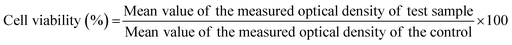In vitro biological evaluation of epigallocatechin gallate (EGCG) release from three-dimensional printed (3DP) calcium phosphate bone scaffolds
Yongdeok
Jo
 ,
Naboneeta
Sarkar
,
Naboneeta
Sarkar
 and
Susmita
Bose
and
Susmita
Bose
 *
*
School of Mechanical and Materials Engineering, Washington State University, Pullman, Washington 99164, USA. E-mail: sbose@wsu.edu
First published on 21st December 2022
Abstract
Three-dimensional printed (3DP) tricalcium phosphate (TCP) scaffolds can guide bone regeneration, especially for patient-specific defect repair applications in low-load bearing sites. Epigallocatechin gallate (EGCG), a green tea compound, has gained attention as a safer alternative treatment for bone disorders. The 3DP TCP scaffold is designed for localized EGCG delivery, which can enhance in vitro osteogenic ability, anti-osteoclastogenic activity, vascularization formation, and chemoprevention. In the cocultures of human bone marrow-derived mesenchymal stem cells (hMSCs) and monocytes (THP-1), EGCG release enhances osteogenic differentiation of hMSCs at day 16 compared to the control; this is indicated by a 2.8- and 4.0-fold upregulation of Runt-related transcription factor 2 (Runx2) and bone gamma-carboxyglutamic acid-containing protein (BGLAP), the early and late osteoblast differentiation marker expressions. However, EGCG significantly downregulates the receptor activator of nuclear factor-κB ligand (RANKL) expression by 7.0-fold, indicating that EGCG suppresses RANKL-induced osteoclast maturation. EGCG also stimulates endothelial tube formation at as early as 3 hours when human umbilical vein endothelial cells (HUVECs) grow on Matrigel. It reduces human osteosarcoma MG-63 cell viability by 66% compared to the control at day 11. An in vitro release kinetics study demonstrates that EGCG shows a ∼64% release within a day followed by a sustained release in the physiological environment (pH 7.4) because its phenolic hydroxyl groups are easily deprotonated at physiological pH. These findings contribute to developing a multifunctional scaffold for the treatment of low load-bearing patient-specific bone defects after trauma and tumor excision.
10th Anniversary StatementOver the past ten years, the Journal of Materials Chemistry B has been an asset to the science community through its consistent dedication to enhancing and expanding scientific discourse. It has published outstanding work in all areas of biology and medicine focusing on ways to serve researchers. The journal's multidisciplinary approach encourages authors to contribute their best work and in doing so, has fostered growth and innovation. We are thrilled to have our work published during the journal's ten-year anniversary and are honored to share our research, as we believe that the concept of natural medicinal drug delivery, like the release of epigallocatechin gallate (EGCG) from binder-jetting-based three-dimensional printed (3DP) calcium phosphate bone tissue engineering scaffolds, is a promising approach for enhancing new bone formation and blood vessel formation in low-load bearing bone defects after trauma and tumor excision. |
1 Introduction
Craniomaxillofacial congenital anomalies constitute a significant cause of death and morbidity among children worldwide. Besides congenital abnormalities, severe craniofacial trauma in adults caused by accidents or injuries also requires defect restoration to protect the brain and recover facial aesthetics.1 Cranial implants are used in more than 6% of annual bone graft procedures.2 Given that the world population is growing, the need for cranial bone grafts will also increase.3 In most cases, traditionally manufactured synthetic bone graft substitutes fail to provide desirable physiological functions based on the patients’ anatomical features and needs.4 However, three-dimensional printed (3DP) biomaterials enable a highly flexible patient-specific therapy because one can create bone scaffolds with controlled chemistry and geometry using bone-like bioceramics.5,6 3DP calcium phosphates (CaPs) have garnered attention due to their superior bioactivity over other biomaterials that are not similar in composition to natural bone tissue.7 The biodegradability of tricalcium phosphate (TCP), one CaP ceramic, enables the scaffolds to be used as bioresorbable implants in low-load or non-load bearing areas of craniofacial bones.8,9Craniofacial defects that require bone grafts can also be caused by several bone-related disorders, one of the more common being osteosarcoma resection surgeries.10 About 6% of the osteosarcomas occur in the jawbones, and those aged between 33 and 36 years suffer craniofacial osteosarcoma more frequently than any other age group.11 Advances in chemotherapy have increased the osteosarcoma survival rate in the past few decades, but the survival rate has remained stagnant in the 21st century.12 Surgical intervention is the ultimate clinical management for osteosarcoma; however, it leaves critical-sized bone defects beyond the bone's self-healing capability.13 Moreover, properly managing chemotherapy-induced toxicities is still a challenge, and the function of chemotherapy in craniofacial osteosarcoma is unclear.14,15
The challenges posed by chemotherapy and the treatment of critical-sized bone defects have reinforced the need for novel multifunctional bone grafts integrated with therapeutic biomolecules to remove residual tumor cells and support bone regeneration. The introduction of natural medicinal compounds (NMCs) provides a potential solution to the risks associated with high dosages of drug.16 NMCs such as epigallocatechin gallate (EGCG) have gained popularity as alternatives to synthetic drugs and growth factors for bone tissue engineering due to their superior safety and efficacy.17 EGCG, the main component extracted from green tea, is one of the most promising candidates for bone tissue engineering because of its wide range of therapeutic efficacy, including anti-inflammation, chemoprevention, and antiresorptive properties. According to a previous study, green tea consumption is linked to improved health outcomes, due to its antibacterial and anti-tumor potency.18,19 The consumption of green tea reduces the incidence of several malignancies, including those in the breast, colon, lung, and stomach.20–22 Furthermore, a study of 1256 women in the United Kingdom found that women aged 65 to 76 who drink the tea made from Camellia sinensis daily, including green tea, had higher bone mineral density than the non-tea drinkers.23 In osteosarcoma and osteoclast cells, EGCG induces apoptosis by inhibiting the nuclear factor kappa B (NF-κB) pathway.24,25 Studies have suggested that EGCG also promotes angiogenesis by enhancing the vascular endothelial growth factor (VEGF) levels and osteoblast proliferation via the bone morphogenetic protein 2 (BMP-2) signaling pathway.26,27
Given the extensive potential of EGCG to promote bone regeneration and reduce osteosarcoma in bone tissue engineering, in vitro single cell studies are needed to evaluate its efficacy in humans. The evaluation of scaffolds in bone tissue engineering requires in vitro cocultures of osteoblasts and osteoclasts because they can more closely mimic the in vivo bone repair process through cell–cell interactions than a monoculture model.28 This technique has become commonplace in biomaterial research, but only a few studies have used in vitro rat studies to demonstrate the effects of EGCG on cell behavior in cocultures.29,30 It necessitates additional in vitro research on EGCG's effects by incorporating human cells in the coculture.
There are gaps in scientific knowledge regarding the development of multifunctional scaffolds with controlled chemistry and natural medicine for the treatment of critical-sized bone defects. Therefore, we will investigate the following two questions: (1) Is it possible to modulate the release behavior of EGCG that mimic an in vivo scaffold microenvironment? (2) Can EGCG loaded 3DP scaffolds induce in vitro bone regeneration, in vitro vasculogenesis, and anticancer activity? To answer these questions, we have investigated the ability of the EGCG-loaded 3DP scaffold to release EGCG in a controlled manner that can induce in vitro osteogenesis, anti-osteoclastogenesis, vasculogenesis, and chemoprevention to provide a potential therapeutic treatment option for postsurgical bone defects in low load-bearing sites.
2 Experimental section
2.1 Scaffold preparation
3DP scaffolds were fabricated using β-tricalcium phosphate (β-TCP) powder prepared by the solid-state synthesis method.31 Commercial hydroxyapatite powder (HA, NEI, USA.) and the synthesized TCP powder were used to fabricate discs by uniaxially pressing at 145 MPa for 2 minutes. The HA discs were used for in vitro coculture study, while the TCP discs were used for in vitro drug release study. A binder jet printer (Innovent+®, ExOne, U.S.A.) was used to fabricate TCP scaffolds with designed porosity. The 3DP TCP scaffolds were employed for in vitro osteoblast, and osteosarcoma cell culture studies, as well as in vitro endothelial cell tube formation assay. The binder was cured for the 3DP scaffolds, and then the green fabricated scaffolds were depowderized. Green parts were then sintered at 1250 °C for 2 h in a conventional furnace.2.2 Drug loading and in vitro release of EGCG
EGCG (97.69%, APExBIO, USA) was dissolved in ethanol (100%, KOPTEC, USA) at a concentration of 2.5 mg mL−1 for the in vitro release study. To understand the drug release behavior of EGCG at different pH, EGCG drug solution was loaded on TCP discs. 40 μL of the drug solution was added dropwise using a pipette on top of each disc to cover its entire surface and the solvent was dried in the dark at room temperature overnight. EGCG drug release was carried out in an acetate buffer solution (pH 5) and phosphate-buffered solution (pH 7.4) to mimic the acidic environment of tumor tissues and the physiological environment of body, respectively. The drug loaded samples were kept in glass vials and immersed in 2 mL of buffer solutions. All vials were placed in a 37 °C shaker rotating at 150 rpm. The buffer medias were collected and replaced with fresh buffers at selected time points; 0.3, 0.6, 1, 1.5, 3, and 9 h, and 1, 4, 7, 14, and 21 days. The concentration of EGCG in the collected solutions was determined using a UV-Vis spectroscopy microplate reader (BioTek Synergy 2 SLFPTAD, USA) at 278 nm. KinetDs (version 3.0) was used to determine the kinetics of EGCG release.322.3 In vitro osteosarcoma and osteoblast cell culture study
3DP scaffolds were sterilized for 60 min at a temperature of 121 °C in an autoclave (Tuttnauer, USA) before the cell culture study. Unloaded 3DP scaffolds were utilized as control. For the treatment group, 0.1 mM of the drug solution was prepared with ethanol and EGCG. In a sterile cell culture hood, 200 μL of the drug solution was loaded on the scaffold to cover its entire surface. The osteosarcoma cells (MG-63, ATCC, USA) were seeded onto the scaffolds at a density of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per scaffold. The Eagle's minimum essential medium (EMEM, ATCC, USA) supplemented with 10% Fetal Bovine Serum (FBS, ATCC, USA) was used as a growth medium. The samples were kept in the incubator at 37 °C under a humidified and 5% CO2 atmosphere, and the media was changed every 2–3 days.
000 cells per scaffold. The Eagle's minimum essential medium (EMEM, ATCC, USA) supplemented with 10% Fetal Bovine Serum (FBS, ATCC, USA) was used as a growth medium. The samples were kept in the incubator at 37 °C under a humidified and 5% CO2 atmosphere, and the media was changed every 2–3 days.
The prepared scaffolds were placed in 24 well plates, and human fetal osteoblastic cells (hFOB 1.19, ATCC, USA) were seeded onto the 3DP scaffolds with a density of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per sample. The cell culture medium was prepared by a 1
000 cells per sample. The cell culture medium was prepared by a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio mixture of Ham's F12 medium and Dulbecco's Modified Eagle's Medium (DMEM/F12, Sigma Aldrich, USA) with 0.3
1 ratio mixture of Ham's F12 medium and Dulbecco's Modified Eagle's Medium (DMEM/F12, Sigma Aldrich, USA) with 0.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mg mL−1 of G418 (Sigma Aldrich, USA), and 1.2 mg mL−1 of sodium bicarbonate (NaHCO3, Fisher Scientific, USA) in filter sterilized deionized (DI) water. The media was then supplemented with 10% FBS and penicillin-streptomycin (ATCC, USA). The samples were kept in the incubator at 34 °C under 5% CO2 atmosphere, and the media was changed every 2–3 days.
mg mL−1 of G418 (Sigma Aldrich, USA), and 1.2 mg mL−1 of sodium bicarbonate (NaHCO3, Fisher Scientific, USA) in filter sterilized deionized (DI) water. The media was then supplemented with 10% FBS and penicillin-streptomycin (ATCC, USA). The samples were kept in the incubator at 34 °C under 5% CO2 atmosphere, and the media was changed every 2–3 days.
2.4 In vitro coculture of hMSCs and monocytes
Human bone marrow-derived mesenchymal stem cells (hMSCs, Lonza, USA) and monocytes (THP-1, ATCC, USA) were used for the coculture. hMSCs cells were grown in mesenchymal stem cell growth medium (Lonza, USA). THP-1 cells were grown in RPMI-1640 growth medium, containing 10% FBS and 0.05 mM 2-mercaptoethanol. Before cell seeding, osteoblast and osteoclast differentiation medias were prepared. Human mesenchymal stem cell (hMSC) osteogenic differentiation media was prepared using hMSC osteogenic differentiation basal medium (Lonza, USA), and hMSC osteogenic differentiation singlequots™ supplements kit (Lonza, USA). Osteoclast differentiation media contained 10 ng mL−1 RANKL (Abcam, USA), 40 ng mL−1 phorbol 12-myristate13-acetate (PMA, Abcam, USA), and 10% FBS (ATCC, USA) in RPMI-1640 medium (ATCC, USA). 2.29 μg of EGCG was loaded on the HA discs, which are denser than the tricalcium phosphate used for the coculture study. hMSC and THP-1 were seeded at a concentration of 1.5 × 104 cells on the HA discs. 1 mL of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of osteoblast and osteoclast differentiation media was used for the coculture. Samples were incubated at 37 °C under a humidified atmosphere containing 5% CO2 and media was changed every 2–3 days.33
1 mixture of osteoblast and osteoclast differentiation media was used for the coculture. Samples were incubated at 37 °C under a humidified atmosphere containing 5% CO2 and media was changed every 2–3 days.33
2.5 The MTT cell viability assay and cellular morphology
The MTT assay was performed at 3, 7, and 11 days for cell cultures. The MTT solution was prepared by adding sterile-filtered phosphate buffered saline (PBS) to the MTT reagent (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) at 5 mg mL−1 concentration. 100 μL of the MTT solution and 900 μL of cell media were added to each sample and incubated for 2 h at 34 °C in a 5% CO2 environment. After 2 h incubation, the media was aspirated. The formed purple formazan crystals were dissolved by adding 600 μL of the MTT solubilizer composed of 10% Triton X-100, 0.1 N HCL and isopropanol. 100 μL of final solution was transferred to 96 well plates and read by a microplate reader (BioTek Synergy 2 SLFPTAD, USA) at 570 nm. The cytotoxicity of the drug to osteosarcoma cells was determined by the percentage cell viability using the following equation.The cellular morphology of all samples was examined by Field Emission Scanning Electron Microscope (FESEM, FEI Quanta 200, FEI Inc., USA). Samples were taken out and fixed with 1 mL of a solution composed of 2% paraformaldehyde (VWR, USA) and 2% glutaraldehyde (Sigma Aldrich, USA) in 0.1 M phosphate buffer (Sigma Aldrich, USA) overnight at 4 °C. After primary fixation, samples were rinsed with 0.1 M phosphate buffer and postfixed in 2% OsO4 at room temperature for 2 h. After the post-fixation, samples were rinsed with 0.1 M phosphate buffer and dehydrated in a 30, 50, 70, 90, and 100% ethanol series for 10 minutes each, followed by hexamethyldisilane (HMDS, EMS, USA) drying. The samples were coated with gold using a sputter coater (Hummer V sputtering system, Hummer, USA) before SEM images were taken.
2.6 Resorption activity analysis
hMSCs and monocytes were cocultured in vitro as described in a previous Section 2.4 and used for the resorption pit assay. The media was removed, and then the sample was rinsed with PBS solution three times. After that, 500 μL of the mixture of the buffer solution and Triton X-100 (500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v ratio) was added to submerge the samples, which were then ultrasonicated for 10 minutes. The samples were washed with PBS solution three times. Later, the samples were dehydrated and dried for SEM imaging. The resorption pit areas were analyzed with ImageJ (ImageJ software, USA).34
1 v/v ratio) was added to submerge the samples, which were then ultrasonicated for 10 minutes. The samples were washed with PBS solution three times. Later, the samples were dehydrated and dried for SEM imaging. The resorption pit areas were analyzed with ImageJ (ImageJ software, USA).34
2.7 Quantitative reverse transcription polymer chain reaction (qRT-PCR)
According to the manufacturer's protocol, the extraction of total RNA was performed with Aurum TM Total RNA Mini Kit (Bio-Rad, USA). After extraction, 15 μL of RNA were reverse transcribed to cDNA using iScript™ cDNA Synthesis Kit (Bio-Rad, USA), according to the manufacturer's protocol. Quantitative RT-PCR (qRT-PCR) was performed to detect expression of Runt-related transcription factor 2 (Runx2), osteocalcin known as bone gamma-carboxyglutamic acid-containing protein (BGLAP), Osteoprotegerin (OPG), receptor activator of nuclear factor-κB ligand (RANKL) with SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad, USA), following the manufacturer's instruction. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used for the internal control, and the relative gene expression was analyzed by the 2−ΔΔCt method.352.8 In vitro vascular tubule formation
Control (3DP TCP scaffold) and treatment (3DP TCP loaded with EGCG) groups were prepared as described in a previous Section 2.3 for the in vitro endothelial cell culture. HUVEC (ATCC, USA) was cultured in a flask with 15 mL vascular cell basal media (ATCC, PCS-100-030, USA) and endothelial cell growth kit-VEGF (ATCC, PCS-100-041, USA). Matrigel was diluted in the complete growth medium (ATCC, USA) in equal proportions. 500 μL of the diluted matrigel was added in the lower chamber of 12 transwell plates (Corning, USA). HUVECs (ATCC, USA) were seeded on Matrigel at a density of 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well. The control 3DP scaffolds and EGCG-loaded 3DP scaffolds were kept in the transwell inserts. Each well was filled with 500 μL of complete growth media. The samples were incubated in a 5% CO2 environment at 37 °C with saturated humidity. The endothelial tube formation was determined 3 h, 12 h, 24 h later by an inverted microscope (ACCU-SCOPE, SERIES 3300, USA) and analyzed by Angiogenesis Analyzer for ImageJ.36
000 cells per well. The control 3DP scaffolds and EGCG-loaded 3DP scaffolds were kept in the transwell inserts. Each well was filled with 500 μL of complete growth media. The samples were incubated in a 5% CO2 environment at 37 °C with saturated humidity. The endothelial tube formation was determined 3 h, 12 h, 24 h later by an inverted microscope (ACCU-SCOPE, SERIES 3300, USA) and analyzed by Angiogenesis Analyzer for ImageJ.36
2.9 Statistical analyses
All statistical analyses were performed using the student's t test. All measurements were made in triplicate (n = 3), and p value ≤ 0.05, is considered as statistically significant.3 Results
3.1 TCP scaffolds show controlled and pH sensitive EGCG release
Binder jetting was used to fabricate 3DP TCP scaffolds (Fig. 1(A)–(D)). Computer aided design (CAD) software was used to 400 μm of highly interconnected designed pore sizes (Fig. 1(A)). The green scaffolds were fabricated with macro pores between 400 and 440 μm (Fig. 1(B)). The sintering process resulted in a densified 3DP TCP scaffold with pore sizes of approximately 340–370 μm and intrinsic residual micropores of 5–20 μm (Fig. 1(C) and (D)). The release of EGCG from TCP discs was measured at pH 5.0 and at pH 7.4 as depicted in Fig. 1(E) and (F). At pH 5.0, EGCG exhibits a cumulative release of 32% in 24 hours and achieves a plateau thereafter (Fig. 1(E)). An initial higher release of 64% of the EGCG is achieved at pH 7.4, followed by a sustained release of the drug for 3 weeks (Fig. 1(F)). After 21 days, a total of 74.8% of the EGCG is released at pH 7.4, while only 33.25% is released at pH 5.0. The acid dissociation constant or pKa value of EGCG is 7.68, indicating EGCG is easily deprotonated at physiological pH (pH = 7.4) (Fig. 1(G)), and thus exhibited higher release. The cumulative release kinetics of EGCG at pH 5.0 and 7.4 are fitted by the Weibull distribution model.373.2 EGCG-loaded 3DP scaffolds promotes hFOB cell proliferation at later time points
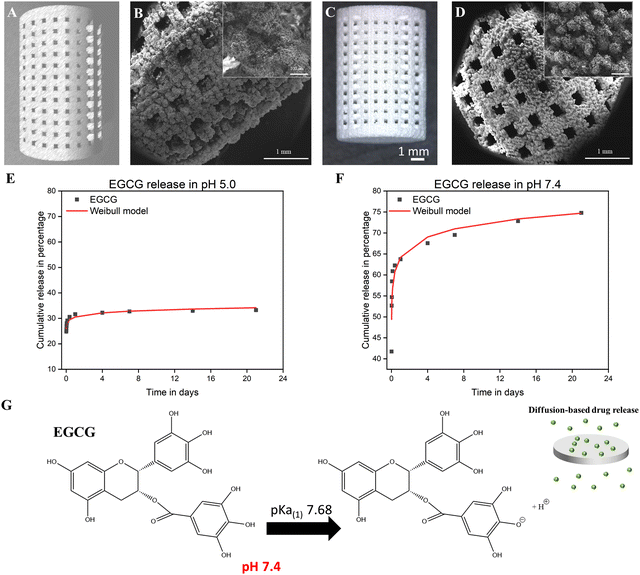
The osteoblast cell culture was used to investigate the cytocompatibility of EGCG with human fetal osteoblastic cells (hFOB). This study found that EGCG did not exhibit any cellular toxicity for hFOB cells at 3, 7, and 11 days. EGCG does not induce early-stage proliferation of osteoblasts, as confirmed by the MTT assay at day 3. However, it does enhance hFOB proliferation at days 7 and 11. The MTT assay indicated that EGCG released from the scaffolds significantly increased cellular viability by 1.3- and 1.2-fold at 7 and 11 days compared to the control (p ≤ 0.05) as shown in Fig. 2(A). Healthy hFOB cell attachment and growth on the surfaces of both the control and EGCG-loaded 3DP scaffolds were examined by FESEM at all time points (Fig. 2(B)). On day 7 and day 11, the cells had more elongated cell morphology and firmer attachments on the surfaces of the EGCG-loaded scaffolds compared to the control.3.3 Controlled EGCG release reduces osteoclast-mediated resorption pit formation
The difference in resorption pits between the control and EGCG treatments was determined. At days 7 and 16, the presence of EGCG resulted in remarkably fewer and smaller lacunae than the control over the entire surface of the HA discs as shown in Fig. 3. The control had a total area of resorption lacunae of approximately 1611 μm2, whereas EGCG had smaller pit formations with an approximate surface area of 195 μm2 at day 7. After 16 days of culture, the pit areas of around 8038 μm2 were measured for the control samples, while the pits formed with an approximate surface area of 476 μm2 on EGCG-loaded samples. EGCG reduced resorption pit formation by 88% and 94% on days 7 and 16, respectively compared to the control. Osteoclast resorption occurs in the sealing zone of mature osteoclasts, which is composed of F-actin.38,39 F-actin binds tightly to the surface, releasing protons that dissolve hydroxyapatite and proteases that digest bone organic compounds. The smaller resorption areas caused by EGCG suggest that the compound inhibits bone resorption.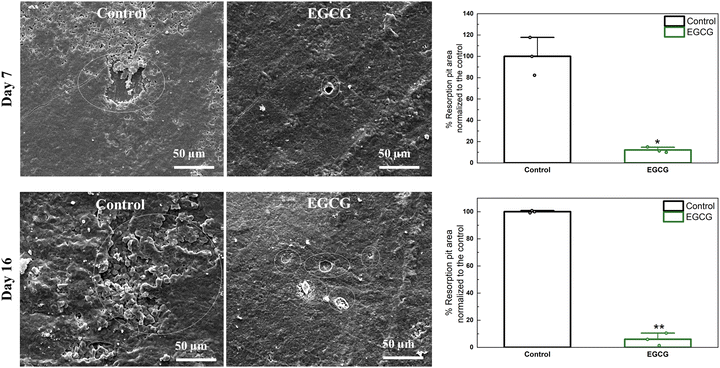 | ||
| Fig. 3 Fewer and smaller resorption pits (white circle) can be found on EGCG-loaded samples than control samples at all time points. The relative resorption pit areas are calculated using ImageJ software.40 The quantification of the resorbed area shows that EGCG significantly reduced osteoclast resorption activity compared to the control. (* denotes p value ≤ 0.05, ** denotes p value ≤ 0.001). | ||
3.4 Controlled EGCG release modulates in vitro osteoblast and osteoclast differentiation markers
The coculture of hMSC-derived osteoblasts and monocyte derived osteoclasts was used to understand the effects of EGCG on in vitro bone remodeling maintained by these cells, as well as their potential signaling pathways. The effects of EGCG on the proliferation and morphology of the cocultured cells were evaluated by SEM after culturing for 7 and 16 days, as shown in Fig. 4(A). The morphology of attached osteoblast cells on the surfaces of samples differed between the EGCG-loaded samples and control after 7 days of cell culture. Compared to the control, EGCG treatment induced a continuous layer of flattened osteoblast-like cells with filopodial prostheses and lamellipodial extensions. At day 16, however, there were no noticeable differences in cell morphology and proliferation between the two groups. Both the EGCG and control showed that osteoblastic cells proliferated uniformly over the surfaces of the samples with high density. After 7 and 16 days, RANKL-stimulated osteoclast cells in the coculture system were examined to observe the morphology and integration of the osteoclasts cells. The control group showed highly networked osteoclast-like cells on the surface of the sample with numerous filopodia and lamellipodia, whereas in the presence of EGCG, these cells had rounder shapes and fewer intercellular attachments, implying EGCG inhibited RANKL-induced osteoclast differentiation between days 7 and 16. To determine the effects of EGCG on in vitro osteogenic potential, relative gene expression analysis of bone gamma-carboxyglutamic acid-containing protein (BGLAP) and runt-related transcription factor 2 (Runx2) were accessed by qRT-PCR following 7 and 16 days of culture. The addition of EGCG did not change the levels of BGLAP and Runx2 gene expression after 7 days.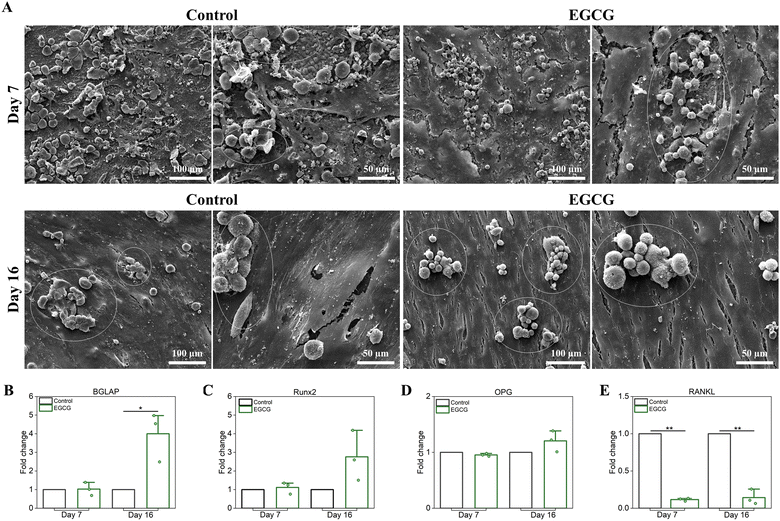 | ||
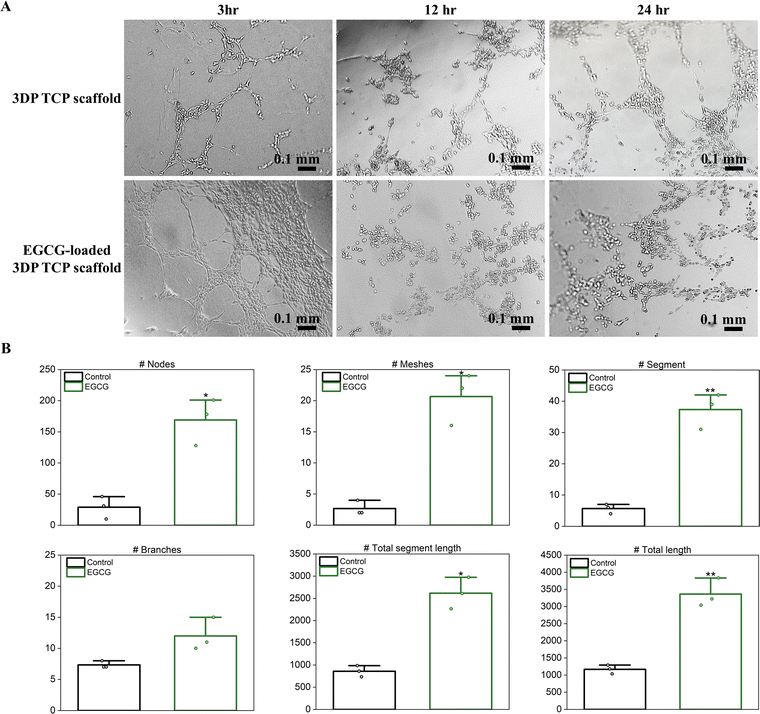
| Fig. 4 Effects of EGCG-loaded HA on coculture of hMSC-derived osteoblasts and monocyte derived osteoclasts were evaluated by SEM microscopy and relative gene expression.35 (A) SEM micrographs showed healthy osteoblast morphology in all samples at all time points. The control showed healthy maturation from differentiated monocytes (white circle) pre-osteoclast cells. The osteoclast cells bonded to form multi-nucleated mature osteoclasts at days 7 and 16. EGCG showed good adhesion and attachment of osteoclast cells, but inhibited maturation compared to the control at that time. (B)–(E) qRT-PCR results were presented as fold change compared to the control, which is standardized to a value of 1. GAPDH was used to normalize all gene expression data. Day 7 was the early time point to observe the effects of EGCG on coculture. The higher expression of (B) bone gamma-carboxyglutamic acid-containing protein (BGLAP) and (C) Runt-related transcription factor 2 (Runx2) in treatment than control implied that EGCG stimulated osteoblast differentiation and maturation on day 16. However, EGCG inhibited osteoclast differentiation and activity by upregulating (D) osteoprotegerin (OPG), a negative regulator of osteoclastogenesis, and downregulating (E) RANKL expression on day 16. (* denotes p value ≤ 0.05, ** denotes p value ≤ 0.001). | ||
However, BGLAP gene expression increased significantly by 4.0-fold with EGCG treatment, whereas Runx2 gene expression increased by 2.8-fold (Fig. 4(B) and (C)). These results indicated that EGCG induced osteoblast differentiation after 16 days. The analysis of expression levels of osteoclastic marker genes, including RANKL and OPG were used to assess the effects of EGCG on in vitro osteoclastogenesis (Fig. 4(D) and (E)). OPG gene expression was slightly downregulated within the error range in EGCG treatment over the control at day 7, whereas the gene expression was increased by 1.2-fold at day 16. This result indicated that EGCG had a minor effect on OPG expression, which is in line with a previous study.41 On the other hand, RANKL expression decreased significantly in the presence of EGCG. RANKL was downregulated by EGCG by 8.4- and 7.0-fold on days 7 and 16, respectively compared to the control. The RANKL/OPG ratio usually determines the balance between bone formation and resorption and thereby also indicates osteoclast activation.42,43 RANKL/OPG ratios lower than 1 indicate bone formation, while levels higher than 1 imply bone destruction.42 The RANK/OPG ratio was 0.12 during EGCG treatment on days 7 and 16. This finding suggested that EGCG inhibited osteoclast activity and differentiation compared to the control, resulting in less hydroxyapatite resorption. Taken together, these findings suggest that EGCG promotes the differentiation of hMSCs into osteoblasts by upregulating Runx2 and BGLAP expressions, while inhibiting differentiation of osteoclast precursors via the RANKL/RANK pathway and bone resorption.
3.5 EGCG release from 3DP TCP inhibits in vitro osteosarcoma cell proliferation
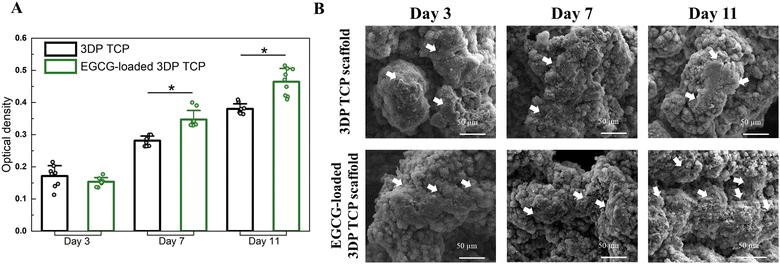
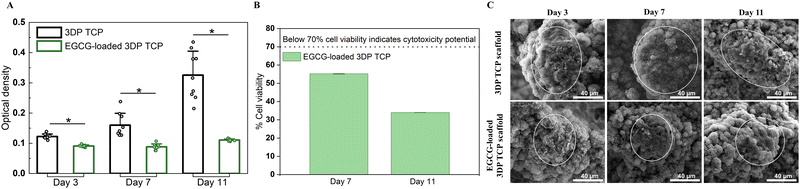
At days 3, 7, and 11, the MTT assays and cellular morphology study of osteosarcoma cells by FESEM were used to assess the anti-osteosarcoma properties of EGCG (Fig. 5). The MTT assay demonstrated that EGCG reduced osteosarcoma cell viability at all time points as shown in Fig. 5(A). In the presence of EGCG, the osteosarcoma cell viability was significantly reduced to 26%, 45%, and 66% of the control value at days 3, 7 and 11, respectively. Biomolecules that reduce cell viability by 70% have the potential to be cytotoxic to the cell.44 This finding implied that EGCG release induces cytotoxicity to osteosarcoma cells at days 7 and 11 (Fig. 5(B)). Further, the cell morphology was examined by FESEM. On days 3, 7 and 11, FESEM images showed that in vitro osteosarcoma cells were well spread on the surface of the control at all time points. In contrast, only a few cells were attached to the EGCG-loaded 3DP scaffolds, indicating EGCG suppresses the proliferation and adhesion of osteosarcoma cells compared to the control (Fig. 5(C)).3.6 Controlled release of EGCG promotes early-stage HUVEC-induced tube formation
To evaluate in vitro angiogenesis of EGCG-loaded 3DP TCP scaffolds, HUVECs were used for the tube formation assay. EGCG release from 3DP TCP scaffolds promoted capillary tube formation of the endothelial cells at as early as 3 hours compared to the control, which showed few tubes and minimal network among the branches (Fig. 6(A)). After 12 hours, the tubes began to break down and were under apoptosis. Most endothelial cells were detached from Matrigel and cell death occurred at less than 24 hours.45 Furthermore, the in vitro angiogenic activities of EGCG were evaluated quantitatively by ImageJ (Fig. 6(B)). In the presence of EGCG for 3 hours, in vitro angiogenesis parameters such as the number of nodes, meshes, segments, branches, total segment length, and total length increased by 5.8-, 7.8-, 6.6-, 1.6-, 3.0- and 2.9-fold, respectively.4 Discussion
Severe bone trauma and tumor resection can result in critical-sized bone defects which do not heal on their own. In fact, following tumor resection, residual cancer cells are frequently left behind, which can move to other tissues and metastasize. These challenges have steered medicine in the direction of natural compounds as alternatives to synthetic drugs to treat bone disorders. To address the challenges of bone defect repair, a 3DP tissue-engineered TCP scaffold has been developed, which enables localized EGCG delivery and significant improvements in in vitro osteogenic, anti-osteoporotic, chemopreventive, and angiogenic properties.To address in vitro osteoblast responses to EGCG-loaded 3DP scaffolds, hFOB cell culture was conducted. EGCG showed no signs of cytotoxicity toward hFOB cells at day 3, followed by an increase in in vitro osteoblast cellular viability and proliferation at days 7 and 11. This result is in line with a previous study, which showed that EGCG increases osteoblast proliferation and decreased osteoblast apoptosis by suppressing oxidative stress.46 To further assess the effect of EGCG on in vitro bone remodeling, a qRT-PCR study was conducted in the in vitro coculture system, mimicking a bone microenvironment.28 The presence of EGCG can promote osteoblast development and osteoblast maturation by increasing the expression of osteoblast-specific target genes like Runx2 and BGLAP. A previous study found that the treatment of EGCG increases mRNA expression of Runx2 in a bone marrow mesenchymal stem cell line.26 Runx2 is a key transcription factor that regulates early-stage osteoblast differentiation.47 It is maximized in the immature osteoblasts and subsequently is decreased in mature osteoblasts.48 EGCG also modulates BGLAP, the most abundant protein in bone and an osteogenic marker at the last stage of osteoblast differentiation, resulting in bone matrix synthesis and mineralization.49 The Runx2 and BGLAP genes are stimulated by EGCG via the Wnt/β-catenin signaling pathway.50 The Wnt/β-catenin pathway is activated by binding between Wnt family member 3a (Wnt3a) and membrane receptor complexes such as the frizzled and low density lipoprotein receptor-related proteins 5/6 (Lrp5/6) receptors. A study confirmed that EGCG increases osteogenic gene expression by stimulating Runx2 and osterix secretion from mesenchymal stem cells (MSCs), which plays an important role in osteogenic differentiation and mineralization of MSCs.51 EGCG also enhanced osteogenesis on stem cells from apical papilla through Smad1/5/9.52 EGCG released from a calcium phosphate matrix upregulated both Runx2 and BGLAP genes, implying that EGCG induces enhanced osteoblast differentiation in vitro after 16 days of culture. It has been reported that EGCG can reduce the activity of bone-resorbing osteoclasts.24 Osteoclasts are multinucleated giant cells that differentiate from monocytes and are responsible for in vivo bone resorption.53 The receptor activator of nuclear factor-κB ligand (RANKL) can influence the production of mature osteoclasts.54 Mature osteoclasts form a firm attachment between the cell membrane and the calcified matrix, resulting in the release of enzymes and the formation of resorption pits.55In vitro study shows that EGCG suppresses RANKL-induced osteoclast differentiation at an early stage when cell fusion and multinucleated cell formation occurs.24 The binding of RANKL to RANK drives the osteoclast differentiation by activating the RANK signaling pathway. However, OPG, a decoy receptor for RANK secreted by osteoblasts, suppresses RANKL–RANK interactions, eventually inhibiting osteoclastic differentiation.56 It has been reported that EGCG suppresses osteoclastogenesis in a coculture of RAW264.7 mouse monocytes and ST2 mouse marrow stromal cells by regulating the RANK/RANKL/OPG pathway.41 By increasing OPG and decreasing RANKL gene expression, EGCG inhibits osteoclast development and mature osteoclast function. Consistent with these studies, EGCG inhibits RANKL-induced osteoclastogenesis in human monocytes by suppressing osteoclast maturation and resorption pit formation, upregulating OPG expression and downregulating RANKL expression compared to the control at day 16. These results suggest that EGCG suppresses osteoclast differentiation by competitively inhibiting RANKL binding to RANK receptors.
After investigating the effects of EGCG release on in vitro osteoblast and osteoclast activities in bone remodeling, we evaluated the in vitro chemopreventive effects of EGCG on osteosarcoma (MG-63) cells. EGCG reduces cell viability in osteosarcoma cells by 55% and 34% after 7 and 11 days, respectively, indicating its in vitro chemoprevention potential. The selective cytotoxicity of EGCG to both osteoclast and osteosarcoma can be explained by the NF-κB pathway (Fig. 7). NF-κB proteins are inactivated in the majority of cells by binding to IκB proteins found in the cytoplasm. However, osteoclast differentiation and osteosarcoma growth can be induced by activated NF-κB signaling caused by various inflammatory stimuli as well as the degradation of IκB proteins.57,58 EGCG can inhibit NF-κB pathway activation by suppressing the proinflammatory gene expressions and phosphorylation of IκB.59,60 In summary, EGCG can suppress proliferation and viability of osteosarcoma cells, as well as osteoclast differentiation by inhibiting activation of the NF-κB pathway.
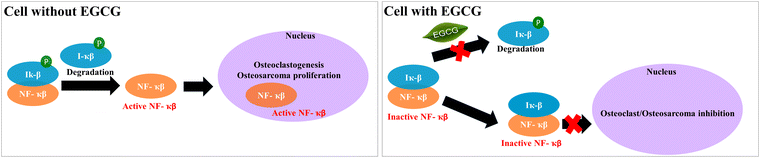 | ||
| Fig. 7 The possible mechanism of action of EGCG towards osteoclast and osteosarcoma cell inhibition is demonstrated through the NF-κB inhibition pathway.57,58 | ||
In the local drug delivery system, we have examined, the sustained release of chemopreventive drugs at the tumor site inhibits osteosarcoma progression and has long-term anticancer effects.61 In the case of the osteogenic compound, however, an initial higher release, followed by a sustained release will promote faster wound healing and bone regeneration.62 EGCG- loaded 3DP scaffolds with chemopreventive and osteogenic properties can be used as local drug delivery systems due to their pH-sensitive release of EGCG. The pH-dependent solubility changes of EGCG in buffer solutions can play a role in different release rates. EGCG exhibits a Weibull distribution with a higher release at pH 7.4, but a sustained release at pH 5.0. Since EGCG has the pKa values of 7.68–7.75 and 8.0, which are close to the pH of phosphate-buffered solution, the phenolic hydroxyl groups of EGCG are easily deprotonated by physiological pH (pH = 7.4). A physiological pH induces EGCG to change from the phenol type to the phenolate ion which is more soluble in water.63 The acidic microenvironment created by surgery and implantation lasts about two weeks before returning to the normal physiological pH level. Therefore, EGCG will slowly be released from the scaffolds until the normal physiological pH level is reached after tumor removal. The remaining tumor cells will be eliminated within this period. When the acidic microenvironment is neutralized to pH 7.4, the scaffold will release the remaining EGCG, which will aid in bone regeneration and wound healing. It will also help in the prevention of tumor cell growth in the surgical region, resulting in inhibition of metastasis.
The healing of critical-sized bone defects is influenced not only by osteogenesis but also by angiogenesis.64 Implant vascularization in critical-sized bone defects can provide a constant supply of oxygen, osteoblast/osteoclast precursors and nutrition, resulting in faster bone remodeling.65 Our findings suggest that EGCG can increase the in vitro angiogenic properties of 3DP TCP scaffolds required for enhancing bone repair. A previous study reported that EGCG enhances vascularization in normal tissues, while inhibiting tumor angiogenesis.27 Results from a recent study are in line with our results where presence of EGCG has been associated with enhanced angiogenesis both in vitro and in vivo. EGCG enhanced in vitro VEGF expression, stimulated HUVEC migration, as well as promoted in vivo angiogenesis by secreting HIF-1α and VEGF expression in a mouse hindlimb ischemic model.66 These results support our finding that EGCG-loaded 3DP scaffolds accelerate in vitro vasculogenesis by enhancing endothelial tube formation of HUVECs at the 3 h time point.
5 Conclusions
A multifunctional EGCG-loaded 3DP tissue engineering TCP scaffold improves in vitro bone remodeling by balancing bone formation and resorption. EGCG increases osteoblast differentiation via increasing Runx2 and BGLAP gene expression over 16 days. However, EGCG also inhibits the differentiation of osteoclasts by decreasing RANKL expression and increasing OPG expression. In addition to its ability to accelerate in vitro bone remodeling, EGCG-loaded 3DP scaffolds promote in vitro osteoblast proliferation and in vitro HUVEC-induced vasculogenesis while inhibiting in vitro osteosarcoma progression. At all time periods, the EGCG-loaded scaffolds do not induce cytotoxicity in hFOB cells while improving cellular proliferation and attachment compared to the control after 7 and 11 days. EGCG release from the scaffolds also shows increased in vitro tubular formation and endothelial vascular network formation. Because of the in vitro chemopreventive potential of EGCG, the scaffolds reduce the cellular viability of the osteosarcoma cells at days 7 and 11. Together, the porous 3DP scaffold incorporated with EGCG can be used as a potential graft material to promote bone formation and wound healing in low load-bearing bone defects caused by trauma or tumor ablation.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors would like to acknowledge financial support from the National Institute of Dental and Craniofacial Research (NIDCR) of the NIH grant number R01 DE029204-01 (PI: Bose) and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of the National Institutes of Health, USA under Award Number R56 AR066361-06 (PI: Bose). A special thanks to the Franceschi Microscopy and Imaging Center at Washington State University.References
- X. Chen, L. Xu, X. Li and J. Egger, Sci. Rep., 2017, 7, 3–12 CrossRef PubMed.
- A. S. Greenwald, S. D. Boden, V. M. Goldberg, Y. Khan, C. T. Laurencin, R. N. Rosier and American Academy of Orthopaedic Surgeons. The Committee on Biological Implants, J Bone Joint Surg Am, 2001, 83A(Suppl 2 Pt 2), 98–103 CrossRef PubMed.
- K. Johnson, Carsey Sch. Public Policy Sch. Repos.
- S. Tarafder, V. K. Balla, N. M. Davies, A. Bandyopadhyay and S. Bose, J. Tissue Eng. Regen. Med., 2013, 7, 631–641 CrossRef CAS PubMed.
- S. Bose, S. Vahabzadeh and A. Bandyopadhyay, Mater. Today, 2013, 16, 496–504 CrossRef CAS.
- W. Chen, L. Tucker, L. Teer and L. Priddy, Biomed. Sci. Instrum., 2021, 56, 1–9 Search PubMed.
- S. Bose, S. Tarafder and A. Bandyopadhyay, Ann. Biomed. Eng., 2017, 45, 261–272 CrossRef PubMed.
- G. Thrivikraman, A. Athirasala, C. Twohig, S. K. Boda and L. E. Bertassoni, Dent. Clin. North Am., 2017, 61, 835–856 CrossRef PubMed.
- S. Bose and S. Tarafder, Acta Biomater., 2012, 8, 1401–1421 CrossRef CAS PubMed.
- M. A. Gentile, A. J. Tellington, W. J. Burke and M. S. Jaskolka, Atlas Oral Maxillofac. Surg. Clin. North Am., 2013, 21, 69–95 CrossRef PubMed.
- D. Baumhoer, P. Brunner, S. Eppenberger-Castori, J. Smida, M. Nathrath and G. Jundt, Oral Oncol., 2014, 50, 147–153 CrossRef PubMed.
- L. Mirabello, R. J. Troisi and S. A. Savage, Cancer, 2009, 115, 1531–1543 CrossRef PubMed.
- L. Ma, X. Feng, H. Liang, K. Wang, Y. Song, L. Tan, B. Wang, R. Luo, Z. Liao, G. Li, X. Liu, S. Wu and C. Yang, Mater. Today, 2020, 36, 48–62 CrossRef CAS.
- J. Thariat, M. Julieron, A. Brouchet, A. Italiano, T. Schouman, P. Y. Marcy, G. Odin, A. Lacout, O. Dassonville, I. Peyrottes-Birstwisles, R. Miller, A. Thyss and N. Isambert, Crit. Rev. Oncol. Hematol., 2012, 82, 280–295 CrossRef PubMed.
- W. M. Mendenhall, R. Fernandes, J. W. Werning, M. Vaysberg, R. S. Malyapa and N. P. Mendenhall, Am. J. Otolaryngol. – Head Neck Med. Surg., 2011, 32, 597–600 Search PubMed.
- S. Bose, N. Sarkar and D. Banerjee, Acta Biomater., 2021, 126, 63–91 CrossRef CAS PubMed.
- S. Bose and N. Sarkar, Trends Biotechnol., 2020, 38, 404–417 CrossRef CAS PubMed.
- Y. Ren, F. Y. Wang, Z. J. Chen, R. T. Lan, R. H. Huang, W. Q. Fu, R. M. Gul, J. Wang, J. Z. Xu and Z. M. Li, J. Mater. Chem. B, 2020, 8, 10428–10438 RSC.
- X. Wang, X. Li, X. Liang, J. Liang, C. Zhang, J. Yang, C. Wang, D. Kong and H. Sun, J. Mater. Chem. B, 2018, 6, 1000–1010 RSC.
- S. Yu, L. Zhu, K. Wang, Y. Yan, J. He and Y. Ren, Medicine, 2019, 98, e1614 Search PubMed.
- Z. Guo, M. Jiang, W. Luo, P. Zheng, H. Huang and B. Sun, Iran. J. Public Health, 2019, 48, 1566–1576 Search PubMed.
- G. P. Yu, C. C. Hsieh, L. Y. Wang, S. Z. Yu, X. L. Li and T. H. Jin, Cancer Causes Control, 1995, 6, 532–538 CrossRef CAS PubMed.
- V. M. Hegarty, H. M. May and K. T. Khaw, Am. J. Clin. Nutr., 2000, 71, 1003–1007 CrossRef CAS PubMed.
- R. W. Lin, C. H. Chen, Y. H. Wang, M. L. Ho, S. H. Hung, I. S. Chen and G. J. Wang, Biochem. Biophys. Res. Commun., 2009, 379, 1033–1037 CrossRef CAS PubMed.
- L. Yang, W. Zhang, S. Chopra, D. Kaur, H. Wang, M. Li, P. Chen and W. Zhang, Curr. Drug Targets, 2020, 21, 1099–1104 CrossRef CAS PubMed.
- S. Y. Lin, L. Kang, C. Z. Wang, H. H. Huang, T. L. Cheng, H. T. Huang, M. J. Lee, Y. S. Lin, M. L. Ho, G. J. Wang and C. H. Chen, Molecules, 2018, 23, 3221 CrossRef PubMed.
- Y. W. Cheon, K. C. Tark and Y. W. Kim, Dermatologic Surg., 2012, 38, 1835–1842 CrossRef CAS PubMed.
- G. Borciani, G. Montalbano, N. Baldini, G. Cerqueni, C. Vitale-Brovarone and G. Ciapetti, Acta Biomater., 2020, 108, 22–45 CrossRef CAS PubMed.
- J. H. Yun, E. K. Pang, C. S. Kim, Y. J. Yoo, K. S. Cho, J. K. Chai, C. K. Kim and S. H. Choi, J. Periodontal Res., 2004, 39, 300–307 CrossRef CAS PubMed.
- M. Kamon, R. Zhao and K. Sakamoto, Cell Biol. Int., 2009, 34, 109–116 Search PubMed.
- A. A. Vu, D. A. Burke, A. Bandyopadhyay and S. Bose, Addit. Manuf., 2021, 39, 101870 CAS.
- A. Mendyk, R. Jachowicz, K. Fijorek, P. Dorozyński, P. Kulinowski and S. Polak, Dissolution Technol., 2012, 19, 6–11 CrossRef.
- A. A. Vu, S. F. Robertson, D. Ke, A. Bandyopadhyay and S. Bose, Acta Biomater., 2019, 92, 325–335 CrossRef CAS PubMed.
- M. Roy and S. Bose, J. Biomed. Mater. Res. A, 2012, 100, 2450–2461 Search PubMed.
- T. D. Schmittgen and K. J. Livak, Nat. Protoc., 2008, 3, 1101–1108 CrossRef CAS PubMed.
- G. Carpentier, S. Berndt, S. Ferratge, W. Rasband, M. Cuendet, G. Uzan and P. Albanese, Sci. Rep., 2020, 10, 1–13 CrossRef PubMed.
- K. Jørgensen and L. Jacobsen, Int. J. Pharm., 1992, 88, 23–29 CrossRef.
- H. K. Vaananen, H. Zhao, M. Mulari and J. M. Halleen, J. Cell Sci., 2000, 113, 377–381 CrossRef CAS PubMed.
- S. Vahabzadeh, M. Roy and S. Bose, J. Mater. Chem. B, 2015, 3, 8973–8982 RSC.
- W. Wang, C. Wu, B. Tian, X. Liu, Z. Zhai, X. Qu, C. Jiang, Z. Ouyang, Y. Mao, T. Tang, A. Qin and Z. Zhu, Int. J. Mol. Sci., 2014, 15, 21913–21934 CrossRef CAS PubMed.
- S. Chen, L. Kang, C. Wang, P. Huang, H. Huang, S. Lin, S. Chou, C. Lu, P. Shen, Y. Lin and C. Chen, Molecules, 2019, 24, 156 CrossRef PubMed.
- L. H. D. Van Tuyl, A. E. Voskuyl, M. Boers, P. Geusens, R. B. M. Landewé, B. A. C. Dijkmans and W. F. Lems, Ann. Rheum. Dis., 2010, 69, 1623–1628 CrossRef CAS PubMed.
- A. E. Kearns, S. Khosla and P. J. Kostenuik, Endocr. Rev., 2008, 29, 155–192 CrossRef CAS PubMed.
- International Organization for Standardization (ISO 10993-5) 2009 Biological evaluation of medical devices—Part 5: tests for in vitro cytotoxicity.
- I. Arnaoutova and H. K. Kleinman, Nat. Protoc., 2010, 5, 628–635 CrossRef CAS PubMed.
- C. L. Shen, J. K. Yeh, J. J. Cao and J. S. Wang, Nutr. Res., 2009, 29, 437–456 CrossRef CAS PubMed.
- S. Bose, S. Vahabzadeh, D. Banerjee and D. Ke, Mater. Today Commun., 2019, 21, 100534 CrossRef CAS.
- G. A. Fielding, W. Smoot and S. Bose, J. Biomed. Mater. Res., Part A, 2014, 102, 2417–2426 CrossRef PubMed.
- X. Huang, X. Guo, L. Qu, Z. Wu, T. Yu, Y. Jiao and C. Zhou, Appl. Mater. Today, 2021, 23, 101067 CrossRef.
- J. Wang, Q. Sun, Y. Wei, M. Hao, W. S. Tan and H. Cai, Int. J. Biol. Macromol., 2021, 176, 96–105 CrossRef CAS PubMed.
- C. H. Chen, M. L. Ho, J. K. Chang, S. H. Hung and G. J. Wang, Osteoporos. Int., 2005, 16, 2039–2045 CrossRef CAS PubMed.
- Z. Liu, Y. Lin, X. Fang, J. Yang and Z. Chen, Molecules, 2021, 26, 1580 CrossRef CAS PubMed.
- M. Roy, G. A. Fielding, A. Bandyopadhyay and S. Bose, Biomater. Sci., 2013, 1, 74–82 RSC.
- G. D. Roodman, Endocr. Rev., 1996, 17, 308–332 CAS.
- S. L. Teitelbaum, Science, 2000, 289, 1504–1508 CrossRef CAS PubMed.
- D. Ke, S. Tarafder, S. Vahabzadeh and S. Bose, Mater. Sci. Eng., C, 2019, 96, 10–19 CrossRef CAS PubMed.
- B. F. Boyce, Y. Xiu, J. Li, L. Xing and Z. Yao, Endocrinol. Metab., 2015, 30, 35–44 CrossRef CAS PubMed.
- R. O. Escárcega, S. Fuentes-Alexandro, M. García-Carrasco, A. Gatica and A. Zamora, Clin. Oncol., 2007, 19, 154–161 CrossRef PubMed.
- S. Y. Joo, Y. A. Song, Y. L. Park, E. Myung, C. Y. Chung, K. J. Park, S. B. Cho, W. S. Lee, H. S. Kim, J. S. Rew, N. S. Kim and Y. E. Joo, Gut Liver, 2012, 6, 188–196 CrossRef CAS PubMed.
- M. Nomura, W. Ma, N. Chen, A. M. Bode and Z. Dong, Carcinogenesis, 2000, 21, 1885–1890 CrossRef CAS PubMed.
- M. Gonçalves, P. Figueira, D. Maciel, J. Rodrigues, X. Qu, C. Liu, H. Tomás and Y. Li, Acta Biomater., 2014, 10, 300–307 CrossRef PubMed.
- N. Sarkar and S. Bose, Acta Biomater., 2020, 114, 407–420 CrossRef CAS PubMed.
- Ö. Dönmez, B. A. Mogol, V. Gökmen, N. Tang, M. L. Andersen and D. E. W. Chatterton, Food Funct., 2020, 11, 6038–6053 RSC.
- F. A. do Monte, N. Ahuja, K. R. Awad, Z. Pan, S. Young, H. K. W. Kim, P. Aswath, M. Brotto and V. G. Varanasi, JBMR Plus, 2021, 5, 1–18 CrossRef PubMed.
- U. Saran, S. Gemini Piperni and S. Chatterjee, Arch. Biochem. Biophys., 2014, 561, 109–117 CrossRef CAS PubMed.
- Z. Chen, J. Duan, Y. Diao, Y. Chen, X. Liang, H. Li, Y. Miao, Q. Gao, L. Gui, X. Wang, J. Yang and Y. Li, Bioact. Mater., 2021, 6, 1–11 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |