + F-Pichia”, resulting in higher reducing sugar production. Additionally, X/F-Pichia produced more ferulic acid than F-Pichia alone or “X-Pichia + F-Pichia”. These findings strongly suggest that the strategy of co-displaying the combination of FAE and XYN enables the utilization of their synergistic and proximity effects for efficiently utilizing lignocellulosic biomass and lays the foundation for sustainable bioprocessing with substantial potential.
2. Materials and methods
2.1 Strains and culture conditions
The yeast and bacterial strains used in this study are summarized in Table 1. P. pastoris strains were cultured in Yeast extract Peptone Dextrose (YPD) agar (20 g L−1 peptone, 10 g L−1 yeast extract, 10 g L−1D-glucose, and 15 g L−1 bacteriological agar) at 30 °C. P. pastoris transformants were screened on YPD supplemented with 100 μg mL−1 Zeocin (Invivogen, USA). For protein expression, P. pastoris transformants were pre-cultivated overnight in YPD at 30 °C with 250 rpm shaking. The overnight cultures were transferred to 100 mL of buffered glycerol complex medium (BMGY) (1% (w/v) yeast extract, 2% (w/v) peptone, 100 mM potassium phosphate, pH 6.0, 1.34% (w/v) yeast nitrogen base (YNB), and 4 × 10−5% (w/v) biotin) supplemented with 1% (v/v) glycerol and grown at 30 °C at 250 rpm for 3 days. The cells were harvested by centrifugation at 8000×g for 5 minutes at room temperature and inoculated into 50 mL of BMMY containing 0.5% methanol. To maintain the induction of protein expression, methanol was added to the yeast culture once daily for 2 days to make its concentration 0.5%.E. coli DH5α (Invitrogen) was used for plasmid propagation. Bacterial cells were grown in Luria–Bertani broth (10 g L−1 peptone, 5 g L−1 yeast extract, and 5 g L−1 NaCl) containing 25 μg mL−1 Zeocin for transformant selection.
2.2 Plasmid construction
PCR amplification was carried out using KOD-plus DNA polymerase (Toyobo, Osaka, Japan) with the primers listed in Table 1. The anchoring domain ScSED1 was amplified from Saccharomyces cerevisiae strain Cen-PK2-1C using SpeI-SED1_F and NotI-SED1_R primers (Table 1). DNAs encoding Thermomyces lanuginosus xylanase (TlXYN; UniProt ID: O43097) and Acremonium alcalophilum feruloyl esterase (AaFaeD; JGI ID: Acral2|1082309) were codon-optimized and synthesized by Thermo Fisher Scientific (Japan). TlXYN, fused with an N-terminal FLAG tag and C-terminal (G4S)3 linker, and AaFaeD, fused with an N-terminal 6× histidine tag and its native C-terminal linker, were amplified using TlXYN-EcoRI_F and TlXYN-G4S-SpeI_R primers, and AaFAE-EcoRI-F and AaFAE-SpeI-R primers, respectively (Table 1). The PCR amplicons were then ligated with the ScSED1 anchoring domain (Fig. 2) and cloned into pPICZαA at the EcoRI and NotI restriction sites, resulting in pPICZαA-XYN-SED and pPICZαA-FAE-SED, respectively (Table 1). To construct the single-displayed strains, X-Pichia and F-Pichia, pPICZαA-XYN-SED and pPICZαA-FAE-SED were linearized using SacI restriction enzyme and integrated into the P. pastoris X-33 genome. The double-displayed strain, X/F-Pichia, was created by co-integrating the linearized pPICZαA-XYN-SED and pPICZαA-FAE-SED into P. pastoris X-33.2.3 Immunofluorescence
To visualize the protein localization of XYN and FAE on the P. pastoris cell surface, induced P. pastoris transformants were collected by centrifugation at 8000×g for 5 minutes and washed twice with 1 × PBS (0.8% NaCl, 0.2% KCl, 0.144% Na2HPO4, and 0.024% KH2PO4). Subsequently, cells were incubated with a 100-fold dilution of His-probe antibody (AD1.1.10) conjugated with Alexa Fluor 488 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and a 100-fold dilution of OctA-Probe antibody (H-5), an IgG1 κ mouse monoclonal anti-FLAG tag antibody, conjugated with Alexa Fluor 594 (Santa Cruz Biotechnology) in 1 × PBS for 1 hour at room temperature. Following incubation, the cells were washed twice with 1 × PBS and mounted on a glass slide using 50% glycerol in 1 × PBS. Fluorescence detection was performed using a fluorescence microscope (FluoView™ FV1000, Olympus, Tokyo, Japan) at excitation wavelengths of 488 nm (argon laser line) and 594 nm (helium/neon laser line). The fluorescent emission channels were set using filters with wavelengths of 510 nm and 610 nm, respectively. Cells were imaged using a 100× oil immersion objective lens, and images were acquired and processed using cellSens Standard software (Olympus, Tokyo, Japan).2.4 Determination of activities of enzymes displayed on the P. pastoris cell surface towards beechwood xylan and methyl ferulate (MFA)
To assess enzyme activities on the yeast cell surface, induced yeast cells were washed three times with 25 mM potassium phosphate at pH 7.0. Following this, the washed cells were adequately diluted (based on OD590 unit) in the washing solution before commencing the assay.The xylanase activity assay was performed using beechwood xylan (Sigma, USA) as the substrate. One mL of reaction mixture contained 1% (w/v) beechwood xylan and 10 OD590 units of P. pastoris cells in 100 mM potassium phosphate, pH 7.0. The reaction was carried out at 30 °C for 30 minutes, and enzymatic activity was stopped by heating the mixture at 100 °C for 10 minutes. The concentration of reducing sugars was measured using the 3,5-dinitrosalicylic acid (DNS; Wako, Japan) method.50 Briefly, a mixture containing 200 μL of the sample and 600 μL of DNS was boiled for 10 minutes and then cooled on ice. The mixture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 1 min to pellet the residual cells and insoluble substrate. The reducing sugar content in the supernatant was then measured as the absorbance at 540 nm against a xylose standard curve (ranging from 0.15 to 1.5 mg mL−1). One unit (U) of enzyme activity was defined as the amount of enzyme that released 1 μmole of reducing sugars per minute.
000×g for 1 min to pellet the residual cells and insoluble substrate. The reducing sugar content in the supernatant was then measured as the absorbance at 540 nm against a xylose standard curve (ranging from 0.15 to 1.5 mg mL−1). One unit (U) of enzyme activity was defined as the amount of enzyme that released 1 μmole of reducing sugars per minute.
FAE activity was assayed using methyl ferulate (MFA; LKT Laboratories, USA) following a modified version of our previous protocol.51 The 250 μL reaction mixture, comprising 0.05 OD590 units of P. pastoris cells in 80 mM potassium phosphate, pH 7.0, and 0.12 mM MFA, was incubated at 30 °C for 30 minutes. The reaction was monitored using a microplate reader (Tecan Infinite 200 Pro, Switzerland) at 340 nm with a 2 minutes interval to obtain the initial reaction rates of hydrolysis. The extinction coefficients of MFA and FA were determined experimentally. One unit of activity was defined as the production of 1 μmol of FA from MFA per minute.
To assess the thermostability and pH stability of X-Pichia and F-Pichia, we measured their activity towards beechwood xylan and MFA, respectively. For thermostability assessment, X-Pichia and F-Pichia were dissolved in 50 mM potassium phosphate, pH 6.5, and incubated for 24 hours at temperatures ranging from 20 to 80 °C. After incubation, the activity was measured at 30 °C using the method described above. To determine the pH stability, P. pastoris cells were dissolved in various solutions with different pH values: 100 mM sodium acetate (pH 4.0–5.0), 100 mM potassium phosphate (pH 6.0–8.0), or 100 mM Tris-HCl (pH 8.0–9.0). The cell suspensions were then incubated at 30 °C for 24 hours, and the activities were measured at 30 °C in 100 mM potassium phosphate, pH 7.0.
To assess the tolerance of the displayed enzymes to some metal ions, inhibitors, and detergents, we measured the activity of X-Pichia and F-Pichia at 30 °C in the presence of these additives under 100 mM Tris-HCl and pH 7.0 conditions. The enzyme activity in the presence of an additive was determined by subtracting the rate of the blank reaction, which contained only the additive, from the individual reaction rate. The activity obtained without the presence of any additives was used as a control and defined as 100%.
2.5 Pretreatment of natural biomass, sugarcane trash
Sugarcane trash was obtained from Eastern Sugar and Industries Ltd. (Sa Kaeo, Thailand). Initially, it was ground to a range of approximately 1–8 mm in length and 0.5–2 mm in thickness. Subsequently, the sugarcane trash was meticulously cut and sieved to obtain particles in the size range of 0.5–1 mm. The sugarcane trash was pretreated using the diluted acid pretreatment protocol of Mkabayi et al. (2020).11 Briefly, the cut sugarcane trash was suspended in a solution of 0.5% (w/w) sulfuric acid at a solid-to-liquid ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10. The suspension was then autoclaved at 121 °C for 20 minutes. Following acid pretreatment, the sugarcane trash was thoroughly washed with Milli-Q water until reaching neutral pH. The washed sugarcane trash was freeze-dried and stored at −20 °C until further used.
10. The suspension was then autoclaved at 121 °C for 20 minutes. Following acid pretreatment, the sugarcane trash was thoroughly washed with Milli-Q water until reaching neutral pH. The washed sugarcane trash was freeze-dried and stored at −20 °C until further used.
2.6 Degradation of natural biomass, sugarcane trash
The degradation of biomass was conducted using P. pastoris cells displaying enzymes on their surface. Acid-pretreated sugarcane trash was used as the substrate. To compare the hydrolysis activity of X/F-Pichia with that of X-Pichia, F-Pichia, and the mixture of X-Pichia and F-Pichia, “X-Pichia + F-Pichia”, we adjusted the amounts of X-Pichia and F-Pichia to match the XYN and/or FAE activities of X/F-Pichia (refer to Table 2). The XYN and/or FAE activities of X-Pichia, F-Pichia, and X/F-Pichia were measured in the previous section. A 1 mL reaction mixture was prepared, comprising 1% (w/v) acid-pretreated sugarcane trash and P. pastoris cells in 100 mM potassium phosphate buffer with a pH of 7.0. As a control, a 1 mL reaction mixture comprising 1% (w/v) acid-pretreated sugarcane trash and P. pastoris X-33 cells was also prepared. The reactions were conducted at 30 °C for 24 and 48 hours, respectively.| P. pastoris strain | Enzyme activity (mU per OD590 unit)a | Amount of P. pastoris (OD590 units) used for a 1 mL hydrolysis of acid-pretreated sugarcane trash | |||||
|---|---|---|---|---|---|---|---|
| XYNb | FAEc | Sample name | |||||
| X-33d | X-Pichiae | F-Pichiaf | “X-Pichia + F-Pichia” | X/F-Pichia | |||
| a Values are the means of three independent replicates. ND: not detected. b XYN activity assay using beechwood xylan as substrate. c FAE activity assay using MFA as substrate. d Reference reaction was carried out using 17.7 OD590 units of P. pastoris X-33. e 8.4 OD590 units of X-Pichia exerts 50 mU XYN activity, which is equivalent to the XYN activity of 10 OD590 units of X/F-Pichia. f 9.3 OD590 units of F-Pichia exerts 18 mU FAE activity, which is equivalent to the FAE activity of 10 OD590 units of X/F-Pichia. | |||||||
| X-33 | ND | ND | 17.7 | 9.3 | 8.4 | — | 7.7 |
| X-Pichia | 6.0 ± 0.1 | ND | — | 8.4 | — | 8.4 | — |
| F-Pichia | ND | 2.0 ± 0.0 | — | — | 9.3 | 9.3 | — |
| X/F-Pichia | 5.0 ± 0.4 | 1.8 ± 0.1 | — | — | — | — | 10 |
2.7 Determination of reducing sugar and ferulic acid contents obtained on the hydrolysis of natural biomass, sugarcane trash
The total reducing sugar content resulting from enzymatic hydrolysis of acid-pretreated sugarcane trash was determined using the DNS method as mentioned above.For analysis of the FA content, 200 μL of a sample was mixed with 600 μL of 100% acetonitrile. The FA content was then determined using an RP-HPLC (Shimadzu, Japan) equipped with a reversed phase column (4.6 mm × 15 cm, TSKgel ODS-80 TM; Tosoh, Tokyo, Japan) and a UV detector (310 nm, SPD-20A UV/VIS detector; Shimadzu, Japan). The binary mobile phase consisted of (A) Milli-Q water + 0.1% trifluoroacetic acid (TFA) and (B) acetonitrile + 0.1% TFA. The elution profile was as follows: isocratic at 5% B for 0.0–5 min, B linearly increased from 5–70% over 5–25 min, isocratic at 70% B for 25–35 min, and isocratic at 5% B for 35–45 min. The flow rate was 1 mL min−1. The FA (MP Biomedicals, USA) content was estimated using standard curves. Data were processed using LabSolution software (Shimadzu, Japan).
The reducing sugar and FA contents that were obtained for the control (a 1 mL reaction mixture comprising 1% (w/v) acid-pretreated sugarcane trash and P. pastoris X-33 cells) were subtracted from the corresponding contents obtained for reaction mixtures with various combinations of P. pastoris described above.
2.8 Statistical analysis
All experiments were performed in triplicate, and the values are reported as means ± standard deviation. Statistical analyses were conducted using Excel Software, employing the t-test for significance testing.3. Results and discussions
3.1 Construction of P. pastoris strains
In this study, we employed wild-type P. pastoris X-33 as the starting strain for displaying fungal XYN and FAE enzymes. Plasmid pPICZαA served as the foundational DNA for constructing pPICZαA-XYN-SED and pPICZαA-FAE-SED (Table 1). The XYN and FAE genes, along with α-factor secretion signal sequences from the plasmid backbone, were expressed under the control of the AOX1 promoter. To anchor the enzymes onto the yeast cell surface, we fused the ScSED1 anchoring protein to the C-terminus of FLAG-tagged XYN and 6× His-tagged FAE (Fig. 2). The enzyme-displaying yeast strains, namely X-Pichia, F-Pichia, and X/F-Pichia, were generated by introducing the gene cassettes of pPICZαA-XYN-SED and/or pPICZαA-FAE-SED into the P. pastoris X-33 genome, as depicted in Fig. 2.3.2 Localization of fusion proteins in P. pastoris
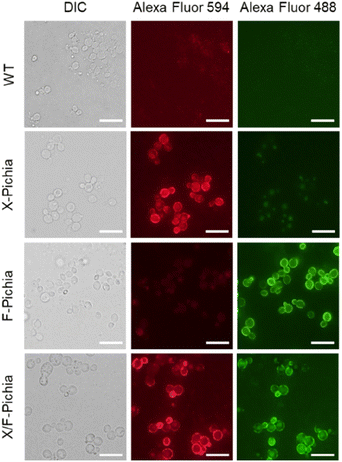
We visualized XYN and FAE displayed on the yeast cell surface via immunofluorescence analysis. We employed a FLAG-tag antibody labeled with Alexa Fluor 594 for XYN and a His-probe antibody labeled with Alexa Fluor 488 for FAE. The results showed strong red fluorescence on the surface of X-Pichia and X/F-Pichia, while F-Pichia and X/F-Pichia displayed intense green fluorescence (Fig. 3). These findings confirm the successful anchoring of XYN and FAE on the cell surface of recombinant P. pastoris X-33.3.3 Enzyme activity of XYN and FAE displayed on the yeast cell surface
It was revealed that P. pastoris X-33 does not exhibit either XYN or FAE activity (Table 2). This is consistent with the previous report that P. pastoris X-33 does not have the XYN activity.52After protein expression, both the P. pastoris cell fraction and the medium fraction were collected. Activity assays were conducted on the cell fraction using beechwood xylan and MFA as substrates to evaluate the functionality of the enzymes displayed on the P. pastoris cell surface. The results revealed XYN activities of 6.0 mU per OD590 unit for X-Pichia and 5.0 mU per OD590 unit for X/F-Pichia, while the FAE activities were 2.0 mU per OD590 unit for F-Pichia and 1.8 mU per OD590 unit for X/F-Pichia (Table 2, left). One OD590 unit represents the amount of enzyme included in 1 mL of the solution whose optical density at 590 nm is 1.0. These findings confirm the functional activity of the enzymes displayed on the cell surface of X-Pichia, F-Pichia and X/F-Pichia.
Further investigation of enzyme activity in the medium fraction indicated that some of the fusion proteins are secreted or released into the medium (data not shown). This suggests that the overexpression of XYN and FAE conjugated with the SED1 anchoring protein might have resulted in the release of these proteins into the medium to some extent.53
3.4 Temperature and pH stability of XYN and FAE on the P. pastoris cell surface
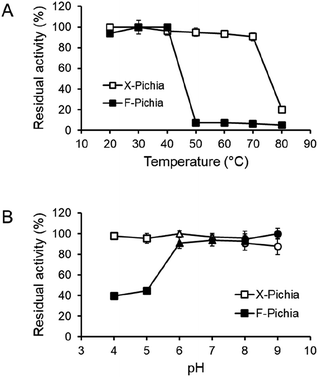
To assess the durability of the immobilized enzymes on the P. pastoris cell surface, we examined the temperature and pH stability of XYN and FAE on X-Pichia and F-Pichia, respectively. Both X-Pichia and F-Pichia were subjected to various temperatures and pH conditions for 24 hours, and afterward, we assessed the remaining XYN and FAE activities.The results indicate that surface-displayed XYN remains stable within a temperature range of 20 to 70 °C, retaining over 90% of its activity (Fig. 4A). However, its activity significantly decreases on incubation at 80 °C for 24 hours (Fig. 4A). Additionally, XYN retains high activity over a broad pH range of 4.0 to 9.0, with residual activity exceeding 85% (Fig. 4B). Notably, the XYN displayed on the cell surface in this study exhibits superior thermal stability compared to surface-displayed Lentinula edodes XYN, which remains stable only within a temperature range of 20 to 30 °C.41 This difference in stability may be attributed to the adaptation of T. lanuginosus to high-temperature environments, such as hot springs.54
Surface-displayed FAE, on the other hand, demonstrates stability within a narrower temperature range, specifically between 20 to 40 °C, and exhibits almost no activity after exposure to temperatures exceeding 50 °C (Fig. 4A). Additionally, it was observed that surface-displayed FAE maintains robust stability at pH levels ranging from 6.0 to 9.0, consistently retaining over 90% of its activity. However, it displays lower stability under mildly acidic conditions, with activity falling below 50% (Fig. 4B). Overall, the temperature and pH stability of surface-displayed XYN and FAE closely align with those of the crude TlXYN55 and secreted AaFaeD (unpublished data), respectively.
3.5 Effect of additives on the enzyme activities of XYN and FAE on the P. pastoris cell surface
In previous studies, it has been reported that certain metal ions and organic reagents can enhance the activities of XYN56,57 and FAE.58 Therefore, in this study, we investigated the impact of additives on the activities of XYN and FAE on the P. pastoris cell surface.The addition of CoCl2, NiCl2, and CuCl2 to X-Pichia led to an increase in XYN activity. Specifically, at a concentration of 5 mM, these salts enhanced the activity by 88%, 23%, and 81%, respectively (Table 3). The increase in XYN activity could be due to the structural change in the active site, resulting in the enlargement of substrate-binding pocket, induced by binding with the metal ion.57 In contrast, the introduction of ethylenediaminetetraacetate (EDTA) had an adverse effect on XYN activity, resulting in a 35% reduction at the same 5 mM concentration. Additionally, the inclusion of 2-mercaptoethanol (2-ME) and dithiothreitol (DTT) showed a slight improvement in XYN activity, with increases of 12% and 16% at the 5 mM concentration, respectively (Table 3). On the other hand, X-Pichia exhibited a 10% decrease in XYN activity when sodium dodecyl sulfate (SDS) and Triton X-100 were added, while the presence of Tween 20 and Tween 80 had no discernible impact on XYN activity.
| Additive | Relative activity (%)a | |||
|---|---|---|---|---|
| XYN activity (X-Pichia)b | FAE activity (F-Pichia)c | |||
| 1 mM | 5 mM | 1 mM | 5 mM | |
| a Reference reactions were carried out for each additive in the absence of X-Pichia or F-Pichia. The values obtained from the reference reactions were subtracted to derive the values presented in the table. The activity of X-Pichia and F-Pichia in the absence of additives was set as 100%. ND: Not Detected. b XYN activity assay was conducted using beechwood xylan as substrate. c FAE activity assay was conducted using MFA as substrate. | ||||
| Metal ions | ||||
| LiCl | 100 ± 3 | 104 ± 3 | 94 ± 9 | 97 ± 4 |
| NaCl | 100 ± 2 | 100 ± 4 | 84 ± 10 | 100 ± 5 |
| KCl | 96 ± 2 | 100 ± 3 | 93 ± 4 | 89 ± 6 |
| MgCl2 | 97 ± 2 | 104 ± 3 | 93 ± 8 | 120 ± 8 |
| CaCl2 | 98 ± 2 | 105 ± 5 | 113 ± 9 | 134 ± 12 |
| MnCl2 | 99 ± 4 | 108 ± 4 | 95 ± 5 | 103 ± 5 |
| CoCl2 | 141 ± 5 | 188 ± 7 | 58 ± 6 | 66 ± 16 |
| NiCl2 | 104 ± 7 | 123 ± 8 | 46 ± 6 | 37 ± 6 |
| CuCl2 | 119 ± 6 | 181 ± 5 | 24 ± 4 | N.D. |
| ZnCl2 | 96 ± 2 | 109 ± 4 | 18 ± 10 | 16 ± 12 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Inhibitors | ||||
| EDTA | 95 ± 8 | 65 ± 4 | 102 ± 5 | 115 ± 9 |
| 2 ME | 111 ± 1 | 112 ± 1 | 112 ± 5 | 101 ± 3 |
| DTT | 106 ± 3 | 116 ± 3 | 102 ± 5 | 106 ± 5 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Detergents | ||||
| SDS | 96 ± 9 | 90 ± 7 | 80 ± 5 | 13 ± 2 |
| Triton X-100 | 96 ± 7 | 93 ± 6 | 84 ± 8 | 56 ± 9 |
| Tween 20 | 100 ± 8 | 100 ± 6 | 34 ± 5 | 34 ± 4 |
| Tween 80 | 106 ± 7 | 100 ± 7 | 88 ± 3 | 59 ± 4 |
The addition of MgCl2 and CaCl2 to F-Pichia resulted in an increase in FAE activity. At a concentration of 5 mM, these salts increased the activity by 20% and 34%, respectively (Table 3). Conversely, the addition of CoCl2, NiCl2, CuCl2, and ZnCl2, had an adverse effect on FAE activity, resulting in 34%, 63%, 100%, and 84% reduction at the same 5 mM concentration, respectively. The addition of EDTA led to a slight improvement in FAE activity, with an increase of 15%. On the other hand, F-Pichia exhibited reductions of 87%, 44%, 66%, and 41% in FAE activity when SDS, Triton X-100, Tween 20, and Tween 80 were added, respectively.
3.6 Sample preparation for investigating synergistic hydrolysis of XYN and FAE displayed on the P. pastoris cell surface
This section outlines the sample preparation process for the subsequent investigation into the hydrolysis of acid-pretreated sugarcane trash by different P. pastoris strains: a single-displaying strain alone (X-Pichia or F-Pichia), a mixture of single-displaying strains (“X-Pichia + F-Pichia”), and the co-displaying strain (X/F-Pichia). Specifically, we examine the hydrolysis of beechwood xylan by cell-surface displayed XYN (referred to as XYN activity) and the hydrolysis of MFA by cell-surface displayed FAE (referred to as FAE activity). In this section, we calculate the relative XYN and FAE activities of each P. pastoris strain to determine the proportions in which they need to be mixed for hydrolysis of natural biomass, sugarcane trash.As established in the previous section, the XYN activities (activities of hydrolysis of beechwood xylan) of X-Pichia and X/F-Pichia were approximately 6.0 and 5.0 mU per OD590 unit, respectively (Table 2, left). Therefore, when one OD590 unit of X/F-Pichia is used, 0.84 OD590 units of X-Pichia are required to equalize their XYN activities. Similarly, the FAE activities (MFA hydrolysis) of F-Pichia and X/F-Pichia were approximately 2.0 and 1.8 mU per OD590 unit, respectively (Table 2, left). In this case, when one OD590 unit of X/F-Pichia is used, 0.93 OD590 units of F-Pichia are needed to equalize their FAE activities.
Firstly, we set the amount of X/F-Pichia to 10 OD590 units (Table 2, right, X/F-Pichia column). Consequently, to achieve equivalent XYN and FAE activities, 8.4 OD590 units of X-Pichia and 9.3 OD590 units of F-Pichia are used. Secondly, the total amount of P. pastoris for “X-Pichia + F-Pichia” becomes 17.7 OD590 units (Table 2, right, “X-Pichia + F-Pichia” column). In order to adjust the total P. pastoris amount to 17.7 OD590 units, original P. pastoris X-33, which contains no XYN or FAE, is added to X-Pichia, F-Pichia, and X/F-Pichia in quantities of 9.3, 8.4, and 7.7 OD590 units, respectively (Table 2, right, X-Pichia, F-Pichia, and X/F-Pichia columns). Finally, a sample of 17.7 OD590 units of X-33 was prepared as a control.
3.7 Synergistic hydrolysis of XYN and FAE on the P. pastoris cell surface
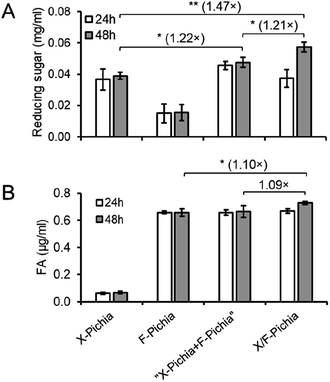
To explore the synergistic action of XYN and FAE displayed on the P. pastoris cell surface during biomass degradation, we conducted activity assays of a single-displaying strain alone (X-Pichia or F-Pichia), a mixture of single-displaying strains (“X-Pichia + F-Pichia”), and the co-displaying strain (X/F-Pichia), using acid-pretreated sugarcane trash as the substrate. Measurements were taken at both 24 and 48 hours, and the product amounts after 48 hours were used for assessment.After 48 hours, the concentration of reducing sugar was ca. 0.04 mg mL−1 for X-Pichia, ca. 0.01 mg mL−1 for F-Pichia, ca. 0.05 mg mL−1 for “X-Pichia + F-Pichia”, and ca. 0.06 mg mL−1 for X/F-Pichia (Fig. 5A). Notably, “X-Pichia + F-Pichia” released 1.22 times more reducing sugar than X-Pichia alone, a clear synergistic action between XYN and FAE in the breakdown of hemicellulose being observed (Fig. 5A). Furthermore, X/F-Pichia released 1.21 times more reducing sugar than “X-Pichia + F-Pichia” and 1.47 times more than X-Pichia (Fig. 5A). In X/F-Pichia, XYN and FAE are co-displayed on the P. pastoris cell surface, bringing these enzymes into close proximity. This physical co-localization enhances the synergistic effect between XYN and FAE. Within hemicellulose, there are numerous sites linked with FA. When FAs at these sites are hydrolyzed and released by FAE, the hemicellulose at these sites becomes exposed, and the local structure of hemicellulose relaxes (Fig. 1). These products have improved accessibility and become better substrates for XYN.6,10–12 Thus, synergistic and proximity effects allow products generated by one enzyme to immediately become substrates for the other, resulting in an observed increase in reducing sugar production.
After 48 hours, the concentration of FA was ca. 0.07 μg mL−1 for X-Pichia, ca. 0.66 μg mL−1 for F-Pichia, ca. 0.66 μg mL−1 for “X-Pichia + F-Pichia”, and ca. 0.73 μg mL−1 for X/F-Pichia (Fig. 5B). The amount of FA produced by X-Pichia (ca. 0.07 μg mL−1) closely matched the initial content in the acid-pretreated sugarcane trash. Interestingly, “X-Pichia + F-Pichia” released the same amount of FA as F-Pichia alone, while X/F-Pichia released 1.10 times more FA than F-Pichia alone (Fig. 5B). This finding underscores the importance of the proximity effect in facilitating the synergistic action of XYN and FAE in producing FA.
4. Conclusions
In this study, we successfully harnessed YSD technology to co-display fungal XYN and FAE enzymes, thereby exploring the intriguing interplay of synergistic and proximity effects in biomass degradation. Our findings shed light on the potential of this innovative strategy for efficient lignocellulosic biomass utilization.We successfully constructed P. pastoris strains, namely X-Pichia, F-Pichia, and X/F-Pichia, expressing FLAG-tagged XYN and 6× His-tagged FAE, anchored to the yeast cell surface using ScSED1. Our investigation demonstrated robust enzyme stability across varying temperatures and pH levels, expanding the versatility of this co-display system. Furthermore, the impact of specific salts and chemicals on XYN and FAE activities provided valuable insights for optimizing their catalytic efficiency in biomass degradation.
The most significant findings emerged from our analysis of synergistic hydrolysis. Activity assays revealed that “X-Pichia + F-Pichia” released more reducing sugar than X-Pichia alone, indicating a clear synergistic effect between XYN and FAE in hemicellulose breakdown. Impressively, X/F-Pichia, displaying both XYN and FAE on the same cell surface, demonstrated even greater enhancements in releasing reducing sugar and FA than “X-Pichia + F-Pichia.” This underscores the amplified synergistic effect achieved through the proximity effect, where the physical co-localization of XYN and FAE facilitated effective substrate turnover, resulting in increased product yields.
In summary, our study has revealed a potent approach to maximize the utilization of lignocellulosic biomass by harnessing the synergistic and proximity effects of co-displayed XYN and FAE on the yeast cell surface. This strategy holds significant promise for sustainable bioprocessing, as it has the potential to increase the production of reducing sugars and valuable phenolic compounds such as ferulic acid.7–9 As we progress, this innovative approach offers the potential to advance biotechnology applications and contribute to a more environmentally friendly and resource-efficient future.
Author contributions
Apisan Phienluphon: conceptualization, methodology, investigation, formal analysis, writing – original draft. Keiko Kondo: methodology, formal analysis, data curation, funding acquisition, writing – review & editing. Hiroyuki Okano: resources, writing – review & editing. Takashi Watanabe: resources, writing – review & editing. Takashi Nagata: conceptualization, supervision, project administration, funding acquisition, writing – review & editing. Masato Katahira: conceptualization, supervision, project administration, funding acquisition, writing – review & editing.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by JSPS KAKENHI to M. K. [22H05596, 23H02419 and 23H04069], T. N. [20K06524, and 23K05664], and K. K. [20K06164, and 23K05334]; the Collaboration Program of the Laboratory for Complex Energy Processes, Institute of Advanced Energy, Kyoto University to TN; JST e-ASIA JRP; the Joint Usage/Research Program on Zero-Emission Energy Research, Institute of Advanced Energy, Kyoto University; collaborative research with DAICEL Co. Ltd.References
- M. Mujtaba, L. Fernandes Fraceto, M. Fazeli, S. Mukherjee, S. M. Savassa, G. Araujo de Medeiros, A. do Espírito Santo Pereira, S. D. Mancini, J. Lipponen and F. Vilaplana, Lignocellulosic biomass from agricultural waste to the circular economy: A review with focus on biofuels, biocomposites and bioplastics, J. Clean. Prod., 2023, 402, 136815 CrossRef CAS.
- A. Zoghlami and G. Paës, Lignocellulosic biomass: Understanding recalcitrance and predicting hydrolysis, Front. Chem., 2019, 7, 874 CrossRef CAS PubMed.
- J. Rao, Z. Lv, G. Chen and F. Peng, Hemicellulose: Structure, chemical modification, and application, Prog. Polym. Sci., 2023, 140, 101675 CrossRef CAS.
- R. Chaudhary, T. Kuthiala, G. Singh, S. Rarotra, A. Kaur, S. K. Arya and P. Kumar, Current status of xylanase for biofuel production: a review on classification and characterization, Biomass Convers. Biorefin., 2021, 13, 8773–8791 CrossRef.
- A. Dilokpimol, M. R. Mäkelä, M. V. Aguilar-Pontes, I. Benoit-Gelber, K. S. Hildén and R. P. De Vries, Diversity of fungal feruloyl esterases: Updated phylogenetic classification, properties, and industrial applications, Biotechnol. Biofuels, 2016, 9, 231 CrossRef PubMed.
- E. Schmitz, S. Leontakianakou, S. Norlander, E. Nordberg Karlsson and P. Adlercreutz, Lignocellulose degradation for the bioeconomy: The potential of enzyme synergies between xylanases, ferulic acid esterase and laccase for the production of arabinoxylo-oligosaccharides, Bioresour. Technol., 2022, 343, 126114 CrossRef CAS PubMed.
- S. Mathew and T. E. Abraham, Ferulic acid: An antioxidant found naturally in plant cell walls and feruloyl esterases involved in its release and their applications, Crit. Rev. Biotechnol., 2004, 24, 59–83 CrossRef CAS PubMed.
- S. Ou and K. C. Kwok, Ferulic acid: Pharmaceutical functions, preparation and applications in foods, J. Sci. Food Agric., 2004, 84, 1261–1269 CrossRef CAS.
- I. Antonopoulou, E. Sapountzaki, U. Rova and P. Christakopoulos, Ferulic acid from plant biomass: A phytochemical with promising antiviral properties, Front. Nutr., 2022, 8, 777576 CrossRef PubMed.
- C. M. P. Braga, P. da S. Delabona, D. J. da S. Lima, D. A. A. Paixão, J. G. da C. Pradella and C. S. Farinas, Addition of feruloyl esterase and xylanase produced on-site improves sugarcane bagasse hydrolysis, Bioresour. Technol., 2014, 170, 316–324 CrossRef CAS PubMed.
- L. Mkabayi, S. Malgas, B. S. Wilhelmi and B. I. Pletschke, Evaluating feruloyl esterase-xylanase synergism for hydroxycinnamic acid and xylo-oligosaccharide production from untreated, hydrothermally pre-treated and dilute-acid pre-treated corn cobs, Agronomy, 2020, 10, 688 CrossRef CAS.
- M. S. Mafa, S. Malgas and B. I. Pletschke, Feruloyl esterase (FAE-1) sourced from a termite hindgut and GH10 xylanases synergy improves degradation of arabinoxylan, AMB Express, 2021, 11, 21 CrossRef CAS PubMed.
- Y. Fujita, S. Takahashi, M. Ueda, A. Tanaka, H. Okada, Y. Morikawa, T. Kawaguchi, M. Arai, H. Fukuda and A. Kondo, Direct and efficient production of ethanol from cellulosic material with a yeast strain displaying cellulolytic enzymes, Appl. Environ. Microbiol., 2002, 68, 5136–5141 CrossRef CAS PubMed.
- K. V. Teymennet-Ramírez, F. Martínez-Morales and M. R. Trejo-Hernández, Yeast surface display system: Strategies for improvement and biotechnological applications, Front. Bioeng. Biotechnol., 2022, 9, 794742 CrossRef PubMed.
- K. Inokuma, T. Hasunuma and A. Kondo, Efficient yeast cell-surface display of exo-and endo-cellulase using the SED1 anchoring region and its original promoter, Biotechnol. Biofuels, 2014, 7, 8 CrossRef PubMed.
- K. Kajiwara, W. Aoki and M. Ueda, Evaluation of the yeast surface display system for screening of functional nanobodies, AMB Express, 2020, 10, 51 CrossRef CAS PubMed.
- A. Phienluphon, W. Mhuantong, K. Boonyapakron, P. Deenarn, V. Champreda, D. Wichadakul and S. Suwannarangsee, Identification and evaluation of novel anchoring proteins for cell surface display on Saccharomyces cerevisiae, Appl. Microbiol. Biotechnol., 2019, 103, 3085–3097 CrossRef CAS PubMed.
- K. Inokuma, Y. Kitada, T. Bamba, Y. Kobayashi, T. Yukawa, R. Den Haan, W. H. Van Zyl, A. Kondo and T. Hasunuma, Improving the functionality of surface-engineered yeast cells by altering the cell wall morphology of the host strain, Appl. Microbiol. Biotechnol., 2021, 105, 5895–5904 CrossRef CAS PubMed.
- J. Arnthong, J. Ponjarat, P. Bussadee, P. Deenarn, P. Prommana, A. Phienluphon, S. Charoensri, V. Champreda, X. Q. Zhao and S. Suwannarangsee, Enhanced surface display efficiency of β-glucosidase in Saccharomyces cerevisiae by disruption of cell wall protein-encoding genes YGP1 and CWP2, Biochem. Eng. J., 2022, 179, 108305 CrossRef CAS.
- H. Tang, M. Song, Y. He, J. Wang, S. Wang, Y. Shen, J. Hou and X. Bao, Engineering vesicle trafficking improves the extracellular activity and surface display efficiency of cellulases in Saccharomyces cerevisiae, Biotechnol. Biofuels, 2017, 10, 53 CrossRef PubMed.
- J. Arnthong, P. Bussadee, A. Phienluphon, P. Deenarn, K. Tulsook, S. N. Plupjeen, C. Siamphan, C. Tachaapaikoon, V. Champreda and S. Suwannarangsee, Overexpression of LAS21 in cellulase-displaying Saccharomyces cerevisiae for high-yield ethanol production from pretreated sugarcane bagasse, Fermentation, 2022, 8, 652 CrossRef CAS.
- T. Sakamoto, T. Hasunuma, Y. Hori, R. Yamada and A. Kondo, Direct ethanol production from hemicellulosic materials of rice straw by use of an engineered yeast strain codisplaying three types of hemicellulolytic enzymes on the surface of xylose-utilizing Saccharomyces cerevisiae cells, J. Biotechnol., 2012, 158, 203–210 CrossRef CAS PubMed.
- Y. Sasaki, T. Takagi, K. Motone, K. Kuroda and M. Ueda, Enhanced direct ethanol production by cofactor optimization of cell surface-displayed xylose isomerase in yeast, Biotechnol. Prog., 2017, 33, 1068–1076 CrossRef CAS PubMed.
- C. Zhang, H. Chen, Y. Zhu, Y. Zhang, X. Li and F. Wang, Saccharomyces cerevisiae cell surface display technology: Strategies for improvement and applications, Front. Bioeng. Biotechnol., 2022, 10, 1056804 CrossRef PubMed.
- S. Shiraga, M. Kawakami, M. Ishiguro and M. Ueda, Enhanced reactivity of Rhizopus oryzae lipase displayed on yeast cell surfaces in organic solvents: Potential as a whole-cell biocatalyst in organic solvents, Appl. Environ. Microbiol., 2005, 71, 4335–4338 CrossRef CAS PubMed.
- X. Li, X. Jin, X. Lu, F. Chu, J. Shen, Y. Ma, M. Liu and J. Zhu, Construction and characterization of a thermostable whole-cell chitinolytic enzyme using yeast surface display, World J. Microbiol. Biotechnol., 2014, 30, 2577–2585 CrossRef CAS PubMed.
- L. H. Fan, Z. J. Zhang, X. Y. Yu, Y. X. Xue and T. W. Tan, Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 13260–13265 CrossRef CAS PubMed.
- Z. Liu, S. H. Ho, K. Sasaki, R. Den Haan, K. Inokuma, C. Ogino, W. H. Van Zyl, T. Hasunuma and A. Kondo, Engineering of a novel cellulose-adherent cellulolytic Saccharomyces cerevisiae for cellulosic biofuel production, Sci. Rep., 2016, 6, 24550 CrossRef CAS PubMed.
- E. Y. Yuzbasheva, T. V. Yuzbashev, I. A. Laptev, T. K. Konstantinova and S. P. Sineoky, Efficient cell surface display of Lip2 lipase using C-domains of glycosylphosphatidylinositol-anchored cell wall proteins of Yarrowia lipolytica, Appl. Microbiol. Biotechnol., 2011, 91, 645–654 CrossRef CAS PubMed.
- Z. Liu, K. Inokuma, S. H. Ho, R. Den Haan, T. Hasunuma, W. H. Van Zyl and A. Kondo, Combined cell-surface display- and secretion-based strategies for production of cellulosic ethanol with Saccharomyces cerevisiae, Biotechnol. Biofuels, 2015, 8, 162 CrossRef PubMed.
- S. Yang, X. Lv, X. Wang, J. Wang, R. Wang and T. Wang, Cell-surface displayed expression of trehalose synthase from Pseudomonas putida ATCC 47054 in Pichia pastoris using Pir1P as an anchor protein, Front. Microbiol., 2017, 8, 2583 CrossRef PubMed.
- Y. Matano, T. Hasunuma and A. Kondo, Display of cellulases on the cell surface of Saccharomyces cerevisiae for high yield ethanol production from high-solid lignocellulosic biomass, Bioresour. Technol., 2012, 108, 128–133 CrossRef CAS PubMed.
- J. Bae, K. Kuroda and M. Ueda, Proximity effect among cellulose-degrading enzymes displayed on the Saccharomyces cerevisiae cell surface, Appl. Environ. Microbiol., 2015, 81, 59–66 CrossRef PubMed.
- M. R. Smith, H. Gao, P. Prabhu, L. F. Bugada, C. Roth, D. Mutukuri, C. M. Yee, L. Lee, R. M. Ziff, J. K. Lee and F. Wen, Elucidating structure–performance relationships in whole-cell cooperative enzyme catalysis, Nat. Catal., 2019, 2, 809–819 CrossRef CAS PubMed.
- S. Fan, B. Liang, X. Xiao, L. Bai, X. Tang, E. Lojou, S. Cosnier and A. Liu, Controllable display of sequential enzymes on yeast surface with enhanced biocatalytic activity toward efficient enzymatic biofuel cells, J. Am. Chem. Soc., 2020, 142, 3222–3230 CrossRef CAS PubMed.
- F. Guo, M. Liu, H. Liu, C. Li and X. Feng, Direct yeast surface codisplay of sequential enzymes with complementary anchor motifs: enabling enhanced glycosylation of natural products, ACS Synth. Biol., 2023, 12, 460–470 CrossRef CAS PubMed.
- Y. Fujita, J. Ito, M. Ueda, H. Fukuda and A. Kondo, Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme, Appl. Environ. Microbiol., 2004, 70, 1207–1212 CrossRef CAS PubMed.
- Z. Liu, K. Inokuma, S.-H. Ho, R. Den Haan, W. H. Van Zyl, T. Hasunuma and A. Kondo, Improvement of ethanol production from crystalline cellulose via optimizing cellulase ratios in cellulolytic Saccharomyces cerevisiae, Biotechnol. Bioeng., 2017, 114, 1201–1207 CrossRef CAS PubMed.
- K. S. K. Teo, K. Kondo, K. Saito, Y. Iseki, T. Watanabe, T. Nagata and M. Katahira, Enhanced depolymerization of beech wood lignin and its removal with peroxidases through continuous separation of lignin fragments, Green Chem., 2023, 25, 7682–7695 RSC.
- J. K. Wang, B. He, W. Du, Y. Luo, Z. Yu and J. X. Liu, Yeast with surface displayed xylanase as a new dual purpose delivery vehicle of xylanase and yeast, Anim. Feed Sci. Technol., 2015, 208, 44–52 CrossRef CAS.
- C. Liu, W. Zhang, Y. Li, K. Pan, K. OuYang, X. Song, X. Xiong, Y. Zang, L. Wang, M. Qu and X. Zhao, Characterization of yeast cell surface displayed Lentinula edodes xylanase and its effects on the hydrolysis of wheat, Int. J. Biol. Macromol., 2022, 199, 341–347 CrossRef CAS PubMed.
- C. Dong, X. Wang, W. Sun, L. Chen, S. Li, K. Wu, K. Wu, L. Ma, L. Ma, Y. Liu and Y. Liu, Engineering Pichia pastoris with surface-display minicellulosomes for carboxymethyl cellulose hydrolysis and ethanol production, Biotechnol. Biofuels, 2020, 13, 108 CrossRef CAS PubMed.
- W. Siripong, P. Wolf, T. P. Kusumoputri, J. J. Downes, K. Kocharin, S. Tanapongpipat and W. Runguphan, Metabolic engineering of Pichia pastoris for production of isobutanol and isobutyl acetate, Biotechnol. Biofuels, 2018, 11, 1 CrossRef PubMed.
- Y. Liu, L. Huang, D. Zheng, Y. Fu, M. Shan, Z. Xu, J. Ma and F. Lu, Development of a: Pichia pastoris whole-cell biocatalyst with overexpression of mutant lipase I PCLG47I from Penicillium cyclopium for biodiesel production, RSC Adv., 2018, 8, 26161–26168 RSC.
- H. Shimoi, H. Kitagaki, H. Ohmori and Y. Iimura, Sed1p Is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance, J. Bacteriol., 1998, 180, 3381–3387 CrossRef CAS PubMed.
- G. Su, X. Zhang and Y. Lin, Surface display of active lipase in Pichia pastoris using Sed1 as an anchor protein, Biotechnol. Lett., 2010, 32, 1131–1136 CrossRef CAS PubMed.
- W. Li, H. Shi, H. Ding, L. Wang, Y. Zhang, X. Li and F. Wang, Cell surface display and characterization of Rhizopus oryzae lipase in Pichia pastoris using Sed1p as an anchor protein, Curr. Microbiol., 2015, 71, 150–155 CrossRef CAS PubMed.
- J. Yang, K. Huang, X. Xu, Y. Miao, Y. Lin and S. Han, Cell surface display of Thermomyces lanuginosus lipase in Pichia pastoris, Front. Bioeng. Biotechnol., 2020, 8, 544058 CrossRef PubMed.
- S. M. R. Khattab, H. Okano, C. Kimura, T. Fujita and T. Watanabe, Efficient integrated production of bioethanol and antiviral glycerolysis lignin from sugarcane trash, Biotechnol. Biofuels Bioprod., 2023, 16, 82 CrossRef CAS PubMed.
- G. L. Miller, Use of dinitrosalicylic acid reagent for determination of reducing sugar, Anal. Chem., 1959, 31, 426–428 CrossRef CAS.
- A. Phienluphon, K. Kondo, B. Mikami, T. Nagata and M. Katahira, Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii, Int. J. Biol. Macromol., 2023, 253, 127188 CrossRef CAS PubMed.
- C. Ouephanit, N. Boonvitthya, S. Bozonnet and W. Chulalaksananukul, High-level heterologous expression of endo-1,4-β-xylanase from Penicillium citrinum in Pichia pastoris X-33 directed through codon optimization and optimized expression, Molecules, 2019, 24, 3515 CrossRef CAS PubMed.
- S. Yang, J. Shen, J. Deng, H. Li, J. Zhao, H. Tang and X. Bao, Engineering cell polarization improves protein production in Saccharomyces cerevisiae, Microorganisms, 2022, 10, 2005 CrossRef CAS PubMed.
- S. Singh, A. M. Madlala and B. A. Prior, Thermomyces lanuginosus: Properties of strains and their hemicellulases, FEMS Microbiol. Rev., 2003, 27, 3–16 CrossRef CAS PubMed.
- M. Adam, I. Gomes, G. Mohiuddint and M. M. Hoq, Production and characterization of thermostable xylanases by Thermomyces lanuginosus and Thermoascus aurantiacus grown on lignocelluloses, Enzyme Microb. Technol., 1994, 16, 298–302 CrossRef.
- M. Gaffney, S. Carberry, S. Doyle and R. Murphy, Purification and characterisation of a xylanase from Thermomyces lanuginosus and its functional expression by Pichia pastoris, Enzyme Microb. Technol., 2009, 45, 348–354 CrossRef CAS.
- M. Hou, C. Liang, Y. Fei, D. Yang, N. Zhang, Y. Lu, L. Wang, Z. Xing and Z. Zhao, Analysis of the effect of metal ions on the ability of xylanase to hydrolyze wheat bran by molecular dynamics simulations, Front. Bioeng. Biotechnol., 2023, 11, 1142873 CrossRef PubMed.
- Y. Zhang, Z. Feng, H. Xiang, X. Zhang and L. Yang, Characterization of feruloyl esterase from Klebsiella oxytoca Z28 and its application in the release of ferulic acid from de-starching wheat bran, Microorganisms, 2023, 11, 989 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |