Open Access Article
Open Access ArticleTissue-specific gelatin bioink as a rheology modifier for high printability and adjustable tissue properties†
Hohyeon
Han‡
a,
Minji
Kim‡
 c,
Uijung
Yong
e,
Yeonggwon
Jo
a,
Yoo-mi
Choi
d,
Hye Jin
Kim
b,
Dong Gyu
Hwang
d,
Dayoon
Kang
c,
Uijung
Yong
e,
Yeonggwon
Jo
a,
Yoo-mi
Choi
d,
Hye Jin
Kim
b,
Dong Gyu
Hwang
d,
Dayoon
Kang
 cd and
Jinah
Jang
cd and
Jinah
Jang
 *bcdf
*bcdf
aSchool of Interdisciplinary Bioscience and Bioengineering, Pohang University of Science and Technology (POSTECH), South Korea
bDepartment of Convergence IT Engineering, Pohang University of Science and Technology (POSTECH), South Korea. E-mail: jinahjang@postech.ac.kr
cDepartment of Mechanical Engineering, Pohang University of Science and Technology (POSTECH), South Korea
dCenter for 3D Organ Printing and Stem Cells, Pohang University of Science and Technology (POSTECH), South Korea
eFuture IT Innovation Laboratory (i-Lab), Pohang University of Science and Technology (POSTECH), South Korea
fInstitute of Convergence Science, Yonsei University, South Korea
First published on 19th March 2024
Abstract
Decellularized extracellular matrix (dECM) has emerged as an exceptional biomaterial that effectively recapitulates the native tissue microenvironment for enhanced regenerative potential. Although various dECM bioinks derived from different tissues have shown promising results, challenges persist in achieving high-resolution printing of flexible tissue constructs because of the inherent limitations of dECM's weak mechanical properties and poor printability. Attempts to enhance mechanical rigidity through chemical modifications, photoinitiators, and nanomaterial reinforcement have often compromised the bioactivity of dECM and mismatched the desired mechanical properties of target tissues. In response, this study proposes a novel method involving a tissue-specific rheological modifier, gelatinized dECM. This modifier autonomously enhances bioink modulus pre-printing, ensuring immediate and precise shape formation upon extrusion. The hybrid bioink with GeldECM undergoes a triple crosslinking system—physical entanglement for pre-printing, visible light photocrosslinking during printing for increased efficiency, and thermal crosslinking post-printing during tissue culture. A meticulous gelatinization process preserves the dECM protein components, and optimal hybrid ratios modify the mechanical properties, tailoring them to specific tissues. The application of this sequential multiple crosslinking designs successfully yielded soft yet resilient tissue constructs capable of withstanding vigorous agitation with high shape fidelity. This innovative method, founded on mechanical modulation by GeldECM, holds promise for the fabrication of flexible tissues with high resilience.
1. Introduction
Decellularized extracellular matrix (dECM) has emerged as a tissue-specific biomaterial recapitulating the microenvironment of native tissue. Compared to conventional single-type extracellular matrix (ECM) materials, such as collagen and laminin, dECM facilitates cell specification and tissue morphogenesis because of its composition with a diverse biochemical milieu.1,2 Recent studies have focused on dECM bioinks derived from various tissues ever since they were formulated into hydrogels.3,4 In addition to highlighting their remarkable regenerative potential across a variety of target tissues in vitro,5,6 the application of dECM bioinks and tissue fabricated with them has shown promising feasibility for its eventual clinical use.1,7 However, dECM's principal thermal crosslinking mechanism is considerably restricted by the heat transport rate within the gel environment, resulting in inadequate structural stability of the printed constructs with unique geometries (e.g., tubular, curved, spherical).8 These limitations pose challenges in printing flexible tissue constructs with high resolution; to date, tissue engineering of hollow organs with dECM bioinks mostly relies on polymer scaffolds to ensure tissue geometry and mechanical stability.9,10Alternatively, exogenous additions and modifications of dECM have been tried in various ways to improve their mechanical rigidity. For example, exogenous additions include chemical crosslinkers,11–14 ionically crosslinkable polymers,15,16 and reinforcing nanomaterials,17 whereas chemical modifications typically involve methacrylation.18–21 Unfortunately, they have critical limitations. Chemical modification methods are often detrimental to the inherent bioactivity of pristine dECM because they affect the physical composition of dECM, undermining the original purpose of using dECM bioinks.22 Furthermore, it is difficult to control the kinetics of the ionic crosslinking, and ionically crosslinked hydrogels tend to swell and dissolve in the aqueous physiological environment.23,24 Integrating nanomaterials increase the stiffness and brittleness of the bioink,25 failing to match the soft yet tough properties of the native soft tissues.26,27
Our recent method enhanced the printability of dECM using a biocompatible visible light photoinitiator system.28 However, as with other photocrosslinking methods, it did not improve the properties of the printing ink itself but rather allowed faster and tighter crosslinking of the printed ink and stiffened the hydrogel matrix. In this study, we developed a rheological modifier derived from dECM that can be tailored to specific tissues. This modifier demonstrates the distinctive ability to autonomously enhance the modulus of the bioink in the pre-printing step, facilitating the immediate and precise formation of the desired shape, following the extrusion of the bioink.
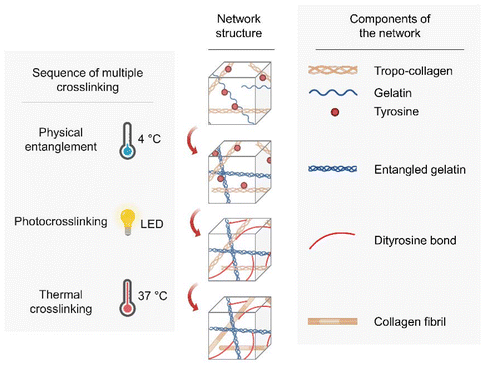
Specifically, we incorporated gelatinized dECM (GeldECM) as a mechanical modifier to dECM and aimed to fabricate a triple crosslinking system for the hybrid bioink (Fig. 1). We expected to employ (i) physical entanglement of the GeldECM portion to enhance printability for pre-printing, (ii) visible light photocrosslinking of the whole hybrid bioink during the printing process to boost crosslinking efficiency, and (iii) thermal crosslinking of the dECM portion after printing to culture the bioprinted tissues. We established a gelatinization process that does not deplete the protein components in dECM; additionally, we determined the best hybridization ratio to modify the mechanical properties of the target tissue. Consequently, we successfully printed cm-scale, tough, and resilient tissue constructs that can withstand vigorous agitation.
We chose small intestine submucosa (SIS) dECM to demonstrate our idea as a promising technique for tissue printing of hollow soft organs. In the future, this sequential multiple crosslinking design based on the mechanical modulatory effect of the GeldECM portion will be useful in fabricating other flexible tissues requiring high resilience.
2. Materials and methods
2.1. Materials
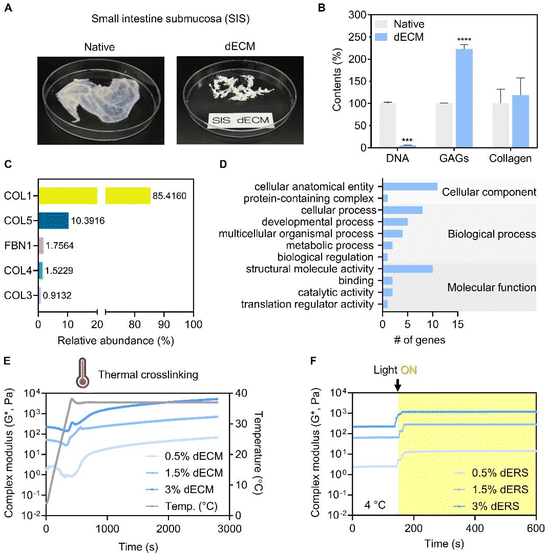
2.1.1.1. Decellularization of the porcine small intestine. Porcine small intestine tissue was obtained from the slaughterhouse. The decellularization process was conducted according to a previously reported protocol, in which the only modification involved the transition from colon to small intestine tissue.29
2.1.1.2. Biochemical analysis of SIS dECM. As described previously, residual DNA, glycosaminoglycans (GAGs), and collagen in both native and decellularized SIS tissues were quantified to validate the efficacy of the decellularization process.30 The obtained results were compared to normalized relative values.
2.1.1.3. Proteomic analysis. Proteomic analysis of SIS dECM and GeldECM was conducted using an ultra-performance liquid chromatography Q-exactive high-resolution mass spectrometer (Q-exactive UHMR hybrid quadrupole-orbitrap; Thermo Fischer Scientific, USA). First, the samples were prepared by in-solution protein digestion into peptides. After lyophilization, the samples were desalted using a C18 micro spin-column. The purified peptide samples were reconstituted in 0.1% formic acid and subsequently injected into the LC-MS/MS system. Mass spectra were automatically analyzed using Proteome Discoverer (version 2.5; Thermo Fischer Scientific, USA). Protein identification was performed by searching the MS/MS data obtained from the peptides against the UniProt database, specifying Sus sucrofa as the species for both SIS dECM and SIS GeldECM.
2.1.1.4. Preparation of SIS dECM bioinks. The lyophilized SIS dECM was digested for 7–10 days using a solution of 0.5 M acetic acid (Merck Millipore, USA) containing pepsin (Sigma-Aldrich, USA). The digested hydrogel was filtered through a 100 μm cell strainer (SPL Life Sciences, Korea) and stored at 4 °C. Before the experiments, the dECM hydrogel was neutralized with 10 N sodium hydroxide solution (NaOH; Biosaesang, Korea) while being maintained on ice and then stored at 4 °C.
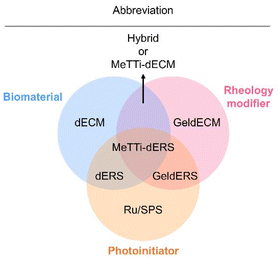
| Abbreviation | Full name |
|---|---|
| dECM | Decellularized extracellular matrix |
| dERS | dECM + Ru/SPS |
| GeldECM | Gelatinized dECM |
| MeTTi–dECM | Mechanically tuned tissue-specific dECM |
| MeTTi–dERS | MeTTi–dECM + Ru/SPS |
| Transmittance (%) = 102−Absorbance |
2.2. Molecular characterization of GeldECM
2.3. Mechanical properties of GeldERS
2.4. Assessment of MeTTi–dECM
2.4.1.1. Time sweep. A time sweep analysis was conducted to measure the complex modulus of the bioinks. To evaluate the photocrosslinking properties of dERS, GeldERS, and MeTTi–dERS, visible light irradiation started at 180 s with an intensity of 10 mW cm−2 and was continued until the experiments finished. The angular frequency was applied in the range of 0.1 to 100 rad s−1 under a 2% strain for all experiments. Oscillation of dECM, dERS, GeldECM, and MeTTi–dERS occurred at 4 °C, whereas GeldERS oscillated at 37 °C.
2.4.1.2. Temperature ramp. A temperature ramp analysis was performed to identify the temperature-dependent crosslinking properties of dERS, GeldERS, and MeTTi–dERS. The temperature was initially held at 4 °C for 180 s, followed by a ramp from 4 to 37 °C at a rate of 5 °C min−1. The temperature was then maintained at 37 °C for 40 min to induce thermal crosslinking in dERS and MeTTi–dERS. Visible light was exposed for 180 s at an intensity of 10 mW cm−2 to photocrosslink MeTTi–dERS. To investigate the physical crosslinking properties of GeldERS, the oscillation temperature was ramped from 37 °C to 4 °C at a rate of 2 °C min−1. A constant angular frequency of 10 rad s−1 and a 2% strain were applied for all experiments.
2.4.1.3. Flow sweep. A steady-state flow sweep analysis was conducted to measure the viscosities of MeTTi–dERS at shear rates ranging from 0.1 to 1000 s−1, at temperatures of 4, 20, and 37 °C.
2.4.2.1. Strand formation test. To evaluate the gelation status of the ink and the uniformity of the strands, extrusion was continued through a 19 G nozzle at 5 kPa until the strands tore off.
2.4.2.2. Shape fidelity analysis. To assess the expansion of bioinks upon extrusion from the nozzle, which results from the release of shear stress, the expansion degree of the filaments was calculated using the equation: α = D/d. To demonstrate the ability to print complex structures, the aspect ratio was calculated as the ratio between the actual height and diameter (H/D). To estimate the collapse of structures in the vertical direction caused by the effect of gravity, the sinking degree was calculated using the equation: γ = H/H0.33 To evaluate the diffusion of strands in the horizontal direction, the ratio between the theoretical diameter and the actual diameter was calculated using the equation: δ = D/D0.
2.5. Assessment of cell viability and maturity
Caco-2 cells (Korea Cell Line Bank, Korea) were encapsulated in dERS and MeTTi–dERS at a density of 5 × 106 cells per mL. Cells were cultured using Dulbecco's modified Eagle's medium high glucose (Hyclone, USA) with media replacement occurring every 2 days.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 ratio in PBS-T and incubated overnight at 4 °C. After three washes with PBS-T, the secondary antibody, Alexa Fluor 488 goat anti-rabbit antibody (Thermo Fischer Scientific, USA), was diluted to obtain a 1
100 ratio in PBS-T and incubated overnight at 4 °C. After three washes with PBS-T, the secondary antibody, Alexa Fluor 488 goat anti-rabbit antibody (Thermo Fischer Scientific, USA), was diluted to obtain a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 ratio with 1% BSA in PBS-T and incubated for 2 h at room temperature. The samples were washed thrice with PBS-T. Finally, VECTASHIELD mounting medium with DAPI (Vector Laboratories, USA) was applied to stain the nucleus and mount the samples. Immunofluorescence imaging was conducted using a spinning disc confocal microscope (Eclipse Ti2-E, Nikon Instruments, Japan).
200 ratio with 1% BSA in PBS-T and incubated for 2 h at room temperature. The samples were washed thrice with PBS-T. Finally, VECTASHIELD mounting medium with DAPI (Vector Laboratories, USA) was applied to stain the nucleus and mount the samples. Immunofluorescence imaging was conducted using a spinning disc confocal microscope (Eclipse Ti2-E, Nikon Instruments, Japan).
For staining the junctional protein markers, the tissues were fixed with 4% paraformaldehyde (Chembio, USA), washed with DPBS for 5 min, and permeabilized with 0.1% Triton X-100 (Biosesang, Korea) in PBS for 1 h at 25 °C. The samples were washed twice using Hank's balanced salt solution (HBSS, Sigma-Aldrich, USA) with 0.05% Tween-20 and DPBS, each for 5 min. Subsequently, they were treated with 5% normal goat serum (NGS; Invitrogen Life Technologies, USA) for blocking for 1 h and washed with PBS for 5 min each. The primary antibody, E-CAD (Cell Signaling, USA), was then prepared as a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution in 1% BSA and incubated at 4 °C overnight. Next, the secondary antibody, Alexa Fluor 594 goat anti-rabbit antibody (Thermo Fischer Scientific, USA), was diluted to obtain a 1
100 dilution in 1% BSA and incubated at 4 °C overnight. Next, the secondary antibody, Alexa Fluor 594 goat anti-rabbit antibody (Thermo Fischer Scientific, USA), was diluted to obtain a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 ratio and incubated at 4 °C overnight. The samples were washed twice with HBSS and 0.05% Tween-20 and with DPBS for 5 min. Finally, the antifade mounting medium, VECTASHIELD (Vector Laboratories, USA), was used for nucleus staining and sample mounting. Immunofluorescence imaging was performed using a fluorescent confocal laser scanning microscope (Nikon Ti Eclipse; Nikon Instruments, Japan).
200 ratio and incubated at 4 °C overnight. The samples were washed twice with HBSS and 0.05% Tween-20 and with DPBS for 5 min. Finally, the antifade mounting medium, VECTASHIELD (Vector Laboratories, USA), was used for nucleus staining and sample mounting. Immunofluorescence imaging was performed using a fluorescent confocal laser scanning microscope (Nikon Ti Eclipse; Nikon Instruments, Japan).
2.6. Statistical analysis
Quantitative results were analyzed with GraphPad Prism 10.1.0 (GraphPad Software, USA), and the data are presented as mean ± standard deviation (S.D.) or standard error of the mean (S.E.M.) as specifically indicated. Student's t-tests were employed to determine the differences between the experimental groups. Statistically significant differences are represented as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.3. Results and discussion
3.1. Preparation of SIS dECM
We evaluated the crosslinking characteristics of SIS dECM and dERS at different concentrations. As previously mentioned, we aimed to develop a triple crosslinking platform where the mechanical properties of dECM bioink can be tuned as desired with the aid of GeldECM. For the designed triple crosslinking strategy to function well, it is important to determine the conditions in which the base hydrogel system, dECM in this study, and the modifier, i.e., GeldECM, have the highest crosslinking efficiency. Increasing the temperature from 4 °C to 37 °C enabled stable thermal crosslinking of the SIS dECMs at all concentrations (Fig. 3E). We assessed the complex modulus under photocrosslinking by supplementing Ru/SPS and thermal crosslinking at physiological temperature (37 °C). All of the dERS concentration ranges (0.5, 1.5, and 3%) exhibited efficient photocrosslinking when visible light was used to irradiate the samples (Fig. 3F). Because the suitable storage modulus range of hydrogel for bioprinting is 102–103 Pa34 and the cell viability in viscous bioinks over 3% can decrease due to high shear stress,18 1.5% was chosen for SIS dECM subsequently (Fig. S1†).
3.2. Preparation of SIS GeldECM
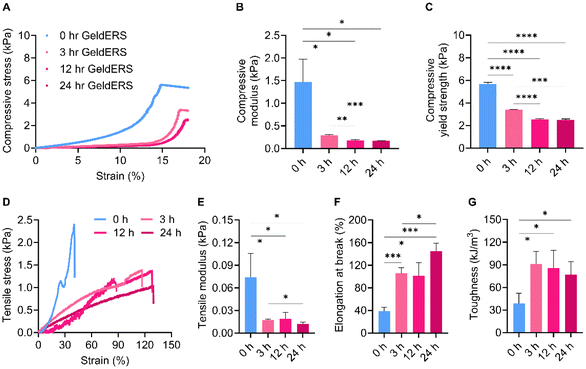
3.2.3.1. Mechanical properties. We identified the characteristics of GeldECM according to the gelatinization time. Because the GeldECM serves as a rheological modifier of dECM, it is important to identify the alterations in its mechanical properties and adapt them as needed. For mechanical testing, GeldECMs with different gelatinization times (0, 3, 12, 24, 72, and 120 h) were supplemented with Ru/SPS and cast into molds to fabricate uniform specimens for compressive and tensile tests. However, samples from 72 and 120 h were excluded because they were not photocrosslinkable.
The compressive and tensile tests showed that gelatinization imparted ductile, stretchable, and tough nature on the stiff and brittle dECM (Fig. 5). Compression testing revealed a decrease in the compressive modulus and yield strength with gelatinization (Fig. 5A–C). The decrease in the compressive modulus indicates that GeldECM became more deformable and less stiff when subjected to compressive forces compared with dECM (Fig. 5B). For a given tensile force, the GeldECM showed higher ductility and stretchability (Fig. 5D–G). The decrease in tensile modulus indicates that the GeldECM became less stiff and experienced more significant elongation (Fig. 5E). Especially, the failure strain of SIS GeldECM increased significantly to approximately 120–130% compared with dECM, which is similar to the failure strain value of native tissue of human small intestine, i.e., 140% (Fig. 5F).35 The toughness increased more than 2-fold after gelatinization (Fig. 5G).
This transformation indicates that GeldECM is soft yet tough, which is largely attributed to physical entanglements in the GeldECM hydrogel network. The physical entanglements function as flexible crosslinking points whereas the chemical crosslinking gives rigidity to the hydrogel.26 Thus, balanced physical and chemical crosslinking in the network could have allowed the energy to dissipate efficiently.36
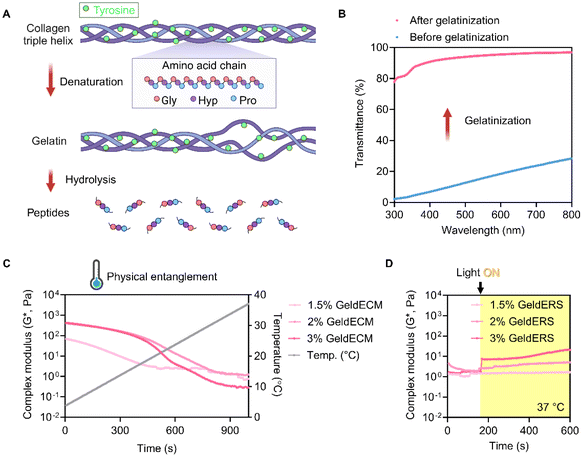
3.2.3.2. Collagen integrity. To reveal the mode of action of gelatinization, we traced the gelatinization-induced changes at the molecular level. Succeeding, we moved on to optimize the gelatinization time for our study. We analyzed the FTIR spectra of the SIS GeldECM by focusing on specific absorbance bands related to collagen structure, such as amides A, I, II, and III (Table 2). FTIR spectroscopy has been employed for the comprehensive examination of protein characteristics, including their secondary structure composition, structural dynamics, and conformational alterations triggered by variations in temperature, pH, and pressure. We identified a red shift in the amide A band peak positions after heat exposure indicating that the structural order of collagen changed due to the disruption of hydrogen bonds (Table 3 and Fig. S2†).37–39 Additionally, a blue shifting in the amide III frequency was observed, which is a typical response to the interruption in the hydrogen bonding of collagen.39 These shifts in both amide A and amide III implied that the hydrogen bonding in collagen was disturbed leading to alteration in its structure during gelatinization.
| Designation | Approximate frequency (cm−1) | Description | Ref. |
|---|---|---|---|
| Amide A | 3300–3500 | Intermolecular hydrogen bonds | 38, 40 and 41 |
| Collagen triple helix structure | |||
| Amide I | 1600–1700 | Secondary structural composition of the peptide | 38 and 42 |
| Intramolecular hydrogen bonds | |||
| Amide II | 1480–1575 | Intact triple helical structure of collagens | 38 and 42 |
| Formation of hydrogen bonds | |||
| Amide III | 1200–1300 | Collagen triple helix structure | 39 and 40 |
| Hydrogen bonding of the collagen peptide bond | |||
| AIII/A1450 | 1.00 for pure collagen | Integrity of collagen triple helical structure | 38 |
| 0.59 for denatured collagen | |||
| Δν (νAI–νAII) | <100 cm−1 | Denaturation of the secondary structure of collagen | 43 |
| Gelatinization time | Parameters | |||||
|---|---|---|---|---|---|---|
| Amide A | Amide I | Amide II | Amide III | AIII/A1450 | Δν | |
| 0 h | 3411.923 | 1637.492 | 1546.842 | 1228.601 | 1.280 | 90.650 |
| 3 h | 3400.351 | 1637.492 | 1546.842 | 1234.384 | 0.997 | 90.650 |
| 12 h | 3408.066 | 1637.492 | 1546.842 | 1235.316 | 0.716 | 90.650 |
| 24 h | 3411.923 | 1637.492 | 1546.842 | 1242.102 | 0.640 | 90.650 |
| 72 h | 3409.995 | 1635.563 | 1544.913 | 1240.173 | 0.683 | 90.650 |
| 120 h | 3408.066 | 1637.492 | 1546.842 | 1240.173 | 0.745 | 90.650 |
Calculation of the intensity ratio of amide III to 1450 cm−1 (AIII/A1450) and the wavenumber difference between amide I and II (Δν) revealed detailed information about where the gelatinization-induced change happened among the three hierarchical levels of collagen's structure (Table 3). The AIII/A1450 ratio of SIS dECM was 1.280, which was similar to that of pure and intact collagen.38 The gelatinization reduced the AIII/A1450 value to 0.997, suggesting a loss of triple helicity of collagen structure.44,45 Meanwhile, the Δν value was consistently under 100 cm−1, disclosing that the secondary structure of the collagen did not denature during gelatinization.43
3.2.3.3. Tyrosine moiety. We confirmed that the tyrosine was abundant in dECM and did not notably decline, which assures efficient photocrosslinking of both dECM and GeldECM as tyrosine is the moiety necessary for photocrosslinking via Ru/SPS (Table 4 and Fig. S3†).
| Gelatinization time | Concentration of tyrosine (μg mL−1) |
|---|---|
| 0 h | 136.871 |
| 3 h | 143.473 |
| 12 h | 124.544 |
| 72 h | 124.148 |
| 120 h | 90.428 |
3.3. Comparison between dECM and GeldECM
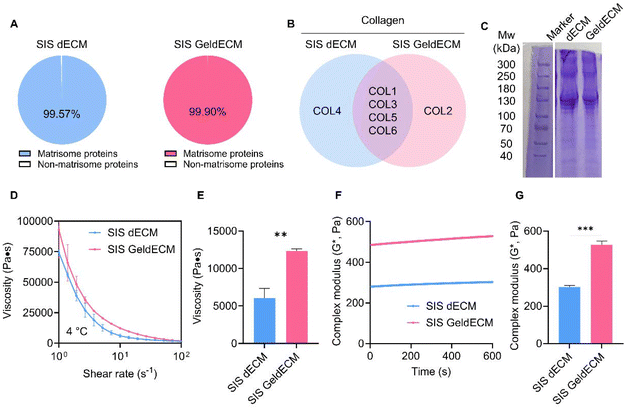
To use GeldECM as a tissue-specific modifier, it is rational to clarify that gelatinization does not damage the bioactivity of dECM. GeldECM should hold tissue-specific microenvironmental cues to support cells and moieties for photocrosslinking. We analyzed and compared the ECM profiles of SIS dECM and 3 h heat-treated GeldECM. The number of proteins that were classified as matrisomes was determined among all proteins identified (Fig. 6A). SIS dECM and GeldECM had a similar ratio of matrisome proteins in their components. We compared the top 5 proteins and collagen types in the SIS dECM and GeldECM. SIS GeldECM presented a significant similarity in the composition of their top 5 proteins and collagens compared with SIS dECM (Fig. S4† and Fig. 6B). They shared three collagen types, I, III, and VI, showing compositional similarity. The SIS dECM and SIS GeldECM have four collagen types in common, indicating that gelatinization did not alter their collagen constitution.35To verify the change in molecular weight, we conducted SDS-PAGE (Fig. 6C and Fig. S5†). Because there were no considerable alternations in their band location and width, the proteins were denatured minimally and the triple helix of collagen unraveled partially after gelatinization. It has been reported that no changes in the position or thickness of the bands were detected on the SDS-PAGE in the case of partial unraveling.46 The dECM and GeldECM showed negligible dispersion of bands in the molecular weight range of the collagen helix, approximately 300 kDa,47 confirming that the partial disentangling of collagen was induced by gelatinization. Overall, regulating the temperature within a range encompassing both collagen denaturation and heat inactivation ensures the preservation of key biological factors of dECM hydrogels.32
Rheological characterization showed instantly enhanced viscosity and modulus in GeldECM (Fig. 6D–G). Gelatinization significantly increased the viscosity and modulus of dECM more than 2-fold (Fig. 6E) and 1.7-fold effortlessly due to the spontaneous physical entanglement at printing temperature (4 °C) (Fig. 6G).
3.4. Rheological properties of hybrid ratio
Because the SIS GeldECM displayed considerably different mechanical properties compared with the SIS dECM, we blended GeldECM with dECM to modulate the rheological properties of the resultant hybrid bioink and the mechanical properties of the printed constructs. We screened various hybrid ratios between SIS dECM and GeldECM to achieve the ultimate mechanical properties through the triple crosslinking strategy.First, we evaluated the suitability of the hybrid bioinks for extrusion-based printing with various blending ratios, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 9
0, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 7
1, 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, and 5
3, and 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
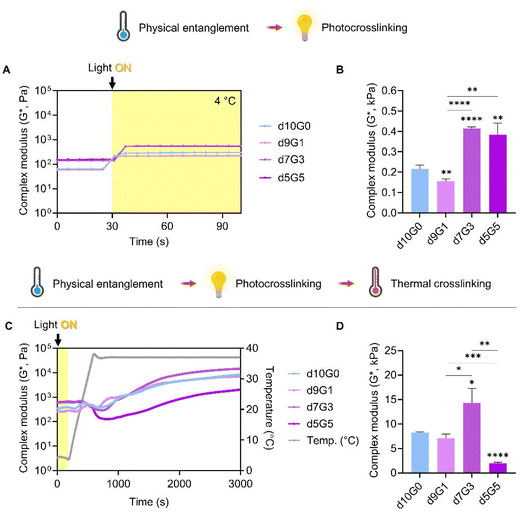
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, (dECM to GeldECM) at different temperatures (Fig. S6†). The viscosity profiles exhibited shear-thinning properties implying that they can be used for extrusion-based printing. We then evaluated the effect of sequential double crosslinking in the hybrid bioinks (Fig. 7A and B). The hybrid bioinks containing more than 30% GeldECM (d7G3 and d5G5) displayed significantly higher modulus compared with dECM (d10G0) because of the high packing density of GeldECM through physical crosslinking (Fig. 7B).
5, (dECM to GeldECM) at different temperatures (Fig. S6†). The viscosity profiles exhibited shear-thinning properties implying that they can be used for extrusion-based printing. We then evaluated the effect of sequential double crosslinking in the hybrid bioinks (Fig. 7A and B). The hybrid bioinks containing more than 30% GeldECM (d7G3 and d5G5) displayed significantly higher modulus compared with dECM (d10G0) because of the high packing density of GeldECM through physical crosslinking (Fig. 7B).
Furthermore, we investigated the impact of successive additions of the thermal crosslinking step (Fig. 7C and D). It was found that the triple crosslinking of bioinks greatly increased the modulus at least 3-fold to a maximum of 32-fold compared to double crosslinking (Fig. 7D). Notably, during the early stages of thermal crosslinking (500 to 1000 s), the modulus of bioinks fluctuates (Fig. 7C). This is because of the thermodynamics during the crosslinking of collagen; the triple helical structure of collagen is partially unwound as the increased temperature gives kinetic energy to overcome the hydrogen bonds maintaining it.48 Subsequently, during the later stages of crosslinking, the collagen molecules reassemble and form covalent bonds, creating a more stabilized and crosslinked structure.
Among the aforementioned three candidate groups (d7G3 and d5G5), the d7G3 hybrid group showed similar depth and span of fluctuations to dECM (d10G0), while having a significantly higher modulus when the crosslinking was complete. Meanwhile, the 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 hybrid group presented a huge decrease in modulus during thermal crosslinking, which resulted in lower stiffness than the dECM group. Crosslinking was hindered at this specific ratio; although each component of the composite hydrogel is stable and crosslinkable, at some ratio they may disrupt the intermolecular crosslinking reaction with one another.49,50 Based on these results, we proceeded to print with the d7G3 hybrid bioink compared with dECM. We named this optimized hybrid condition a mechanically tuned tissue-specific bioink (MeTTi–dECM).
5 hybrid group presented a huge decrease in modulus during thermal crosslinking, which resulted in lower stiffness than the dECM group. Crosslinking was hindered at this specific ratio; although each component of the composite hydrogel is stable and crosslinkable, at some ratio they may disrupt the intermolecular crosslinking reaction with one another.49,50 Based on these results, we proceeded to print with the d7G3 hybrid bioink compared with dECM. We named this optimized hybrid condition a mechanically tuned tissue-specific bioink (MeTTi–dECM).
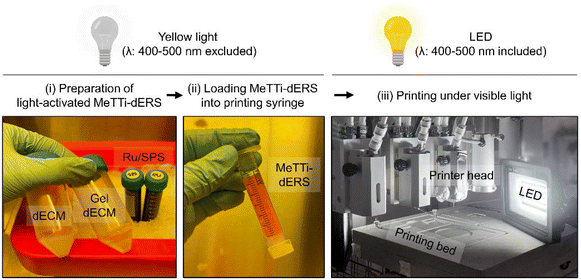
3.5. Printability assessment of the MeTTi–dERS
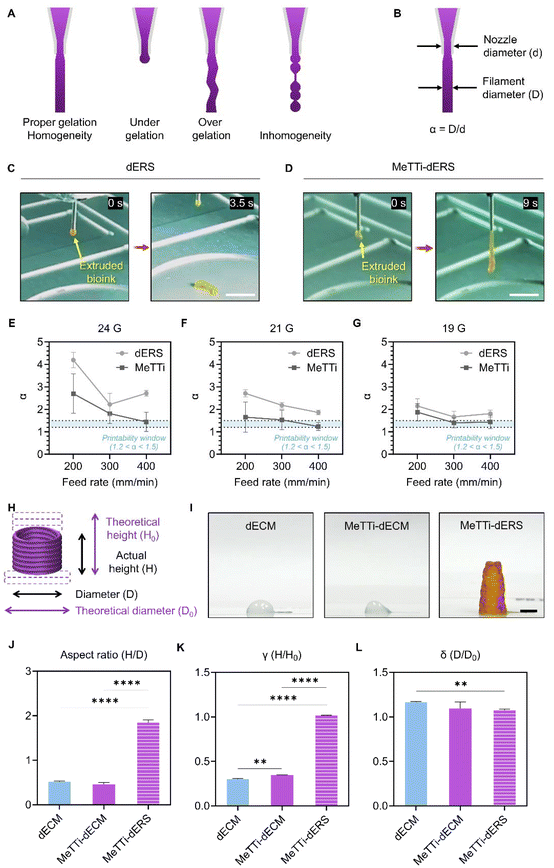
Next, we evaluated whether the MeTTi–dECM can enhance the printability of the dECM. We established a printing environment that does not activate the photocrosslinking of dERS or MeTTi–dERS during bioink preparation, but instantly activates photocrosslinking during printing by adjusting illumination (Fig. 8).Extrusion-based printing offers an appealing feature in its ability to freely draw intricate details with high resolution using finely extruded ink lines. The increased adjustability of the width of the printed line directly correlates with the enhanced shape fidelity of the intended structure.14 To demonstrate line adjustability, we printed dERS and MeTTi–dERS under various conditions, while manipulating parameters, such as nozzle size, pneumatic pressure during dispensing, and motion speed of the printer head (feed rate; abbreviated as F) (Fig. 9A–G). We chose 24, 21, and 19 gauges (abbreviated as G) as representatives for thin, moderate, and thick nozzles, respectively.
The strand formation test assesses static extrusion pressure, ink gelation status, and strand uniformity, providing a brief insight into the hydrogel's 3D printing capabilities (Fig. 9A).51 Because the MeTTi–dERS is a composite hydrogel consisting of two different gels with opposite thermodynamic behavior, it may cause inhomogeneity in the ink, hindering the formation of straight strands. We extruded the two bioinks through the 19 G nozzle at 5 kPa for the longest time until the strands were removed (Fig. 9C and D). With the strand formation test, we confirmed the homogeneity of the bioinks from their uniform strands. Remarkably, the MeTTi–dERS produced a much longer strand compared with dECM, indicating that the MeTTi–dERS is more strongly gelatinized to withstand the weight of the strand.
When the ink is extruded out of the nozzle, it expands because of the release of shear stress, increasing the filament diameter. Previously, an α value, i.e., the ratio between the filament diameter and the nozzle diameter (Fig. 9B), was introduced to describe the spread of the printed filament.33 Quantifying the α values from the line structures printed using dECM and MeTTi–dERS reveals that more α values fell within the printable range, 1.2 < α < 1.5, in the MeTTi–dERS group under the same printing conditions (Fig. 9E–G).
To address the imperative need in regenerative medicine for fabricating elongated tissues that are commensurate with the size of defects, we evaluated the aspect ratio, γ, and δ values (Fig. 9H–L, ESI movies 1 and 2†). The aspect ratio is expressed as H/D, where H is the actual height and D is the diameter of the stacked constructs (Fig. 9H and I). Quantification of aspect ratios revealed significantly increased stacked height when using MeTTi–dERS, underscoring the effectiveness of the sequentially added-up triple crosslinking strategy (Fig. 9J). The γ value, which describes the sinking degree of the stacked constructs and is denoted as H/H0, is the ratio between the actual and theoretical heights (Fig. 9H). The MeTTi–dERS showed a significantly higher γ value compared with the other groups, indicating that it accurately matches the initially designed dimensions (Fig. 9K). Lastly, stacking the hydrogel structure often encounters the problem of filament diffusion in the bottom layers. This degree of diffusion can be expressed as δ, which is the ratio of the actual and theoretical diameters (Fig. 9H). The MeTTi–dECM had a smaller δ value, indicating less horizontal diffusion of the printed filaments when stacked (Fig. 9L).
3.6. Structural integrity assessment of printed tissues
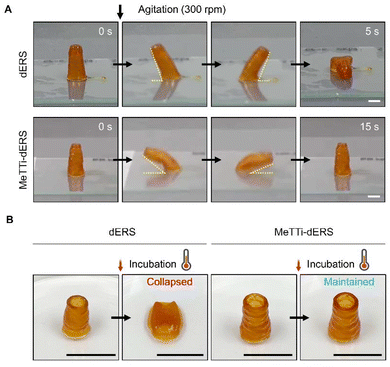
Tissue resilience was remarkably different across the samples (Fig. 10A, ESI movies 3 and 4†). We agitated the dERS and MeTTi–dERS tissues with the same aspect ratio on an orbital shaker at 300 rpm before thermal crosslinking. The dERS collapsed 5 s after agitation, whereas the MeTTi–dERS withstood agitation for 15 s and recovered to its original shape. This clearly shows their capability to dissipate energy through flexible physical crosslinking points within the hydrogel network. In addition, thermal crosslinking made the dERS tissue unstable, as previously predicted by rheology data, which obstructs the steady culture of tissues with a high aspect ratio or structural complexity (Fig. 10B). Notably, the MeTTi–dERS tissues maintained their shape during incubation, which is possibly attributed to the high modulus obtained from triple crosslinking. Prolonged culture dissolved the unreacted Ru/SPS in the printed tissues without disturbing tissue integrity (Fig. S8†).3.7. Biological assessment of printed tissues
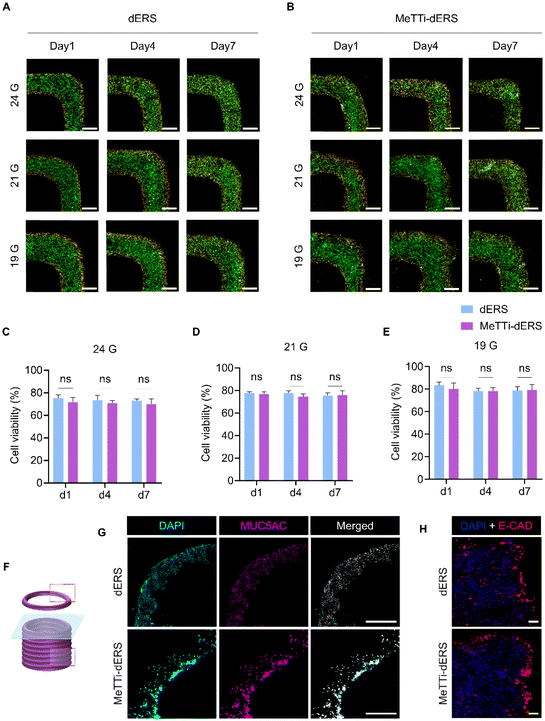
As observed in the rheological analysis, bioinks have high viscosity. Therefore, it is essential to establish the conditions suitable for high printability and cellular assessments. Before the fabrication of tissues, we checked the viability of cells printed with the bioinks using Caco-2 cells (Fig. 11A and B). Using a 19 G nozzle, both dERS and MeTTi–dERS demonstrated a high viability of more than 80%, and there was no significant difference between the groups (Fig. 11C). They also showed good viability of over 70% in the 21 G and 24 G conditions (Fig. 11D and E).Lastly, we evaluated the maturation of printed tissues (Fig. 11F–H). IF staining of mucin revealed that the dERS and MeTTi–dERS tissues exhibited comparable levels of mucin expression (Fig. 11G). Furthermore, we evaluated the expression of junctional protein markers, E-CAD, in the tissues as Caco-2 is an enterocyte that is responsible for forming the intestinal epithelium (Fig. 11H). We observed comparable expression of this junctional protein and a barrier-like morphology in both groups. Overall, intestinal tissues bioprinted with MeTTi–dERS denoted excellent printability, structural integrity, and biological function.
4. Conclusions
In summary, we used GeldECM as a rheological modifier and developed MeTTi–dERS, which has superior mechanical properties because of its inherent physical entanglement. To the best of our knowledge, this is the first report on a tissue-specific rheological modifier. The tissues printed with MeTTi–dERS showed improved printability, shape fidelity, and structural integrity compared with other bioinks based on dECM with single or double crosslinking sequences and excellent biological function. Taken together, the proposed strategy employing GeldECM as a rheology modifier of dECM opens new avenues for fabricating soft and flexible in vitro tissues.Author contributions
Hohyeon Han, Minji Kim: conceptualization, methodology, validation, formal analysis, investigation, writing – original draft, writing – review & editing, and visualization. Uijung Yong: methodology, validation, formal analysis, and investigation. Yeonggwon Jo: validation and investigation. Yoo-mi Choi, Hye Jin Kim, and Dong Gyu Hwang: investigation. Dayoon Kang: investigation and resources. Jinah Jang: conceptualization, methodology, writing – review & editing, supervision, project administration, and funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2021R1A2C2004981). This work was also supported by the Alchemist Project 1415180884 (20012378, Development of Meta Soft Organ Module Manufacturing Technology without Immunity Rejection and Module Assembly Robot System) and the Technology Development Program (S3318933) funded by the Ministry of Trade, Industry & Energy (MOTIE) and the Ministry of SMEs and Startups (MSS) in Korea, respectively.References
- B. S. Kim, S. Das, J. Jang and D.-W. Cho, Chem. Rev., 2020, 120, 10608–10661 CrossRef CAS PubMed.
- W. Han, N. K. Singh, J. J. Kim, H. Kim, B. S. Kim, J. Y. Park, J. Jang and D. W. Cho, Biomaterials, 2019, 224, 119496 CrossRef CAS PubMed.
- D. O. Freytes, J. Martin, S. S. Velankar, A. S. Lee and S. F. Badylak, Biomaterials, 2008, 29, 1630–1637 CrossRef CAS PubMed.
- J. Yoon and J. Jang, Decellularized Tissue-Derived Materials as Advanced Bioinks, in Handbook of the Extracellular Matrix, ed. F. R. A. Maia, J. M. Oliveira and R. L. Reis, Springer International Publishing, Cham, 2023, pp. 1–43 Search PubMed.
- A. A. Golebiowska, J. T. Intravaia, V. M. Sathe, S. G. Kumbar and S. P. Nukavarapu, Bioact. Mater., 2024, 32, 98–123 CAS.
- M. T. Spang and K. L. Christman, Acta Biomater., 2018, 68, 1–14 CrossRef CAS PubMed.
- J. H. Traverse, T. D. Henry, N. Dib, A. N. Patel, C. Pepine, G. L. Schaer, J. A. DeQuach, A. M. Kinsey, P. Chamberlin and K. L. Christman, JACC: Basic Transl. Sci., 2019, 4, 659–669 Search PubMed.
- J. A. Serna, L. Rueda-gensini, D. N. Céspedes-valenzuela, J. Cifuentes, J. C. Cruz and C. Muñoz-camargo, Polymers, 2021, 13, 3263 CrossRef CAS PubMed.
- Z. Galliger, C. D. Vogt and A. Panoskaltsis-Mortari, Transl. Res., 2019, 211, 19–34 CrossRef PubMed.
- H. Nam, H. J. Jeong, Y. Jo, J. Y. Lee, D. H. Ha, J. H. Kim, J. H. Chung, Y. S. Cho, D. W. Cho, S. J. Lee and J. Jang, Sci. Rep., 2020, 10, 1–14 CrossRef PubMed.
- K. Esmaeili Pourfarhangi, S. Mashayekhan, S. G. Asl and Z. Hajebrahimi, Biologicals, 2018, 53, 10–18 CrossRef CAS PubMed.
- B. Nyambat, Y. B. Manga, C.-H. Chen, U. Gankhuyag, A. Pratomo WP, M. Kumar Satapathy and E.-Y. Chuang, Int. J. Mol. Sci., 2020, 21, 4864 CrossRef CAS PubMed.
- Y. E. Arslan, Y. F. Galata, T. Sezgin Arslan and B. Derkus, J. Mater. Sci. Mater. Med., 2018, 29, 127 CrossRef PubMed.
- J. Jang, T. G. Kim, B. S. Kim, S. W. Kim, S. M. Kwon and D. W. Cho, Acta Biomater., 2016, 33, 88–95 CrossRef CAS PubMed.
- G. Gao, H. Kim, B. S. Kim, J. S. Kong, J. Y. Lee, B. W. Park, S. Chae, J. Kim, K. Ban, J. Jang, H. J. Park and D. W. Cho, Appl. Phys. Rev., 2019, 6, 041402 Search PubMed.
- X. Ma, C. Yu, P. Wang, W. Xu, X. Wan, C. S. E. Lai, J. Liu, A. Koroleva-Maharajh and S. Chen, Biomaterials, 2018, 185, 310–321 CrossRef CAS PubMed.
- J. Yoon, H. Han and J. Jang, Nano Convergence, 2023, 10, 52 CrossRef CAS PubMed.
- J. Lee, J. Hong, W. J. Kim and G. H. Kim, Carbohydr. Polym., 2020, 250, 116914 CrossRef CAS PubMed.
- D. O. Visscher, H. Lee, P. P. M. van Zuijlen, M. N. Helder, A. Atala, J. J. Yoo and S. J. Lee, Acta Biomater., 2021, 121, 193–203 CrossRef CAS PubMed.
- M. Ali, A. K. PR, J. J. Yoo, F. Zahran, A. Atala and S. J. Lee, Adv. Healthc. Mater., 2019, 8, 1800992 CrossRef PubMed.
- H. Kim, B. Kang, X. Cui, S. H. Lee, K. Lee, D. W. Cho, W. Hwang, T. B. F. Woodfield, K. S. Lim and J. Jang, Adv. Funct. Mater., 2021, 31, 2011252 CrossRef CAS.
- H. Zhang, Y. Wang, Z. Zheng, X. Wei, L. Chen, Y. Wu, W. Huang and L. Yang, Theranostics, 2023, 13, 2562–2587 CrossRef CAS PubMed.
- C. K. Kuo and P. X. Ma, J. Biomed. Mater. Res., Part A, 2008, 84, 899–907 CrossRef PubMed.
- D. C. Bassett, A. G. Håti, T. B. Melø, B. T. Stokke and P. Sikorski, J. Mater. Chem. B, 2016, 4, 6175–6182 RSC.
- Y. J. Shin, R. T. Shafranek, J. H. Tsui, J. Walcott, A. Nelson and D. H. Kim, Acta Biomater., 2021, 119, 75–88 CrossRef CAS PubMed.
- M. Wang, S. Sun, G. Dong, F. Long and J. T. Butcher, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2213030120 CrossRef CAS PubMed.
- G. Su, Y. Zhang, X. Zhang, J. Feng, J. Cao, X. Zhang and T. Zhou, Chem. Mater., 2022, 34, 1392–1402 CrossRef CAS.
- H. Kim, B. Kang, X. Cui, S. H. Lee, K. Lee, D. W. Cho, W. Hwang, T. B. F. Woodfield, K. S. Lim and J. Jang, Adv. Funct. Mater., 2021, 31, 2011252 CrossRef CAS.
- H. Han, Y. Park, Y. mi Choi, U. Yong, B. Kang, W. Shin, S. Min, H. J. Kim and J. Jang, Adv. Healthc. Mater., 2022, 11, 2101768 CrossRef CAS PubMed.
- F. Pati, J. Jang, D. H. Ha, S. Won Kim, J. W. Rhie, J. H. Shim, D. H. Kim and D. W. Cho, Nat. Commun., 2014, 5, 3935 CrossRef CAS PubMed.
- K. S. Lim, R. Levato, P. F. Costa, M. D. Castilho, C. R. Alcala-Orozco, K. M. A. Van Dorenmalen, F. P. W. Melchels, D. Gawlitta, G. J. Hooper, J. Malda and T. B. F. Woodfield, Biofabrication, 2018, 10, 034101 CrossRef PubMed.
- H. Kim, J. H. Jang, W. Han, H. J. Hwang, J. Jang, J. Y. Kim and D. W. Cho, Biomaterials, 2023, 292, 121941 CrossRef CAS PubMed.
- Q. Gao, X. Niu, L. Shao, L. Zhou, Z. Lin, A. Sun, J. Fu, Z. Chen, J. Hu, Y. Liu and Y. He, Biofabrication, 2019, 11, 035006 CrossRef CAS PubMed.
- N. Ashammakhi, S. Ahadian, C. Xu, H. Montazerian, H. Ko, R. Nasiri, N. Barros and A. Khademhosseini, Mater. Today Bio, 2019, 1, 100008 CrossRef CAS PubMed.
- V. I. Egorov, I. V. Schastlivtsev, E. V. Prut, A. O. Baranov and R. A. Turusov, Mechanical properties of the human gastrointestinal tract $, 2002, vol. 35 Search PubMed.
- C. Norioka, Y. Inamoto, C. Hajime, A. Kawamura and T. Miyata, NPG Asia Mater., 2021, 13, 34 CrossRef CAS.
- O. S. Rabotyagova, P. Cebe and D. L. Kaplan, Mater. Sci. Eng., C, 2008, 28, 1420–1429 CrossRef CAS PubMed.
- T. Riaz, R. Zeeshan, F. Zarif, K. Ilyas, N. Muhammad, S. Z. Safi, A. Rahim, S. A. A. Rizvi and I. U. Rehman, Appl. Spectrosc. Rev., 2018, 53, 703–746 CrossRef CAS.
- N. S. Myshakina, Z. Ahmed and S. A. Asher, J. Phys. Chem. B, 2008, 112, 11873–11877 CrossRef CAS PubMed.
- J. H. Muyonga, C. G. B. Cole and K. G. Duodu, Food Chem., 2004, 86, 325–332 CrossRef CAS.
- Y. Ji, X. Yang, Z. Ji, L. Zhu, N. Ma, D. Chen, X. Jia, J. Tang and Y. Cao, ACS Omega, 2020, 5, 8572–8578 CrossRef CAS PubMed.
- J. Kong and S. Yu, Acta Biochim. Biophys. Sin., 2007, 39, 549–559 CrossRef CAS PubMed.
- M. G. Albu, M. V. Ghica, M. Giurginca, V. Trandafir, L. Popa and C. Cotrut, Rev. Chim., 2009, 60, 666–672 CAS.
- B. F. D. Zaelani, M. Safithri, K. Tarman, I. Setyaningsih and M. Meydia, IOP Conf Ser Earth Environ Sci, DOI:10.1088/1755-1315/241/1/012033.
- A. Pielesz, Spectrochim. Acta, Part A, 2014, 118, 287–293 CrossRef CAS PubMed.
- M. Sun, X. Wei, H. Wang, C. Xu, B. Wei, J. Zhang, L. He, Y. Xu and S. Li, Food Bioprocess Technol., 2020, 13, 367–378 CrossRef CAS.
- A. León-López, A. Morales-Peñaloza, V. M. Martínez-Juárez, A. Vargas-Torres, D. I. Zeugolis and G. Aguirre-Álvarez, Molecules, 2019, 24, 4031 CrossRef PubMed.
- T. L. Willett, R. S. Labow, N. C. Avery and J. M. Lee, Ann. Biomed. Eng., 2007, 35, 1961–1972 CrossRef PubMed.
- J. E. Torres, F. Meng, S. Bhattacharya, K. P. Buno, A. Ahmadzadegan, S. Madduri, P. M. Babiak, P. P. Vlachos, L. Solorio, Y. Yeo and J. C. Liu, Biomacromolecules, 2023, 24, 4718–4730 CrossRef CAS PubMed.
- Y. He, Z. Hou, J. Wang, Z. Wang, X. Li, J. Liu, X. Yang, Q. Liang and J. Zhao, Int. J. Biol. Macromol., 2020, 149, 1275–1284 CrossRef CAS PubMed.
- F. F. Cai, S. Heid and A. R. Boccaccini, J. Biomed. Mater. Res., Part B, 2021, 109, 1090–1104 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3bm02111d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |