Next-generation materials for RNA–lipid nanoparticles: lyophilization and targeted transfection
Ting
Wang†
 a,
Tzu-Cheng
Sung†
a,
Tao
Yu†
a,
Hui-Yu
Lin
b,
Yen-Hung
Chen
b,
Zhe-Wei
Zhu
b,
Jian
Gong
c,
Jiandong
Pan
a and
Akon
Higuchi
a,
Tzu-Cheng
Sung†
a,
Tao
Yu†
a,
Hui-Yu
Lin
b,
Yen-Hung
Chen
b,
Zhe-Wei
Zhu
b,
Jian
Gong
c,
Jiandong
Pan
a and
Akon
Higuchi
 *abde
*abde
aState Key Laboratory of Ophthalmology, Optometry and Visual Science, Eye Hospital, Wenzhou Medical University, No. 270, Xueyuan Road, Wenzhou, Zhejiang 325027, China. E-mail: higuchi@wmu.edu.cn
bDepartment of Chemical and Materials Engineering, National Central University, No. 300, Jhongda RD., Jhongli, Taoyuan 32001, Taiwan. E-mail: higuchi@ncu.edu.tw
cDepartment of Laboratory Medicine, The Second Affiliated Hospital and Yuying Children's Hospital of Wenzhou Medical University, No. 109, Xueyuan West Road, Lucheng District, Wenzhou City, Zhejiang Province 325027, China
dR&D Center for Membrane Technology, Chung Yuan Christian University, Chungli, Taoyuan 32001, Taiwan
eDepartment of Reproduction, National Center for Child Health and Development, 2-10-1 Okura, Setagaya-ku, Tokyo 157-8535, Japan
First published on 4th April 2023
Abstract
RNA, including mRNA, siRNA and miRNA, is part of a new class of patient treatments that prevent and treat several diseases. As an alternative to DNA therapy using plasmid DNA, RNA functions in the cellular cytosol, avoiding the potential risks of insertion into patient genomes. RNA drugs, including mRNA vaccines, need carrier materials for delivery into the patient's body. Several delivery carriers of mRNA, such as cationic polymers, lipoplexes, lipid–polymer nanoparticles and lipid nanoparticles (LNPs), have been investigated. For clinical applications, one of the most commonly selected types of RNA delivery carrier is LNPs, which are typically formed with (a) ionizable lipids, which bind to RNA; (b) cholesterol for stabilization; (c) phospholipids to form the LNPs; and (d) polyethylene glycol-conjugated lipids to prevent aggregation and provide stealth characteristics. Most RNA-LNP research has been devoted to achieving highly efficient RNA expression in vitro and in vivo. It is also necessary to study the extended storage of RNA-LNPs under mild conditions. One of the most efficient methods to store RNA-LNPs for a long time is to prepare freeze-dried (lyophilized) RNA-LNPs. Future research should include investigating LNP materials for the development of freeze-dried RNA-LNPs using optimal lipid components and compositions with optimal cryoprotectants. Furthermore, the development of sophisticated RNA-LNP materials for targeted transfection into specific tissues, organs or cells will be a future direction in the development RNA therapeutics. We will discuss the prospects for the development of next-generation RNA-LNP materials.
Introduction
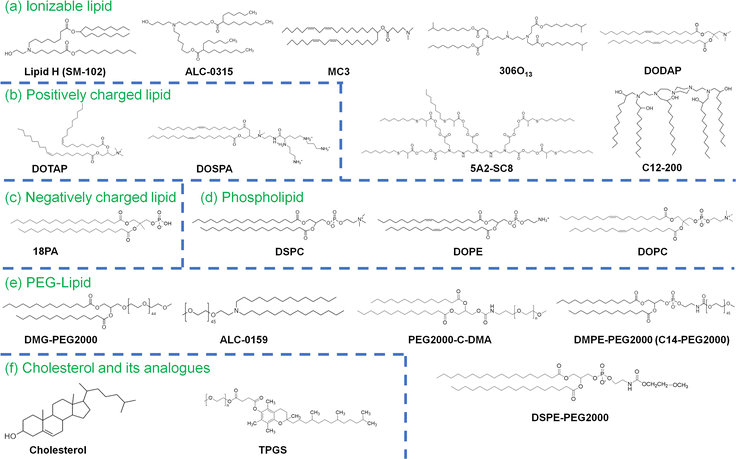
RNA, including messenger RNA (mRNA), siRNA (small interfering RNA) and microRNA (miRNA), contributes to a new class of patient treatments that mitigate, prevent or treat several diseases. As an alternative to DNA therapy using plasmid DNA, RNA functions in the cellular cytosol, preventing the possible risks of insertion into patient genomes.1,2 RNA drugs can be developed very quickly in response to new unknown diseases, including biohazards from bioweapons or unknown viruses. Only mRNA vaccines were approved in the USA, Japan and European countries in December 2020, when coronavirus disease 2019 (COVID-19) spread worldwide in late 2019.2–4 However, RNA drugs, including mRNA vaccines, need carriers for delivery into the patient body. Several delivery carriers of mRNA, such as cationic polymers, lipoplexes, lipid–polymer nanoparticles and lipid nanoparticles (LNPs), have been investigated.5 In research on RNA delivery into cells or animals, cationic materials such as polyethylene imine or lipids such as lipofectamine (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPW)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2,3-dioleoyloxy-N-[2(sperminecarboxamido)ethyl]-N,N-dimethyl-1-propaniminium trifluoroacetate (DOSPA) = 1
2,3-dioleoyloxy-N-[2(sperminecarboxamido)ethyl]-N,N-dimethyl-1-propaniminium trifluoroacetate (DOSPA) = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) (Fig. 1) can be used as transfection agents for RNA. In the clinical application of RNA delivery, a commonly used type of RNA delivery carrier is LNPs, which are typically formed with (a) ionizable lipids, which bind to RNA; (b) cholesterol for stabilization; (c) phospholipids such as 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) and DOPE to form the LNPs; (d) polyethylene glycol (PEG)-conjugated lipids to prevent aggregation and provide stealth characteristics (Fig. 1); and (e) mRNA to generate proteins to treat or mitigate diseases in patients or miRNA and siRNA to regulate specific protein expression and/or signals in the cells (Fig. 2).6 The BNT162b2 (Comirnaty) vaccine produced by BioNtech/Pfizer is formed with ALC-0159 (PEGylated lipid), cholesterol, DSPC, and ALC-0315 (((4-hydroxybutyl)azanediyl)bis(hexane-6,1-diyl)bis(2-hexyldecanoate)) (Fig. 1) in a weight ratio of 1.6
3) (Fig. 1) can be used as transfection agents for RNA. In the clinical application of RNA delivery, a commonly used type of RNA delivery carrier is LNPs, which are typically formed with (a) ionizable lipids, which bind to RNA; (b) cholesterol for stabilization; (c) phospholipids such as 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) and DOPE to form the LNPs; (d) polyethylene glycol (PEG)-conjugated lipids to prevent aggregation and provide stealth characteristics (Fig. 1); and (e) mRNA to generate proteins to treat or mitigate diseases in patients or miRNA and siRNA to regulate specific protein expression and/or signals in the cells (Fig. 2).6 The BNT162b2 (Comirnaty) vaccine produced by BioNtech/Pfizer is formed with ALC-0159 (PEGylated lipid), cholesterol, DSPC, and ALC-0315 (((4-hydroxybutyl)azanediyl)bis(hexane-6,1-diyl)bis(2-hexyldecanoate)) (Fig. 1) in a weight ratio of 1.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 42.7
42.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9.4
9.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 46.3.7 The mRNA-1273 (Spikevax) vaccine produced by Moderna for SARS-CoV-2 is formed with DMG-PEG2000, cholesterol, DSPC and Lipid H (SM-102, heptadecan-9-yl-8-((2-hydroxyethyl)(6-oxo-6-(undecyloxy)hexyl)amino) octanoate)8 in a molar ratio of 1.5
46.3.7 The mRNA-1273 (Spikevax) vaccine produced by Moderna for SARS-CoV-2 is formed with DMG-PEG2000, cholesterol, DSPC and Lipid H (SM-102, heptadecan-9-yl-8-((2-hydroxyethyl)(6-oxo-6-(undecyloxy)hexyl)amino) octanoate)8 in a molar ratio of 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.5
38.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 (Table 1).7 However, these mRNA vaccines (mRNA-LNP) should be frozen and stored at −20 or −70 °C in a cold chain to protect mRNA-LNP efficiency during storage and transportation, whereas most conventional (classical) vaccines can be stored at approximately 4 °C in a refrigerator for more than half a year. The cold chain problem of mRNA-LNP vaccines generates a critical obstacle to vaccine delivery in developing countries with poor infrastructure.2 Most research on mRNA-LNPs, miRNA-LNPs or siRNA-LNPs has been devoted to pursuing highly efficient RNA expression in vitro and in vivo.8–22 Only a few studies have investigated the long-term storage of RNA-LNPs.1,23–25 One of the most efficient methods to store RNA-LNPs for a long time is freeze-drying (lyophilization). It is necessary to investigate LNP materials to develop freeze-dried RNA-LNPs using (i) optimal lipid components and their optimal concentrations and (ii) compositions of cryoprotectants in the future. Furthermore, the development of sophisticated RNA-LNP materials for targeted transfection into specific tissues, organs or cells will be a future direction in RNA therapeutics. We will discuss these prospects for the development of next-generation RNA-LNP materials together with the current situation in the following sections.
50 (Table 1).7 However, these mRNA vaccines (mRNA-LNP) should be frozen and stored at −20 or −70 °C in a cold chain to protect mRNA-LNP efficiency during storage and transportation, whereas most conventional (classical) vaccines can be stored at approximately 4 °C in a refrigerator for more than half a year. The cold chain problem of mRNA-LNP vaccines generates a critical obstacle to vaccine delivery in developing countries with poor infrastructure.2 Most research on mRNA-LNPs, miRNA-LNPs or siRNA-LNPs has been devoted to pursuing highly efficient RNA expression in vitro and in vivo.8–22 Only a few studies have investigated the long-term storage of RNA-LNPs.1,23–25 One of the most efficient methods to store RNA-LNPs for a long time is freeze-drying (lyophilization). It is necessary to investigate LNP materials to develop freeze-dried RNA-LNPs using (i) optimal lipid components and their optimal concentrations and (ii) compositions of cryoprotectants in the future. Furthermore, the development of sophisticated RNA-LNP materials for targeted transfection into specific tissues, organs or cells will be a future direction in RNA therapeutics. We will discuss these prospects for the development of next-generation RNA-LNP materials together with the current situation in the following sections.
| Chemical name | Type | Abbreviations | Molar ratios | Sucrose concen. |
|---|---|---|---|---|
| a Sucrose concen. represents for sucrose concentration (w/v, g mL−1). | ||||
| COMIRNATY® (COVID-19 Vaccine, mRNA) | ||||
| ((4-Hydroxybutyl)azanediyl)bis(hexane-6,1-diyl)bis(2-hexyldecanoate) | Ionizable lipids | ALC-0315 | 46.3 | 2% |
| 2-[(Polyethylene glycol)-2000]-N,N-ditetradecylacetamide | PEG-lipids | ALC-0159 | 1.6 | |
| 1,2-Distearoyl-sn-glycero-3-phosphocholine | Help lipids | DSPC | 9.4 | |
| Cholesterol | Cholesterol | Chol. | 42.7 | |
| SPIKEVAX (COVID-19 Vaccine, mRNA) | ||||
| Heptadecan-9-yl 8-((2-hydroxyethyl)(6-oxo-6-(undecyloxy)hexyl)amino)octanoate | Ionizable lipids | SM-102 | 50 | 8.7% |
| 1,2-Dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 | PEG-lipids | DMG-PEG2000 | 1.5 | |
| 1,2-Distearoyl-sn-glycero-3-phosphocholine | Neutral lipids | DSPC | 10 | |
| Cholesterol | Cholesterol | Chol. | 38.5 | |
| ONPATTRO (polyneuropathy of hereditary transthyretin-mediated amyloidosis, siRNA) | ||||
| (6Z,9Z,28Z,31Z)-Heptatriaconta-6,9,28,31-tetraen-19-yl 4-(dimethylamino)butanoate | Ionizable lipids | DLin-MC3-DMA | 50 | None |
| 1,2-Dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 | PEG-lipids | DMG-PEG2000 | 1.5 | |
| 1,2-Distearoyl-sn-glycero-3-phosphocholine | Neutral lipids | DSPC | 10 | |
| Cholesterol | Cholesterol | Chol. | 38.5 | |
Structure of RNA-LNPs
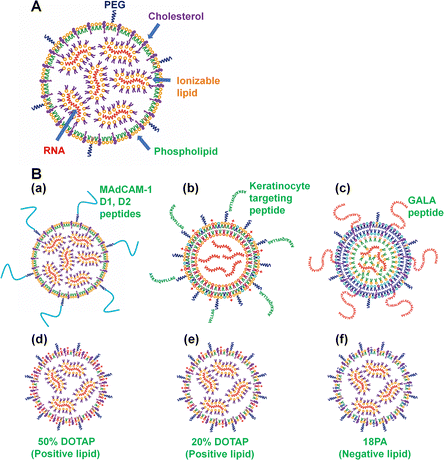
RNA-LNPs are typically composed of (a) ionizable lipid; (b) cholesterol or cholesterol derivatives, which can serve as a stabilizing agent for the RNA-LNPs, (c) phospholipids such as DSPC and DOPE to form the LNPs; (d) PEG–lipid or derivatives; and (e) mRNA, as described in the previous section. Other lipids, such as positively or negatively charged lipids, are sometimes used as additional components of LNPs. Furthermore, in some cases, lipids are conjugated with peptides or nucleotides, such as RNA or DNA aptamers, to guide RNA-LNPs into specific cell types, tissues or organs. A schematic representation of RNA-LNPs is shown in Fig. 2. Ionizable lipids, but not cationic lipids, are extensively utilized for LNP compositions that are ionized (protonated) at the pH in endosomes (low pH) but neutral at pH 7. The pH-dependent properties of ionizable lipids favor RNA transfection into cells in vivo because neutral lipids are biocompatible with the negatively charged membranes of blood cells. The positive charge of the ionizable lipids at low pH facilitates the endosomal escape of mRNA. Most researchers use DLin-MC3-DMA (MC3) as an ionizable lipid (Fig. 1).2,8–20 The ionizable lipid SM-102 (Lipid H) is utilized in the mRNA-1273 (Spikevax) vaccine, and the ionizable lipid ALC-0315 is used in the BNT162b2 (Comirnaty) vaccine (Fig. 1).PEG-lipids inhibit LNP aggregation as well as nonspecific proteins adhesion, conferring stealth properties on LNPs. Several PEG chain lengths in PEG-lipids were investigated, including C14-PEG350 (232 nm), C14-PEG1000 (121 nm), C14-PEG2000 (67 nm), 1,2-distearoylsn-glycero-3-phosphoethanolamine-N-[methoxy-PEG2000]) (C18-PEG2000 (DSPE-PEG2000); 110 nm), and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy-PEG3000] (C14-PEG3000; LNP diameter = 96 nm),26 and C14-PEG2000 (the smallest LNP) elicited the strongest immune response as represented by the CD8+ T-cell count in mouse blood. In general, many investigators have chosen PEG2000 lipids, such as DMPE-PEG2000 (C14-PEG2000),19,26–28 C18-PEG2000 (DSPE-PEG2000 or 18:00 PEG2000-PE),29,30 and DMG-PEG2000.1,12,15,31–34
Development of freeze-dried RNA-LNPs for long-term storage of RNA-LNPs under mild conditions
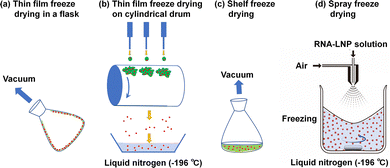
In general, the biological efficiency (expression of RNA) of RNA-LNPs in solution is decreased and the size of mRNA-LNPs increased after one week at room temperature. It is difficult to maintain the biological efficiency of mRNA-LNPs in solution under mild conditions (4 °C or room temperature) for more than one week. Several methods have been developed for the long-term storage of liposomes or LNPs, such as freezing-thawing, spray drying, supercritical fluid technology and freeze-drying (lyophilization) processes.35–37 The lyophilization method is typically utilized in pharmaceuticals to enhance the shelf life and stability of various drugs by eliminating water from the drugs.23,38,39 Some freeze-drying methods are schematically illustrated in Fig. 3. In a freeze-drying state, RNA-LNPs in a dry powder form can be conveniently delivered worldwide with no need for cold storage at −20 or −70 °C. The freeze-drying process of RNA-LNPs is currently the most reliable and stable method to maintain the biological efficiency (expression of RNA) of RNA-LNPs for long-term storage at 4 °C or room temperature for 3 or 6 months. Cryoprotectant (5–20%) is typically added to the RNA-LNP solution before freeze-drying.The biological efficiency and size of RNA-LNPs depend on the concentration and selection of the cryoprotectants. There are two major considerations for the mechanisms of cryoprotectants to avoid RNA-LNPs from their freezing damage. Some cryoprotectants function by lowering the glass transition temperature of the RNA-LNP solution. Therefore, the cryoprotectant prevents actual freezing of RNA-LNP solution. Another mechanism is that the cryoprotectants replace the water molecules in the RNA-LNP solution. Because the hydrogen bonding in the aqueous solutions is important for the proper function of RNA-LNPs. Therefore, RNA-LNPs can retain their native physiological structure and function. As for adverse effect of cryoprotectant usage for storage of RNA-LNPs, the size of RNA-LNPs enhances more or less after reconstruction of freeze-dried RNA-LNPs. The cryoprotectants that have been evaluated for the freeze-drying of LNPs include monosaccharides (glucose,40–44 fructose,41–43,45 mannitol,1,44,46–53 mannose,40,43–45 sorbitol,41,44,46,47 and xylose45), disaccharides (sucrose,1,23,24,40,42,45,52,54–57 lactose,44,45 maltose,23,40,42,44,54 and trehalose1,24,40,44,46,47,53,54,56,58–60), polysaccharides (raffinose),45 hydrophilic polymers (polyvinylpyrrolidone (PVP),44,46,47 polyvinyl alcohol (PVA),44,51 and gelatin44), and hydrophilic chemicals (glycine,44,46,47) (Table 2).
| Monosaccharides | Disaccharides | Polysaccharides | Hydrophilic polymers | Amino acid |
|---|---|---|---|---|
| a PVA, polyvinylalcohol; PVP, polyvinylpyrrolidone. | ||||
| Glucose | Sucrose | Raffinose | PVP | Glycine |
| Fructose | Lactose | PVA | ||
| Mannitol | Maltose | Gelatin | ||
| Mannose | Trehalose | |||
| Sorbitol | ||||
| Xylose | ||||
Ball et al. prepared freeze-dried siRNA-LNPs with 0, 1, 5, 10, or 20% trehalose or sucrose, where the LNPs were composed of DSPC, PEG2000-PE (DSPE-PEG2000, 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000]), cholesterol, and lipidoid 306O13 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5
1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.5
38.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) (Fig. 1).24 In a freeze-thaw process rather than a freeze-drying process, their results showed very low efficacy of siRNA., i.e., gene silencing in cells after reconstitution of siRNA-LNP in water.24 The effects of pH, temperature and lyophilization on siRNA-LNP efficacy in HeLa cells were also evaluated in their study. Their results suggested that siRNA-LNPs in aqueous solution were stable over 150 days at 2 °C compared to frozen siRNA-LNPs at −20 °C. The stability of siRNA-LNPs showed good stability at 2 °C, which were stored in any pH of the storage buffer at pH 3, 7.4 and 9 in their study.
50) (Fig. 1).24 In a freeze-thaw process rather than a freeze-drying process, their results showed very low efficacy of siRNA., i.e., gene silencing in cells after reconstitution of siRNA-LNP in water.24 The effects of pH, temperature and lyophilization on siRNA-LNP efficacy in HeLa cells were also evaluated in their study. Their results suggested that siRNA-LNPs in aqueous solution were stable over 150 days at 2 °C compared to frozen siRNA-LNPs at −20 °C. The stability of siRNA-LNPs showed good stability at 2 °C, which were stored in any pH of the storage buffer at pH 3, 7.4 and 9 in their study.
Lyophilized siRNA-LNPs with 20% sucrose or trehalose maintained 80–87% gene silencing compared to that of fresh siRNA-LNPs. Typically, the RNA bioactivity of freeze-thawed RNA-LNPs with a cryoprotectant is less easy to maintain than that of freeze-dried RNA-LNPs. The aggregation of siRNA-LNPs and loss of siRNA efficacy were observed during freeze-thaw cycles, and the stability of siRNA-LNPs could be improved using the cryoprotectants trehalose or sucrose. Trehalose and sucrose are also lyoprotectants used in commercially available lyophilized products such as Avastin (bevacizumab, a monoclonal antibody used as a cancer treatment drug and an angiogenesis inhibitor), Herceptin (trastuzumab, a monoclonal antibody used as a cancer treatment drug), LEP-ETU (liposome-entrapped paclitaxel easy-to-use formulation), and AmBisome (amphotericin B, an antifugal medication) for enhanced long-term stability.24,61,62
Zhao et al. prepared lyophilized firefly luciferase-encoding mRNA-LNPs and found that the reconstituted mRNA-LNPs maintained the efficiency of mRNA expression in mice based on in vivo bioluminescence imaging data.1 They evaluated the stability of lyophilized firefly luciferase-encoding mRNA-LNPs with different concentrations of cryoprotectants (mannitol, trehalose, or sucrose) under freezing or lyophilization.1 By performing mRNA delivery studies in vitro and in vivo, they investigated the optimal storage conditions and found that the addition of 5% (w/v) trehalose or sucrose to mRNA-LNPs maintained mRNA efficacy for at least 3 months in liquid nitrogen storage. Our preliminary data also suggest that the addition of 5% (w/v) sucrose or trehalose to mRNA-LNPs enables the long-term storage of freeze-dried mRNA-LNPs under mild conditions (4 °C storage), whereas the storage of freeze-thawed mRNA-LNPs requires the addition of a higher concentration of sucrose, such as 20% sucrose, to the mRNA-LNP solution (unpublished data).
Stitz et al. developed mRNA-LNPs using CureVac's RNActive technology, incorporating mRNA that encoded full-length rabies virus glycoprotein, and then freeze-dried the mRNA-LNPs.25,63 Their mRNA-LNPs could be stored at temperatures as high as 70 °C for several months, after which mice injected with the mRNA-LNPs could produce neutralizing antibodies and showed protection against lethal rabies infection. Thus, their mRNA-LNPs showed high thermal stability.25,63 However, it is unknown whether their mRNA-LNPs were freeze-dried with a specific cryoprotectant.
Muramatsu et al. prepared mRNA-LNPs and freeze-dried mRNA-LNPs,23 in which the LNPs consisted of PEG lipid (PEG2000-C-DMA), cholesterol, DSPC and an ionizable lipid ((6Z,16Z)-12-((Z)-dec-4-en-1-yl)docosa-6,16-dien-11-yl 5-(dimethylamino)pentanoate) in a molar ratio of 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.5
38.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50. mRNA-LNPs with 10% sucrose and 10% maltose (cryoprotectants) were freeze-dried and stored at −80, −20, 4, 25 (room temperature), and 42 °C for one, three, and six months. The physicochemical properties (size and polydispersity index (PDI)) did not change greatly after storage at 4 °C for six months or at 25 °C for 3 months. Firefly luciferase-encoding mRNA-LNPs showed high bioluminescence imaging (high production of luciferase) in mice when the mRNA-LNPs were injected into the mice intramuscularly or intradermally after storage at 4 °C for six months or 25 °C for 3 months.23 Furthermore, when an mRNA (a nucleoside-modified mRNA for hemagglutinin from the A/Puerto Rico/8/1934 influenza virus strain, PR8 HA)-LNP vaccine was injected into mice intramuscularly or intradermally, mRNA-LNPs could generate an immune response in mice even after 6 months of storage at 4 °C or 3 months of storage at room temperature in a lyophilized form.23 Their study indicated that the freeze-dried form of mRNA-LNPs is useful for long-term storage under mild conditions such as 4 °C or room temperature.
50. mRNA-LNPs with 10% sucrose and 10% maltose (cryoprotectants) were freeze-dried and stored at −80, −20, 4, 25 (room temperature), and 42 °C for one, three, and six months. The physicochemical properties (size and polydispersity index (PDI)) did not change greatly after storage at 4 °C for six months or at 25 °C for 3 months. Firefly luciferase-encoding mRNA-LNPs showed high bioluminescence imaging (high production of luciferase) in mice when the mRNA-LNPs were injected into the mice intramuscularly or intradermally after storage at 4 °C for six months or 25 °C for 3 months.23 Furthermore, when an mRNA (a nucleoside-modified mRNA for hemagglutinin from the A/Puerto Rico/8/1934 influenza virus strain, PR8 HA)-LNP vaccine was injected into mice intramuscularly or intradermally, mRNA-LNPs could generate an immune response in mice even after 6 months of storage at 4 °C or 3 months of storage at room temperature in a lyophilized form.23 Their study indicated that the freeze-dried form of mRNA-LNPs is useful for long-term storage under mild conditions such as 4 °C or room temperature.
Most researchers use a single-component cryoprotectant. However, Muramatsu et al. used 10% sucrose and 10% maltose as cryoprotectants of mRNA-LNPs.23 Unfortunately, they did not investigate the effect of single-component or multicomponent cryoprotectants on the physicochemical characteristics and/or bioassay (in vivo expression of proteins in mice by injection of mRNA-LNP intramuscularly or intradermally) results of mRNA-LNPs, such as observing the luciferase expression or immune response in mice after the injection of freeze-dried mRNA-LNPs with (a) 10% or 20% sucrose, (b) 10% or 20% maltose, or (c) 10% sucrose plus 10% maltose. It is necessary to evaluate whether the excellent data reported in this study are based on the multicomponent cryoprotectant usage of 10% sucrose plus 10% maltose.
Kamiya et al. prepared LNPs loaded with griscofulvin (an antifungal medication) instead of mRNA using a high-pressure homogenization method.54 When LNPs were not freeze-dried together with a cryoprotectant, the size of LNPs in the solution increased from the native size of approximately 50 nm to 300 nm after half a year of storage at 25 °C. When the nanoparticles were freeze-dried with 5% cryoprotectant, the particle size of LNPs was decreased to 160 nm using a cryoprotectant of monosaccharide (galactose or glucose), but freeze-dried LNPs with a disaccharide cryoprotectant (trehalose, maltose, or sucrose) maintained their size at 55–65 nm even after half a year of storage at 25 °C.54 The powder X-ray diffraction pattern of freeze-dried LNPs with monosaccharide showed a crystal state of monosaccharide. However, the X-ray diffraction pattern of freeze-dried LNPs with disaccharide indicated an amorphous state of disaccharide. From these X-ray diffraction results, the high viscosity of an amorphous disaccharide should prevent LNP aggregation by retarding molecular movement. It was typically found that freeze-dried LNPs containing disaccharide were stable for longer periods than LNPs in solution or freeze-dried LNPs containing monosaccharide. The disaccharides directly contact the polar groups of the lipids to inhibit particle fusion during freezing and can also inhibit ice generation by increasing the glass transition temperature of the solution.24,64,65 The vitrification explanation indicates that the addition of sugar keeps the RNA-LNP solution in a glassy matrix state, which inhibits both aggregation and ice crystal attack on the lipid layer.24,38
This finding explains why disaccharides such as sucrose or trehalose are better cryoprotectants than monosaccharides to maintain the original diameter of LNPs containing not only drugs but also RNA during and after freeze-drying. The development of freeze-dried RNA-LNPs, especially mRNA-LNP vaccines for COVID-19, is currently an urgent societal need.
Targeted therapy using RNA-LNPs
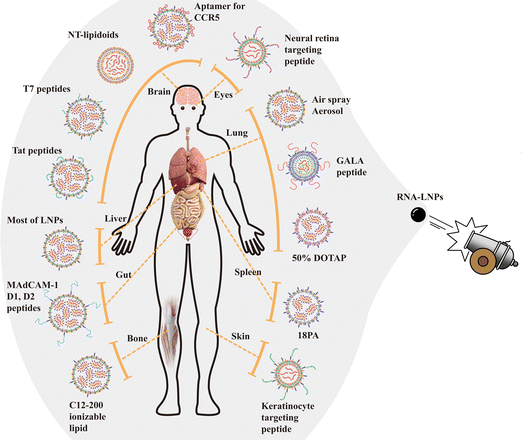
Targeting the transfection of RNA into specific cells or organs using RNA-LNPs will be one of the future directions in the development of sophisticated RNA-LNP materials. RNA-LNPs and other drugs typically accumulate in liver cells upon intravenous administration.66 Several LNPs have been developed for the delivery of LNPs into targeted tissues by different administration routes in vivo,48,67–72 by adapting the specific lipid composition,73,74 and by the addition of specific peptides,55,74–76 aptamers,77 and ligands (Fig. 4).66,78 Some recent examples of the targeted delivery of RNA-LNPs into specific organs or tissues are described below.Aerosolizable siRNA-LNPs,15 which were prepared using thin-film freeze-drying, spray-drying or conventional freeze-drying (Fig. 3), were developed by Wang et al.48 for the direct delivery of siRNA into the patient's lung in LNPs composed of PEG–lipid, cholesterol and lecithin. The zeta potential, PDI, and size of the siRNA-LNPs were maintained when the siRNA-LNPs were thin-film freeze-dried and reconstituted into siRNA-LNP solution.48 However, the characteristics of the original siRNA-LNPs were not reproduced after the siRNA-LNPs were subjected to conventional freeze-drying and reconstituted into siRNA-LNP solution. The dry powders generated by thin-film freeze-drying exhibited better aerosol characteristics than those generated by spray drying.48 The thin-film freeze-drying of siRNA-LNPs is a promising way to generate aerosolizable dry powder siRNA-LNPs for future patient treatment.
Nanoparticles targeted to a specific cell type are expected to show high uptake efficiency in the desired cell type. Therefore, Li et al. prepared freeze-dried keratinocyte-targeted LNPs (nanocarriers) of locked nucleic acid (LNA)-modified anti-miR-107 (Fig. 2(B)), where the LNPs consisted of 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP, positively charged lipid)/1,2-dioleoyl-3-dimethylammonium propane (DODAP, an ionizable lipid), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, a phospholipid), D-α-tocopherol polyethylene glycol 1000 succinate (TPGS) (Fig. 1), and DSPE-PEG2000-amine-keratinocyte-targeting peptide sequence (ASKAIQVFLLAG) (Table 3).55 The anti-miRNA (miR)-107 oligopeptide neutralizes miR-107. Freeze-drying of the LNPs was performed with 20% sucrose to enhance the shelf life of the LNPs.
| Peptide name | Sequences | Targeting tissue or cell |
|---|---|---|
| Keratinocyte-targeting peptide | ASKAIQVFLLAG | Keratinocyte |
| GALA | WEAALAEALAEALAEHLAEALAEALEALAA | Lung endothelium |
| MH43 | SPALHFL | Neural retina |
| TaT | YGRKKRRQRRR | BBB penetrating |
| T7 | HAIYPRH | BBB penetrating |
Twice-weekly treatment of burn wounds on the skin of mice or rats with hydrogel-dispersed freeze-dried LNPs with anti-miR107 greatly enhanced wound closure and the restoration of skin barrier characteristics.55 Freeze-dried LNPs with anti-miR107 depleted miR-107 and upregulated Dicer expression, which enhanced keratinocyte differentiation.55 These LNPs hold promise as treatment drugs for the management of burn injury of the skin. In the presence of 10% serum, the LNPs showed 90% uptake by keratinocytes within the first 4 h, which demonstrated targeting efficiency and specificity. Rapid uptake by targeted cells should minimize clearance by immune cells at the wound site.
Hagino et al. developed double-coated LNPs decorated with the GALA peptide (WEAALAEALAEALAEHLAEALAEALEALAA, α-helical peptide79)75 because the GALA peptide has a high affinity for the lung endothelium (Table 3)80 and was therefore utilized as a ligand conjugated with cholesterol to target the lung endothelium as a component of LNPs. They successively delivered pDNA of luciferase into the lung tissue of mice using pDNA–LNPs at approximately 75 ng of luciferase protein per mg of protein.75
The targeted delivery of LNPs across the blood-brain barrier (BBB) is currently a challenge. For example, there is effective anti-retroviral medicine for the treatment of human immunodeficiency virus 1 (HIV-1). However, HIV-associated neurological disorders persist because of the poor uptake of anti-retroviral medicine across the BBB.81–83 Therefore, Ray et al. used two strategies to develop LNPs that can permeate across the BBB.77 The LNPs in this study consisted of DMG-PEG2000, DSPC, cholesterol, and DLin-MC3-DMA (Fig. 1) as well as lipid-conjugated cell-penetrating peptides (TaT and T7 peptides) (Table 3).77 One strategy is that the BBB-penetrating trans-activator of transcription (TaT, YGRKKRRQRRR) peptide or the T7 peptide (HAIYPRH), an arginine-rich short cell-penetrating peptide obtained from the natural nuclear Tat protein of HIV-1, was immobilized on the LNP surface.77 Several researchers have reported that HIV-1 Tat peptides can penetrate the BBB by acting as cell-penetrating peptides (Table 3).74,84 The T7 peptide has a high affinity for the transferrin receptor.85,86 Transferrin receptors are present in large numbers in brain capillaries, nucleated cells, and rapidly dividing cells,87 and transferrin is known to enhance the transport of oligonucleotides or small molecules across the BBB.87–90 The T7 peptide is reported to support drug delivery into brain tissue.77,85,86,88,90,91 These peptides were conjugated to the N-terminal lipid anchor of LNPs.
Currently, RNA-LNP delivery is restricted to the Müller glia or retinal pigment epithelium (RPE).76 RNA-LNPs need to pass ocular barriers for transfection of neuronal cells, which are important for visual phototransduction of the photoreceptors (PRs). Herrera-Barrera et al. identified peptides, which target PRs by utilizing a combinatorial M13 bacteriophage–based heptameric peptide phage display library.76 The peptide, they found, SPALHFL, showed immediate localization to the PRs. Therefore, they prepared RNA-LNPs conjugated with the peptide, which successfully could deliver mRNA into RPE, Müller glia, and PRs in mice and nonhuman primate, where LNPs were consisted of DMG-PEG2000, DSPC, cholesterol, and DLin-MC3-DMA as well as DSPE-PEG2000-conjugated MH43 peptide (SPALHFL-GGGSC).76 The peptide, GGGSC, was used as a joint segment to bind DSPE-PEG2000 and MH43 peptide. They developed specific peptide-conjugated RNA-LNPs that can enable mRNA delivery to the neural retina (PRs), which can expand the utility of mRNA-LNP therapies for inherited blindness of patients in future.
The delivery of siRNA using LNPs specific to activation-sensitive receptors expressed on gut-homing leukocytes in a mouse model of colitis was investigated by Dammes et al.66 with a targeting site consisting of a specific protein conformation, namely, the high-affinity conformation of integrin α4β7. This is because gut-homing leukocytes use this pivotal intestinal homing receptor to adhere to the intestinal endothelium, where α4β7 integrin is a key protein in homing leukocytes to the gut during intestinal inflammation. Integrin α4β7 has the potential to bind both vascular cell adhesion molecule-1 (VCAM-1) for homing to peripheral tissues and mucosal vascular addressin cell adhesion molecule-1 (MAdCAM-1) for homing to intestinal tissues but not for simultaneous binding. Only leukocytes that actively home to the intestinal tissue possess α4β7 integrin in the high-affinity conformation. Therefore, LNPs that target high-affinity α4β7-expressing cells were prepared in this study.66 Domain D1 and D2 peptides of MAdCAM-1, which can specifically bind to α4β7 integrin with the high-affinity conformation, was immobilized on a PEG lipid to form one of the components of LNPs in this study. By targeting the high-affinity conformation of α4β7 integrin, which is a hallmark of inflammatory gut-homing leukocytes, siRNA-LNPs loaded with siRNA of interferon-γ extensively silenced interferon-γ in the gut, resulting in improved therapeutic results in a mouse model of colitis. The siRNA-LNPs did not show liver toxicities or adverse immune activation. This study suggests that RNA-LNPs targeting specific tissues or organs could be valuable for the selective delivery of payloads to other conformation-sensitive targets.
In another strategy, an RNA aptamer targeted to glycoprotein gp160 derived from human immunodeficiency virus (HIV) or C–C chemokine receptor 5 (CCR5, an HIV-1 coreceptor) was conjugated on siRNA through a stick bridge motif.77 An aptamer is an artificial oligonucleotide (DNA or RNA) binding to one or more specific target molecules. The molecular weight of an aptamer is approximately 5–30 kDa, which is much smaller than that of antibodies at approximately and above 150 kDa, making aptamers suitable for conjugation to lipids or RNA as one of the components of RNA-LNPs. A CCR5-selective RNA aptamer contributed to facilitating the entry of LNPs immobilized into a BBB model (brain model) and the uptake of these LNPs into CCR5-expressing cells (HIV-1 infected cell models) without immune reaction in primary human monocyte-derived macrophages. However, the gp160 aptamer-immobilized LNP could not permeate into a BBB model and could not be taken up into CCR5-expressing cells.77 The addition of cell-penetrating peptides, T7 and Tat, on the surface of LNPs with CCR5-selective RNA aptamer did not enhance the penetration into a BBB model.77 This research revealed the ability of aptamer-loaded LNPs to enhance the specific targeting of cells and delivery of central-nervous-system-active RNAi (RNA interference)-LNP therapy across the BBB.
In addition to specific peptides or aptamers, RNA-LNPs can be passively delivered into specific tissues or organs by the selection of specific lipids for use in the LNPs.69,73,75,92–95 LNPs containing the ionizable lipid C12-200 (1,10-[[2-[4-[2-[[2-[bis(2-hydroxydodecyl)amino]ethyl](2-hydroxydodecyl)amino]ethyl]-1-piperazinyl]ethyl]imino]bis-2-dodecanol) exhibit improved specific delivery of LNPs into muscles.73,94,95 Ma et al. prepared neurotransmitter-derived lipidoids (NT-lipidoids), which can cross the BBB and deliver antisense oligonucleotides (ASOs) against tau when administered by intravenous injection.73,93
Cheng et al.69 and Wei et al.96 reported that the addition of DOTAP (a positively charged lipid) to LNPs allowed LNPs to target specific organs such as the lung. RNA-LNPs composed of 50% DOTAP, namely, DOTAP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMG-PEG2000
DMG-PEG2000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol
cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE
DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5A2-SC8 = 50
5A2-SC8 = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.4
2.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 23.8
23.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11.9
11.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11.9, and loaded with Cas9 mRNA/single-guide RNA were intravenously injected into mice and successfully edited phosphatase and tensin homolog (PTEN) exclusively in the lungs with 15.1% indels.69
11.9, and loaded with Cas9 mRNA/single-guide RNA were intravenously injected into mice and successfully edited phosphatase and tensin homolog (PTEN) exclusively in the lungs with 15.1% indels.69
RNA-LNPs composed of 20% DOTAP, namely, DOTAP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMG-PEG2000
DMG-PEG2000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol
cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE
DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5A2-SC8 = 20
5A2-SC8 = 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.8
3.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.1
38.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19.05
19.05![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19.05, successfully edited PTEN exclusively in the liver of mice with 13.9% gene editing.69 Furthermore, Cheng et al. prepared LNPs with the addition of 1,2-dioleoyl-sn-glycero-3-phosphate (18PA, a negatively charged lipid) to enhance the delivery of RNA-LNPs into the spleen.69 Their LNPs consisted of 18PA
19.05, successfully edited PTEN exclusively in the liver of mice with 13.9% gene editing.69 Furthermore, Cheng et al. prepared LNPs with the addition of 1,2-dioleoyl-sn-glycero-3-phosphate (18PA, a negatively charged lipid) to enhance the delivery of RNA-LNPs into the spleen.69 Their LNPs consisted of 18PA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMG-PEG2000
DMG-PEG2000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol
cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE
DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5A2-SC8 = 30
5A2-SC8 = 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.3
3.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33.3
33.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 16.7
16.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 16.7. These LNPs containing the Cas9 protein and an sgRNA targeting PTEN were intravenously injected into mice, and successful gene editing of PTEN in the spleen was observed.69
16.7. These LNPs containing the Cas9 protein and an sgRNA targeting PTEN were intravenously injected into mice, and successful gene editing of PTEN in the spleen was observed.69
However, in most cases, we do not currently know why some specific lipids in RNA-LNPs facilitate specific delivery into the targeted tissue or organs (Fig. 4). It should be an interesting study to investigate the reason and mechanism by which specific lipids in RNA-LNPs promote the specific delivery of RNA-LNPs into the targeted tissue or organs.
Current RNA-LNP products
The BNT162b2 (Comirnaty) vaccine produced by BioNtech/Pfizer and the mRNA-1273 (Spikevax) vaccine produced by Moderna for SARS-CoV-2 are RNA-LNP products that were already approved by the FDA, but they should be stored at −70 or −20 °C for long-term storage.2 Some freeze-dried RNA-LNP products are also in clinical trials or on the market. TKM-100802, which was developed by Tekmira Pharmaceuticals, is a freeze-dried RNA-LNP containing siRNA for the treatment of Zaire Ebola virus (ZEBOV) infection that targets the L polymerase proteins VP35 and VP24.23,97,98 Onpattro (patisiran), composed of DLin-MC3-DMA, PEG2000-C-DMG ((α-(3′-((1,2-di(myristyloxy)propanoxy) carbonylamino)propyl)-ω-methoxy, polyoxyethylene), DSPC, cholesterol, and siRNA, which was developed by Alnylam Pharmaceuticals and is now on the market for the treatment of hereditary transthyretin-mediated amyloidosis,48,99 can be stored in solution form at 2 or 8 °C for 3 years. Currently, commercially available RNA-LNPs are not designed to be delivered into specific tissues or organs. It is necessary to develop RNA-LNPs with characteristics enabling targeted delivery into specific organs or tissues, such as the muscle, brain, lung, heart, spleen, bone, or eyes, in the future.Conclusions
The freeze-drying of RNA-LNPs with optimal cooling programs and optimal cryoprotectants is important for the development of RNA-LNP products, including mRNA-LNP vaccines, for storage under mild conditions (room temperature or 4 °C) and delivery to remote areas. To achieve therapeutic effects, the RNA in RNA-LNPs should be delivered into specific target organs and cells to perform functions such as the production of proteins or the inhibition of specific signaling pathways. However, the targeted delivery and endosomal escape of RNA-LNPs remain challenging for RNA delivery systems, which highlights the need for the development of safe, effective and specific targeted delivery methods for RNA-LNPs in patient bodies. The conjugation and expression of specific peptides or aptamers on RNA-LNPs is likely to be the most reliable strategy to recognize and deliver RNA-LNPs into specific organs and/or cells. Furthermore, the development and discovery of specific lipids components that can target the delivery of RNA-LNPs into specific organs or tissues will be important for the safe delivery of RNA-LNPs into the body of patients.Author contributions
Conceptualization: A. H. Funding acquisition: A. H. Investigation: T. W., T. Y., H. L., Y. C., Z. Z., J. G., J. P. Supervision: A. H. Writing – original draft: T. S., A. H. writing – review & editing: A. H.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (52250710155), the National Key Research and Development Program of China (2022YFA1105501), and the Project of the State Key Laboratory of Ophthalmology, Optometry and Vision Science, Wenzhou Medical University (J02-20210201). This study was also supported by the Wenzhou Municipal Science and Technology Bureau (Y2020227, Y2020920).Notes and references
- P. Zhao, X. Hou, J. Yan, S. Du, Y. Xue, W. Li, G. Xiang and Y. Dong, Bioact. Mater., 2020, 5, 358–363 CrossRef PubMed.
- A. Higuchi, T. C. Sung, T. Wang, Q. D. Ling, S. S. Kumar, S. T. Hsu and A. Umezawa, Polym. Rev., 2023, 63, 394–436 CrossRef CAS.
- L. A. Jackson, E. J. Anderson, N. G. Rouphael, P. C. Roberts, M. Makhene, R. N. Coler, M. P. McCullough, J. D. Chappell, M. R. Denison, L. J. Stevens, A. J. Pruijssers, A. McDermott, B. Flach, N. A. Doria-Rose, K. S. Corbett, K. M. Morabito, S. O’Dell, S. D. Schmidt, P. A. Swanson, 2nd, M. Padilla, J. R. Mascola, K. M. Neuzil, H. Bennett, W. Sun, E. Peters, M. Makowski, J. Albert, K. Cross, W. Buchanan, R. Pikaart-Tautges, J. E. Ledgerwood, B. S. Graham, J. H. Beigel and mRNA-1273 Study Group, N. Engl. J. Med., 2020, 383, 1920–1931 CrossRef CAS PubMed.
- A. Wadhwa, A. Aljabbari, A. Lokras, C. Foged and A. Thakur, Pharmaceutics, 2020, 12, 102 CrossRef CAS PubMed.
- X. Hou, T. Zaks, R. Langer and Y. Dong, Nat. Rev. Mater., 2021, 6, 1078–1094 CrossRef CAS PubMed.
- M. L. Guevara, F. Persano and S. Persano, Front. Chem., 2020, 8, 589959 CrossRef CAS PubMed.
- L. Schoenmaker, D. Witzigmann, J. A. Kulkarni, R. Verbeke, G. Kersten, W. Jiskoot and D. J. A. Crommelin, Int. J. Pharm., 2021, 601, 120586 CrossRef CAS PubMed.
- K. J. Hassett, K. E. Benenato, E. Jacquinet, A. Lee, A. Woods, O. Yuzhakov, S. Himansu, J. Deterling, B. M. Geilich, T. Ketova, C. Mihai, A. Lynn, I. McFadyen, M. J. Moore, J. J. Senn, M. G. Stanton, O. Almarsson, G. Ciaramella and L. A. Brito, Mol. Ther.–Nucleic Acids, 2019, 15, 1–11 CrossRef CAS PubMed.
- J. F. Nabhan, K. M. Wood, V. P. Rao, J. Morin, S. Bhamidipaty, T. P. LaBranche, R. L. Gooch, F. Bozal, C. E. Bulawa and B. C. Guild, Sci. Rep., 2016, 6, 20019 CrossRef CAS PubMed.
- E. Robinson, K. D. MacDonald, K. Slaughter, M. McKinney, S. Patel, C. Sun and G. Sahay, Mol. Ther., 2018, 26, 2034–2046 CrossRef CAS PubMed.
- M. Sedic, J. J. Senn, A. Lynn, M. Laska, M. Smith, S. J. Platz, J. Bolen, S. Hoge, A. Bulychev, E. Jacquinet, V. Bartlett and P. F. Smith, Vet. Pathol., 2018, 55, 341–354 CrossRef CAS PubMed.
- S. Patel, N. Ashwanikumar, E. Robinson, Y. Xia, C. Mihai, J. P. Griffith, 3rd, S. Hou, A. A. Esposito, T. Ketova, K. Welsher, J. L. Joyal, O. Almarsson and G. Sahay, Nat. Commun., 2020, 11, 983 CrossRef CAS PubMed.
- E. Kon, I. Hazan-Halevy, D. Rosenblum, N. Cohen, S. Chatterjee, N. Veiga, P. Raanani, O. Bairey, O. Benjamini, A. Nagler and D. Peer, Pharmaceutics, 2020, 12, 520 CrossRef CAS PubMed.
- A. Mukherjee, K. D. MacDonald, J. Kim, M. I. Henderson, Y. Eygeris and G. Sahay, Sci. Adv., 2020, 6, eabc5911 CrossRef CAS PubMed.
- H. Zhang, J. Leal, M. R. Soto, H. D. C. Smyth and D. Ghosh, Pharmaceutics, 2020, 12, 1042 CrossRef CAS PubMed.
- D. Ross-Thriepland, A. Bornot, L. Butler, A. Desai, H. Jaiswal, S. Peel, M. R. Hunter, U. Odunze, B. Isherwood and D. Gianni, SLAS Discovery, 2020, 25, 605–617 CrossRef CAS PubMed.
- Q. Li, C. Chan, N. Peterson, R. N. Hanna, A. Alfaro, K. L. Allen, H. Wu, W. F. Dall’Acqua, M. J. Borrok and J. L. Santos, ACS Chem. Biol., 2020, 15, 830–836 CrossRef CAS PubMed.
- M. Y. Arteta, T. Kjellman, S. Bartesaghi, S. Wallin, X. Q. Wu, A. J. Kvist, A. Dabkowska, N. Szekely, A. Radulescu, J. Bergenholtz and L. Lindfors, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E3351–E3360 CAS.
- R. S. Riley, M. V. Kashyap, M. M. Billingsley, B. White, M. G. Alameh, S. K. Bose, P. W. Zoltick, H. Li, R. Zhang, A. Y. Cheng, D. Weissman, W. H. Peranteau and M. J. Mitchell, Sci. Adv., 2021, 7, eaba1028 CrossRef CAS PubMed.
- M. I. Henderson, Y. Eygeris, A. Jozic, M. Herrera and G. Sahay, Mol. Pharmaceutics, 2022, 19, 4275–4285 CrossRef CAS PubMed.
- K. J. Kauffman, J. R. Dorkin, J. H. Yang, M. W. Heartlein, F. DeRosa, F. F. Mir, O. S. Fenton and D. G. Anderson, Nano Lett., 2015, 15, 7300–7306 CrossRef CAS PubMed.
- K. J. Kauffman, F. F. Mir, S. Jhunjhunwala, J. C. Kaczmarek, J. E. Hurtado, J. H. Yang, M. J. Webber, P. S. Kowalski, M. W. Heartlein, F. DeRosa and D. G. Anderson, Biomaterials, 2016, 109, 78–87 CrossRef CAS PubMed.
- H. Muramatsu, K. Lam, C. Bajusz, D. Laczko, K. Kariko, P. Schreiner, A. Martin, P. Lutwyche, J. Heyes and N. Pardi, Mol. Ther., 2022, 30, 1941–1951 CrossRef CAS PubMed.
- R. L. Ball, P. Bajaj and K. A. Whitehead, Int. J. Nanomed., 2017, 12, 305–315 CrossRef CAS PubMed.
- L. Stitz, A. Vogel, M. Schnee, D. Voss, S. Rauch, T. Mutzke, T. Ketterer, T. Kramps and B. Petsch, PLoS Neglected Trop. Dis., 2017, 11, e0006108 CrossRef PubMed.
- M. A. Oberli, A. M. Reichmuth, J. R. Dorkin, M. J. Mitchell, O. S. Fenton, A. Jaklenec, D. G. Anderson, R. Langer and D. Blankschtein, Nano Lett., 2017, 17, 1326–1335 CrossRef CAS PubMed.
- M. M. Billingsley, N. Singh, P. Ravikumar, R. Zhang, C. H. June and M. J. Mitchell, Nano Lett., 2020, 20, 1578–1589 CrossRef CAS PubMed.
- K. A. Hajj, R. L. Ball, S. B. Deluty, S. R. Singh, D. Strelkova, C. M. Knapp and K. A. Whitehead, Small, 2019, 15, e1805097 CrossRef PubMed.
- J. C. Kaczmarek, K. J. Kauffman, O. S. Fenton, K. Sadtler, A. K. Patel, M. W. Heartlein, F. DeRosa and D. G. Anderson, Nano Lett., 2018, 18, 6449–6454 CrossRef CAS PubMed.
- X. Su, J. Fricke, D. G. Kavanagh and D. J. Irvine, Mol. Pharmaceutics, 2011, 8, 774–787 CrossRef CAS PubMed.
- Q. Cheng, T. Wei, Y. Jia, L. Farbiak, K. Zhou, S. Zhang, Y. Wei, H. Zhu and D. J. Siegwart, Adv. Mater., 2018, 30, e1805308 CrossRef PubMed.
- X. Zhang, W. Zhao, G. N. Nguyen, C. Zhang, C. Zeng, J. Yan, S. Du, X. Hou, W. Li, J. Jiang, B. Deng, D. W. McComb, R. Dorkin, A. Shah, L. Barrera, F. Gregoire, M. Singh, D. Chen, D. E. Sabatino and Y. Dong, Sci. Adv., 2020, 6, eabc2315 CrossRef CAS PubMed.
- T. Miwa, H. Saito and H. Akita, Exp. Brain Res., 2021, 239, 425–433 CrossRef CAS PubMed.
- S. S. Nogueira, A. Schlegel, K. Maxeiner, B. Weber, M. Barz, M. A. Schroer, C. E. Blanchet, D. I. Svergun, S. Ramishetti, D. Peer, P. Langguth, U. Sahin and H. Haas, ACS Appl. Nano Mater., 2020, 3, 10634–10645 CrossRef CAS.
- L. J. Wang, X. W. Hu, B. D. Shen, Y. C. Xie, C. Y. Shen, Y. Lu, J. P. Qi, H. L. Yuan and W. Wu, J. Drug Delivery Sci. Technol., 2015, 30, 163–170 CrossRef CAS.
- F. Yin, S. Guo, Y. Gan and X. Zhang, Int. J. Nanomed., 2014, 9, 1665–1676 CrossRef PubMed.
- R. Campardelli, P. Trucillo and E. Reverchon, Ind. Eng. Chem. Res., 2016, 55, 5359–5365 CrossRef CAS.
- C. Chen, D. Han, C. Cai and X. Tang, J. Controlled Release, 2010, 142, 299–311 CrossRef CAS PubMed.
- J. C. Kasper, G. Winter and W. Friess, Eur. J. Pharm. Biopharm., 2013, 85, 162–169 CrossRef CAS PubMed.
- S. Doktorovova, R. Shegokar, L. Fernandes, P. Martins-Lopes, A. M. Silva, R. H. Muller and E. B. Souto, Pharm. Dev. Technol., 2014, 19, 922–929 CrossRef CAS PubMed.
- S. Soares, P. Fonte, A. Costa, J. Andrade, V. Seabra, D. Ferreira, S. Reis and B. Sarmento, Int. J. Pharm., 2013, 456, 370–381 CrossRef CAS PubMed.
- H. Ohshima, A. Miyagishima, T. Kurita, Y. Makino, Y. Iwao, T. Sonobe and S. Itai, Int. J. Pharm., 2009, 377, 180–184 CrossRef CAS PubMed.
- P. Shahgaldian, J. Gualbert, K. Aissa and A. W. Coleman, Eur. J. Pharm. Biopharm., 2003, 55, 181–184 CrossRef CAS PubMed.
- C. Schwarz and W. Mehnert, Int. J. Pharm., 1997, 157, 171–179 CrossRef CAS PubMed.
- S. Kamiya, T. Kurita, A. Miyagishima, S. Itai and M. Arakawa, Eur. J. Pharm. Biopharm., 2010, 74, 461–466 CrossRef CAS PubMed.
- M. E. Ali and A. Lamprecht, Int. J. Pharm., 2017, 516, 170–177 CrossRef CAS PubMed.
- L. Kumar, M. S. Reddy, R. Verma and K. B. Koteshwara, Lat. Am. J. Pharm., 2016, 35, 284–290 CAS.
- J. L. Wang, M. S. Hanafy, H. Xu, J. Leal, Y. Zhai, D. Ghosh, R. O. Williams Iii, H. David Charles Smyth and Z. Cui, Int. J. Pharm., 2021, 596, 120215 CrossRef CAS PubMed.
- F. Alihosseini, S. Ghaffari, A. R. Dabirsiaghi and S. Haghighat, Braz. J. Pharm. Sci., 2015, 51, 797–802 CrossRef CAS.
- M. Burra, R. Jukanti, K. Y. Janga, S. Sunkavalli, A. Velpula, S. Ampati and K. N. Jayaveera, Adv. Powder Technol., 2013, 24, 393–402 CrossRef CAS.
- Y. J. Wang, K. Kho, W. S. Cheow and K. Hadinoto, Int. J. Pharm., 2012, 424, 98–106 CrossRef CAS PubMed.
- J. Varshosaz, S. Ghaffari, M. R. Khoshayand, F. Atyabi, A. J. Dehkordi and F. Kobarfard, Pharm Dev Technol, 2012, 17, 187–194 CrossRef CAS PubMed.
- H. Zhang, F. M. Zhang and S. J. Yan, Int. J. Nanomed., 2012, 7, 2901–2910 CrossRef CAS PubMed.
- S. Kamiya, Y. Nozawa, A. Miyagishima, T. Kurita, Y. Sadzuka and T. Sonobe, Chem. Pharm. Bull., 2006, 54, 181–184 CrossRef CAS PubMed.
- J. Li, S. Ghatak, M. S. El Masry, A. Das, Y. Liu, S. Roy, R. J. Lee and C. K. Sen, Mol. Ther., 2018, 26, 2178–2188 CrossRef CAS PubMed.
- K. L. Aves, C. M. Janitzek, C. E. Fougeroux, T. G. Theander and A. F. Sander, Pharmaceutics, 2022, 14, 1301 CrossRef CAS PubMed.
- L. Zhang, P. Li, D. Li, S. Guo and E. Wang, Langmuir, 2008, 24, 3407–3411 CrossRef CAS PubMed.
- H. Amekyeh and N. Billa, Molecules, 2021, 26, 908 CrossRef CAS PubMed.
- G. Mancini, R. M. Lopes, P. Clemente, S. Raposo, L. M. D. Goncalves, A. Bica, H. M. Ribeiro and A. J. Almeida, Eur. J. Lipid Sci. Technol., 2015, 117, 1947–1959 CrossRef CAS.
- A. del Pozo-Rodriguez, M. A. Solinis, A. R. Gascon and J. L. Pedraz, Eur. J. Pharm. Biopharm., 2009, 71, 181–189 CrossRef CAS PubMed.
- S. Ohtake and Y. J. Wang, J. Pharm. Sci., 2011, 100, 2020–2053 CrossRef CAS PubMed.
- H. I. Chang and M. K. Yeh, Int. J. Nanomed., 2012, 7, 49–60 CAS.
- M. Schnee, A. B. Vogel, D. Voss, B. Petsch, P. Baumhof, T. Kramps and L. Stitz, PLoS Neglected Trop. Dis., 2016, 10, e0004746 CrossRef PubMed.
- P. V. Date, A. Samad and P. V. Devarajan, AAPS PharmSciTech, 2010, 11, 304–313 CrossRef CAS PubMed.
- N. Khatri, D. Baradia, I. Vhora, M. Rathi and A. Misra, AAPS PharmSciTech, 2014, 15, 1630–1643 CrossRef CAS PubMed.
- N. Dammes, M. Goldsmith, S. Ramishetti, J. L. J. Dearling, N. Veiga, A. B. Packard and D. Peer, Nat. Nanotechnol., 2021, 16, 1030–1038 CrossRef CAS PubMed.
- C. D. Sago, M. P. Lokugamage, K. Paunovska, D. A. Vanover, C. M. Monaco, N. N. Shah, M. Gamboa Castro, S. E. Anderson, T. G. Rudoltz, G. N. Lando, P. Munnilal Tiwari, J. L. Kirschman, N. Willett, Y. C. Jang, P. J. Santangelo, A. V. Bryksin and J. E. Dahlman, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E9944–E9952 CrossRef CAS PubMed.
- N. Pardi, S. Tuyishime, H. Muramatsu, K. Kariko, B. L. Mui, Y. K. Tam, T. D. Madden, M. J. Hope and D. Weissman, J. Controlled Release, 2015, 217, 345–351 CrossRef CAS PubMed.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson, S. A. Dilliard and D. J. Siegwart, Nat. Nanotechnol., 2020, 15, 313–320 CrossRef CAS PubMed.
- K. A. Whitehead, J. R. Dorkin, A. J. Vegas, P. H. Chang, O. Veiseh, J. Matthews, O. S. Fenton, Y. Zhang, K. T. Olejnik, V. Yesilyurt, D. Chen, S. Barros, B. Klebanov, T. Novobrantseva, R. Langer and D. G. Anderson, Nat. Commun., 2014, 5, 4277 CrossRef CAS PubMed.
- M. Jayaraman, S. M. Ansell, B. L. Mui, Y. K. Tam, J. Chen, X. Du, D. Butler, L. Eltepu, S. Matsuda, J. K. Narayanannair, K. G. Rajeev, I. M. Hafez, A. Akinc, M. A. Maier, M. A. Tracy, P. R. Cullis, T. D. Madden, M. Manoharan and M. J. Hope, Angew. Chem., Int. Ed., 2012, 51, 8529–8533 CrossRef CAS PubMed.
- A. Tam, J. Kulkarni, K. An, L. Li, D. R. Dorscheid, G. K. Singhera, P. Bernatchez, G. Reid, K. Chan, D. Witzigmann, P. R. Cullis, D. D. Sin and C. J. Lim, Eur. J. Pharm. Sci., 2022, 176, 106234 CrossRef CAS PubMed.
- K. Godbout and J. P. Tremblay, Pharmaceutics, 2022, 14, 2129 CrossRef CAS.
- Y. Qin, H. Chen, W. Yuan, R. Kuai, Q. Zhang, F. Xie, L. Zhang, Z. Zhang, J. Liu and Q. He, Int. J. Pharm., 2011, 419, 85–95 CrossRef CAS PubMed.
- Y. Hagino, I. A. Khalil, S. Kimura, K. Kusumoto and H. Harashima, Mol. Pharmaceutics, 2021, 18, 878–888 CrossRef CAS PubMed.
- M. Herrera-Barrera, R. C. Ryals, M. Gautam, A. Jozic, M. Landry, T. Korzun, M. Gupta, C. Acosta, J. Stoddard, R. Reynaga, W. Tschetter, N. Jacomino, O. Taratula, C. Sun, A. K. Lauer, M. Neuringer and G. Sahay, Sci. Adv., 2023, 9, eadd4623 CrossRef.
- R. M. Ray, A. H. Hansen, M. Taskova, B. Jandl, J. Hansen, C. Soemardy, K. V. Morris and K. Astakhova, Beilstein J. Org. Chem., 2021, 17, 891–907 CrossRef CAS.
- D. Gonzalez-Carter, X. Liu, T. A. Tockary, A. Dirisala, K. Toh, Y. Anraku and K. Kataoka, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 19141–19150 CrossRef CAS PubMed.
- S. Santiwarangkool, H. Akita, T. Nakatani, K. Kusumoto, H. Kimura, M. Suzuki, M. Nishimura, Y. Sato and H. Harashima, J. Pharm. Sci., 2017, 106, 2420–2427 CrossRef CAS PubMed.
- K. Kusumoto, H. Akita, T. Ishitsuka, Y. Matsumoto, T. Nomoto, R. Furukawa, A. El-Sayed, H. Hatakeyama, K. Kajimoto, Y. Yamada, K. Kataoka and H. Harashima, ACS Nano, 2013, 7, 7534–7541 CrossRef CAS PubMed.
- K. S. Rao, A. Ghorpade and V. Labhasetwar, Expert Opin. Drug Delivery, 2009, 6, 771–784 CrossRef CAS PubMed.
- O. Osborne, N. Peyravian, M. Nair, S. Daunert and M. Toborek, Trends Neurosci., 2020, 43, 695–708 CrossRef CAS PubMed.
- L. Bertrand, M. Velichkovska and M. Toborek, J. Neuroimmune Pharmacol., 2021, 16, 74–89 CrossRef PubMed.
- E. Vives, P. Brodin and B. Lebleu, J. Biol. Chem., 1997, 272, 16010–16017 CrossRef CAS.
- M. Liang, C. Gao, Y. Wang, W. Gong, S. Fu, L. Cui, Z. Zhou, X. Chu, Y. Zhang, Q. Liu, X. Zhao, B. Zhao, M. Yang, Z. Li, C. Yang, X. Xie, Y. Yang and C. Gao, Drug Delivery, 2018, 25, 1652–1663 CrossRef CAS PubMed.
- J. H. Lee, J. A. Engler, J. F. Collawn and B. A. Moore, Eur. J. Biochem., 2001, 268, 2004–2012 CrossRef CAS PubMed.
- Z. M. Qian, H. Li, H. Sun and K. Ho, Pharmacol. Rev., 2002, 54, 561–587 CrossRef CAS PubMed.
- Z. Pang, H. Gao, Y. Yu, L. Guo, J. Chen, S. Pan, J. Ren, Z. Wen and X. Jiang, Bioconjugate Chem., 2011, 22, 1171–1180 CrossRef CAS PubMed.
- Z. R. Crook, E. Girard, G. P. Sevilla, M. Merrill, D. Friend, P. B. Rupert, F. Pakiam, E. Nguyen, C. Yin, R. O. Ruff, G. Hopping, A. D. Strand, K. A. K. Finton, M. Coxon, A. J. Mhyre, R. K. Strong and J. M. Olson, J. Mol. Biol., 2020, 432, 3989–4009 CrossRef CAS PubMed.
- G. Sharma, S. Lakkadwala, A. Modgil and J. Singh, Int. J. Mol. Sci., 2016, 17, 806 CrossRef PubMed.
- Z. Pang, H. Gao, Y. Yu, J. Chen, L. Guo, J. Ren, Z. Wen, J. Su and X. Jiang, Int. J. Pharm., 2011, 415, 284–292 CrossRef CAS PubMed.
- S. Liu, Q. Cheng, T. Wei, X. Yu, L. T. Johnson, L. Farbiak and D. J. Siegwart, Nat. Mater., 2021, 20, 701–710 CrossRef CAS PubMed.
- F. H. Ma, L. Yang, Z. R. Sun, J. J. Chen, X. H. Rui, Z. Glass and Q. B. Xu, Sci. Adv., 2020, 6, eabb4429 CrossRef CAS PubMed.
- P. P. G. Guimaraes, R. Zhang, R. Spektor, M. Tan, A. Chung, M. M. Billingsley, R. El-Mayta, R. S. Riley, L. Wang, J. M. Wilson and M. J. Mitchell, J. Controlled Release, 2019, 316, 404–417 CrossRef CAS.
- E. Kenjo, H. Hozumi, Y. Makita, K. A. Iwabuchi, N. Fujimoto, S. Matsumoto, M. Kimura, Y. Amano, M. Ifuku, Y. Naoe, N. Inukai and A. Hotta, Nat. Commun., 2021, 12, 245007094 Search PubMed.
- T. Wei, Q. Cheng, Y. L. Min, E. N. Olson and D. J. Siegwart, Nat. Commun., 2020, 11, 3232 CrossRef CAS PubMed.
- T. W. Geisbert, A. C. Lee, M. Robbins, J. B. Geisbert, A. N. Honko, V. Sood, J. C. Johnson, S. de Jong, I. Tavakoli, A. Judge, L. E. Hensley and I. Maclachlan, Lancet, 2010, 375, 1896–1905 CrossRef CAS.
- T. W. Geisbert, L. E. Hensley, E. Kagan, E. Y. Z. Yu, J. B. Geisbert, K. Daddario-DiCaprio, E. A. Fritz, P. B. Jahrling, K. McClintock, J. R. Phelps, A. C. H. Lee, A. Judge, L. B. Jeffs and I. MacLachlan, J. Infect. Dis., 2006, 193, 1650–1657 CrossRef CAS PubMed.
- Y. Dong, D. J. Siegwart and D. G. Anderson, Adv. Drug Delivery Rev., 2019, 144, 133–147 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |