Open Access Article
Open Access ArticleApplication of quantum dots in brain diseases and their neurotoxic mechanism
Yuanyuan
Hu
 a,
Xiaoli
Wang
a,
Yiru
Niu
a,
Keyu
He
*b and
Meng
Tang
a,
Xiaoli
Wang
a,
Yiru
Niu
a,
Keyu
He
*b and
Meng
Tang
 *a
*a
aKey Laboratory of Environmental Medicine & Engineering, Ministry of Education, School of Public Health, Southeast University, Nanjing, Jiangsu 210009, China. E-mail: tm@seu.edu.cn
bBlood Transfusion Department, Clinical Laboratory, Zhongda Hospital, Southeast University, Nanjing, Jiangsu 210009, China. E-mail: hky@seu.edu.cn
First published on 5th June 2024
Abstract
The early-stage diagnosis and therapy of brain diseases pose a persistent challenge in the field of biomedicine. Quantum dots (QDs), nano-luminescent materials known for their small size and fluorescence imaging capabilities, present promising capabilities for diagnosing, monitoring, and treating brain diseases. Although some investigations about QDs have been conducted in clinical trials, the concerns about the toxicity of QDs have continued. In addition, the lack of effective toxicity evaluation methods and systems and the difference between in vivo and in vitro toxicity evaluation hinder QDs application. The primary objective of this paper is to introduce the neurotoxic effects and mechanisms attributable to QDs. First, we elucidate the utilization of QDs in brain disorders. Second, we sketch out three pathways through which QDs traverse into brain tissue. Ultimately, expound upon the adverse consequences of QDs on the brain and the mechanism of neurotoxicity in depth. Finally, we provide a comprehensive summary and outlook on the potential development of quantum dots in neurotoxicity and the difficulties to be overcome.
1. Introduction
Quantum dots (QDs) technology dates back to the mid-1970s when Dr Brus accidentally discovered that cadmium sulfide displays different colors as the particle size changes.1 After this, in-depth research on quantum dots was started, and it was found that they have excellent optical properties, such as tunable fluorescence properties,2 good photostability,3 small size,4 and simple synthesis processes.5 Until P. Alivisatos et al.6 used quantum dots to label fibroblasts in 1998, quantum dots had come a long way in their synthesis, modification, and applications.Quantum dots exhibit various varieties and predominantly manifest as core–shell structures. Initially, binary quantum dots predominantly consisted of heavy metal ions, such as cadmium-based7 and silver-based QDs.8 However, advancements in technology have led to the emergence of non-metallic quantum dots, including silicon-containing QDs (SiQDs),9 carbon-containing QDs (CQDs),10 graphene-containing QDs (GQDs),11etc. There has been a growing development of ternary quantum dots, such as CuInS212 and CdZnSe.13 In addition to altering the composition of QDs, researchers also modified diverse ligands on their surfaces, aiming to improve bioavailability and diminish side effects. These modifications are tailored to meet the specific demands of different applications, such as cell labelling and tracking,14 deep tissue imaging,15 theranostics,16,17 biosensors,4 and drug and gene delivery.18 Specifically, the small size of QDs, typically ranging from 2 to 20 nm, allows them to penetrate the Blood–Brain Barrier (BBB),19 thereby presenting a novel avenue for the manufacture of functional brain probes utilizing QDs.
Despite the proven advantages of quantum dots, concerns about potential toxic effects have always accompanied them. QDs can be transported to different tissues and organs upon exposure, resulting in adverse effects in organisms.20 This review aims to gather information on recent advancements in applying QDs for identifying and treating brain diseases. It also provides an overview of the pathways through which QDs enter the brain and their interactions with the nervous system. Additionally, it elucidates the mechanisms underlying the toxic effects of QDs within the cerebral nervous system.
2. Application of quantum dots in brain diseases
With the rapid aging of the population and lifestyle change, the incidence of neurological diseases, such as brain tumours, cerebrovascular diseases, and neurodegenerative diseases, continues to rise, placing a heavy burden on health and socio-economic development.21,22 However, due to the brain's complex structure, almost all neurological diseases lack effective diagnosis and therapy methods in the clinic. Medical imaging techniques or pathological methods commonly used in clinical practice are mainly focused on anatomical and functional changes in organs, with limited help in the early diagnosis of diseases. Various physiological barriers prevent drugs from entering the brain. There is an unmet need to formulate new methods for early diagnosis and real-time tracking of the prognosis of CNS disorders. The unparalleled ability of tissue penetration (a few centimeters), spatial resolution (∼10 mm), as well as the capability to act as drug carriers make QDs ideal candidates to break through the above dilemma.2.1. Brain tumours
The overall survival of patients with brain tumours is usually less than 5 years.23 The earlier diagnosis always indicates a better prognosis. Therefore, developing a highly sensitive and selective diagnostic reagent for detecting brain tumours is urgently needed. QDs can emit iridescent fluorescence and target different cells by modifying various molecules and proteins, making them ideal for visualizing and tracking molecular processes within the cell.24 More importantly, the feature of QDs penetrating the blood–brain barrier25 makes it possible to monitor tumour development and neovascularization in real time. Some studies have confirmed the potential of QDs in tumour diagnosis and treatment. Ganganboina et al.26 well-crafted a dual functional probe, S-GQDs@Au–CNS nanocomposites decorated with Angiopep-2, for glioma cell detection. The probe has an excellent linearity range, from 100 cell per mL to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cell per mL, and the threshold detection limit is 40 cell per mL for detecting glial cells in human serum in vitro. In addition to creating a new diagnostic tool, QDs can also be coupled with existing diagnostic methods to improve the accuracy and sensitivity of brain tumor diagnosis. Chunyan Li et al.27 synthesized a unique Gd-DOTA doped Ag2S QD nanoprobe, which showed an outstanding contrast enhancement in deep tissue imaging, and moreover revealed a high signal-to-background ratio and high spatiotemporal resolution of fluorescence imaging in the NIR-II of Ag2S. Due to concerns about the toxicity of Gd, the boron-doped graphene quantum dot was a choice for Gd-containing contrast agents. It showed a better T1 contrast enhancement in imaging in vivo and was demonstrated to be transported to the brain.28
000 cell per mL, and the threshold detection limit is 40 cell per mL for detecting glial cells in human serum in vitro. In addition to creating a new diagnostic tool, QDs can also be coupled with existing diagnostic methods to improve the accuracy and sensitivity of brain tumor diagnosis. Chunyan Li et al.27 synthesized a unique Gd-DOTA doped Ag2S QD nanoprobe, which showed an outstanding contrast enhancement in deep tissue imaging, and moreover revealed a high signal-to-background ratio and high spatiotemporal resolution of fluorescence imaging in the NIR-II of Ag2S. Due to concerns about the toxicity of Gd, the boron-doped graphene quantum dot was a choice for Gd-containing contrast agents. It showed a better T1 contrast enhancement in imaging in vivo and was demonstrated to be transported to the brain.28
Over and above being a nano-diagnostic tool, the potential of QDs in brain tumour therapy is equally fascinating. The most effective treatment for brain tumours is surgery. Traditional organic fluorophores have limitations distinguishing tumours adjacent to eloquent brain regions, leading to incomplete surgical resection and residual micro lesions.29 Remnants of microscopic lesions postoperatively constitute a significant cause of tumour recurrence and poor prognosis. Thus, accurate determination of the boundary between malignant neoplasm and normal tissue and identification of micrometastatic tumor cells are essential considerations for complete resection and a critical factor in preventing glioma recurrence.30 Donna et al. employed QDs-EGF and QDs-EGFR as staining agents for glioma cells, tumour bearing mice, and human brain glioma sections. They found a visible distinction between normal and tumour tissue.31
Due to the obstruction of the BBB, how to deliver drugs across the BBB and target the tumor tissue are two obstacles to glioma treatment. Due to their small size, quantum dots can serve as vehicles delivering anti-tumor drugs, such as epirubicin, temozolomide, and gemcitabine.32,33 At the same time, they can control the drug release34 and increase the concentration of anti-tumour drugs at the tumour site while reducing their concentration in the systemic circulation. Li Z. et al.35 synthesized an Nd3+ ion-ligated black phosphorus quantum dot that could penetrate the BBB, monitor the growth of glioblastoma in real-time, and participate in tumour treatment as a photodynamic chemotherapy synergist under specific X-ray guidance. In addition to directly killing tumour cells, quantum dots also have a synergistic effect with chemotherapy, so chemotherapy can play an anti-tumour role at a lower dose level, which is conducive to reducing chemotherapy toxicity.36 Here, we summarized the applications of QDs in brain tumours in Table 1.
| No. | Core | Shell | Modification | Size | Model | Uses | References |
|---|---|---|---|---|---|---|---|
| a Abbreviations: MIONs, magnetic iron oxide nanoparticles; PEGNIO, polyethylene glycol niosome; CMCel, carboxymethylcellulose; CMCelCys, carboxymethylcellulose with L-cysteine; CMCelPolyArg, poly-L-arginine with carboxymethylcellulose; BSA-DTPAGd, DTPA-coupled BSA with Gd3+ chelation; c(RGDyk), cyclic arginine–glycine–aspartic acid short chain polypeptide. | |||||||
| 1 | InP | ZnS | PEGNIO | 178.5 nm | U87 cell line | Enhanced MRI imaging effect of glioma | 37 |
| MIONs | |||||||
| Tf | |||||||
| 2 | CdSe | ZnS | Interleukin-13 | 15–20 nm | U251, T3691, and T387 | Facilitated the early detection of tumor recurrence, distinguished recurrent tumors from pseudoprogress, and potentially enabled the diagnosis of patients in high-risk populations | 38 |
| Brain tumour patients' cerebrospinal fluid | |||||||
| 3 | CuInS2 | ZnS | BSA-DTPAGd | 45 nm | SU2 stem cells | Specifically target the tumor cells and enhance the efficacy of contrast agents of MRI imaging | 39 |
| Anti-CD133 monoclonal antibody | Nude mice | ||||||
| 4 | QD800 | — | EG2-hFc | U87MG.EGFRvIII | Non-invasive imaging of brain tumours in vivo, detection of tumour aggressiveness and resistance | 40 | |
| EG2-Cys | U87MG.EGFRvIII bearing mice | ||||||
| 5 | AgInS2 | CMCel | AIS-CMCel: 3.5 ± 0.5 nm | HEK 293T | Bioimaging and biolabeling in glioma cells | 41 | |
| CMCel-Cys | AIS-CMCel-Cys: 3.3 ± 0.4 nm | ||||||
| CMCel-PolyArg | AIS-CMCel-PolyArg: 3.7 ± 0.5 nm | U-87 MG cells | |||||
| 6 | Carbon dots | — | Nitrogen | ∼2.6 nm | 293T cells | Intracellular bioimaging and transfer of the BBB | 42 |
| BBB model | |||||||
| 7 | AgInS2 | CMC-KLA | — | U87 MG | Imaging and cellular tracking in U-87 MG cells | 43 | |
| CMC-cysteine | |||||||
| CMC-CYS-KLA | Chicken eggs | Inhibited the formation of the new blood vessels in CAM assays | |||||
| 8 | Gold QDs | — | <10 nm | T98G cells, U87 cells, U373 cells, SNU-80 cells, H460 cells, MRC5 cells, and HEK293 cells | Inhibit self-renewal of tumour cells and reduce tumour metastasis | 44 | |
| 9 | Graphene QDs | NH2– | 10 nm | U87 cells | Green-QDs and COOH-GQDs serve as synergistic agents with doxorubicin on U87 cells | 45 | |
| COOH– | |||||||
| Green– | Mouse cortical neurons | ||||||
| 10 | CdSe | ZnS | Biotinylated aptamer | 20 nm | U87 cells, HUVEC cells, and U87-EGFRvIII cells | Brain tumour imaging in cells and surgical guidance in tumour-bearing mice | 46 |
| Nude mouse | |||||||
| 11 | CdSe | ZnS | Liposomes, superparamagnetic iron oxide nanoparticles | CdSe/ZnS: ∼8 nm | C6 cells | Surgical guidance and delivery of antineoplastic drugs | 47 |
| Cilengitide | QSC-Lip: 100 ± 1.24 nm | Glioma-bearing rats | |||||
| 12 | ZnCdSe | ZnS | c(RGDyk)-poloxamer-188 | 212.4 nm | C6 cells, HUVECs, and PC12 cells | Targeting and biolabeling C6 cells in vitro, and accumulation in glioma tissue in orthotropic tumour rats | 48 |
| Orthotropic tumour rats | Surgical guidance of glioma | ||||||
| 13 | Graphene QDs | — | Nitrogen and boron | ∼4.7 nm | SF-763 cells, 4T1 cells, and B16F10 cells | Visualization of blood vessels and organs in vivo. Used as a photothermal therapy agent in the treatment of tumours | 49 |
| Nude mice | |||||||
| Glioma-bearing nude mice | |||||||
2.2. Cerebrovascular diseases
Cerebrovascular disease refers to cerebrovascular lesions caused by various reasons or brain dysfunction caused by blood flow disorders, including ischemic cerebrovascular disease and hemorrhagic cerebrovascular disease, which is one of the major factors leading to death.50 Real-time visualization of the cerebral vasculature in situ is essential for curing cerebrovascular disease. QDs have a range of superiorities in fluorescence imaging, high tissue penetration depth, good resolution, and rapid data acquisition,51 which can be applied to non-invasive vascular visualization and continuous monitoring. Jun Q. et al.52 performed bilayer modification of PbS/CdS QDs, which showed high biostability and pH stability, and were successfully used for deep cerebral angiography in mice. In addition to their fluorescence effects, QDs have a sensitization effect to enhance the luminescence efficiency of other fluorescent nanomaterials. Lanthanide-doped nanomaterials (LnNCs) are some of the NIR-II nanoprobes. The sensitizing effect of QDs can increase the brightness of LnNCs by more than 100 times, and their penetration depth reaches 11 mm.53 The probe can be used for cerebral vascular structure imaging of ischemic stroke and traumatic brain injury and to monitor cerebral vascular hemodynamics. The material is expected to be helpful in pre-hospital diagnosis or early detection of ischemic stroke and in-hospital monitoring of this condition. Here, we summarized the applications of QDs in cerebrovascular diseases in Table 2.| No. | Core | Shell | Modification | Size | Model | Uses | Disease name | References |
|---|---|---|---|---|---|---|---|---|
| 1 | PbS | Ag2Se | V&C | ∼150 nm | HUVECs cells | Early detection of ischemia stroke | Stroke | 54 |
| BALB/c mouse | ||||||||
| 2 | Ag2S | — | V&A | 39.4 nm | Brain-injured mice | Early real-time assessment of traumatic brain injury | Traumatic brain injury | 55 |
| 3 | CdSe | ZnS/ZnS | mPEG-PAEA-DDA | — | Cerebral ischemia rats | Detect cerebral ischemic disease in rats | Cerebral ischemia | 56 |
2.3. Neurodegenerative disorders
Neurodegenerative disorders (NDs) are a class of diseases in the brain and spinal cord which are characterized by loss of synapses and neurons. Most research focuses on AD and PD. Some hypothesis had been proposed to explain the pathogenesis of NDs, like oxidative stress,57 mitochondrial dysfunction,58,59 immune inflammation,60 metal ion disorder,61 and so on, and among them the amyloid cascade hypothesis is the most famous. Due to the complex pathogenesis of NDs, there are very few approaches that have been approved.With the development of nanotechnology, many studies have found that quantum dots seem to have potential for the diagnosis and treatment of NDs. QDs can scavenge free radicals effectively, such as superoxide anions, hydrogen peroxide, and hydroxyl radicals. GOQDs exert broad-spectrum antioxidant activity to relieve apoptosis and alleviate α-synuclein and mitochondrial damage in MPP+-treated zebrafish.62 Once inside the brain, SeQDs can play a similar catalase activity; they can rapidly attenuate AD, significantly ameliorate memory impairment, and improve the learning and memory capacity of AD mice.63 Beyond that, QDs also achieve some progress in the diagnosis and treatment of Parkinson's disease. GQDs inhibit α-syn fiber formation and trigger fiber depolymerization, promote neuronal and synaptic regeneration, reduce the formation of Lewy bodies and Lewy synapses, promote mitochondrial repair, and prevent interneuronal transmission of α-syn pathology.64 GQDs could enter neural stem cells via endocytosis without altered viability, metabolic activity, proliferation, and differentiation potential in human neural progenitor cells.65 These studies provide new possibilities for the treatment of neurodegenerative disorders using QDs. Here, we summarized the applications of QDs in neurodegenerative disorders in Table 3.
| No. | Core | Shell | Modification | Size | Dosage | Model | Uses | Disease name | Hypothesis | References |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CQD | — | GSH | 4.37 ± 0.87 nm | 250 μg mL−1 | Human serum | Monitoring treatment efficacy and prognosis of PD by testing levodopa | Parkinson's disease | Dopamine theory | 66 |
| 2 | GQDs | — | PEGylated biotin | — | 1 μg mL−1 in cells | Primary cortical neuron cells and C57BL/6 mice | Treat PD by inhibiting α-synuclein (α-syn) fibrillization and disaggregating fibrils | Parkinson's disease | α-syn amyloid aggregation | 67 |
| 50 μg in mice | ||||||||||
| 3 | CQD | — | Sodium citrate, phenylboronic acid, and 4-aminophenylboronic acid | 2–9 nm, 12–22 nm, 5–13 nm | 80 μg mL−1 | SH-SY5Y cells | Treat PD by scavenging reactive oxygen species | Parkinson's disease | Oxidative stress | 68 |
| 4 | CdSe | — | Mito-metformin | — | 30 μg mL−1 | N27 cells | Treat PD by reducing mitochondrial dysfunction | Parkinson's disease | Mitochondrial dysfunction | 69 |
| 5 | SeQD | — | — | 5 nm | 20 μg mL−1 | SH-SY5Y cells and AD model mice | Treat PD by scavenging reactive oxygen species | Alzheimer's disorder | Inflammatory, mitochondrial dysfunctions, and oxidative stress | 63 |
| 1 mg kg−1 | ||||||||||
| 6 | CuInS2 | ZnS | Dopamine | 3–5 nm | 400 μM | Human serum | Diagnosis of AD by detect ion of tau protein | Alzheimer's disorder | Aβ accumulation, tau protein hyperphosphorylation | 70 |
| 7 | CdSe | ZnS | Biphenyl ethers | 5 nm | 5–10 mM | PSEN1dE9 transgenic mouse | Treat AD by inhibiting the formation of Aβ fibrils | Alzheimer's disorder | Amyloid hypothesis | 71 |
| 8 | GQDs | — | Glycine–proline–glutamate | 18 nm | 200 μg mL−1 | APP/PS1 transgenic mouse | Treat AD by inhibiting Aβ plaque production and stimulating new neural precursor cells and neuron generation | Alzheimer's disorder | Amyloid hypothesis | 72 |
| 9 | MoS2 QDs | — | TPP | 50 nm | 100 μg mL−1 | hCMEC/D3 cells, BV-2 cells, PC12 cells, and APP/PS1 transgenic mouse | Treat AD by converting M1 microglia to M2 microglia and clearing Aβ aggregates | Alzheimer's disorder | Amyloid hypothesis | 73 |
| 10 | CNDs | Memantine | <5 nm | 50 mg mL−1 | Zebrafish | Treat AD by inhibiting the aggregation of tau proteins | Alzheimer's disorder | Amyloid hypothesis | 74 | |
| B-CDs |
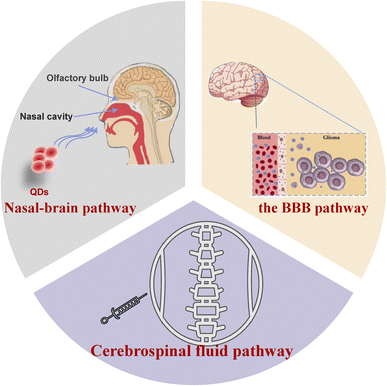
3. The primary way for QDs to enter brain tissue
Due to the brain's complex structure, traditional delivery methods make it difficult to transport drugs to lesion locations. Nevertheless, the ultra-small size of quantum dots makes them uniquely useful for drug delivery. Studies have shown that QDs can enter the central nervous system through various pathways, e.g., blood–brain barrier, nasal–brain transport, cerebrospinal fluid pathway, etc. Some quantum dots are synthesized using heavy metals, and brain tissue is susceptible to these substances. Therefore, an in-depth understanding of how quantum dots enter the brain is conducive to developing new diagnoses and treatments of brain diseases. The different ways for QDs to enter the brain are outlined in Fig. 1.3.1. Transport through the BBB pathway
The blood–brain barrier is a dynamic interface comprised of blood vessels regulating the passage of compounds both into and out of the brain, from internal and external sources. It can self-regulate in response to subtle changes in the brain or blood circulation to maintain the homeostatic environment of the central nervous system.75,76 Only lipid-soluble molecules with molecular weight <400 Da can break through the BBB.77 This property is a double-edged sword, protecting the brain from damage while also making it difficult for most therapeutic drugs to work. It seems to be the crux that has hindered the treatments for neurological diseases.Due to the small size, biocompatibility, long-term stability, and drug delivery capacity, QDs are expected to solve this dilemma. The following pathways endow QDs with the possibility to go through the BBB: (1) endocytosis, (2) passive diffusion, (3) inhibition of efflux pump, and (4) opening of tight junctions between BBB cells. All of these pathways can also exist simultaneously. S. Kato et al. intraperitoneally injected CdSe/ZnS QDs coated with captopril into ICR mice. The results show that quantum dots can cross the BBB and reach the brain and brain parenchyma through transcytosis-mediated patterns. At the same time, they determined cadmium concentrations in different brain regions by ICP-MS. The result showed that cadmium levels significantly increased within the olfactory bulb, cerebral cortex, hippocampus, thalamus, and brainstem of mice.78 Moreover, quantum dots have demonstrated the ability to traverse the BBB model via CD13 receptor-mediated transport channels.79 QDs destroyed the BBB structure by over-produced ROS and damaged membrane phosphatidylserine.80
Zhang et al. found that CdSe/CdS-PEG-OH quantum dots clustered in various brain regions can be internalized by microglia.81 After entering cells, most QDs were distributed in vesicles. In contrast, a few were distributed in tubular structures and polyvesicles.82 CdS QDs induce neuronal and Purkinje cell degeneration and inflammation in rats.83
3.2. Nasal–brain pathway
Intranasal administration is a viable, non-invasive method of intracranial administration. Instilled or inhaled drugs spread across the nasal mucosa and reach the central nervous system through the respiratory epithelium pathway or olfactory epithelium pathway.84 Studies have shown that ultrafine nanoparticles (<50 nm) are more likely to invade the brain in this way.85 CdTe QDs went into the nasal cavity of mice and diffused into the olfactory bulb, and nasal fluorescence intensity gradually declined after 2 h. At 24 h, the fluorescence in the olfactory bulb decreased, but the fluorescence was observed in the brain tissue.86 These results indicate that QDs were transported across the nasal mucosa via the axons of the olfactory neurons to the olfactory bulb and distributed throughout the olfactory bulb and the brain. The transportation mode depends on the dimensions and surface modification of quantum dots. When bovine nasal mucosal explants were stained with QDs-COOH and QDs-PEG, respectively, it was found that only QDs-COOH could be detected in nasal mucosal explants.87 The nasal–brain route can avoid general circulation and reduce systemic adverse events. However, its delivery efficiency is not high, and the drugs that can reach the target lesions are extremely limited, so overdose or repeated doses are required to achieve therapeutic effects. The direction of future research will be how to improve transportation efficiency and reduce the local toxicity of the nasal cavity.3.3. Cerebrospinal fluid pathway
In addition to the BBB pathway and the nasal–brain pathway, drugs can also reach brain tissue through the cerebrospinal fluid pathway. Moreover, the cerebrospinal fluid pathway allows the rapid distribution of drugs in the cerebrospinal fluid88 to achieve effective concentrations while simultaneously avoiding the adverse effects of large intravenous doses and effectively increasing the intracerebral concentration of the drug. In order to validate the targeting of brain tissue and the imaging role of QDs in a nude mouse model, Liang et al.89 injected folic acid-modified CdSeTe/Zinc Sulfide (ZnS)QDs into the orthotopic U87MG nude mice transplanted model via intrathecal injection. After coupling with folic acid, CdSeTe/ZnS QDs penetrate the tumour interior and potentially contribute to the detection and management of cancer, including image-guided surgery. Juan A. Varela et al.90 performed two types of injections in different cerebral hemispheres of the same rat. At 3 hours after QD administration, massive activation of microglia in cerebral hemispheres injected with localized brain tissue indicated phagocytosis of microglia within the brain within the QDs, and few QDs were observed outside of microglia. In contrast, brain tissue injected into the ventricles did not show microglia activation, and most of the QDs were scattered throughout the tissue. These results suggest that utilization of the cerebrospinal fluid pathway is more conducive to quantum dot distribution within intracerebral tissues.4. Neurotoxic effects and the mechanism of QDs
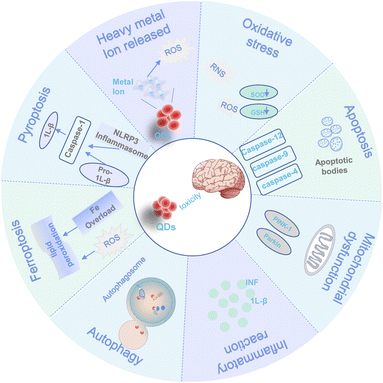
With the gradual use of quantum dots, the controversy about the toxicity of quantum dots has never stopped. The toxicity of QDs depends on the size, surface charge, concentration, surface modification, character, and solubility of the materials, as well as the purity.91–93 There needs to be more comprehensive analyses of the mechanism of QD-induced neurotoxicity. The different ways of interaction between quantum dots and the brain are outlined in Fig. 2.4.1. Neurobehavioural impairments and brain tissue damage
Neurotoxicity can impair neurological functions and endanger human motor coordination, learning ability, and cognitive functions. Exposure to CdSe/ZnS QDs reduced hatch and survival rates and delayed long development with fertilized zebrafish germ cells.94 It allows zebrafish to spend more time in dark areas while reducing motor activity in the dark. These changes may result from the alteration of dopaminergic in developing larvae.95 Prolonged exposure to 0.1–1 g per L CdTe QDs can cause entry of REM motor neurons via the intestinal barrier, leading to altered REM development and abnormal foraging behavior in nematodes.96 In addition, it has been found that QDs decreased the body bends and head trash frequency and negatively impacted learning and memory ability in Caenorhabditis elegans.97 The same results were observed in the rodent model. Wu et al.98 found that CdTe QDs can decrease Wistar rats' spatial memory capacity; moreover, the short-term memory impairment was dose and time-dependent.After entering the body via the gastrointestinal tract, skin, and respiratory tract, QDs are rapidly distributed throughout the kidneys, liver, lung, gastrointestinal tract, and other tissues and organs with blood circulation.99–101 They can also cross the blood–brain barrier and enter the cerebral cortex, hippocampus, cerebral nuclei, cerebellu, and other brain regions.102 The nasal drip of N-GQDs caused decreased organ coefficients and pathological structural damage in rat brain tissue.103 Prasad et al.104 found that after long-term exposure of CdTe QDs to differentiated PC12 cells, quantum dots were deposited in the cytoplasm. CdSe/ZnS QDs induce autophagosome formation in primary neuronal cells, which triggers autophagy in neuronal cells and results in impaired synaptic function in area CA1 of the hippocampus.105 CdS QDs injected intraperitoneally into mice for one week caused vascular atrophy, congestion, and degeneration in the brain and cerebellar tissue, necrotic degeneration, and DNA breaks in neurons and astrocytes in cortical regions, and pathologic features such as reduced Purkinje cell number and size, cellular degeneration and apoptosis.83
4.2. Mechanism of neurotoxicity induced by QDs
Several mechanisms contribute to the neurotoxicity of QDs. They mainly include non-neurological specific mechanisms (e.g., such as oxidative stress, heavy metal ion release, apoptosis, mitochondrial dysfunction, inflammation, autophagy, ferroptosis, pyroptosis, and genomic instability.) and neurological specific mechanisms of action (e.g., intervention in GABA metabolic pathways and neurotransmitter receptor-mediated). An overview of the non-neurological mechanisms of QDs is provided in this review. Table 4 demonstrates some neurotoxicity mechanisms in QDs.| No. | QDs | Dose | Exposure time | Model | Toxicity mechanism | Reference |
|---|---|---|---|---|---|---|
| 1 | CdSe | 40 μg mL−1 | 72 h | T98G human glioma cells | Regulated neuroinflammation via hormonal control of the hypothalamus via gonadotropin-releasing hormone receptors. Downregulated the pro-inflammatory interleukin gene | 106 |
| 2 | CdS | In vitro: 0.01–100 μg mL−1 | 24 h | Cerebellar cell culture and male SD rats | CdS at 0.01 μg mL−1 had no significant toxic effect. The expression of 8-OHdG and GFAP was significantly up-regulated at high doses | 83 |
| In vivo: 0.1–25 mg kg−1 | ||||||
| 3 | MPA-CdTe | 0, 20 μM, 40 μM, and 80 μM | 24 h | RSC96 cells | The induction of RSC96 apoptosis and autophagy by CdTe QDs was concentration-dependent and could lead to apoptosis and autophagy through the mitochondrial and endoplasmic reticulum pathways | 107 |
| 4 | Ag2Se | 0, 0.01 μM, 0.1 μM, and 1 μM | 24 h and 72 h | C. elegans | The accumulation of Ag2Se QDS in C. elegans shortens its lifespan and disrupts its normal neural behavior. In addition, Ag2Se QDs caused an overproduction of ROS and altered the expression of genes involved in REDOX balance | 108 |
| 5 | Graphene QDs | U251cells | ROS generation and singlet linear oxygen kill U251 human glioma cells via autophagy and apoptosis | 109 | ||
| 6 | CuInS2/ZnS | 12.5–100 μg mL−1 | 24 h | U87 cells | CuInS2/ZnS QDs can enter cells with low cytotoxicity, primarily in the cytoplasm. However, they can cause significant alterations in gene expression patterns, including ribosomes, DNA-chromosome binding, and chromosome assembly | 110 |
| 7 | MPA-CdTe, CdTe@ZnS | BV2 cells:1.25 nM and 5 nM | BV2 cells: 12 h | BV2 cells | In C. elegans, exposure to QDs induced immune responses and neurobehavioural deficits. Neuroinflammatory responses to QD exposure, including activation of microglia and release of IL-1β, were observed in the hippocampal region of QD-treated mice. QDs containing Cd activate the NLRP3 inflammasome by inducing excessive ROS production and releasing IL-1β | 111 |
| C. elegans: 0.01 μM and 1 μM | C. elegans: 24 h, 72 h | C. elegans | ||||
| Mice: 0, 0.25 mM, and 2.5 mM | Mice: 24 h, 28 days | Mice | ||||
| 8 | MPA-CdTe | 10 nM, 20 nM, and 40 nM | 24 h | BV2 cells | Exposure to high levels of MPA-CdTe QDs activated microglia, promoted the secretion of IL-1β, and was strongly correlated with activation of the TLR2/MyD88/NF-κB pathway and NLRP3 inflammasome induced by ROS | 112 |
| 9 | CdSe/ZnS-LPS | 10 μg mL−1 | 24 h | N9 cells | LPS QDs induce toll-like receptor 4 (TLR-4) activation and subsequent NLRP3 inflammasome vesicle activation through p38 and JNK signaling. In the inflammasome, pro-caspase 1 is cleaved, and caspase-1 is released from pre-cleaved IL-1β, ultimately releasing the pro-inflammatory cytokine (IL-1β) | 113 |
| 10 | Gold QDs | 25 nM | 48 h | U373 cells and U87 cells | Gold QDs and cold atmospheric plasma (CAP) induce different glioma cell death through the Fas/TRAIL signal pathway | 114 |
| 11 | CdTe and CdTe@ZnS | 1.25 nM, 2.5 nM and 5 nM | 12 h | BV2 cells | Both QDs could trigger NLRP3 priming and pro-IL-1β expression, up-regulation in IL-1 β release, ultimately causing cell death by pyroptosis | 115 |
| 12 | Ag2Se QDs | 1.25 nM, 2.5 nM and 5 nM | 12 h | BV2 cells | Ag2Se QDs triggered NLRP3 priming and pro-IL-1β expression through the activated NF-κB pathway and ROS generation; these factors all up-regulated in IL-1 β release and then led to pyroptosis in microglia | 116 |
| 13 | MoS2 QDs | 50 μg mL−1 | 0 h, 4 h, 6 h, and 12 h | Primary microglia | MoS2 QDs promote NLRP3 inflammasome priming, resulting in microglia cell pyroptosis through the caspase-1 signal pathway | 117 |
| Primary astrocytes |
Laurie E. Hopkins et al.125 exposed mice to aerosolized CdSe/ZnS QDs for 1 h. No acute cytotoxicity of the nasal epithelium or olfactory bulb was found, but increased microglia activation in the olfactory bulb of mice exposed to QDs was observed. Microglia are the intrinsic immune cells of the central nervous system and constitute only 5–20% of glial cells. Under stressful conditions, microglia are activated and release anti-inflammatory cytokines and chemokines that reduce the inflammatory response while producing reactive oxygen and nitrogen species. Ren et al.,73 in their exploration of QDs for AD, found that TPP-MoS2 QDs attenuated Aβ aggregation-mediated neurotoxicity by switching microglia from a pro-inflammatory M1 phenotype to an anti-inflammatory M2 phenotype, resulting in the elimination of Aβ aggregation in mice with AD. On the other hand, if overactivated or chronically activated, microglia can lead to further brain injury caused by the release of pro-inflammatory cytokines. After entering microglia, CdTe/ZnS QDs are capable of polarizing microglia towards the M1 phenotype through activation of the mammalian target of the rapamycin (mTOR) protein pathway and activation of Nod-like receptor 3 (NLRP3) inflammatory vesicles to release inflammatory factors TNF-α, IL-1β, and NO, resulting in secondary inflammatory damage.126 Fuster et al.106 assessed the effects of CdSe-QDs on human T98G glioblastoma cells using RNAseq at maximum noncytotoxic dose. They found that after 72 h of CdSe-QDs exposure, altered signaling pathways primarily involved regulating inflammatory and immune responses and regulating the gonadotropin-releasing hormone receptor pathway, including the NF-κB and MAPK signaling pathways.127 In addition, CuInS2/ZnS-PE negatively regulates the downstream MAPK cascade, which significantly downregulates the expression of the NGF receptor (p75NTR) and inhibits NGF-induced synaptic growth through the NGF/p75NTR/MAPK pathway,128 causing neurotoxic damage.
The concentration of heavy metal ions is a critical factor in the cytotoxicity of QDs.126 Studies have shown that Cd2+ dissociated from CdTe QDs can promote the adverse effects of CdTe QDs.133 We speculated that the following reactions would occur in cells with QD exposure: CdTe QDs were disassembled by intracellular enzymes and released Cd2+. Cd2+ and CdTe QDs induced cells to produce excess ROS together;121 the excess ROS produced could chemically degrade the CdTe QDs, which resulted in more Cd2+ being released from the CdTe QDs.134,135 Besides the toxic effects caused by heavy metals, studies have shown that Cd2+ can affect the metabolism of other metal ions, like Cu2+ (ref. 136) in organisms, which in turn causes further damage to the body.
4.2.3.1 Cell-surface death receptors. In nucleated cells, the extrinsic pathway of apoptosis is triggered by the interaction of death ligands of the tumour necrosis factor superfamily with the death receptors on the external cell surface membrane, including FAS ligand-FAS/APO1, TNF–TNF receptors, and TRAIL–TRAIL receptors. Co-treatment with AuQDs and CAP induces reactive oxygen/nitrogen species (RONS) that promote apoptosis in tumour cells by positively regulating the expression of the apoptosis-associated Fas signaling pathway in both U373 and U87 cells.114 NAC and Trolox are two common antioxidants, and Trolox does not have a significant protective effect against the toxicity of the CdTe QDs on PC12 cells. However, it is still being determined how to protect against these effects while NAC can withstand active quantum dot damage on PC12 cells.124 Thus, NAC may activate the Ras-ERK pathway in PC12 cells,143 promoting the activation of PC12 cell proliferation and producing an anti-apoptotic effect.
4.2.3.2 Mitochondrial apoptotic pathway. Mitochondria are the main sites of ATP synthesis, providing energy for cellular activities, maintaining intracellular Ca2+ homeostasis, controlling aerobic metabolism, and maintaining cell survival.144–146 Chan et al.147 found that QDs up-regulated BAX, p-MAPK/JNK expression in human neuroblastoma cells and down-regulated BCL2, p-MAPK/ERK, HSP90, RAF1, and RAS protein expression, leading to a loss of mitochondrial membrane potential, mitochondrial release of cytochrome c, as well as apoptosis-inducing effects of caspase-9 and caspase-3 leading to apoptosis. Based on these findings, structural disruption and functional impairment of mitochondria are considered two major factors of quantum dot toxicity. The leading cause of the endogenous apoptotic pathway is mitochondrial damage, where exogenous or endogenous toxicants enter the cell and block the transmission of the mitochondrial respiratory chain, causing mitochondrial swelling, lowering mitochondrial membrane potential, altering mitochondrial membrane fluidity, inducing the opening of the permeability transition pore (MPT) of the mitochondrial inner membrane, and the release of pro-apoptotic proteins.148,149 Both pro-apoptotic and anti-apoptotic Bcl-2 family members have been shown to regulate apoptosis by regulating the release of mitochondrial factors, including cytochrome c; the quantum dot-induced apoptosis signaling pathway has also been shown in human neuroblastoma.
5. Conclusions
Quantum dots have obvious biomedical value. On the one hand, there is a thirst for this advanced technology. On the other hand, there are severe concerns about toxic effects. What lies ahead of the widespread use of quantum dots is a path of continued exploration. QDs cause neurological damage through multiple pathways, and these toxic effects may involve various mechanisms and numerous neural factors. However, the existing studies cannot fully grasp the transformation process and the existing form of quantum dots in the nervous system. Therefore, further studies are needed to focus on the molecular mechanisms of quantum dot-induced neurotoxicity.Meanwhile, due to the complex structures and functions of the nervous system, most of the studies had been carried out in vitro. More in vivo studies still need to be performed to predict the toxic effects of quantum dots on humans more accurately. At the same time, evaluating the toxicity and safety of QDs has yet to catch up with their applications. Further understanding of the mechanism of neurotoxicity caused by quantum dots is expected to accelerate the application of quantum dots.
Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82173545, 32101116, 31671034, and 21876026) and the Fundamental Research Funds for the Central Universities (No. 2242023k30020).References
- R. Rossetti, S. Nakahara and L. E. Brus, J. Chem. Phys., 1983, 79, 1086–1088 CrossRef CAS.
- Y. Zhang, H. Yang, X. An, Z. Wang, X. Yang, M. Yu, R. Zhang, Z. Sun and Q. Wang, Small, 2020, 16, e2001003 CrossRef.
- Isnaeni, L. H. Jin and Y. H. Cho, J. Colloid Interface Sci., 2013, 395, 45–49 CrossRef CAS.
- S. Chung, R. A. Revia and M. Zhang, Adv. Mater., 2021, 33, e1904362 CrossRef.
- S. Iravani and R. S. Varma, Environ. Chem. Lett., 2020, 18, 703–727 CrossRef CAS.
- M. Bruchez, M. Moronne, P. Gin, S. Weiss and A. P. Alivisatos, Science, 1998, 281, 2013–2016 CrossRef CAS.
- T. V. Duncan, A. Bajaj and P. J. Gray, J. Hazard. Mater., 2022, 439, 129687 CrossRef CAS.
- C. Ding, Y. Huang, Z. Shen and X. Chen, Adv. Mater., 2021, 33, e2007768 CrossRef.
- B. J. Furey, B. J. Stacy, T. Shah, R. M. Barba-Barba, R. Carriles, A. Bernal, B. S. Mendoza, B. A. Korgel and M. C. Downer, ACS Nano, 2022, 16, 6023–6033 CrossRef CAS.
- G. Calabrese, G. De Luca, G. Nocito, M. G. Rizzo, S. P. Lombardo, G. Chisari, S. Forte, E. L. Sciuto and S. Conoci, Int. J. Mol. Sci., 2021, 22(21), 11783 CrossRef CAS.
- H. Yan, Q. Wang, J. Wang, W. Shang, Z. Xiong, L. Zhao, X. Sun, J. Tian, F. Kang and S.-H. Yun, Adv. Mater., 2023, 35, e2210809 CrossRef.
- J. Ning, Z. Duan, S. V. Kershaw and A. L. Rogach, ACS Nano, 2020, 14, 11799–11808 CrossRef CAS.
- G. Mao, G. Wu, M. Chen, C. Yan, J. Tang, Y. Ma and X.-E. Zhang, Anal. Chem., 2022, 94, 6665–6671 CrossRef CAS.
- M. U. Zahid, L. Ma, S. J. Lim and A. M. Smith, Nat. Commun., 2018, 9, 1830 CrossRef.
- Y. Yang, Y. Xie and F. Zhang, Adv. Drug Delivery Rev., 2023, 193, 114697 CrossRef CAS.
- R. M. Abdelgalil, S. N. Khattab, S. Ebrahim, K. A. Elkhodairy, M. Teleb, A. A. Bekhit, M. A. Sallam and A. O. Elzoghby, ACS Omega, 2023, 8, 5655–5671 CrossRef CAS.
- Z. Lei and F. Zhang, Angew. Chem., Int. Ed. Engl., 2021, 60, 16294–16308 CrossRef CAS.
- P. S. Yasaswi, K. Shetty and K. S. Yadav, J. Controlled Release, 2021, 336, 549–571 CrossRef CAS.
- E. S. Seven, Y. B. Seven, Y. Zhou, S. Poudel-Sharma, J. J. Diaz-Rucco, E. Kirbas Cilingir, G. S. Mitchell, J. D. Van Dyken and R. M. Leblanc, Nanoscale Adv., 2021, 3, 3942–3953 RSC.
- B. B. Manshian, J. Jiménez, U. Himmelreich and S. J. Soenen, Biomaterials, 2017, 127, 1–12 CrossRef CAS.
- F. J. Charlson, A. J. Baxter, H. G. Cheng, R. Shidhaye and H. A. Whiteford, Lancet, 2016, 388, 376–389 CrossRef.
- M. Naghavi, BMJ, 2019, 364, l94 CrossRef.
- R. Ma, M. J. B. Taphoorn and P. Plaha, J. Neurol., Neurosurg. Psychiatry, 2021, 92, 1103–1111 CrossRef.
- O. Semyachkina-Glushkovskaya, I. Fedosov, A. Shirokov, E. Vodovozova, A. Alekseeva, A. Khorovodov, I. Blokhina, A. Terskov, A. Mamedova, M. Klimova, A. Dubrovsky, V. Ageev, I. Agranovich, V. Vinnik, A. Tsven, S. Sokolovski, E. Rafailov, T. Penzel and J. Kurths, Nanophotonics, 2021, 10, 3215–3227 CrossRef CAS.
- Y. Zhou, Z. Peng, E. S. Seven and R. M. Leblanc, J. Controlled Release, 2018, 270, 290–303 CrossRef CAS.
- A. B. Ganganboina, N. K. Dega, T. Hai Linh, W. Darmonto and R.-A. Doong, Biosens. Bioelectron., 2021, 181, 113151 CrossRef CAS.
- C. Li, L. Cao, Y. Zhang, P. Yi, M. Wang, B. Tan, Z. Deng, D. Wu and Q. Wang, Small, 2015, 11, 4517–4525 CrossRef CAS.
- H. Wang, R. Revia, Q. Mu, G. Lin, C. Yen and M. Zhang, Nanoscale Horiz., 2020, 5, 573–579 RSC.
- D. Li, J. Zhang, C. Chi, X. Xiao, J. Wang, L. Lang, I. Ali, G. Niu, L. Zhang, J. Tian, N. Ji, Z. Zhu and X. Chen, Theranostics, 2018, 8, 2508–2520 CrossRef CAS.
- N. L. Martirosyan, D. D. Cavalcanti, J. M. Eschbacher, P. M. Delaney, A. C. Scheck, M. G. Abdelwahab, P. Nakaji, R. F. Spetzler and M. C. Preul, J. Neurosurg., 2011, 115, 1131–1138 Search PubMed.
- D. J. Arndt-Jovin, S. R. Kantelhardt, W. Caarls, A. H. de Vries, A. Giese and T. M. Jovin Ast, IEEE Trans. NanoBiosci., 2009, 8, 65–71 Search PubMed.
- S. D. Hettiarachchi, R. M. Graham, K. J. Mintz, Y. Zhou, S. Vanni, Z. Peng and R. M. Leblanc, Nanoscale, 2019, 11, 6192–6205 RSC.
- P. Y. Liyanage, Y. Zhou, A. O. Al-Youbi, A. S. Bashammakh, M. S. El-Shahawi, S. Vanni, R. M. Graham and R. M. Leblanc, Nanoscale, 2020, 12, 7927–7938 RSC.
- M. Shamsipour, A. M. Mansouri and P. Moradipour, AAPS PharmSciTech, 2019, 20, 259 CrossRef.
- Z. Li, C. Zhao, Q. Fu, J. Ye, L. Su, X. Ge, L. Chen, J. Song and H. Yang, Small, 2022, 18, e2105160 CrossRef.
- G. Perini, V. Palmieri, G. Ciasca, M. D’Ascenzo, A. Primiano, J. Gervasoni, F. De Maio, M. De Spirito and M. Papi, Materials, 2020, 13(18), 4139 CrossRef CAS.
- D. Ag Seleci, V. Maurer, F. B. Barlas, J. C. Porsiel, B. Temel, E. Ceylan, S. Timur, F. Stahl, T. Scheper and G. Garnweitner, Int. J. Mol. Sci., 2021, 22(9), 4556 CrossRef.
- A. B. Madhankumar, O. D. Mrowczynski, S. R. Patel, C. L. Weston, B. E. Zacharia, M. J. Glantz, C. A. Siedlecki, L.-C. Xu and J. R. Connor, Acta Biomater., 2017, 58, 205–213 CrossRef CAS.
- J. Zhang, G. Hao, C. Yao, S. Hu, C. Hu and B. Zhang, J. Mater. Chem. B, 2016, 4, 4110–4118 RSC.
- D. Fatehi, T. N. Baral and A. Abulrob, J. Nanosci. Nanotechnol., 2014, 14, 5355–5362 CrossRef CAS.
- I. C. Carvalho, A. A. P. Mansur, S. M. Carvalho, R. M. Florentino and H. S. Mansur, Int. J. Biol. Macromol., 2019, 133, 739–753 CrossRef CAS.
- S. Lu, S. Guo, P. Xu, X. Li, Y. Zhao, W. Gu and M. Xue, Int. J. Nanomed., 2016, 11, 6325–6336 CrossRef CAS.
- A. A. P. Mansur, M. R. B. Paiva, O. A. L. Cotta, L. M. Silva, I. C. Carvalho, N. S. V. Capanema, S. M. Carvalho, E. A. Costa, N. R. Martin, R. Ecco, B. S. Santos, S. L. Fialho, Z. I. P. Lobato and H. S. Mansur, Int. J. Biol. Macromol., 2022, 210, 530–544 CrossRef CAS.
- R. Wahab, N. Kaushik, F. Khan, N. K. Kaushik, S.-J. Lee, E. H. Choi and A. A. Al-Khedhairy, Int. J. Nanomed., 2019, 14, 1131–1148 CrossRef CAS.
- G. Perini, V. Palmieri, G. Ciasca, M. D’Ascenzo, J. Gervasoni, A. Primiano, M. Rinaldi, D. Fioretti, C. Prampolini, F. Tiberio, W. Lattanzi, O. Parolini, M. De Spirito and M. Papi, Int. J. Mol. Sci., 2020, 21, 6301 CrossRef CAS.
- J. Tang, N. Huang, X. Zhang, T. Zhou, Y. Tan, J. Pi, L. Pi, S. Cheng, H. Zheng and Y. Cheng, Int. J. Nanomed., 2017, 12, 3899–3911 CrossRef CAS.
- H.-L. Xu, J.-J. Yang, D.-L. Zhuge, M.-T. Lin, Q.-Y. Zhu, B.-H. Jin, M.-Q. Tong, B.-X. Shen, J. Xiao and Y.-Z. Zhao, Adv. Healthcare Mater., 2018, 7, 1701130 CrossRef.
- Q.-L. Wu, H.-L. Xu, C. Xiong, Q.-H. Lan, M.-L. Fang, J.-H. Cai, H. Li, S.-T. Zhu, J.-H. Xu, F.-Y. Tao, C.-T. Lu, Y.-Z. Zhao and B. Chen, Artif. Cells, Nanomed., Biotechnol., 2020, 48, 143–158 CrossRef CAS.
- H. Wang, Q. Mu, K. Wang, R. A. Revia, C. Yen, X. Gu, B. Tian, J. Liu and M. Zhang, Appl. Mater. Today, 2019, 14, 108–117 CrossRef.
- P. W. Yoon, B. Bastian, R. N. Anderson, J. L. Collins and H. W. Jaffe, MMWR Morb. Mortal. Wkly. Rep., 2014, 63, 369–374 Search PubMed.
- S. G. Ryan, M. N. Butler, S. S. Adeyemi, T. Kalber, P. S. Patrick, M. Z. Thin, I. F. Harrison, D. J. Stuckey, M. Pule and M. F. Lythgoe, Sci. Rep., 2019, 9, 19223 CrossRef CAS.
- A. Zebibula, N. Alifu, L. Xia, C. Sun, X. Yu, D. Xue, L. Liu, G. Li and J. Qian, Adv. Funct. Mater., 2018, 28, 1703451 CrossRef.
- D. Song, M. Zhu, S. Chi, L. Xia, Z. Li and Z. Liu, Anal. Chem., 2021, 93, 7949–7957 CrossRef CAS.
- X. Yang, Z. Wang, H. Huang, S. Ling, R. Zhang, Y. Zhang, G. Chen, C. Li and Q. Wang, Adv. Healthcare Mater., 2021, 10, e2001544 CrossRef.
- C. Li, W. Li, H. Liu, Y. Zhang, G. Chen, Z. Li and Q. Wang, Angew Chem. Int. Ed. Engl., 2020, 59, 247–252 CrossRef CAS.
- H. Y. Yang, Y. Fu, M.-S. Jang, Y. Li, W. P. Yin, T. K. Ahn, J. H. Lee, H. Chae and D. S. Lee, Colloids Surf., B, 2017, 155, 497–506 CrossRef CAS.
- K. J. Barnham, C. L. Masters and A. I. Bush, Nat. Rev. Drug Discovery, 2004, 3, 205–214 CrossRef CAS.
- M. T. Lin and M. F. Beal, Nature, 2006, 443, 787–795 CrossRef CAS.
- T. Ashleigh, R. H. Swerdlow and M. F. Beal, Alzheimer's Dementia, 2023, 19, 333–342 CrossRef CAS.
- E.-K. Tan, Y.-X. Chao, A. West, L.-L. Chan, W. Poewe and J. Jankovic, Nat. Rev. Neurol., 2020, 16, 303–318 CrossRef.
- J.-J. Guo, F. Yue, D.-Y. Song, L. Bousset, X. Liang, J. Tang, L. Yuan, W. Li, R. Melki, Y. Tang, P. Chan, C. Guo and J.-Y. Li, Cell Death Dis., 2021, 12, 81 CrossRef CAS.
- C. Ren, X. Hu and Q. Zhou, Adv. Sci., 2018, 5, 1700595 CrossRef.
- X. Guo, Q. Lie, Y. Liu, Z. Jia, Y. Gong, X. Yuan and J. Liu, ACS Appl. Mater. Interfaces, 2021, 13, 30261–30273 CrossRef CAS.
- J. Liu, F. Erogbogbo, K.-T. Yong, L. Ye, J. Liu, R. Hu, H. Chen, Y. Hu, Y. Yang, J. Yang, I. Roy, N. A. Karker, M. T. Swihart and P. N. Prasad, ACS Nano, 2013, 7, 7303–7310 CrossRef CAS.
- W. Shang, X. Zhang, M. Zhang, Z. Fan, Y. Sun, M. Han and L. Fan, Nanoscale, 2014, 6, 5799–5806 RSC.
- S. W. Park, T. E. Kim and Y. K. Jung, Anal. Chim. Acta, 2021, 1165, 338513 CrossRef CAS.
- D. Kim, J. M. Yoo, H. Hwang, J. Lee, S. H. Lee, S. P. Yun, M. J. Park, M. Lee, S. Choi, S. H. Kwon, S. Lee, S.-H. Kwon, S. Kim, Y. J. Park, M. Kinoshita, Y.-H. Lee, S. Shin, S. R. Paik, S. J. Lee, S. Lee, B. H. Hong and H. S. Ko, Nat. Nanotechnol., 2018, 13, 812–818 CrossRef CAS.
- J. Ahlawat and M. Narayan, ACS Sustain. Chem. Eng., 2022, 10, 4610–4622 CrossRef CAS.
- B. W. Schlichtmann, B. Kalyanaraman, R. L. Schlichtmann, M. G. Panthani, V. Anantharam, A. G. Kanthasamy, S. K. Mallapragada and B. Narasimhan, J. Biomed. Mater. Res. B Appl. Biomater., 2022, 110, 450–459 CrossRef CAS.
- L. Chen, J. Lin, J. Yi, Q. Weng, Y. Zhou, Z. Han, C. Li, J. Chen and Q. Zhang, Anal. Bioanal. Chem., 2019, 411, 5277–5285 CrossRef CAS.
- S. Gupta, P. Babu and A. Surolia, Biomaterials, 2010, 31, 6809–6822 CrossRef CAS.
- S. Xiao, D. Zhou, P. Luan, B. Gu, L. Feng, S. Fan, W. Liao, W. Fang, L. Yang, E. Tao, R. Guo and J. Liu, Biomaterials, 2016, 106, 98–110 CrossRef CAS.
- C. Ren, D. Li, Q. Zhou and X. Hu, Biomaterials, 2020, 232, 119752 CrossRef CAS.
- W. Zhang, N. Kandel, Y. Zhou, N. Smith, B. C. L. B Ferreira, M. Perez, M. L. Claure, K. J. Mintz, C. Wang and R. M. Leblanc, J. Colloid Interface Sci., 2022, 617, 20–31 CrossRef CAS.
- N. J. Abbott, L. Rönnbäck and E. Hansson, Nat. Rev. Neurosci., 2006, 7, 41–53 CrossRef CAS.
- B. Obermeier, R. Daneman and R. M. Ransohoff, Nat. Med., 2013, 19, 1584–1596 CrossRef CAS.
- W. M. Pardridge, J. Cereb. Blood Flow Metab., 2012, 32, 1959–1972 CrossRef CAS.
- S. Kato, K. Itoh, T. Yaoi, T. Tozawa, Y. Yoshikawa, H. Yasui, N. Kanamura, A. Hoshino, N. Manabe, K. Yamamoto and S. Fushiki, Nanotechnology, 2010, 21, 335103 CrossRef.
- N. Huang, S. Cheng, X. Zhang, Q. Tian, J. Pi, J. Tang, Q. Huang, F. Wang, J. Chen, Z. Xie, Z. Xu, W. Chen, H. Zheng and Y. Cheng, Nanomedicine, 2017, 13, 83–93 CrossRef CAS.
- K. D. Kania, W. Wagner and L. Pulaski, Int. J. Mol. Sci., 2021, 22, 1068 CrossRef CAS.
- M. Zhang, B. P. Bishop, N. L. Thompson, K. Hildahl, D. Binh, O. Mironchuk, N. Chen, R. Aoki, V. C. Holmberg and E. Nance, Nanoscale Adv., 2019, 1, 3424–3442 RSC.
- S. Paris-Robidas, D. Brouard, V. Emond, M. Parent and F. Calon, J. Cereb. Blood Flow Metab., 2016, 36, 731–742 CrossRef CAS.
- A. Varmazyari, A. Taghizadehghalehjoughi, C. Sevim, O. Baris, G. Eser, S. Yildirim, A. Hacimuftuoglu, A. Buha, D. R. Wallace, A. Tsatsakis, M. Aschner and Y. Mezhuev, Toxicol. Rep., 2020, 7, 637–648 CrossRef CAS.
- T. P. Crowe, M. H. W. Greenlee, A. G. Kanthasamy and W. H. Hsu, Life Sci., 2018, 195, 44–52 CrossRef CAS.
- K. Sarlo, K. L. Blackburn, E. D. Clark, J. Grothaus, J. Chaney, S. Neu, J. Flood, D. Abbott, C. Bohne, K. Casey, C. Fryer and M. Kuhn, Toxicology, 2009, 263, 117–126 CrossRef CAS.
- F. R. Yin, C. Y. Chen, Y. X. Dong, W. U. Gang and Y. X. Gao, Chin. Sci. Bull., 2010, 55, 547–552 Search PubMed.
- B. C. Bejgum and M. D. Donovan, Mol. Pharm., 2021, 18, 429–440 CrossRef CAS.
- G. Naseri Kouzehgarani, T. Feldsien, H. H. Engelhard, K. K. Mirakhur, C. Phipps, V. Nimmrich, D. Clausznitzer and D. R. Lefebvre, Adv. Drug Delivery Rev., 2021, 173, 20–59 CrossRef CAS.
- Z. Liang, Y. Yang, F. Jia, K. Sai, S. Ullah, C. Fidelis, Z. Lin and F. Li, ACS Appl. Bio Mater., 2019, 2, 1432–1439 CrossRef CAS.
- J. A. Varela, J. P. Dupuis, L. Etchepare, A. Espana, L. Cognet and L. Groc, Nat. Commun., 2016, 7, 10947 CrossRef CAS.
- A. Nagy, A. Steinbrück, J. Gao, N. Doggett, J. A. Hollingsworth and R. Iyer, ACS Nano, 2012, 6, 4748–4762 CrossRef CAS.
- X. Han, J. Lei, K. Chen, Q. Li, H. Hao, T. Zhou, F.-L. Jiang, M. Li and Y. Liu, Ecotoxicol. Environ. Saf., 2019, 174, 467–474 CrossRef CAS.
- J. G. Paithankar, S. Kushalan, N. S, S. Hegde, S. Kini and A. Sharma, Chemosphere, 2022, 295, 133836 CrossRef CAS.
- N. Zheng, J. Yan, W. Qian, C. Song, Z. Zuo and C. He, J. Environ. Sci., 2021, 100, 240–249 CrossRef CAS.
- M. Zonouzi-Marand, M. Naderi and R. W. M. Kwong, Aquat. Toxicol., 2022, 247, 106157 CrossRef CAS.
- Y. Zhao, X. Wang, Q. Wu, Y. Li and D. Wang, J. Hazard. Mater., 2015, 283, 480–489 CrossRef CAS.
- H. Xu, X. Wang, X. Zhang, J. Cheng, J. Zhang, M. Chen and T. Wu, Nanomaterials, 2021, 11, 3314 CrossRef CAS.
- T. Wu, K. He, S. Ang, J. Ying, S. Zhang, T. Zhang, Y. Xue and M. Tang, Int. J. Nanomed., 2016, 11, 2737–2755 CrossRef CAS.
- G. Lin, T. Chen, Y. Pan, Z. Yang, L. Li, K.-T. Yong, X. Wang, J. Wang, Y. Chen, W. Jiang, S. Weng, X. Huang, J. Kuang and G. Xu, Nanotheranostics, 2020, 4, 173–183 CrossRef.
- G. Amiri, A. Valipoor, K. Parivar, M. Modaresi, A. Noori, H. Gharamaleki, J. Taheri and A. Kazemi, Int. J. Fertil. Steril., 2016, 9, 512–520 CAS.
- W. Zou, L. Li, Y. Chen, T. Chen, Z. Yang, J. Wang, D. Liu, G. Lin and X. Wang, Front. Pharmacol., 2019, 10, 437 CrossRef CAS.
- I. Kang, J. M. Yoo, D. Kim, J. Kim, M. K. Cho, S.-E. Lee, D. J. Kim, B.-C. Lee, J. Y. Lee, J.-J. Kim, N. Shin, S. W. Choi, Y.-H. Lee, H. S. Ko, S. Shin, B. H. Hong and K.-S. Kang, Nano Lett., 2021, 21, 2339–2346 CrossRef CAS.
- T. Wu, X. Wang, J. Cheng, X. Liang, Y. Li, M. Chen, L. Kong and M. Tang, Part. Fibre Toxicol., 2022, 19, 22 CrossRef CAS.
- B. R. Prasad, N. Nikolskaya, D. Connolly, T. J. Smith, S. J. Byrne, V. A. Gérard, Y. K. Gun'ko and Y. Rochev, J. Nanobiotechnol., 2010, 8, 7 CrossRef.
- L. Chen, Y. Miao, L. Chen, P. Jin, Y. Zha, Y. Chai, F. Zheng, Y. Zhang, W. Zhou, J. Zhang, L. Wen and M. Wang, Biomaterials, 2013, 34, 10172–10181 CrossRef CAS.
- E. Fuster, H. Candela, J. Estevez, E. Vilanova and M. A. Sogorb, Int. J. Mol. Sci., 2022, 23, 2267 CrossRef CAS.
- C. Bai, Y. Yao, Z. Wang, X. Huang, T. Wei, L. Zou, N. Liu, T. Zhang and M. Tang, J. Appl. Toxicol., 2022, 42, 1962–1977 CrossRef CAS.
- X. Liang, X. Wang, J. Cheng, X. Zhang and T. Wu, Bull. Environ. Contam. Toxicol., 2022, 109, 279–285 CrossRef CAS.
- Z. M. Markovic, B. Z. Ristic, K. M. Arsikin, D. G. Klisic, L. M. Harhaji-Trajkovic, B. M. Todorovic-Markovic, D. P. Kepic, T. K. Kravic-Stevovic, S. P. Jovanovic, M. M. Milenkovic, D. D. Milivojevic, V. Z. Bumbasirevic, M. D. Dramicanin and V. S. Trajkovic, Biomaterials, 2012, 33, 7084–7092 CrossRef CAS.
- D. Xue, W. Zou, D. Liu, L. Li, T. Chen, Z. Yang, Y. Chen, X. Wang, W. Lu and G. Lin, Neurotoxicology, 2022, 88, 134–143 CrossRef CAS.
- T. Wu, X. Liang, K. He, X. Liu, Y. Li, Y. Wang, L. Kong and M. Tang, Int. J. Nanomed., 2020, 15, 3217–3233 CrossRef CAS.
- T. Wu, X. Liang, K. He, T. Wei, Y. Wang, J. Lu, Y. Yao, T. Zhang, Y. Xue and M. Tang, Toxicol. In Vitro, 2018, 52, 41–51 CrossRef CAS.
- A. Moquin, E. Hutter, A. O. Choi, A. Khatchadourian, A. Castonguay, F. M. Winnik and D. Maysinger, ACS Nano, 2013, 7, 9585–9598 CrossRef CAS.
- N. K. Kaushik, N. Kaushik, R. Wahab, P. Bhartiya, N. N. Linh, F. Khan, A. A. Al-Khedhairy and E. H. Choi, Cancers, 2020, 12, 457 CrossRef CAS.
- X. Liang, T. Wu, Y. Wang, T. Wei, L. Zou, C. Bai, N. Liu, T. Zhang, Y. Xue and M. Tang, Toxicol. In Vitro, 2020, 65, 104827 CrossRef CAS.
- T. Wu, X. Liang, K. He, T. Wei, Y. Wang, L. Zou, C. Bai, N. Liu, T. Zhang, Y. Xue and M. Tang, Nanoscale, 2019, 11, 20820–20836 RSC.
- P. Yang, S. Ke, L. Tu, Y. Wang, S. Ye, S. Kou and L. Ren, ACS Biomater. Sci. Eng., 2020, 6, 1764–1775 CrossRef CAS.
- M. C. Dos Santos, W. R. Algar, I. L. Medintz and N. Hildebrandt, TrAC, Trends Anal. Chem., 2020, 125, 115819 CrossRef.
- A. Anas, H. Akita, H. Harashima, T. Itoh, M. Ishikawa and V. Biju, J. Phys. Chem. B, 2008, 112, 10005–10011 CrossRef CAS.
- R. Bai, J. Guo, X.-Y. Ye, Y. Xie and T. Xie, Ageing Res. Rev., 2022, 77, 101619 CrossRef CAS.
- A. Cuypers, M. Plusquin, T. Remans, M. Jozefczak, E. Keunen, H. Gielen, K. Opdenakker, A. R. Nair, E. Munters, T. J. Artois, T. Nawrot, J. Vangronsveld and K. Smeets, BioMetals, 2010, 23, 927–940 CrossRef CAS.
- W. Hong, H. Kuang, X. He, L. Yang, P. Yang, B. Chen, Z. P. Aguilar and H. Xu, Nanomaterials, 2019, 9, 257 CrossRef CAS.
- Y. Wang and M. Tang, Sci. Total Environ., 2018, 625, 940–962 CrossRef CAS.
- J. Lovrić, H. S. Bazzi, Y. Cuie, G. R. A. Fortin, F. M. Winnik and D. Maysinger, J. Mol. Med., 2005, 83, 377–385 CrossRef.
- L. E. Hopkins, E. S. Patchin, P.-L. Chiu, C. Brandenberger, S. Smiley-Jewell and K. E. Pinkerton, Nanotoxicology, 2014, 8, 885–893 CrossRef CAS.
- K. He, X. Liang, T. Wei, N. Liu, Y. Wang, L. Zou, C. Bai, Y. Yao, T. Wu, L. Kong, T. Zhang, Y. Xue and M. Tang, Chemosphere, 2020, 246, 125629 CrossRef CAS.
- J. A. DiDonato, F. Mercurio and M. Karin, Immunol. Rev., 2012, 246, 379–400 CrossRef.
- Z. Yang, W. Zou, Y. Pan, K.-T. Yong, L. Li, X. Wang, D. Liu, T. Chen, D. Xue and G. Lin, Ecotoxicol. Environ. Saf., 2021, 207, 111378 CrossRef CAS.
- D. D. Vilela, A. B. Justino, D. C. Caixeta, A. V. de Souza, R. R. Teixeira, R. R. Franco, A. L. Saraiva, B. B. Fonseca, N. O. Dantas, A. C. Almeida Silva and F. S. Espindola, J. Biomed. Mater. Res. B Appl. Biomater., 2022, 110, 1140–1150 CrossRef CAS.
- A. M. Derfus, W. C. W. Chan and S. N. Bhatia, Nano Lett., 2004, 4, 11–18 CrossRef CAS.
- Y.-F. Liu and J.-S. Yu, J. Colloid Interface Sci., 2010, 351, 1–9 CrossRef CAS.
- K. He, X. Liang, T. Wei, N. Liu, Y. Wang, L. Zou, J. Lu, Y. Yao, L. Kong, T. Zhang, Y. Xue, T. Wu and M. Tang, J. Appl. Toxicol., 2019, 39, 525–539 CrossRef CAS.
- Y. Su, M. Hu, C. Fan, Y. He, Q. Li, W. Li, L.-h. Wang, P. Shen and Q. Huang, Biomaterials, 2010, 31, 4829–4834 CrossRef CAS.
- Y.-H. Luo, S.-B. Wu, Y.-H. Wei, Y.-C. Chen, M.-H. Tsai, C.-C. Ho, S.-Y. Lin, C.-S. Yang and P. Lin, Chem. Res. Toxicol., 2013, 26, 662–673 Search PubMed.
- Z. A. Shaikh, T. T. Vu and K. Zaman, Toxicol. Appl. Pharmacol., 1999, 154, 256–263 CrossRef CAS.
- W. Zhang, X. Sun, L. Chen, K.-F. Lin, Q.-X. Dong, C.-J. Huang, R.-B. Fu and J. Zhu, Environ. Toxicol. Chem., 2012, 31, 2117–2123 CrossRef CAS.
- T. Ku, Z. Ren, R. Yang, Q. S. Liu, N. Sang, F. Faiola, Q. Zhou and G. Jiang, Environ. Int., 2022, 170, 107572 CrossRef CAS.
- S. Elmore, Toxicol. Pathol., 2007, 35, 495–516 CrossRef CAS.
- N. N. Danial and S. J. Korsmeyer, Cell, 2004, 116, 205–219 CrossRef CAS.
- E. Garcia-Calvo, P. Cabezas-Sanchez and J. L. Luque-Garcia, Chemosphere, 2021, 263, 128170 CrossRef CAS.
- R. A. Lockshin and Z. Zakeri, Nat. Rev. Mol. Cell Biol., 2001, 2, 545–550 CrossRef CAS.
- N. Morishima, K. Nakanishi, H. Takenouchi, T. Shibata and Y. Yasuhiko, J. Biol. Chem., 2002, 277, 34287–34294 CrossRef CAS.
- C. Y. Yan and L. A. Greene, J. Neurosci., 1998, 18, 4042–4049 CrossRef CAS.
- L. Galluzzi, O. Kepp, C. Trojel-Hansen and G. Kroemer, Circ. Res., 2012, 111, 1198–1207 CrossRef CAS.
- E. Murphy and C. Steenbergen, Annu. Rev. Physiol., 2021, 83, 107–126 CrossRef CAS.
- K. Salin, S. K. Auer, A. M. Rudolf, G. J. Anderson, C. Selman and N. B. Metcalfe, Physiol. Biochem. Zool., 2016, 89, 511–523 CrossRef.
- W.-H. Chan, N.-H. Shiao and P.-Z. Lu, Toxicol. Lett., 2006, 167, 191–200 CrossRef CAS.
- P. Meier and K. H. Vousden, Mol. Cell, 2007, 28, 746–754 CrossRef CAS.
- J. Li, Y. Zhang, Q. Xiao, F. Tian, X. Liu, R. Li, G. Zhao, F. Jiang and Y. Liu, J. Hazard. Mater., 2011, 194, 440–444 CrossRef CAS.
- J. Yi, A. M. Minikes and X. Jiang, Cell Chem. Biol., 2019, 26, 621–622 CrossRef CAS.
- N. Liu, Y. Liang, T. Wei, L. Zou, X. Huang, L. Kong, M. Tang and T. Zhang, J. Hazard. Mater., 2022, 436, 129043 CrossRef CAS.
- J. Zhou, Y. Jin, Y. Lei, T. Liu, Z. Wan, H. Meng and H. Wang, Neurodegener. Dis., 2020, 20, 20–34 CrossRef CAS.
- X. Song and D. Long, Front. Neurosci., 2020, 14, 267 CrossRef.
- T. Wu, X. Liang, X. Liu, Y. Li, Y. Wang, L. Kong and M. Tang, Part. Fibre Toxicol., 2020, 17, 30 CrossRef CAS.
- L. Yao, M.-M. Zhao, Q.-W. Luo, Y.-C. Zhang, T.-T. Liu, Z. Yang, M. Liao, P. Tu and K.-W. Zeng, ACS Nano, 2022, 16, 9228–9239 CrossRef CAS.
- Y. Lu, S. Xu, H. Chen, M. He, Y. Deng, Z. Cao, H. Pi, C. Chen, M. Li, Q. Ma, P. Gao, Y. Ji, L. Zhang, Z. Yu and Z. Zhou, Biomaterials, 2016, 90, 27–39 CrossRef CAS.
- M. A. Brennan and B. T. Cookson, Mol. Microbiol., 2000, 38, 31–40 CrossRef CAS.
- L. Jiang, H. Cai, W. Qin, Z. Li, L. Zhang and H. Bi, Bioconjugate Chem., 2023, 34, 1387–1397 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |