 Open Access Article
Open Access ArticlePaper-based sustainable biosensors
Anuj
Kumar
* and
Pralay
Maiti

School of Materials Science and Technology, Indian Institute of Technology (BHU), Varanasi 221005, Uttar Pradesh, India. E-mail: anuj.mst@itbhu.ac.in; anuj.budhera@gmail.com
First published on 21st March 2024
Abstract
Sustainability is the priority of the research community in designing and manufacturing value-added products, including biosensing devices. In this case, the utilization of paper and cellulose-based fibrous materials has been widely explored for developing biosensing devices in the past decade due to their high versatility, adaptability, biodegradability, and low cost. Recently, paper-based multifunctional biosensing devices have been developed for a sustainable future in a variety of applications such as the healthcare, agriculture, food safety and security, and environmental fields. However, there are still various technological and environmental issues that need to be overcome for manufacturing the desired biosensors. In this review, we provide the precise fundamentals of biosensing concepts, sustainable materials, and their utilization in developing paper-based sustainable biosensing devices for different healthcare, agriculture, food industry, environmental, and other applications. Moreover, the challenges, opportunities, and future perspectives regarding the use of these sustainable materials, together with their economic and advanced technological features are presented.
1. Introduction
Sensing devices are analytical tools employed to monitor analytes linked to environmental pollution, human diseases, food safety and security, and human–machine interactions. The extensive research and utilization of these devices have remarkably increased in the past few decades1,2 owing to their simplicity, cheap reagents, and flexibility for on-site detection.3,4 Herein, compared to micro-materials, the use of nanoengineered materials can enhance the recognition of distinct analytes at a very low content due to their high surface area-to-volume ratio and specific functional mechanisms to facilitate the efficient recognition of chemical and/or biological species. Among the variety of available materials for developing sensing devices, great efforts have been devoted to employing natural polymers as substrates or even as components of the active layer in sensors5–8 to realize sustainable and/or biocompatible sensing devices by avoiding non-renewable and hazardous/toxic elements. In this case, cellulose materials and/or nanomaterials have been considered as potential components of sensors and biosensors. Therefore, the use of paper and nanopaper as substrates to fabricate electrodes and substituting conventional glass- or plastic-based materials with low-cost and disposable materials is highly desirable.In this trend, standard paper has widely been utilized in the field of printed sensors, pursuing the upgrades advanced by the simple loading and delivery of reagents in the 3D cellulosic-network, ability to operate using very low volume of reagents and sample amounts, pores and channels of the substrate for avoiding interference of the components, and microfluidic patterns with no requirement of micropumps for flow initiation and avoiding bubble formation.9,10 Moreover, natural materials including nanocellulose materials provide interesting benefits in developing nanopaper-based sensors and biosensors, resulting in inexpensive, consistent, sustainable, disposable, and reagent-free tools for use in the monitoring of food quality and safety.9,11 Also, they provide the opportunity for the successful immobilization of various biological components to develop biocompatible and nontoxic biosensors.10–12 Moreover, paper (cellulose and nitrocellulose) is an appropriate substrate for a range of applications, including sensors for environmental and healthcare monitoring.13 Paper has been utilized passively in various analytical applications (e.g., pH indicator strips, pregnancy tests, and rapid Covid-19 antigen tests).14 Accordingly, it has been utilized in many other applications including electronic and electrochemical devices due to its inexpensive nature, flexibility, fluid conduct (capillary action due to rough fibrous-network), significant sensitivity and easy disposal.15,16 Therefore, different electroactive elements can easily be detected by using paper-based sensor devices such as ions,17 dopamine,18 and biomarkers,19 together with their easily disposal. In this advancement, the fabrication of multifunctional paper-based biosensors to detect multiple parameters using only one physical input is highly desirable.
Presently, artificial intelligence (AI) and wearable sensors/biosensors have become two important areas to realize the tailoring of the best precision medicine therapies for individual patients. Therefore, the integration of AI and biosensing devices can potentially enable the better acquisition of patient data and enhance the design of wearable and implantable biosensing devices to monitor the fitness, health, and surroundings of patients. However, AI-biosensors with suitable practical characteristics encounter new opportunities and challenges. In this case, the innovations in materials, biorecognition components, signal acquisition and transport, data processing and intelligent decision system are the essential factors. Further, integration, slimness, miniaturization, and low energy utilization are the upcoming prospects of AI-biosensors for human healthcare. To this end, more technological advances are needed to detect low levels of specific biomarkers in biofluids and the achieve system integration. Herein, biomarkers must channel the cavity between biophysical and biochemical markers to prepare all-inclusive prototypes for individuals. Moreover, great efforts need to be devoted to developing flexible electronic materials integrated with chip technology, the Internet of Things, huge data, and AI to accomplish the collective functioning of AI-biosensors, wherein the accumulated data is consumed by machine learning algorithms to monitor crucial signs, spot abnormalities and track therapies for further correlated interactions at the human–machine interface.20
2. Biosensors
Sensors are devices that respond to stimuli and detect the events or changes in a specific environment and communicate the information to other interconnected technical devices (e.g., computer system). Various physical and chemical characteristics (e.g., odor, pressure, force, temperature, and pH) of any compound are measured and detected by these sensors and their associated interfaces in the presence of special chemical moieties. The moieties are solely sensitive to the targeted physical or chemical quantity for measurement (insensitive to all other parameters) and do not influence the characteristics of the input physical and/or chemical quantities. Therefore, important features such as selectivity, sensitivity, accuracy, calibration scope, resolution, repeatability, and cost-effectiveness are important factors in the selection of sensor systems.21,22 Moreover, sensors can be grouped broadly into two types, physical and chemical sensors. Physical sensor devices detect and/or quantify responses such as force, temperature, absorbance, refractive index, mass change, conductivity, and magnetic field.23 Alternatively, chemical sensing devices have a chemically selective interface (layer) that responds selectively to a distinct analyte24 and provides chemical reaction information (with analyte) or physical characteristic of the system being probed. For example, the content of certain constituents or probing the total composition (being transformed into signals such as changes in conductance, light, current, voltage, and sound effects). The operating principles of physical and chemical sensors are remarkably different. Most physical sensors are direct and only affected by a few parameters, whereas the signals from chemical sensors may be affected by various parameters. In this way, chemical sensors have attracted greater attention in broad commercial applications such as industrial, environmental, agricultural, and clinical fields.22,25Biosensors are sensors or a type of chemical sensor composed of a biological molecule as the recognition component. Therefore, biosensors are classified as analytical devices containing biological or biological-derived sensing components either unified within or closely linked with physicochemical transducers for the recognition of analytes.26,27 In the past, different biologically derived substances such as enzymes, nucleic acids, antigen-antibodies, cells, and microorganisms were included in various mechanical, electrochemical, optical, and acoustic detectors to develop biosensing devices with high selectivity and sensitivity.28,29 The first biosensor, as an amperometric oxygen electrochemical sensor, was originally established in 196230 using glucose oxidase to detect the content of glucose. However, the term “biosensor” was proposed in 1977 by Rechnitz et al.31 using an arginine selective electrode (living organisms) as the recognition element.
Biosensors should be highly responsive, free of physical constraints (e.g., pH and temperature), and reusable. Biosensors are composed of three modules, as follows: (1) bioreceptor, (2) transducer, and (3) electric circuit (Fig. 1). In this case, the bioreceptor senses and responds to the analyte to generate a signal, the transducer transforms this biologically recognized action into an assessable signal, and then the electric circuit quantifies the transduced signal and collects it for display and examination, leading to diagnostic assessments.32–34 However, the lack of biorecognition elements impedes the development of diverse biosensors.35 The transducer-based output signals include optical (e.g., absorbance, luminescence, chemiluminescence, and surface plasmon),36 mass (e.g., piezoelectric and magnetoelectric),37 thermometric,38 and electrochemical signals.39 Electrochemical biosensors can be categorized into amperometric, potentiometric, voltammetric, impedimetric/conductometric, and capacitive sensors.22 By using electronics and signal processors, biosensing and bioelectronic devices have been effectively utilized in several fields such as environmental monitoring, drug screening, clinical diagnostics, and governing of food quality. Recent research has mainly been focused on the miniaturization of biosensing and bioelectronic devices to enable the real-time, POC, and easy-to-use detection of analytes, especially in clinical and environmental trials.40,41
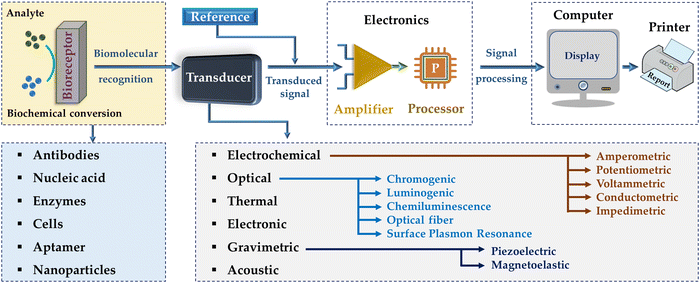 | ||
| Fig. 1 Schematic illustration of a biosensing device, which is comprised of a bioreceptor, transducer, and amplifier.42 | ||
Moreover, biosensor platforms are typically categorized into three ways, i.e., dipstick tests, lateral flow assays (LFAs), and microfluidic paper-based analytical devices (μPADs), among which, μPADs are the most versatile owing to their pattern-paper made using hydrophobic polymers around designated areas in the paper. These barrier patterns are designed for accommodating small sample volumes for multiple reactions. However, the techniques used to create these hydrophobic barrier patterns and layers in inexpensive μPADs require a long manufacturing time and very expensive.
2.1. Materials used for sensing devices
Sensors or biosensor devices are products that respond to stimuli and utilized to detect and quantify light, temperature, sound, chemicals, motion, and biological elements. Based on the material type, design approaches, and fabrication technologies, the structures of these devices can be constructed into robust and sophisticated apparatus. There Various types of sensing devices are utilized based on their working principles (e.g., force, temperature, gas, humidity, proximity, chemical, optical, motion, and imaging) and applications (e.g., environmental monitoring, agriculture, health industry, pharma industries, medical imaging, space, paint and coating industry, and human–machine interfaces). Various types of materials have been utilized to develop and manufacture different types of sensors. The area of sensing materials utilized for sensing devices can be divided into two groups, as follows: (1) direct sensing devices (i.e., a change in electrical signals) and (2) complex sensing devices (i.e., no change in electrical features, need a conversion process).43Various types of materials have been used for fabricating these biosensing devices such as metals, metal oxides, metal organic frameworks (MOFs), sol–gel materials, graphene and carbon nanomaterials, synthetic polymers, and petroleum-based materials. However, there are still various challenges in the real-world applications of these devices, wherein these material types are associated with some difficulties owing to their bulkiness, lack of affinity between material and the substrate, vulnerability to oxidation, and shortcomings considering robustness and waste management (disposal).44 In this regard, most of the sensors are comprised of petroleum-based materials from non-renewable resources and exhibit long-term stability in the soil (difficult to be degraded) when discarded, causing severe environmental pollution. Further, the application of these devices in the biological or biomedical field is limited due to various challenges including their biocompatibility and biodegradability. Therefore, numerous research efforts have been devoted to searching for sustainable materials for the development of biocompatible, biodegradable, and renewable sensing and biosensing devices45–47 and still in progress for achieving the desired outcomes for their diverse applications in environmental pollution, agriculture, food industry, soft-robotics, human–machine interaction, bioelectronics, etc.
Among the biopolymers, cellulose as the most abundant polysaccharide has been promisingly considered for the fabrication of biodegradable sensors with various potential applications due to their chemical distinctiveness, remarkable biocompatibility, appealing degradation profile under physiological conditions, high-temperature stability, transparency, flexibility, and easy processing. Also, cellulosic materials can be transformed into electronically conductive carbon materials with abundant pore networks and high specific surface areas using a straightforward carbonization process.52 However, it is difficult to control the mechanical properties and degradation rate of naturally derived substrates. Thus, biodegradable synthetic polymers with predictable and repeatable mechanical and degradable characteristics have been synthesized in controlled manner and utilized with or without biopolymers to fabricate sensor substrates.48 Overall, renewable and sustainable materials have received significant attention over the past few years in basic research and real-world applications owing to their ability to minimize environmental, consumption, and cost-related issues. In this way, these materials have widely been utilized in pristine form or a combination to manufacture sensing and biosensing devices for suitable and targeted utilization and outcomes with low cost, satisfactory biodegradability, high flexibility, and low-energy consumption.
The use of composite materials based on renewable resources has increased remarkably and continues in the development of new biosensing devices. In this case, various basic and technological methods have been applied to manufacture desired composite sensing and biosensing devices partially based on organic and renewable materials and their properties can be controlled by modifying the combination of components of renewable, inorganic, and organic nanomaterials. For example, different research groups developed various sensing devices by using different composite materials, for example, conductive paper was manufactured using carbonized paper for sensing human activity,53 cellulose/CNT aerogels for gas sensing,54 nanocellulose with silver nanowires (AgNWs)55 and/or carbon nanotubes (CNTs),55–57 and bacterial cellulose/AgNPs/molybdenum trioxide (MoO3) for gas sensing,58 and for different applications including wearable sensing fibres, CNT/cellulose-based humidity sensor59 and ammonia gas sensor,60 leaves and wood,61 carbonized wood,62 wood cellulose fibres/barium titanate (BaTiO3),63 paper substrate/zinc oxide nanowires (ZnO NWs),64 and sodium lignosulfonate/graphene.65 Overall, renewable resources (e.g., wood, plant, bio-based carbon, and cellulose) in various forms and from various origins have greatly established their significance by using them in combination with other materials to manufacture low-cost, biodegradable, and sustainable composites. Therefore, depending on their combination and fabrication process, these composite sensing materials can be manufactured into different forms such as films, fibres, paper, and aerogels for a variety of potential applications (e.g., gas sensing, supercapacitors, photovoltaic cells, wearable and flexible sensing devices, and human–machine interface).66 Throughout history, paper has been utilized in a broad variety of applications, and therefore, cellulose-based paper, which is a porous and green material, has been utilized to manufacture different biosensors owing to its low cost, flexibility, portability, biocompatibility, degradability and capillary force-assisting fluid transfer characteristics.67
3. Paper-based biosensors
3.1. Paper and its properties
The use of paper as a flexible, porous, and degradable cellulosic material with an environment-friendly nature has shown great potential to be applied in sustainable biosensors, bioelectronics and other areas.68,69 Herein, paper-based devices provide multipurpose, inexpensive, and accurate systems for sensing humidity, gases, chemicals, strain, and biological species.10,69–72 However, it is difficult to simplify a set of characteristics of papers due to the differences in their material compositions, structures, and fabrication methods. For example, printing paper is composed of a high filler content, which can be either natural (e.g., limestone, talc, and clay) or synthetic alternatives (e.g., precipitated calcium carbonate, gypsum, and titanium dioxide) and distinguish the cost, thickness, structure, pore size, strength, and exterior form of the paper, and other characteristics based on its type and quantity.73–76 In addition, due to the highly hydrophilic nature of paper, the presence of moisture can soften the materials, thereby decreasing their elastic modulus with a poor thermal performance,77 and therefore, it is processed under low-temperature conditions. Moreover, due to the flexible nature of paper, it has been broadly utilized as a substrate for actuators, sensors, and energy storage components78–83 despite the diverse set of characteristics across various types of paper.In addition, smartphones have also been integrated with sensors (e.g., strips, chips, and hand-held detectors) for biochemical detection owing to their portability and global accessibility. Smartphones are often utilized as the controller, analyzer, and display for fast, real-time, and point-of-care monitoring, which can remarkably simplify the design and reduce the cost of detecting devices. Paper-based biosensors and bioelectronics integrated with smartphones have greatly emerged for portable and disposable devices that can be applied in environmental monitoring, healthcare diagnosis, and food safety.84 The search for renewable and sustainable power sources is crucial for developing green electronics and sensor networks. For example, paper-based origami triboelectric nanogenerators (TENGs) with high flexibility, light weight, low cost, and recyclability were developed. These TENGs were efficient to harvest ambient mechanical energy from different types of human motions (e.g., stretching, lifting, and twisting) and could directly light LEDs. Additionally, these TENGs could also work as self-powered pressure sensors.85 In a study, an inexpensive turnip tissue paper-based mediated amperometric H2O2 biosensing device was fabricated using screen-printed carbon electrodes (SPCEs). Herein, raw turnip peroxidase and potassium hexacyanoferrate(II) as a mediator were immobilized on a cellulose paper matrix. This device required a low amount of the sample (0.5 mL) for analysis and exhibited a linear detection range of 0.02–0.50 mM with an LOD of 4.1 mM H2O2. Moreover, the biosensing device maintained 70% of its activity after a storage period of 25 days at 4 °C.86 Most recently, some innovative paper-based biosensors have been reported for renal surveillance in fingerprint blood,87 visual detection of microbially contaminated food,88 nucleic acid amplification-fee and label-free detection of micro-RNA,89 controlling the pandemic situation (e.g., Covid-19),90etc.
3.2. Processing and manufacturing of paper-based biosensing devices
A significant advancement in the development of portable and disposable paper-based biosensor (PBB) and microfluidic paper-based analytical devices (μPADs) has recently been realized. In this case, paper, which is composed of cellulose microfibers, provides both a huge microporous network for storing analytical elements and capillary force to drive liquid samples to a devoted reaction zone for the rapid detection of the desired analytes. Due to their inexpensive nature and ultra-high sensitivity, these biosensors have shown great potential for medical emergencies, point-of-care health diagnosis, and wide early-detection of cancer compared to the conventional advanced analytical devices.10 Additionally, the development of inexpensive wearable flexible biosensing devices has attracted significant attention for the monitoring of health issues and chronic diseases in the human body. Herein, conventional wearable devices uninterruptedly record signals such as stress, pressure, and temperature for the real-time monitoring of human actions. However, the uninterrupted monitoring of different chemicals in body fluids and respired breath using wearable sensors still faces certain challenges such as accuracy, sensitivity, flexibility, and fitting of the sensor to human skin.91,92 Paper is a favorable substrate to manufacture POC in-vitro diagnostic devices due to its abundant availability, affordability, chemical stability, biodegradability, prominent porous fibrous network with high surface area for interactions, and natural capillary-driven mass transport of liquid samples.93–97 Various well-established technologies for the manufacture and processing of paper have been applied for the fabrication of paper-based microfluidic devices, including folding, cutting,98,99 masking,100,101 laser writing,102 and printing.103–105 However, paper can vary immensely based on its composition, internal structure, and wettability owing to its sources and manufacturing method,106 for example, filter paper,107,108 chromatography paper,109 common printer paper,110 and paper towels. The performance of diagnostic sensing devices for various target analytes is largely affected by the internal structure of paper. In this case, nanomaterials have shown a considerable prospect in developing paper-based sensors. In one study, CNT-based electrodes were fabricated for use in paper-based electrochemical sensing devices via a series of laser-cutting, drop-casting, and origami. Herein, a range of filter papers with varying porosities and different 3D cellulose-networks was utilized with this process to analyze the effect of the cellulose scaffold on the ultimate CNT network and resulting electrochemical detection of glucose.111 The synergistic blending of materials and interfaces is a decisive strategy in developing novel functional sensing devices for different potential applications, including inexpensive paper-based POC systems. Surface-functionalized polypyrrole (PPy) constructs were integrated monolithically into a 3D multilayered paper-based microfluidic biosensing device to monitor local humidity variations. The PPy constructs exhibited remarkable mechanical stability, which is relevant to flexible electronics and the electrical resistance connects with local relative humidity (RH) interior of the sealed microfluidic channels, and their sensing response was entirely reversible. In addition, the results showed that RH changes of 5–10% could affect the flow of the prolonged channels (>5 cm) even when they were fully surrounded.1124. Applications
Biosensors have been used in various fields, including food, environment, and medicine. These developed biosensor devices are categorized into three types for human healthcare, as follows: (1) biosensors for in vitro diagnosis with urine, saliva, and blood samples, (2) continuous monitoring biosensors, and (3) wearable biosensors. Among them, in vitro diagnosis biosensors have received been widely developed via various approaches, and also used in integrated devices, such as lateral flow assays (LFAs) and microfluidic/electrochemical paper-based analytical devices (μPADs/ePADs). Moreover, high-quality and inexpensive smartphone-based POC devices for basic healthcare monitoring can be developed using paper-based platforms. Exclusively, μPADs have shown immense potential in using this inexpensive matrix for detecting blood glucose, urine metabolites, pH level, liver function, and infectious agents.113 Also, μPADs have various benefits such as low cost, simple and rapid manufacturing, and disposability compared to the alternative traditional lab-on-chip devices.114 The integration of PADs with paper-microfluidics has empowered various studies for detecting different enzymatic and biological reactions with an electronic readout.83 However, recently, continuous monitoring and wearable biosensors have attracted significant attention in terms of commercialization. A schematic illustration of these biosensors devices and their data processing and interpretation with the concept and implementation of artificial intelligence (AI) is provided in Fig. 2.115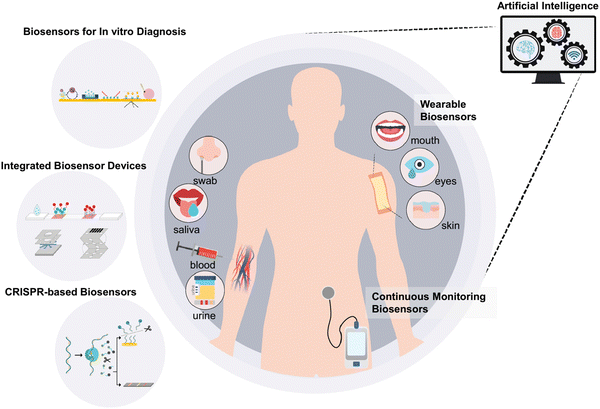 | ||
| Fig. 2 Schematic illustration of conceptual implementation of AI to process and interpret the received data. Reproduced with permission from ref. 115 Copyright 2022, Elsevier. | ||
4.1. Healthcare
Worldwide, human healthcare is the priority and requires sophisticated and affordable medical care products and devices. Among the various healthcare products and devices, paper-based biosensors are affordable diagnostic tools, but their wide utilization is impeded owing to the scarcity of sensitive detection means to be implemented on paper substrates.A paper-based cholesterol biosensing device was developed by modifying an electrode through electro-spraying using a nanocomposite of graphene (G), polyvinylpyrrolidone (PVP), and PANI (GPP). Herein, in this nanocomposite, the existence of a small quantity of PVP (2 mg mL−1) enhanced the dispersibility of G and improved the electrochemical conductivity of the electrode, resulting in improved sensitivity in the biosensor. Compared to the unmodified electrode, a 3-times increased current signal was observed including remarkable electrocatalytic action to the oxidation of H2O2. Further, cholesterol oxidase was fixed to the GPP-modified electrode for the amperometric determination of cholesterol. Under the optimal situations, it exhibited a linear range of 50 μM to 10 mM for cholesterol with an LOD of 1 μM.118 In a study, a portable paper-based biosensor with a specific enzymatic calorimetric detection was developed for blood alcohol sensing (direct quantification of ethanol in whole blood) for on-site toxicological tests.119 Similarly, the detection of diverse biomarker types with zero background was achieved by using a paper-based electrochemical biosensor (paper functionalized with single molecule-labeled DNA and a screen-printed electrode along with target recognition solutions). It enabled high sensitivity and selectivity for the target via high specific target-triggered polymerization/nicking and DNAzyme-catalyzed signal amplification. These assays could be used to identify the target in spiked serum trials for the point-of-care analysis of clinical samples.120
Paper-based microfluidic devices are portable, rapid, inexpensive, and disposable and only require small volumes of regents and samples. The combination of these devices with the electrochemical detection mechanism extends the extra advantages of high selectivity, sensitivity, simplicity, and portability, and low-cost. In one study, an integrated paper-based screen-printed electrochemical biosensing device to detect and quantify a nerve agent contamination in situ. The strategy of this reagent-free method was the dual electrochemical analysis of butyrylcholinesterase (BChE) enzymatic action towards butyrylcholine with or without exposure to polluted samples, wherein the sensitivity was enhanced by using a nanocomposite of carbon black/Prussian Blue as a working electrode modifier. Herein, a nitrocellulose membrane strip was integrated with a paper-based test area holding a screen-printed electrode and BChE and detected paraoxon as a nerve agent stimulant linearly as low as 3 μg L−1.121 In addition, a paper-based microfluidic electrochemical DNA biosensor was developed to detect EGFR mutations sensitively in patients using their saliva. In this study, oligonucleotides were functionalized on the surface of the electrodes and the outcome of the electrochemical signals were received by investigating the hybridization of DNA, where the indicator-labeled on DNA was recognized by horseradish peroxidase (HRP), showing a remarkable electrocatalytic response to H2O2. Under the optimum situations, it exhibited a linear association between current value and the logarithm of the target DNA content (5–500.0 nM) with a limit of detection as low as 0.167 nM and showed satisfactory stability and high specificity in distinguishing the single nucleotide polymorphism of the target DNA.122 In the case of progesterone monitoring for in vitro fertilization therapy of dairy cows, it depends on antibodies, making this procedure expensive. Thus, an alternative transcription factor-based progesterone sensor was utilized with a portable and affordable paper fluidic arrangement, where fluorescent dye-labeled oligonucleotides were immobilized on nitrocellulose through a biotin–streptavidin interaction. In the absence and presence of progesterone, these oligonucleotides bind and unbind with the fluorescent transcription factor, respectively. Also, the LOD of the device was 27 nm, which is the clinically significant level of progesterone.35
The use of nanomaterials in combination improves the efficiency of biosensors, for example, a disposable paper-based chemiresistive biosensor with manifold detection capability and microfluidic controls using single-walled carbon nanotubes (SWCNTs) was developed for label-free immunosensing. In this study, a stable water-based ink composed of pyrene carboxylic acid (PCA) and SWCNTs was synthesized and deposited on paper in a facile approach, which did not require extra covers or patterns. Specific antibodies were functionalized on the PCA/SWCNTs, and then human serum albumin (HSA) was rapidly, sensitively, and selectively detected and quantified with a limit of detection of 1 pM due to the porous-network of the paper surface, wherein more SWCNTs were accommodated and formed an electric network of SWCNTs.123 Micro RNAs (miRNAs), which are 19–23 nucleotide-long non-coding RNA, have been identified as important biomarkers for various diseases including cancer. This type of biomarker (miRNA) was detected using a rapid, simple, ultrasensitive, greatly specific, and label-free paper-based SWCNT field-effect transistor (FET) biosensor, without nucleic acid amplification. This approach involved a two-step process, as follows: (1) direct hybridization of the targeted miRNA with a certain RNA probe immobilized on SWCNT system deposited on paper substrate to produce the first signal, and then (2) recognition of the resulting RNA/miRNA duplexes with the Carnation Italian ringspot virus p19 protein (p19) in a size-dependent mode to provide the second electrical response. For demonstration, the miRNA-122a biomarker (lowest detection: 0.1 aM and broad dynamic range: 0.1 aM–1 fM) was detected for the diagnosis of early-stage hepatocellular carcinoma in various samples, including human serum, phosphate buffer, and synthetic saliva. Overall, without any sample preparation, the biosensor successfully detected miRNA spiked in serum and artificial saliva.89 In another study, multi-walled CNTs (MWCNTs) were coated on a paper substrate (15–20 nm average diameter) using stencil-free wax deposition and vacuum filtration to create hydrophilic and hydrophobic channels for the label-free, highly selective and sensitive detection of cholesterol owing to the high affinity of cholesterol oxidase towards MWCNTs through the electrostatic gating effect and direct electron transfer mechanism between cholesterol and MWCNTs. The disposable biosensor showed stability, reproducibility, and high specificity towards cholesterol (response to a concentration range of 10 nM–75 μM to 100 μM–8 mM and an LOD of 3.2 nM (3 S m−1)), while negligible interaction from interfering analytes.124 In another study, a paper-based biosensor was developed via the electropolymerization of 5-amino-1,3,4-thiadiazole-2-thiol (ATT) on the surface of graphene oxide (GO) and showed high sensitivity by requiring only 200 μL of analyte solution for the cyclic voltammetry (CV) analysis. ATT showed good selectivity for specific binding with uric acid and the interference from nitrites in human blood and blood fluids could be prevented in the CV analysis owing to their different oxidation potentials.125 In another study, a robust, user-friendly, and portable smartphone-aided biomimetic metal–organic framework (MOF) nanoreactor colorimetric paper (SBMCP) device was fabricated for the on-demand POC recognition of endogenous biomolecules. In this study, the single/multiple enzyme constituents trapped within the micropores network of ZIF-8 (exoskeleton) facilitated the sensitive and selective detection of the target analyte, and then relocated the detection event to a visual color signal based on the cascade effect. In the SBMCP assay, only a small volume (5 μL) of sample was needed, and it provided the efficient and real-time colorimetric recognition of glucose and uric acid in diabetes and gout events.126
Due to the changes in dietary habits and living standards, the occurrence of chronic kidney disease (CKD) is a rapidly rising global health concern. Thus, the early detection of CKD is critical in avoiding progression to final-stage renal disease and preventing expensive therapies. For this, a series of biomarkers, including blood urea nitrogen (BUN), has been proposed for this specific detection. Clinically, BUN is utilized as an indicator for renal function analysis and considered the gold standard for urea bioassay. However, the testing of BUN requires blood sampling, which is generally inconvenient, painful, time-utilizing, and expensive method. Therefore, inexpensive, simple, fast, and stable point-of-care biosensors are greatly desirable for evaluating the BUN levels with the minimum sample requirements. However, there are various challenges in developing novel sensing biosensing devices. In this regard, some biosensors such as origami paper-based sensors and smartphone-assisted paper-based biosensors have been developed for colorimetric readouts. Nevertheless, there are several challenges due to variations in camera quality and software, and contamination issues. In this case, the demand for electrochemical point-of-care biosensors is increasing due to their inexpensive system, high sensitivity, robust miniaturization potential and ease of integration for the fabrication of compact devices. In 2023, Cheng et al. developed an inexpensive handheld optical biosensor with multilayer paper-based photochemical disposable test strips (Fig. 3), wherein these strips can help in separating and filtering whole blood samples and the colorimetric reaction uses urease immobilized on a membrane to hydrolyze urea into NH3, exhibiting intense color changes with a pH indicator. Further, this color change was detected using an optical reader via the reflection of light, and then BUN quantified via a unique photodetection algorithm. This process could be accomplished within 2 min using only 20 μL of fingertip blood sample for high accuracy and consistency with a broad concentration range (BUN: 2.46–38.14 mM) and LOD of 0.03 mM in different samples. Additionally, Bluetooth was employed in the reader and connected to a smartphone for active health monitoring or remote diagnosis for renal diseases.87
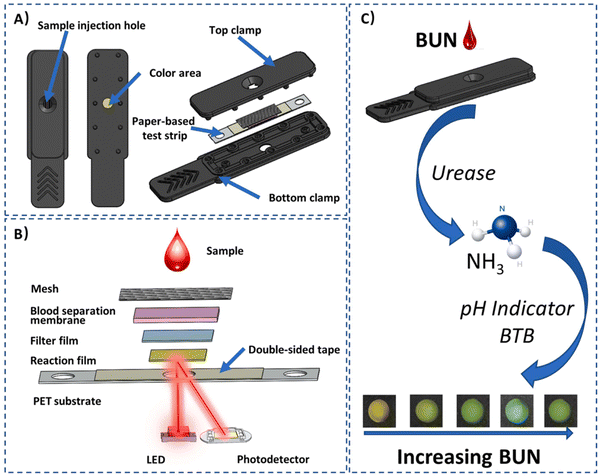 | ||
| Fig. 3 (A) Schematic illustration of a disposable photochemical test strip, (B) stacking of multilayer film strips with reflectance photometry, and (C) principle of enzymatic colorimetric response. Reproduced with permission from ref. 87 Copyright 2024, Elsevier. | ||
In a study, an anthraquinone-labeled pyrrolidinyl peptide nucleic acid probe (AQ-PNA) and graphene-polyaniline (G-PANI)-functionalized electrode were utilized in developing a paper-based electrochemical DNA biosensing device to detect human papillomavirus (HPV) type 16 DNA (corresponding sequence of synthetic 14-base oligonucleotide target). Under the optimal conditions, the limit of detection of HPV type 16 DNA was observed to be 2.3 nM with a linear range of 10–200 nM. In addition, the real-time screening and monitoring of HPV-DNA type 16 were evaluated with the detection of PCR-intensified DNA samples from SiHa cells for the identification of the primary phases of cervical cancer.127 In another study, a hand-operated paper-based rotational vertical-flow immunosensor (r-VFI) platform was designed, which required fewer pipetting steps, for the electrochemical detection of α-fetoprotein with numerous and time-progression steps without any interference from the convective factor of fluid motion. In this case, free transfer-switch-stop fluid flow could be achieved by rotating the paper disk. Overall, the assay duration was shortened to 9 min with the linear range (LR) of 0.01–500 ng mL−1 and LOD of 1.65 pg mL−1, and the sensitivity could be improved remarkably by switching off the sample cascade, which could extend the LR to 0.5 pg mL−1 with LOD of 3.54 fg mL−1. This is the lowest detectable level reported to date for paper-based sensors.128
The rapid detection of deoxyribonuclease I (DNase I) activity as a biomarker based on the variations in the viscosity of DNA mucus was performed, exhibiting a linear range of 0.01–10 U mL−1, remarkable sensitivity, specificity, and reproducibility. In addition, the limit of detection achieved was as low as 0.003 U mL−1. This sensing device provided great potential in detecting DNase I and effective and rapid analysis of nucleic acid scavenger.129 Further, in the case of investigating various biological actions and events of enantiomers, chiral examination is of crucial importance in different fields. For this, a paper-based chiral biosensor device was developed for detecting lactate enantiomers in human serum samples. For the purpose of achieving the sensitive and real-time detection of lactate enantiomers, an alginate hydrogel was utilized to construct enantiomer-selective sensing zones and dual enzyme (i.e., lactate dehydrogenase (LDH) and glutamic pyruvic transaminase (GPT))-linked synergistic catalytic interactions with substrate recycling were used to improve the sensor response. The well-blended solutions including the components for gelation and sensing (e.g., alginate, Ca2+, GPT, coenzymes, NAD+, L-Glu, L-LDH and D-LDH) were supplemented in the sensing domains. The fluorescence signals from nicotinamide adenine dinucleotide were acquired using a smartphone with color evaluation software. The results exhibited remarkable sensitivity with an LOD of 30.0 ± 0.7 μM and 3.0 ± 0.2 μM and broad linear range of detection of 0.1–3.0 mM and 0.01–0.5 mM for the concentrations of L-lactate and D-lactate, respectively, with reasonable accuracy.130
A paper-based microfluidic electrochemical DNA biosensor was developed to detect EGFR mutations sensitively in patients with saliva. In this study, oligonucleotides were functionalized on the surface of electrodes and the outcome of the electrochemical signals were received by investigating the hybridization of DNA, where the indicator-labeled DNA was recognized by horseradish peroxidase (HRP), showing a remarkable electrocatalytic response to H2O2. Under the optimum conditions, it exhibited a linear association between current value and the logarithm of the target DNA content (5–500.0 nM) with a limit of detection as low as 0.167 nM and showed satisfactory stability and high specificity in distinguishing single nucleotide polymorphism of target DNA.122 Bhattacharya et al. 2022 developed high-quality laser-induced graphene (LIG) electrodes, which were integrated into a paper-based electrochemical biosensor device for the detection of uric acid. Mechanistically, the enhanced charge-transfer kinetics in the paper-based two-electrode chronoamperometric biosensor led to improved functioning for sensing uric acid (i.e., sensitivity: 24.35 1.55 μA μM−1 and LOD: 41 nM).131 Further, the storage of very concentrated reagents and their release with high proficiency are vital in developing very sensitive biosensors, while the performance of the reservoirs in paper-based biosensors is restricted due to the intrinsic physicochemical characteristics of non-treated cellulose. Therefore, in a study, some polymeric water-soluble reservoirs (e.g., carboxymethyl cellulose, chitosan, carrageenan, and poly(vinyl alcohol)) were created via the drop-casting method with nine different modifications to store and release enzymes and nanoparticles in paper-based biosensors. Exclusively, the release of the enzyme from the reservoir composed of carboxymethyl cellulose rapidly at a high concentration exhibited an ultralow LOD of 0.005 mM for colorimetric glucose sensing, while the reservoirs composed of polymer blends focused antibody-patterned nanoparticles and reduced the LOD of the model immunosensor by 10 times. Moreover, the pathogen Klebsiella pneumoniae in urine samples was detected rapidly with high specificity at the infectious dose threshold.132
In a study, the polylactic acid (PLA) filament and wax filament were used to create hydrophobic patterns on paper for μPADs using the 3D printing method and combined with a fluidic chip in a prototype biosensor, wherein the barrier patterns held cell-free reactions and the fluidic chip facilitated the sample delivery to the reactions in the device. Overall, this rapid prototyping device type could differentiate Dengue virus serotypes considering the differences in their small nucleotide sequences.133 In another study, a CO2 laser was used to generate etched grooves (a pitch of 0.75 mm) on paper to hasten the speed of the wicking channels in μPADs. This study exhibited the influence of various adhesive sealed grooved channels on wicking, where tape sealing demonstrated faster wicking and use of lamination showed a negative effect on wicking. Moreover, this chemiresistive paper-based biosensor efficiently detected human serum albumin (15 mM) spiked in phosphate buffer, artificial urine, and artificial saliva.134 A fullerene C60-functionalized disposable graphite paper-based electrode was utilized to develop a highly sensitive electrochemical immunosensor for determining suppression of tumorigenicity 2 (ST2) in human serum. It showed remarkable repeatability and reproducibility with a broad detection range (0.1–100 fg mL−1), low LOD (0.124 fg mL−1), and limit of quantification (0.414 fg mL−1).135 In a study, an innovative phenylalanine electrochemical paper-based biosensor was manufactured using a hybrid of zinc oxide (ZnO) nanorods and gold (Au) nanoparticles together with the enzyme phenylalanine hydroxylase (from the extract of moss leaf-like tissue). The immobilization of the enzyme was performed by dropping ZnO@Au hybrids and moss extract respectively onto a filter paper and the functionalized paper was positioned on the top of a graphite screen-printed electrode (GSPE). Further, it was effectively utilized in determining phenylalanine in a sample of human blood serum and exhibited the phenylalanine concentration in the range of 5.0 nM and 100 μM with an LOD (S/N: 3) and quantification limit of 3.0 nM and 10.0 nM, respectively.136 In another study, AuNPs were utilized to develop a colorimetric paper-based biosensor device to detect phenylalanine, wherein phenylalanine dehydrogenase enzyme reacted with phenylalanine selectively in the presence of NAD+ to form NADH, which reacts with MTT to form purple-colored formazan in the presence of diaphorase enzyme. Also, AuNPs interact with MTT as a cationic dye and increased the color intensity. Under the optimal circumstances, it exhibited a linear detection range of 0.8 to 33 mg dL−1 and LOD of 0.57 mg dL−1.137
Recently, Covid-19 triggered by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) appeared as a worldwide pandemic outbreak and in China. During the Covid-19 pandemic, the global demand for fast, inexpensive, broad distributable, and point-of-care nucleic acid diagnostic tools tremendously increased to stop widespread infections, preserve health/lives, and sustain economies.138 For rapid and accurate testing, a paper-based electrochemical biosensor with a specific and sensitive immunosensor for the detection of immunoglobulins produced against SARS-CoV-2 was developed. Unlike other LFAs that involve multiple antibodies, this label-free paper-based electrochemical platform targeted SARS-CoV-2 antibodies without the definite obligation of an antibody. Additionally, this method was extended to detect an antigen (the spike protein of SARS-CoV-2) for the possible diagnosis of Covid-19 symptoms.139 In another study, lyophilized cell-free protein synthesis (CFPS) and toehold switch riboregulators (TSRs) were used to develop a paper-based nucleic acid diagnostic template activated by only adding saliva. Herein, TSRs were engineered to express the bioluminescent reporter NanoLuc upon reaction with the SARS-CoV-2 RNA sequences present in saliva tests. This biosensor system produced a visible signal within 7 min after administrating 15 μL saliva and costed less than 0.50 USD roughly. However, additional research to reduce the LOD of this device is desirable for an effective outcome for society.138
Nucleic acid amplification testing (NAAT) is a highly sensitive and specific technique, but it is not appropriate for rapid point-of-care testing. Thus, a 3D microfluidic paper-based electrochemical NAAT biosensor was developed using off-the-shelf gold plasma-coated threads (i.e., electrodes) to combine electrochemical readouts via the ex situ construction of self-assembled monolayers on the threads. Then, movable stacks of filter papers were integrated with incubation, rinsing, and detection steps via a sandwich hybridization assay for time-sequenced responses (Fig. 4). This biosensor device utilized glass fibre substrates for loading the recombinase polymerase amplification components and conducting isothermal intensification. This paper-based biosensor device was used to detect the toxic microalgae Ostreopsis cf. ovata and the detection of 1 ng μL−1 of O. cf. ovata genomic DNA was reported with an LOD of 0.06 pM for targeted synthetic DNA. Also, no significant cross-reactivity was observed from closely related microalgae species.140
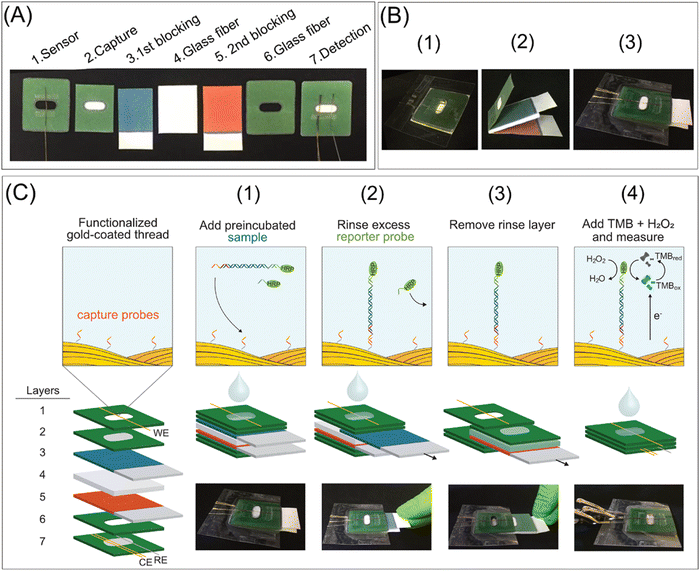 | ||
| Fig. 4 Digital images and schematic representation of the elements, manufacturing, and operation of the device: (A) elements and functions, (B) manufacturing (1: detection, spacer, and sensor layers are aligned and laminated; 2: movable layers are created), 3: movable layers are incorporated between the laminated detection and space layers, and then the modified working electrode is employed in the detection layer, and (C) operational steps from sample incubation to chronoamperometric measurement. Reproduced with permission from ref. 140 Copyright 2021, the American Chemical Society. | ||
Pancreatic ductal adenocarcinoma is the leading neoplastic disease of the pancreas, which is fourth most common reason for death in cancer-related disease with only 8% survival rate after five years of the diagnosis. Therefore, an inexpensive office paper-based biosensor device was fabricated for detecting miRNA-492 for the early diagnosis of pancreatic ductal adenocarcinoma. In this design, an electrochemical sensor was screen-printed on paper using wax-printing, and then it was organized with highly specific peptide nucleic acid (PNA) as the biorecognition component. The construction of the PNA/miRNA-492 duplex was measured by monitoring the reaction between positively-charged ruthenium(III) hexamine with uncharged PNA and/or negatively-charged PNA/miRNA-492 complex. It showed a linear range of up to 100 nM with an LOD of 6 nM.141
Porcine pseudorabies, which is one of the most acute infectious diseases, influences the pig-breeding industry and causes massive losses, highlighting the vital requirement for developing clinical techniques for the exact and rapid diagnosis of wild-type PRV infection. PRV-gE antibodies are a notable indicator of PRV source only in the serum of infected individuals (wild-type PRV), not in injected PRV-gE-deleted vaccines, and enables the distinction of wild-type PRV infection from vaccine immunization. In this case, in 2021, Huang et al. developed a paper-based smartphone biosensing device based on an immunoassay for the distinct diagnosis of wild-type pseudorabies virus infection against vaccination immunization. The test of swine clinical samples exhibited the conformity of 98% between this system and the most commonly utilized commercial gE-ELISA kit. Overall, this biosensing device was fast (15 min), inexpensive, and easy-to-operate, and served as an on-site differential diagnosis method for PRV infection.142
Recently, in 2021, Li et al. developed an inexpensive, miniaturized, and greatly combined sensing paper using Ti3C2Tx MXene and foldable layered wholly-paper substrates for the real-time monitoring of lactate and glucose in human sweat. In this study, the working were screen-printed carbon electrodes (SPCPEs) and functionalized with Ti3C2Tx MXene and methylene blue (MB), and the reference electrode was created using silver paste. Further, a drop of glucose oxidase was placed on the electrode surface for detecting glucose, whereas lactate oxidase was utilized for the detection of lactate. This device showed a linear chronoamperometric current response with a concentration in the range of 0.08–1.25 mM and LOD of 17.05 mM. At low and high contents, the approximate detection sensitivity of glucose was observed to be 2.4 nA mM−1 and 1.0 nA mM−1, respectively. Additionally, the amperometric lactate response was also observed in the linear range of 0.3–20.3 mM with an LOD of 3.73 mM, including a sensitivity of 0.49 mA mM−1. Moreover, in the presence of non-target metabolites and analytes, no obvious interference signals were recognized, the selectivity was good, and the response hardly altered after 14 days.149 In another study, in 2022, Singh et al. developed a highly sensitive enzyme-free human sweat glucose sensor on a Whatman filter paper substrate by utilizing a dual-step pencil and pen strategy (Fig. 5). In this case, two pencil drawn electrodes (PDE) were made on the filter paper, with one of them as the working electrode was drawn with a sensitive layer of copper nanoparticle ink (CuNPs ink, 43.5%) pen and the second one as the reference electrode was drawn with silver conductive ink (Ag ink) pen. The developed CuNPs/PDE promoted rapid electron transfer during the electro-oxidation of glucose in a strong alkaline medium due to its highly conductive and crystalline nature and offered a rapid response time of ∼1.5 s with a sensitivity of 2691.7 μA mM−1 cm−2 and LOD of 0.5 μM in a linear range of 1.2–40 μM. Moreover, the size of the sensor (25 mm × 10 mm) provides the opportunity to develop a miniaturized and wearable electronic system for uninterrupted glucose monitoring.150
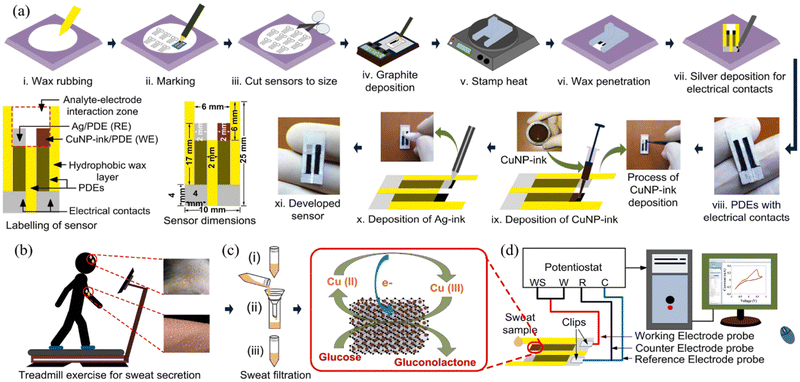 | ||
| Fig. 5 (a) Schematic of the design and manufacture of two-electrode enzyme-free human sweat glucose sensor (i–ix) with the sensor dimensions and labelling, (b) secretion of sweat during treadmill exercise at room temperature, (c) filtration of sweat, and (d) set-up for developed sensor connected to a potentiostat with a computer system for detecting the level of sweat glucose in a human sweat sample. Reproduced with permission from ref. 150 Copyright 2022, Elsevier. | ||
In 2023, Fiore et al. developed a sustainable paper-based microfluidic device for the delivery of capillary-driven microfluidics for a reagent-free competitive magnetic-bead based immunosensor for cortisol determination in sweat samples. The presence of magnetic beads modified with monoclonal antibodies for recognizing cortisol in the reaction zone allowed the measurement of a specific analyte. The competitive interaction between the target cortisol and acetylcholinesterase enzyme-labeled cortisol demonstrated a reaction inversely proportional to the target cortisol (ranging from 10 to 140 ng mL−1) by merely folding the pad loaded with the enzymatic substrate. Further, this paper-based microfluidic device was merged with a near-field communication wireless module to fabricate a flexible integrated wearable analytical device for detecting cortisol in sweat samples (Fig. 6).151
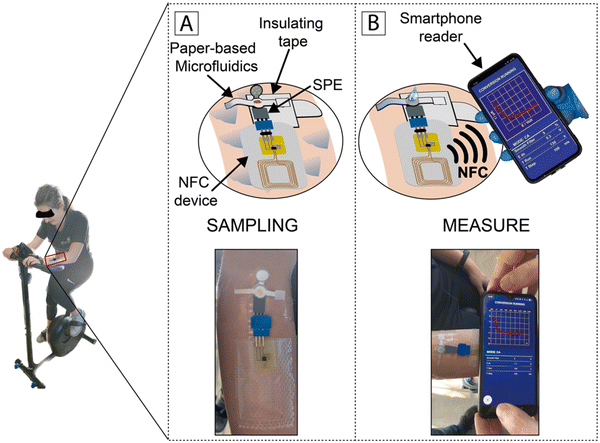 | ||
| Fig. 6 Schematic illustration of monitoring cortisol by chasing the level of the subject during physical activity: (A) processing of samples and (B) assessing data and wireless transmission to a smartphone through NFC. Reproduced with permission from ref. 151 Copyright 2018, Elsevier. | ||
4.2. Agriculture
Agriculture generates food for the world and great revenue with technological innovations. In this case, to realize the better production of food and supplies, new effective strategies are required to identify the on-site yield limiting factors compared to conventional testing, which are mostly non-green, lack precision, and highly non-accessible to farmers. Therefore, biosensors for precision agriculture as a technology-assisted strategy can be applied as real-time, sensitive, and quantitative diagnostic devices when there is a high global food demand. Also, as POC devices, they can be utilized for the protection of crops, monitoring of soil health, and assessment of crop productivity.152Pesticides are utilized in agriculture to control pests, weeds, and diseases in plants with an excellent contribution to food security and availability.153 However, the extensive use of pesticides in the atmosphere causes pollution, which has a negative impact on the ecosystem (e.g., produced food, soil, and groundwater), and thereby human health.154,155 Thus, the extensive use of pesticides should be reduced to prevent or minimize the contamination of unwanted areas (other than targeted species). In this regard, effective electrochemical biosensors have been developed to detect and quantify the over-utilization of pesticides for precision agriculture with long-term food production efficiency and profitability, reducing the unwanted impact on wildlife and the atmosphere using real-time monitoring. Also, the usage of paper as a sustainable substrate reduces the environmental influence of these biosensors owing to its good biodegradability, porosity, and capillary forces.156,157 Recently, some paper-based sustainable biosensors have been developed to detect pesticides.158–160 For example, in 2019, Arduini et al. developed an origami paper-based enzymatic electrochemical biosensor for detecting three different types of pesticides in river water trials158 and an origami wearable paper-based enzymatic biosensor to detect mustard agents in aerosol form.161 In addition, a flower-like origami paper-based enzymatic electrochemical biosensor was developed to detect three classes of analytes such as paraoxon (PX), 2,4-dichlorophenoxyacetic acid (DCPA) and glyphosate (GP) in the aerosol phase at the ppb level by quantifying their inhibitory action towards three different enzymes (i.e., butyrylcholinesterase, alkaline phosphatase, and peroxidase), respectively, for precision agriculture. This integrated electrochemical biosensor was made of three office paper-based screen-printed electrodes and filter paper-based pads laden with enzymes and enzymatic substrates (Fig. 7). Herein, the percentage of inhibition in relation to the quantity of aerosolized pesticides was evaluated by using a smartphone-potentiostat (with initial and residual enzyme activity through chronoamperometric method).162
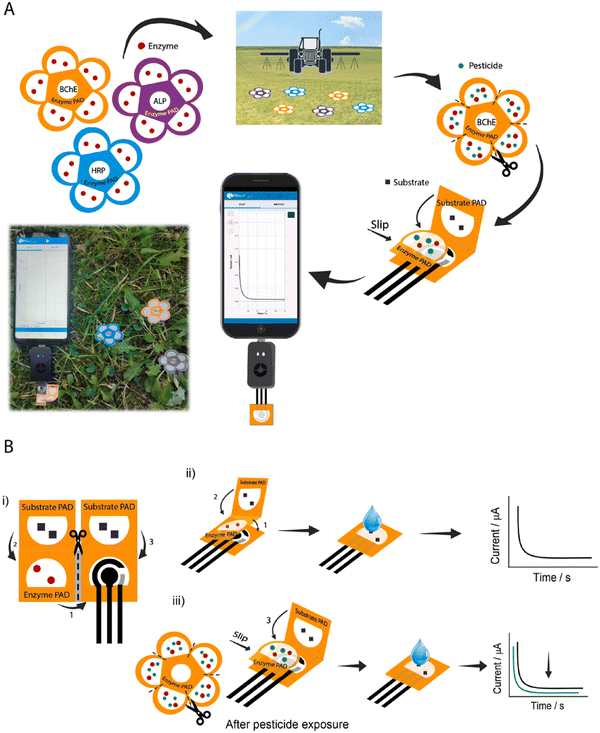 | ||
| Fig. 7 Schematic representation of (A) flower-like origami paper-based electrochemical biosensor for detecting pesticides using smartphone-assisted potentiostat and (B) device composed of office paper-based screen-printed electrode and three filter pads (i), evaluation procedure of initial enzymatic activity in the absence of pesticide (ii), and residual enzymatic activity after exposure to pesticide solution (iii). Reproduced with permission from ref. 162 Copyright 2022, Elsevier. | ||
A wide range of insecticides acts and initiates physiological responses when combined with nicotinic acetylcholine receptors (nAChRs), leading to paralysis and the death of pest organisms.163–166 Neonicotinoids is one of the most commonly utilized class of insecticides worldwide.167–169 The adverse effect of neonicotinoid pesticide residues on human health and the environment via contamination is a major concern, and thus sensitive, rapid, and on-site devices are highly desirable for detecting these residues for food security and sustainable development of environment. Various effective and highly sensitive methods have been developed and utilized for these purposes, but their usage is limited due to their high cost and time-consuming sample provision. Therefore, for food safety and public health, it is imperative to develop modest, fast, and low-cost diagnostic techniques for the real-time monitoring of these pesticides and insecticides. In this case, a paper-based surface-enhanced Raman scattering (SERS) intensified strategy was proposed by integrating multilayered plasmonic coupling using hybrids of 3D silver dendrite (SD), electropolymerized molecular identifier (EMI), and silver nanoparticles (AgNPs). Due to the distinct design of the multilayered coupling amplification approach, the SERS paper chips exhibited remarkable specificity and ultra-high sensitivity in detecting imidacloprid (IMI), with a low limit of detection of 0.02811 ng mL−1. Moreover, this approach showed good prospects for the automated screening of a diverse range of pollutants.170 In another study, an eco-designed paper-based algal biosensor was developed for optical detection of nano-encapsulated-atrazine (a nanoformulated herbicide). In this study, a paper substrate soaked with an agar thin film was immobilized with a unicellular green photosynthetic algae Chlamydomonas reinhardtii and placed in a glass optical analysis. The detection of nano-encapsulated-atrazine demonstrated a range of 0.5 to 200 nM, demonstrating a relationship in reasonable dose–response curves and limit of detection of 4 pM. Further, the interference analyses showed a minor interference in the presence of 2 ppm copper and 10 ppb at safety bounds, a suitable recovery value of 96 ± 5% for 75 nM nano-encapsulated-atrazine, and good storage stability for up to 3 weeks.171
Soil pollution, caused by organophosphorus pesticides and their main metabolites, is a major concern worldwide. In this case, the on-site screening of these pollutants for determining soil bioavailability is still a challenge. Therefore, whole-cell biosensors are effective devices to distinguish the interaction mechanisms between pollutants and microbes. In this way, a novel, inexpensive and portable paper-strip biosensor was developed, which could precisely detect methyl parathion (MP) and its prime metabolite p-nitrophenol elements (Fig. 8). In this study, the Escherichia coli BL21/pNP-LacZ system was fixed to filter paper using an alginate biogel and polymyxin B sensitizer. The color intensity of this paper-strip biosensor recorded by a mobile app could provide the content of MP and p-nitrophenol. This method could detect and quantify the content of 10–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg kg−1 for p-nitrophenol and 25–7500 for μg kg−1 MP, wherein the investigation of soil samples showed the detection limit of 5.41 μg kg−1 for p-nitrophenol and 9.57 μg kg−1 for MP. Also, it could detect and quantify the content of 10–10
000 μg kg−1 for p-nitrophenol and 25–7500 for μg kg−1 MP, wherein the investigation of soil samples showed the detection limit of 5.41 μg kg−1 for p-nitrophenol and 9.57 μg kg−1 for MP. Also, it could detect and quantify the content of 10–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg kg−1 for p-nitrophenol and 25–7500 for μg kg−1 MP. Moreover, this method could also be utilized to study the gene expression in complex environments.172
000 μg kg−1 for p-nitrophenol and 25–7500 for μg kg−1 MP. Moreover, this method could also be utilized to study the gene expression in complex environments.172
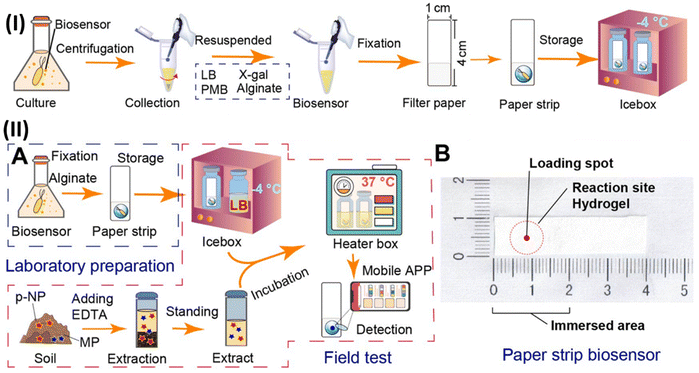 | ||
| Fig. 8 (I) Schematic illustration of manufacturing of paper-strip biosensor. (II) Diagram of whole-cell biosensor device for detecting MP and p-nitrophenol (p-NP): flow-diagram of the fabrication and utilization (A) and objective drawing (B) of whole-cell paper-strip biosensor. Reproduced with permission from ref. 172 Copyright 2023, Elsevier. | ||
Further, the extensive use of organophosphorus pesticides (OPs) in agriculture has reduced the possible threats of residue complications. Therefore, it is highly essential and desirable to develop operative diagnostic methods (e.g., rapid and reliable biosensor). In general, conventional biosensors majorly depend on biorecognition components, which limit their real-world applications. Thus, to address these issues, nanozymes are broadly utilized in developing nanozyme-enabled biosensors owing to their inexpensive nature, remarkable stability, and promising catalytic action.173 In this advancement, in 2022, Song et al. developed an AChE-nanozyme bioactive paper together with a 3D-printed system for the fast detection of different pesticides. After serially incorporating AChE, acetylcholine (ACh), and TMB, and H2O2 solutions via various syringes on the 3D printed system, the SAzyme (SACe-N-C) with peroxidase-like action exhibited continuous responses to AChe. However, OPs may impede the action of AChE and yield less or no choline (no reduction of blue oxTMB, oxidized by SACe-N-C) to the colorless redTMB successively, leading to the release of the action of SACe-N-C and generation of the color of bioactive paper positively according to content of OPs. Moreover, this biosensing device could monitor the concentration of OP residues in fruits and vegetables within 30 min with an LOD of 55.83 ng mL−1 and 77.51 ng mL−1 for omethoate and methamidophos, respectively.174
The occurrence of antibiotic-resistant bacteria due to the presence of antibiotics in environmental water is a serious issue. In this case, rapid and sensitive colorimetric paper-based biosensing devices can be utilized for detecting antibiotics the inhibit bacterial protein synthesis. By considering the concept of color change induced by β-galactosidase synthesis in real water samples without antibiotics, it exhibited the approximate limit of detection by the naked eye of 0.5, 2.1, 0.8, and 6.1 mg mL−1 for paromomycin, tetracycline, chloramphenicol, and erythromycin, respectively, by hampering the inhibition of β-galactosidase synthesis with antibiotics.175
4.3. Food industry
Microorganisms such as bacteria, fungi, and viruses play a vital role in our life, wherein some microorganisms (e.g., Penicillium (2), Saccharomyces (1), and Lactobacillus (3)) are beneficial and broadly utilized in the food and medicinal industries. However, in many cases, the microbial contamination of food causes severe health problems, leading to significant economic loss annually. Therefore, the fast, sensitive, specific, and visual detection of microbials in food items is highly desirable. In this regard, various microbial detection devices have been developed, but they are complex and time-consuming methods, and are not effective in growing food testing demands. The climate change and increasing global population, which affect agriculture and food safety, are major challenges worldwide, wherein the agricultural and food industries are the major support of our society and essential to be developed uninterruptedly to realize improved productivity and quality. For this purpose, fertilizers and pesticides have been extensively utilized in modern times for tremendous profits to agricultural productivity; however, the harmful impact of their residues in food products and environmental contamination cannot be ignored.176 Some chemicals hazards (e.g., pesticides and veterinary drug residues), illegal or excessive additives, environmental impurities, natural toxins, and biological hazards (e.g., microbial impurities) can have adverse effects on human health even at low levels.177,178 Thus, suitable, effective, selective, and sensitive diagnostic devices are greatly desirable. In recent times, sensors as diagnostic devices showed an advantage of straightforward operation, high proficiency, and sensitivity. These devices include sensitive elements and conversion elements, which can quantify specific targets by converting them into usable signals according to certain laws.178,179 Herein, paper-based biosensors are very effective for the efficient and visual detection of microorganisms for food safety and environmental hygiene owing to their simple, low-cost, high speed, and all-in-one device.88 In a study, an inexpensive and dual-step pen-on-paper strategy was used to fabricate a fully drawn fluidic patterned electrochemical paper-based biosensor for the amperometric enzymatic determination of glucose, wherein glucose oxidase and potassium ferricyanide (as the mediator) were immobilized in the test area. The results exhibited the limit of quantification of 0.08 mmol L−1, reproducibility of <12% for 6 different devices, and the assay time of 120 s. This device was applied for the determination of glucose in food samples with 94–106% recovery.180 Some paper-based biosensing devices exhibited low sensitivity, and in this case, aptamers as bioreceptors are used to improve the selectivity and sensitivity of the sensors, specifically paper-based biosensors. Therefore, a paper-based electrochemical aptasensor was developed to detect ractopamine (RAC) with good sensitivity and selectively under the optimal conditions in the range of 0.001 μM to 100 mM (lower limit of quantification: 0.01 μM). Further, the monitoring of RAC in non-treated human plasma samples was observed with the lower limit of quantification of 0.01 μM and linear range of 0.01 μM–10 mM.181Contamination of foods is the greatest concern to consumers,182 wherein food contamination occurs in various ways (e.g., physical, chemical, and biological processes). Among them, infectious microorganisms are the most serious contaminants of food, especially in packaged foods.183 In this situation, the development of active and sensitive food packaging is highly desirable to protect and preserve foods or communicate between packaging foods and consumers.184,185 Therefore, the integration of biopolymer-based smart food packaging with biosensing entities has provided an attractive opportunity for the real-time monitoring of the microbial breakdown of packaged foods for food quality and safety.186
The trustworthy detection of pathogenic bacteria in complex biological specimens using simple assays or sensing devices remains a key challenge. A simple colorimeter paper biosensing device was developed and used to detect Helicobacter pylori (H. pylori) specifically and sensitively. H. pylori is a pathogen intensely connected to gastric carcinoma, gastric ulcers, and duodenal ulcers in stool samples. The sensor device was designed based on the RNA-cleaving characteristic of the DNAzyme, which is activated by a protein biomarker from H. pylori, enabling the sensitive and rapid detection of H. pylori in a human stool specimen within minutes. Moreover, the diagnostic device maintained its functionality under storage for at least 130 days at ambient temperature.187 A multiplex lateral flow immunoassay (LFA) was developed to detect primary marine biotoxins (e.g., amnesic, paralytic, and diarrhetic shellfish poisoning toxins) with a single sample isolation method and diluted volume and showed no matrix effect on its detection performance.188
Infections triggered by Gram-positive bacteria cause several severe diseases (e.g., bacteremia, sepsis, pneumonia, osteomyelitis, and endocarditis).189,190 Therefore, the rapid inspection of Gram-positive bacteria-contaminated food stuffs is highly required to prevent and control the epidemic situations induced by them.191 Compared to the lengthy traditional approach, nucleic acid-, immunological-, and biosensing-based strategies have been developed to reduce the time for detecting bacteria.192–194 However, the possible interference of a complex matrix on these approaches may generate false-positive or negative outcomes with no cultural supplementation. Thus, the development of pre-processing approaches to overcome matrix-linked inhibition and reach the low content of bacteria in foodstuffs is crucial to improve the accuracy of these approaches. Herein, an on-site colorimeter biosensing device was developed for ultrasensitive and fast recognition by using polydopamine and polyethyleneimine (PDA–PEI)-functionalized papers (PDA–PEI@paper) for isolating bacteria and peptidoglycan-targeting carbon dots for the selective colorimetric recognition of Gram-positive bacteria. Moreover, this device could detect S. aureus up to 1 cfU mL−1, L. monocytogenes up to 5 cfU mL−1, and B. subtilis up to 9 cfU mL−1 within 55 min. Also, the recognition of Gram-positive bacteria was performed in the samples of eggshell and sausage with recoveries in the range of 91.2–110%.195 In another study, AuNPs were utilized in a paper-based DNA biosensing device for the visual detection of dairy products for their food authenticity, wherein species-specific DNA sequences were separated, intensified and recognized using specific DNA probes in the biosensing device. In this case, three species (e.g., cow, sheep, and goat) were analyzed as low as 1.6 fmol for cow and goat, and 3.1 fmol for sheep PCR result. In addition, contamination down to 0.01% could also be detected based on binary mixtures of milk yogurts of cow, goat, and sheep having 0.01% to 5% of cow yogurt in the yogurt of the sheep and goat, respectively. Overall, this biosensing device exhibited good specificity, reproducibility, and 10 fold detectability compared to other methods.196
Salmonella causes millions of foodborne illnesses annually and result in higher death rates compared to other foodborne bacterial pathogens. A colorimeter paper-based analytical device (PAD) linked with immunomagnetic separation (IMS) was used for the specific detection of Salmonella typhimurium (S. typhimurium) without any interference from other pathogenic bacteria (e.g., Escherichia coli). The LOD for S. typhimurium was observed to be 102 CFU mL−1, and also this device was utilized for inoculated Starling bird fecal and whole milk samples with an LOD of 105 CFU g−1 and 103 CFU mL−1, respectively.197 The analysis of complex matrices in food samples is tedious due to the interference among them. In this case, optical biosensors with chemiluminescence and fluorescence detection mechanisms have been developed and applied over traditional methods due to their good sensitivity and selectivity. For example, phenolic compounds (PCs), which are key plant ingredients having redox characteristics responsible for antioxidant action, were detected by a paper-based chemiluminescence smartphone biosensing device. In this study, molasses, honey, and tea as food samples were analyzed and showed the LOD for PCs of 0.12, 0.28, 0.46, 0.85, and 1.23 μg mL−1 for gallic acid, quercetin, catechin, kaempferol, and caffeic acid, respectively.198 For the detection of procymidone in vegetables, three paper-based biosensor devices using a core biological immune scaffold (CBIS) were fabricated as time-resolved fluorescence immunochromatography strips with europium oxide (Eu-TRFICS). Herein, CBIS was designed using secondary fluorescent probes (of goat anti-mouse IgG and europium oxide time-resolved fluorescent microspheres) and procymidone monoclonal antibody (PCM-Ab). Three paper-based devices of Eu-TRFICS-1 (fixing of secondary fluorescent probes onto conjugated pad and mixing of PCM-Ab in a sample solution), Eu-TRFICS-2 (fixing of CBIS onto conjugate pad), and Eu-TRFICS-3 (direct mixing of CBIS with sample solution). These systems demonstrated multidimensional labeling and directional-coupling (by resolving the difficulties of steric interruption of antibody labeling, inadequate exposure of antigen-recognition zone and easy loss of action in conventional methods). Among the three types, Eu-TRFICS-1 exhibited the best detection ability with the reduction of antibody usage by 25%, increased sensitivity by 3 times, detection of range of 1–800 ng mL−1, and LOD of 0.12 ng mL−1 with a visible LOD of 5 ng mL−1.199
The freshness of fresh meat can be evaluated by determining the Hx level (a metabolic product of ATP metabolism) and it can be utilized as a biomarker for detecting the early stage of spoilage.200 In a study, a pattern-free paper enzyme biosensing device was fabricated for one-step fish-freshness detection by indicating hypoxanthine (Hx). For this development, chitosan oligosaccharide lactate-functionalized nitrocellulose (NC) membranes were immobilized with a dual-enzyme system (xanthine oxidase and horseradish peroxidase) and showed a remarkable microfluidic accumulation effect. The as-developed biosensing device showed a color within 3 min and exhibited a linear response of 0.01–0.16 mmol L−1 with an LOD of 8.22 μmol L−1, and selective recognition of Hx with satisfactory recoveries of 96.13–103.11% for fish specimens. Herein, the biochemical signal of Hx level was converted to a colorimetric signal via a two-step enzymatic interaction for visual detection. Also, a remarkable color distinction between the negative specimen (colorless) and positive specimen (green) with the naked eyes could be observed by attaching it directly to the surface of fish meat. Also, quantitative analysis could be achieved using a smartphone.201 Most recently, in 2024, Li et al. reported the fabrication of a pomegranate-structured film-based sensing device using PANI, multi-branched cellulose nanofibers (MCNFs), and multi-walled carbon nanotubes (MWCNTs) for the detection of human-physiology and food spoilage gas signals for real-time monitoring by integrating it with a wireless heart-rate display and smart packaging censoring technique.202
4.4. Environmental pollutants
Various environmental pollutants pose a significant threat to human health and the global climate (atmosphere). Among them, the contamination of heavy metal ions in water is gaining considerable interest due to their serious harmful consequences to human health and adverse ecological effects. Mercury (Hg2+) and lead (Pb2+) are toxic metals present in water, posing a remarkable hazard to the environment and aquatic life via biological accumulation in the food chain, ultimately affecting human health. Exposure to even a trace amount of Hg2+ can trigger severe damage to human organs.203 Also, the accumulation of Pb2+ beyond a threshold value causes permanent damage.204 In this case, in vitro transcription (IVT) technology with an allosteric transcription factor (aTF)-based cell-free paper-based biosensor device was developed for the on-site detection of Hg2+ and Pb2+ simultaneously in a consistent and sensitive mode with just one drop of water (Fig. 9). It could detect Hg2+ and Pb2+ quantitatively in a linear range of 0.5–500 nM and 1–250 nM within a period of one hour, with an LOD of 0.5 nM and 0.1 nM for Hg2+ and Pb2+, respectively. Moreover, the recoveries ranged from 91.09% to 123.24% for the detection of real water models.205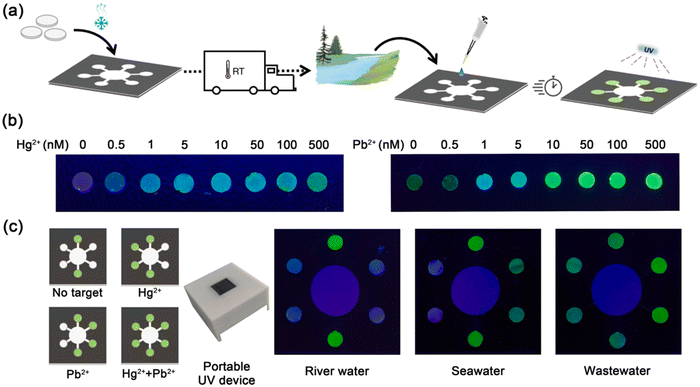 | ||
| Fig. 9 (a) Schematic illustration of the development and application of paper-based biosensing device, (b) fluorescence output of paper-based biosensing device at different contents of Hg2+ and Pb2+, and (c) detection of real water models using this inexpensive and portable paper-based biosensing device. Reproduced with permission from ref. 205 Copyright 2021, Elsevier. | ||
Water-borne pathogens are also causing a remarkable increment in mortality rate due to issues such as poor sanitation, sewage sludge, and industrial effluents. Thus, to prevent this, easy, sensitive, user-friendly, and rapid on-site biosensor devices for detecting pathogens are needed. In this advancement, an inexpensive, label-free, and eco-friendly paper-based screen-printed graphene oxide (GO)-functionalized electrochemical impedimetric biosensing device immobilized with lectin Concanavalin A (ConA) as the biorecognition unit was developed for the detection of a water-borne bacterial contamination, Escherichia coli, in water. Mechanistically, the high surface area and chemical action of GO on graphene led to the remarkably specific immobilization of ConA in bulk quantity onto the surface of GO owing to its active functional groups (–COOH) for site-specific reaction in the pH range of 4.5–6.0. This device achieved ultra-high sensitivity for detecting bacterial contamination over a wide linear range of 102–108 CFU mL−1 and ultra-low LOD of 0.1 × 102 CFU mL−1 without any chemical intensification.206 In another study, a ‘turn-off’ fluorescent biosensing device (mCherry L199C) for sensing Hg2+ was designed and developed via the direct modification of the chromophore environment of the fluorescent protein mCherry. The designed variant of mCherry L199C was expressed directly on the outer-membrane of E. coli cells via the cell surface display approach. The fluorescent biosensing device exhibited a favorable response to Hg2+ at the micromole level among other metal ions and over a wide pH range. The cells of this biosensor were incorporated in an alginate hydrogel to develop cell-alginate hydrogel-based paper. This system was simple and inexpensive, which could detect mercury pollution within 5 min and keep stable fluorescence and activity at 4 °C for 24 h.207
4.5. Other applications
There are various other mixed applications, wherein paper-based sustainable biosensing devices have been utilized for desired outcomes as efficient diagnostic platforms. For example, a biosensor composed of a hydrophilic cellulosic paper disk with immobilized GOX placed on SPCE was developed and this biosensing device, using 0.1 M phosphate buffer solution (PBS, pH 7.0) and 10 mM soluble ferrocene monocarboxylic acid mediator, detected a low amount of glucose (5 μL) and showed a linear range of 1–5 mM glucose with an LOD of 0.18 mM, and maintained 98% of its activity after a period of 4 months. It was successfully utilized for the selective and sensitive assay of glucose in various commercial soda beverages.208 In another study, the level of glucose in beverages (e.g., commercial orange juice) via potentiometric detection was monitored using a developed novel paper-based biosensor device. In this case, the electrodes were composed of platinized paper as a support and a biocompatible polymeric membrane composed of a blend of chitosan and poly(vinyl alcohol) holding glucose oxidase as the recognition layer. This biosensing device selectively and sensitively detected H2O2 produced by an enzymatic response. It exhibited rapid and accurate results, presenting the sensitivity of 119 ± 6.4 mV dec−1 in the range of 0.03–1.0 mM with an LOD of 0.02 mM.209 Presently, the use of volatile toxic complexes as chemical weapons as a large-scale destructive system has posed a new threat worldwide. In this case, the development of effective wearable sensing devices can be a good candidate for security monitoring. Therefore, a wearable electrochemical biosensor was fabricated for the rapid and on the spot detection of mustard agents directly in the aerosol phase. In this case, the electrodes were screen-printed onto a filter paper to create an origami-like and reagent-free device. The detection of mustard agents was conducted by monitoring their inhibitory effects via amperometric analysis of an enzymatic by-product (hydrogen peroxide, H2O2). In addition, the nanocomposite of carbon black/Prussian blue was utilized to modify the conductive graphite ink forming the working electrode for the electrocatalysis of H2O2 reduction. Moreover, after verification with a mustard agent, the developed origami-like biosensor was used to detect real sulfur mustard rapidly and in real time, demonstrating the LOD of 1 mM and 0.019 g min m−3 for the liquid and aerosol phases, respectively (Fig. 10).161 | ||
| Fig. 10 Schematic of the working principle of the developed wearable origami-like ePAD. Reproduced with permission from ref. 161 Copyright 2019, Elsevier. | ||
5. Conclusion, challenges and future research directions
In this review, the fundamentals of sensing or biosensing devices and their utilized materials were precisely described. To guide the future research on the design of biosensors or bioelectronics, we focused on discussing the efficacy of the use of renewable and sustainable materials for biosensing devices as an alternative to petroleum-based polymers. Further, the recent advances in emerging paper-based sustainable materials and high-performance devices in biosensing, such as healthcare, agriculture, food industry, environmental pollutants, and others were discussed.As discussed above, renewable and sustainable materials have received significant attention in the past few years in terms of basic research and real-world applications owing to their ability to minimize environmental, consumption, and cost-related issues. Accordingly, these materials have widely been utilized in pristine form or in combination to manufacture sensing and biosensing devices for suitable and targeted utilization and outcomes with low cost, satisfactory biodegradability, high flexibility, and low-energy consumption. Herein, paper is a workable alternative to traditional materials utilized in biosensing devices for various applications due to its inexpensive nature, flexibility, fluid conduct (capillary action due to rough fibrous-network), significant sensitivity and easy disposal. In addition, the combination of cellulose with a diverse range of additives (e.g., conductive polymers and nanomaterials) produces composite materials with remarkable electrical conductivity, while preserving the sustainability and biocompatibility of cellulose. Therefore, extensive research activities on paper- and cellulose-based biosensing devices have widely been reported and still on-going for effective utilization in various potential applications. However, there are still various challenges in using renewable materials in developing inexpensive sensing devices with satisfactory sensitivity, selectivity, response time, steadiness, and operational durability. Furthermore, they require an enhanced operational timing, low maintenance and installation cost, and data management.
For an independent measurement of environmental factors, calibrations, numerical corrections, and doping or coating methods are used for precise and effective operations. Further, with the development of communication technologies, high-frequency microprocessors, the Internet of things, real-time detection, and storage capacity, the successful integration of these technologies into the developed biosensing devices for on-spot real-time data is still another challenge. Various design approaches and fabrication technologies should be used to enhance or convert renewable materials into value-added inexpensive products with desirable properties such as mechanical strength, flexibility, thermal stability, electrical characteristics, and recyclability. Significant functioning and advances in computer science with machine learning algorithms are beneficial in achieving optimal parameters and reduction in operational time and associated costs.210–212 In addition, artificial intelligence methods have provided a great opportunity in all sections of the world by allowing machines to mimic imagination, decision-making process, and manufacturing new materials.213,214 In this case, the use of microfabrication technologies (e.g., additive manufacturing) will decrease the time of manufacturing with anticipated structural design and reduction in wastage of materials, thereby decreasing the overall cost of the product.66,215 Moreover, the economic and technological challenges must be considered and encouraged by scientific research to convince decision-makers to develop the ideas for new sensing and biosensing devices.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. K. acknowledges the support from the Ramalingaswami Re-entry Fellowship (Ref: BT/HRD/35/02/2006) of Department of Biotechnology, Government of India.References
- H. Teymourian, M. Parrilla, J. R. Sempionatto, N. F. Montiel, A. Barfidokht, R. Van Echelpoel, K. De Wael and J. Wang, ACS Sens., 2020, 5, 2679–2700 CrossRef CAS PubMed.
- J. van der Zalm, S. Chen, W. Huang and A. Chen, J. Electrochem. Soc., 2020, 167, 037532 CrossRef CAS.
- D. Li, H. Duan, Y. Ma and W. Deng, Anal. Chem., 2018, 90, 5719–5727 CrossRef CAS PubMed.
- H. Nawaz, J. Zhang, W. Tian, K. Jin, R. Jia, T. Yang and J. Zhang, J. Hazard. Mater., 2020, 387, 121719 CrossRef CAS PubMed.
- A. Pavinatto, L. A. Mercante, C. S. Leandro, L. H. Mattoso and D. S. Correa, J. Electroanal. Chem., 2015, 755, 215–220 CrossRef CAS.
- J. Fan, S. Zhang, F. Li and J. Shi, Cellulose, 2020, 27, 5477–5507 CrossRef CAS.
- G. Barandun, M. Soprani, S. Naficy, M. Grell, M. Kasimatis, K. L. Chiu, A. Ponzoni and F. Güder, ACS sensors, 2019, 4, 1662–1669 Search PubMed.
- R. Prathapan, R. F. Tabor, G. Garnier and J. Hu, ACS Appl. Bio Mater., 2020, 3, 1828–1844 CrossRef CAS PubMed.
- F. Arduini, L. Micheli, V. Scognamiglio, V. Mazzaracchio and D. Moscone, TrAC, Trends Anal. Chem., 2020, 128, 115909 CrossRef CAS.
- K. Ratajczak and M. Stobiecka, Carbohydr. Polym., 2020, 229, 115463 CrossRef CAS PubMed.
- K. B. Teodoro, R. C. Sanfelice, F. L. Migliorini, A. Pavinatto, M. H. Facure and D. S. Correa, ACS Sens., 2021, 6, 2473–2496 CrossRef CAS PubMed.
- S. Kamel and T. A. Khattab, Biosensors, 2020, 10, 67 CrossRef CAS PubMed.
- V. B. C. Lee, N. F. Mohd-Naim, E. Tamiya and M. U. Ahmed, Anal. Sci., 2018, 34, 7–18 CrossRef CAS PubMed.
- B. G. Andryukov, AIMS Microbiol., 2020, 6, 280 CAS.
- E. Solhi, M. Hasanzadeh and P. Babaie, Anal. Methods, 2020, 12, 1398–1414 RSC.
- A. Antonacci, V. Scognamiglio, V. Mazzaracchio, V. Caratelli, L. Fiore, D. Moscone and F. Arduini, Front. Bioeng. Biotechnol., 2020, 8, 339 CrossRef PubMed.
- W.-J. Lan, X. U. Zou, M. M. Hamedi, J. Hu, C. Parolo, E. J. Maxwell, P. Bühlmann and G. M. Whitesides, Anal. Chem., 2014, 86, 9548–9553 CrossRef CAS PubMed.
- N. F. Santos, S. O. Pereira, A. Moreira, A. V. Girão, A. F. Carvalho, A. J. Fernandes and F. M. Costa, Adv. Mater. Technol., 2021, 6, 2100007 CrossRef CAS.
- N. Nontawong, M. Amatatongchai, W. Wuepchaiyaphum, S. Chairam, S. Pimmongkol, S. Panich, S. Tamuang and P. Jarujamrus, Int. J. Electrochem. Sci., 2018, 13, 6940–6957 CrossRef CAS.
- X. Jin, C. Liu, T. Xu, L. Su and X. Zhang, Biosens. Bioelectron., 2020, 165, 112412 CrossRef CAS PubMed.
- P. Gründler, Chemical Sensors: An Introduction for Scientists and Engineers, Springer Science & Business Media, 2007 Search PubMed.
- A. A. Ensafi, Electrochemical Biosensors, Elsevier, 2019, pp. 1–10 Search PubMed.
- S. Li, Y. Zhang, Y. Wang, K. Xia, Z. Yin, H. Wang, M. Zhang, X. Liang, H. Lu and M. Zhu, InfoMat, 2020, 2, 184–211 CrossRef CAS.
- J. Janata, Principles of chemical sensors, Springer Science & Business Media, 2010 Search PubMed.
- M. S. H. Ador, P. Bhattacharjee, S. Kabir, M. T. Ahmed, F. Ahmed and I. A. Choudhury, Ref. Module Mater. Sci. Mater. Eng., 2023 DOI:10.1016/B978-0-323-96020-5.00043-1.
- F.-G. Banica, Chemical sensors and biosensors: fundamentals and applications, John Wiley & Sons, 2012 Search PubMed.
- A. P. Turner, Chem. Soc. Rev., 2013, 42, 3184–3196 RSC.
- A. Qureshi, Y. Gurbuz and J. H. Niazi, Sens. Actuators, B, 2012, 171, 62–76 CrossRef.
- S. Saha, M. Tomar and V. Gupta, J. Appl. Phys., 2012, 111, 102804 CrossRef.
- L. C. Clark Jr and C. Lyons, Ann. N. Y. Acad. Sci., 1962, 102, 29–45 CrossRef CAS PubMed.
- G. A. Rechnitz, R. Kobos, S. Riechel and C. Gebauer, Anal. Chim. Acta, 1977, 94, 357–365 CrossRef CAS PubMed.
- N. Bhattacharyya, S. Jain, S. Bhattacharyya, S. Pal, A. Jana and S. Mukherjee, Ref. Module Mater. Sci. Mater. Eng., 2023 DOI:10.1016/B978-0-323-96020-5.00058-3.
- D. G. Buerk, Biosensors: Theory and applications, CRC Press, 2014 DOI:10.1201/9781498710770.
- C. Karunakaran, K. Bhargava and R. Benjamin, Biosensors and Bioelectronics, Elsevier, 2015 Search PubMed.
- M. Zamani, J. Dupaty, R. Baer, U. Kuzmanovic, A. Fan, M. W. Grinstaff, J. E. Galagan and C. M. Klapperich, ACS Omega, 2022, 7, 5804–5808 CrossRef CAS PubMed.
- F. S. Ligler and C. R. Taitt, Optical biosensors: today and tomorrow, Elsevier, 2011 Search PubMed.
- C. Steinem and A. Janshoff, Piezoelectric Sensors, Springer Science & Business Media, 2007 Search PubMed.
- S. Zhou, Y. Zhao, M. Mecklenburg, D. Yang and B. Xie, Biosens. Bioelectron., 2013, 49, 99–104 CrossRef CAS PubMed.
- S. Cosnier, Electrochemical biosensors, CRC Press, 2015 Search PubMed.
- O. Tokel, F. Inci and U. Demirci, Chem. Rev., 2014, 114, 5728–5752 CrossRef CAS PubMed.
- S. Sang, Q. Feng, X. Tang, T. Wang, X. Huang, A. Jian, Z. Ma and W. Zhang, Microelectron. Eng., 2015, 134, 33–37 CrossRef CAS.
- V. Naresh and N. Lee, Sensors, 2021, 21, 1109 CrossRef CAS PubMed.
- S. Varshney, P. K. Rajput, A. Singhal and G. Varshney, Mater. Today: Proc., 2022, 62, 6679–6683 Search PubMed.
- G. Dandegaonkar, A. Ahmed, L. Sun, B. Adak and S. Mukhopadhyay, Mater. Adv., 2022, 3, 3766–3783 RSC.
- H. Ma, Z. Cheng, X. Li, B. Li, Y. Fu and J. Jiang, J. Bioresour. Bioprod., 2023, 8, 15–32 CrossRef CAS.
- L. Wang, D. Chen, K. Jiang and G. Shen, Chem. Soc. Rev., 2017, 46, 6764–6815 RSC.
- H. Zhu, W. Luo, P. N. Ciesielski, Z. Fang, J. Zhu, G. Henriksson, M. E. Himmel and L. Hu, Chem. Rev., 2016, 116, 9305–9374 CrossRef CAS PubMed.
- S. Sreejith, L. L. Joseph, S. Kollem, V. Vijumon and J. Ajayan, Measurement, 2023, 113261 CrossRef.
- Q. Wang, T. Liu, X. Xu, H. Chen and S. Chen, Int. J. Biol. Macromol., 2021, 169, 60–66 CrossRef CAS PubMed.
- D. Molinnus, A. Drinic, H. Iken, N. Kröger, M. Zinser, R. Smeets, M. Köpf, A. Kopp and M. J. Schöning, Biosens. Bioelectron., 2021, 183, 113204 CrossRef CAS PubMed.
- J. Park, J.-K. Kim, S. A. Park and D.-W. Lee, Microelectron. Eng., 2019, 206, 1–5 CrossRef CAS.
- D. Zhao, Y. Zhu, W. Cheng, W. Chen, Y. Wu and H. Yu, Adv. Mater., 2021, 33, 2000619 CrossRef CAS PubMed.
- K. Xia, X. Chen, X. Shen, S. Li, Z. Yin, M. Zhang, X. Liang and Y. Zhang, ACS Appl. Electron. Mater., 2019, 1, 2415–2421 CrossRef CAS.
- H. Qi, J. Liu, J. Pionteck, P. Pötschke and E. Mäder, Sens. Actuators, B, 2015, 213, 20–26 CrossRef CAS.
- H. Koga, M. Nogi, N. Komoda, T. T. Nge, T. Sugahara and K. Suganuma, NPG Asia Mater., 2014, 6, e93–e93 CrossRef CAS.
- R. K. Mishra, P. Mishra, K. Verma, A. Mondal, R. G. Chaudhary, M. M. Abolhasani and S. Loganathan, Environ. Chem. Lett., 2019, 17, 767–800 CrossRef CAS.
- S.-Y. Cho, H. Yu, J. Choi, H. Kang, S. Park, J.-S. Jang, H.-J. Hong, I.-D. Kim, S.-K. Lee and H. S. Jeong, ACS Nano, 2019, 13, 9332–9341 CrossRef CAS PubMed.
- P. Sukhavattanakul and H. Manuspiya, Carbohydr. Polym., 2020, 230, 115566 CrossRef CAS PubMed.
- J.-W. Han, B. Kim, J. Li and M. Meyyappan, J. Phys. Chem. C, 2012, 116, 22094–22097 CrossRef CAS.
- J.-W. Han, B. Kim, J. Li and M. Meyyappan, Rsc Adv., 2014, 4, 549–553 RSC.
- T. S. D. Le, S. Park, J. An, P. S. Lee and Y. J. Kim, Adv. Funct. Mater., 2019, 29, 1902771 CrossRef.
- S. Teng, G. Siegel, W. Wang and A. Tiwari, ECS Solid State Lett., 2014, 3, M25 CrossRef CAS.
- S. K. Mahadeva, K. Walus and B. Stoeber, ACS Appl. Mater. Interfaces, 2014, 6, 7547–7553 CrossRef CAS PubMed.
- H. Koga, K. Nagashima, Y. Huang, G. Zhang, C. Wang, T. Takahashi, A. Inoue, H. Yan, M. Kanai and Y. He, ACS Appl. Mater. Interfaces, 2019, 11, 15044–15050 CrossRef CAS PubMed.
- M. Niu, Y. Yao, Y. Shi, J. Luo, X. Duan, T. Liu and X. Guo, Ind. Eng. Chem. Res., 2019, 58, 10364–10372 CrossRef CAS.
- A. Nasri, M. Petrissans, V. Fierro and A. Celzard, Mater. Sci. Semicond. Process., 2021, 128, 105744 CrossRef CAS.
- R. Tang, M. Y. Xie, M. Li, L. Cao, S. Feng, Z. Li and F. Xu, Appl. Mater. Today, 2022, 26, 101305 CrossRef.
- A. Singh, D. Lantigua, A. Meka, S. Taing, M. Pandher and G. Camci-Unal, Sensors, 2018, 18, 2838 CrossRef PubMed.
- A. Kumar, K. Chen, T. Thundat and M. T. Swihart, ACS Appl. Mater. Interfaces, 2023, 15, 5439–5448 CrossRef CAS PubMed.
- A. Quddious, S. Yang, M. M. Khan, F. A. Tahir, A. Shamim, K. N. Salama and H. M. Cheema, Sensors, 2016, 16, 2073 CrossRef PubMed.
- E. Noviana, D. B. Carrão, R. Pratiwi and C. S. Henry, Anal. Chim. Acta, 2020, 1116, 70–90 CrossRef CAS PubMed.
- C.-M. Wang, C.-Y. Chen and W.-S. Liao, Anal. Chim. Acta, 2021, 1144, 158–174 CrossRef CAS PubMed.
- M. A. Hubbe and R. A. Gill, BioResources, 2016, 11, 2886–2963 CAS.
- K. J. Hipolit, Chemical Processing Aids in Papermaking: A Practical Guide, Tappi press, 1992 Search PubMed.
- R. Bown, Pap. Technol., 1998, 39, 44–48 CAS.
- R. Davidson, Pap. Technol., 1965, 6, 107–114 CAS.
- N. L. Salmen, Tappi, 1980, 63, 117–120 CAS.
- F. Brunetti, A. Operamolla, S. Castro-Hermosa, G. Lucarelli, V. Manca, G. M. Farinola and T. M. Brown, Adv. Funct. Mater., 2019, 29, 1806798 CrossRef.
- L. Chen, J. Lv, L. Ding, G. Yang, Z. Mao, B. Wang, X. Feng, S. Zapotoczny and X. Sui, Chem. Eng. J., 2020, 400, 125950 CrossRef CAS.
- J. H. Kim, D. Lee, Y. H. Lee, W. Chen and S. Y. Lee, Adv. Mater., 2019, 31, 1804826 CrossRef PubMed.
- X. Zhao, W. Han, Y. Jiang, C. Zhao, X. Ji, F. Kong, W. Xu and X. Zhang, Nanoscale, 2019, 11, 17725–17735 RSC.
- Z. Wang, Y. H. Lee, S. W. Kim, J. Y. Seo, S. Y. Lee and L. Nyholm, Adv. Mater., 2021, 33, 2000892 CrossRef CAS PubMed.
- S. M. Khan, J. M. Nassar and M. M. Hussain, ACS Appl. Electron. Mater., 2020, 3, 30–52 CrossRef.
- D. Zhang and Q. Liu, Biosens. Bioelectron., 2016, 75, 273–284 CrossRef CAS PubMed.
- P. K. Yang, Z. H. Lin, K. C. Pradel, L. Lin, X. Li, X. Wen, J. H. He and Z. L. Wang, ACS Nano, 2015, 9, 901–907 CrossRef CAS PubMed.
- N. C. Sekar, L. Ge, S. A. M. Shaegh, S. H. Ng and S. N. Tan, Sens. Actuators, B, 2015, 210, 336–342 CrossRef CAS.
- J. Cheng, Y. Fu, J. Guo and J. Guo, Sens. Actuators, B, 2023, 387, 133795 CrossRef CAS.
- J. Li, K. Chen, Y. Su, L. Zhu, H. Zhang, W. Xu and X. Li, Journal of Future Foods, 2024, 4, 61–70 CrossRef.
- Y. Shen and A. Mulchandani, Biosens. Bioelectron.: X, 2023, 100364 Search PubMed.
- T. Pinheiro, A. R. Cardoso, C. E. Sousa, A. C. Marques, A. P. Tavares, A. M. Matos, M. T. Cruz, F. T. Moreira, R. Martins and E. Fortunato, ACS Omega, 2021, 6, 29268–29290 CrossRef CAS PubMed.
- L. Xu, Z. Zhou, M. Fan and X. Fang, Int. J. Electrochem. Sci., 2023, 18, 13–19 CrossRef CAS.
- Z. Ouyang, S. Li, J. Liu, H.-Y. Yu, L. Peng, S. Zheng, D. Xu and K. C. Tam, Nano Energy, 2022, 104, 107963 CrossRef CAS.
- M. Yence, A. Cetinkaya, S. I. Kaya, G. Ozcelikay and S. A. Ozkan, in Recent Developments in Green Electrochemical Sensors: Design, Performance, and Applications, ACS Publications, 2023, pp. 411–439 Search PubMed.
- K. Beaver, A. Dantanarayana and S. D. Minteer, J. Phys. Chem. B, 2021, 125, 11820–11834 CrossRef CAS PubMed.
- N. Postulka, T. Meckel and M. Biesalski, Langmuir, 2021, 37, 8746–8752 CrossRef CAS PubMed.
- W. Heng, L. Weihua and K. Bachagha, Carbohydr. Polym., 2023, 121306 CrossRef CAS PubMed.
- R. Gerulskis and S. D. Minteer, ECS Adv., 2023, 2, 035501 CrossRef.
- S. Selvaraj and D. R. Shankaran, ChemistrySelect, 2023, 8, e202302027 CrossRef CAS.
- X. Li, H. Yang, S. Lu and R. Cai, Adv. Mater. Technol., 2023, 2300079 CrossRef CAS.
- A. Uçar, G. A. Tığ and E. Er, TrAC, Trends Anal. Chem., 2023, 117027 CrossRef.
- R. Moscoso, S. Abarca, C. Yáñez and J. Squella, Electrochim. Acta, 2023, 443, 141984 CrossRef CAS.
- P. B. Deroco, D. Wachholz Junior and L. T. Kubota, Electroanalysis, 2023, 35, e202200177 CrossRef CAS.
- S. R. Benjamin, F. de Lima, V. A. D. Nascimento, G. M. de Andrade and R. B. Oriá, Biosensors, 2023, 13, 689 CrossRef CAS PubMed.
- T. S. Martins, S. A. Machado, O. N. Oliveira Jr and J. L. Bott-Neto, Food Chem., 2023, 410, 135429 CrossRef CAS PubMed.
- X. Ji, X. Zhao, Z. Zhang, Y. Si, W. Qian, H. Fu, Z. Chen, Z. Wang, H. Jin and Z. Yang, Nano Res., 2023, 1–8 Search PubMed.
- H. Liu, J. Quan, J. Wang, Q. Hou, R. Wang, B. Cui, T. Lang, Y. Chen, H. Pan and J. Xu, Cellulose, 2023, 30, 1225–1244 CrossRef CAS.
- H. Manisha, J. Sonia, S. Shashikiran, S. Yuvarajan, P. Rekha and K. S. Prasad, Electrochem. Commun., 2022, 137, 107259 CrossRef CAS.
- G. Maduraiveeran, Curr. Anal. Chem., 2022, 18, 509–517 CrossRef CAS.
- Y. Zheng, Y. Li, L. Fan, H. Yao and Z. Zhang, Anal. Methods, 2022, 14, 1268–1278 RSC.
- H. Zhang, X. Li, Q. Zhu and Z. Wang, J. Electroanal. Chem., 2022, 909, 116140 CrossRef CAS.
- C. J. Valentine, K. Takagishi, S. Umezu, R. Daly and M. De Volder, ACS Appl. Mater. Interfaces, 2020, 12, 30680–30685 CrossRef CAS PubMed.
- M. Santhiago, P. G. da Costa, M. P. Pereira, C. C. Correa, V. R. B. de Morais and C. C. Bufon, ACS Appl. Mater. Interfaces, 2018, 10, 35631–35638 CrossRef CAS PubMed.
- R. S. Khan, Z. Khurshid and F. Yahya Ibrahim Asiri, Diagnostics, 2017, 7, 39 CrossRef PubMed.
- L. Cai, C. Xu, S. Lin, J. Luo, M. Wu and F. Yang, Biomicrofluidics, 2014, 8, 056504 CrossRef PubMed.
- E. R. Kim, C. Joe, R. J. Mitchell and M. B. Gu, Trends Biotechnol., 2023, 41, 374–395 CrossRef CAS PubMed.
- X. Luo, J. Xia, X. Jiang, M. Yang and S. Liu, Anal. Chem., 2019, 91, 15461–15468 CrossRef CAS PubMed.
- N. C. Sekar, S. A. M. Shaegh, S. H. Ng, L. Ge and S. N. Tan, Sens. Actuators, B, 2014, 204, 414–420 CrossRef.
- N. Ruecha, R. Rangkupan, N. Rodthongkum and O. Chailapakul, Biosens. Bioelectron., 2014, 52, 13–19 CrossRef CAS PubMed.
- Y. Thepchuay, T. Sonsa-Ard, N. Ratanawimarnwong, S. Auparakkitanon, J. Sitanurak and D. Nacapricha, Anal. Chim. Acta, 2020, 1103, 115–121 CrossRef CAS PubMed.
- X. Liu, X. Li, X. Gao, L. Ge, X. Sun and F. Li, ACS Appl. Mater. Interfaces, 2019, 11, 15381–15388 CrossRef CAS PubMed.
- S. Cinti, C. Minotti, D. Moscone, G. Palleschi and F. Arduini, Biosens. Bioelectron., 2017, 93, 46–51 CrossRef CAS PubMed.
- T. Tian, H. Liu, L. Li, J. Yu, S. Ge, X. Song and M. Yan, Sens. Actuators, B, 2017, 251, 440–445 CrossRef CAS.
- Y. Shen, T.-T. Tran, S. Modha, H. Tsutsui and A. Mulchandani, Biosens. Bioelectron., 2019, 130, 367–373 CrossRef CAS PubMed.
- S. Veeralingam and S. Badhulika, J. Ind. Eng. Chem., 2022, 113, 401–410 CrossRef CAS.
- Y.-J. Chang, M.-C. Lee and Y.-C. Chien, SLAS Technol., 2022, 27, 54–62 CrossRef CAS PubMed.
- X. Kou, L. Tong, Y. Shen, W. Zhu, L. Yin, S. Huang, F. Zhu, G. Chen and G. Ouyang, Biosens. Bioelectron., 2020, 156, 112095 CrossRef CAS PubMed.
- P. Teengam, W. Siangproh, A. Tuantranont, C. S. Henry, T. Vilaivan and O. Chailapakul, Anal. Chim. Acta, 2017, 952, 32–40 CrossRef CAS PubMed.
- A. Yakoh, E. Mehmeti, K. Kalcher and S. Chaiyo, Anal. Chem., 2022, 94, 5893–5900 CrossRef CAS PubMed.
- X. Li, Q. Duan, M. Khan, D. Yang, Q. Liu, F. Yin, Q. Hu and L. Yu, Talanta, 2024, 266, 124994 CrossRef CAS PubMed.
- J. Xu, M. Wang, M. Li, J. Yang and L. Yang, Anal. Chim. Acta, 2023, 1279, 341834 CrossRef CAS PubMed.
- G. Bhattacharya, S. J. Fishlock, S. Hussain, S. Choudhury, A. Xiang, B. Kandola, A. Pritam, N. Soin, S. S. Roy and J. A. McLaughlin, ACS Appl. Mater. Interfaces, 2022, 14, 31109–31120 CrossRef CAS PubMed.
- M. d M. G. del Campo, A. Alba-Patino, C. Palomino, M. Bauzá, E. Rojo-Molinero, A. Oliver, G. Turnes and R. de la Rica, Sens. Actuators, B, 2022, 354, 131214 CrossRef.
- R. Suvanasuthi, S. Chimnaronk and C. Promptmas, Talanta, 2022, 237, 122962 CrossRef CAS PubMed.
- S. Modha, Y. Shen, H. Chamouni, A. Mulchandani and H. Tsutsui, Biosens. Bioelectron., 2021, 180, 113090 CrossRef CAS PubMed.
- B. Demirbakan and M. K. Sezgintürk, Anal. Chim. Acta, 2021, 1144, 43–52 CrossRef CAS PubMed.
- M. Rahimi-Mohseni, J. B. Raoof, T. A. Aghajanzadeh and R. Ojani, Microchem. J., 2021, 160, 105739 CrossRef CAS.
- P. Jafari, S. M. Beigi, F. Yousefi, S. Aghabalazadeh, M. Mousavizadegan, M. Hosseini, S. Hosseinkhani and M. R. Ganjali, Microchem. J., 2021, 163, 105909 CrossRef CAS.
- J. P. Hunt, E. L. Zhao, T. J. Free, M. Soltani, C. A. Warr, A. B. Benedict, M. K. Takahashi, J. S. Griffitts, W. G. Pitt and B. C. Bundy, New Biotechnol., 2022, 66, 53–60 CrossRef CAS PubMed.
- A. Yakoh, U. Pimpitak, S. Rengpipat, N. Hirankarn, O. Chailapakul and S. Chaiyo, Biosens. Bioelectron., 2021, 176, 112912 CrossRef CAS PubMed.
- S. Khaliliazar, A. Toldrà, G. Chondrogiannis and M. M. Hamedi, Anal. Chem., 2021, 93, 14187–14195 CrossRef CAS PubMed.
- M. Moccia, V. Caratelli, S. Cinti, B. Pede, C. Avitabile, M. Saviano, A. L. Imbriani, D. Moscone and F. Arduini, Biosens. Bioelectron., 2020, 165, 112371 CrossRef CAS PubMed.
- L. Huang, W. Xiao, T. Xu, H. Chen, Z. Jin, Z. Zhang, Q. Song and Y. Tang, Sens. Actuators, B, 2021, 327, 128893 CrossRef CAS.
- J. Heikenfeld, A. Jajack, B. Feldman, S. W. Granger, S. Gaitonde, G. Begtrup and B. A. Katchman, Nat. Biotechnol., 2019, 37, 407–419 CrossRef CAS PubMed.
- Y. J. Hong, H. Lee, J. Kim, M. Lee, H. J. Choi, T. Hyeon and D. H. Kim, Adv. Funct. Mater., 2018, 28, 1805754 CrossRef.
- P. J. Derbyshire, H. Barr, F. Davis and S. P. Higson, J. Physiol. Sci., 2012, 62, 429–440 CrossRef CAS PubMed.
- J. Moyer, D. Wilson, I. Finkelshtein, B. Wong and R. Potts, Diabetes Technol. Ther., 2012, 14, 398–402 CrossRef CAS PubMed.
- A. Alizadeh, A. Burns, R. Lenigk, R. Gettings, J. Ashe, A. Porter, M. McCaul, R. Barrett, D. Diamond and P. White, Lab Chip, 2018, 18, 2632–2641 RSC.
- A. Singh, A. Hazarika, C. Talukdar, M. Bhuyan, L. Dutta, M. Barthakur and K. Chakraborty, IEEE Trans. Ind. inf., 2021, 17, 7546–7553 Search PubMed.
- M. Li, L. Wang, R. Liu, J. Li, Q. Zhang, G. Shi, Y. Li, C. Hou and H. Wang, Biosens. Bioelectron., 2021, 174, 112828 CrossRef CAS PubMed.
- A. Singh, A. Hazarika, L. Dutta, A. Bhuyan and M. Bhuyan, Anal. Chim. Acta, 2022, 1227, 340257 CrossRef CAS PubMed.
- L. Fiore, V. Mazzaracchio, A. Serani, G. Fabiani, L. Fabiani, G. Volpe, D. Moscone, G. M. Bianco, C. Occhiuzzi and G. Marrocco, Sens. Actuators, B, 2023, 379, 133258 CrossRef CAS.
- S. Mandal and G. C. Banik, Chem. Sci. Rev. Lett., 2021, 10, 116–112 Search PubMed.
- E.-C. Oerke, J. Agric. Sci., 2006, 144, 31–43 CrossRef.
- P. Nicolopoulou-Stamati, S. Maipas, C. Kotampasi, P. Stamatis and L. Hens, Public Health Front., 2016, 4, 148 Search PubMed.
- M. A. Beketov, B. J. Kefford, R. B. Schäfer and M. Liess, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 11039–11043 CrossRef CAS PubMed.
- N. E. Pollok, C. Rabin, C. T. Walgama, L. Smith, I. Richards and R. M. Crooks, ACS Sens., 2020, 5, 853–860 CrossRef CAS PubMed.
- E. Noviana and C. S. Henry, Curr. Opin. Electrochem., 2020, 23, 1–6 CrossRef CAS.
- F. Arduini, S. Cinti, V. Caratelli, L. Amendola, G. Palleschi and D. Moscone, Biosens. Bioelectron., 2019, 126, 346–354 CrossRef CAS PubMed.
- A. Cioffi, M. Mancini, V. Gioia and S. Cinti, Environ. Sci. Technol., 2021, 55, 8859–8865 CrossRef CAS PubMed.
- X. Ruan, Y. Wang, E. Y. Kwon, L. Wang, N. Cheng, X. Niu, S. Ding, B. J. Van Wie, Y. Lin and D. Du, Biosens. Bioelectron., 2021, 184, 113238 CrossRef CAS PubMed.
- N. Colozza, K. Kehe, G. Dionisi, T. Popp, A. Tsoutsoulopoulos, D. Steinritz, D. Moscone and F. Arduini, Biosens. Bioelectron., 2019, 129, 15–23 CrossRef CAS PubMed.
- V. Caratelli, G. Fegatelli, D. Moscone and F. Arduini, Biosens. Bioelectron., 2022, 205, 114119 CrossRef CAS PubMed.
- C. Botías, A. David, J. Horwood, A. Abdul-Sada, E. Nicholls, E. Hill and D. Goulson, Environ. Sci. Technol., 2015, 49, 12731–12740 CrossRef PubMed.
- F. Si, R. Zou, S. Jiao, X. Qiao, Y. Guo and G. Zhu, Ecotoxicol. Environ. Saf., 2018, 148, 862–868 CrossRef CAS.
- H. Li, R. Jin, D. Kong, X. Zhao, F. Liu, X. Yan, Y. Lin and G. Lu, Sens. Actuators, B, 2019, 283, 207–214 CrossRef CAS.
- K.-L. Lee, M.-L. You, C.-H. Tsai, E.-H. Lin, S.-Y. Hsieh, M.-H. Ho, J.-C. Hsu and P.-K. Wei, Biosens. Bioelectron., 2016, 75, 88–95 CrossRef CAS PubMed.
- D. Englert, J. P. Zubrod, M. Link, S. Mertins, R. Schulz and M. Bundschuh, Environ. Sci. Technol., 2017, 51, 5793–5802 CrossRef CAS PubMed.
- C.-H. Chang, D. MacIntosh, B. Lemos, Q. Zhang and C. Lu, J. Agric. Food Chem., 2018, 66, 10097–10105 CrossRef CAS PubMed.
- T. Liu, D. Chen, Y. Li, X. Wang and F. Wang, J. Agric. Food Chem., 2018, 66, 4531–4540 CrossRef CAS PubMed.
- P. Zhao, H. Liu, L. Zhang, P. Zhu, S. Ge and J. Yu, ACS Appl. Mater. Interfaces, 2020, 12, 8845–8854 CrossRef CAS PubMed.
- V. Scognamiglio, A. Antonacci, F. Arduini, D. Moscone, E. V. Campos, L. F. Fraceto and G. Palleschi, J. Hazard. Mater., 2019, 373, 483–492 CrossRef CAS PubMed.
- Z. Ma, Y. Li, C. Lu and M. Li, J. Hazard. Mater., 2023, 131725 CrossRef CAS PubMed.
- F. Zhao, L. Wang, M. Li, M. Wang, G. Liu and J. Ping, TrAC, Trends Anal. Chem., 2023, 117152 CrossRef CAS.
- G. Song, J. Zhang, H. Huang, X. Wang, X. He, Y. Luo, J.-C. Li, K. Huang and N. Cheng, Food Chem., 2022, 387, 132896 CrossRef CAS PubMed.
- T. T. M. Duyen, H. Matsuura, K. Ujiie, M. Muraoka, K. Harada and K. Hirata, J. Biosci. Bioeng., 2017, 123, 96–100 CrossRef CAS PubMed.
- X. Wang, Y. Luo, K. Huang and N. Cheng, Adv. Agrochem, 2022, 1, 3–6 CrossRef.
- G. Bayramoglu, M. Kilic and M. Y. Arica, Food Chem., 2022, 382, 132353 CrossRef CAS PubMed.
- Y. Li, Q. Wu, Z. Wu, Y. Zhuang, L. Sun, X. Fan, T. Zhao, L. Yi and Y. Gu, Food Chem., 2023, 405, 134974 CrossRef CAS PubMed.
- C. Griesche and A. J. Baeumner, TrAC, Trends Anal. Chem., 2020, 128, 115906 CrossRef CAS.
- V. Pagkali, D. Soulis, C. Kokkinos and A. Economou, Sens. Actuators, B, 2022, 358, 131546 CrossRef CAS.
- H. K. Kordasht, A. Saadati and M. Hasanzadeh, Food Chem., 2022, 373, 131411 CrossRef CAS PubMed.
- C. Nerín, M. Aznar and D. Carrizo, Trends Food Sci. Technol., 2016, 48, 63–68 CrossRef.
- B. Kuswandi, Y. Wicaksono, Jayus, A. Abdullah, L. Y. Heng and M. Ahmad, Sens. Instrum. Food Qual. Saf., 2011, 5, 137–146 CrossRef.
- H.-z Chen, M. Zhang, B. Bhandari and C.-h Yang, LWT, 2019, 99, 43–49 CrossRef CAS.
- S. Y. Lee, S. J. Lee, D. S. Choi and S. J. Hur, J. Sci. Food Agric., 2015, 95, 2799–2810 CrossRef CAS PubMed.
- A. Sobhan, K. Muthukumarappan and L. Wei, Food Packag. Shelf Life, 2021, 30, 100745 CrossRef CAS.
- M. M. Ali, M. Wolfe, K. Tram, J. Gu, C. D. Filipe, Y. Li and J. D. Brennan, Angew. Chem., 2019, 131, 10012–10016 CrossRef.
- C. Mills, M. J. Dillon, P. K. Kulabhusan, D. Senovilla-Herrero and K. Campbell, Environ. Sci. Technol., 2022, 56, 12210–12217 CrossRef CAS PubMed.
- S. K. Choi, A. Myc, J. E. Silpe, M. Sumit, P. T. Wong, K. McCarthy, A. M. Desai, T. P. Thomas, A. Kotlyar and M. M. B. Holl, ACS Nano, 2013, 7, 214–228 CrossRef CAS PubMed.
- H. J. Chung, T. Reiner, G. Budin, C. Min, M. Liong, D. Issadore, H. Lee and R. Weissleder, ACS Nano, 2011, 5, 8834–8841 CrossRef CAS PubMed.
- N. Verdoodt, C. R. Basso, B. F. Rossi and V. A. Pedrosa, Food Chem., 2017, 221, 1792–1796 CrossRef CAS PubMed.
- Y. Mao, X. Huang, S. Xiong, H. Xu, Z. P. Aguilar and Y. Xiong, Food Control, 2016, 59, 601–608 CrossRef CAS.
- B. H. Park, S. J. Oh, J. H. Jung, G. Choi, J. H. Seo, E. Y. Lee and T. S. Seo, Biosens. Bioelectron., 2017, 91, 334–340 CrossRef CAS PubMed.
- T. Wang, P. Li, Q. Zhang, W. Zhang, Z. Zhang, T. Wang and T. He, Sci. Rep., 2017, 7, 4348 CrossRef PubMed.
- L. Fu, S. Deng, Y. Luo, Q. Fu, Y. Fan and L. Jia, Talanta, 2023, 265, 124920 CrossRef CAS PubMed.
- E. T. Bougadi and D. P. Kalogianni, Food Chem., 2020, 322, 126758 CrossRef CAS PubMed.
- M. Srisa-Art, K. E. Boehle, B. J. Geiss and C. S. Henry, Anal. Chem., 2018, 90, 1035–1043 CrossRef CAS PubMed.
- H. A. Al Lawati, J. Hassanzadeh, N. Bagheri and I. A. Lawati, Talanta, 2021, 234, 122648 CrossRef CAS PubMed.
- G. Li, J. Sun, J. Li, Y. Zhang, J. Huang, F. Yue, H. Dong, F. Li, H. Xu and Y. Guo, Talanta, 2023, 124843 CrossRef CAS PubMed.
- P. K. Prabhakar, S. Vatsa, P. P. Srivastav and S. S. Pathak, Food Res. Int., 2020, 133, 109157 CrossRef CAS PubMed.
- X. Wang, Y. Wang, C. Guo, X. Zhang, Y. Wang, L. Lv, X. Wang and M. Wei, Food Chem., 2023, 405, 134811 CrossRef CAS PubMed.
- S. Li, S. Wang, B. Wu, M. Jiang, H.-Y. Yu, D. Ge, Y. Dong, W. Xu and K. C. Tam, Nano Energy, 2024, 120, 109148 CrossRef CAS.
- O. Pourret and A. Hursthouse, Int. J. Environ. Res. Public Health, 2019, 16, 4446 CrossRef CAS PubMed.
- S. Zhu, M. Xia, Y. Chu, M. A. Khan, W. Lei, F. Wang, T. Muhmood and A. Wang, Appl. Clay Sci., 2019, 169, 40–47 CrossRef CAS.
- Y. Zhang, C. Zhao, H. Bi, X. Zhang, B. Xue, C. Li, S. Wang, X. Yang, Z. Qiu and J. Wang, J. Hazard. Mater., 2022, 438, 129499 CrossRef CAS PubMed.
- S. Karuppiah, N. C. Mishra, W.-C. Tsai, W.-S. Liao and C.-F. Chou, ACS Sens., 2021, 6, 3214–3223 CrossRef CAS PubMed.
- Y. Zheng, L. Wei, L. Duan, F. Yang, G. Huang, T. Xiao, M. Wei, Y. Liang, H. Yang and Z. Li, J. Environ. Sci., 2021, 106, 161–170 CrossRef CAS PubMed.
- C. S. K. Lawrence, S. N. Tan and C. Z. Floresca, Sens. Actuators, B, 2014, 193, 536–541 CrossRef.
- L. Guadarrama-Fernández, M. Novell, P. Blondeau and F. J. Andrade, Food Chem., 2018, 265, 64–69 CrossRef PubMed.
- A. D. Clayton, A. M. Schweidtmann, G. Clemens, J. A. Manson, C. J. Taylor, C. G. Niño, T. W. Chamberlain, N. Kapur, A. J. Blacker and A. A. Lapkin, Chem. Eng. J., 2020, 384, 123340 CrossRef CAS.
- C. W. Coley, R. Barzilay, T. S. Jaakkola, W. H. Green and K. F. Jensen, ACS Cent. Sci., 2017, 3, 434–443 CrossRef CAS PubMed.
- G. H. Gu, J. Noh, I. Kim and Y. Jung, J. Mater. Chem. A, 2019, 7, 17096–17117 RSC.
- W. Sha, Y. Guo, Q. Yuan, S. Tang, X. Zhang, S. Lu, X. Guo, Y.-C. Cao and S. Cheng, Adv. Intell. Syst., 2020, 2, 1900143 CrossRef.
- Z. Chen, Z. Chen, Z. Song, W. Ye and Z. Fan, J. Semicond., 2019, 40, 111601 CrossRef CAS.
- C. Mendes-Felipe, J. Oliveira, I. Etxebarria, J. L. Vilas-Vilela and S. Lanceros-Mendez, Adv. Mater. Technol., 2019, 4, 1800618 CrossRef.
| This journal is © The Royal Society of Chemistry 2024 |


