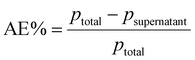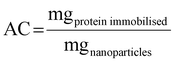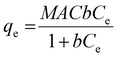Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Adsorption of immunomodulatory proteins over silica nanoparticles and the in vitro effect
Exequiel David
Giorgi
ab,
Sofía
Genovés
ab,
María Eugenia
Díaz
ab,
Sofía
Municoy
c,
Martin Federico
Desimone†
 *c and
Mauricio César
De Marzi†
*ab
*c and
Mauricio César
De Marzi†
*ab
aCONICET-Universidad Nacional de Luján, Instituto de Ecología y Desarrollo Sustentable (INEDES), Grupo de Investigaciones Básicas y Aplicadas en Inmunología y Bioactivos (GIBAIB), Av. Constitución y Ruta 5, Luján, Buenos Aires, Argentina. E-mail: desimone@ffyb.uba.ar
bUniversidad Nacional de Lujan, Departamento de Ciencias Básicas, Laboratorio de Inmunología, Av. Constitución y Ruta 5, Luján, Buenos Aires, Argentina
cUniversidad de Buenos Aires, Facultad de Farmacia y Bioquímica, CONICET, Instituto de Química y Metabolismo del Fármaco (IQUIMEFA), Buenos Aires, Argentina
First published on 5th December 2023
Abstract
Silica NPs (SiNPs) used as a platform to deliver molecules have huge potential for biomedical applications. In order to generate new immunomodulatory tools, 2 variants of SiNPs were synthesized and 3 proteins were adsorbed over their surface: bovine serum albumin and the cytokines IL-1β and TGF-β. Protein adsorption was analyzed according to Langmuir and Freundlich models. The adsorption isotherm of IL-1β on both SiNP variants had a good fit to the Freundlich model, indicating the formation of a protein multilayer around the NPs. For BSA and TGF-β isotherms, the fit to the Langmuir model was better, evidencing the presence of a protein monolayer on the NPs. SiNPs@TGF-β complexes were tested in THP-1 cells (human monocytes cells). The complexes reduced cellular metabolic activity and did not cause an increase in nitric oxide expression, which is related to the immunosuppressive activity of TGF-β, but was potentiated and prolonged over time compared to the cytokine alone. These nanocomplexes could be important tools for use as nanoinmunomodulators (NIMs) for the therapeutic treatment of inflammatory diseases.
Introduction
Metal oxide nanoparticles (NPs) have been used in several industries and consumer products. There is evidence showing that NPs could generate reactive oxygen species (ROS), which endow them with antimicrobial activity but, on the other hand, can induce oxidative stress which could be harmful to the cell membrane and cause DNA damage.1–4 Moreover, when proteins interact with the NP surface, the adsorption forces can disrupt non-covalent interactions in the protein structure, so a denaturation process could occur.5 In biological systems, a layer of proteins formed on the NP surface is called the protein corona (which includes proteins, lipids, and ions that are immediately adsorbed onto the NP surface),6,7 and is an entity that can change over time.5,8Silica NPs (SiNPs) are among the most widely used NPs for the development of potential delivery tools4,9,10 and they can be found in several food and cosmetic products.11 Indeed, SiNPs have been used in clinical trials and clinical studies. They are mostly employed for oral delivery or bioimaging studies.4,12
Although the biological effects of SiNPs in vivo are not fully understood, it is clear that high specific surface areas are associated with increased cytotoxicity in most cases.11,13 Orally ingested SiNPs can be tolerated at high doses, but when inhaled they could lead to chronic inflammation.14 There is evidence of degradation of silica particles into silicic acid in biological media, which can be excreted through the urine.15 However, it is not possible to establish a general degradation kinetics for different SiNPs because it is a process that not only depends on the particle characteristics, such as surface area, functional groups, pore size, and others, but also on the degradation medium characteristics (i.e., temperature, pH, and concentration).15,16 Moreover, SiNPs’ biocompatibility depends on their properties such as size and surface properties.17–19
It has been reported that the presence of a protein corona could mitigate the toxicity of these NPs and also change the way they interact with cells, altering their internalization efficiency compared to SiNPs without the protein corona.6,20,21
Bovine serum albumin (BSA) is a common protein model used in NP adsorption studies.22,23 Some studies have been carried out on the adsorption of BSA on various surfaces of inorganic and polymeric NPs in biological media and the pH effects on this process.24–27 Since oxide NPs are postulated as transporters for therapeutic drugs, immunomodulatory molecules, or antigens, it is necessary to understand the characteristics of protein corona formation under different conditions and for different proteins of biological interest. Herein lies the importance of the biological effect of NPs, because the interaction between cells and NPs depends on the proteins immobilized on their surface, which will determine the biological action and fate of the NPs.28 In this sense, since BSA has similar properties, molecular weight, and amino acid sequence to its human variant, human serum albumin (HSA), it is an excellent study model since HSA is the most abundant protein in the blood and is present in different body fluids.5,22
In addition to the importance of using BSA as a protein model, several proteins of high interest with immunomodulatory activity have been identified that could be adsorbed on NPs to obtain NIMs (nanoimmunomodulators) with potential for use in the treatment of various pathologies. Interleukin 1 beta (IL-1β) is a pro-inflammatory cytokine produced by several cells, but in particular, by macrophages in tissues or lymphoid organs in response to inflammatory signals.29 Moreover, IL-1β secreted by activated Antigen Presenting Cells (APCs) induces T Helper 1 immune response demonstrating its beneficial effect against infections.30 On the other hand, Transforming Growth Factor beta (TGF-β) is a regulatory molecule with pleiotropic effects over cell proliferation, differentiation, migration, and survival and it is implicated in several biological processes such as tissue development, carcinogenesis, fibrosis, cicatrization, and immune response. It has 3 different isoforms (TGF-β1, TGF-β2, and TGF-β3) but TGF-β1 is the most expressed by immune cells. However, all isoforms present similar in vitro properties.31 TGF-β has different effects on each type of immune cells, sometimes stimulating their proliferation but in other cases inhibiting them. TGF-β inhibits the activation and differentiation of macrophages. The inhibitory capacity of TGF-β on macrophages can resolve the inflammatory process and prevent the development of immunopathologies (as seen in autoimmune diseases). It is also a key protein in the cicatrization process because of its anti-inflammatory and chemoattractant capacity of monocytes in the context of tissue damage. Not less important, it can promote cellular proliferation of chondrocytes, osteoblasts, fibroblasts, and endothelial cells, among others, and it can also favor extracellular matrix deposition.31
Due to all the above mentioned reasons, the immobilization of different proteins on NPs could result in the generation of nanocomplexes capable of reproducing or improving their immunomodulatory activity on human immune cells such as monocytes. These nanocomplexes, named NIMs, could offer new options for the treatment of different pathologies. Particularly, NPs have shown high potential as immunomodulators, stimulating the immune system in the context of immunosuppression or regulating the exacerbated immune response in processes such as infections, allergies, or hypersensitivity reactions.32 There are also great advances in nanomaterial-based drug delivery of immunomodulatory factors for bone and cartilage tissue engineering.33,34 The inclusion of anti-inflammatory drugs and cytokines in nanomaterials is one of the approaches used. These would favor M2-type macrophage development, promoting tissue regeneration.35
In this work, to develop NIMs for future use in targeted therapies, different proteins of immunological interest were adsorbed on conventional and modified SiNPs, obtaining stable complexes and testing some of them in vitro as possible immunomodulators in human monocytes.
Experimental
Synthesis of silica nanoparticles
Bare SiNPs were obtained by the Stöber method.36 In this method, when the concentration of the silica precursor is higher than solubility, the nucleation phenomenon takes place and generates homogeneous particles. Briefly, as was described by Baudou et al.,17,18,37 tetraethyl orthosilicate 98% w/w (TEOS, Sigma-Aldrich) was added dropwise to a solution of ultrapure water, absolute ethanol (Biopack) and ammonium hydroxide (25% w/v) under stirring. The solution was stirred overnight at room temperature. Particles were washed twice with absolute ethanol, once with deionized water, and then centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g). The obtained pellet (SiOHNPs) was dried in a vacuum, resuspended, and stored in 10 mM KCl at 4 °C. Half of these NPs were mixed with 3-aminopropyltriethoxysilane (APTES) at 500 rpm for 24 h. This reagent chemically modified the SiOHNP surface by adding amine groups to obtain the other variant (SiNH2NPs). Finally, they were analyzed by dynamic light scattering, zeta potential measurements, and transmission electron microscopy.
000g). The obtained pellet (SiOHNPs) was dried in a vacuum, resuspended, and stored in 10 mM KCl at 4 °C. Half of these NPs were mixed with 3-aminopropyltriethoxysilane (APTES) at 500 rpm for 24 h. This reagent chemically modified the SiOHNP surface by adding amine groups to obtain the other variant (SiNH2NPs). Finally, they were analyzed by dynamic light scattering, zeta potential measurements, and transmission electron microscopy.
Hydrodynamic diameter and surface charge
Dynamic light scattering (DLS, Brookhaven Instruments) was used to determine the hydrodynamic diameter of SiNPs. Using the same instrument, the zeta potential was also measured on the same samples to determine the surface charge at pH 7. SiNPs were suspended in a 10 mM KCl aqueous solution; the suspensions obtained were sonicated in an ultrasonic bath for 10 min and then the measurements were performed. Samples were analyzed in triplicate/quadruplicate for media and standard deviation determination.Transmission electron microscopy
TEM was performed on a Zeiss EM109T at 200 keV. For the analysis, SiNP suspensions were diluted 1:20 in milliQ water, and 10 μL were deposited in carried grids (S147-4, Plano GmbH, Germany) and allowed to dry under ambient conditions before imaging. The particle size was determined using ImageJ software.38Scanning electron microscopy
SiNPs were evaluated by SEM using a Zeiss Crossbeam 340 at 3.00 kV. For the analysis, SiNP suspensions were diluted 1:20 in milliQ water, and 10 μL were deposited in carried grids and allowed to dry under ambient conditions before imaging.Surface area (BET) measurements
The surface area of SiNPs (BET measurements) was determined by N2 adsorption at 77 K (AUTOSORB-1, Quantachrome); before analysis, the samples (ca. 0.45 g) were outgassed at 120 °C for 12 h.39Fourier transform infrared spectroscopy
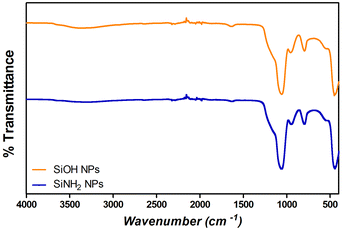
Fourier transform infrared spectroscopy (FT-IR) analysis was performed on different samples using a Nicolet iS 50 FT-IR spectrophotometer, with a KBr beam splitter. Spectra were obtained with a resolution of 2 cm−1 using a DTGS detector. The samples were measured using the attenuated total reflectance (ATR) technique. For this, 150 μL of the samples were dried under a nitrogen flow and the powder was then placed on the attenuated total reflection accessory of the spectrometer, and the spectra were recorded. FT-IR spectra were conducted to confirm the presence of Si–O–Si bonds (with absorption bands at 1100 cm−1) and silanol groups (Si–OH) (absorption bands at 960 cm−1) in SiNPs, and S–S bonds and amide I, II, and III (characteristic absorption bands for proteins) to corroborate the adsorption of proteins over SiNPs.23,37,40Protein adsorption studies
Different proteins of immunological interest such as BSA, TGF-β and IL-1β were used for immobilization on NPs. A protein stock solution (2 mg mL−1 in the case of BSA and 0.50 mg mL−1 for the other proteins tested) was prepared in phosphate buffer solution (PBS). Serial dilutions of this protein stock solution were made (until 0.05 mg mL−1) for subsequent use. SiOHNPs and SiNH2NPs (8 mg mL−1) were dispersed in the same buffer. Equal volumes of protein solution and SiOHNPs or SiNH2NPs suspension were mixed and incubated at 25 °C in 25 mM PBS pH 7.4 overnight under stirring to favor protein adsorption on the NP surfaces. Finally, the mix was centrifuged and supernatants (SNs) were removed and stored. The pellet (NPs@protein) was washed and centrifuged 3 times with 25 mM PBS pH 7 and stored in 300 μL of the same work buffer for characterization. SNs with non-adsorbed protein were isolated and stored at −20 °C for later protein quantification by Bradford determination. The NPs@protein was resuspended in 1 mL of PBS 1X. Data on the adsorption of different amounts of proteins onto a fixed amount of NPs were collected.The protein content adsorbed on NPs was calculated as the difference in concentration of the protein in the SN before and after adsorption. Protein concentration was determined using a standard calibration curve of BSA by the Bradford micro-method.41,42 These results were corroborated by estimating the protein content using SDS-PAGE and comparing it with BSA samples of known concentrations. The percentage of adsorption efficiency was calculated as shown below:
Also, adsorption capacity was calculated as follows:
From AC and Psupernatant data, adsorption isotherms were composed and their adjustment to Langmuir and Freundlich models was calculated.43,44
Langmuir adsorption model assumes that there are a finite number of interaction points in the surface of NPs where proteins can bind. For that, there is a maximum amount of protein that could be adsorbed, which is represented by the maximum adsorption capacity (MAC) forming a monolayer on the NP surface.43,45 The model equation can be represented as follows:
From this model the Km value can be obtained, which represents the equilibrium concentration necessary to reach 50% of MAC.43
The Freundlich adsorption model assumes a heterogeneous surface with high and low affinity regions but with a progressive reduction of adsorption affinity due to lateral repulsion between the adsorbed molecules.45 The model is represented by the following equation:
| qe = KfC1/ne |
Protein release studies
In vitro release of adsorbed proteins was determined by suspending the protein-loaded NPs in 1 mL of PBS solution at a final NP concentration of 4 mg mL−1. All suspensions were placed in an Eppendorf mixer (100 rpm, 4 °C). The amount of released protein was determined by removing the SN after centrifugation (6800g, 10 min) and replacing it with buffer (1 mL) at specified time points (0.5; 1; 2; 4; 8; 24; 48; 96; 192; 384 and 720 h). Desorbed proteins were measured in the SNs by the Bradford method.Effect of nanoparticles on cell proliferation
Human monocyte cell line THP-1 (TIB-202™) from American Type Culture Collection (ATCC) were cultivated in complete RPMI 1640 medium (10% fetal bovine serum) at 37 °C and 5% CO2 atmosphere. For assays, cells were placed in 24-well plates at a final concentration of 1 × 105 cells per mL with 1 mL as the final volume. Cell proliferation in the presence of NPs alone or NPs carrying different proteins was measured for 24–72 h. For this purpose, a concentration of 300 μg mL−1 of NPs or nanocomplexes was employed.Metabolic activity as an indicator of cellular proliferation was determined with MTT assay as described previously.17,37 Results were expressed as mean ± standard deviation for triplicate experiments.
Nitric oxide expression
Secreted nitric oxide by monocytes is quickly oxidized to nitrites and nitrates by oxygen. Due to this, it is possible to use nitrite concentration as an indicator of NO production. Nitrite determination was performed by the Griess reaction as described by De Marzi et al.17 Cells cultured without particles were used as controls.Results and discussion
Characterization of silica nanoparticles
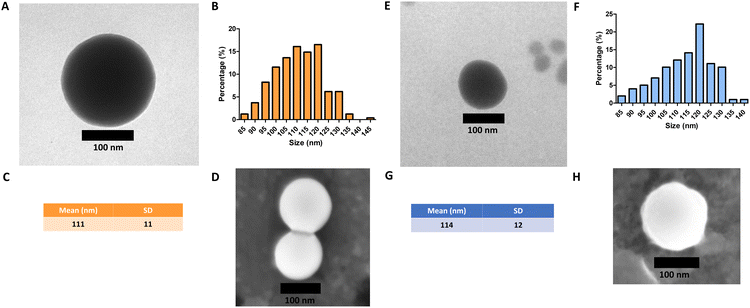
Monodisperse spherical bare SiNPs were obtained by the Stöber method. Samples were analyzed by TEM. Spherical NPs were observed, shown in Fig. 1(A) and (D). Particle diameters were measured from microimages obtained by employing ImageJ Software. The mean diameters for SiNPs were 111 ± 11 nm (SiOHNPs) and 114 ± 12 nm (SiNH2NPs) (Fig. 1(B), (C), (F), and (G)). TEM images reveal that the particle mean diameter was not significantly affected by surface modification. Surface area (BET) measurements were 15 m2 g−1 for SiOHNPs and 11 m2 g−1 for SiNH2NPs. These results are in accordance with what was described by Ali et al. (2022) who reported surface areas in the same range for commercial Stöber nanoparticles.46 On the other hand, it is important to highlight the importance of the surface area since, as Zych et al. described, nanoparticles of similar size can present quite different specific surface areas. In these cases, the high specific surface area is related to the presence of a relatively high number of micropores (pores with sizes less than 2 nm).47Further analysis by SEM confirms the spherical shape with smooth surface for both types of SiNPs (Fig. 1(D) and (H)). FTIR spectra of SiOHNPs and SiNH2NPs show the characteristic absorption bands for Si–O–Si bonds at 1100 cm−1 and for silanol groups at 960 cm−1 (Fig. 2).37,40 For SiOHNPs and SiNH2NPs, zeta potentials (ζ) were −23.6 mV and 13.3 mV, respectively, and their hydrodynamic diameters were also measured by DLS (Table 1). In this way, highly homogeneous negative and positive charged SiNPs were obtained at physiological pH (7.4).
| Hydrodynamic diameter (nm) | Z potential (mV) | |
|---|---|---|
| SiOHNPs | 151 ± 9 | −23.7 |
| SiNH2NPs | 154 ± 8 | +13.3 |
Protein adsorption and release studies
After obtaining NPs with a homogeneous size but different Z potential due to the chemical modification performed (as corroborated by physicochemical characterization), the adsorption of different proteins of interest on SiNPs was evaluated.To evaluate the ability of SiNPs to adsorb and release proteins, different proteins were mixed with SiOHNPs and SiNH2NPs in different proportions, maintaining a fixed amount of NPs. This allowed us to obtain the values of adsorption capacity and equilibrium concentration required to draft adsorption isotherms and to analyze how well they fit the Langmuir and Freundlich models. For the interpretation of this analysis, some properties of the proteins immobilized such as their pI and molecular weight were considered (Table 2).
| Protein | Molecular weight | Isoelectric point | Net charge at pH 7.4 |
|---|---|---|---|
| BSA | 66.46 kDa | 4.7 | (−) |
| IL-1β | 17.5 kDa | 6.1 | (−) |
| TGF-β1 | 25.62 kDa | 8.8 | (+) |
It was observed that the adsorption process was quick and it reached the equilibrium with a constant amount of adsorbed protein after 2 hours of incubation at room temperature and constant stirring. To better comprehend these phenomena, adsorption isotherms graphics were analyzed according to the adjustment of the data to Langmuir and Freundlich models.43 Based on these models, it is possible to obtain theoretical values such as Maximum Adsorption Capacity (MAC), Km, and other parameters.
The Langmuir adsorption model assumes the formation of a protein monolayer, which indicates a finite number of sites of immobilization. The MAC obtained by this model indicates when all these sites are occupied by molecules. Km values indicate the equilibrium concentration necessary to reach an AC of 50% MAC. The Freundlich model assumes the formation of multiple layers, which can indicate that not all the proteins are in direct interaction with NP surfaces. From this model were obtained the Kf and n values, related to the AC and the adsorption intensity, respectively.43
One of the characteristics of the proteins that is essential for adsorption is the net charge at the medium pH. It is considered that when proteins present an opposite charge to NPs, the adsorption could be favored by electrostatic interactions. It is also important how large the difference is between the isoelectric point of the proteins and the pH of the medium, since the greater this difference, the more charge the proteins will have and therefore the greater the electrostatic repulsion between them, which makes the process of adsorption on the surfaces of NPs. Choosing a pH medium near pI could boost the adsorption efficiency, because protein–protein repulsions are minimized, allowing high compaction over NPs.5,48 Moreover, there is evidence that not only electrostatic interaction is important in protein–NP interaction.49 It has also been reported that BSA adsorption is optimal at isoelectric point 4.7 at room temperature.26 According to R2 values presented in Table 3 and to the adsorption curve shown in Fig. 3, BSA immobilization on SiOHNPs had a better adjustment to the Langmuir model (R2 = 0.9802) than to the Freundlich model (R2 = 0.9281). The MAC calculated from the Langmuir isotherm model was 285.8 mg g−1 SiOHNPs. In the case of BSA adsorption on SiNH2NPs a similar adjustment to Langmuir model was obtained (R2 = 0.9780) but for the Freundlich model a higher R2 value was calculated (R2 = 0.9652). Also, according to the Langmuir model, almost double MAC was obtained (540.2 mg BSA per g SiNH2NPs) in comparison with SiOHNPs, which clearly indicates a higher cargo capacity. In both cases, protein release was not detected after several washes. This is a sign of the great stability of the nanocomplexes formed.
| Protein adsorbed | SiOH NPs | SiNH2 NPs | ||
|---|---|---|---|---|
| Langmuir model | Freundlich model | Langmuir model | Freundlich model | |
| BSA | MAC = 285.8 mg g−1 NPs | K f = 267.4 | MAC = 540.2 mg g−1 NPs | K f = 433.0 |
| K m = 0.1265 mg mL−1 | n = 0.3636 | K m = 0.4181 mg mL−1 | n = 0.6151 | |
| R 2 = 0.9802 | R 2 = 0.9281 | R 2 = 0.9780 | R 2 = 0.9652 | |
| Protein percentage release = no detected | Protein percentage release = no detected | |||
| IL-1β | MAC = exceeds the range | K f = 30380 | MAC = exceeds the range | K f = 10790 |
| K m = exceeds the range | n = 1.891 | K m = exceeds the range | n = 1.870 | |
| R 2 = 0.7996 | R 2 = 0.9350 | R 2 = 0.8265 | R 2 = 0.9210 | |
| Protein percentage release = 4.74% | Protein percentage release = 6.18% | |||
| TGF-β1 | MAC = 94.62 mg per cg NPs | K f = 215.2 | MAC = 37.61 mg g−1 NPs | K f = 100.4 |
| K m = 0.1998 mg mL−1 | n = 0.8219 | K m = 0.05415 mg mL−1 | n = 0.5978 | |
| R 2 = 0.9530 | R 2 = 0.9548 | R 2 = 0.7890 | R 2 = 0.8208 | |
| Protein percentage release = 22.53% | Protein percentage release = 40.54% | |||
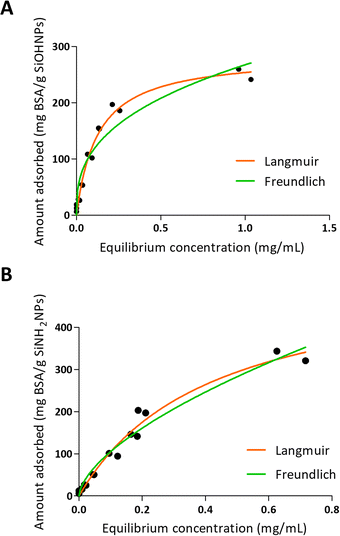 | ||
| Fig. 3 Adsorption isotherms of BSA over SiOHNPs (A) and SiNH2NPs (B) with representation of Langmuir and Freundlich curves in orange and green, respectively. | ||
When IL-1β was absorbed into NPs, a different pattern was presented as can be seen in Fig. 4. On one hand, the values obtained in the adsorption of IL-1β over SiOHNPs presented a poor adjustment for the Langmuir model (R2 = 0.7996) but a better one for the Freundlich model (R2 = 0.9350). MAC and km values were out of range. On the other hand, the Kf and n values of the Freundlich model were very high, showing a high interaction of the protein over the surface. Due to the absence of highly heterogeneous surface, the formation of multilayer could be correlated with protein–protein interactions. When IL-1β adsorption was performed over SiNH2NPs, similar R2 values were obtained for the Langmuir and Freundlich models (0.8265 and 0.9210 respectively). The Kf value was lower than the one presented in SiOHNP adsorption, but the n value was similar. IL-1β pI is 6.1, so it is possible that at pH 7.4 the repulsion between proteins is not too strong to make the adsorption process difficult. Furthermore, IL-1β presents a lower molecular weight (17.5 kDa) than BSA (66.46 kDa), which can explain the differences in the adsorption pattern.
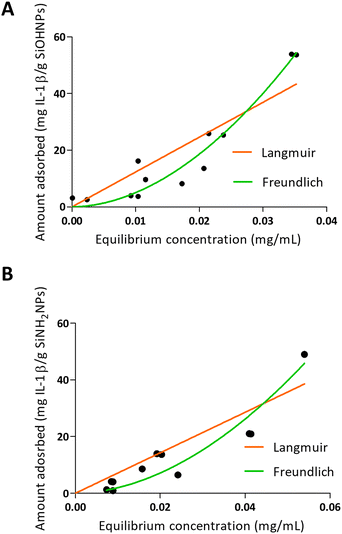 | ||
| Fig. 4 Adsorption isotherms of IL-1β over SiOHNPs (A) and SiNH2NPs (B) with representation of Langmuir and Freundlich curves in orange and green, respectively. | ||
In both NP variants, a low protein release was found over time, that was observed at 30 minutes with no further releases in all other times measured (as far as a month). The protein release percentages for SiOHNPs and SiNH2NPs were 4.74% and 6.18% respectively (Table 3). Once again, it seems that the obtained nanocomplexes presented great stability despite the several washes with PBS through time.
In another sense, it was previously reported that the presence of BSA on the surface of NPs alters their charge state and improves their dissolution kinetics. Perhaps this reduces its useful life but improves protein delivery.46
When TGF-β adsorption was analyzed over SiOHNPs (Fig. 5), R2 = 0.9530 was obtained for the Langmuir model and R2 = 0.9548 for the Freundlich model. The MAC was 94.62 mg TGF-β per g SiOHNPs. On the other hand, the data obtained for TGF-β adsorption on SiNH2NPs presented lower adjustment to the Langmuir and Freundlich models (R2 = 0.7890 and R2 = 0.8280 respectively). MAC was lower, presenting a value of 37.61 mg TGF-β per g SiNH2NPs. The protein release percentages for SiOHNPs and SiNH2NPs were 22.53% and 40.54% respectively and it was detected in the first hour, with no more detectable released protein after that (Table 3). Taking all this data together it is clear that not only MAC was lower for SiNH2NPs, but also a higher percentage of protein was lost in the first washes with PBS. Despite that, both nanocomplexes seem to present good stability at least for a month at 4 °C.
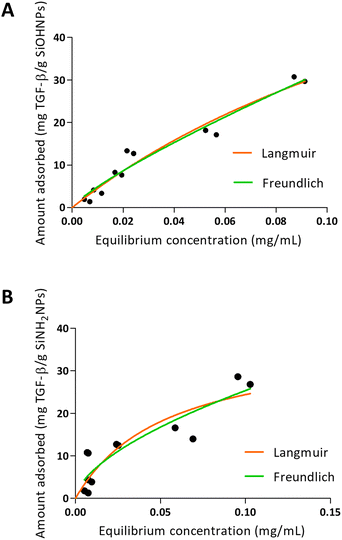 | ||
| Fig. 5 Adsorption isotherms of TGF-β over SiOHNPs (A) and SiNH2NPs (B) with representation of Langmuir and Freundlich curves in orange and green, respectively. | ||
The differences of adjustment to the models between the adsorption in TGF-β and the adsorption of IL-1β over the two types of NPs can be related to the protein corona formed and the interactions presented. When the data fit better to the Langmuir model, as seen for the adsorption of TGF-β, a protein monolayer is probably formed around the NPs.
Although in some combinations, NPs and proteins did not have opposite charges, successful adsorption was achieved. This was also reported in previous studies. Indeed, Villarruel et al. reported the adsorption of recombinant human growth hormone (hGH), which presents an isoelectric point around 5, over silica NPs (synthesized by the Stöber method) in phosphate buffer.50 At this point, both NPs and hGH should have presented a net negative charge, but the immobilization was still accomplished.
Effect of nanoparticles on cell proliferation
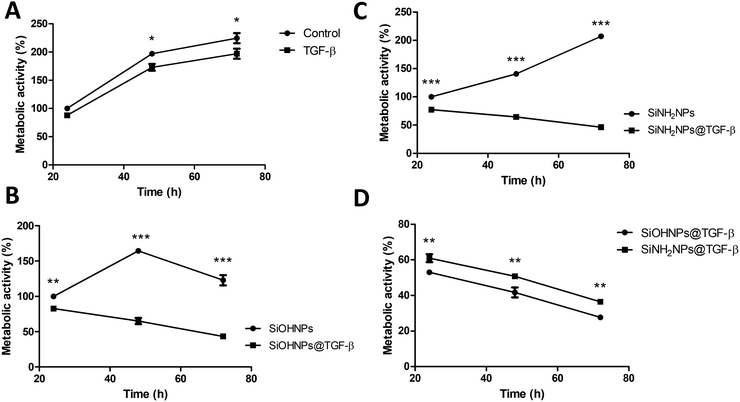
The effect of bare SiOHNPs and SiNH2NPs of similar sizes over the metabolic activity of THP-1 cells at a concentration of 0.6 mg mL−1 (600 μg NPs mL−1) was previously analyzed.18 It was observed that SiOHNPs generated a decrease in cell metabolic activity while SiNH2NPs did not. Given the precedent, the effect of nanocomplexes on cell metabolic activity was analyzed in comparison with the effect generated by bare NPs to identify differences due to the protein coating. SiOHNPs@TGF-β and SiNH2NPs@TGF-β were tested as model NIMs because TGF-β activity is related to a wide variety of immunological processes and has been reported to be associated with immunosuppression and immunotolerance.31,51,52The nanocomplexes used in THP-1 culture were produced from a mix of NPs and TGF-β in a proportion of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 because at this point the AE% was still near 50%. At higher proportions of TGF-β:NPs, an important decrease of AE% was observed. The treatment with NPs and nanocomplexes was made with a final concentration of 300 μg NPs per mL. At this concentration of nanocomplexes, the concentration of TGF-β adsorbed was around 8 μg mL−1. Initially, the effect of TGF-β was analyzed at that concentration (8 μg mL−1). Fig. 6(A) shows the curves of proliferation considering the value of metabolic activity of unexposed cells, at 24 h as 100%. A decrease in metabolic activity of cells treated with TGF-β at 48 and 72 h can be observed, reaching at the last time a decrease of ∼25%. In this way, the TGF-β biological activity over human monocytes was corroborated.
20 because at this point the AE% was still near 50%. At higher proportions of TGF-β:NPs, an important decrease of AE% was observed. The treatment with NPs and nanocomplexes was made with a final concentration of 300 μg NPs per mL. At this concentration of nanocomplexes, the concentration of TGF-β adsorbed was around 8 μg mL−1. Initially, the effect of TGF-β was analyzed at that concentration (8 μg mL−1). Fig. 6(A) shows the curves of proliferation considering the value of metabolic activity of unexposed cells, at 24 h as 100%. A decrease in metabolic activity of cells treated with TGF-β at 48 and 72 h can be observed, reaching at the last time a decrease of ∼25%. In this way, the TGF-β biological activity over human monocytes was corroborated.
When the effect of SiOHNPs and SiOHNPs@TGF-β over THP-1 cells was analyzed (Fig. 6(B)), SiOHNPs@TGF-β showed a higher effect in the reduction of the metabolic activity. At 24 h, cells treated with SiOHNPs@TGF-β presented an activity ∼15% below the activity of the ones treated with bare SiOHNPs. At 48 h, the difference was higher, with an activity of 65% for the nanocomplex (taking the 100% as the value of activity of cells treated with SiOHNPs at 24 h). At 72 h, cells treated with SiOHNPs@TGF-β showed even less activity, showing 3-fold lower activity than cells treated with SiOHNPs. The inhibitory effect of TGF-β was enhanced and maintained through the time when it was immobilized over SiOHNPs.
SiOHNPs’ effect over metabolic activity at 24 h is similar to what was observed in previous studies by De Marzi et al. and Mitarotonda et al., where SiOHNPs generated a decrease in THP-1 cell proliferation.17,18 In particular, for NPs around ∼150 nm, this effect was evident but not as important as in bigger particles (∼500 nm).
In Fig. 6(C) the proliferation curves of cells treated with SiNH2NPs and SiNH2NPs@TGF-β are presented. While SiNH2NPs did not affect the growth rate of THP-1 cells, SiNH2NPs@TGF-β generated a decrease in metabolic activity. In comparison with the metabolic activity of cells treated with SiNH2NPs at 24 h (100%), SiNH2NPs@TGF-β presented an activity reduction of ∼20%, ∼36%, and ∼54% at 24, 48, and 72 h respectively. The good biocompatibility presented by SiNH2NPs was also previously reported by Mitarotonda et al and Baudou et al.18,37
As the tendency seemed to be similar for both complexes, the metabolic activities of cells treated were compared with control cells’ activity. At all the times tested a difference in metabolic cell activity of around 8% between cells treated with SiNH2NPs@TGF-β and the ones treated with SiOHNPs@TGF-β was found (Fig. 6(D)). As the amount of TGF-β adsorbed over NPs was similar in both cases, the only attributable difference between the complexes was the protein percentage release (∼20% for SiOHNPs@TGF-β and ∼45% SiNH2NPs@TGF-β), which could explain the slighter increased effect of the SiOHNPs@TGF-β over monocytes.
These results could indicate that NPs-TGF-β complexes maintain their biological activity over time, even more than TGF-β alone, not only because of the slow release that the NPs would perform but also because of the stability that the NPs provide to the protein. Another point of great interest is that nanocomplexes presented a similar tendency over cells despite the Z potential of the original NPs (Table 1) used for immobilization. As can be seen in the comparison with NPs alone, their typical effects of them are completely changed by the presence of TGF-β on the surface of the nanocomplexes. So, the use of NPs previously characterized as adsorption platforms of different proteins of interest can have a big impact on their biological effect and fate.
Nitric oxide expression
It has been described that TGF-β (in conjunction with IL-10) regulates arginase expression on several immune cells, so it also has an important role in the suppression of NO production mediated by arginase.53,54 This is why NIMs transporting TGF-β could have a crucial effect on macrophage polarization M1/M2.55 Nitrite concentration in monocyte cell cultures was analyzed as an indicator of cell activation.No significant differences were observed when comparing nitrite concentration in exposed and non-exposed cells to TGF-β at all times (Fig. 7). Only cells treated with SiOHNPs presented high concentrations of nitrites (∼4.5 μM) at 24 h. A similar tendency in the induction of nitric oxide expression in THP-1 cells by SiOHNPs was reported in a previous study.18 In the rest of the cases, even when significant differences were found, it did not reach higher values than 2 μM. Therefore, it is considered that nanocomplexes did not generate cell activation, despite the effect that bare NPs could generate. Furthermore, cell cultures treated with SiOHNPs@TGF-β and SiNH2NPs@TGF-β did not present significant differences in nitrite concentration between them at any time, which indicates a similar behavior for both nanocomplexes.
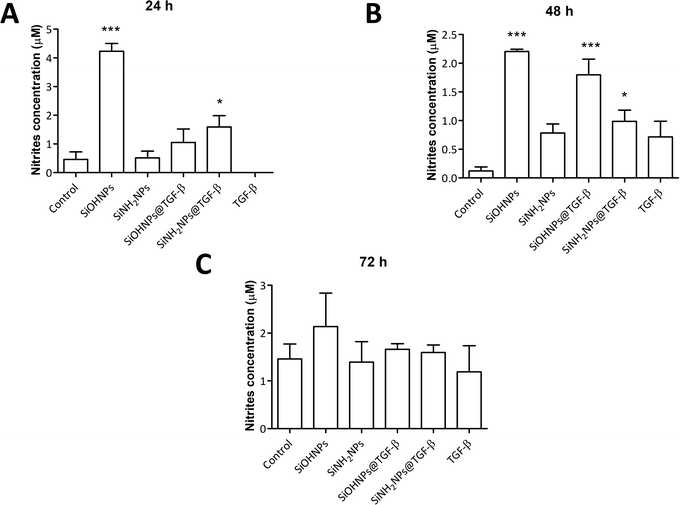 | ||
| Fig. 7 Nitric oxide expression: (A) nitrite concentration in THP-1 cell cultures with different treatments at 24 h; (B) 48 h; (C) 72 h. All comparisons are with control cells. | ||
The evidence collected shows that the effect of nanocomplexes over monocytes metabolic activity and their expression of NO seems to be correlated with the effect of the TGF-β adsorbed and presented on their surface, determining the interaction between NPs and cells. TGF-β activity could also be altered by conformational changes suffered in the adsorption process.5 Nanocomplexes obtained demonstrate that immobilized TGFβ not only maintains biological activity but that it is better than soluble TGFβ.
These nanocomplexes could be the basis for a new generation of immunomodulators, called NIMs. However, it is still necessary to have a wider comprehension of the effects of these NIMs over THP-1 cells and to test them in another model that better emulates the natural physiological environment and cell phenotypes such as 3D models and PMBCs.
As far as we know, this is the first time that TGF-β is adsorbed over silica NPs to provide a potential therapeutic tool. Two studies from the last years incorporate different isoforms of TGF-β in a 3D scaffold to achieve a controlled release in the context of cartilage damage. One of them presented a hydrogel with TGF-β1 and bone marrow-derived mesenchymal stem cells encapsulated with cartilage regeneration properties, demonstrating the high potential of TGF-β as part of complex therapeutic systems for regeneration and different disease treatments.56 On the other hand, Shen et al. demonstrated that the incorporation of a nanomaterial with TGF-β3 adsorbed in a 3D scaffold improved the long-term release and therefore its in vivo effect.57
Herein, the obtained NIMs with an apparent immunosuppressive activity represent true candidates to be tested in a chronic inflammation model and analyze their immunoregulatory capacity. Moreover, these NIMs could be also incorporated into hydrogels and applied to damaged articular cartilage.
Conclusions
In this work, negatively and positively charged SiNPs capable of efficiently adsorbing different proteins on their surface are described. In this sense, all the proteins under study were able to be adsorbed on the surface of both synthesized NPs, but in the case of BSA, SiNH2NPs showed greater adsorption capacity. The results obtained with IL-1β suggest the formation of a protein multilayer around the NPs while with TGF-β a higher MAC was obtained for the SiOHNPs. This indicates that the adsorption, as expected, was more favored when the NPs presented an opposite charge to the proteins.The SiOHNPs@TGF-β and SiNH2NPs@TGF-β nanocomplexes similarly inhibited monocyte metabolic activity, with SiNH2NPs@TGF-β having a slightly smaller initial effect. Both nanocomplexes did not induce important increases in NO, indicating the absence of cell activation.
These observations differ from the effect generated by bare NPs, demonstrating that nanocomplexes have novel properties regardless of the original chemical nature of the NP surface.
Therefore, SiNPs are proposed as potential platforms for delivering immunoregulatory molecules to increase their effects on immune cells. Thus, SiNPs present a potential immunomodulatory tool (NIMs) for future use in treating immunological diseases such as hypersensitivity reactions and autoimmune diseases.
Author contributions
Exequiel Giorgi contributed to the synthesis of NPs and nanocomplexes, analysed the effect of nanocomplexes on cell proliferation and activation and performed statistical analysis. He actively participated in the writing of the article. Sofía Genovés contributed with the cell cultures and metabolic activity determinations as well as statistical analysis. She also actively participated in the writing of the article. María Eugenia Díaz contributed with cell cultures, with the analysis of the effect of nanoparticles on cell proliferation and with statistical analysis. She participated in the writing of the article. Sofía Municoy contributed to the physicochemical characterization of NPs and statistical analysis. She participated in the writing of the article. Martín F. Desimone contributed to the design of experiments and data interpretation. Mauricio C. De Marzi contributed to the conception, design of experiments and acquisition and interpretation of data. Both, Martín F. Desimone and Mauricio C. De Marzi led the writing of the article and grants acquisition. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Exequiel Giorgi is grateful for his doctoral fellowship granted by CONICET. This work was supported by the under grant from the Universidad de Buenos Aires PIDAE2022, PIP 0826 and UBACYT 20020190100083BA (to MFD), and from UNLu-RESREC-LUJ 37/22, UNLu- CDDCB-No 326-19 and PICT-2021-GRF-TI-00507 (to MCDM).Notes and references
- T. Jaswal and J. Gupta, Mater. Today Proc., 2023, 81, 859–863 CrossRef CAS.
- D. B. Warheit, C. M. Sayes, K. L. Reed and K. A. Swain, Pharmacol. Ther., 2008, 120, 35–42 CrossRef CAS.
- V. Stone, H. Johnston and M. J. D. Clift, IEEE Trans Nanobioscience, 2007, 6, 331–340 Search PubMed.
- S. Paul, S. Mukherjee and P. Banerjee, Mater. Adv., 2023, 4, 2042–2061 RSC.
- G. Bashiri, M. S. Padilla, K. L. Swingle, S. J. Shepherd, M. J. Mitchell and K. Wang, Lab Chip, 2023, 23, 1432–1466 RSC.
- A. Lesniak, F. Fenaroli, M. P. Monopoli, C. Åberg, K. A. Dawson and A. Salvati, ACS Nano, 2012, 6, 5845–5857 CrossRef PubMed.
- I. A. Mudunkotuwa, A. Al Minshid and V. H. Grassian, Analyst, 2014, 139, 870–881 RSC.
- M. P. Monopoli, C. Aberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef.
- H. Zhang, Z. Ji, T. Xia, H. Meng, C. Low-Kam, R. Liu, S. Pokhrel, S. Lin, X. Wang, Y. P. Liao, M. Wang, L. Li, R. Rallo, R. Damoiseaux, D. Telesca, L. Mädler, Y. Cohen, J. I. Zink and A. E. Nel, ACS Nano, 2012, 6, 4349–4368 CrossRef PubMed.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CrossRef CAS.
- A. M. Mebert, C. J. Baglole, M. F. Desimone and D. Maysinger, Food Chem. Toxicol., 2017, 109, 753–770 CrossRef CAS PubMed.
- T. I. Janjua, Y. Cao, C. Yu and A. Popat, Nat. Rev. Mater., 2021, 6, 1072–1074 CrossRef CAS PubMed.
- G. Oberdörster, E. Oberdörster and J. Oberdörster, Environ. Health Perspect., 2005, 113, 823–839 CrossRef.
- J. G. Croissant, K. S. Butler, J. I. Zink and C. J. Brinker, Nat. Rev. Mater., 2020, 5, 886–909 CrossRef CAS.
- J. G. Croissant, Y. Fatieiev, A. Almalik and N. M. Khashab, Adv. Healthcare Mater., 2018, 7, 1–75 Search PubMed.
- A. Ale, M. F. Gutierrez, A. S. Rossi, C. Bacchetta, M. F. Desimone and J. Cazenave, Environ. Toxicol. Pharmacol., 2021, 87, 103689 CrossRef CAS PubMed.
- M. C. De Marzi, M. Saraceno, R. Mitarotonda, M. Todone, M. Fernandez, E. L. Malchiodi and M. F. Desimone, Ther. Delivery, 2017, 8, 1035–1049 CrossRef CAS PubMed.
- R. Mitarotonda, M. Saraceno, M. Todone, E. Giorgi, E. L. Malchiodi, M. F. Desimone and M. C. De Marzi, Ther. Delivery, 2021, 12, 443–459 CrossRef CAS.
- V. S. Andrade, A. Ale, S. Municoy, C. Bacchetta, M. F. Desimone, M. F. Gutierrez and J. Cazenave, Environ. Toxicol. Pharmacol., 2023, 102, 104238 CrossRef CAS.
- J. Saikia, M. Yazdimamaghani, S. P. Hadipour Moghaddam and H. Ghandehari, ACS Appl. Mater. Interfaces, 2016, 8, 34820–34832 CrossRef CAS.
- K. Saha, D. F. Moyano and V. M. Rotello, Mater. Horiz., 2014, 1, 102–105 RSC.
- S. Dominguez-Medina, J. Blankenburg, J. Olson, C. F. Landes and S. Link, ACS Sustainable Chem. Eng., 2013, 1, 833–842 CrossRef CAS.
- B. E. Givens, Z. Xu, J. Fiegel and V. H. Grassian, J. Colloid Interface Sci., 2017, 493, 334–341 CrossRef CAS.
- N. El Kadi, N. Taulier, J. Y. Le Huérou, M. Gindre, W. Urbach, I. Nwigwe, P. C. Kahn and M. Waks, Biophys. J., 2006, 91, 3397–3404 CrossRef CAS.
- H. Larsericsdotter, S. Oscarsson and J. Buijs, J. Colloid Interface Sci., 2005, 289, 26–35 CrossRef CAS.
- M. Wïniewska, K. Szewczuk-Karpisz and D. Sternik, J. Therm. Anal. Calorim., 2015, 120, 1355–1364 CrossRef.
- S. J. McClellan and E. I. Franses, Colloids Surf., A, 2005, 260, 265–275 CrossRef CAS.
- V. H. Nguyen and B. J. Lee, Int. J. Nanomed., 2017, 12, 3137–3151 CrossRef CAS.
- N. Kaneko, M. Kurata, T. Yamamoto, S. Morikawa and J. Masumoto, Inflammation Regener., 2019, 39, 1–16 CrossRef CAS.
- R. Bent, L. Moll, S. Grabbe and M. Bros, Int. J. Mol. Sci., 2018, 19, 2155 CrossRef.
- M. Morikawa, R. Derynck and K. Miyazono, Cold Spring Harbor Perspect. Biol., 2016, 8, a021873 CrossRef PubMed.
- R. Mitarotonda, E. Giorgi, T. Eufrasio-da-Silva, A. Dolatshahi-Pirouz, Y. K. Mishra, A. Khademhosseini, M. F. Desimone, M. De Marzi and G. Orive, Biomater. Adv., 2022, 135, 212726 CrossRef CAS PubMed.
- K. Schuhladen, L. Stich, J. Schmidt, A. Steinkasserer, A. R. Boccaccini and E. Zinser, Biomater. Sci., 2020, 8, 2143–2155 RSC.
- K. Zheng, W. Niu, B. Lei and A. R. Boccaccini, Acta Biomater., 2021, 133, 168–186 CrossRef PubMed.
- I. Lukin, I. Erezuma, M. F. Desimone, Y. S. Zhang, A. Dolatshahi-Pirouz and G. Orive, Biomater. Adv., 2023, 154, 213637 CrossRef.
- W. Stöber, A. Fink and E. Bohn, J. Colloid Interface Sci., 1968, 26, 62–69 CrossRef.
- F. G. Baudou, L. Fusco, E. Giorgi, E. Diaz, S. Municoy, M. F. Desimone, L. Leiva and M. C. De Marzi, Colloids Surf., B, 2020, 193, 111128 CrossRef.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef PubMed.
- B. Ledesma, E. Sabio, C. M. González-García, S. Román, M. E. Fernandez, P. Bonelli and A. L. Cukierman, Processes, 2023, 11, 1–22 CrossRef.
- A. Beganskienė, V. Sirutkaitis, A. Kareiva, M. Kurtinaitienė and R. Juškėnas, Mater. Sci., 2004, 10, 287–290 Search PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS.
- T. Zor and Z. Selinger, Anal. Biochem., 1996, 236, 302–308 CrossRef CAS PubMed.
- K. Y. Foo and B. H. Hameed, Chem. Eng. J., 2010, 156, 2–10 CrossRef CAS.
- I. Langmuir, J. Am. Chem. Soc., 1918, 60, 1361–1403 CrossRef.
- M. Naderi, Prog. Filtr. Sep., 2015, 585–608 Search PubMed.
- M. S. Ali, M. J. Uttinger, S. Romeis, J. Schmidt and W. Peukert, Colloids Surf., B, 2022, 214, 112466 CrossRef CAS PubMed.
- Ł. Zych, A. M. Osyczka, A. Łacz, A. Różycka, W. Niemiec, A. Rapacz-Kmita, E. Dzierzkowska and E. Stodolak-Zych, Materials, 2021, 14, 1–17 Search PubMed.
- B. E. Givens, N. D. Diklich, J. Fiegel and V. H. Grassian, Biointerphases, 2017, 12, 02D404 CrossRef.
- S. Tenzer, D. Docter, S. Rosfa, A. Wlodarski, A. Rekik, S. K. Knauer, C. Bantz, T. Nawroth, C. Bier, J. Sirirattanapan, W. Mann, L. Treuel, R. Zellner, M. Maskos and R. H. Stauber, Am. Chem. Soc. NANO, 2011, 5, 7155–7167 CAS.
- L. A. Villarruel, B. Brie, S. Municoy, D. Becú-Villalobos, M. F. Desimone and P. N. Catalano, Int. J. Pharm., 2023, 634, 122662 CrossRef CAS PubMed.
- K. Fujio, T. Komai, M. Inoue, K. Morita, T. Okamura and K. Yamamoto, Autoimmun. Rev., 2016, 15, 917–922 CrossRef CAS.
- M. O. Li, Y. Y. Wan, S. Sanjabi, A. K. Robertson and R. A. Flavell, Annu. Rev. Immunol., 2006, 24, 99–146 CrossRef CAS.
- M. Kupani, S. Sharma, R. K. Pandey, R. Kumar, S. Sundar and S. Mehrotra, Front. Cell. Infect. Microbiol., 2021, 10, 1–9 Search PubMed.
- Y. Vodovotz and C. Bogdan, Prog. Growth Factor Res., 1994, 5, 341–351 CrossRef CAS PubMed.
- C. D. Mills, K. Kincaid, J. M. Alt, M. J. Heilman and A. M. Hill, J. Immunol., 2000, 164, 6166–6173 CrossRef CAS.
- D. Zheng, T. Chen, L. Han, S. Lv, J. Yin, K. Yang, Y. Wang and N. Xu, Int. Wound J., 2022, 19, 1023–1038 CrossRef.
- H. Shen, H. Lin, A. X. Sun, S. Song, B. Wang, Y. Yang, J. Dai and R. S. Tuan, Acta Biomater., 2020, 105, 44–55 CrossRef CAS.
Footnote |
| † Both authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |