Innate and engineered attributes of bacterial microcompartments for applications in bio-materials science
S. M.
Rose†
 ,
Aarcha
Radhakrishnan†
,
Aarcha
Radhakrishnan†
 and
Sharmistha
Sinha
and
Sharmistha
Sinha
 *
*
Chemical Biology Unit, Institute of Nano Science and Technology, Sector-81, Knowledge City, Sahibzada Ajit Singh Nagar, Punjab 140306, India. E-mail: sinhas@inst.ac.in
First published on 27th April 2023
Abstract
Bacterial microcompartments (BMCs) are sophisticated all-protein bionanoreactors widely spread in bacterial phyla. BMCs facilitate diverse metabolic reactions, which assist bacterial survivability in normal (by fixing carbon dioxide) and energy dearth conditions. The past seven decades have uncovered numerous intrinsic features of BMCs, which have attracted researchers to tailor them for customised applications, including synthetic nanoreactors, scaffold nano-materials for catalysis or electron conduction, and delivery vehicles for drug molecules or RNA/DNA. In addition, BMCs provide a competitive advantage to pathogenic bacteria and this can pave a new path for antimicrobial drug design. In this review, we discuss different structural and functional aspects of BMCs. We also highlight the potential employment of BMCs for novel applications in bio-material science.
1. Introduction
A cell, the tiniest living unit, is a compartment of selected biomolecules in a complex metabolic environment. In the course of evolution, the cell itself has gradually developed membrane-enclosed sub-compartments within it to provide spatiotemporal regulation by assorting substrates and intermediates of interconnected metabolic processes. The physical barriers of all biological compartments are composed of either lipid or protein. While lipid membrane-bound compartments are present exclusively in eukaryotes, protein-based compartments exist in both prokaryotes and eukaryotes.1 Ferritin and its homologs, bacterial microcompartments (BMCs), lumazine synthase, vault complex, and encapsulin are some examples of lipid-free proteinaceous compartments involved in various cell functions.2–7 Ferritin is the first discovered protein compartment and is involved in the iron storage and homeostasis of iron.8 It is present in both eukaryotic and prokaryotic cells. On the other hand, BMCs and encapsulins are two distinct lineages of all protein compartments present only in prokaryotes. These compartments are reported to segregate and orchestrate metabolic reactions inside them.9,10 Another class of protein-based compartments is vault complexes, which are highly conserved among eukaryotes. These ribonucleoprotein complexes take part in nuclear-cytoplasmic transport, drug resistance, and various cell signalling processes inside cells.11 These protein-based compartments can serve as localized nano-reactors or nano-containers in vivo or in vitro and hence have attracted the attention of several researchers in the field. In past few years, they have been utilized to fabricate novel materials with a wide range of applications.2,12–17Fig. 1 chronologically summarizes the discovery and the first attempt towards the application of these naturally occurring protein compartments.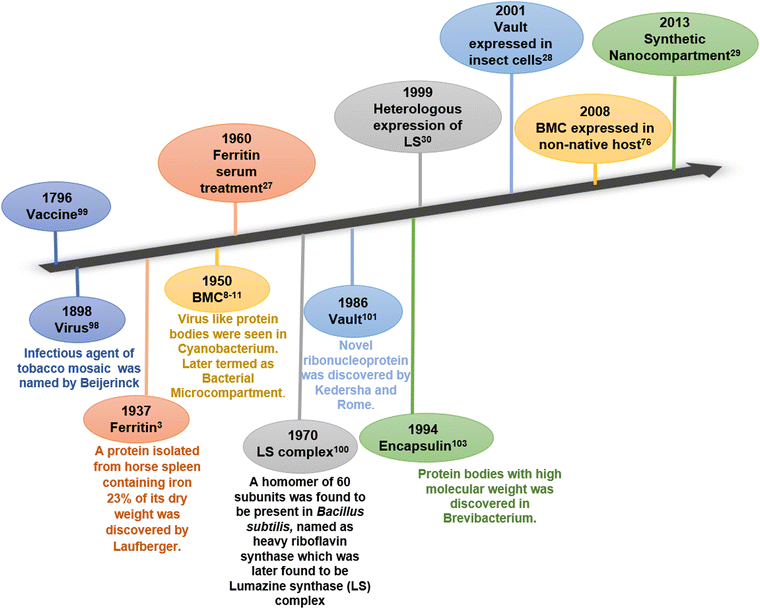 | ||
| Fig. 1 Timeline of discovery (bottom) and first attempts towards applications (top) of various protein compartments. | ||
Among these protein compartments, bacterial microcompartments stand unique in terms of their shell architecture and diverse functions. They evolved transiently and conditionally in several bacterial and algal species to carry out certain conditional metabolic reactions.18 In the 1950s, BMCs were first reported in the cytoplasm of cyanobacteria Phormidium uncinatum through Transmission Electron Microscopy (TEM).19–22 In the beginning, these polyhedral bodies were thought of as protein inclusions and were later considered viruses due to their morphological similarity to phages. Follow-up studies revealed that they play an important role in enhancing CO2 fixation during photosynthesis in cyanobacteria and some chemoautotrophic bacteria and were accordingly termed carboxysomes. In 1973, carboxysomes were first purified from the bacterium Halothiobacillis neapolitanus.23,24 After 40 years of carboxysome discovery, homologs of carboxysomes were found to be present in other bacteria performing special catabolic reactions and were termed catabolosomes (or metabolosomes).25,26 To date, BMC-related genes are clustered in 7000 loci in 45 bacterial phyla that can induce 68 BMC types and subtypes.27 Studies followed by the isolation and purification of BMCs revealed that these structures are entirely composed of protein and these protein bodies (BMCs) have an enzymatic core within a protein shell.28–30 Although the core enzyme cluster varies for different types of microcompartments, the protein subunits that make up the outer shell (shell proteins) are similar throughout several genres of BMCs.31 The protein shell of all BMCs is made up of self-assembling protein subunits containing the two types of BMC domains.32–35 These shell proteins have similar and superimposable structures and tile together to make up the heterogeneously self-assembled compartment envelope.12,13 Entrapment of the metabolic machineries inside the small, confined semipermeable protein envelope of BMC increases the catalytic efficiency, orchestrates multi-enzyme pathways, and sequesters any toxic intermediates.31 Typically, the genes that encode the BMCs are present in a single operon or clustered together.36 These BMCs offer an open palette to design and fabricate customized biomaterials with desirable structure and function. Several efforts have already been employed to exploit the BMCs to construct synthetic bioreactors. In this review, we discuss in detail the natural and engineered properties of bacterial microcompartments towards the development of novel biomaterials for various applications. This review updates our current understanding of bacterial microcompartment applications and provides new insights into the future applications of these prokaryotic nano-organelles.
2. Innate features of BMC
2.1. Functional features of BMC
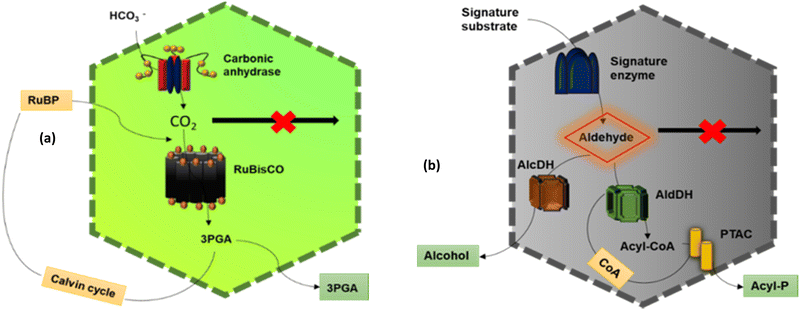
The main function of BMCs is to carry out different metabolic reactions in an efficient way inside bacterial cells, thus helping in their survival. According to these reactions, BMCs have two lineages – anabolic BMCs (carboxysomes) and catabolic BMCs (metabolosomes).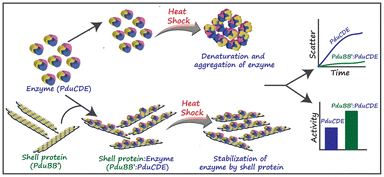 | ||
| Fig. 3 The chaperone-like activity of the PduBB′ shell protein of Salmonella enterica on the enzyme, PduCDE, providing thermal stability to the enzyme. (Adapted from Kumar et al.51). | ||
2.1.2.1. Propanediol Utilization Bacterial Microcompartment- Pdu BMC. PduBMC is one of the most studied metabolosomes. It is found mainly in Salmonella and other bacteria that contribute to the gut and soil microbiota.52,53 The fermentation of fucose and rhamnose, common carbohydrates found in plant cell walls, bacterial capsules and glycoconjugates in eukaryotic cells, produces 1,2 propanediol. Pdu BMC catabolizes the 1,2 propanediol using vitamin B12-dependent diol dehydratase. The reaction intermediate (propanaldehyde), being toxic and volatile, is prevented from moving out of the shell. Next, propanol dehydrogenase and propionaldehyde dehydrogenase convert propanaldehyde into propanol and propionyl CoA, respectively. Phosphotransacylase converts this propionyl-CoA into the propionyl-PO4 ion; this reaction happens at the junction of the shell and cytoplasm, releasing the final product into the cytoplasm.46,54
2.1.2.2. Ethanolamine utilization bacterial microcompartment – Eut BMC. Eut BMC is involved in the degradation of the ethanolamine obtained from the phosphodiesterase-mediated degradation of phosphatidylethanolamine.55 Here, the substrate-signature enzyme, ethanolamine ammonia lyase, converts ethanolamine into ammonia and acetaldehyde in the presence of vitamin B12 cofactor. Acetaldehyde as a volatile and lipophilic intermediate is sequestrated inside the shell and some of these molecules are converted into acetyl-CoA at first and then to acetyl phosphate, which is further converted to acetate.56 Other molecules of acetaldehyde produce ethanol to maintain NADP/NAD+ levels. Two different alcohol dehydrogenases are the key enzymes for this process and on completion of this metabolic circuit, one molecule of ATP is gained. A part of acetyl-CoA is released into the cytoplasm while another part is converted to acetal-P by phosphotransacetylase present at the border of the shell and cytoplasm.57
2.1.2.3. Ethanol utilization bacterial microcompartment – Etu BMC. This microcompartment was discovered in Clostridium Kluyveri, when it was grown on ethanol and acetate.45 Before its discovery, researchers predicted the presence of a BMC involved in ethanol degradation from genome studies of Clostridium Kluyveri, in which the etu operon is located near the BMC loci.45 The etu operon is considered to have of genes for two shell proteins, two ethanol dehydrogenases and three aldehyde dehydrogenases. These enzymes are involved in the conversion of ethanol generated from the fermentation of sugars or as a side metabolic product of Eut BMC, to acetyl-CoA.58
2.1.2.4. Glycyl radical enzyme containing microcompartments – Grm BMC. Grm BMCs represent a group of vitamin B12-independent metabolosomes. They are involved in the degradation of propanediol, fucose and rhamnose, choline, etc.49,50 The enzymes are commonly called glycyl radical enzymes. GRMs are subdivided into many groups, GRM1, GRM2, GRM3, GRM4, GRM5, GRM6, according to the substrate they metabolize. GRM1 and GRM2 are involved in choline metabolism. GRM3, 4 and 6 are similar to Pdu BMC and are involved in the degradation of 1,2-propanediol. GRM5 takes fuculose as the substrate.27
2.1.2.5. Other BMCs. Genome sequencing data of different bacteria done by many groups revealed the likelihood of the presence of more types of BMCs in the bacterial phyla.59 Some of these are as follows. Sugar-phosphate utilization BMC (SPU BMC) is involved in the degradation of exogenous DNA and RNA in bacteria to produce energy. RMM BMC (Rhodococcus and Mycobacterium Microcompartemnt) is involved in the metabolism of amino acetone to propionyl-CoA. PVM BMC is involved in the metabolism of rhamnose and fucose, encapsulating a class-II aldolase enzyme.44
2.2. Structural features
Bacterial microcompartments can be viewed as polyhedral nano-reactors having a size range of 40–200 nm.18 Although they are involved in diverse metabolic functions, they share some similarities in their structural features. This section presents a comparative analysis of the phenotypic and genotypic makeup of the different BMCs.The central axes of BMC-H and BMC-T form pores having average size ranges of 4 Å–7 Å and 12 Å–14 Å, respectively.64 The size and the charge around the pore and its conformation decide the selectivity for the passage of the substrates, cofactors and products in and out of the BMCs.64 For example, EutL (pseudohexamer) of Eut BMC has an average negative charge and is reported to open upon exposure to zinc ions.65,66 Moreover, in the double trimer (BMC-TD) one trimer faces the lumen and the other faces the cytoplasm. The pore of the cytoplasm-facing trimer is open and the other is obstructed by pore-surrounding amino acids, thus forming gated pores that open and close according to the substrate and amino acid interaction.64,67 Thus, this semipermeable shell sequestrates the toxic intermediates of the reaction inside the compartment and protects the cell. Further, these pores can be engineered to tune their selectivity either by increasing or decreasing their size and altering the charge of amino acids around the pores. The permeability of different substances is gated based on the size and charges around each pore, which differs from bacteria to bacteria.
The shell proteins thus function as a semi-permeable physical barrier between the cytoplasm and the catalytic core. Some reports suggest that the shell proteins can protect the catalysts from thermal and pH stress. A recent study by Kumar et al. (Fig. 3) showed that the major shell protein of Pdu BMC shows chaperone-like activity by conserving the structure and activity of its signature enzyme under thermal stress.51
2.3. BMC provides competitive advantages to pathogenic bacteria
BMC enables bacteria to use alternative carbon sources other than glucose. Such alternative carbon sources like ethanolamine and 1,2-propanediol are constantly produced in the human gut through the metabolism of different food constitutents.57,76 Enteric bacteria living in the animal gut can express different types of catabolic BMCs that utilize these substrates, thus providing a nutritional advantage over those bacteria that cannot express BMC.77,78 Enterohaemorrhagic E. coli, for example, has a nutritional advantage over commensal E. coli because of ethanolamine metabolism when grown in bovine intestinal contents.79 From these observations, researchers speculated that there was a connection between BMC and pathogenicity. It has been shown that the invading pathogen (Salmonella enterica) splits into two life cycle patterns; one directly attacks the gut epithelium causing inflammation, leading to anaerobic conditions and thus helping the other bacteria to colonize within the gut by functional BMC induction.80 Studies on S. typhimurium suggest the upregulation of the Eut and Pdu operons during colonization of the bacteria in the chicken caecum.81 Another study shows that deletion of the regulatory unit of Pdu and Eut operons reduces the proliferation of S. typhimurium in an animal model.82 Although a direct connection between BMC and pathogenicity is yet to be found, it is evident that BMC provides growth advantages to bacteria irrespective of pathogenicity. Following these directions, drug molecules that can inhibit the assembly of BMC can be designed to restrict the growth of bacteria.3. Engineering BMC for biomaterial applications
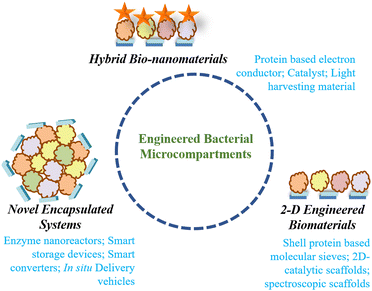
The confinement of enzymatic machinery inside the small volume of a protein shell commendably increases the catalytic efficiency. Further, the protein shell also protects the encapsulated enzymatic cargo with high thermal and pH stability.51 Another important property of bacterial microcompartments is that they can self-assemble heterogeneously, and each shell protein can self-assemble, homogenously forming different nanostructures based on the concentration in both in vivo and in vitro environments. All these features of BMCs, the high catalytic efficiency, multifunctionality, semi-permeability and self-assembling nature of the protein shell make BMCs superior to other protein compartments for exploitation as biosynthetic nano-reactors. Enticed by these special features, scientists have tailored BMCs for different bio-material applications.The engineering of bacterial microcompartments was initially considered for use in expressing heterologous enzymes inside the shell to increase their catalytic efficiency. Later, inorganic catalysts like nanoparticles, and organic polymers like DNA were incorporated within or on the shell for industrial and biological applications. The self-assembly of the shell proteins has been used to attach desired reactive moieties nearby for electron conduction, charge transfer, etc. and the core with diverse functionality.
Any engineered BMCs or their components need to be supported by in vitro physical and biochemical characterization, following the expression and purification of these structures as a whole or as components. Further purification will be needed for the covalent attachment of any moieties to these proteins. To date, four different types of BMCs have been purified from different organisms by three main methods: cell lysis, followed by ultracentrifugation, sucrose gradient separation and affinity chromatography.83 Purification of BMCs can be carried out from host organisms as well as heterologous expression systems.83
3.1. The fate of heterologous expressions of BMC and its components
An initial step in engineering bacterial microcompartments is the expression of BMC proteins in non-native hosts. This step is important as native BMC proteins will be expressed along with the engineered ones otherwise. For the first time, Parson et al. demonstrated the heterologous expression of Pdu BMC of Citrobacter freundii in E. coli, which was a fully functional and complete recombinant metabolosome.84 Similarly, the α-carboxysome of Halothiobacillus neapolitanus was also successfully expressed in E. coli, which was observed to be fully intact and operative.68 In 2018, β-carboxysomes of Syncechococcus elongates were expressed in E. coli, which was also functional.85 These are some examples of the first few successful attempts at the heterologous expression of different bacterial microcompartments. Since the BMC operon can be expressed heterologously, engineering the same for synthetic compartments and structures was considered. Although the BMC operon codes for many proteins, all proteins are not important in forming the complete polyhedral structure of BMC. This was identified in the year 2010 when Parsons et al. expressed all the shell proteins86 of the Pdu operon heterologously in E. coli and later deleted individual shell proteins to study the interaction and importance of different shell proteins for forming Pdu BMC. Following this, they were successful in expressing an empty minimal microcompartment in vivo. While conducting these experiments by deleting certain shell protein genes, researchers observed that with certain combinations of shell proteins, novel architectures that were different from those of the polyhedral organelles were formed. A combination of PduA and PduB formed axial filaments that showed motility when tagged with PduV (enzyme).87 Cheng et al. showed that PduA, PduBB′, PduJ and PduN are only required to form a complete functional Pdu BMC. PduJ and PduN deletion resulted in the formation of an elongated BMC, while the deletion of PduA resulted in an enlarged BMC.88 Choudhary et al. showed that the EutS shell protein alone can form a compartment-like structure by heterologously expressing the recombinant EutS of Salmonella enterica in E. coli.89 In 2018, Hagen et al. formed an in vitro minimal microcompartment using a physical mixture of Haliangium ochraceum BMC-H, BMC-T and BMC-P in a ratio of 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12, respectively. Another example of in vivo nanoarchitecture formation is the tubular Pdu BMC observed as a result of PduN deletion from the Pdu operon.63 A recent report suggests the formation of pleiotropic structures including nanotubes and nanocones when six shell proteins of a GRM from Rhodopseudomonas palustris were expressed in E. coli.90 The self-assembly of shell proteins into such diverse types of structures disclosed the wide plasticity of BMC in bio-material applications.
12, respectively. Another example of in vivo nanoarchitecture formation is the tubular Pdu BMC observed as a result of PduN deletion from the Pdu operon.63 A recent report suggests the formation of pleiotropic structures including nanotubes and nanocones when six shell proteins of a GRM from Rhodopseudomonas palustris were expressed in E. coli.90 The self-assembly of shell proteins into such diverse types of structures disclosed the wide plasticity of BMC in bio-material applications.
3.2. Repurposing BMC as a nanoreactor
Researchers have so far explored both anabolic and catabolic bacterial microcompartments for various biotechnology applications. BMC as a nanoreactor is prominent among all the other applications. Being such a well-orchestrated system, BMCs can carry out complex engineered pathways in the most efficient manner resulting in enhanced yield. The encapsulation of non-native cargo with encapsulation peptide (EP: described in the previous section) inside the BMC shell was the first step taken to fabricate the nanoreactor. Green Fluorescent Protein (GFP) fused with either aldehyde dehydrogenase or the first 34 amino acids from the N-terminal of the enzyme was targeted to the compartment. Although the efficiency of targeting was not high, this study demonstrated how cargo could be targeted to BMC.83 In the same year, pyruvate decarboxylase and alcohol dehydrogenase fused with EP were targeted to Pdu BMC to produce a BMC that was efficient in producing ethanol from pyruvate91 (Fig. 4). Wagner et al. showed that 3 different non-native enzymes, beta-galactosidase, esterase Est 5 and glycerol dehydrogenase, with 19 amino acids from PduP can be targeted to BMC without altering the enzyme efficiency.92 When the BMC operon from Citrobacter freundii is co-expressed with polyphosphate kinase (PPK 1) fused with EP, the enzyme became encapsulated in BMC. PPK 1 loaded in BMC was able to sequester inorganic phosphate (Pi), a common water pollutant with more efficiency than free PPK 1. BMC can thus be repurposed to fabricate a reactor that can be utilized in wastewater treatment.31 In another attempt, a hydrogen-producing nanoreactor was created using shell proteins of α-carboxysomes and directing hydrogen-producing machineries, [Fe–Fe] hydrogenase, ferredoxin (Fd) and ferredoxin:NADP+ oxidoreductase (FNR), into the protein shell. The developed reactor was not only producing hydrogen more efficiently than the free hydrogenase enzyme but also protecting the hydrogenase from oxygen.93 To enhance CO2 fixation by Rubisco in carboxysomes, the components of Rubisco activase (CbbO and CbbQ) were installed inside the carboxysome shell.94 Prentice et al. invented recombinant bacteria expressing BMC with heterologous enzymes that were used for the accumulation of metabolites. In this case, lower molecular weight substrates were converted to polymeric or higher molecular weight products by the encapsulated enzymes. These polymeric metabolites were stored within the synthetic BMC.95 In the context of developing nanoreactors by engineering BMC, these are some versatile paradigms for using EP to target non-native cargo into the BMC lumen. Just like EP, a heterodimeric coiled-coil system can also be used to target cargo in the BMC lumen, as described by Lee et al.96 It was also reported that the SpyTag/SpyCatcher system was utilized to incorporate various fluorescent cargo inside the compartment.97 Using this method, recent reports proclaimed the development of a synthetic formate utilizing microcompartments (sFUT).98,99 This recombinant microcompartment was loaded with pyruvate formate lyase (PFL) and phosphate acetyl transferase and thus it was able to utilize formate and acetyl-phosphate to produce pyruvate. Zhang et al. covalently attached the SpyTag/SpyCatcher system to EutM, the hexameric sheet-forming shell protein of Eut BMC, for conducting augmented cascade reactions.100 These studies revealed the calibre of BMC for developing a bio-reactor for various applications.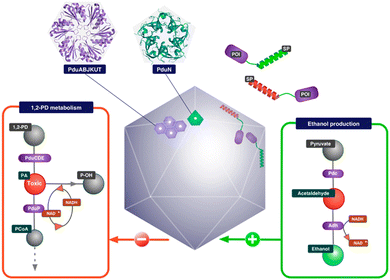 | ||
| Fig. 4 Schematic illustration of an ethanol bioreactor, repurposing Pdu BMC. (Adapted from Wagner et al.92). | ||
3.3. Delivery vehicle derived from BMC
One of the challenges addressed by many researchers is to develop a vehicle for delivering cargo molecules like drugs, DNA, RNA, metal nanoparticles, peptides or proteins inside cells. These cargo-loaded systems could have potential applications in cancer diagnosis and therapy.101,102 In this context, very few attempts have been made with BMC. The possibility of using BMC as a cargo carrier was reported elsewhere.103 The shell protein of BMC could be engineered to have cysteine at the desired position and with that, a drug molecule of specific function can be attached.103 Recently, a protein shell (PS) was fabricated from a sheet-forming shell protein of Pdu BMC (Fig. 5). Here, anticancer drugs like doxorubicin and curcumin were loaded in the lumen of the protein shell and their release was checked.104 Moreover, the non-native enzyme, Cyt C, was also able to be encapsulated inside the protein shell and this enzyme-loaded protein shell successfully converted pyrogallol to phloroglucinol.104 In a recent report, DNA was targeted to the lumen of BMC for the first time. In this work, a DNA segment with the Lac repressor binding site was used and the Lac repressor was fused with an enhanced green fluorescent protein and Pdu BMC targeting peptide to target the DNA assembled inside the BMC lumen.105 The use of BMCs has been also proposed in the field of phytonanotechnology using the nanostructures derived from the BMC shells for the controlled uptake and release of fertilizers, pesticides, or other agrochemicals that can interact directly to benefit the plant.106 An advantage of BMCs as drug delivery vehicles is that they can shelter the cargo from thermal and pH stress.51,107 However, the biocompatibility of BMC needs to be explored.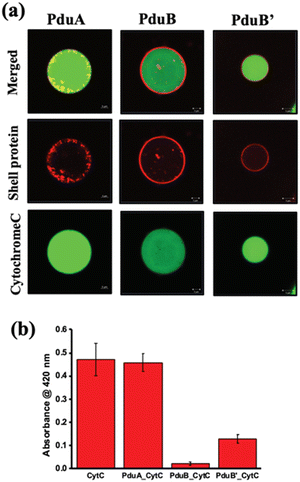 | ||
| Fig. 5 (a) Confocal images of alexa 488-labelled cytochrome C loaded in a constructed protein shell. (b) Enzymatic conversion of pyrogallol to phloroglucinol by Cyt C inside the protein shell. (Adapted from Bari et al.).104 | ||
3.4. Developing protein-based hybrid materials
The symbiotic combination of biomolecules with inorganic nanoparticles or organic molecules produces hybrid materials, which are known to have better functionality and elaborate applications. To fabricate such materials, proteins or peptides have been used widely since their shape and size can be precisely controlled through genetic engineering. Whole Pdu BMC was used to reduce auric chloride to gold nanoparticles and after reduction, the gold nanoparticles were found to be present on the outer protein shell of BMC.108 The developed hybrid system was able to perform standard gold nanoparticle-catalyzed reactions without losing the internal enzyme cascade activity of the BMC core. Another work from the same group demonstrated how the quaternary structure of the protein can affect the morphology and catalytic efficiency of nanocatalysts. In this work, they used a globular, non-self-assembling protein i.e., BSA and a self-assembling and sheet-forming shell protein of Pdu BMC, i.e., Pdu BB′, to develop a hybrid copper nanoflower (Cu NF). The Cu NF developed with BMC protein was found to have a different morphology and higher catalytic activity in comparison to BSA Cu NF.109 In another study, Ccm (Cytochrome c maturation) system 1 (Ccm 1) was used to covalently attach heme with BMC-H to develop a long-distance electron transport system.110 After the attachment of the heme moiety, shell proteins retained their property of self-assembling into hexameric structures (Fig. 6). As a proof of concept, the pore of a BMC shell protein was functionalized to bind inorganic redox counterparts such as copper or Fe–S clusters. The redox potential of these hybrid shell proteins was checked against the Standard Hydrogen Electrode (SHE). Inorganic counterparts bound to the concave side of the shell protein were found to have more positive formal potential in comparison to those variants having the same counterpart at the convex face.110 Kaur et al. showed that a 2D sheet-forming shell protein, PduA (from Pdu BMC), when mutated to PduA [K26A] formed a 3D scaffold. Then, both 2D and 3D structures were used to synthesize a protein-gold nanoparticle hybrid material. In the case of a 3D scaffold, the size of the gold nanoparticle was found to be lower as this scaffold has a higher surface area that provides more nucleation sites for nanoparticle formation. Moreover, gold nanoparticles synthesized in the presence of a 3D scaffold have greater efficiency in reducing para-nitrophenol.111 These attempts at combining BMC shell proteins with organic or inorganic counterparts open many future directions to have novel hybrid material with diverse applications.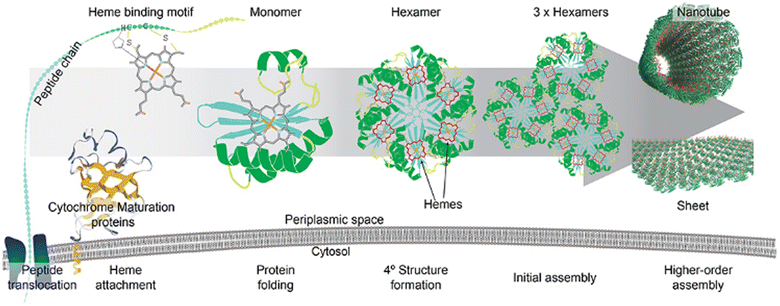 | ||
| Fig. 6 The attachment of a heme moiety with a hexameric shell protein of BMC to construct a hybrid electron transporter system. (Adapted from Huang et al., 2020).110 | ||
4. Conclusions and future outlook
The bacterial microcompartments are unique paradigms of biomolecular compartmentalization that can be exploited for myriads of applications in biology and biomaterial sciences (Fig. 7). To date, the literature has ample resources on the techniques that can be used for the genetic engineering of these nano-architectures leading to reactors with different shapes and properties. Further studies have also delineated the properties of the individual components. However, certain innate aspects of microcompartments still need to be explored and studied. The most relevant question is concerned with what determines the size and shape of these nano-organelles. The answer to this question will aid in the bio-synthetic production of microcompartment-like engineered nanoreactors. The next issue that needs attention is the mechanism of biogenesis of such a complex system. Although recent studies show that the biogenesis of different BMC paradigms occurs via liquid–liquid phase separation, more studies in vivo and in vitro need to be performed for conclusive outcomes.112 Another aspect that remains to be deciphered is the role of BMCs in the pathogenesis of the micro-organism it is associated with. Such studies will help in the identification of alternative targets for microbial management.Further, the components of BMCs can be exploited in permutations and combinations to form different protein architectures in 2- and 3-dimensions. These scaffolds will have high thermal and pH stability as indicated earlier and can be used in catalysis, Raman spectroscopic probes, etc. The 3-dimensional structures such as protein vesicles derived from BMC components can be employed to carry bioactive molecules such as anticancer drugs, nucleic acids or peptides. This area needs the attention of researchers since self-assembling BMC shell proteins have remarkable potential for use as delivery vehicles.
Author contributions
SS conceived the theme of the review. SMR and AR performed the literature survey and wrote the manuscript. SS wrote and edited the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors acknowledge Institute of Nano Science and Technology for financial support.Notes and references
- Y. Diekmann and J. B. Pereira-Leal, Evolution of intracellular compartmentalization, Biochem. J., 2012, 449(2), 319–331 CrossRef PubMed
.
- D. Goel and S. Sinha, Naturally occurring protein nano compartments: basic structure, function, and genetic engineering, Nano Express, 2021, 2(4), 042001 CrossRef
.
- H. Lecoq, Discovery of the first virus, the tobacco mosaic virus: 1892 or 1898?, C. R. Acad. Sci., Ser. III, 2001, 324(10), 929 CAS
.
- S. Riedel, Edward Jenner and the history of smallpox and vaccination. Proc (Bayl Univ Med Cent), 2005, 18(1), 21.
- R. Ladenstein, M. Fischer and A. Bacher, The lumazine synthase/riboflavin synthase complex: shapes and functions of a highly variable enzyme system, FEBS J., 2013, 280(11), 2537 CrossRef CAS PubMed
.
- N. L. Kedersha and L. H. Rome, Preparative agarose gel electrophoresis for the purification of small organelles and particles, Anal. Biochem., 1986, 156(1), 161 CrossRef CAS PubMed
.
- N. Valdés-Stauber and S. Scherer, Isolation and characterization of Linocin M18, a bacteriocin produced by Brevibacterium linens, Appl. Environ. Microbiol., 1994, 60(10), 3809 CrossRef
.
- W. Wang, M. A. Knovich, L. G. Coffman, F. M. Torti and S. V. Torti, Serum Ferritin: Past, Present and Future, Biochim Biophys Acta, 2010, 1800(8), 760 CrossRef CAS
.
- J. A. Jones and T. W. Giessen, Advances in encapsulin nanocompartment biology and engineering, Biotechnol. Bioeng., 2021, 118(1), 491–505 CrossRef CAS PubMed
.
- C. A. Kerfeld and O. Erbilgin, Bacterial microcompartments and the modular construction of microbial metabolism, Trends Microbiol., 2015, 23(1), 22–34 CrossRef CAS PubMed
.
- A. van Zon, M. H. Mossink, R. J. Scheper, P. Sonneveld and E. A. C. Wiemer, The vault complex, Cell. Mol. Life Sci., 2003, 60(9), 1828 CrossRef CAS PubMed
.
- A. M. Demchuk and T. R. Patel, The biomedical and bioengineering potential of protein nanocompartments, Biotechnol. Adv., 2020, 41, 107547 CrossRef CAS PubMed
.
- T. G. W. Edwardson, M. D. Levasseur, S. Tetter, A. Steinauer, M. Hori and D. Hilvert, Protein Cages: From Fundamentals to Advanced Applications, Chem. Rev., 2022, 122(9), 9145 CrossRef CAS PubMed
.
- D. A. Rothstein, S. Rosen, A. Markowitz and J. B. Fuller, Ferritin and antiferritin serum treatment of dogs in irreversible hemorrhagic shock, Am. J. Physiol., 1960, 198, 844 CrossRef CAS PubMed
.
- A. G. Stephen, S. Raval-Fernandes, T. Huynh, M. Torres, V. A. Kickhoefer and L. H. Rome, Assembly of Vault-like Particles in Insect Cells Expressing Only the Major Vault Protein, J. Biol. Chem., 2001, 276(26), 23217 CrossRef CAS PubMed
.
- R. Rahmanpour and T. D. H. Bugg, Assembly in vitro of Rhodococcus jostii RHA1 encapsulin and peroxidase DypB to form a nanocompartment, FEBS J., 2013, 280(9), 2097 CrossRef CAS PubMed
.
- D. B. Jordan, K. O. Bacot, T. J. Carlson, M. Kessel and P. V. Viitanen, Plant riboflavin biosynthesis. Cloning, chloroplast localization, expression, purification, and partial characterization of spinach lumazine synthase, J. Biol. Chem., 1999, 274(31), 22114 CrossRef CAS
.
- C. Chowdhury, S. Sinha, S. Chun, T. O. Yeates and T. A. Bobik, Diverse bacterial microcompartment organelles, Microbiol. Mol. Biol. Rev., 2014, 78(3), 438 CrossRef PubMed
.
- G. Drews and W. Niklowitz, Beiträge zur Cytologie der Blaualgen, Arch. Mikrobiol., 1956, 24(2), 147 CAS
.
- T. O. Yeates, C. A. Kerfeld, S. Heinhorst, G. C. Cannon and J. M. Shively, Protein-based organelles in bacteria: carboxysomes and related microcompartments, Nat. Rev. Microbiol., 2008, 6(9), 681 CrossRef CAS
.
- E. Gantt and S. F. Conti, Ultrastructure of Blue-Green Algae1, J. Bacteriol., 1969, 97(3), 1486 CrossRef CAS
.
- J. M. Shively, G. L. Decker and J. W. Greenawalt, Comparative Ultrastructure of the Thiobacilli, J. Bacteriol., 1970, 101(2), 618 CrossRef CAS PubMed
.
- J. M. Shively, F. L. Ball and B. W. Kline, Electron Microscopy of the Carboxysomes (Polyhedral Bodies) of Thiobacillus neapolitanus, J. Bacteriol., 1973, 116(3), 1405 CrossRef CAS PubMed
.
- J. M. Shively, F. Ball, D. H. Brown and R. E. Saunders, Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus, Science, 1973, 182(4112), 584 CrossRef CAS PubMed
.
- I. Stojiljkovic, A. J. Bäumler and F. Heffron, Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster, J. Bacteriol., 1995, 177(5), 1357 CrossRef CAS PubMed
.
- P. Chen, D. I. Andersson and J. R. Roth, The control region of the pdu/cob regulon in Salmonella typhimurium., J. Bacteriol., 1994, 176(17), 5474 CrossRef CAS
.
- M. Sutter, M. R. Melnicki, F. Schulz, T. Woyke and C. A. Kerfeld, A catalog of the diversity and ubiquity of bacterial microcompartments, Nat. Commun., 2021, 12(1), 3809 CrossRef CAS PubMed
.
- G. D. Havemann and T. A. Bobik, Protein Content of Polyhedral Organelles Involved in Coenzyme B12-Dependent Degradation of 1,2-Propanediol in Salmonella enterica Serovar Typhimurium LT2, J. Bacteriol., 2003, 185(17), 5086 CrossRef CAS PubMed
.
- Y. A. Holthuijzen, J. F. L. van Breemen, J. G. Kuenen and W. N. Konings, Protein composition of the carboxysomes of Thiobacillus neapolitanus, Arch. Microbiol., 1986, 144(4), 398 CrossRef CAS
.
- G. D. Price, S. M. Howitt, K. Harrison and M. R. Badger, Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function, J. Bacteriol., 1993, 175(10), 2871 CrossRef CAS PubMed
.
- M. Liang, S. Frank, H. Lünsdorf, M. J. Warren and M. B. Prentice, Bacterial microcompartment-directed polyphosphate kinase promotes stable polyphosphate accumulation in E. coli, Biotechnol. J., 2017, 12(3), 1600415 CrossRef PubMed
.
- T. O. Yeates, M. C. Thompson and T. A. Bobik, The protein shells of bacterial microcompartment organelles, Curr. Opin. Struct. Biol., 2011, 21(2), 223 CrossRef CAS PubMed
.
- T. O. Yeates, C. S. Crowley and S. Tanaka, Bacterial Microcompartment Organelles: Protein Shell Structure and Evolution, Annu. Rev. Biophys., 2010, 39, 185 CrossRef CAS PubMed
.
- M. Sutter, B. Greber, C. Aussignargues and C. A. Kerfeld, Assembly principles and structure of a 6.5 MDa bacterial microcompartment shell, Science, 2017, 356(6344), 1293 CrossRef CAS PubMed
.
- M. Sutter, M. Faulkner, C. Aussignargues, B. C. Paasch, S. Barrett and C. A. Kerfeld,
et al., Visualization of Bacterial Microcompartment Facet Assembly Using High-Speed Atomic Force Microscopy, Nano Lett., 2016, 16(3), 1590 CrossRef CAS PubMed
.
- C. A. Kerfeld, C. Aussignargues, J. Zarzycki, F. Cai and M. Sutter, Bacterial microcompartments, Nat. Rev. Microbiol., 2018, 16(5), 277 CrossRef CAS PubMed
.
- M. R. Badger, D. Hanson and G. D. Price, Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria, Funct. Plant Biol., 2002, 29(3), 161 CrossRef CAS PubMed
.
- B. D. Rae, B. M. Long, M. R. Badger and G. D. Price, Functions, Compositions, and Evolution of the Two Types of Carboxysomes: Polyhedral Microcompartments That Facilitate CO2 Fixation in Cyanobacteria and Some Proteobacteria, Microbiol. Mol. Biol. Rev., 2013, 77(3), 357 CrossRef CAS PubMed
.
- M. R. Badger and G. D. Price, CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution, J. Exp. Bot., 2003, 54(383), 609 CrossRef CAS PubMed
.
- Z. Dou, S. Heinhorst, E. B. Williams, C. D. Murin, J. M. Shively and G. C. Cannon, CO2 Fixation Kinetics of Halothiobacillus neapolitanus Mutant Carboxysomes Lacking Carbonic Anhydrase Suggest the Shell Acts as a Diffusional Barrier for CO2, J. Biol. Chem., 2008, 283(16), 10377 CrossRef CAS
.
- F. Cai, B. B. Menon, G. C. Cannon, K. J. Curry, J. M. Shively and S. Heinhorst, The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier, PLoS One, 2009, 4(10), e7521 CrossRef
.
- T. A. Bobik, G. D. Havemann, R. J. Busch, D. S. Williams and H. C. Aldrich, The Propanediol Utilization (pdu) Operon of Salmonella enterica Serovar Typhimurium LT2 Includes Genes Necessary for Formation of Polyhedral Organelles Involved in Coenzyme B12-Dependent 1,2-Propanediol Degradation, J. Bacteriol., 1999, 181(19), 5967 CrossRef CAS PubMed
.
- I. Stojiljkovic, A. J. Bäumler and F. Heffron, Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster, J. Bacteriol., 1995, 177(5), 1357 CrossRef CAS PubMed
.
- O. Erbilgin, K. L. McDonald and C. A. Kerfeld, Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides, Appl. Environ. Microbiol., 2014, 80(7), 2193 CrossRef PubMed
.
- H. Seedorf, W. F. Fricke, B. Veith, H. Brüggemann, H. Liesegang and A. Strittmatter,
et al., The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(6), 2128 CrossRef CAS PubMed
.
- E. M. Sampson and T. A. Bobik, Microcompartments for B12-Dependent 1,2-Propanediol Degradation Provide Protection from DNA and Cellular Damage by a Reactive Metabolic Intermediate, J. Bacteriol., 2008, 190(8), 2966 CrossRef CAS PubMed
.
- S. D. Axen, O. Erbilgin and C. A. Kerfeld, A Taxonomy of Bacterial Microcompartment Loci Constructed by a Novel Scoring Method, PLoS Comput. Biol., 2014, 10(10), e1003898 CrossRef PubMed
.
- O. Erbilgin, M. Sutter and C. A. Kerfeld, The Structural Basis of Coenzyme A Recycling in a Bacterial Organelle, PLoS Biol., 2016, 14(3), e1002399 CrossRef PubMed
.
- B. Ferlez, M. Sutter and C. A. Kerfeld, Glycyl Radical Enzyme-Associated Microcompartments: Redox-Replete Bacterial Organelles, mBio, 2019, 10(1), e023272-18 CrossRef PubMed
.
- J. Zarzycki, O. Erbilgin and C. A. Kerfeld, Bioinformatic Characterization of Glycyl Radical Enzyme-Associated Bacterial Microcompartments, Appl. Environ. Microbiol., 2015, 81(24), 8315 CrossRef CAS PubMed
.
- G. Kumar, N. K. Bari, J. P. Hazra and S. Sinha, A Major Shell Protein of 1,2-Propanediol Utilization Microcompartment Conserves the Activity of Its Signature Enzyme at Higher Temperatures, ChemBioChem, 2022, 23(9), e202100694 CrossRef CAS PubMed
.
- T. A. Bobik, Polyhedral organelles compartmenting bacterial metabolic processes, Appl. Microbiol. Biotech., 2006, 70, 517–525 CrossRef CAS PubMed
.
- S. Cheng, Y. Liu, C. S. Crowley, T. O. Yeates and T. A. Bobik, Bacterial microcompartments: their properties and paradoxes, BioEssays, 2008, 30(11–12), 1084 CrossRef CAS PubMed
.
- R. M. Jeter, Cobalamin-dependent 1,2-propanediol utilization by Salmonella typhimurium, Microbiol. Soc., 1990, 136(5), 887–896 CAS
.
- D. M. Roof and J. R. Roth, Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium, J. Bacteriol., 1989, 171(6), 3316 CrossRef CAS
.
- J. T. Penrod and J. R. Roth, Conserving a Volatile Metabolite: a Role for Carboxysome-Like Organelles in Salmonella enterica, J. Bacteriol., 2006, 188(8), 2865 CrossRef CAS PubMed
.
- D. A. Garsin, Ethanolamine Utilization in Bacterial Pathogens: Roles and Regulation, Nat. Rev. Microbiol., 2010, 8(4), 290 CrossRef CAS PubMed
.
- D. Heldt, S. Frank, A. Seyedarabi, D. Ladikis, J. B. Parsons and M. J. Warren,
et al., Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri, Biochem. J., 2009, 423(2), 199–207 CrossRef CAS PubMed
.
- M. Sutter and C. A. Kerfeld, BMC Caller: a webtool to identify and analyze bacterial microcompartment types in sequence data, Biol. Direct, 2022, 17(1), 9 CrossRef CAS PubMed
.
- S. Cheng, Y. Liu, C. S. Crowley, T. O. Yeates and T. A. Bobik, Bacterial microcompartments: their properties and paradoxes, BioEssays, 2008, 30(11–12), 1084 CrossRef CAS PubMed
.
- E. Mallette and M. S. Kimber, A Complete Structural Inventory of the Mycobacterial Microcompartment Shell Proteins Constrains Models of Global Architecture and Transport, J. Biol. Chem., 2017, 292(4), 1197 CrossRef CAS PubMed
.
- F. Cai, M. Sutter, J. C. Cameron, D. N. Stanley, J. N. Kinney and C. A. Kerfeld, The Structure of CcmP, a Tandem Bacterial Microcompartment Domain Protein from the β-Carboxysome, Forms a Subcompartment Within a Microcompartment, J. Biol. Chem., 2013, 288(22), 16055 CrossRef CAS
.
- C. E. Milla, C. Waltmann, A. G. Archer, N. W. Kennedy, C. H. Abrahamson, A. D. Jackson, E. W. Roth, S. Shirman, M. C. Jewett, N. M. Mangan, M. O. Cruz and D. T. Ereck, Vertex protein PduN tunes encapsulated pathway performance by dictating bacterial metabolosome morphology, Nat. Commun., 2022, 13(1), 3746 CrossRef PubMed
.
- M. G. Klein, P. Zwart, S. C. Bagby, F. Cai, S. W. Chisholm and S. Heinhorst,
et al., Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport, J. Mol. Biol., 2009, 392(2), 319 CrossRef CAS PubMed
.
- M. Sagermann, A. Ohtaki and K. Nikolakakis, Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(22), 8883–8887 CrossRef CAS PubMed
.
- M. Takenoya, K. Nikolakakis and M. Sagermann, Crystallographic Insights into the Pore Structures and Mechanisms of the EutL and EutM Shell Proteins of the Ethanolamine-Utilizing Microcompartment of Escherichia coli., J. Bacteriol., 2010, 192(22), 6056 CrossRef CAS PubMed
.
- F. Cai, M. Sutter, J. C. Cameron, D. N. Stanley, J. N. Kinney and C. A. Kerfeld, The structure of CcmP, a tandem bacterial microcompartment domain protein from the β-carboxysome, forms a subcompartment within a microcompartment, J. Biol. Chem., 2013, 288(22), 16055 CrossRef CAS PubMed
.
- S. Heinhorst, E. B. Williams, F. Cai, C. D. Murin, J. M. Shively and G. C. Cannon, Characterization of the Carboxysomal Carbonic Anhydrase CsoSCA from Halothiobacillus neapolitanus, J. Bacteriol., 2006, 188(23), 8087 CrossRef CAS PubMed
.
- S. Cheng and T. A. Bobik, Characterization of the PduS Cobalamin Reductase of Salmonella enterica and Its Role in the Pdu Microcompartment, J. Bacteriol., 2010, 192(19), 5071 CrossRef CAS PubMed
.
- N. K. Bari, J. P. Hazra, G. Kumar, S. Kaur and S. Sinha, Probe into a multi-protein prokaryotic organelle using thermal scanning assay reveals distinct properties of the core and the shell. Biochimica et Biophysica Acta (BBA) - General Subjects. 2020, 1864(10) 129680.
- R. Daniel, T. A. Bobik and G. Gottschalk, Biochemistry of coenzyme B12-dependent glycerol and diol dehydratases and organization of the encoding genes, FEMS Microbiol. Rev., 1998, 22(5), 553 CrossRef CAS PubMed
.
- C. Fan, S. Cheng, Y. Liu, C. M. Escobar, C. S. Crowley, R. E. Jefferson, T. O. Yeates and T. A. Bobik, Short N-terminal sequences package proteins into bacterial microcompartments, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(16), 7509–7514 CrossRef CAS PubMed
.
- C. Fan, S. Cheng, S. Sinha and T. A. Bobik, Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(37), 14995 CrossRef CAS PubMed
.
- C. Fan and T. A. Bobik, The N-Terminal Region of the Medium Subunit (PduD) Packages Adenosylcobalamin-Dependent Diol Dehydratase (PduCDE) into the Pdu Microcompartment, J. Bacteriol., 2011, 193(20), 5623–5628 CrossRef CAS PubMed
.
- T. M. Nichols, N. W. Kennedy and D. Tullman-Ercek, Cargo encapsulation in bacterial microcompartments: Methods and analysis, Methods Enzymol., 2019, 617, 155 CAS
.
- A. Dank, Z. Zeng, S. Boeren, R. A. Notebaart, E. J. Smid and T. Abee, Bacterial Microcompartment-Dependent 1,2-Propanediol Utilization of Propionibacterium freudenreichii, Front. Microbiol., 2021, 12, 679827 CrossRef PubMed
.
- M. B. Prentice, Bacterial microcompartments and their role in pathogenicity, Curr. Opin. Microbiol., 2021, 63, 19–28 CrossRef CAS PubMed
.
- P. Thiennimitr, S. E. Winter, M. G. Winter, M. N. Xavier, V. Tolstikov and D. L. Huseby,
et al., Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(42), 17480 CrossRef CAS PubMed
.
- Y. Bertin, J. P. Girardeau, F. Chaucheyras-Durand, B. Lyan, E. Pujos-Guillot and J. Harel,
et al., Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content, Environ. Microbiol., 2011, 13(2), 365 CrossRef CAS PubMed
.
- C. M. Jakobson and D. Tullman-Ercek, Dumpster Diving in the Gut: Bacterial Microcompartments as Part of a Host-Associated Lifestyle, PLoS Pathog., 2016, 12(5), e1005558 CrossRef PubMed
.
- P. C. Harvey, M. Watson, S. Hulme, M. A. Jones, M. Lovell and A. Berchieri,
et al., Salmonella enterica Serovar Typhimurium Colonizing the Lumen of the Chicken Intestine Grows Slowly and Upregulates a Unique Set of Virulence and Metabolism Genes, Infect. Immun., 2011, 79(10), 4105 CrossRef CAS PubMed
.
- S. Srikumar and T. M. Fuchs, Ethanolamine Utilization Contributes to Proliferation of Salmonella enterica Serovar Typhimurium in Food and in Nematodes, Appl. Environ. Microbiol., 2011, 77(1), 281 CrossRef CAS PubMed
.
- J. K. Lassila, S. L. Bernstein, J. N. Kinney, S. D. Axen and C. A. Kerfeld, Assembly of Robust Bacterial Microcompartment Shells Using Building Blocks from an Organelle of Unknown Function, J. Mol. Biol., 2014, 426(11), 2217 CrossRef CAS PubMed
.
- J. B. Parsons, S. D. Dinesh, E. Deery, H. K. Leech, A. A. Brindley and D. Heldt,
et al., Biochemical and Structural Insights into Bacterial Organelle Form and Biogenesis, J. Biol. Chem., 2008, 283(21), 14366 CrossRef CAS PubMed
.
- Y. Fang, F. Huang, M. Faulkner, Q. Jiang, G. F. Dykes and M. Yang,
et al., Engineering and Modulating Functional Cyanobacterial CO2-Fixing Organelles, Front. Plant Sci., 2018, 9, 739 CrossRef PubMed
.
- J. B. Parsons, S. Frank, D. Bhella, M. Liang, M. B. Prentice and D. P. Mulvihill,
et al., Synthesis of Empty Bacterial Microcompartments, Directed Organelle Protein Incorporation, and Evidence of Filament-Associated Organelle Movement, Mol. Cell, 2010, 38(2), 305 CrossRef CAS PubMed
.
- J. B. Parsons, S. Frank, D. Bhella, M. Liang, M. B. Prentice and D. P. Mulvihill,
et al., Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement, Mol. Cell, 2010, 38(2), 305 CrossRef CAS PubMed
.
- S. Cheng, S. Sinha, C. Fan, Y. Liu and T. A. Bobik, Genetic analysis of the protein shell of the microcompartments involved in coenzyme B12-dependent 1,2-propanediol degradation by Salmonella, J. Bacteriol., 2011, 193(6), 1385–1392 CrossRef CAS PubMed
.
- S. Choudhary, M. B. Quin, M. A. Sanders, E. T. Johnson and C. Schmidt-Dannert, Engineered Protein Nano-Compartments for Targeted Enzyme Localization, PLoS One, 2012, 7(3), e33342 CrossRef CAS
.
- B. H. Ferlez, H. Kirst, B. J. Greber, E. Nogales, M. Sutter and C. A. Kerfeld, Heterologous Assembly of Pleomorphic Bacterial Microcompartment Shell Architectures Spanning the Nano- to Microscale, Adv Mater., 2023, 2212065 CrossRef PubMed
.
- A. D. Lawrence, S. Frank, S. Newnham, M. J. Lee, I. R. Brown and W. F. Xue,
et al., Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor, ACS Synth. Biol., 2014, 3(7), 454 CrossRef CAS PubMed
.
- H. J. Wagner, C. C. Capitain, K. Richter, M. Nessling and J. Mampel, Engineering bacterial microcompartments with heterologous enzyme cargos, Eng. Life Sci., 2017, 17(1), 36 CrossRef CAS
.
- T. Li, Q. Jiang, J. Huang, C. M. Aitchison, F. Huang and M. Yang,
et al., Reprogramming bacterial protein organelles as a nanoreactor for hydrogen production, Nat. Commun., 2020, 11(1), 5448 CrossRef CAS
.
- T. Chen, Y. Fang, Q. Jiang, G. F. Dykes, Y. Lin and G. D. Price,
et al., Incorporation of Functional Rubisco Activases into Engineered Carboxysomes to Enhance Carbon Fixation, ACS Synth. Biol., 2022, 11(1), 154 CrossRef CAS PubMed
.
- M. Prentice, M. Warren and M. Liang, Accumulation of metabolic products in bacterial microcompartments, University of Kent at Canterbury, University College Cork, US Pat. Search PubMed
, US9187766, 2015.
- M. J. Lee, J. Mantell, I. R. Brown, J. M. Fletcher, P. Verkade and R. W. Pickersgill,
et al., De novo targeting to the cytoplasmic and luminal side of bacterial microcompartments, Nat. Commun., 2018, 9, 3413 CrossRef PubMed
.
- A. Hagen, M. Sutter, N. Sloan and C. A. Kerfeld, Programmed loading and rapid purification of engineered bacterial microcompartment shells, Nat. Commun., 2018, 9(1), 2881 CrossRef PubMed
.
- V. Müller, A synthetic bacterial microcompartment as production platform for pyruvate from formate and acetate, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(9), e2201330119 CrossRef PubMed
.
- H. Kirst, B. H. Ferlez, S. N. Lindner, C. A. R. Cotton, A. Bar-Even and C. A. Kerfeld, Toward a glycyl radical enzyme containing synthetic bacterial microcompartment to produce pyruvate from formate and acetate, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(8), e2116871119 CrossRef CAS PubMed
.
- G. Zhang, M. B. Quin and C. Schmidt-Dannert, Self-Assembling Protein Scaffold System for Easy in Vitro Coimmobilization of Biocatalytic Cascade Enzymes, ACS Catal., 2018, 8(6), 5611 CrossRef CAS
.
- K. Xue, L. Wang and J. Liu, Bacterial outer membrane vesicles and their functionalization as vehicles for bioimaging, diagnosis and therapy, Mater. Adv., 2022, 3(19), 7185 RSC
.
- Z. Cao and J. Liu, Bacteria and bacterial derivatives as drug carriers for cancer therapy, J. Controlled Release, 2020, 326, 396–407 CrossRef CAS PubMed
.
-
S. J. Tsai and T. O. Yeates, in Bacterial Microcompartments: Insights into the Structure, Mechanism, and Engineering Applications, Progress in Molecular Biology and Translational Science, ed. S. Howorka, Academic Press, 2011, vol. 103, p. 1–20 Search PubMed
.
- N. K. Bari, G. Kumar, J. P. Hazra, S. Kaur and S. Sinha, Functional protein shells fabricated from the self-assembling protein sheets of prokaryotic organelles, J. Mater. Chem. B, 2020, 8(3), 523 RSC
.
- J. Otoničar, M. Hostnik, M. Grundner, R. Kostanjšek, T. Gredar and M. Garvas,
et al., A method for targeting a specified segment of DNA to a bacterial microorganelle, Nucleic Acids Res., 2022, 50(19), e113 CrossRef PubMed
.
- D. A. Raba and C. A. Kerfeld, The potential of bacterial microcompartment architectures for phytonanotechnology, Environ. Microbiol. Rep., 2022, 14(5), 700 CrossRef CAS PubMed
.
- M. Faulkner, L. S. Zhao, S. Barrett and L. N. Liu, Self-Assembly Stability and Variability of Bacterial Microcompartment Shell Proteins in Response to the Environmental Change, Nanoscale Res. Lett., 2019, 14(1), 54 CrossRef PubMed
.
- N. K. Bari, G. Kumar, A. Bhatt, J. P. Hazra, A. Garg and Md. E. Ali,
et al., Nanoparticle Fabrication on Bacterial Microcompartment Surface for the Development of Hybrid Enzyme-Inorganic Catalyst, ACS Catal., 2018, 8(9), 7742 CrossRef CAS
.
- H. Kaur, N. K. Bari, A. Garg and S. Sinha, Protein morphology drives the structure and catalytic activity of bio-inorganic hybrids, Int. J. Biol. Macromol., 2021, 176, 106 CrossRef CAS PubMed
.
- J. Huang, B. H. Ferlez, E. J. Young, C. A. Kerfeld, D. M. Kramer and D. C. Ducat, Functionalization of Bacterial Microcompartment Shell Proteins With Covalently Attached Heme, Front. Bioeng. Biotechnol., 2020, 7, 432 CrossRef PubMed
.
- S. Kaur, N. K. Bari and S. Sinha, Varying protein architectures in 3-dimensions for scaffolding and modulating properties of catalytic gold nanoparticles, Amino Acids, 2022, 54(3), 441 CrossRef CAS PubMed
.
- H. Wang and M. Hayer-Hartl, Phase Separation of Rubisco by the Folded SSUL Domains of CcmM in Beta-Carboxysome Biogenesis, Methods Mol. Biol., 2022, 2563, 269 CrossRef PubMed
.
Footnote |
| † Equal contribution. |
| This journal is © The Royal Society of Chemistry 2023 |