Open Access Article
Open Access ArticleRecent advances and prospects in nanomaterials for bacterial sepsis management
Chaoyang
Zhou
*a,
Yong
Liu
 *ac,
Yuanfeng
Li
*ab and
Linqi
Shi
*ac,
Yuanfeng
Li
*ab and
Linqi
Shi
 d
d
aDepartment of Critical Care Medicine, The People's Hospital of Yuhuan, Taizhou, Zhejiang 317600, China. E-mail: zcy2018666@163.com
bTranslational Medicine Laboratory, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang 325035, China. E-mail: yuanfengli@wmu.edu.cn
cWenzhou Institute, University of Chinese Academy of Sciences, Wenzhou, Zhejiang 325001, China. E-mail: y.liu@ucas.ac.cn
dKey Laboratory of Functional Polymer Materials of Ministry of Education, Institute of Polymer Chemistry, College of Chemistry, Nankai University, Tianjin 300071, China
First published on 25th October 2023
Abstract
Bacterial sepsis is a life-threatening condition caused by bacteria entering the bloodstream and triggering an immune response, underscoring the importance of early recognition and prompt treatment. Nanomedicine holds promise for addressing sepsis through improved diagnostics, nanoparticle biosensors for detection and imaging, enhanced antibiotic delivery, combating resistance, and immune modulation. However, challenges remain in ensuring safety, regulatory compliance, scalability, and cost-effectiveness before clinical implementation. Further research is needed to optimize design, efficacy, safety, and regulatory strategies for effective utilization of nanomedicines in bacterial sepsis diagnosis and treatment. This review highlights the significant potential of nanomedicines, including improved drug delivery, enhanced diagnostics, and immunomodulation for bacterial sepsis. It also emphasizes the need for further research to optimize design, efficacy, safety profiles, and address regulatory challenges to facilitate clinical translation.
1. Introduction
Bacterial sepsis, also known as septicemia or blood poisoning, is a serious medical condition caused by the presence of bacteria and their toxins in the bloodstream.1,2 Sepsis can occur when an infection in one part of the body, such as the lungs, urinary tract, or skin, spreads to the bloodstream and triggers an immune response. The consequences of untreated or severe bacterial sepsis can be life-threatening. It can damage multiple organs, including the heart, brain, kidneys, and liver, leading to septic shock and organ failure.3 Bacterial sepsis is a leading cause of death in hospitals and intensive care units, with mortality rates ranging from 10% to 50% depending on the severity of the infection and the patient's overall health status.4 Early recognition and prompt treatment are critical for improving outcomes in sepsis. Common signs and symptoms of sepsis include fever, chills, rapid heartbeat, rapid breathing, low blood pressure, and confusion. Treatment typically involves antibiotics, fluids, oxygen therapy, and other supportive measures to stabilize the patient's vital signs and prevent complications.5In bacterial sepsis, as shown in Fig. 1, the body experiences a severe response to a bacterial infection. This condition is characterized by the presence of bacteria in the bloodstream, leading to widespread infection throughout the body.6 The immune system, which normally acts to defend against such infections, becomes compromised or suppressed, making it harder for the body to fight off the invading bacteria. Additionally, bacterial sepsis triggers an exaggerated and dysregulated inflammatory response known as hyperinflammation.7–10 This excessive inflammation can further contribute to tissue damage and organ dysfunction, potentially leading to life-threatening complications if not effectively managed.
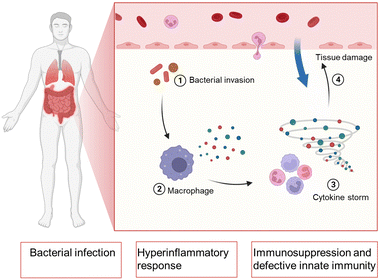 | ||
| Fig. 1 Schematic illustration of the procedure in bacterial sepsis and the physiological parameters during bacterial sepsis. Created with BioRender.com. | ||
The treatment and management of sepsis including the initial diagnosis, timely anti-microbial therapy and attenuating the host response and accompanying organ dysfunction.11 The appropriate cultures should be obtained, to determine the use of antibiotics. The hypotension commonly treated by intravenous fluids or vasopressors. Low-dose steroid is suggested to administer, and plateau airway pressure should be maintained. The deep vein thrombosis and upper gastrointestinal bleeding should be carefully prevented.
Nanomedicine is a rapidly growing field that involves the use of nanotechnology in medicine. It offers many advantages and has great potential in various applications, including the treatment of bacterial sepsis. In the context of sepsis therapy, an advantage of nanomedicine is its ability to enhance the diagnostic capabilities of medical practitioners.12 Several nanomaterials, such as gold nanoparticles (Au NPs), quantum dots, and magnetic nanoparticles (MNPs), have been explored for their potential as biosensors for the detection of bacterial toxins, cytokines, and other biomarkers of sepsis.13,14 These sensors offer high sensitivity, specificity, and accuracy, which could enable earlier detection and diagnosis of sepsis, leading to faster treatment and better outcomes. Moreover, nanomedicine also provides unique opportunities for imaging and monitoring of sepsis. Nanoparticles (NPs) can be designed to be detectable by imaging techniques such as magnetic resonance imaging (MRI) or computed tomography (CT). This could enable real-time monitoring of the spread and severity of sepsis, allowing for more targeted and personalized treatment approaches.
Besides, nanotechnology could be particularly valuable because it could help to deliver antibiotics directly to the site of infection, allowing for more effective and efficient treatment.15 Nanomedicine also offers potential solutions for overcoming the problem of antibiotic resistance.16 The emergence of drug-resistant bacteria is a major concern in the treatment of bacterial infections, including sepsis. NPs can increase the efficacy of antibiotics against drug-resistant bacteria by enhancing their uptake at the site of infection, reducing the risk of resistance development and minimizing damage to healthy tissues.17–19 Another promising application of nanomedicine in sepsis therapy is the modulation of the immune response. Sepsis is characterized by an excessive and dysregulated immune response that can cause tissue damage and organ dysfunction. NPs can be designed to target specific cells or molecules involved in the immune response and modulate their activity.20,21 Despite these advantages, there are also challenges associated with the development and translation of nanomedicine into clinical practice. One of the biggest challenges is ensuring safety and regulatory compliance. Due to their unique properties and potential risks, NPs require extensive testing and evaluation before they can be approved for clinical use. Furthermore, the manufacturing and scalability of NPs may be challenging, and cost-effectiveness needs to be evaluated carefully.
In this review, we highlight the significant potential of nanomedicines in the treatment and diagnosis of bacterial sepsis (Fig. 2). In this regard, nanomedicines offer many advantages, including improved drug delivery, enhanced diagnostic capabilities, and opportunities for immunomodulation and combating antibiotic resistance. Also, further research is discussed that needed to optimize the design and efficacy of nanomedicines, improve their safety profile, and address regulatory challenges for their clinical translation.
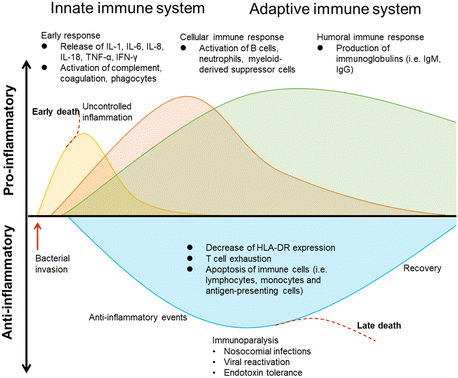
2. Host response in bacterial infection
As summarized in Fig. 3, in bacterial sepsis, both innate and adaptive immune responses play crucial roles in combating the infection.During the proinflammatory response in bacterial infections,22 there is an early immune reaction characterized by the release of various proinflammatory cytokines such as interleukin-1 (IL-1), interleukin-6 (IL-6), interleukin-8 (IL-8), interleukin-18 (IL-18), tumor necrosis factor-alpha (TNF-α), and interferon-gamma (IFN-γ). These cytokines contribute to the activation of complement, coagulation pathways, and phagocytes,23,24 which are important for eliminating the invading bacteria.
The cellular immune response is also triggered during bacterial infections. B cells, neutrophils, and myeloid-derived suppressor cells (MDSCs) become activated.25 B cells play a role in producing immunoglobulins, including IgM and IgG, which can specifically bind to bacterial antigens and aid in their clearance. Neutrophils, a type of phagocyte, are mobilized to the site of infection to engulf and destroy bacteria. MDSCs, on the other hand, have an immunosuppressive function and can inhibit T cell responses.
In contrast to the proinflammatory response, the anti-inflammatory response emerges as an attempt to regulate the excessive inflammation.26 This response is characterized by a decrease in the expression of human leukocyte antigen-DR (HLA-DR), resulting in impaired immune cell function. T cell exhaustion occurs, leading to reduced T cell activity and responsiveness. Moreover, immune cells, such as lymphocytes, monocytes, and antigen-presenting cells, undergo apoptosis, further compromising the immune response.
Immunoparalysis is a state of immune dysfunction that occurs during the anti-inflammatory response, leaving individuals susceptible to nosocomial infections (hospital-acquired infections) and reactivation of latent viral infections.6,27,28 Endotoxin tolerance, where the body becomes less responsive to bacterial endotoxins, can also contribute to immunoparalysis. Ultimately, if the anti-inflammatory response is not controlled, it can lead to late death in bacterial sepsis.
In summary, both the proinflammatory and anti-inflammatory responses are involved in the immune response during bacterial sepsis. While the proinflammatory response aims to eliminate the bacteria, the anti-inflammatory response acts as a regulatory mechanism. However, an uncontrolled anti-inflammatory response can lead to immunoparalysis and negative outcomes such as late death.
In bacterial sepsis, SIRS (systemic inflammatory response syndrome),29 sepsis, severe sepsis, and septic shock are different stages in the continuum of infection-related conditions. SIRS is a systemic inflammatory response that can be triggered by various factors, including infection, pancreatitis, burns, trauma, etc.30 Sepsis occurs when there is evidence of infection along with the presence of SIRS criteria. Severe sepsis refers to sepsis with evidence of organ dysfunction, while septic shock is a life-threatening condition characterized by persistent hypotension despite treatment.31 Their relationship was summarized in Fig. 4. Timely recognition and appropriate management of these conditions are critical to improve patient outcomes.
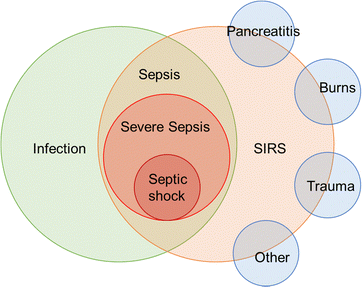 | ||
| Fig. 4 Schematic showing of the relationship between SIRS, sepsis, severe sepsis, and septic shock can be described as a progressive continuum of infection-related conditions. | ||
3. Nanodiagnostic techniques for early detection of bacterial sepsis
3.1. Nanomaterials for detecting early bacterial infections
Early detection and diagnosis of bacterial infections are critical for effective treatment and prevention of disease progression. Nanomaterials have emerged as promising platforms for the development of sensitive and specific diagnostic tools for detecting early bacterial infections. In this section, we will explore the different types of nanomaterials that have been used for early detection of bacterial infections.AuNPs have been widely explored for their potential in bacterial detection. AuNPs can be functionalized with antibodies or peptides that specifically bind to bacterial cell wall components, such as lipopolysaccharides (LPS)32,33 or peptidoglycan.34 The binding of AuNPs to bacterial cells results in a color change or shift in the surface plasmon resonance, which can be detected using various techniques, such as UV-visible spectroscopy or colorimetry.35 AuNPs-based assays have been developed for the detection of a variety of bacterial pathogens, including Escherichia coli (E. coli), Pseudomonas aeruginosa (P. aeruginosa), and Staphylococcus aureus (S. aureus).36 For example, aptamers for ethanolamine and E. coli O111:B4 LPS were used to modify AuNPs. The aptamers for ethanolamine-modified AuNPs (G-probes) can bind to ethanolamine and identify any type of LPS due to the presence of ethanolamine in all LPS variants. This interaction causes the AuNPs to aggregate in a sandwich-like manner, resulting in a color change from red to blue. Additionally, specific probes (S-probes) were created by functionalizing the AuNPs with an aptamer specific to E. coli O111:B4 LPS. These two probes were utilized to develop a logical typing method that can detect LPS within the concentration range of 2.5 to 20 μg mL−1, with a detection limit of 1 μg mL−1 (Fig. 5).32 Additionally, a colorimetric assay using an LPS-binding peptide (LBP) and unmodified AuNPs was developed for the label-free detection of LPS. When cationic LBP probes with a C-terminal cysteine thiol anchor to the surface of anionic AuNPs, it causes the AuNPs to aggregate, leading to a red-to-purple color change and the emergence of a new absorption peak at 586 nm. However, in the presence of LPS, the LPS and LBP form LPS–LBP complexes that prevent the aggregation of AuNPs, which causes prominent color and absorption spectrum differences. This assay allows for quantitative determination of LPS concentration through UV-vis spectroscopy and the detection range is between 10–1000 nM with a detection limit of 2.0 nM.37
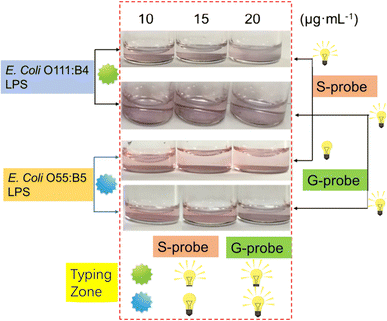 | ||
| Fig. 5 The signal output configuration oflogic typing sensor based on G- probe & S-probe. Reproduced from ref. 32 with permission from Springer Nature, copyright 2019. | ||
Magnetic nanoparticles (MNPs) have also been investigated for their potential in bacterial detection.38,39 MNPs can be modified with antibodies40 or peptides41 that selectively attach to specific components of bacterial cell walls. When MNPs bind to bacterial cells, they generate a magnetic signal that can be identified through techniques such as MRI42 or magnetoresistive sensors.43 For instance, a low-field magnetic resonance imaging (LF-MRI) aptasensor that enables fast detection of P. aeruginosa was developed. This aptasensor relies on the disparity in magnetic behavior between two magnetic nanoparticles: MN10, which has a diameter of 10 nm, and MN400, which has a diameter of 400 nm. To capture the targeted bacteria, specific anti-P. aeruginosa aptamers were chemically immobilized onto the magnetic nanoparticles. When P. aeruginosa is present, the aptamers situated on the magnetic nanoparticles bind to the P. aeruginosa cells, resulting in the formation of a complex called MN10–bacteria–MN400 (MBM).44 By applying a magnetic field, the MBM complex along with the free MN400 particles can be swiftly separated magnetically. Meanwhile, the unbound MN10 particles remain in the solution and serve as a single readout for T2 (transverse relaxation time) in MRI measurements. The LF-MRI platform not only allows for image analysis but also facilitates quantitative detection of P. aeruginosa. Under optimized conditions, it exhibits a detection limit of 100 CFU mL−1 (Fig. 6).
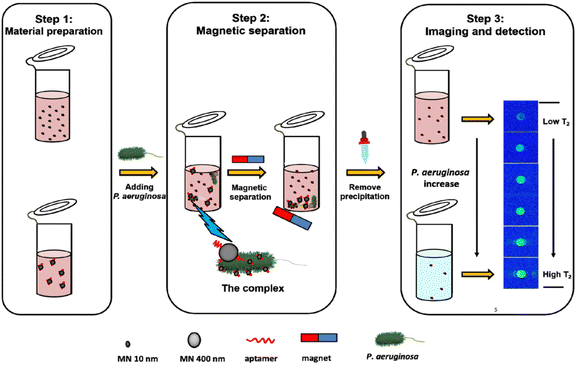 | ||
| Fig. 6 LF-MRI aptasensor schematic and the detection of P. aeruginosa. Reproduced from ref. 44 with permission from American Chemical Society, copyright 2021. | ||
In another example, the sensitivity of magnetoresistive sensors, the portability of a lab-on-chip platform, and the specificity of phage receptor binding proteins (RBPs) as probes were integrated for the rapid and multiplex detection of Enterococcus and Staphylococcus. To achieve this, bacterial cells were initially labeled with RBPs-functionalized MNPs, and subsequently, they were measured using MR sensors. The results clearly demonstrate that the RBP-MNPs enable the specific capture of over 70% of both Enterococcus and Staphylococcus cells, both individually and simultaneously. Moreover, the MR sensors exhibited robust signals for these samples, thereby enabling the detection of both pathogens even at low concentrations (10 CFU mL−1) in less than 2 hours.45
Carbon nanotubes (CNTs) have also shown great potential for bacterial detection.46 CNTs can be functionalized with antibodies or peptides that specifically bind to bacterial cell wall components.47 The binding of bacteria to CNTs results in changes in the electrical conductivity or fluorescence intensity of the CNTs, which can be detected using various techniques, such as field-effect transistor (FET) or fluorescence spectroscopy.48 CNTs-based assays have been developed for the detection of several bacterial pathogens, including E. coli and S. aureus.49 A carbon nanotube field-effect transistor (CNT-FET) is an electronic device that utilizes carbon nanotubes as the conducting channel, offering high speed, low power consumption, and compatibility with flexible electronics. By applying a gate voltage to control the flow of current through the carbon nanotube channel, CNT-FETs can modulate their electrical characteristics, making them promising for various applications such as digital logic circuits, sensors, and biomedical devices.50 For example, a CNT-FET was fabricated by assembling high-purity semiconductor carbon nanotube films onto the sensing channel. To enhance detection sensitivity, carboxylated graphene quantum dots (cGQDs) were coupled to the CNT surface using poly-L-lysine (PLL). Covalent binding of polymyxin B (PMB), a highly specific endotoxin binder, to cGQDs enabled efficient capture and detection of endotoxin. This method exhibited a low limit of detection in both PBS (4.6 fg mL−1) and serum (30.3 fg mL−1) with excellent resistance to interference. It also allowed for rapid analysis of Gram-negative bacterial infections in blood samples, demonstrating superior diagnostic accuracy (AUC = 0.990) based on the receiver operating characteristic curve (ROC).51
In addition to their sensitivity and specificity, nanomaterial-based assays offer several advantages over traditional bacterial detection methods, such as culture-based methods or polymerase chain reaction (PCR).52 Nanomaterial-based assays are rapid, requiring only minutes to hours for detection, compared to days for culture-based methods or PCR. They also require minimal sample preparation and can be performed using portable or handheld devices, making them attractive for point-of-care applications.
3.2. Nanobiosensors for detecting biomarkers of sepsis
Biomarkers of sepsis, such as procalcitonin,53 C-reactive protein,54 and IL-6,55 have been identified and used in clinical practice for the diagnosis and management of sepsis.6 However, these biomarkers have limited sensitivity and specificity, highlighting the need for more sensitive and specific diagnostic tools. Nanobiosensors have emerged as promising platforms for the detection of sepsis biomarkers.56 In this section, we will describe the different types of nanobiosensors that have been used for detecting biomarkers of sepsis.CNTs have emerged as promising tools in the field of sepsis biomarker sensing due to their unique properties. Extensive research has been conducted to explore the potential of CNTs in effectively detecting sepsis biomarkers. By functionalizing CNTs with antibodies or peptides that specifically recognize sepsis biomarkers, such as procalcitonin or C-reactive protein,57 the binding interaction between the biomarker and the CNTs triggers noticeable changes in the electrical conductivity or fluorescence intensity of the CNTs. These changes serve as valuable indicators that enable the detection and quantification of sepsis biomarkers.58 Various advanced techniques, including differential pulse voltammetry (DPV) or fluorescence spectroscopy,59 are employed to accurately measure these alterations and provide reliable detection results. Significantly, CNT-based biosensors have been successfully developed and optimized for the specific identification and measurement of several sepsis biomarkers, including procalcitonin and C-reactive protein.60 This breakthrough opens up new possibilities for early sepsis diagnosis, allowing for timely intervention and improved patient outcomes.61 Continued advancements in CNT-based biosensor technology hold great promise for enhancing sepsis management and contributing to the development of personalized medicine approaches in the future.
In addition to carbon nanotubes, AuNPs have garnered significant attention and investigation for their potential in sepsis biomarker detection. The unique properties of AuNPs make them suitable candidates for functionalization with antibodies or peptides that selectively target sepsis biomarkers, including interleukin-6.62 When the sepsis biomarker binds to the AuNPs, it induces a color change or shift in the surface plasmon resonance of the NPs. This optical phenomenon can be effectively detected using techniques like UV-visible spectroscopy or colorimetry. By measuring the changes in absorbance or color intensity, the presence and concentration of sepsis biomarkers can be accurately determined. The use of AuNP-based biosensors offers several advantages, including their high stability, biocompatibility, and ease of synthesis and modification.63 These properties make them valuable tools for rapid and sensitive detection of sepsis biomarkers. Ongoing research and development in this field aim to optimize AuNP-based sensing platforms, leading to advancements in early sepsis diagnosis, monitoring of treatment effectiveness, and personalized patient care.64
MNPs have also emerged as a promising approach for the detection of sepsis biomarkers, showcasing their significant potential in this field. By functionalizing MNPs with antibodies or peptides that possess high specificity towards sepsis biomarkers like procalcitonin, they can selectively bind to these biomolecules. The binding interaction between the sepsis biomarker and the MNPs generates a magnetic signal, which can be harnessed for detection purposes. Techniques such as MRI or magnetoresistive sensors can effectively capture and analyze this magnetic signal.
The use of MNPs in sepsis biomarker detection offers distinct advantages, including their biocompatibility, stability, and the possibility of multiplexed detection with different biomarkers simultaneously. This technology holds immense promise for improving sepsis diagnosis, facilitating early intervention, and monitoring treatment response. Continued research and development efforts are focused on optimizing MNP-based sensing platforms, advancing their sensitivity, specificity, and applicability in clinical settings. In addition to their sensitivity and specificity, nanobiosensors offer several advantages over traditional diagnostic tools for sepsis biomarkers, such as enzyme-linked immunosorbent assays (ELISA). Nanobiosensors are rapid, requiring only minutes to hours for detection, compared to hours for ELISAs. They also require minimal sample preparation and can be performed using portable or handheld devices, making them attractive for point-of-care applications.
3.3. Nanoparticle imaging techniques for diagnosing sepsis
Nanoparticle imaging techniques have emerged as promising tools for the diagnosis and monitoring of sepsis. In this article, we will explore the different types of nanoparticle imaging techniques that have been used for diagnosing sepsis.MRI is a non-invasive imaging technique that uses magnetic fields and radio waves to create images of internal organs and tissues. Superparamagnetic iron oxide nanoparticles (SPIONs) have been widely explored as contrast agents for MRI in sepsis imaging. SPIONs can be functionalized with antibodies or peptides that specifically bind to sepsis biomarkers, such as LPS or microbial cell wall components.65 The binding of SPIONs to bacterial cells results in changes in magnetic susceptibility, which can be detected using MRI. SPION-based MRI has been shown to improve the sensitivity and specificity of sepsis detection compared to traditional MRI methods.
Optical imaging is another non-invasive imaging technique that uses light to visualize organs and tissues. Near-infrared fluorescence (NIRF) imaging is a type of optical imaging that takes advantage of the transparency of biological tissues to near-infrared light. NIRF imaging can be used to detect fluorescently labeled NPs that specifically bind to sepsis biomarkers. Au NPs and quantum dots have both been investigated as fluorescent contrast agents for NIRF imaging in sepsis. For example, Au NPs can be functionalized with antibodies or peptides that specifically bind to LPS, resulting in enhanced fluorescence signals in regions of sepsis.
Positron emission tomography (PET) is a nuclear imaging technique that uses radioactive tracers to visualize metabolic processes in the body.66 Radiolabeled NPs, such as iron oxide nanoparticles or Au NPs, have been investigated as contrast agents for PET imaging in sepsis.67 Radiolabeled NPs can be functionalized with targeting moieties that specifically bind to sepsis biomarkers, allowing for targeted imaging of septic foci. For example, radiolabeled iron oxide nanoparticles have been used to detect neutrophils by PET in LPS-induced lung injury (Fig. 7).68,69
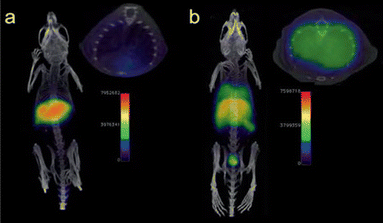 | ||
| Fig. 7 PET/CT imaging of a C57BL/6 mouse. No 68Ga signal was observed in the lungs without LPS-treated mouse (a), but appeared in liver and spleen. The 68Ga signal was showed in lungs with LPS-induced lung infection (b). Reproduced from ref. 64 with permission from Royal Chemical Society, copyright 2023. | ||
In addition to their sensitivity and specificity, nanoparticle imaging techniques offer several advantages over traditional diagnostic tools for sepsis, such as blood cultures or biomarker assays. Nanoparticle imaging techniques provide real-time information on the location and extent of septic foci, allowing for precise localization and monitoring of sepsis. They are also non-invasive, reducing the risk of complications associated with invasive procedures.67,70
4. Nanomedicine in antibiotic therapy
4.1. NPs to enhance stability, bioavailability, and efficacy of antibiotics
NPs have shown great potential in enhancing the stability, bioavailability, and efficacy of antibiotics. They offer several advantages over traditional antibiotic formulations, including increased drug solubility, targeted delivery, prolonged circulation time, and reduced toxicity.16,71 In this section, we will describe the different types of NPs that have been used to improve antibiotic therapy and their mechanisms of action (Fig. 8). The mechanisms by which NPs enhance antibiotic delivery and efficacy are multifactorial. One of the primary mechanisms is through increased drug solubility and stability. NPs can encapsulate hydrophobic drugs, such as some antibiotics, and protect them from degradation or aggregation.72,73 This allows for increased circulation time and improved availability of the drug at the site of infection. Targeted delivery is another mechanism by which NPs improve antibiotic therapy. By functionalizing the surface of NPs with targeting ligands,74 they can selectively bind to bacterial cells and deliver the drug directly to the site of infection.75,76 This reduces the risk of off-target effects and minimizes damage to healthy tissues.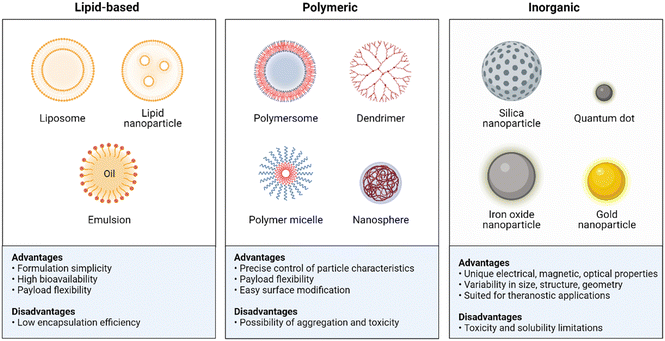 | ||
| Fig. 8 Summary advantages and disadvantages of the NPs used in antibiotic delivery. Created with BioRender.com. | ||
One of the most common types of NPs used for antibiotic delivery is lipid-based NPs, including liposomes, lipid NPs, and emulsions. Liposomes are spherical particles composed of a phospholipid bilayer that can encapsulate hydrophilic or hydrophobic drugs.77 The phospholipid bilayer provides a protective barrier around the drug, reducing its degradation and increasing its circulation time in the bloodstream. Liposomes can also be functionalized with targeting ligands, such as antibodies or peptides, to enhance their specificity and selectivity towards bacterial pathogens. For instance, compared to non-targeted liposomes, these targeted liposomes showed higher binding affinity to P. aeruginosa and were found in greater numbers in the circulation and at the site of infection in mice. In a mouse model, treatment with ciprofloxacin-loaded, targeted liposomes resulted in increased survival time compared to other liposome formulations and free ciprofloxacin.78 Lipid NPs are a type of nanoparticle that are composed primarily of lipids, which are naturally occurring molecules such as phospholipids and cholesterol. These NPs have gained significant attention in the field of drug delivery due to their ability to encapsulate and protect various types of therapeutic agents, including small molecules, proteins, nucleic acids, and vaccines. For example, a novel ciprofloxacin-conjugated lipid nanoparticle has been developed for bacterial biofilm eradication.79,80 By conjugating boronic acid moieties on drugs with a lipid derived from α-aminophosphonate containing catechol through dynamic covalent bonding (Fig. 9), we obtained lipid NPs with desirable properties. Efficient drug loading is ensured by this conjugation, achieving effective encapsulation and eliminating the need for purification as water is generated. Charge reversal is achieved due to lowered pKa of adjacent amines to phosphonates, enabling adaptive targeting in acidic microenvironments. Specific drug release occurs via the responsive nature of lipid NPs to acidic/oxidative conditions in cancer cells or infection sites, allowing for in situ release of cargoes and therapeutic efficacy.79
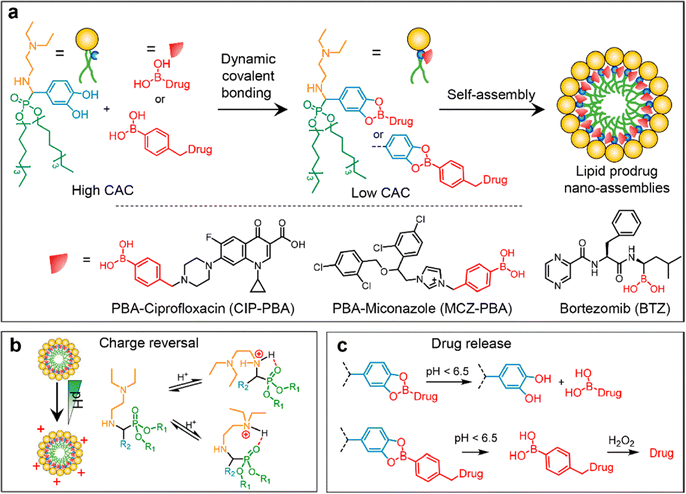 | ||
| Fig. 9 The schematic diagram depicts (a) the formation of lipid prodrug nanoassemblies (LPNAs), (b) a hypothetical stereochemical scheme illustrating the protonation of amines adjacent to phosphonates, and (c) the specific release of drugs in an acidic/oxidative microenvironment. Reproduced from ref. 79 with permission from American Chemical Society, copyright 2023. | ||
Polymeric NPs are another type of nanoparticle used for antibiotic delivery. Polymeric NPs encompass a range of structures, including polymersomes, polymeric micelles, dendrimers, and nanospheres. Polymersomes are vesicular structures formed by self-assembly of amphiphilic block copolymers. These large, hollow structures provide an excellent platform for encapsulating hydrophilic and hydrophobic antibiotics within their aqueous core and lipid bilayer membrane,81 respectively. Polymeric micelles are formed through the self-assembly of amphiphilic block copolymers in an aqueous solution. They consist of a hydrophobic core surrounded by a hydrophilic shell, allowing for the encapsulation of hydrophobic drugs within the core.82 These NPs are made from amphiphilic polymers, such as poly(lactic-co-glycolic acid) (PLGA), and can encapsulate both hydrophilic and hydrophobic drugs.73,83 Polymeric NPs provide sustained release of the drug and increased bioavailability compared to free drug formulations.84 They also allow for targeted delivery by functionalizing the surface of the NPs with targeting moieties.85 For example, a pH/enzyme-responsive amphiphilic block copolymer consisting of biotinylated poly(ethylene glycol)-b-poly(β-amino ester)-b-poly(ethylene glycol) grafted with PEGylated lipid (Biotin-PEG-b-PAE(-g-PEG-b-DSPE)-b-PEG-Biotin) was applied to loaded an antibiotic (ciprofloxacin, CIP) and an anti-inflammatory agent (2-[(amino-carbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide, TPCA-1), followed by coating of anti-mouse ICAM-1 antibody via the binding of biotin to avidin. The resulting polymeric micelles could the highly expresses ICAM-1 molecules in infectious sites, which the local acidic and enzymatic microenvironment could trigger the release of the loaded drugs to eliminate bacteria and alleviate inflammation.86
Dendrimers are highly branched and symmetrical macromolecules with multiple branches emanating from a central core.87 Their unique architecture allows for precise control over size, shape, and surface functionality. Dendrimers can encapsulate hydrophilic and hydrophobic drugs and can be functionalized with targeting moieties.88 Their small size allows for easy penetration into tissues and efficient cellular uptake, making them attractive for intracellular infections.89 Nanospheres refer to solid NPs composed of polymeric materials. They are typically spherical in shape and can be engineered to have controlled sizes and surface properties. Nanospheres can be used for encapsulating drugs or imaging agents, and their release kinetics can be finely tuned based on the polymer choice and formulation parameters.
Inorganic NPs, such as silica NPs, iron oxide NPs, Au NPs, and quantum dots, have been explored for antibiotic delivery purposes. Silica NPs are composed of silicon dioxide and can be easily synthesized with controlled sizes and surface properties.90 They have a high loading capacity for antibiotics and can protect the drugs from degradation. Additionally, their porous structure allows for sustained release of antibiotics over time.91 Iron oxide NPs, particularly magnetic iron oxide NPs, offer unique advantages for antibiotic delivery. They can be guided and concentrated to infection sites using external magnetic fields, enabling localized drug delivery.92 Furthermore, they possess inherent imaging properties, allowing for real-time monitoring of drug distribution and therapeutic efficacy.93 Au NPs have excellent biocompatibility and can serve as carriers for antibiotics. They can be functionalized with targeting ligands to selectively deliver antibiotics to specific cells or pathogens.94 Moreover, Au NPs have plasmonic properties that can enable controlled drug release through photothermal or photochemical methods.95 Quantum dots are semiconductor NPs with tunable optical properties. Although commonly used for imaging applications, they can also be employed for antibiotic delivery. Quantum dot-based drug carriers can provide controlled release of antibiotics, and their fluorescence properties can facilitate tracking and monitoring of the drug distribution in biological systems.96,97 The use of these inorganic NPs holds promise for antibiotic delivery, offering unique features and capabilities that can enhance drug efficacy, improve targeted therapy, and provide valuable insights into drug release and distribution. However, it is important to note that further research is ongoing to fully understand their safety, toxicity, and long-term effects in biomedical applications.
4.2. Targeted drug delivery using size, shape, and surface modification of NPs
Targeted drug delivery is a critical area of research in nanomedicine. Using the size, shape, and surface modification of NPs, it is possible to create drug delivery systems that selectively target specific tissues or cells while minimizing damage to healthy tissues.One approach to targeted drug delivery is through size selection. NPs can be designed to have a specific size range, allowing them to accumulate in certain tissues or organs preferentially. For example, NPs with a diameter of 10–100 nm are ideal for passive targeting of solid tumors due to their enhanced permeability and retention effect (EPR).98 This phenomenon is caused by the leaky vasculature and poor lymphatic drainage of infections, which allows NPs to accumulate within the infection microenvironment.99
Another approach to targeted drug delivery is through shape selection. The shape of an object can have an impact on its ability to kill bacteria, primarily in terms of surface area and physical structure. Surface area: the shape determines the surface area of an object, and the surface area is related to the contact area between the object and the surrounding environment. Microorganisms such as bacteria often adhere to surfaces, so a larger surface area can provide more contact area, increasing the chances of coming into contact with bacteria and effectively killing them. For example, shapes with many small pores or uneven surfaces can provide more space for accommodating bacteria, making it easier for disinfectants or antimicrobial agents to come into contact with them.100 Physical structure: different shapes have different physical structures, which can also affect their efficacy in killing bacteria. For instance, certain shapes may create crowded environments that make it difficult for bacteria to escape or cause mechanical damage, leading to bacterial injury or death. Additionally, specific shapes can influence the dispersion of light or chemical sterilizers, enhancing the antimicrobial effects. It is important to note that shape is just one factor that influences the efficacy of antibacterial actions. Other factors, such as the type of disinfectant, concentration, contact time, etc., also play a role in determining the effectiveness. Therefore, in practical applications, multiple factors need to be considered when selecting the appropriate shape or method for killing bacteria.
Surface modification is another essential strategy for targeted drug delivery using NPs. By functionalizing the surface of NPs with targeting ligands, such as antibodies, peptides, or aptamers, it can selectively bind to bacteria.101 This approach is beneficial for actively targeting the infection sites. The targeting ligands can recognize specific receptors or markers on the cell surface, allowing for selective uptake of the NPs into the target cells. For instance, mannose has the ability to bind to specific receptors on the surface of macrophages. These receptors are called mannose receptors and are mainly present on the surface of macrophages.102 Therefore, NPs modified with mannose are able to target macrophages and efficiently mediate the delivery of antibiotics for killing intracellular bacteria.103,104
Surface charge is another parameter that can be modified to improve targeted drug delivery using NPs.105 The surface charge of NPs can affect their interaction with bacteria. For example, positively charged NPs have been shown to have improved cellular uptake compared to negatively charged NPs due to their electrostatic interaction with the negatively charged bacterial cell membrane.79 However, positively charged NPs can also cause toxicity and non-specific binding to healthy tissues,16 so carefully considering the surface charge is necessary.
In conclusion, targeted drug delivery using size, shape, and surface modification of NPs has great potential for improving the efficacy and safety of drug therapy. The design of NPs can be optimized to enhance passive or active targeting of specific tissues or cells while minimizing off-target effects. However, additional research is needed to optimize the design and efficacy of nanoparticle formulations, evaluate their safety profile, and address regulatory challenges for clinical translation.
4.3. NPs to modulate the host immune system
To treat sepsis, immunomodulators such as nicotinamide adenine dinucleotide (NAD+) have been loaded into NPs in order to modulate the host immune system. For instance, NAD+,106 which is an immunomodulator with potential sepsis treatment capabilities, has recently been loaded into various NPs like liposomes and zeolitic imidazolate frameworks. While NAD+ is recognized as an immune modulator, its exact relationship with inflammation is still not completely understood, and its clinical application is hindered by its inability to be directly taken up by cells. However, when NAD+ or its reduced form (NADH) is loaded into NPs, it improves cellular energy supply, suppresses inflammation, and prevents inflammation-induced cell pyroptosis and apoptosis. In vivo experiments have shown that these NPs can prevent multiorgan injury caused by caecal ligation and puncture, as well as improve outcomes of secondary P. aeruginosa infections following caecal ligation and puncture. Hence, this innovative and translational approach holds promise for efficient and safe sepsis treatment.107NPs can also modulate the immune response to improve antibiotic therapy. For example, polymeric NPs can reduce the production of proinflammatory cytokines,108,109 which contribute to tissue damage and organ dysfunction in sepsis. For instance, a nanoparticle that mimics the behavior of macrophage cells can be used as a potential therapy for controlling sepsis. The nanoparticle works through a two-step process where it first neutralizes endotoxins and then sequesters cytokines. These biomimetic NPs have an exterior that resembles macrophage cells, which allows them to bind to endotoxins and proinflammatory cytokines.110 In another work, a new approach to treating sepsis has been developed using a nanotrap called a telodendrimer (TD) to capture various biomolecules via multivalent, hybrid and synergistic interactions. The TD-NTs adsorb septic molecules with high efficiency. Different charges displayed on proinflammatory and anti-inflammatory cytokines allow for selective capture. The efficacy of the therapy is time and charge dependent. When combined with moderate antibiotic treatment, the therapy resulted in 100% survival in severe septic mice by controlling infection and hyperinflammation, whereas individual therapies only resulted in 50–60% survival.7 Cytokine analysis, inflammatory gene activation and tissue histopathology support the benefits of the treatment.7 In another example, macrophage biomimetic NPs were created by encapsulating polymer cores with cell membranes derived from macrophages. These NPs possess an antigenic exterior identical to the source cells. By mimicking macrophages, these NPs act as decoys and effectively bind and neutralize endotoxins that would otherwise trigger an immune response (Fig. 10). Additionally, these macrophage-like NPs have the capability to sequester proinflammatory cytokines, preventing them from exacerbating the sepsis cascade. In a mouse model of E. coli bacteremia, treatment with these macrophage-mimicking NPs, known as MΦ-NPs, resulted in decreased levels of proinflammatory cytokines, inhibited bacterial dissemination, and ultimately conferred a significant survival advantage to infected mice.110 By inhibiting the inflammatory response, NPs can improve the outcome of antibiotic therapy and reduce the risk of complications.
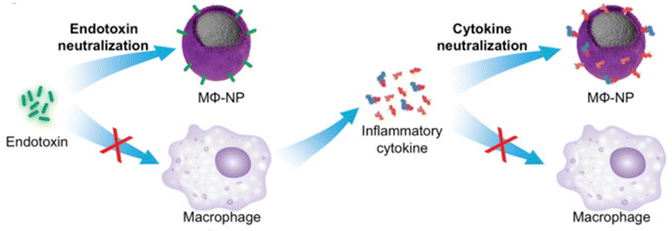 | ||
| Fig. 10 Schematic representation of using MΦ-NPs to neutralize endotoxins and proinflammatory cytokines as a two-step process for sepsis management. Reproduced from ref. 110 under Creative Commons licenses: CC BY-NC-ND. Copyright at the authors. | ||
4.4. Nanoparticle-mediated killing of drug-resistant bacteria
Antibiotic resistance is a major global health threat, and new strategies to combat drug-resistant bacteria are urgently needed. In treatment and management of sepsis, administration of broad-spectrum antibiotics is essential. However, the abuse or overuse antibiotics is prevalent in clinic, especially in low-income and middle-income countries,111 resulting the development of drug-resistant bacteria. It is reported that more than 67% Gram-negtive isolates from neonates with sepsis were resistant to at least one β-lactam and one aminoglycoside in 26 countries in Africa.112 The drug-resistant bacterial induced sepsis is harder to treatment by empirical use of combined antibiotics, which increased the mortality of sepsis. NPs have emerged as a promising approach for addressing this problem by enhancing the killing efficacy of antibiotics against resistant strains. In this regard, many non-antibiotic nanomedicines have been developed, including metal and positively charged NPs.Other types of NPs, such as zinc oxide NPs (ZnO NPs) and titanium dioxide NPs (TiO2 NPs), have also been investigated for their antimicrobial properties. ZnO NPs have been shown to be effective against both Gram-positive and Gram-negative bacteria, including antibiotic-resistant strains. The mechanism of action involves the release of zinc ions, which causes damage to the bacterial cell membrane and intracellular components. TiO2 NPs can exhibit antibacterial activity when activated by ultraviolet light. They can produce reactive oxygen species (ROS), which can damage bacterial cell membranes and intracellular components.
5. Future trends and challenges
5.1. Preclinical progress and current obstacles
Nanotechnology-based approaches have shown promise for the treatment of sepsis,129,130 but there are still several obstacles that need to be addressed before these approaches can be translated into clinical practice.Preclinical studies have shown promising results for the use of nanotechnology-based approaches in treating bacterial sepsis. For example, liposomes loaded with vancomycin have been shown to improve survival rates and reduce bacterial load in a mouse model of sepsis caused by methicillin-resistant Staphylococcus aureus (MRSA).131 Polymeric NPs loaded with dexamethasone have been shown to reduce proinflammatory cytokine levels in a rat model of sepsis induced by the injection of LPS.132 Dendrimers functionalized with Toll-like receptor ligands have been shown to enhance the phagocytic activity of macrophages and improve survival rates in a mouse model of sepsis caused by E. coli.133
Despite these promising results, there are still several obstacles that need to be addressed before nanotechnology-based approaches can be translated into clinical practice. One challenge is optimizing the design and formulation of NPs to maximize drug delivery and efficacy while minimizing off-target effects. Another challenge is evaluating the safety profile of nanoparticle-based approaches, particularly in terms of long-term toxicity and potential for accumulation in the body. Moreover, regulatory approval for nanotechnology-based therapies can be challenging due to the complex nature of NPs and the lack of standardized manufacturing processes.
In conclusion, nanotechnology-based approaches offer promising strategies for treating bacterial sepsis through targeted drug delivery or modulation of immune responses. Preclinical studies have shown promising results for liposomes, polymeric NPs, and dendrimers in treating sepsis caused by a variety of bacterial pathogens. However, there are still several obstacles that need to be addressed before these approaches can be translated into clinical practice. Nonetheless, the use of nanotechnology-based approaches represents a promising strategy for improving the treatment and outcomes of bacterial sepsis.
5.2. Safety considerations and regulatory requirements for nanomedicine
As with any new technology, safety considerations and regulatory requirements are essential to ensure the safe and effective translation of nanomedicine from the laboratory to clinical practice. In this article, we will explore the safety considerations and regulatory requirements for nanomedicine.Toxicity. NPs can interact with biological systems in unpredictable ways due to their unique physicochemical properties. The toxicity of NPs depends on various factors, such as size, shape, surface charge, and chemical composition. Safety assessments of nanomedicines should include comprehensive toxicity studies to evaluate potential adverse effects on human health.
Immunogenicity. NPs can also elicit immune responses in the body that may affect their efficacy and safety. The immunogenicity of nanomedicines should be evaluated to determine their potential to induce allergic reactions or other immune-related adverse events.
Biodistribution. The biodistribution of nanomedicines is another important safety consideration, as NPs can accumulate in certain organs or tissues and cause toxicity. Imaging techniques, such as MRI or positron emission tomography (PET), can be used to track the distribution and clearance of NPs in vivo.
Manufacturing. Nanomedicines must be manufactured according to current good manufacturing practices (cGMP) to ensure their quality and consistency. The manufacturing process should be well-controlled and validated to minimize batch-to-batch variability and ensure reproducibility.
Preclinical testing. Preclinical testing of nanomedicines should follow established guidelines, such as those provided by the International Conference on Harmonization (ICH). Preclinical studies should include comprehensive toxicity testing, pharmacokinetics, and efficacy evaluations in relevant animal models.
Clinical trials. Clinical trials of nanomedicines must be conducted according to the same regulatory requirements as traditional drugs, such as those provided by the US Food and Drug Administration (FDA) or the European Medicines Agency (EMA). Clinical trials should follow established guidelines for safety and efficacy evaluations, including phase I, II, and III trials.
Labeling. Nanomedicines must be properly labeled to ensure their safe and effective use by healthcare providers and patients. The labeling should include information on the composition, dosage, administration, and potential adverse events associated with the nanomedicine.
In conclusion, safety considerations and regulatory requirements are critical for the safe and effective translation of nanomedicine from the laboratory to clinical practice. Safety considerations include toxicity, immunogenicity, and biodistribution, while regulatory requirements include manufacturing, preclinical testing, clinical trials, and labeling. As the field of nanomedicine continues to advance, efforts to ensure its safety and efficacy will remain essential to improving patient outcomes.
Author contributions
Conceptualization, literature analysis and collection of references, writing and preparing final version of schemes and figures – original draft preparation, writing – review and editing, Y. L. and Y. F. L.; conceptualization, methodology, project administration, resources, writing – review and editing, supervision, data curation, C. Y. Z. and L. Q. S. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Grant No. 22275043 and 52203184), Startup Funding of Wenzhou Institute, University of Chinese Academy of Sciences (Grant No. WIUCASQD2021022), and Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (Grant No. 2023KY1348 and 2023RC200).References
- J. Cohen, Nature, 2002, 420, 885–891 CrossRef CAS PubMed.
- J. Cohen, J.-L. Vincent, N. K. J. Adhikari, F. R. Machado, D. C. Angus, T. Calandra, K. Jaton, S. Giulieri, J. Delaloye and S. Opal, Lancet Infect. Dis., 2015, 15, 581–614 CrossRef.
- J. Fernández, J. Acevedo, R. Wiest, T. Gustot, A. Amoros, C. Deulofeu, E. Reverter, J. Martínez, F. Saliba and R. Jalan, Gut, 2018, 67, 1870–1880 CrossRef PubMed.
- H. Cao, Y. Gao, H. Jia, L. Zhang, J. Liu, G. Mu, H. Gui, Y. Wang, C. Yang and J. Liu, Nano Lett., 2022, 22, 7882–7891 CrossRef CAS PubMed.
- N. C. Riedemann, R.-F. Guo and P. A. Ward, Nat. Med., 2003, 9, 517–524 CrossRef CAS PubMed.
- T. Van Der Poll, F. L. Van De Veerdonk, B. P. Scicluna and M. G. Netea, Nat. Rev. Immunol., 2017, 17, 407–420 CrossRef CAS PubMed.
- C. Shi, X. Wang, L. Wang, Q. Meng, D. Guo, L. Chen, M. Dai, G. Wang, R. Cooney and J. Luo, Nat. Commun., 2020, 11, 3384 CrossRef CAS PubMed.
- M. Ciszek-Lenda, M. Strus, M. Walczewska, G. Majka, A. Machul-Żwirbla, D. Mikołajczyk, S. Górska, A. Gamian, B. Chain and J. Marcinkiewicz, Inflammation Res., 2019, 68, 397–413 CrossRef CAS PubMed.
- V. Castiglia, A. Piersigilli, F. Ebner, M. Janos, O. Goldmann, U. Damböck, A. Kröger, S. Weiss, S. Knapp and A. M. Jamieson, Cell Host Microbe, 2016, 19, 375–387 CrossRef CAS PubMed.
- T. D. Y. Reijnders, A. Saris, M. J. Schultz and T. van der Poll, Lancet Respir. Med., 2020, 8, 619–630 CrossRef CAS PubMed.
- J. Rello, F. Valenzuela-Sánchez, M. Ruiz-Rodriguez and S. Moyano, Adv. Ther., 2017, 34, 2393–2411 CrossRef CAS PubMed.
- B. Pelaz, C. Alexiou, R. A. Alvarez-Puebla, F. Alves, A. M. Andrews, S. Ashraf, L. P. Balogh, L. Ballerini, A. Bestetti, C. Brendel, S. Bosi, M. Carril, W. C. W. Chan, C. Chen, X. Chen, X. Chen, Z. Cheng, D. Cui, J. Du, C. Dullin, A. Escudero, N. Feliu, M. Gao, M. George, Y. Gogotsi, A. Grünweller, Z. Gu, N. J. Halas, N. Hampp, R. K. Hartmann, M. C. Hersam, P. Hunziker, J. Jian, X. Jiang, P. Jungebluth, P. Kadhiresan, K. Kataoka, A. Khademhosseini, J. Kopeček, N. A. Kotov, H. F. Krug, D. S. Lee, C. M. Lehr, K. W. Leong, X. J. Liang, M. Ling Lim, L. M. Liz-Marzán, X. Ma, P. Macchiarini, H. Meng, H. Möhwald, P. Mulvaney, A. E. Nel, S. Nie, P. Nordlander, T. Okano, J. Oliveira, T. H. Park, R. M. Penner, M. Prato, V. Puntes, V. M. Rotello, A. Samarakoon, R. E. Schaak, Y. Shen, S. Sjöqvist, A. G. Skirtach, M. G. Soliman, M. M. Stevens, H. W. Sung, B. Z. Tang, R. Tietze, B. N. Udugama, J. S. VanEpps, T. Weil, P. S. Weiss, I. Willner, Y. Wu, L. Yang, Z. Yue, Q. Zhang, Q. Zhang, X. E. Zhang, Y. Zhao, X. Zhou and W. J. Parak, ACS Nano, 2017, 11, 2313–2381 CrossRef CAS PubMed.
- L. Papafilippou, A. Claxton, P. Dark, K. Kostarelos and M. Hadjidemetriou, Adv. Healthcare Mater., 2021, 10, 2001378 CrossRef CAS PubMed.
- A. Pant, I. Mackraj and T. Govender, J. Biomed. Sci., 2021, 28, 1–30 CrossRef PubMed.
- K. Forier, K. Raemdonck, S. C. De Smedt, J. Demeester, T. Coenye and K. Braeckmans, J. Controlled Release, 2014, 190, 607–623 CrossRef CAS PubMed.
- Y. Liu, L. Q. Shi, L. Z. Su, H. C. van der Mei, P. C. Jutte, Y. J. Ren and H. J. Busscher, Chem. Soc. Rev., 2019, 48, 428–446 RSC.
- Y.-Z. Piao, Y. Qi, X.-W. Hu, Y. Wang, Y. Li, T. Zhou, L. Shi, Y. Liu and C. Zhou, J. Controlled Release, 2022, 352, 1–14 CrossRef CAS.
- Y. Li, Y. Liu, R. Ma, Y. Xu, Y. Zhang, B. Li, Y. An and L. Shi, ACS Appl. Mater. Interfaces, 2017, 9, 13056–13067 CrossRef CAS PubMed.
- Y. Liu, Y. Li and L. Shi, J. Controlled Release, 2021, 329, 1102–1116 CrossRef CAS PubMed.
- D. F. Moyano, Y. Liu, D. Peer and V. M. Rotello, Small, 2016, 12, 76–82 CrossRef CAS PubMed.
- Y. Yang, K. Wang, Y. Pan, L. Rao and G. Luo, Adv. Sci., 2021, 8, 2102330 CrossRef.
- H. K. De Jong, T. Van Der Poll and W. J. Wiersinga, J. Innate Immun., 2010, 2, 422–430 CrossRef CAS PubMed.
- A. Oberholzer, C. Oberholzer and L. L. Moldawer, Shock, 2001, 16, 83–96 CrossRef CAS PubMed.
- A. Mantovani, M. A. Cassatella, C. Costantini and S. Jaillon, Nat. Rev. Immunol., 2011, 11, 519–531 CrossRef CAS PubMed.
- J. Pillay, T. Tak, V. M. Kamp and L. Koenderman, Cell. Mol. Life Sci., 2013, 70, 3813–3827 CrossRef CAS PubMed.
- N. J. Shubin, S. F. Monaghan and A. Ayala, Sepsis-Pro-Inflammatory Anti-inflamm. Responses, 2011, 17, 108–124 CAS.
- R. S. Hotchkiss, G. Monneret and D. Payen, Lancet Infect. Dis., 2013, 13, 260–268 CrossRef CAS.
- F. Venet and G. Monneret, Nat. Rev. Nephrol., 2018, 14, 121–137 CrossRef CAS.
- M. G. Davies and P. Hagen, Br. J. Surg., 1997, 84, 920–935 CrossRef CAS.
- C. M. Robertson and C. M. Coopersmith, Microbes Infect., 2006, 8, 1382–1389 CrossRef CAS.
- D. C. Angus and T. Van der Poll, N. Engl. J. Med., 2013, 369, 840–851 CrossRef CAS PubMed.
- L. Zhu, S. Li, X. Shao, Y. Feng, P. Xie, Y. Luo, K. Huang and W. Xu, Microchim. Acta, 2019, 186, 1–6 CrossRef CAS PubMed.
- W. Wang, L. Liu, S. Song, L. Xu, H. Kuang, J. Zhu and C. Xu, Sci. China Mater., 2016, 59, 665–674 CrossRef CAS.
- T. Lee, J. Lim, K. Park, E.-K. Lim and J.-J. Lee, ACS Sens., 2020, 5, 3099–3108 CrossRef CAS PubMed.
- F. Fu, L. Li, Q. Luo, Q. Li, T. Guo, M. Yu, Y. Song and E. Song, Analyst, 2018, 143, 1133–1140 RSC.
- C. N. Elliott, M. C. Becerra, J. C. Bennett, L. Graham and G. L. Hallett-Tapley, RSC Adv., 2021, 11, 14161–14168 RSC.
- C. Lei, Z. Qiao, Y. Fu and Y. Li, Anal. Methods, 2016, 8, 8079–8083 RSC.
- Y. W. Chu, D. A. Engebretson and J. R. Carey, J. Biomed. Nanotechnol., 2013, 9, 1951–1961 CrossRef CAS PubMed.
- C. Xu, O. U. Akakuru, J. Zheng and A. Wu, Front. Bioeng. Biotechnol., 2019, 7, 141 CrossRef PubMed.
- T. Yoneyama, A. Kuwahata, T. Murayama, L. Tonthat, S. Yabukami, Y. Sato, Y. Teramura, W. Ikeda-Ohtsubo and T. Ogawa, IEEE Trans. Magn., 2022, 58, 1–6 Search PubMed.
- C. Wang, B. Gu, Q. Liu, Y. Pang, R. Xiao and S. Wang, Int. J. Nanomed., 2018, 1159–1178 CrossRef CAS PubMed.
- H. Shao, C. Min, D. Issadore, M. Liong, T.-J. Yoon, R. Weissleder and H. Lee, Theranostics, 2012, 2, 55 CrossRef CAS PubMed.
- T. A. P. Rocha-Santos, TrAC, Trends Anal. Chem., 2014, 62, 28–36 CrossRef CAS.
- F. Jia, X. Bai, X. Zhang, Y. Fu, Y. Li, X. Li and J. L. Kokini, Anal. Chem., 2021, 93, 8631–8637 CrossRef CAS PubMed.
- A. P. Cunha, R. Henriques, S. Cardoso, P. P. Freitas and C. M. Carvalho, Biotechnol. Bioeng., 2021, 118, 3164–3174 CrossRef CAS PubMed.
- B. Prieto-Simón, N. M. Bandaru, C. Saint and N. H. Voelcker, Biosens. Bioelectron., 2015, 67, 642–648 CrossRef PubMed.
- A. M. Münzer, Z. P. Michael and A. Star, ACS Nano, 2013, 7, 7448–7453 CrossRef PubMed.
- C. A. S. Andrade, J. M. Nascimento, I. S. Oliveira, C. V. J. de Oliveira, C. P. de Melo, O. L. Franco and M. D. L. Oliveira, Colloids Surf., B, 2015, 135, 833–839 CrossRef CAS PubMed.
- C. García-Aljaro, L. N. Cella, D. J. Shirale, M. Park, F. J. Muñoz, M. V. Yates and A. Mulchandani, Biosens. Bioelectron., 2010, 26, 1437–1441 CrossRef PubMed.
- M. D. Bishop, G. Hills, T. Srimani, C. Lau, D. Murphy, S. Fuller, J. Humes, A. Ratkovich, M. Nelson and M. M. Shulaker, Nat. Electron., 2020, 3, 492–501 CrossRef CAS.
- Q. Cui, J. Li, Y. Li, L. Tang, K. Li, T. Li, X. Chen, Z. Zhang and G. J. Zhang, Talanta, 2024, 266, 125035 CrossRef CAS PubMed.
- C. Picard, C. Ponsonnet, E. Paget, X. Nesme and P. Simonet, Appl. Environ. Microbiol., 1992, 58, 2717–2722 CrossRef CAS PubMed.
- K. Reinhart and M. Meisner, Crit. Care Clin., 2011, 27, 253–263 CrossRef CAS PubMed.
- S. Eschborn and J.-H. Weitkamp, J. Perinatol., 2019, 39, 893–903 CrossRef PubMed.
- J. Eichberger and B. Resch, Front. Pediatr., 2022, 10, 840778 CrossRef PubMed.
- A. Alba-Patiño, A. Vaquer, E. Barón, S. M. Russell, M. Borges and R. de la Rica, Microchim. Acta, 2022, 189, 74 CrossRef PubMed.
- D. Kumar and B. B. Prasad, Sens. Actuators, B, 2012, 171, 1141–1150 CrossRef.
- A. Schuck, H. E. Kim, M. Kang and Y.-S. Kim, BioChip J., 2023, 17, 274–283 CrossRef CAS.
- Y.-S. Fang, H.-Y. Wang, L.-S. Wang and J.-F. Wang, Biosens. Bioelectron., 2014, 51, 310–316 CrossRef CAS PubMed.
- M. Buch and J. Rishpon, Electroanalysis, 2008, 20, 2592–2594 CrossRef CAS.
- S. Balayan, N. Chauhan, W. Rosario and U. Jain, Appl. Surf. Sci. Adv., 2022, 12, 100343 CrossRef.
- B. S. Munge, C. E. Krause, R. Malhotra, V. Patel, J. S. Gutkind and J. F. Rusling, Electrochem. Commun., 2009, 11, 1009–1012 CrossRef CAS PubMed.
- M. A. Khan and M. Mujahid, Sensors, 2020, 20, 646 CrossRef PubMed.
- S. Singh, P. S. Podder, M. Russo, C. Henry and S. Cinti, Lab Chip, 2023, 23, 44–61 RSC.
- A. Popov, B. Brasiunas, A. Kausaite-Minkstimiene and A. Ramanaviciene, Chemosensors, 2021, 9, 85 CrossRef CAS.
- S. Basu, T. Chryssikos, S. Moghadam-Kia, H. Zhuang, D. A. Torigian and A. Alavi, Seminars in nuclear medicine, Elsevier, 2009, vol. 39, pp. 36–51 Search PubMed.
- L. Zhang, W.-F. Dong and H.-B. Sun, Nanoscale, 2013, 5, 7664–7684 RSC.
- M. Krekorian, K. R. G. Cortenbach, M. Boswinkel, A. Kip, G. M. Franssen, A. Veltien, T. W. J. Scheenen, R. Raavé, N. K. van Riessen and M. Srinivas, Cancers, 2021, 13, 5069 CrossRef CAS PubMed.
- J. Pellico, A. V. Lechuga-Vieco, E. Almarza, A. Hidalgo, C. Mesa-Nuñez, I. Fernández-Barahona, J. A. Quintana, J. Bueren, J. A. Enríquez, J. Ruiz-Cabello and F. Herranz, Sci. Rep., 2017, 7, 13242 CrossRef PubMed.
- P. Padmanabhan, A. Kumar, S. Kumar, R. K. Chaudhary and B. Gulyás, Acta Biomater., 2016, 41, 1–16 CrossRef CAS PubMed.
- A. Gupta, S. Mumtaz, C.-H. Li, I. Hussain and V. M. Rotello, Chem. Soc. Rev., 2019, 48, 415–427 RSC.
- Y. Liu, H. C. van der Mei, B. Zhao, Y. Zhai, T. Cheng, Y. Li, Z. Zhang, H. J. Busscher, Y. Ren and L. Shi, Adv. Funct. Mater., 2017, 27, 1701974 CrossRef.
- Y. Liu, H. J. Busscher, B. Zhao, Y. Li, Z. Zhang, H. C. Van Der Mei, Y. Ren and L. Shi, ACS Nano, 2016, 10, 4779–4789 CrossRef CAS PubMed.
- R. Molinaro, C. Corbo, J. O. Martinez, F. Taraballi, M. Evangelopoulos, S. Minardi, I. K. Yazdi, P. Zhao, E. De Rosa, M. B. Sherman, A. De Vita, N. E. Toledano Furman, X. Wang, A. Parodi and E. Tasciotti, Nat. Mater., 2016, 15, 1037–1046 CrossRef CAS PubMed.
- H. Koo, R. N. Allan, R. P. Howlin, P. Stoodley and L. Hall-Stoodley, Nat. Rev. Microbiol., 2017, 15, 740 CrossRef CAS PubMed.
- M. Van Oosten, T. Schäfer, J. A. C. Gazendam, K. Ohlsen, E. Tsompanidou, M. C. De Goffau, H. J. M. Harmsen, L. M. A. Crane, E. Lim, K. P. Francis, L. Cheung, M. Olive, V. Ntziachristos, J. M. Van Dijl and G. M. Van Dam, Nat. Commun., 2013, 4, 2584 CrossRef PubMed.
- Z. Drulis-Kawa and A. Dorotkiewicz-Jach, Int. J. Pharm., 2010, 387, 187–198 CrossRef CAS PubMed.
- A. Singla, S. B. Simbassa, B. Chirra, A. Gairola, M. R. Southerland, K. N. Shah, R. E. Rose, Q. Chen, A. Basharat, J. Baeza, R. Raina, M. J. Chapman, A. M. Hassan, I. Ivanov, A. Sen, H.-J. Wu and C. L. Cannon, ACS Appl. Mater. Interfaces, 2022, 14, 40724–40737 CrossRef CAS PubMed.
- Y. Ding, X. Hu, Y. Piao, R. Huang, L. Xie, X. Yan, H. Sun, Y. Li, L. Shi and Y. Liu, ACS Nano, 2023, 17, 6601–6614 CrossRef CAS PubMed.
- Y. Zhan, X. Hu, Y. Li, Y. Wang, H. Chen, C. A. Omolo, T. Govender, H. Li, F. Huang, L. Shi, X. Hu and Y. Liu, Adv. Funct. Mater., 2023, 33, 2214299 CrossRef CAS.
- Y. Xi, Y. Wang, J. Gao, Y. Xiao and J. Du, ACS Nano, 2019, 13, 13645–13657 CrossRef CAS PubMed.
- Y. Liu, Y. Ren, Y. Li, L. Su, Y. Zhang, F. Huang, J. Liu, J. Liu, T. G. van Kooten, Y. An, L. Shi, H. C. van der Mei and H. J. Busscher, Acta Biomater., 2018, 79, 331–343 CrossRef CAS PubMed.
- W. Cai, J. Wu, C. Xi and M. E. Meyerhoff, Biomaterials, 2012, 33, 7933–7944 CrossRef CAS.
- T. Wang, Y. Li, E. J. Cornel, C. Li and J. Du, ACS Nano, 2021, 15, 9027–9038 CrossRef CAS PubMed.
- A. F. Radovic-Moreno, T. K. T. K. Lu, V. A. Puscasu, C. J. Yoon, R. Langer and O. C. Farokhzad, ACS Nano, 2012, 6, 4279–4287 CrossRef CAS.
- C. Y. Zhang, J. Gao and Z. Wang, Adv. Mater., 2018, 30, 1803618 CrossRef.
- E. R. Gillies and J. M. J. Frechet, Drug Discovery Today, 2005, 10, 35–43 CrossRef CAS PubMed.
- S. K. Choi, A. Myc, J. E. Silpe, M. Sumit, P. T. Wong, K. McCarthy, A. M. Desai, T. P. Thomas, A. Kotlyar, M. M. B. Holl, B. G. Orr and J. R. Baker, ACS Nano, 2013, 7, 214–228 CrossRef CAS PubMed.
- Y. Gao, J. Wang, M. Chai, X. Li, Y. Deng, Q. Jin and J. Ji, ACS Nano, 2020, 14, 5686–5699 CrossRef CAS PubMed.
- A. Bernardos, E. Piacenza, F. Sancenón, M. Hamidi, A. Maleki, R. J. Turner and R. Martínez-Máñez, Small, 2019, 15, 1900669 CrossRef PubMed.
- L. B. Capeletti, J. F. A. de Oliveira, L. M. D. Loiola, F. E. Galdino, D. E. da Silva Santos, T. A. Soares, R. de Oliveira Freitas and M. B. Cardoso, Adv. Funct. Mater., 2019, 29, 1904216 CrossRef CAS.
- Y. Liu, P. C. Naha, G. Hwang, D. Kim, Y. Huang, A. Simon-Soro, H.-I. Jung, Z. Ren, Y. Li, S. Gubara, F. Alawi, D. Zero, A. T. Hara, D. P. Cormode and H. Koo, Nat. Commun., 2018, 9, 2920 CrossRef PubMed.
- B. M. Geilich, I. Gelfat, S. Sridhar, A. L. Van De Ven and T. J. Webster, Biomaterials, 2017, 119, 78–85 CrossRef CAS PubMed.
- P. Yang, P. Pageni, A. Rahman, M. Bam, T. Zhu, Y. P. Chen, M. Nagarkatti, A. W. Decho and C. Tang, Adv. Healthcare Mater., 2018, 1800854 Search PubMed.
- D. Hu, H. Li, B. Wang, Z. Ye, W. Lei, F. Jia, Q. Jin, K.-F. Ren and J. Ji, ACS Nano, 2017, 11, 9330–9339 CrossRef CAS PubMed.
- H.-J. Jian, R.-S. Wu, T.-Y. Lin, Y.-J. Li, H.-J. Lin, S. G. Harroun, J.-Y. Lai and C.-C. Huang, ACS Nano, 2017, 11, 6703–6716 CrossRef CAS PubMed.
- Y. Li, S. G. Harroun, Y. Su, C. Huang, B. Unnikrishnan, H. Lin, C. Lin and C. Huang, Adv. Healthcare Mater., 2016, 5, 2545–2554 CrossRef CAS PubMed.
- Y. Ding, Y. Xu, W. Yang, P. Niu, X. Li, Y. Chen, Z. Li, Y. Liu, Y. An, Y. Liu, W. Shen and L. Shi, Nano Today, 2020, 35, 100970 CrossRef CAS.
- L. Su, Y. Li, Y. Liu, Y. An and L. Shi, Macromol. Biosci., 2019, 19, 1900289 CrossRef CAS PubMed.
- Y. Cheng, G. Feng and C. I. Moraru, Front. Microbiol., 2019, 10, 191 CrossRef PubMed.
- H. R. Jia, Y. X. Zhu, Z. Chen and F. G. Wu, ACS Appl. Mater. Interfaces, 2017, 9, 15943–15951 CrossRef CAS PubMed.
- F.-Y. Su, S. Srinivasan, B. Lee, J. Chen, A. J. Convertine, T. E. West, D. M. Ratner, S. J. Skerrett and P. S. Stayton, J. Controlled Release, 2018, 287, 1–11 CrossRef CAS PubMed.
- C. Yang, S. Krishnamurthy, J. Liu, S. Liu, X. Lu, D. J. Coady, W. Cheng, G. De Libero, A. Singhal, J. L. Hedrick and Y. Y. Yang, Adv. Healthcare Mater., 2016, 5, 1272–1281 CrossRef CAS PubMed.
- X. Yang, B. Xie, H. Peng, G. Shi, B. Sreenivas, J. Guo, C. Wang and Y. He, J. Controlled Release, 2021, 329, 454–467 CrossRef CAS PubMed.
- T. Wei, Q. Yu, W. Zhan and H. Chen, Adv. Healthcare Mater., 2016, 5, 449–456 CrossRef CAS PubMed.
- A. Grahnert, A. Grahnert, C. Klein, E. Schilling, J. Wehrhahn and S. Hauschildt, Innate Immun., 2011, 17, 212–233 CrossRef CAS PubMed.
- M. Ye, Y. Zhao, Y. Wang, R. Xie, Y. Tong, J.-D. Sauer and S. Gong, Nat. Nanotechnol., 2022, 17, 880–890 CrossRef CAS PubMed.
- F. Liu, S. Sheng, D. Shao, Y. Xiao, Y. Zhong, J. Zhou, C. H. Quek, Y. Wang, J. Dawulieti, C. Yang, H. Tian, X. Chen and K. W. Leong, Matter, 2021, 4, 3677–3695 CrossRef CAS.
- Y. Li, Y.-Z. Piao, H. Chen, K. Shi, J. Dai, S. Wang, T. Zhou, A.-T. Le, Y. Wang, F. Wu, R. Ma, L. Shi and Y. Liu, Bioact. Mater., 2023, 27, 288–302 CAS.
- S. Thamphiwatana, P. Angsantikul, T. Escajadillo, Q. Zhang, J. Olson, B. T. Luk, S. Zhang, R. H. Fang, W. Gao and V. Nizet, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 11488–11493 CrossRef CAS PubMed.
- K. M. Thomson, C. Dyer, F. Liu, K. Sands, E. Portal, M. J. Carvalho, M. Barrell, I. Boostrom, S. Dunachie and R. Farzana, Lancet Infect. Dis., 2021, 21, 1677–1688 CrossRef PubMed.
- K. Sands, M. J. Carvalho, E. Portal, K. Thomson, C. Dyer, C. Akpulu, R. Andrews, A. Ferreira, D. Gillespie and T. Hender, Nat. Microbiol., 2021, 6, 512–523 CrossRef CAS PubMed.
- G. Qing, X. Zhao, N. Gong, J. Chen, X. Li, Y. Gan, Y. Wang, Z. Zhang, Y. Zhang, W. Guo, Y. Luo and X. J. Liang, Nat. Commun., 2019, 10, 1–12 CrossRef CAS PubMed.
- Y. Huang, J. Ren and X. Qu, Chem. Rev., 2019, 119, 4357–4412 CrossRef CAS PubMed.
- L. Zhang, L. Zhang, H. Deng, H. Li, W. Tang, L. Guan, Y. Qiu, M. J. Donovan, Z. Chen and W. Tan, Nat. Commun., 2021, 12, 2002 CrossRef CAS PubMed.
- P. C. Naha, Y. Liu, G. Hwang, Y. Huang, S. Gubara, V. Jonnakuti, A. Simon-Soro, D. Kim, L. Gao, H. Koo and D. P. Cormode, ACS Nano, 2019, 13, 4960–4971 CrossRef CAS PubMed.
- F. Wu, J. Ma, Y. Wang, L. Xie, X. Yan, L. Shi, Y. Li and Y. Liu, ACS Nano, 2023, 17, 2980–2991 CrossRef CAS PubMed.
- L. Chen, M. Peng, J. Zhou, X. Hu, Y. Piao, H. Li, R. Hu, Y. Li, L. Shi and Y. Liu, Adv. Mater., 2023, 35, 2301664 CrossRef CAS PubMed.
- X. Du, M. Zhang, H. Zhou, W. Wang, C. Zhang, L. Zhang, Y. Qu, W. Li, X. Liu, M. Zhao, K. Tu and Y.-Q. Li, Research, 2022, 2022, 9767643 CAS.
- Y. Wang, J. Zhou, L. Yuan, F. Wu, L. Xie, X. Yan, H. Li, Y. Li, L. Shi, R. Hu and Y. Liu, Small, 2023, 19, 2206657 CrossRef CAS PubMed.
- M. Liang and X. Yan, Acc. Chem. Res., 2019, 52, 2190–2200 CrossRef CAS PubMed.
- L. Gao, K. Fan and X. Yan, Theranostics, 2017, 7, 3207–3227 CrossRef CAS PubMed.
- H. Ding, Y. Cai, L. Gao, M. Liang, B. Miao, H. Wu, Y. Liu, N. Xie, A. Tang, K. Fan, X. Yan and G. Nie, Nano Lett., 2019, 19, 203–209 CrossRef CAS PubMed.
- B. Xu, H. Wang, W. Wang, L. Gao, S. Li, X. Pan, H. Wang, H. Yang, X. Meng, Q. Wu, L. Zheng, S. Chen, X. Shi, K. Fan, X. Yan and H. Liu, Angew. Chem., Int. Ed., 2019, 58, 4911–4916 CrossRef CAS PubMed.
- P. Makvandi, C. Wang, E. N. Zare, A. Borzacchiello, L. Niu and F. R. Tay, Adv. Funct. Mater., 2020, 30, 1910021 CrossRef CAS.
- S. J. Lam, N. M. O’Brien-Simpson, N. Pantarat, A. Sulistio, E. H. H. Wong, Y.-Y. Chen, J. C. Lenzo, J. A. Holden, A. Blencowe, E. C. Reynolds and G. G. Qiao, Nat. Microbiol., 2016, 1, 16162 CrossRef CAS PubMed.
- J. Xie, M. Zhou, Y. Qian, Z. Cong, S. Chen, W. Zhang, W. Jiang, C. Dai, N. Shao, Z. Ji, J. Zou, X. Xiao, L. Liu, M. Chen, J. Li and R. Liu, Nat. Commun., 2021, 12, 5898 CrossRef CAS PubMed.
- Y. Jiang, W. Zheng, K. Tran, E. Kamilar, J. Bariwal, H. Ma and H. Liang, Nat. Commun., 2022, 13, 197 CrossRef CAS PubMed.
- X. Hu, Y. Li, Y. Piao, M. Karimi, Y. Wang, F. Wen, H. Li, L. Shi and Y. Liu, Adv. Mater., 2023, 35, 2301623 CrossRef CAS PubMed.
- Y. Qian, X. Hu, J. Wang, Y. Li, Y. Liu and L. Xie, Colloids Surf., B, 2023, 231, 113542 CrossRef CAS PubMed.
- J. C. Nwabuife, A. M. Pant and T. Govender, Adv. Drug Delivery Rev., 2021, 178, 113861 CrossRef CAS PubMed.
- Z. Zhai, W. Ouyang, Y. Yao, Y. Zhang, H. Zhang, F. Xu and C. Gao, Bioact. Mater., 2022, 14, 430–442 CAS.
- B. A. J. Hoffman, E. A. Pumford, A. I. Enueme, K. L. Fetah, O. M. Friedl and A. M. Kasko, Trends Biotechnol., 2023, 41, 1139–1154 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2023 |