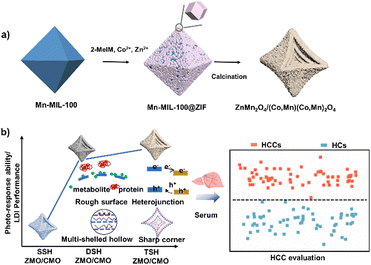
Hollow multishelled heterostructures with enhanced performance for laser desorption/ionization mass spectrometry based metabolic diagnosis†
Congcong
Pei‡
a,
Rui
Su‡
bc,
Songting
Lu‡
a,
Xiaonan
Chen
a,
Yajie
Ding
a,
Rongxin
Li
a,
Weikang
Shu
a,
Yu
Zeng
a,
Yingying
Lin
a,
Liang
Xu
b,
Yuqiang
Mi
*bc and
Jingjing
Wan
 *a
*a
aSchool of Chemistry and Molecular Engineering, East China Normal University, Shanghai, 200241, P. R. China. E-mail: jjwan@chem.ecnu.edu.cn
bTianjin Second People's Hospital, Tianjin Medical University, Tianjin 300192, China. E-mail: miyuqiang@tj.gov.cn
cTianjin Institute of Hepatology, Tianjin 300192, China
First published on 9th August 2023
Abstract
High-performance metabolic diagnosis-based laser desorption/ionization mass spectrometry (LDI-MS) improves the precision diagnosis of diseases and subsequent treatment. Inorganic matrices are promising for the detection of metabolites by LDI-MS, while the structure and component impacts of the matrices on the LDI process are still under investigation. Here, we designed a multiple-shelled ZnMn2O4/(Co, Mn)(Co, Mn)2O4 (ZMO/CMO) as the matrix from calcined MOF-on-MOF for detecting metabolites in LDI-MS and clarified the synergistic impacts of multiple-shells and the heterostructure on LDI efficiency. The ZMO/CMO heterostructure allowed 3–5 fold signal enhancement compared with ZMO and CMO with the same morphology. Furthermore, the ZMO/CMO heterostructure with a triple-shelled hollow structure displayed a 3-fold signal enhancement compared to its nanoparticle counterpart. Taken together, the triple-shelled hollow ZMO/CMO exhibits 102-fold signal enhancement compared to the commercial matrix products (e.g., DHB and DHAP), allowing for sensitive metabolic profiling in bio-detection. We directly extracted metabolic patterns by the optimized triple-shelled hollow ZMO/CMO particle-assisted LDI-MS within 1 s using 100 nL of serum and used machine learning as the readout to distinguish hepatocellular carcinoma from healthy controls with the area under the curve value of 0.984. Our approach guides us in matrix design for LDI-MS metabolic analysis and drives the development of a nanomaterial-based LDI-MS platform toward precision diagnosis.
1. Introduction
Metabolic profiling is more distal than proteomic and genomic analysis,1–3 and is thereby widely applied in molecular diagnosis for clinical practice.4–6 Mass spectrometry (MS), particularly laser desorption/ionization (LDI) MS with easy operation and high throughput, has attracted intense attention in metabolic-based diagnosis.7–9 Of note, the rational design of an effective matrix is essential for metabolic profiling using LDI-MS, due to extensive molecular fragmentation in the low mass range and decreased LDI efficiency arising from the conventional organic matrix.10 Various inorganic nanoparticles with light absorption and heat stability, such as noble metals,11 metal oxides,12 and carbon,13 have been customized as matrices for LDI-MS detection of metabolites, while the structure and component impacts of the matrices on the particle-assisted desorption/ionization process are still under investigation. The deep study revealing the key elements in matrix design could enhance the metabolic profiling capability of LDI-MS for clinical use.Global efforts,14–16 including ours, have been devoted to elucidating the relationship between the physical effects of an inorganic matrix and LDI efficiency. For example, the physical effect affecting photo-absorption, charge, and thermal transfer from matrices determines the ion-generation process in LDI-MS.17 Meanwhile, the physical effect of controllable surface roughness in the matrix selectively traps small-size molecules in the crevices,18 realizing an in situ large protein separation in serum for selective ionization. Our group recently reported a semiconductor metal oxide material that combined heterojunctions, hollow interiors, sharp corners, and surface roughness for LDI-MS detection.19 The four physical effects endow the matrix with excellent photo-response under laser irradiation and size-exclusive effect towards the metabolite simultaneously. Compared with hollow interiors, multi-shelled hollow structures display better performance in photocatalysis owing to the improved light absorption from the scattering and reflection effects.20,21 We anticipated that integrating the multi-shelled hollow structure into the semiconductor heterojunction structure would bring in further enhancement in LDI efficiency.
Various approaches have been established for multi-shelled hollow semiconductor heterojunction structures, such as soft-/hard-template and metal–organic framework (MOF)-derived methods.22–25 Due to the easy preparation of the MOF precursor, the MOF-derived method overcomes the shortcoming of uncontrolled dispersity and complicated procedures (e.g., etching) of soft- or hard-templating methods, thus considered an emerging strategy for nanomaterials with complex structures and tailored compositions.26–28 Of note, the new family of MOF-on-MOF precursor is conjugated with two or more distinct MOF units.29–31 A large amount of organic linker in the MOF-on-MOF precursor is decomposed and shrinks inward with calcination, and a small crystal nucleus on the outside will contract and grow with precursors. Whether the combustion rate matches the nucleation rate will determine the formation of multiple-shelled metal oxides. The competitive oxidation and different nucleation rates of pre-organized different MOFs during the heat treatment are expected to provide a delicate composition and structural control easily.32 Despite the high diversity of structure and composition, the development of MOF-on-MOF chemistry is still in its infancy, and the conversion of MOF-on-MOF hybrids via calcination to multi-shelled hollow metal oxide composite remains largely unexplored.29,33 Thus, the establishment of a MOF-on-MOF based strategy for a multiple-shelled semiconductor heterojunction provides a superior matrix for metabolic detection and deepens the understanding of MOF-derived nanomaterials.
Herein, we reported the synthesis of triple-shelled hollow ZnMn2O4/(Co, Mn)(Co, Mn)2O4 (TSH ZMO/CMO) concave octahedrons with improved LDI performance. TSH ZMO/CMO was prepared using a prearranged MOF-on-MOF hybrid, as illustrated in Scheme 1. It is demonstrated that the shell number of ZMO/CMO can be controlled by changing the heating rate of calcined MOF-on-MOF. The unique triple-shelled structure ZMO/CMO enabled greatly enhanced performance (3–5-fold signal enhancement compared to single-shelled ZMO/CMO, TSH ZMO, or TSH CMO, 102-fold signal enhancement compared to the commercial matrix products) for direct LDI-MS analysis of metabolites. For cancer detection, we demonstrated in vitro metabolic diagnosis of hepatocellular carcinoma patients using a serum with an area under the curve (AUC) value of 0.984. This work contributed to designing nanomaterial-based platforms for high-performance metabolic analysis and large-scale diagnostic use.
2. Materials and methods
2.1. Reagents
Mn(NO3)2·4H2O (98%, Adamas), Zn(NO3)2·6H2O (98%, Adamas), Co(NO3)2·6H2O (98%, Adamas), 2-methylimidazole (2-MeIM, Aldrich), lauric acid (99%+), trimesic acid (99%+, Adamas), ethanol (AR), methanol (AR), potassium chloride (99.5%), sodium chloride (99.5%), bovine serum albumin (BSA, 98%), L-alanine (Ala) (99%), L-valine (Val) (98%), L-lysine (Lys) (98%), D-mannitol (Man) (99%), D-aspartic acid (Asp) (99%), histidine (His) (99%), proline (Pro) (99%), glutamic acid (Glu) (99%), arginine (Arg) (99%), glucose (Gluc) (96%), juglone (97%, Adamas), rhodamine B (RhB) (BS, Adamas), verapamil hydrochloride (Ver), D-sucrose (Suc) (99%), L(+)-ascorbic acid (99%), 4-chloro-α-cyanocinnamic acid (Cl-CCA, 95.0%, Sigma Aldrich), α-cyano-4-hydroxycinnamic acid (CHCA, 99%), and 2,5-dihydroxybenzoic acid (DHB, 99%) obtained from Adamas Reagent Ltd and 2′,6′-dihydroxyacetophenone (DHAP, 98%) obtained from Accela were used. Millipore water (18.2 MΩ·cm) was used in all experiments.2.2. Characterization
Scanning electron microscopy (SEM) images were obtained from an S4800 field emission scanning electron microscope (Hitachi, Japan). Transmission electron microscopy (TEM) images were obtained with a JEM-2100F (JEOL, Japan) operating at 200 kV equipped with an X-ray energy dispersive spectrometer (EDX: X-Max 80T, Oxford, UK) for chemical composition analyses. X-ray diffraction (XRD) patterns were recorded using a Bruker D8 Advanced X-Ray diffractometer with Cu Kα radiation (λ = 0.154 nm). X-ray photoelectron spectroscopy (XPS) measurement was carried out on a Thermo ESCALAB 250 using an Al Kα radiation and C 1s (284.8 eV) as a reference to correct the binding energy. UV/vis diffuse reflectance spectra were collected on a Lambda950 spectrophotometer and BaSO4 was chosen as the reflectance standard. Nitrogen sorption–desorption isotherms were measured using an Autochem 2920 Micromeritics. The samples were degassed in a vacuum at 460 K for 12 h before analysis. The photocurrent was determined using a standard three-electrode system on a CHI 660E electrochemistry workstation (ChenHua Instruments Co., China). The platinum wire was chosen as a counter electrode, the material as the working electrode, Ag/AgCl as a reference electrode, and 0.5 M Na2SO4 as the electrolyte solution. Room temperature photoluminescence (PL) spectra were recorded on an Edinburgh FS5 spectrofluorometer in the 390–600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm range. Time-resolved photoluminescence (TRPL) spectra were recorded on an FSL980 transient fluorescence spectrometer.
nm range. Time-resolved photoluminescence (TRPL) spectra were recorded on an FSL980 transient fluorescence spectrometer.
2.3. Synthesis of Mn-MIL-100 and Mn-MIL-100/ZIF
To synthesize Mn-MIL-100, 0.240 mmol Mn(NO3)2·4H2O and 90 mg lauric acid were dissolved in 30 mL methanol containing 1.00 mmol trimesic acid by stirring for 15 min. The obtained solution was transferred into a Teflon-lined stainless steel autoclave and heated at 125 °C for 7 h. After cooling to 25 °C naturally, the product was collected by centrifugation, washed thrice with methanol, and dried overnight.To synthesize the Mn-MIL-100/ZIF, 5.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mg of Mn-MIL-100, 5
mg of Mn-MIL-100, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mL of 50
mL of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM 2-MeIM methanol solution, and 3
mM 2-MeIM methanol solution, and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mL of 25
mL of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM Zn(NO3)2·6H2O/Co(NO3)2·6H2O (Zn/Co molar ratio of 2/3) were mixed under stirring. The mixture was allowed to react at room temperature for 12 h. The final product was collected by centrifugation, washed with methanol three times, and dried for further use.
mM Zn(NO3)2·6H2O/Co(NO3)2·6H2O (Zn/Co molar ratio of 2/3) were mixed under stirring. The mixture was allowed to react at room temperature for 12 h. The final product was collected by centrifugation, washed with methanol three times, and dried for further use.
2.4. Synthesis of TSH ZMO/CMO
TSH ZMO/CMO was obtained by thermal treatment of as-synthesized Mn-MIL-100/ZIF at 500 °C in the air for 5 h (0.5 °C min−1). TSH ZMO and TSH CMO synthesis procedures are the same as that for composites except for the molar ratio of Zn and Co in the Mn-MIL-100/ZIF. The molar ratio of Zn![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Co = 0
Co = 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 affords TSH CMO. The molar ratio of Zn
25 affords TSH CMO. The molar ratio of Zn![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Co = 25
Co = 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 affords TSH ZMO. The shell number of ZMO/CMO can be controlled by changing the heating rate.
0 affords TSH ZMO. The shell number of ZMO/CMO can be controlled by changing the heating rate.
2.5. Bio-sample harvesting
All investigation protocols in this study were approved by the Second People's Hospital Ethics Committee Affiliated with Tianjin Medical University (Ethics No.: [2023] 10/[2021] 39). Written informed consent from patients has been obtained since the project started. Patients donated blood samples. All recruited patients were eligible and precisely diagnosed based on histological confirmation and laboratory and imaging findings. Hepatocellular carcinoma serum samples were collected from patients who were diagnosed. As previously reported,34 serum samples were prepared from the blood for LDI-MS analysis. The bio-samples were stored at −80 °C before use.2.6. Mass spectrometry analysis
For the matrix preparation, the matrices were dispersed in water at a concentration of 1 mg mL−1. The Cl-CCA (4.00 mg mL−1), CHCA (4.00 mg mL−1), DHB (10.00 mg mL−1), and DHAP (10.00 mg mL−1) were prepared by dissolving in the solvent (water/ACN/TFA, 70/30/0.1, v/v/v). Small molecule standards (Ala, Man, Asp, Lys, Val, Suc, RhB, and Ver) and serum were dissolved in deionized water. The salt and protein tolerance effects were analyzed by testing a mixture of standard molecules (Ala, Lys, Man, and Suc) within salts (0.50 M NaCl, 0.50 M KCl,) and proteins (BSA, 5.00 mg mL−1). A typical LDI-MS experiment mixed 1 μL analyte solution (small molecules/serum) with 1 μL matrix suspension for LDI-MS detection. LDI-MS was performed using an Autoflex MALDI-TOF/TOF (Bruker) equipped with a 355 nm Nd: YAG solid-state smart beam. The MS spectra were operated in the positive ion reflector mode, using delayed extraction with a repetition rate of 1 kHz, an acceleration voltage of 20 kV, a delay time of 150 ns, and 2000 per laser shots analysis for all LDI-MS detection. The program was pre-calibrated by standard molecules. Accurate mass measurements were employed for the peak assignments of typical metabolites. All spectra were directly used without any smoothing procedures.2.7. Theoretical simulation of the electromagnetic (EM) field enhancement
To compare the differences in the optical response of the synthesized nanoparticles, the finite element method (FEM) provided by the Wave Optical Module of Multiphysics software (COMSOL Inc., USA) was used to solve the Helmholtz equation about the time-harmonic electric field (E)35| ∇ × (μr−1∇ × E) − k02εrE = 0 | (1) |
2.8. Statistical analysis for diagnosis
Five independent spectra were recorded for each bio-sample for the serum metabolic patterns. Peak extraction, alignment, normalization, and mass spectra standardization were performed using Python's “home-built” code (version 3.7.0). For machine learning, one-way ANOVA analysis was conducted to compare the sex and age significance of hepatocellular carcinoma patients and healthy controls. The least absolute shrinkage and selection operator (Lasso) was performed using Python (version 3.7.0).36 Lasso represented the typical linear model defined as follows:| y = Xβ | (2) |
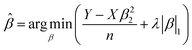 | (3) |
3. Results and discussion
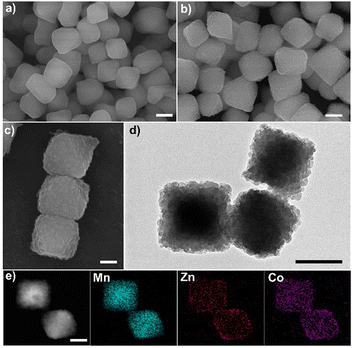
Mn-MIL-100 was first synthesized as a host MOF by a solvothermal method. The SEM image (Fig. 1a) shows that Mn-MIL-100 possessed a smooth surface and a solid octahedron morphology with an average length of ∼250 nm. The XRD pattern shows the high crystallinity of Mn-MIL-100 (Fig. S1, ESI†). After the growth of ZIF, solid octahedrons were found densely coated on the surfaces of Mn-MIL-100 (Fig. 1b). The formation process was divided into two steps, including etching and re-growth. After immersing Mn-MIL-100 in methanol solutions of Zn(NO3)2·6(H2O), Co(NO3)2·6H2O, and 2-MeIM, weakly alkaline 2-MeIM slightly etched Mn-MIL-100 and resulted in Mn2+ ion release. From Fig. S2 (ESI†), although the morphology of Mn-MIL-100 has changed slightly, the crystalline structure has not changed. Surplus 2-MeIM reacted with Mn2+, Zn2+, and Co2+ for 24 h leading to highly dispersed particles Mn-MIL-100@ZIF with a uniform octahedral and rough surface morphology (Fig. 1c). The STEM and line scan images (Fig. S3, ESI†) show the distribution of Mn, Zn, and Co elements in the ZIF of the outer layer, confirming the process of etching and re-growth. The successful growth of ZIF was further confirmed by Fourier transform infrared spectroscopy (FTIR) spectra, in which characteristic peaks at 1578 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1421 cm−1 (C–N), and 997 cm−1 (C–N) are observed (Fig. S4, ESI†). The TEM image (Fig. 1d) further displayed the solid nature of Mn-MIL-100@ZIF, in which core-satellite architecture is further evidenced. The high-angle annular dark-field scanning TEM (HAADF STEM) image and corresponding elemental mapping images (Fig. 1e) show the inner Mn core and external Mn, Zn, and Co satellite. XRD studies revealed the compositions of both MOFs in the Mn-MIL-100@ZIF heterostructure (Fig. S5, ESI†).
N), 1421 cm−1 (C–N), and 997 cm−1 (C–N) are observed (Fig. S4, ESI†). The TEM image (Fig. 1d) further displayed the solid nature of Mn-MIL-100@ZIF, in which core-satellite architecture is further evidenced. The high-angle annular dark-field scanning TEM (HAADF STEM) image and corresponding elemental mapping images (Fig. 1e) show the inner Mn core and external Mn, Zn, and Co satellite. XRD studies revealed the compositions of both MOFs in the Mn-MIL-100@ZIF heterostructure (Fig. S5, ESI†).
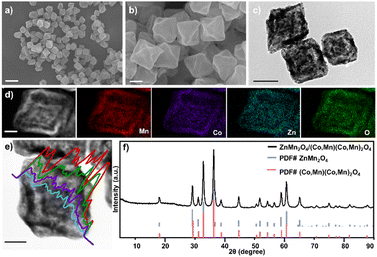
The synthesized Mn-MIL-100@ZIF hybrids were transformed to TSH ZMO/CMO via gradient heating calcination. The SEM image (Fig. 2a) shows highly dispersed particles with a uniform octahedral and concave morphology, indicating that the MOF-on-MOF-derived synthetic route is easy and capable of large-scale and quality-control matrix synthesis. Compared to the MOF precursors, the TSH ZMO/CMO particles that underwent a nanoscale combustion and contraction process had rougher surfaces and sharper corners derived from the concave surface and curly edges (average edge length of ∼300 nm, Fig. 2b). Well-defined triple-shelled structures can be observed in the TEM image (Fig. 2c). To further characterize the triple-shelled structure of ZMO/CMO, the material was embedded in epoxy resin and then cut into ultrathin slices by ultramicrotomy for electron microscopy analysis (Fig. S6, ESI†). The STEM image clearly shows the triple-shelled hollow structures. The size of the shells from the outermost to the innermost is 300 nm, 180 nm, and 90 nm, respectively. The corresponding elemental mapping images (Fig. 2d) and line scan spectrum (Fig. 2e) show that the Mn, Zn, Co, and O elements were evenly distributed in the TSH ZMO/CMO shells. On the other hand, selected area elemental analysis (Fig. S7, ESI†) showed that the molar ratios of Zn/Co/Mn are 9.34/11.34/14.52, 8.05/12.71/14.66 and 10.45/9.64/16.03 from the outermost to the innermost, further proving that TSH ZMO/CMO was evenly distributed in the shells from the outer to the inner. The XRD pattern of TSH ZMO/CMO (Fig. 2f) exhibits typical diffraction peaks for both ZMO and CMO, indicating the formation of the ZMO–CMO heterojunction. XPS (Fig. S8, ESI†) was carried out to investigate the surface chemical states of ZMO/CMO. The detection of Zn2+, Co2+, Co3+, Mn2+, Mn3+, and Mn4+ ions was consistent with metal oxide ZMO/CMO species, confirming the co-existence of ZMO and CMO in the composites.
As for TSH ZMO/CMO formation, whether the combustion rate matches the nucleation rate will determine the formation of multiple-shelled metal oxides. And this matching could be achieved by controlling the reaction temperature and the heating rate. In our work, the shell number of ZMO/CMO can be controlled by changing the heating rate. The single-shelled hollow structure (SSH), double-shelled hollow structure (DSH), and triple-shelled hollow (TSH) can be synthesized with a heating rate of 2.0 °C min−1, 1.0 °C min−1, and 0.5 °C min−1, respectively. From Fig. S9 (ESI†), we determined that all ZMO/CMO hollow octahedrons had a similar size. The ZMO/CMO crystalline structures of the prepared single-shelled hollow octahedrons and double-shelled hollow octahedrons were also proven by XRD (Fig. S9c, ESI†). The TSH ZMO and TSH CMO can be obtained by changing the MOF-on-MOF precursors. Therefore, multiple-shelled metal oxide with tunable compositions and shell numbers was successfully synthesized and can be utilized as a matrix for metabolic analysis.
Studying the shell numbers of the hollow structure is critical to understanding the fundamental properties of the matrix and designing the high-performance matrix. A crucial prerequisite is a facile approach for a multiple-shelled metal oxide heterojunction matrix. Metal oxides with multi-shelled hollow structures have been synthesized using soft-/hard-template methods. However, the synthetic process leads to morphology variation (Fig. S10a–f, ESI†), hampering the detection performance (3-fold lower compared to TSH ZMO/CMO) and reproducibility (the relative standard deviation (RSD) = 23.3% for similar metal oxides; RSD = 9.6% for TSH ZMO/CMO) (Fig. S10, ESI†). We also summarized the previous reports on the cost of raw materials, the synthesis cycle, and hazardous raw materials for synthesizing similar morphologies/compositions without the MOF template process (Table S1, ESI†). In contrast, we demonstrated a feasible strategy via simple calcination of the MOF template to form the nanoparticle not only with a flexible hollow structure but also with expected physical effects, paving the way for advanced matrix synthesis and large-scale clinical use.37 As a new family of MOF material, MOF-on-MOF holds excellent potential in efficiently fabricating a multishelled hollow metal oxide composite using competitive oxidation of multiple MOFs.38 In our study, TSH ZMO/CMO composite can be synthesized via simple calcination of solid Mn-MIL-100@ZIF, leading to uniform, well-dispersed, and stable metal oxide nanoparticles with heterojunctions, rough surface, and sharp corners. Furthermore, the shell number of ZMO/CMO can be controlled by changing the heating rate. The established MOF-on-MOF-based strategy paves the way for advanced matrix synthesis and drives us to understand the fundamental properties of the matrix. From the perspective of large-scale commercial applications, the TSH ZMO/CMO precursors are (€ 0.16 g−1, counts with Zn, Mn, and Co as the raw materials) much cheaper than the noble metal matrices (e.g., Au (€ 42.72 g−1) and Ag (€ 0.49 g−1)). Consequently, the TSH ZMO/CMO composites would advance high-performance applications due to easy operation and low costs, including but not limited to LDI-MS.
Favorable optical properties are among the preconditions for the MALDI matrix, which can promote subsequent energy transfer and charge transfer.39,40 It has been reported that complex multiple-shelled hollow structures show enhanced performance over conventional simple hollow structures owing to the improved light absorption properties from the scattering and reflection effects.41,42 The influence of shell number on the LDI performance of ZMO/CMO was further investigated. We prepared single-shelled hollow ZMO/CMO (SSH ZMO/CMO), double-shelled hollow ZMO/CMO (DSH ZMO/CMO), and TSH ZMO/CMO with the same component for performance comparison.
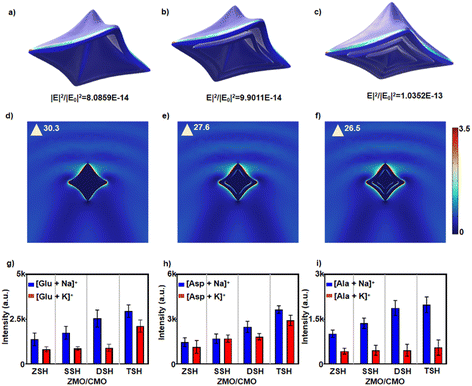
We stimulated the surface electric field enhancement using the finite element method to understand the light absorption of ZMO/CMO with different shell numbers. For the multiple-shelled hollow structure (Fig. 3a), the TSH structure exhibited a higher total relative enhancement of 1.0352 × 10−13 in the EM field, higher than those in SSH (8.0859 × 10−14) and DSH (9.9011 × 10−14) structures. The triple-shelled hollow (TSH) and double-shelled hollow (DSH) structures displayed a 27% and 22.5% increase in field enhancement compared with the single-shelled hollow (SSH). The TSH structure had been slightly enhanced, possibly due to the limited multiple light reflections in the interior cavity of the small-size hollow nanoparticle. The EM field enhancement of the matrix structure correlates with its capability to trap light (e.g., scattering and reflection) for affecting the LDI efficiency.35 The TSH structure afforded a higher scattering/reflection effect domain for enhancing the EM field and facilitating the LDI process when used as the matrix compared with the SSH and DSH. The multiple-shelled hollow structure exhibited a larger local electric field enhancement (up to 26.5–30.3) in the two-dimensional cross-section compared to the reported nanosphere (1.66) and nanocube (16.9) structures (Fig. 3d–f).19 This result suggested that it not only facilitated the LDI performance of the metabolites adsorbed on itself but also benefited the metabolites adsorbed on the adjacent nanoparticles.
To further understand the light trapping of multi-shelled hollow ZMO/CMO, We ground the triple-shelled hollow ZMO/CMO (TSH ZMO/CMO) into particles without the hollow structure (ZSH-ZMO/CMO). We used an ultraviolet-visible (UV-Vis) spectrophotometer to study the absorption spectra. As shown in Fig. S11 (ESI†), TSH ZMO/CMO exhibited enhanced absorption compared to ZSH-ZMO/CMO in UV-Vis spectroscopy and the absorption increased with the increase of the shell number, signifying that TSH ZMO/CMO increased the light trapping by the multi-shelled hollow morphology. We compared the specific surface areas of ZSH-ZMO/CMO nanoparticle with those of TSH ZMO/CMO. The N2 adsorption isotherms of ZSH ZMO/CMO and TSH ZMO/CMO are recorded. The Brunauer–Emmett–Teller (BET) specific surface areas are calculated to be 46.244 m2 g−1 and 45.507 m2 g−1, respectively. The ZSH ZMO/CMO nanoparticle shows similar BET specific surface areas to TSH ZMO/CMO. (Fig. S12, ESI†). We tested four analytes (Glu, Ala, Asp, and Arg) using ZSH, SSH, DSH, and TSH ZMO/CMO as the matrices, respectively. As shown in Fig. 3g–i and Fig. S13 (ESI†), TSH ZMO/CMO displayed nearly 3-fold mean signal intensities than ZSH-ZMO/CMO and 2-fold than SSH ZMO/CMO, confirming the advantage of the multiple-shelled hollow structure of ZMO/CMO.
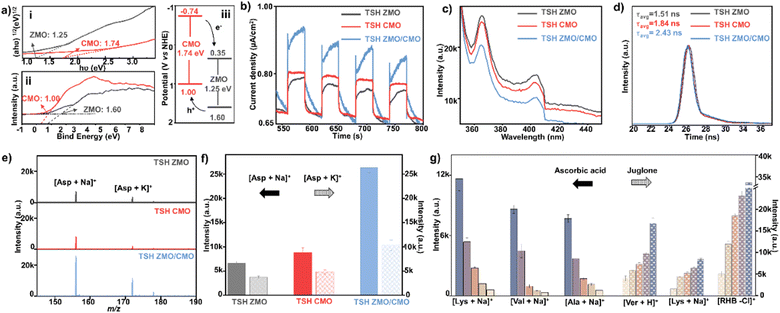
The excellent charge transfer ability that roots in good photo-response properties of the obtained heterostructure samples was investigated in comparison to TSH ZMO (Fig. S14a and b, ESI†) and TSH CMO (Fig. S14c and d, ESI†) with the same morphology. First, UV-vis DRS was used to explore the optical absorption properties. As shown in Fig. S15 (ESI†), TSH ZMO and TSH CMO exhibit light absorption in the region of 355 nm, which is consistent with the laser wavelength. The bandgaps of TSH ZMO and TSH CMO (Fig. 4a(i)) were calculated to be 1.25 and 1.74 eV, respectively. In addition, the valence band values of TSH ZMO and TSH CMO were measured to be 1.60 and 1.00 by the valence band (VB)-XPS (Fig. 4a(ii)). The bandgaps derived from UV-vis DRS calculated the conduction band values of TSH ZMO and TSH CMO as 0.35 and −0.74 eV, respectively. For the TSH ZMO/CMO heterojunction with a staggered band structure, there are heterojunction type II. The photogenerated electrons can migrate from TSH CMO to TSH ZMO while the holes transfer in the opposite direction, suggesting that a type-II heterojunction may improve electron–hole separation.
To explore the effect of the heterojunction on charge separation efficiency, photocurrent response, photoluminescence (PL) emission spectroscopy, and time-resolved photoluminescence (TRPL) spectroscopy analyses were performed. Among all samples, TSH ZMO/CMO displayed the strongest photocurrent intensity compared to the single TSH ZMO and TSH CMO (Fig. 4b), initially indicating the enhanced electron–hole separation. TRPL spectra further surveyed the photogenerated charge carrier transfer dynamics. The average PL lifetime (τavg) of TSH ZMO/CMO was calculated to be 2.43 ns, longer than TSH ZMO (1.51 ns) and TSH CMO (1.84 ns) (Fig. 4d), suggesting longer lived electron–hole pairs in TSH ZMO/CMO. The TSH ZMO/CMO exhibited the weakest PL emission peak intensity, further evidencing charge transferability (Fig. 4c). The separated electron–hole in TSH ZMO/CMO was anticipated to facilitate ionization by enhanced charge transfer in analytes. We observed the intensity enhancement of ∼3–5 folds for the sodium adduction peaks in detecting Asp, His, and Pro using the TSH ZMO/CMO matrix (Fig. 4e, f, and Fig. S16, ESI†). To verify the effect of the charge separation on TSH ZMO/CMO, juglone and ascorbic acid (AA) were chosen as electron and hole scavengers in the positive ion model, respectively. From Fig. 4g, the addition of the juglone will improve the performance of the matrix in detecting metabolites, in contrast to the addition of AA. These results demonstrated the necessity of holes in the charge transfer process in the positive ion model since the analytes had to be protonated to form cations. Therefore, the enhanced electron–hole separation in TSH ZMO/CMO can produce more active holes for charger transfer in LDI-MS detection. The TSH ZMO/CMO matrix enabled the metabolic detection with desired performance.
With enhanced photo-absorption and charge transfer, we further verified the sensitivity and selectivity of TSH ZMO/CMO. The concentration effect of the matrix on the sensitivity of small analytes was further investigated (Fig. S17, ESI†), and the composite with a concentration of 1 mg mL−1 displayed the best performance in detecting standard metabolites. We evaluated the limit of detection (LOD). The LOD of TSH ZMO/CMO was lower, by more than an order of magnitude (0.6 μmol L−1–2.5 μmol L−1 for creatinine, Gluc, and Asp), than in single composition of TSH ZMO, TSH CMO, SSH ZMO/CMO, and DSH ZMO/CMO (Table S2, ESI†). These results indicated that enhanced photo-response and electron–hole separation in the TSH ZMO/CMO could improve detection sensitivity and IN VIVO metabolite detection. The lowered LOD would aid the discovery of the low-abundance metabolites. The detection performance of TSH ZMO/CMO was further evaluated compared with four commonly used organic matrices, including CHCA, Cl-CCA, DHB, and DHAP.
TSH ZMO/CMO led to the enhancement of ∼9–102-fold compared with organic matrices for LDI-MS detection (Fig. S18, including Val, Lys, and Man, ESI†). Fig. S19 (ESI†) shows that the matrix-sample co-crystallization using TSH ZMO/CMO had a uniform surface roughness compared with CHCA, Cl-CCA, DHB, and DHAP.
Given that metabolite abundance and sample complexity affect MS analysis, pre-treatment steps are required to enrich and separate metabolites from complex body fluids. The nanoscale surface roughness can enrich small-sized metabolites in the nanogaps for in situ protein separation in bio-samples, achieving selective LDI-MS towards metabolites. Experimentally, in the presence of high salt (0.5 M KCl or 0.5M NaCl) and protein (5.00 mg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mL−1 BSA) concentrations, TSH ZMO/CMO with surface roughness successfully detected the signal of the metabolite mixture with low signal interference and high signal intensity (Fig. S20, ESI†). The selectivity of TSH ZMO/CMO has been further demonstrated for serum metabolic fingerprints (SMFs) direct acquisition from the interference of proteins and salts in serum with minimum sample treatment, shown in Fig. S21 (ESI†). We also included a standard sample during the fingerprinting process to validate the selectivity and signal reproducibility. In detail, a mixture of 4 analytes was loaded in serum for validation (5 replicates per sample), including Man, Ala, Lys, and Suc. The intra-batch and inter-batch coefficient of variance for the intensity of molecule peaks distributed at 5.4–20.1% (Fig. S22, ESI†), demonstrating a desirable signal reproducibility of TSH ZMO/CMO.
mL−1 BSA) concentrations, TSH ZMO/CMO with surface roughness successfully detected the signal of the metabolite mixture with low signal interference and high signal intensity (Fig. S20, ESI†). The selectivity of TSH ZMO/CMO has been further demonstrated for serum metabolic fingerprints (SMFs) direct acquisition from the interference of proteins and salts in serum with minimum sample treatment, shown in Fig. S21 (ESI†). We also included a standard sample during the fingerprinting process to validate the selectivity and signal reproducibility. In detail, a mixture of 4 analytes was loaded in serum for validation (5 replicates per sample), including Man, Ala, Lys, and Suc. The intra-batch and inter-batch coefficient of variance for the intensity of molecule peaks distributed at 5.4–20.1% (Fig. S22, ESI†), demonstrating a desirable signal reproducibility of TSH ZMO/CMO.
Favorable optical properties are among the preconditions for the MALDI matrix. The thin-shelled topologies in the hollow structures exhibit enhanced photo-response, which improves the performance of LDI-MS. Investigating the shell numbers of hollow structures is crucial for understanding the fundamental properties in matrix design, yet none of the studies have been reported in LDI. We established a simple MOF-based strategy to regulate the shell number of ZMO/CMO. The triple-shelled ZMO/CMO structure exhibited enhanced photo-response, with a ∼27% increase in the signal compared to its single-shelled counterparts, resulting in a surprising ∼3-fold signal enhancement. On further integration with other physical effects (surface roughness, sharp corners, and heterojunction), the optimized TSH ZMO/CMO can enhance the signal by up to 9–102-fold compared to commercial matrices. Therefore, well-designed inorganic matrices are required for understanding the mechanism of LDI-MS for small metabolite detection, which are also expected to offer future guidance for the matrix design for intensive MS detection.
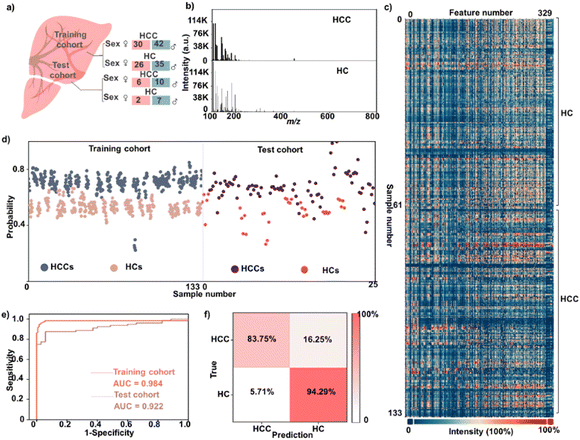
We differentiated 72 hepatocellular carcinoma patients (HCCs) from 61 healthy controls (HCs) by TSH ZMO/CMO metabolic analysis of serum (Fig. 5a), and there was no significant difference in age and gender distribution between the two cohorts (P > 0.05, Table S3). With no time-consuming pre-treatments, we directly recorded the metabolic m/z signals of HCCs and HCs in the low mass range (m/z of 100–800 Da) using TSH ZMO/CMO. The typical LDI-MS spectra of the HCC patient and HC are shown in Fig. 5b, in which multiple peaks with distinct differences can be observed in both cases. After data pre-treatment, we extracted serum metabolic fingerprints (SMFs) from the above HCs and HCCs from each serum sample (No. 1-61 for HCs and No. 62-133 for HCCs, Fig. 5c), serving as the database for building a diagnostic model. The slight difference can be directly observed in the two groups' blueprints of serum metabolic patterns, indicative of the necessity of efficient big data processing.
To better elucidate these data, the distinction between HCCs and HCs was performed based on machine learning of the obtained metabolic fingerprints. The LASSO (a method that involves only a subset of m/z features) constructed the sparse classification model based on SMFs. The training cohort included 72 HCCs and 61 HCs for model building. Fig. 5d showed the stratification of HCCs and HCs in the training cohort from LDI-MS data. A clear separation of these two groups can be observed, suggesting the apparent metabolic fluctuations during the disease progression. A receiver operating characteristic (ROC) curve was plotted to demonstrate the sparse learning performance (Fig. 5e), yielding an averaged AUC of 0.984 with 95% confidence interval (CI) from 0.971 to 0.997. We then obtained consistent results in the test cohort (9 HCs and 17 HCCs) with an AUC value of 0.922 for the metabolic patterns (95% CI of 0.854 to 0.990), validating the advanced diagnostic power of metabolic patterns based on the TSH ZMO/CMO for distinguishing between HCCs and HCs. The performance of the sparse classification model was further tested using a confusion matrix, which obtained an accuracy of 85.5% between HCCs and HCs (Fig. 5f). As a result, the high-performance metabolic patterns based on TSH ZMO/CMO allowed for diagnosis with HCCs.
HCC is the third most common cause of cancer-related death worldwide, causing approximately 7 million deaths worldwide. According to epidemiological investigations, the 5 year relative survival rate of HCC is only about 18.4%.43,44 Current diagnostic methods of ultrasound and α-fetoprotein (AFP) are expensive and lack accuracy in tumor detection.45 The sensitivity of AFP is limited to 65% for clinical HCC diagnosis and <40% for preclinical prediction.44 Metabolic profiles, which are affected by many physiological and pathological processes, can provide a solution to cancer precision diagnosis. Liquid chromatography-mass spectrometry (LC-MS)-based metabolomics is a powerful tool for diagnosing HCC.46–48 However, LC-MS-based metabolic analysis usually requires pre-treatment to remove the proteins and salts.49 The tedious sample pre-treatments (at least 1–3 hours for each sample) are indispensable. These time-consuming procedures may cause sample information loss and high overhead, hindering their clinical application. By comparison, our approach directly profiled serum metabolites (∼0.1 μL) with enhanced LDI efficacy in seconds, exhibiting high efficiency and feasibility for real clinic use. The average time spent on all steps before MS analysis is calculated to be ∼30 s, based on a high-throughput analysis (i.e., a batch of 384 samples). We reported the TSH ZMO/CMO matrix for LDI-MS-based serum metabolic patterns analysis toward HCC detection, having high accuracy with an AUC value of 0.984. Therefore, our diagnostic protocol is high throughput, simple, accurate, and cost-effective for HCC detection.
4. Conclusions
In conclusion, the TSH ZMO/CMO heterostructure with pre-designed physiochemical parameters was chosen as the LDI-MS matrix to afford efficient ionization of LDI-MS for detecting metabolites. The triple-shelled ZMO/CMO structure exhibited enhanced photo-response ability, resulting in a surprising signal enhancement for detecting metabolites in LDI-MS. Serum metabolic fingerprints directly extracted from TSH ZMO/CMO allowed for the high throughput discrimination of HCCs from HCs. There are still several limitations of our study. First, further regulating the matrix's single physiochemical effect is required to reveal the mass spectrometry ionization mechanism. Second, an elaborated cohort design and increased sample size are necessary to draw more in-depth conclusions. Our work makes solid contributions to designing nano-material-based LDI-MS platforms, accelerating advanced metabolic analysis toward medicine in the near future.Author contributions
J. W. and Y. M. foresaw this work and designed the overall approach. C. P., R. S., and S. L. carried out the experiments and contributed equally to this work. C. P. and J. W. wrote the manuscript. S. L., X. C., Y. D., W. S., R. L., Y. L, and Y. Z. contributed to the characterization and synthesis of the materials and analyzed the data. R. S., Y. M., and L. X. helped with the clinical sample collection and preliminary sample analysis. All authors joined in the critical discussion and edited the manuscript.Conflicts of interest
The authors declare competing financial interests and have filed patents for this approach.Acknowledgements
The authors gratefully acknowledge the financial support from Project 22074044 and 22122404 of the National Natural Science Foundation of China (NSFC) and Project KF2105 by the State Key Laboratory of Oncogenes and Related Genes, Project TJYXZDXK-059B of the Tianjin Key Medical Discipline (Specialty) Construction Project, Project TJWJ2022XK034 of the Tianjin Health Science and Technology Project Key Discipline Special and Project 2021022 of the Research Project of Chinese Traditional Medicine and Chinese Traditional Medicine Combined With Western Medicine of Tianjin Municipal Health Commission.Notes and references
- C. Guijas, J. R. Montenegro-Burke, B. Warth, M. E. Spilker and G. Siuzdak, Nat. Biotechnol., 2018, 36, 316–320 CrossRef CAS PubMed.
- D. S. Wishart, Nat. Rev. Drug Discovery, 2016, 15, 473–484 CrossRef CAS PubMed.
- A. J. Grooms, B. J. Burris and A. K. Badu-Tawiah, Mass Spectrom. Rev., 2022, e21826, DOI:10.1002/mas.21826.
- H. Chen, C. Huang, Y. Wu, N. Sun and C. Deng, ACS Nano, 2022, 16, 12952–12963 CrossRef CAS PubMed.
- J. Yang, X. Yin, L. Zhang, X. Zhang, Y. Lin, L. Zhuang, W. Liu, R. Zhang, X. Yan, L. Shi, W. Di, L. Feng, Y. Jia, J. Wang, K. Qian and X. Yao, Adv. Mater., 2022, 34, e2201422 CrossRef PubMed.
- V. Ruiz-Rodado, A. Lita and M. Larion, Nat. Methods, 2022, 19, 1048–1063 CrossRef CAS PubMed.
- L. Huang, J. Wan, X. Wei, Y. Liu, J. Huang, X. Sun, R. Zhang, D. D. Gurav, V. Vedarethinam, Y. Li, R. Chen and K. Qian, Nat. Commun., 2017, 8, 220 CrossRef PubMed.
- J. Wu, M. Xu, W. Liu, Y. Huang, R. Wang, W. Chen, L. Feng, N. Liu, X. Sun, M. Zhou and K. Qian, Small Methods, 2022, 6, e2200264 CrossRef PubMed.
- J. Su, J. Cao, H. Yang, W. Xu, W. Liu, R. Wang, Y. Huang, J. Wu, X. Gao, R. Weng, J. Pu, N. Liu, Y. Gu, K. Qian and W. Ni, Small Methods, 2023, 7, e2201486 CrossRef PubMed.
- Y. Ding, C. Pei, W. Shu and J. Wan, Chem. – Asian J., 2022, 17, e202101310 CAS.
- A. Palermo, E. M. Forsberg, B. Warth, A. E. Aisporna, E. Billings, E. Kuang, H. P. Benton, D. Berry and G. Siuzdak, ACS Nano, 2018, 12, 6938–6948 CrossRef CAS PubMed.
- Z. Li, Y. W. Zhang, Y. L. Xin, Y. Bai, H. H. Zhou and H. W. Liu, Chem. Commun., 2014, 50, 15397–15399 RSC.
- Q. Min, X. Zhang, X. Chen, S. Li and J. J. Zhu, Anal. Chem., 2014, 86, 9122–9130 CrossRef CAS PubMed.
- V. Vedarethinam, L. Huang, M. Zhang, H. Su, H. Hu, H. Xia, Y. Liu, B. Wu, X. Wan, J. Shen, L. Xu, W. Liu, J. Ma and K. Qian, Adv. Funct. Mater., 2020, 30, 2002791 CrossRef CAS.
- X. Li, A. S. Kulkarni, X. Liu, W. Q. Gao, L. Huang, Z. Hu and K. Qian, Small Methods, 2021, 5, e2001001 CrossRef PubMed.
- R. Li, Y. Zhou, C. Liu, C. Pei, W. Shu, C. Zhang, L. Liu, L. Zhou and J. Wan, Angew. Chem., Int. Ed., 2021, 60, 12504–12512 CrossRef CAS PubMed.
- A. S. Kulkarni, L. Huang and K. Qian, J. Mater. Chem. B, 2021, 9, 3622–3639 RSC.
- H. Su, X. Li, L. Huang, J. Cao, M. Zhang, V. Vedarethinam, W. Di, Z. Hu and K. Qian, Adv. Mater., 2021, 33, e2007978 CrossRef PubMed.
- C. Pei, Y. Wang, Y. Ding, R. Li, W. Shu, Y. Zeng, X. Yin and J. Wan, Adv. Mater., 2023, 35, 2209083 CrossRef CAS PubMed.
- J. Wang, N. Yang, H. Tang, Z. Dong, Q. Jin, M. Yang, D. Kisailus, H. Zhao, Z. Tang and D. Wang, Angew. Chem., Int. Ed., 2013, 52, 6417–6420 CrossRef CAS PubMed.
- C. Jiao, Z. Wang, X. Zhao, H. Wang, J. Wang, R. Yu and D. Wang, Angew. Chem., Int. Ed., 2019, 58, 996–1001 CrossRef CAS PubMed.
- L. Zhou, Z. Zhuang, H. Zhao, M. Lin, D. Zhao and L. Mai, Adv. Mater., 2017, 29, 1602914 CrossRef PubMed.
- J. Wang, Y. Cui and D. Wang, Adv. Mater., 2019, 31, e1801993 CrossRef PubMed.
- H. Ren and R. Yu, Inorg. Chem. Front., 2019, 6, 2239–2259 RSC.
- L. Zong, Z. Wang and R. Yu, Small, 2019, 15, e1804510 CrossRef PubMed.
- K. Sun, Y. Qian and H. L. Jiang, Angew. Chem., Int. Ed., 2023, e202217565, DOI:10.1002/anie.202217565.
- Y. Xue, S. Zheng, H. Xue and H. Pang, J. Mater. Chem. A, 2019, 7, 7301–7327 RSC.
- Q. Kong, L. Zhang, M. Wang, M. Li, H. Yao and J. Shi, Sci. Bull., 2016, 61, 1195–1201 CrossRef CAS.
- C. Liu, J. Wang, J. Wan and C. Yu, Coord. Chem. Rev., 2021, 432, 213743 CrossRef CAS.
- L. Yuan, C. Zhang, Y. Zou, T. Bao, J. Wang, C. Tang, A. Du, C. Yu and C. Liu, Adv. Funct. Mater., 2023, 33, 2214627 CrossRef CAS.
- C. Liu, L. Lin, Q. Sun, J. Wang, R. Huang, W. Chen, S. Li, J. Wan, J. Zou and C. Yu, Chem. Sci., 2020, 11, 3680–3686 RSC.
- L. Chai, J. Pan, Y. Hu, J. Qian and M. Hong, Small, 2021, 17, e2100607 CrossRef PubMed.
- T. Bao, Y. Xia, J. Lu, C. Zhang, J. Wang, L. Yuan, Y. Zhang, C. Liu and C. Yu, Small, 2022, 18, e2103106 CrossRef PubMed.
- W. Shu, M. Zhang, C. Zhang, R. Li, C. Pei, Y. Zeng, L. Zhao, J. Zhao and J. Wan, Adv. Funct. Mater., 2022, 33, 2210267 CrossRef.
- X. Yin, J. Yang, M. Zhang, X. Wang, W. Xu, C. H. Price, L. Huang, W. Liu, H. Su, W. Wang, H. Chen, G. Hou, M. Walker, Y. Zhou, Z. Shen, J. Liu, K. Qian and W. Di, ACS Nano, 2022, 16, 2852–2865 CrossRef CAS PubMed.
- J. Yang, R. Wang, L. Huang, M. Zhang, J. Niu, C. Bao, N. Shen, M. Dai, Q. Guo, Q. Wang, Q. Wang, Q. Fu and K. Qian, Angew. Chem., Int. Ed., 2020, 59, 1703–1710 CrossRef CAS PubMed.
- X.-C. Xie, K.-J. Huang and X. Wu, J. Mater. Chem. A, 2018, 6, 6754–6771 RSC.
- J. Shi, J. Xiong, L. Qiao, C. Liu and Y. Zeng, Appl. Surf. Sci., 2023, 609, 155271 CrossRef CAS.
- R. Li, Y. Zhou, C. Liu, C. Pei, W. Shu, C. Zhang, L. Liu, L. Zhou and J. Wan, Angew. Chem., Int. Ed., 2021, 60, 12504–12512 CrossRef CAS PubMed.
- Y. Wang, W. Shu, S. Lin, J. Wu, M. Jiang, S. Li, C. Liu, R. Li, C. Pei, Y. Ding, J. Wan and W. Di, Small, 2022, 18, e2106412 CrossRef PubMed.
- W. Liu, J. Huang, Q. Yang, S. Wang, X. Sun, W. Zhang, J. Liu and F. Huo, Angew. Chem., Int. Ed., 2017, 56, 5631 CrossRef CAS.
- L. Li, X. Dai, D. L. Chen, Y. Zeng, Y. Hu and X. W. D. Lou, Angew. Chem., Int. Ed., 2022, 61, e202205839 CrossRef CAS PubMed.
- Q. Huang, Y. Tan, P. Yin, G. Ye, P. Gao, X. Lu, H. Wang and G. Xu, Cancer Res., 2013, 73, 4992–5002 CrossRef CAS PubMed.
- P. Luo, P. Yin, R. Hua, Y. Tan, Z. Li, G. Qiu, Z. Yin, X. Xie, X. Wang, W. Chen, L. Zhou, X. Wang, Y. Li, H. Chen, L. Gao, X. Lu, T. Wu, H. Wang, J. Niu and G. Xu, Hepatology, 2018, 67, 662–675 CrossRef CAS PubMed.
- X. Wang, A. Zhang and H. Sun, Hepatology, 2013, 57, 2072–2077 CrossRef CAS PubMed.
- H. Yin, J. Zhu, M. Wang, Z. P. Yao and D. M. Lubman, Anal. Chem., 2020, 92, 8201–8208 CrossRef CAS PubMed.
- H. Y. Pan, Q. Q. Wu, Q. Q. Yin, Y. N. Dai, Y. C. Huang, W. Zheng, T. C. Hui, M. J. Chen, M. S. Wang, J. J. Zhang, H. J. Huang and Y. X. Tong, ACS Omega, 2021, 6, 1160–1170 CrossRef CAS PubMed.
- Y. C. Yang, M. H. Chien, T. C. Lai, M. C. Tung, Y. H. Jan, W. M. Chang, S. M. Jung, M. H. Chen, C. N. Yeh and M. Hsiao, J. Biomed. Sci., 2021, 28, 29 CrossRef CAS PubMed.
- S. W. Hyung, S. Lee, J. Han, J. Lee, S. Y. Beak, B. Kim, K. Choi and S. Ahn, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2021, 1179, 122725 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb00766a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |