Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Complementary charge-driven encapsulation of functional protein by engineered protein cages in cellulo†
Daniel
Zakaszewski
 ab,
Lukasz
Koziej
a,
Jędrzej
Pankowski
ab,
Lukasz
Koziej
a,
Jędrzej
Pankowski
 ac,
V. Vishal
Malolan
ab,
Nina
Gämperli
ac,
V. Vishal
Malolan
ab,
Nina
Gämperli
 d,
Jonathan G.
Heddle
d,
Jonathan G.
Heddle
 a,
Donald
Hilvert
a,
Donald
Hilvert
 d and
Yusuke
Azuma
d and
Yusuke
Azuma
 *ad
*ad
aMalopolska Centre of Biotechnology (MCB), Jagiellonian University, Gronostajowa 7A, 30387 Krakow, Poland. E-mail: yusuke.azuma@uj.edu.pl
bDoctoral School of Exact and Natural Sciences, Jagiellonian University, Prof. S. Łojasiewicza 11, 30348 Krakow, Poland
cFaculty of Biochemistry, Biophysics, sand Biotechnology, Jagiellonian University, Gronostajowa 7, 30387 Krakow, Poland
dLaboratory of Organic Chemistry, ETH Zurich, 8093 Zurich, Switzerland
First published on 10th July 2023
Abstract
Charge-driven inclusion complex formation in live cells was examined using a degradation-prone fluorescent protein and a series of protein cages. The results show that sufficiently strong host–guest ionic interaction and an intact shell-like structure are crucial for the protective guest encapsulation.
Compartmentalization is a fundamental feature underlying complex cellular systems. Such spatial separation, typically through physical barriers, enables otherwise incompatible biochemical processes to occur in parallel. While eukaryotic membrane-bound organelles remain the most prominent representatives of this concept, compartmentalization can also be achieved with proteins that self-assemble into regular, cage-like arrangements.1,2 Such proteinaceous compartments are ubiquitous throughout all domains of life and play a role in many crucial biological processes, including mineralization (ferritins),3 proteostasis (molecular chaperones and proteasomes),4,5 as well as harbouring short metabolic pathways (bacterial microcompartments).6 The broad range of morphologies and functionalities exhibited by naturally occurring protein cages has inspired researchers to reengineer these structures into customized organelle-like subcellular compartments with potential applications in biomedicine, metabolic engineering, and synthetic biology.2
Many functions of protein cages rely on their ability to sequester cargo molecules within their lumenal cavity. This is typically achieved by employing specific binding strategies, including charge complementarity,7–9 bioaffinity,10–12 genetic fusion,13–15 or bioconjugation.16–18 Charge complementarity, in particular, offers a simple and general approach for encapsulation: negatively charged guests bind to a positively charged cage lumen and vice versa. Notable examples found in nature are viral capsids that display arginine-rich motifs on their lumenal surface to recognize and package their genomic material.19 To facilitate complex formation between two naturally unrelated molecules, such a propensity for ionic interaction-based encapsulation can also be introduced by engineering. This strategy has been successfully applied to the cage-forming enzyme, Aquifex aeolicus lumazine synthase (AaLS) (Fig. 1(a)). The lumenal surface of the wild-type protein (AaLS-wt) assembly was endowed with a high negative charge by site-directed mutagenesis23 and subsequently optimized by directed evolution,24 yielding variants called AaLS-neg and AaLS-13, respectively. While AaLS-wt assembles into a 16 nm dodecahedral structure, the engineered variants adopt expanded cage-like assemblies with keyhole-shaped pores (Fig. 1(a)).25 Their structural characteristics allow efficient loading of positively charged cargo simply by mixing the guests and host cages in aqueous solution.26–29 However, most experiments to date have been performed in vitro, leaving questions about the utility of these complementary charge-based packaging systems for forming inclusion complexes in living cells. Here we describe the possibility of charge complementarity-based formation of a protein cage–cargo complex, and how these systems can rescue proteins that are rapidly degraded in the bacterial cytosol.
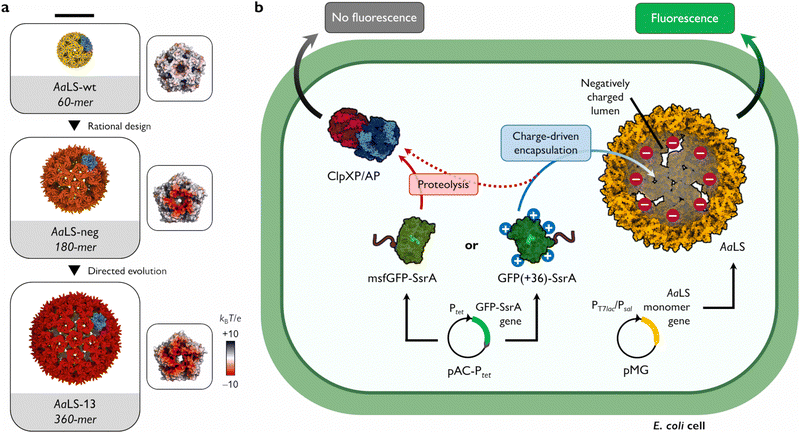 | ||
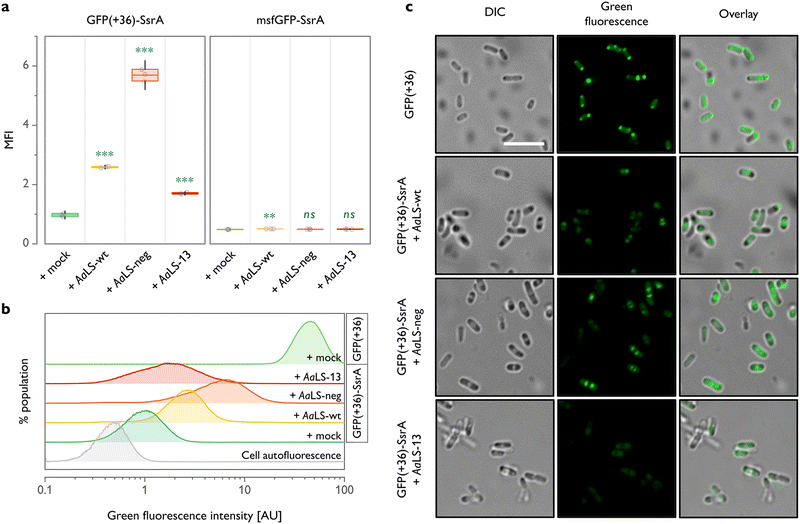
| Fig. 1 Shield-or-shred system: in cellulo ionic interaction-based guest encapsulation of otherwise degradation-prone fluorescent protein by engineered Aquifex aeolicus lumazine synthase (AaLS) protein cages. (a) Structures of AaLS-wt 60-mer (PDB: 5MPP), AaLS-neg 180-mer (5MQ3), AaLS-13 360-mer (5MQ7). Scale bar – 20 nm. Electrostatic potential molecular surfaces (Connolly representation, upscaled 3×) of the pentameric subassemblies, projected perpendicularly to the five-fold symmetry axis, viewed from the lumenal face, are shown next to the respective cages.20–22 (b) Conceptual scheme of the in cellulo guest encapsulation experiments. A GFP fused to SsrA degron peptide, either non-supercharged (msfGFP-SsrA) or positively supercharged (GFP(+36)-SsrA), is coproduced in E. coli cells with a selected protein cage and its fluorescence is analysed. Models are not to scale. | ||
As a cargo molecule we chose a positively supercharged variant of green fluorescent protein30 equipped with a C-terminal SsrA bacterial degradation signal peptide31 (GFP(+36)-SsrA). This undecapeptide is recognized by the bacterial housekeeping proteases ClpXP and ClpAP, rendering the tagged protein rapidly degradable (Fig. 1(b)).32 Successful encapsulation of GFP(+36)-SsrA would be expected to prevent proteolysis by obstructing its access to the proteolytic machinery, named “shield-or-shred” system, which would afford a distinctive fluorescence signature to host cells capable of forming such complexes. In contrast, if GFP were produced without the SsrA tag, the cytoplasmic and encapsulated fractions would be indistinguishable in fluorescence assays. Thus, such a “rescue effect” can serve as a simple tool to probe the formation of host–guest complexes in the cytoplasmic environment of intact cells, which has been exploited in both bacterial33–35 and eukaryotic36–38 systems. In this method, successful encapsulation should lead to an increase in fluorescence stemming from functional guest protein – a notable difference from the sequestration of a cytotoxic protease previously used as a selection pressure to obtain AaLS-13, where the cell survival relied on the guest inactivation by protein cages.24
To test the “shield-or-shred” approach for a charge complementarity system (Fig. 1(b)), we coproduced GFP(+36)-SsrA with different AaLS variants equipped with a C-terminal Strep-tag for isolation purposes in Escherichia coli cells. Expression of GFP and AaLS genes was controlled by a tetracycline operon promoter (Ptet) and a T7 promoter coupled with lactose operon operator (PT7lac), respectively. A monomeric variant of superfolder GFP, msfGFP,39,40 with a net theoretical charge of −6 was used in parallel as a non-supercharged analogue of GFP(+36). Analytical flow cytometry was performed to compare the distributions of fluorescence intensity among cell populations producing the SsrA-tagged GFPs alone or together with an AaLS variant.
For all three AaLS proteins, coproduction with GFP(+36)-SsrA resulted in a significantly higher mean fluorescence intensity when compared to GFP(+36)-SsrA produced alone (Fig. 2(a) and (b)). This was the case even for AaLS-wt, which we thought would have little to no propensity for interaction with GFP(+36)-SsrA due to its near-neutral charge. As we proved later, this observation is likely not a consequence of encapsulation but increased molecular crowding or weak association of GFP(+36)-SsrA with the protein cage exterior, potentially inhibiting proteolysis. A high negative charge of the protein cage lumen appears to aid the rescue effect, as evidenced by a >2-fold higher fluorescence signal for AaLS-neg compared to that for AaLS-wt. However, to our big surprise, AaLS-13 which possesses the most negatively charged lumen showed the smallest rescue effect among the tested variants.
Through the previously performed directed evolution experiments,24AaLS-13 had apparently gained a high net negative charge in exchange for instability or slow assembly kinetics. It has been known that the protein is obtained as a mixture of fully assembled cages and its fragments when produced in E. coli cells.26 Although immature cage fragments might be sufficient to reduce the toxicity of the enzyme, this structural feature is likely not beneficial for protection of guests from proteases. Meanwhile, AaLS-neg cages,23 as well as AaLS-wt,41 seem to be more stable, and unassembled fragments have not been observed. Combined with the prior knowledge, these flow cytometry data suggest that not only lumenal charge but also assembling characteristics of a given protein cage are crucial for efficient guest protection in cells.
In analogous experiments, it was confirmed that msfGFP-SsrA, which is not positively supercharged, undergoes nearly complete proteolysis regardless of the choice of coproduced AaLS (Fig. 2(a)). These results highlight the importance of cargo surface charge to be recognized and protected by AaLS proteins.
Although the different rescue effect observed could conceivably reflect the variation in the amounts of AaLS proteins produced, this seems unlikely for the experiments shown in Fig. 2(a) and (b), as the production levels of all three AaLS proteins were found to be similar by SDS-PAGE analysis (Fig. S1(a), ESI†). Nevertheless, the intracellular concentration of AaLS proteins is an important factor in the absolute fluorescence intensity of GFP(+36)-SsrA-producing cells. When the AaLS genes were expressed under the control of a weaker salicylate operon promoter (Psal), no substantial rescue effect was observed for any of the samples, presumably because the concentration of the protein cages was too low for efficient capture of the guest protein (Fig. S1(b) and (c), ESI†).
Cells were next analysed by confocal microscopy to examine the subcellular localization of GFP. Overproduction of heterologous proteins can lead to the formation of inclusion bodies where the constituent proteins could be protected from proteolysis and still preserve their functionality.42 Indeed, cells producing GFP(+36) lacking the degradation tag showed a strong fluorescence signal in the polar regions (Fig. 2(c)), indicating partial accumulation of the functional protein in inclusion bodies. We were concerned that direction of GFP(+36)-SsrA to inclusion bodies, promoted by overproduction of AaLS proteins, could therefore be an additional, or even alternative, mechanism to encapsulation in protein cages for achieving the rescue effect. This is, however, unlikely the case. When GFP(+36)-SsrA was produced alone (Fig. S2, ESI†), fluorescence was mostly scattered throughout the whole cytoplasm; its coproduction with AaLS proteins confined it to the area between the polar and central regions, presumably inclusion bodies and nucleoid, respectively, featured most prominently for AaLS-neg (Fig. 2(c)). While no notable accumulation in the inclusion bodies was confirmed, these observations suggested that AaLS protein cages limit GFP(+36)-SsrA distribution to a certain area of cells.
To gain further insight into host–guest complex formation, AaLS proteins were isolated using StrepTactin affinity chromatography and characterized outside of the cellular context. Cryogenic transmission electron microscopy (cryo-EM) analysis of AaLS-wt showed the presence of monodisperse ∼16 nm particles, while AaLS-neg and AaLS-13 were found to be more diverse in size and morphology (Fig. 3(a), top, Fig. S3, ESI†), forming pseudospherical assemblies of ∼27 nm and ∼35 nm mean diameter, respectively. For AaLS-13 samples, immature cages and unassembled fragments were also visible as expected.26 Coproduction of GFP(+36)-SsrA did not detectably alter their morphology (Fig. 3(a), bottom).
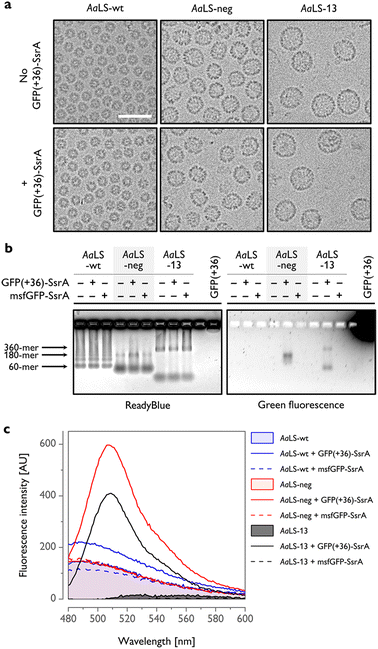 | ||
| Fig. 3 In vitro characterization of AaLS proteins coproduced with SsrA-tagged GFPs. (a) Cryo-EM micrographs of AaLS cages produced with or without GFP(+36)-SsrA. Scale bar – 50 nm. (b) Native agarose gel electrophoresis (AGE) of the AaLS proteins produced in cells alone or coproduced with a GFP-SsrA variant, visualized using ReadyBlue (left) and fluorescence (right). Possible locations of bands corresponding to the most typical AaLS cage assemblies are indicated by arrows. (c) Representative fluorescence emission spectra of isolated AaLS proteins produced in cells alone or coproduced with a GFP-SsrA variant. Spectra for AaLS-neg and AaLS-13 coproduced with GFP(+36)-SsrA exhibit a major peak at 509 nm, corresponding to GFP emission. The background signal observed for AaLS-wt and AaLS-neg is probably attributable to copurified lumazine.43 Spectra of AaLS-wt and AaLS-neg produced alone overlap. | ||
The bands in native agarose gel electrophoresis (AGE) gels corresponding to assembled AaLS-neg and AaLS-13 particles coproduced with GFP(+36)-SsrA exhibited a green fluorescence signal, indicating that GFP(+36)-SsrA comigrated with the cages (Fig. 3(b)). For the AaLS-13 samples, a similar signal was also observed for the incomplete subassemblies, indicating that cage fragments are also capable of binding GFP(+36)-SsrA, likely due to their extraordinarily high net negative charge. The lack of a fluorescence signal for the AaLS-wt samples confirmed little complex formation as we assumed. In the control experiments, AaLS proteins were coproduced with msfGFP-SsrA, yet without any detectable output fluorescence signal. Together, these results confirm that charge complementarity is required for driving the formation of host–guest inclusion complexes in cells.
The number of GFP(+36)-SsrA copurified with AaLS proteins was estimated from the fluorescence spectra to be ∼0.1 molecules per 180-meric AaLS-neg and 360-meric AaLS-13 (Fig. 3(c) and Fig. S4, ESI†). Low complex formation efficiency was also confirmed by SDS-PAGE analysis showing bands corresponding to the expected molecular mass of AaLS monomers and no clear bands corresponding to GFPs (Fig. S5, ESI†). Multiple factors could be in play, including the concentration of the SsrA-tagged GFP in the cytosol, as well as kinetic competition between irreversible proteolysis and potentially reversible encapsulation processes due to the pores of these AaLS cages allowing in- and outflux of GFP-sized proteins (Fig. 1(a)). The observed results likely reflect the overall system ultimately reaching a steady state that depends on the rates of GFP formation, degradation, and encapsulation.
Despite the relatively low rescue efficiency, AaLS-13 proteins were able to form a complex with GFP(+36)-SsrA to a similar extent as AaLS-neg (Fig. S4, ESI†). This may indicate that there is potentially another mechanism, in addition to SsrA-mediated proteolysis, to decrease GFP fluorescence by AaLS-13 in the cytosolic environment. Importantly, such putative inactivation of a guest protein would be even preferable for the sequestration of a toxic enzyme.24 On the other hand, the difference in net negative charge between AaLS-neg and AaLS-13 is probably not crucial for encapsulation of GFP(+36)-SsrA, likely due to the stiff structure with an unusually high net positive charge allowing strong binding to the lumen of both AaLS cages.44 Taken together, our results illustrate how the optimal structures and chemical properties of artificial subcellular compartments may diverge depending on the desired outputs.
Conclusions and perspectives
With appropriate assembling characteristics and binding affinity towards guests based on charge complementarity, the AaLS cages could protectively encapsulate and store an otherwise degradation-prone fluorescent protein in bacterial cells. This finding, together with a previously reported RNA packaging study,45 highlights potential applications of protein cages as novel biotechnological tools for capturing intracellular substances that are homeostatically rare or transient due to rapid degradation or conversion to other forms. Notably, the anionic nature of the host cages, as opposed to commonly employed viral capsids possessing positively charged interiors,2,19 could be useful to deter undesired encapsulation of nucleic acids. Furthermore, encapsulation systems that sequester proteins based on their net surface charges could provide an ideal screening approach for engineering supercharged proteins.46 As shown in this study, flow cytometry constitutes a powerful method for assessing the function of fluorescent proteins and enzymes producing fluorogenic molecules.47 Owing to genetic encodability and evolvability, protein cages hold a great potential to gain tailored properties by reengineering.48–51 This makes our simple charge complementarity system a promising platform for the construction of artificial organelle-like compartments16,37,38 that fulfil specific roles in living cells.Author contributions
Y. A. conceptualized the project, oversaw the research and acquired the funding. D. Z. analysed data, prepared the figures, and wrote the original draft. D. Z., L. K., J. P., V. V. M., N. G. and Y. A. designed and performed experiments. D. Z., L. K., J. G. H., D. H. and Y. A. reviewed and edited the manuscript. All authors read and approved the final version of the manuscript.Conflicts of interest
The authors assert that the research was conducted without any involvement that could be construed as a conflict of interest.Acknowledgements
This work is generously supported by the National Science Centre of Poland (SONATA 14 K/PBD/000307, OPUS 18 K/PBO/000754). We are grateful to the Structural Biology Core Facility (supported by the TEAM TECH CORE FACILITY/2017-4/6 grant from Foundation for Polish Science) for their support in cryo-EM experiments. This publication was developed under the provision of the Polish Ministry of Education and Science project: support for research and development with the use of research infrastructure of the National Synchrotron Radiation Centre SOLARIS under contract nr 1/SOL/2021/2. We acknowledge SOLARIS Centre for their support in cryo-EM measurements on the Glacios-Falcon 4 microscope. We would like to thank Dr Michał Bochenek (Malopolska Centre of Biotechnology, Jagiellonian University) and Anette Schütz (Flow Cytometry Core Facility, ETH Zurich) for their help with the flow cytometry experiments, and Dr Paweł Hermanowicz (Malopolska Centre of Biotechnology, Jagiellonian University) with confocal microscopy imaging. Open-access publication of this article was funded by the Priority Research Area BioS under the program “Initiative of Excellence—Research University” at Jagiellonian University in Krakow.References
- Y. Diekmann and J. B. Pereira-Leal, Evolution of intracellular compartmentalization, Biochem. J., 2013, 449, 319–331 CrossRef CAS PubMed.
- T. G. W. Edwardson, M. D. Levasseur, S. Tetter, A. Steinauer, M. Hori and D. Hilvert, Protein cages: from fundamentals to advanced applications, Chem. Rev., 2022, 122, 9145–9197 CrossRef CAS PubMed.
- N. D. Chasteen and P. M. Harrison, Mineralization in ferritin: an efficient means of iron storage, J. Struct. Biol., 1999, 126, 182–194 CrossRef CAS PubMed.
- F. U. Hartl, A. Bracher and M. Hayer-Hartl, Molecular chaperones in protein folding and proteostasis, Nature, 2011, 475, 324–332 CrossRef CAS PubMed.
- R. T. Sauer and T. A. Baker, AAA+ Proteases: ATP-fueled machines of protein destruction, Annu. Rev. Biochem., 2011, 80, 587–612 CrossRef CAS PubMed.
- C. A. Kerfeld, C. Aussignargues, J. Zarzycki, F. Cai and M. Sutter, Bacterial microcompartments, Nat. Rev. Microbiol., 2018, 16, 277–290 CrossRef CAS PubMed.
- M. Brasch, R. M. Putri, M. V. de Ruiter, D. Luque, M. S. T. Koay, J. R. Castón and J. J. L. M. Cornelissen, Assembling enzymatic cascade pathways inside virus-based nanocages using dual-tasking nucleic acid tags, J. Am. Chem. Soc., 2017, 139, 1512–1519 CrossRef CAS PubMed.
- Y. Azuma, D. L. V. Bader and D. Hilvert, Substrate sorting by a supercharged nanoreactor, J. Am. Chem. Soc., 2018, 140, 860–863 CrossRef CAS PubMed.
- J. A. Bulos, R. Guo, Z. Wang, M. A. Delessio, J. G. Saven and I. J. Dmochowski, Design of a superpositively charged enzyme: human carbonic anhydrase II variant with ferritin encapsulation and immobilization, Biochemistry, 2021, 60, 3596–3609 CrossRef CAS PubMed.
- M. B. Quin, S. A. Perdue, S. Y. Hsu and C. Schmidt-Dannert, Encapsulation of multiple cargo proteins within recombinant Eut nanocompartments, Appl. Microbiol. Biotechnol., 2016, 100, 9187–9200 CrossRef CAS PubMed.
- Y. Azuma, R. Zschoche and D. Hilvert, The C-terminal peptide of Aquifex aeolicus riboflavin synthase directs encapsulation of native and foreign guests by a cage-forming lumazine synthase, J. Biol. Chem., 2017, 292, 10321–10327 CrossRef CAS PubMed.
- R. Lizatović, M. Assent, A. Barendregt, J. Dahlin, A. Bille, K. Satzinger, D. Tupina, A. J. R. Heck, S. Wennmalm and I. André, A protein-based encapsulation system with calcium-controlled cargo loading and detachment, Angew. Chem., Int. Ed., 2018, 57, 11334–11338 CrossRef PubMed.
- A. O’Neil, C. Reichhardt, B. Johnson, P. E. Prevelige and T. Douglas, Genetically programmed in vivo packaging of protein cargo and its controlled release from bacteriophage P22, Angew. Chem., Int. Ed., 2011, 50, 7425–7428 CrossRef PubMed.
- M. Wang, D. Abad, V. A. Kickhoefer, L. H. Rome and S. Mahendra, Vault nanoparticles packaged with enzymes as an efficient pollutant biodegradation technology, ACS Nano, 2015, 9, 10931–10940 CrossRef CAS PubMed.
- S. Deshpande, N. D. Masurkar, V. M. Girish, M. Desai, G. Chakraborty, J. M. Chan and C. L. Drum, Thermostable exoshells fold and stabilize recombinant proteins, Nat. Commun., 2017, 8, 1442 CrossRef PubMed.
- T. W. Giessen and P. A. Silver, A catalytic nanoreactor based on in vivo encapsulation of multiple enzymes in an engineered protein nanocompartment, ChemBioChem, 2016, 17, 1931–1935 CrossRef CAS PubMed.
- L. Schoonen, R. J. M. Nolte and J. C. M. van Hest, Highly efficient enzyme encapsulation in a protein nanocage: towards enzyme catalysis in a cellular nanocompartment mimic, Nanoscale, 2016, 8, 14467–14472 RSC.
- V. V. Shuvaev, M. Khoshnejad, K. W. Pulsipher, R. Y. Kiseleva, E. Arguiri, J. C. Cheung-Lau, K. M. LeFort, M. Christofidou-Solomidou, R. V. Stan, I. J. Dmochowski and V. R. Muzykantov, Spatially controlled assembly of affinity ligand and enzyme cargo enables targeting ferritin nanocarriers to caveolae, Biomaterials, 2018, 185, 348–359 CrossRef CAS PubMed.
- R. D. Requião, R. L. Carneiro, M. H. Moreira, M. Ribeiro-Alves, S. Rossetto, F. L. Palhano and T. Domitrovic, Viruses with different genome types adopt a similar strategy to pack nucleic acids based on positively charged protein domains, Sci. Rep., 2020, 10, 5470 CrossRef PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, E. C. Meng, G. S. Couch, T. I. Croll, J. H. Morris and T. E. Ferrin, UCSF ChimeraX: Structure visualization for researchers, educators, and developers, Protein Sci., 2021, 30, 70–82 CrossRef CAS PubMed.
- M. V. Shapovalov and R. L. Dunbrack, A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions, Structure, 2011, 19, 844–858 CrossRef CAS PubMed.
- E. Jurrus, et al., Improvements to the APBS biomolecular solvation software suite, Protein Sci., 2018, 27, 112–128 CrossRef CAS PubMed.
- F. P. Seebeck, K. J. Woycechowsky, W. Zhuang, J. P. Rabe and D. Hilvert, A simple tagging system for protein encapsulation, J. Am. Chem. Soc., 2006, 128, 4516–4517 CrossRef CAS PubMed.
- B. Wörsdörfer, K. J. Woycechowsky and D. Hilvert, Directed evolution of a protein container, Science, 2011, 331, 589–592 CrossRef PubMed.
- E. Sasaki, D. Böhringer, M. van de Waterbeemd, M. Leibundgut, R. Zschoche, A. J. R. Heck, N. Ban and D. Hilvert, Structure and assembly of scalable porous protein cages, Nat. Commun., 2017, 8, 14663 CrossRef PubMed.
- B. Wörsdörfer, Z. Pianowski and D. Hilvert, Efficient in vitro encapsulation of protein cargo by an engineered protein container, J. Am. Chem. Soc., 2012, 134, 909–911 CrossRef PubMed.
- T. Beck, S. Tetter, M. Künzle and D. Hilvert, Construction of Matryoshka-type structures from supercharged protein nanocages, Angew. Chem., Int. Ed., 2015, 54, 937–940 CrossRef CAS PubMed.
- E. Sasaki, R. M. Dragoman, S. Mantri, D. N. Dirin, M. V. Kovalenko and D. Hilvert, Self-assembly of proteinaceous shells around positively charged gold nanomaterials enhances colloidal stability in high-ionic-strength buffers, ChemBioChem, 2020, 21, 74–79 CrossRef CAS PubMed.
- Y. Azuma, R. Zschoche, M. Tinzl and D. Hilvert, Quantitative packaging of active enzymes into a protein cage, Angew. Chem., Int. Ed., 2016, 55, 1531–1534 CrossRef CAS PubMed.
- M. S. Lawrence, K. J. Phillips and D. R. Liu, Supercharging proteins can impart unusual resilience, J. Am. Chem. Soc., 2007, 129, 10110–10112 CrossRef CAS PubMed.
- G.-F. Tu, G. E. Reid, J.-G. Zhang, R. L. Moritz and R. J. Simpson, C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide, J. Biol. Chem., 1995, 270, 9322–9326 CrossRef CAS PubMed.
- M. Lies and M. R. Maurizi, Turnover of endogenous SsrA-tagged proteins mediated by ATP-dependent proteases in Escherichia coli, J. Biol. Chem., 2008, 283, 22918–22929 CrossRef CAS PubMed.
- E. Y. Kim and D. Tullman-Ercek, A rapid flow cytometry assay for the relative quantification of protein encapsulation into bacterial microcompartments, Biotechnol. J., 2014, 9, 348–354 CrossRef CAS PubMed.
- C. M. Jakobson, E. Y. Kim, M. F. Slininger, A. Chien and D. Tullman-Ercek, Localization of proteins to the 1,2-propanediol utilization microcompartment by non-native signal sequences is mediated by a common hydrophobic motif, J. Biol. Chem., 2015, 290, 24519–24533 CrossRef CAS PubMed.
- M. J. Lee, I. R. Brown, R. Juodeikis, S. Frank and M. J. Warren, Employing bacterial microcompartment technology to engineer a shell-free enzyme-aggregate for enhanced 1,2-propanediol production in Escherichia coli, Metab. Eng., 2016, 36, 48–56 CrossRef CAS PubMed.
- Y. H. Lau, T. W. Giessen, W. J. Altenburg and P. A. Silver, Prokaryotic nanocompartments form synthetic organelles in a eukaryote, Nat. Commun., 2018, 9, 1311 CrossRef PubMed.
- F. Sigmund, C. Massner, P. Erdmann, A. Stelzl, H. Rolbieski, M. Desai, S. Bricault, T. P. Wörner, J. Snijder, A. Geerlof, H. Fuchs, M. H. de Angelis, A. J. R. Heck, A. Jasanoff, V. Ntziachristos, J. Plitzko and G. G. Westmeyer, Bacterial encapsulins as orthogonal compartments for mammalian cell engineering, Nat. Commun., 2018, 9, 1990 CrossRef PubMed.
- L. C. Cheah, T. Stark, L. S. R. Adamson, R. S. Abidin, Y. H. Lau, F. Sainsbury and C. E. Vickers, Artificial self-assembling nanocompartment for organizing metabolic pathways in yeast, ACS Synth. Biol., 2021, 10, 3251–3263 CrossRef CAS PubMed.
- J. D. Pédelacq, S. Cabantous, T. Tran, T. C. Terwilliger and G. S. Waldo, Engineering and characterization of a superfolder green fluorescent protein, Nat. Biotechnol., 2006, 24, 79–88 CrossRef PubMed.
- D. A. Zacharias, J. D. Violin, A. C. Newton and R. Y. Tsien, Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells, Science, 2002, 296, 913–916 CrossRef CAS PubMed.
- L. Koziej, A. Gawin and Y. Azuma, Lumazine synthase nanocompartments, in Microbial Production of High-Value Products, ed. B. H. A. Rehm and D. Wibowo, Microbiology Monographs, 2022, vol. 37, pp. 335–355 Search PubMed.
- E. García-Fruitós, N. González-Montalbán, M. Morell, A. Vera, R. M. Ferraz, A. Arís, S. Ventura and A. Villaverde, Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins, Microb. Cell Fact., 2005, 4, 27 CrossRef PubMed.
- J. Lee, Lumazine protein and the excitation mechanism in bacterial bioluminescence, Biophys. Chem., 1993, 48, 149–158 CrossRef CAS PubMed.
- Y. Azuma and D. Hilvert, Enzyme Encapsulation in an Engineered Lumazine Synthase Protein Cage, in Protein Scaffolds, ed. A. Udit, Methods in Molecular Biology, Humana Press, New York, NY, 2018, vol. 1798 DOI:10.1007/978-1-4939-7893-9_4.
- Y. Azuma, T. G. W. Edwardson, N. Terasaka and D. Hilvert, Modular protein cages for size-selective RNA packaging in vivo, J. Am. Chem. Soc., 2018, 140, 566–569 CrossRef CAS PubMed.
- C. Ma, A. Malessa, A. J. Boersma, K. Liu and A. Herrmann, Supercharged proteins and polypeptides, Adv. Mater., 2020, 32, 1905309 CrossRef CAS PubMed.
- G. Yang and S. G. Withers, Ultrahigh-throughput FACS-based screening for directed enzyme evolution, ChemBioChem, 2009, 10, 2704–2715 CrossRef CAS PubMed.
- G. L. Butterfield, M. J. Lajoie, H. H. Gustafson, D. L. Sellers, U. Nattermann, D. Ellis, J. B. Bale, S. Ke, G. H. Lenz, A. Yehdego, R. Ravichandran, S. H. Pun, N. P. King and D. Baker, Evolution of a designed protein assembly encapsulating its own RNA genome, Nature, 2017, 552, 415–420 CrossRef CAS PubMed.
- E. C. Hartman, C. M. Jakobson, A. H. Favor, M. J. Lobba, E. Álvarez-Benedicto, M. B. Francis and D. Tullman-Ercek, Quantitative characterization of all single amino acid variants of a viral capsid-based drug delivery vehicle, Nat. Commun., 2018, 9, 1385 CrossRef PubMed.
- N. Terasaka, Y. Azuma and D. Hilvert, Laboratory evolution of virus-like nucleocapsids from nonviral protein cages, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 5432–5437 CrossRef CAS PubMed.
- S. Tetter, N. Terasaka, A. Steinauer, R. J. Bingham, S. Clark, A. J. P. Scott, N. Patel, M. Leibundgut, E. Wroblewski, N. Ban, P. G. Stockley, R. Twarock and D. Hilvert, Evolution of a virus-like architecture and packaging mechanism in a repurposed bacterial protein, Science, 2021, 372, 1220–1224 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb00754e |
| This journal is © The Royal Society of Chemistry 2023 |