Self-assembly of a fluorescent virus-like particle for imaging in tissues with high autofluorescence†
Ikeda
Trashi
a,
Mateusz Z.
Durbacz
b,
Orikeda
Trashi
a,
Yalini H.
Wijesundara
a,
Ryanne N.
Ehrman
a,
Alyssa C.
Chiev
a,
Cary B.
Darwin
a,
Fabian C.
Herbert
a,
Jashkaran
Gadhvi
 c,
Nicole J.
De Nisco
c,
Steven O.
Nielsen
c,
Nicole J.
De Nisco
c,
Steven O.
Nielsen
 a and
Jeremiah J.
Gassensmith
a and
Jeremiah J.
Gassensmith
 *ad
*ad
aDepartment of Chemistry and Biochemistry, The University of Texas at Dallas, Richardson, Texas 75080, USA. E-mail: gassensmith@utdallas.edu
bDepartment of Molecular Biology and Hamon Center for Regenerative Science and Medicine, University of Texas Southwestern Medical Center, Dallas, Texas, USA
cDepartment of Biological Science, The University of Texas at Dallas, Richardson, Texas 75080, USA
dDepartment of Bioengineering, The University of Texas at Dallas, Richardson, Texas 75080, USA
First published on 25th April 2023
Abstract
Virus-like particles (VLPs) are engineered nanoparticles that mimic the properties of viruses-like high tolerance to heat and proteases-but lack a viral genome, making them non-infectious. They are easily modified chemically and genetically, making them useful in drug delivery, enhancing vaccine efficacy, gene delivery, and cancer immunotherapy. One such VLP is Qβ, which has an affinity towards an RNA hairpin structure found in its viral RNA that drives the self-assembly of the capsid. It is possible to usurp the native way infectious Qβ self-assembles to encapsidate its RNA to place enzymes inside the VLP's lumen as a protease-resistant cage. Further, using RNA templates that mimic the natural self-assembly of the native capsid, fluorescent proteins (FPs) have been placed inside VLPs in a “one pot” expression system. Autofluorescence in tissues can lead to misinterpretation of results and unreliable science, so we created a single-pot expression system that uses the fluorescent protein smURFP, which avoids autofluorescence and has spectral properties compatible with standard commercial filter sets on confocal microscopes. In this work, we were able to simplify the existing “one-pot” expression system while creating high-yielding fluorescent VLP nanoparticles that could easily be imaged inside lung epithelial tissue.
Introduction
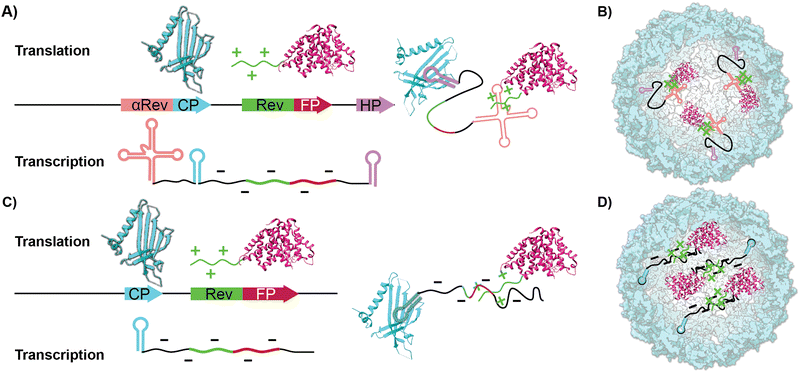
Virus-like particles (VLPs) are genetically engineered nanoparticles that resemble infectious viruses but lack a viral genome and are not infectious.1–3 VLPs typically comprise many identical coat proteins whose self-assembly is templated by the mRNA generated in their recombinant expression. This assembly mimics the properties of the native virus, starting with the nucleation phase and the production of oligomers around the nucleic acids host.4–6 The exterior of VLPs displays various amino acids that can be used to fuse peptides or chemically conjugate different molecules through amide coupling, diazonium addition, and click chemistry.7–11 Their packing ability has been used to protect RNA by expressing and packing it inside or delivering the scaffold proteins using ‘’one pot’’ expression systems.12,13 Their biocompatibility and easy chemical and genetic modification have made them useful in drug delivery, enhancing vaccine efficacy,14–17 gene delivery,18–20 and cancer immunotherapy.21–27One such representative VLP is Qβ, derived from a bacteriophage in the Leviviridae family. The VLP forms an icosahedral capsid comprising 180 identical coat protein (CP) subunits.28 Self-assembly of the natural Qβ has been thought to be driven by a specific RNA hairpin structure that has an affinity toward the native CPs and is found in the Qβ's viral RNA, positioned at the 5’ end, making possible its interaction with CPs during protein synthesis on the bacterial ribosome.29,30
In addition to chemical and genetic modification to the CP-which leads to new covalently bound surface functionality-work on VLPs has focused on using the ample void space as a haven for biomacromolecular cargo. For instance, encapsidating enzymes within the interior lumen of VLPs helps serve as a protease-resistant cage that protects the more sensitive enzymes.31,32 Two common ways to pack proteinaceous cargo inside VLPs are through an iterative disassembly-reassembly process or a “one pot” expression system.33 The former disassembly-reassembly process is best understood as a “supramolecular assembly” process, where self-assembly to reform the capsid is aided by proteins and RNAs that act as templates. For instance, we have previously shown it is possible to encapsidate the small Ultra-Red Fluorescent Protein (smURFP) within Qβ by reducing the capsid's disulfide bonds and forcing the disassembly of the capsid at low pH and high salt concentration. This is followed by a supramolecular assembly process aided by a 10× fold excess of the smURFP cargo at neutral pH.34 In contrast to previous reports, we found that the charge matching of the interior of the Qβ coat protein, which is positively charged, with the anionic charge of smURFP promotes assembly without the need for osmolytes. The 68% yield proved sufficient for small-scale experiments but was cumbersome when significant quantities-i.e., hundreds of milligrams-were required. We were thus drawn to the encapsidation of fluorescent proteins (FPs) inside VLPs via cloning, which allows the bacteria to do the time-consuming work of placing cargo within the capsid in high yields producing larger quantities of material.35,36 Previous work by Finn and colleagues have shown that proteins or enzymes can be packaged inside VLP Qβ using an RNA template that mimics the putative natural way the native capsid self-assembles. This requires engineering recognition elements into the transcribed mRNA to direct fluorescent proteins inside the capsid.37,38 As illustrated in Scheme 1A and B, proteinaceous cargo is tagged with an arginine-rich rev peptide tag that recognizes and binds to an RNA aptamer upstream of the ribosome binding site.39 An RNA hairpin downstream of the stop codon binds to the interior of the CP monomers directing the rev-tagged protein in the interior of VLP. Conceptually, this expression system uses the mRNA as a rope that electrostatically ties cargo to the CP. To the best of our knowledge, this method yielded two fluorescent VLPs-one containing the green fluorescent protein (GFP) and one containing the near-infrared proteins (iRFP and mIFP). GFP and iRFPs have outstanding photophysical properties but have some practical and physical limitations in the lab. Most fluorescent microscopes do not have filter sets to image NIR proteins, and GFP has considerable spectral overlap with biomolecules typically found in tissues.40,41 The kidney, lung, liver, and spleen are notorious for intense autofluorescence, which makes imaging them with green fluorophores challenging.42–44 This often leads to misinterpretation of the results and unreliable science. The solution to overcoming autofluorescence is selecting a spectrally shifted FP that fluoresces in the red channel.
smURFP was evolved from light-harvesting phycobilisomes yielding a photostable protein.45,46 The photostability of this protein is higher than other proteins like eGFP, mCherry, and iRFP 1.4. Red fluorescent proteins like iRFPs, iFPs, and smURFP need the fluorophore biliverdin (Bv), which binds to the protein via carboxylate groups on Bv to fluoresce. The inside of the smURFP is positively charged, allowing attraction and interaction with the carboxylase groups of fluorophore Bv. Following binding, the fluorescence increases 1400× and shows spectral properties similar to Cy5; however, Bv hydrochloride is expensive, and incorporating Bv post synthetically can produce inconsistent results (vide infra).36,47–50 Heme oxygenase 1 (HO-1) is an enzyme that breaks heme down into biliverdin, carbon monoxide (CO), and ferrous (Fe2+) ions.51 HO-1 is found in humans but not E. coli: however, a recombinant version that can be expressed in bacteria exists.52–54
In this report, we show that the co-expression of HO-1, along with the VLP coat protein and the FP, smURFP, produces a very bright encapsidation complex (s@Qβ) both efficiently and in high yield. We demonstrate that the self-assembly of s@Qβ is sufficiently robust that neither hairpin recognition motifs nor RNA-based aptamers are necessary to induce high-yielding s@Qβ (Scheme 1C and D). Finally, we prove that we can administer s@Qβ via pulmonary lavage, extract lung tissue, and subsequently image s@Qβ within the cells of the lung without autofluorescence as a confounding issue.
Results and discussion
Cloning
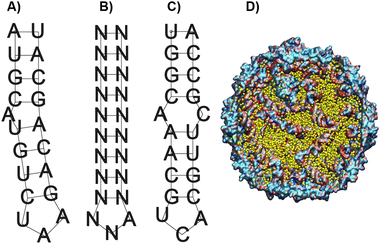
As a starting point in the cloning process, we used the plasmid pJF51-CP-mIFP (gifted by the Finn laboratory), which contains the sequence for both the CP of Qβ and the near-infrared fluorophore mIFP-the mIFP will be digested and replaced with smURFP in this work. The induction of proteins on this plasmid is triggered by the addition of the isopropylthio-β-galactoside (IPTG)- a molecular mimic of lactose. The plasmid was digested with two enzymes XhoI and EagI (New England Biolabs), allowing us to remove the mIFP. The digested backbone was isolated and cleaned from the gel using a NucleosSpin gel and PCR clean-up kit (Macherey-Nagel). Co-expression of the s@Qβ with the HO-1 gene was done based on prior reports36 that incorporation of exogenous commercially obtained Bv during expression produces inconsistent results. We decided to test if the co-expression of HO-1, CP, and smURFP could overcome this issue. To eliminate the need for synthetic biliverdin, we used the pBAD-smURFP-HO-1 plasmid, which expresses smURFP and HO-1 under the L-arabinose operon. smURFP and HO-1 expression induction is triggered by adding L-arabinose to the media. The only restriction enzymes we could use would cause the removal of the smURFP sequence and the HO-1 sequence. Thus, we decided to digest both and reinsert the HO-1 fragment by first amplifying the HO-1 sequence from the pBAD plasmid. Next, the pBAD vector was digested by BamHI and PmeI (New England Biolabs) restriction enzymes. The digested backbone of pBAD was isolated and cleaned with the Macherey-Nagel, NucleoSpin gel, and PCR clean up. The smURFP sequence was ligated with the digested backbone of pJF51-CP (Fig. S1, ESI†). The pBAD backbone and amplified HO-1 were ligated using Gibson assembly (Fig. S2, ESI†). We confirmed the proper ligation by Sanger sequencing. Finally, a double transformation of both plasmids in BL21(DE3) E. coli yielded fluorescent s@Qβ VLPs.Previous papers have shown that the wild-type hairpin (Fig. 1A) is necessary to induce a conformational change of the MS2 CP dimers to form the capsid. We can infer that Qβ-and VLPs derived from the same family Leviviridae-use a similar assembly mechanism.55 Other data have shown that the Qβ hairpin can tolerate significant change and still induce self-assembly, requiring only an eight-base pair stem and unpaired adenosine to induce a CP dimer conformational change, shown as the minimal sequence in Fig. 1B.56 Using RNAfold software, which returns several low free energy structures for a given RNA sequence, we discovered that the RNA that codes for the CP always contains hairpin structures (Fig. 1C) consistent with the minimal sequence known to require assembly. This suggests that as long as the CP is expressed, the capsid should be able to assemble. It has been shown through cryo-electron microscopy experiments on Qβ that RNA hairpin interactions are needed for more than just creating the structure; they also stabilize the capsid.57 Cryogenic electron microscope EM data shows that fifty-nine stem-loops in the Qβ viral capsid appear to contribute to its stability; however, it is not known what the minimum amount of stem-loops are needed for a Qβ VLP to form. Another change in our strategy is the removal of the αREV aptamer, which is putatively needed to capture the rev-tag fused to the N-terminus of the FP. In our system, we retained the rev tag-which is strongly cationic but removed the αREV aptamer. We hypothesize that the transcribed anionic mRNAs that direct CP self-assembly would also interact strongly with the rev tag's cationic surface. We also considered the total void volume to determine how many proteins we could load within the VLP. Since the expression strategy (Scheme 1) mainly involves RNA, with the smURFP protein accounting for less than 5% by mass, we used the space occupied by the genomic RNA in the Qβ virion (pdb entry 7LHD, see Fig. 1D) to estimate how much RNA would fill the VLP. This estimate predicts the maximum number of smURFPs complexes that fit inside the Qβ as 10–17 (computational details in ESI†).
Expression
The dual system with two different operons carries with it the possibility of inhibiting the L-arabinose operon by the presence of IPTG. It has been found that at low concentrations of arabinose, the pBAD can be repressed by IPTG-influenced expression. However, oversaturation of the system with a high amount of arabinose can inhibit the repressor effect of IPTG.58 To overcome the inhibition effect of IPTG, we increased the amount of added L-arabinose to a total of 1% w/v. For protein expression, bacteria were grown in Luria broth (LB) and induced when the OD450∼1. To test if our co-expression system produces superior results, we prepared two samples-one where Bv was added externally (s@Qβ-Bv) and one where they were co-expressed (s@Qβ-HO). For the system s@Qβ-Bv, induction was carried out with 1 mM IPTG and 15 mM Bv as optimized previously36 in the presence of 50 μg mL−1 streptomycin.For the co-expression system, smURFP@Qβ-HO induction was performed with 1 mM IPTG and 1% w/v L-arabinose in LB media supplemented with 50 μg mL−1 streptomycin and 50 μg mL−1 ampicillin. Further purification includes pelleting down the bacteria, sonication, salting out, 40% w/v sucrose cushion, and again pelleting down to obtain the blue protein pellet. Protein concentration was determined by Coomassie Plus Protein Reagent, and from this, we determined that we obtained 40–60 mg L−1 of smURFP@Qβ-HO. This is comparable to the 50–70 mg L−1 yield obtained using the αREV aptamer approach (Scheme 1A) reported by Finn et al.36
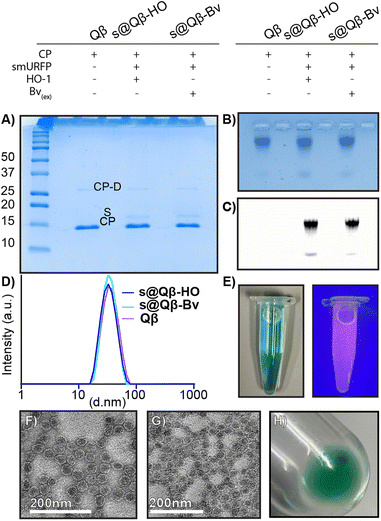
Characterization to compare our different preparations of s@Qβ VLPs with native Qβ VLP was performed. A non-reducing 18% SDS (Fig. 2A) gel showed the CP monomer at 14.2 kDa and a faint CP dimer at 28 kDa for Qβ, s@Qβ-HO, s@Qβ-Bv, and the smURFP at 15 kDa, as expected. Agarose electrophoresis (Fig. 2B) shows that Qβ, s@Qβ-Bv, and s@Qβ-HO do not significantly change in mobility, suggesting that both size and overall charge are unaffected by protein encapsidation. Fluorescence gel imaging (Fig. 2C) showed a signal corresponding to smURFP in the Cy5 channels for both s@Qβ-HO and s@Qβ-Bv but not for our control, Qβ, further proving we have encapsidated smURFP within the VLP in both systems. Dynamic light scattering (DLS) was used to measure the size of the Qβ VLPs. We found the encapsidation did not alter nanoparticle size much: Qβ(∼34 nm, PDI= 0.077), s@Qβ-Bv(∼32 nm, PDI = 0.087) and s@Qβ-HO (∼33 nm, PDI = 0.053) (Fig. 2D). Autocorrelation graphs are reported in Fig. S3 (ESI†). s@Qβ VLP solution in light is blue to the eye and under UV shows a pink luminescence (Fig. 2E). To observe the VLPs morphology, transmission electron microscopy (TEM) was used for s@Qβ-HO (Fig. 2F), and Qβ (Fig. 2G) confirming our DLS results, which found no differences in size or changes in morphology as shown in (Fig. S4, ESI†) Following centrifugation, the s@Qβ VLP pellet appears blue (Fig. 2H). Using gel densitometry, we determined that ∼12 smURFPs were loaded in s@Qβ-HO, larger than the ∼8 smURFPs inside s@Qβ-Bv (Fig. S4 and S5, ESI†). We obtained similar results when the amount was calculated with fluorescence intensity-∼10 smURFPs were packed in s@Qβ-HO and ∼8 smURFPs inside s@Qβ-Bv (Fig. S7, ESI†).
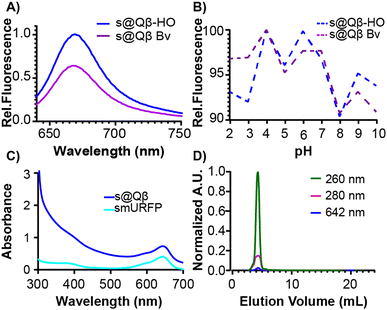
To further compare these different expression methods, more analyses were performed. The fluorescence intensity was measured at the same concentration of VLPs, and s@Qβ-HO exhibited a higher fluorescence intensity than s@Qβ-Bv (Fig. 3A). We speculate that this might be because bacteria cells uptake small molecules less efficiently when introduced externally rather than the ones the cells make themselves. It is worth mentioning that the fluorescence of s@Qβ-Bv declined significantly after some days, which we did not observe for s@Qβ-HO. The VLPs were incubated for 30 min at a pH of 2–10 to evaluate the photophysical properties of the smURFP. While this experiment says nothing of the stability of the VLP at these pH's we can say that we observed minimal loss of fluorescence, with the maximum being a 10% reduction in intensity at pH 8 meaning that the cargo (fluorescent protein) is still stable (Fig. 3B). The spectral properties of the encapsidated and free smURFPs showed identical meaningmax; however, considerable broadening at lower wavelengths was noted (Fig. 3C and Fig. S8, ESI†) To verify the purity of the VLP, size exclusion chromatography (SEC) was used. It showed coelution of the VLPs indicative of the absorbance at 280 and 260 nm for the protein and at 642 nm for smURFP (Fig. 3D). It is worth mentioning that if smURFP had not been encapsidated post-expression, we would observe the free smURFP as a separate peak in the SEC chromatogram.
From these experiments and in agreement with the literature, we conclude that the co-expression system reliably produced a brightly fluorescent encapsidated complex. An added benefit is that the synthesis does not require adding expensive Bv and produces consistent results. Based on these results, we recommend co-expression whenever possible and will use the s@Qβ-HO as we advance in this manuscript. To further explore if removing the REV tag would allow encapsidation of smURFP in Qβ VLP, we digested the REV tag out, performed ligation, and then started expression. No blue color was observed during expression, and the protein solution appeared white/transparent in color indicating smURFP was not packed inside the VLP. These results demonstrate the necessity of the REV tag in the cargo encapsulation. SDS page following purification confirmed no smURFP was encapsidated (Fig. S9, ESI†).
Cell studies
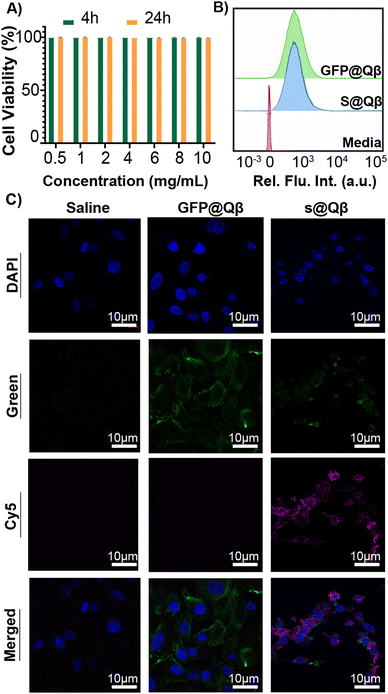
GFP is a well-known FP widely used in imaging owing to its great brightness and stability. However, imaging with GFP can be very challenging as specific molecules found in cells and tissue are bioluminescent at similar wavelengths that GFP emits. This autofluorescence makes data interpretation a difficult process. For this reason, we have benchmarked our system against the previously reported construct GFP@Qβ and studied them both in vitro and in vivo.To observe their behavior first in cells, we performed cell viability studi es and cellular uptake in the A549 human lung adenocarcinoma cell line. Using a lactate dehydrogenase (LDH) assay, no toxicity was observed at any concentration from 0.5 mg mL−1 to 10 mg mL−1 (Fig. 4A). Cellular uptake was quantified using flow cytometry (Fig. 4B and Fig. S10, ESI†), and we found the VLP uptake to be the same regardless of the FP used. Additionally, laser scanning microscopy (CLSM) images were acquired for cells treated with saline, GFP@Qβ s@Qβ, and, as shown in Fig. 4C, we saw similar localizations within cells regardless of the FP used.
Fluorescence imaging in vivo
In tissues, molecules and endogenous fluorophores become fluorescent and absorb at a specific wavelength. The most well-known fluorophores in tissues are flavins, pyridinic acid, and collagen. Imaging experiments must be performed to distinguish the fluorescence when treating tissue with exogenous fluorophores. In this experiment, we attempted to observe if s@Qβ VLP would be sufficiently spectrally shifted and bright to be observed in lung tissue. Three groups of BALB/c mice (n = 3) were administered with saline, GFP@Qβ, and s@Qβ endotracheally, as shown in Fig. 5A. Four hours post-delivery, the mice were sacrificed, and their lungs were extracted, fixed, paraffin-embedded and thin sectioned using a microtome. Tissue slides were dehydrated and cleaned, and nuclei were stained with DAPI and imaged by CSLM. As illustrated in the micrograph in Fig. 5B, when GFP is used, it is difficult to determine which signal arises because of autofluorescence and what arises from GFP@Qβ. Indeed, green fluorescence is obvious in all the samples, even if they were not given GFP@Qβ. In contrast, the micrographs showing samples where s@Qβ was used is very clear in the Cy5 channel-imaged samples stained with s@Qβ using a standard Cy5 filter set produced unambiguous results, clearly showing the lung tissue had taken in the VLPs.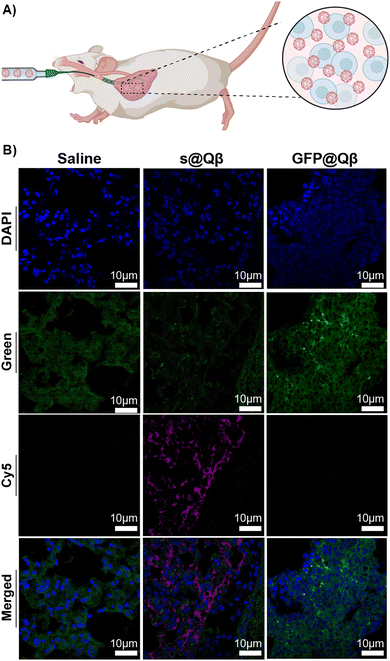 | ||
| Fig. 5 (A) Schematic representation of the delivery. (B) CSLM images of BALB/c lung tissues ex vivo for saline (left), GFP@Qβ (center), and s@Qβ (right). | ||
Conclusions
This study shows that the mRNA used in translating CPs for the Qβ capsid contains hairpin structures sufficient to nucleate capsid formation while also encapsulating cargo. We have also shown that the encapsidation of cargo featuring a REV tag can occur without the complementary αREV aptamer; however, no encapsidation occurs in the absence of the REV tag. We suspect this assembly is driven purely by electrostatic interactions between the cationic REV tag and the anionic backbone of the mRNA. We demonstrated that the genetic co-expression of s@Qβ and HO-1 yields consistent results and lowers the cost s@Qβ expression compared to the expression of s@Qβ with exogenous addition of BV during induction. Finally, we demonstrated that encapsidation of the smURFP protein inside the Qβ VLP drastically allowed for imaging in the Cy5 channel, reducing background auto-fluorescence compared to the spectral window used to image GFP fluorescence.Ethical statement
Female BALB/c mice were obtained from Charles River Lab (Wilmington, Ma). All animal studies used for endotracheal delivery were performed in strict accordance with the protocol #18-17 approved by the University of Texas at Dallas Institutional Animal Care and Use Committee (IACUC).Author contributions
Conceptualization: JJG, IT, methodology: IT, OT, YHW, MD, FCH, investigation: IT, OT, YHW, MD, RNE, ACC, CBD, FCH, JG, NDN, SON, JJG, supervision JJG, writing – original draft: IT, JJG, writing review & editing IT, JJG.Data availability
The raw data associated with this work can be accessed at the Open Science Foundation (OSF) via this link: https://osf.io/cj9r7/?view_only=d06e0579eeb4422f8173663daeb6f706.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Finn lab for providing the pJF51-CP-mIFP plasmid, which was used as a starting point for our project. J. J. G. acknowledges support from Welch Foundation (AT-198920190330) and the National Science Foundation (DMR-2003534). University of Texas at Dallas lab animal resource center (LARC) for their assistance in animal care.References
- A. Zeltins, Construction and Characterization of Virus-Like Particles: A Review, Mol. Biotechnol., 2013, 53(1), 92–107, DOI:10.1007/s12033-012-9598-4.
- B. C. Bundy and J. R. Swartz, Efficient disulfide bond formation in virus-like particles, J. Biotechnol., 2011, 154(4), 230–239, DOI:10.1016/j.jbiotec.2011.04.011.
- T. Bhat, A. Cao and J. Yin, Virus-like particles: Measures and biological functions, Viruses., 2022, 14(2), 383 CrossRef CAS PubMed.
- D. T. Le and K. M. Müller, In Vitro Assembly of Virus-Like Particles and Their Applications, Life, 2021, 11(4), 334, DOI:10.3390/life11040334.
- M. Marchetti, D. Kamsma, E. Cazares Vargas, A. Hernandez García, P. Van Der Schoot, R. De Vries, G. J. Wuite and W. H. Roos, Real-time assembly of viruslike nucleocapsids elucidated at the single-particle level, Nano Lett., 2019, 19(8), 5746–5753 CrossRef CAS PubMed.
- L. Zhang, L. H. Lua, A. P. Middelberg, Y. Sun and N. K. Connors, Biomolecular engineering of virus-like particles aided by computational chemistry methods, Chem. Soc. Rev., 2015, 44(23), 8608–8618 RSC.
- Y. H. Wijesundara, F. C. Herbert, S. Kumari, T. Howlett, S. Koirala, O. Trashi, I. Trashi, N. M. Al-Kharji and J. J. Gassensmith, Rip it, stitch it, click it: A Chemist’s guide to VLP manipulation, Virology, 2022, 577, 105–123, DOI:10.1016/j.virol.2022.10.008.
- Z. Chen, N. Li, S. Li, M. Dharmarwardana, A. Schlimme and J. J. Gassensmith, Viral chemistry: the chemical functionalization of viral architectures to create new technology, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol, 2016, 8(4), 512–534 CrossRef PubMed.
- R. P. Welch, H. Lee, M. A. Luzuriaga, O. R. Brohlin and J. J. Gassensmith, Protein–polymer delivery: Chemistry from the cold chain to the clinic, Bioconjugate Chem., 2018, 29(9), 2867–2883 CrossRef CAS PubMed.
- M. Dharmarwardana, A. F. Martins, Z. Chen, P. M. Palacios, C. M. Nowak, R. P. Welch, S. Li, M. A. Luzuriaga, L. Bleris and B. S. Pierce, Nitroxyl modified tobacco mosaic virus as a metal-free high-relaxivity MRI and EPR active superoxide sensor, Mol. Pharmaceutics, 2018, 15(8), 2973–2983 CrossRef CAS PubMed.
- H. Lee, A. Shahrivarkevishahi, J. L. Lumata, M. A. Luzuriaga, L. M. Hagge, C. E. Benjamin, O. R. Brohlin, C. R. Parish, H. R. Firouzi and S. O. Nielsen, Supramolecular and biomacromolecular enhancement of metal-free magnetic resonance imaging contrast agents, Chem. Sci., 2020, 11(8), 2045–2050 RSC.
- P.-Y. Fang, J. C. Bowman, L. M. G. Ramos, C. Hsiao and L. D. Williams, RNA: packaged and protected by VLPs, RSC Adv., 2018, 8(38), 21399–21406 RSC.
- J. Sharma, M. Uchida, H. M. Miettinen and T. Douglas, Modular interior loading and exterior decoration of a virus-like particle, Nanoscale, 2017, 9(29), 10420–10430 RSC.
- M. A. Luzuriaga, R. P. Welch, M. Dharmarwardana, C. E. Benjamin, S. Li, A. Shahrivarkevishahi, S. Popal, L. H. Tuong, C. T. Creswell and J. J. Gassensmith, Enhanced Stability and Controlled Delivery of MOF-Encapsulated Vaccines and Their Immunogenic Response In Vivo, ACS Appl. Mater. Interfaces, 2019, 11(10), 9740–9746, DOI:10.1021/acsami.8b20504.
- L. X. Doan, M. Li, C. Chen and Q. Yao, Virus-like particles as HIV-1 vaccines, Rev. Med. Virol., 2005, 15(2), 75–88 CrossRef PubMed.
- J. W. Wang and R. B. Roden, Virus-like particles for the prevention of human papillomavirus-associated malignancies, Expert Rev. Vaccines, 2013, 12(2), 129–141 CrossRef CAS PubMed.
- L. H. Lua, N. K. Connors, F. Sainsbury, Y. P. Chuan, N. Wibowo and A. P. Middelberg, Bioengineering virus-like particles as vaccines, Biotechnol. Bioeng., 2014, 111(3), 425–440 CrossRef CAS PubMed.
- P. Lam and N. F. Steinmetz, Delivery of siRNA therapeutics using cowpea chlorotic mottle virus-like particles, Biomater. Sci., 2019, 7(8), 3138–3142 RSC.
- P. Erbacher, J.-S. Remy and J.-P. Behr, Gene transfer with synthetic virus-like particles via the integrin-mediated endocytosis pathway, Gene Ther., 1999, 6(1), 138–145 CrossRef CAS PubMed.
- A. Touze and P. Coursaget, In vitro gene transfer using human papillomavirus-like particles, Nucleic Acids Res., 1998, 26(5), 1317–1323 CrossRef CAS PubMed.
- W. Li, Z. Jing, S. Wang, Q. Li, Y. Xing, H. Shi, S. Li and Z. Hong, P22 virus-like particles as an effective antigen delivery nanoplatform for cancer immunotherapy, Biomaterials, 2021, 271, 120726 CrossRef CAS PubMed.
- K. Cheng, T. Du, Y. Li, Y. Qi, H. Min, Y. Wang, Q. Zhang, C. Wang, Y. Zhou and L. Li, Dual-antigen-loaded hepatitis B virus core antigen virus-like particles stimulate efficient immunotherapy against melanoma, ACS Appl. Mater. Interfaces, 2020, 12(48), 53682–53690 CrossRef CAS PubMed.
- A. Shahrivarkevishahi, L. M. Hagge, O. R. Brohlin, S. Kumari, R. Ehrman, C. Benjamin and J. Gassensmith, Virus-like particles: a self-assembled toolbox for cancer therapy. Materials Today, Chemistry, 2022, 24, 100808 CAS.
- A. Shahrivarkevishahi, M. A. Luzuriaga, F. C. Herbert, A. C. Tumac, O. R. Brohlin, Y. H. Wijesundara, A. V. Adlooru, C. Benjamin, H. Lee and P. Parsamian, PhotothermalPhage: a virus-based photothermal therapeutic agent, J. Am. Chem. Soc., 2021, 143(40), 16428–16438 CrossRef CAS PubMed.
- C. E. Benjamin, Z. Chen, P. Kang, B. A. Wilson, N. Li, S. O. Nielsen, Z. Qin and J. J. Gassensmith, Site-selective nucleation and size control of gold nanoparticle photothermal antennae on the pore structures of a virus, J. Am. Chem. Soc., 2018, 140(49), 17226–17233 CrossRef CAS PubMed.
- P. Parsamian, Y. Liu, C. Xie, Z. Chen, P. Kang, Y. H. Wijesundara, N. M. Al-Kharji, R. N. Ehrman, O. Trashi and J. Randrianalisoa, Enhanced Nanobubble Formation: Gold Nanoparticle Conjugation to Qβ Virus-like Particles, ACS Nano, 2023, 17(8), 7797–7805, DOI:10.1021/acsnano.3c00638.
- C. E. Benjamin, Z. Chen, O. R. Brohlin, H. Lee, A. Shahrivarkevishahi, S. Boyd, D. D. Winkler and J. J. Gassensmith, Using FRET to measure the time it takes for a cell to destroy a virus, Nanoscale, 2020, 12(16), 9124–9132, 10.1039/C9NR09816J.
- S. D. Brown, J. D. Fiedler and M. Finn, Assembly of hybrid bacteriophage Qβ virus-like particles, Biochemistry, 2009, 48(47), 11155–11157 CrossRef CAS PubMed.
- R. Golmohammadi, K. Fridborg, M. Bundule, K. Valegård and L. Liljas, The crystal structure of bacteriophage Qβ at 3.5 Å resolution, Structure, 1996, 4(5), 543–554 CrossRef CAS PubMed.
- T. M. Kozlovska, I. Cielēns, D. Dreilinņa, A. Dišlers, V. Baumanis, V. Ose and P. Pumpēns, Recombinant RNA phage Qβ capsid particles synthesized and self-assembled in Escherichia coli, Gene, 1993, 137(1), 133–137 CrossRef CAS PubMed.
- J. D. Fiedler, S. D. Brown, J. L. Lau and M. Finn, RNA-directed packaging of enzymes within virus-like particles, Angew. Chem., Int. Ed., 2010, 49(50), 9648–9651 CrossRef CAS PubMed.
- H. K. Waghwani, M. Uchida, C.-Y. Fu, B. LaFrance, J. Sharma, K. McCoy and T. Douglas, Virus-like particles (VLPs) as a platform for hierarchical compartmentalization, Biomacromolecules, 2020, 21(6), 2060–2072 CrossRef CAS PubMed.
- K.-I. Ishizu, H. Watanabe, S.-I. Han, S.-N. Kanesashi, M. Hoque, H. Yajima, K. Kataoka and H. Handa, Roles of disulfide linkage and calcium ion-mediated interactions in assembly and disassembly of virus-like particles composed of simian virus 40 VP1 capsid protein, J. Virol., 2001, 75(1), 61–72 CrossRef CAS PubMed.
- F. C. Herbert, O. R. Brohlin, T. Galbraith, C. Benjamin, C. A. Reyes, M. A. Luzuriaga, A. Shahrivarkevishahi and J. J. Gassensmith, Supramolecular Encapsulation of Small-Ultrared Fluorescent Proteins in Virus-Like Nanoparticles for Noninvasive In Vivo Imaging Agents, Bioconjugate Chem., 2020, 31(5), 1529–1536, DOI:10.1021/acs.bioconjchem.0c00190.
- J. D. Fiedler, M. R. Fishman, S. D. Brown, J. Lau and M. Finn, Multifunctional enzyme packaging and catalysis in the Qβ protein nanoparticle, Biomacromolecules, 2018, 19(10), 3945–3957 CrossRef CAS PubMed.
- S. Das, L. Zhao, S. N. Crooke, L. Tran, S. Bhattacharya, E. A. Gaucher and M. G. Finn, Stabilization of Near-Infrared Fluorescent Proteins by Packaging in Virus-like Particles, Biomacromolecules, 2020, 21(6), 2432–2439, DOI:10.1021/acs.biomac.0c00362.
- J.-K. Rhee, M. Hovlid, J. D. Fiedler, S. D. Brown, F. Manzenrieder, H. Kitagishi, C. Nycholat, J. C. Paulson and M. G. Finn, Colorful Virus-like Particles: Fluorescent Protein Packaging by the Qβ Capsid, Biomacromolecules, 2011, 12(11), 3977–3981, DOI:10.1021/bm200983k.
- M. E. Matlashov, D. M. Shcherbakova, J. Alvelid, M. Baloban, F. Pennacchietti, A. A. Shemetov, I. Testa and V. V. Verkhusha, A set of monomeric near-infrared fluorescent proteins for multicolor imaging across scales, Nat. Commun., 2020, 11(1), 239 CrossRef CAS PubMed.
- W. Xu and A. D. Ellington, Anti-peptide aptamers recognize amino acid sequence and bind a protein epitope, Proc. Natl. Acad. Sci. U. S. A., 1996, 93(15), 7475–7480 CrossRef CAS PubMed.
- J. I. García-Plazaola, B. Fernández-Marín, S. O. Duke, A. Hernández, F. López-Arbeloa and J. M. Becerril, Autofluorescence: biological functions and technical applications, Plant Sci., 2015, 236, 136–145 CrossRef PubMed.
- M. Z. Lin, M. R. McKeown, H.-L. Ng, T. A. Aguilera, N. C. Shaner, R. E. Campbell, S. R. Adams, L. A. Gross, W. Ma and T. Alber, Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals, Chem. Biol., 2009, 16(11), 1169–1179 CrossRef CAS PubMed.
- Y. W. Jun, H. R. Kim, Y. J. Reo, M. Dai and K. H. Ahn, Addressing the autofluorescence issue in deep tissue imaging by two-photon microscopy: the significance of far-red emitting dyes, Chem. Sci., 2017, 8(11), 7696–7704 RSC.
- N. Billinton and A. W. Knight, Seeing the wood through the trees: a review of techniques for distinguishing green fluorescent protein from endogenous autofluorescence, Anal. Biochem., 2001, 291(2), 175–197 CrossRef CAS PubMed.
- M. Monici, Cell and tissue autofluorescence research and diagnostic applications, Biotechnol. Annu. Rev., 2005, 11, 227–256 CAS.
- E. A. Rodriguez, G. N. Tran, L. A. Gross, J. L. Crisp, X. Shu, J. Y. Lin and R. Y. Tsien, A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein, Nat. Methods, 2016, 13(9), 763–769, DOI:10.1038/nmeth.3935.
- J.-H. Machado, R. Ting, J. Y. Lin and E. A. Rodriguez, A self-labeling protein based on the small ultra-red fluorescent protein, smURFP, RSC Chem. Biol., 2021, 2(4), 1221–1226 RSC.
- J. P. Fuenzalida-Werner, R. Janowski, K. Mishra, I. Weidenfeld, D. Niessing, V. Ntziachristos and A. C. Stiel, Crystal structure of a biliverdin-bound phycobiliprotein: Interdependence of oligomerization and chromophorylation, J. Struct. Biol., 2018, 204(3), 519–522 CrossRef CAS PubMed.
- F. Montecinos-Franjola, J. Y. Lin and E. A. Rodriguez, Fluorescent proteins for in vivo imaging, where's the biliverdin?, Biochem. Soc. Trans., 2020, 48(6), 2657–2667 CrossRef CAS PubMed.
- I. V. Polyakov, B. L. Grigorenko, V. A. Mironov and A. V. Nemukhin, Modeling structure and excitation of biliverdin-binding domains in infrared fluorescent proteins, Chem. Phys. Lett., 2018, 710, 59–63 CrossRef CAS.
- D. M. Shcherbakova, Near-infrared and far-red genetically encoded indicators of neuronal activity, J. Neurosci. Methods, 2021, 362, 109314 CrossRef PubMed.
- M. Unno, T. Matsui and M. Ikeda-Saito, Structure and catalytic mechanism of heme oxygenase, Nat. Prod. Rep., 2007, 24(3), 553–570 RSC.
- G. Kikuchi, T. Yoshida and M. Noguchi, Heme oxygenase and heme degradation, Biochem. Biophys. Res. Commun., 2005, 338(1), 558–567, DOI:10.1016/j.bbrc.2005.08.020.
- T. Yoshida and C. T. Migita, Mechanism of heme degradation by heme oxygenase, J. Inorg. Biochem., 2000, 82(1–4), 33–41 CrossRef CAS PubMed.
- N. Frankenberg-Dinkel, Bacterial heme oxygenases, Antioxid. Redox Signaling, 2004, 6(5), 825–834 CAS.
- J. Rumnieks and K. Tars, Crystal structure of the bacteriophage Qβ coat protein in complex with the RNA operator of the replicase gene, J. Mol. Biol., 2014, 426(5), 1039–1049 CrossRef CAS PubMed.
- V. L. Morton, W. Burkitt, G. O'Connor, N. J. Stonehouse, P. G. Stockley and A. E. Ashcroft, RNA-induced conformational changes in a viral coat protein studied by hydrogen/deuterium exchange mass spectrometry, Phys. Chem. Chem. Phys., 2010, 12(41), 13468–13475 RSC.
- J.-Y. Chang, K. V. Gorzelnik, J. Thongchol and J. Zhang, Structural assembly of Qβ virion and its diverse forms of virus-like particles, Viruses., 2022, 14(2), 225 CrossRef CAS PubMed.
- P. Stargardt, G. Striedner and J. Mairhofer, Tunable expression rate control of a growth-decoupled T7 expression system by L-arabinose only, Microb. Cell Fact., 2021, 20, 1–17 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb00469d |
| This journal is © The Royal Society of Chemistry 2023 |