microRNA-181a silencing by antisense oligonucleotides delivered by virus-like particles
Soo Khim
Chan
a and
Nicole F.
Steinmetz
 *abcdefg
*abcdefg
aDepartment of NanoEngineering, University of California San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA. E-mail: nsteinmetz@ucsd.edu
bDepartment of Bioengineering, University of California San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA
cDepartment of Radiology, University of California San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA
dCenter for Nano-ImmunoEngineering, University of California San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA
eCenter for Engineering in Cancer, Institute for Engineering in Medicine, University of California San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA
fMoores Cancer Center, University of California San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA
gInstitute for Materials Discovery and Design, University of California San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA
First published on 4th January 2023
Abstract
Cowpea chlorotic mottle virus (CCMV) is a positive-sense RNA virus that can be repurposed for gene delivery applications. Understanding the self-assembly process of the virus enabled to remove its genome and replace it with desired nucleic acids, and we and others have previously reported using CCMV virus-like particle (VLP) to encapsulate siRNA, mRNA, as well as CpG oligodeoxynucleotides. In this study, the CCMV VLP was applied to encapsulate two different formats of anti-miR-181a oligonucleotides: naked RNA and chemically stabilized RNA to knockdown highly regulated miR-181a in ovarian cancer cells. miR-181a expression in ovarian tumors is associated with high aggressiveness, invasiveness, resistance to chemotherapy, and overall poor prognosis. Therefore, miR-181a is an important target for ovarian cancer therapy. qPCR data and cancer cell migration assays demonstrated higher knockdown efficacy when anti-miR-181a oligonucleotides were encapsulated and delivered using the VLPs resulting in reduced cancer cell invasiveness. Importantly, delivery of anti-miR-181a oligonucleotide into cells could be achieved without the aid of a transfection agent or surface modification. These results highlight the opportunity of plant-derived VLPs as nucleic acid carriers.
Introduction
Ovarian cancer is the most common cause of death among gynecological malignancies, with more than 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fatalities worldwide every year. More than 90% of all patients with ovarian carcinoma will succumb to the disease.1 This high mortality rate mainly reflects the fact that the majority (>75%) of patients present at an advanced stage, with widely disseminated disease within the peritoneal cavity.1 Treatment options include surgical cytoreduction and adjuvant chemotherapy; nevertheless, most patients relapse and overall prognosis is poor.2 There is an urgent need for targeted therapies. Toward this goal, we focused on a nucleic acid delivery strategy targeting microRNA miR-181a.
000 fatalities worldwide every year. More than 90% of all patients with ovarian carcinoma will succumb to the disease.1 This high mortality rate mainly reflects the fact that the majority (>75%) of patients present at an advanced stage, with widely disseminated disease within the peritoneal cavity.1 Treatment options include surgical cytoreduction and adjuvant chemotherapy; nevertheless, most patients relapse and overall prognosis is poor.2 There is an urgent need for targeted therapies. Toward this goal, we focused on a nucleic acid delivery strategy targeting microRNA miR-181a.
microRNAs (miRNAs) are small, non-coding RNA molecules that negatively regulate gene expression at the post-transcriptional level in a sequence-specific manner, primarily via base pairing to the 3′-UTR of the target messenger RNA transcripts. Targeting miRNAs to regulate protein expression has shown great promise in oncology. Here we target miR181a which plays a critical role in ovarian cancer.3 The miR181a gene resides in the 1q ‘hotspot’ region which is overexpressed in ovarian carcinomas.4 miR181a promotes epithelial-to-mesenchymal transition (EMT) and correlates with increased cellular survival, migration, invasion, and metastasis. Importantly, miR-181a expression is associated with shorter time to recurrence and poor prognosis in women with epithelial/serous ovarian cancer.3 Data further indicate that miR-181a high cells from primary ovarian tumors exhibit cancer stem cell properties.5 Lastly, a growing body of data has linked miR-181a expression to chemo-resistance in ovarian cancer,6–8 and other tumor types including T cell malignancies, colorectal cancer, as well as prostate cancer,9–11 thus highlighting the clinical importance of the proposed therapeutic target.
To enable knockdown of miR-181a, we turned toward a nanotechnology-based approach to deliver corresponding anti-miRs. Recombinant viruses such as lentivirus, adenovirus, adeno-associated virus (AAV), etc. have been widely utilized as vectors for gene delivery.12–14 Viral-based vectors evolved mechanisms for efficient cell entry and trafficking, therefore affording efficient gene or nucleic acid delivery.15 On the other hand, because of their efficient integration machinery, mammalian vectors bear risks of insertional mutagenesis leading to oncogene activation and onset of malignancies.16 Non-viral gene delivery systems offer safety and are generally less immunogenic.17–25 The COVID-19 pandemic has highlighted the power of nanotechnology platform technologies with lipid nanoparticles being amongst the first entering clinical trials and gaining approvals with vaccinations occurring worldwide.26,27 For this study, we turned toward plant viruses, namely virus-like particles (VLPs) from cowpea chlorotic mottle virus (CCMV) as an alternative nanotechnology platform compared to synthetic nanoparticles or mammalian viral vectors. The plant virus-based approach offers advantages over contemporary viral and non-viral systems: compared to mammalian viral vector systems, plant viruses are not virulent in animal cells and therefore offer a better safety profile.28 Plant viruses do not replicate in or infect mammals;29 the system is non-integrating and therefore does not bear the risk insertional mutagenesis.16 Manufacture in plants produces yields of up 1–2 grams plant virus-based particles per kg of leaf tissue, and production in bacteria or yeast gives rise to similar yields.30 This is in stark contrast to production of mammalian viral vectors, which are produced in tissue culture. While mammalian viruses grow to high titers in tissue culture, typical yields obtained are only a few milligrams per 1 L cell culture.31 Therefore, expression of plant viruses gives raise to yields that are 10–100 times higher. The approach is scalable; for example, Medicago Inc. already produces a product line of virus-like particles in plants.32–35 The plant virus-based approach also provides advantages compared to synthetic systems: a barrier to the widespread use of these novel pharmaceutical agents is their high instability in aqueous suspensions and/or need to store nanoparticles in ultralow freezers.36–39 While several advances have been made to counter the storage instability of non-viral systems,40 the synthetic platforms do not reach the level of stability that can be achieved using the viral counterparts, which are stable in biological media and can be stored as purified solutions or in infected leaf tissue.
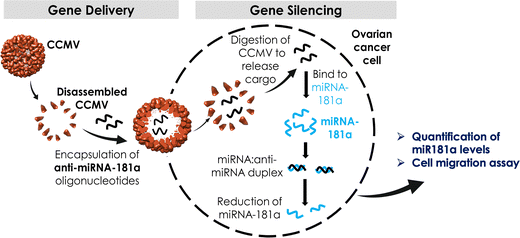
In this work, we focused on CCMV as a platform for anti-miR delivery. CCMV is a plant virus from the family Bromoviridae and it infects legumes including cowpeas and soybeans. The structure of CCMV is known to near-atomic resolution and CCMV forms a T = 3 capsid measuring 30 nm in diameter composed of 180 identical copies of a single coat protein; the capsid carries a tri-partite positive-sense, single-stranded RNA genome.41 The self-assembly process of CCMV is well understood; in fact CCMV particles are highly dynamic platforms that undergo reversible pH- and metal ion-dependent structural transitions,41,42 which can be exploited for cargo loading and shape-shifting.43 For biomedical research, CCMV has been developed by us and others as a nucleic acid carrier for delivery of siRNA,44–46 mRNA,47,48 as well as CpG oligodeoxynucleotides49 for applications in vaccines and cancer therapy. Here we exploited CCMV VLPs to encapsulate two different formats of anti-miR-181a oligonucleotides: naked RNA and chemically stabilized RNA to knockdown highly regulated miR-181a in ovarian cancer cells (the overall workflow is outlined in Scheme 1).
Results and discussion
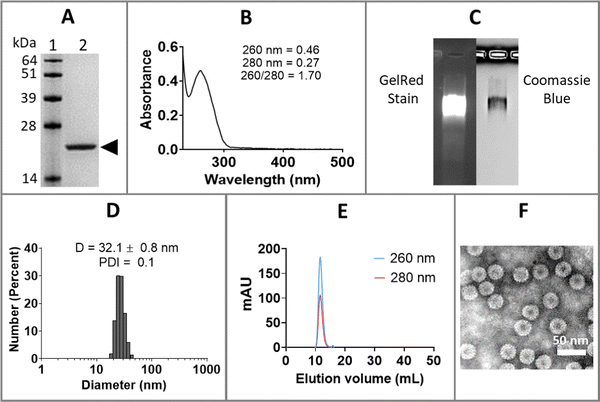
Purified CCMV was characterized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), ultraviolet-visible (UV-Vis) spectrophotometry, dynamic light scattering (DLS), size exclusion chromatography (SEC), and transmission electron microscopy (TEM), as shown in Fig. 1. SDS-PAGE of CCMV revealed a single 21 kDa coat protein band (Fig. 1(A)); protein contaminants were not detectable. The ratio of 260/280 of ∼1.7 determined by UV-Vis spectrophotometry is also indicative of intact and pure CCMV (Fig. 1(B)). The intactness and purity of CCMV were further validated by native agarose gel electrophoresis (Fig. 1(C)) showing colocalization of a single band after GelRed (nucleic acids stain) and Coomassie blue (protein stain) staining – indicating the RNA and protein co-migrate. No free nucleic acids or proteins were observed. DLS (Fig. 1(D)) confirmed the monodispersity of CCMV with polydispersity index (PDI) around 0.1 and diameter around 32 nm. SEC profiles (Fig. 1(E)) were in agreement and confirmed the purity of CCMV by showing single peak eluting at 10–12 mL using a Superose6 Increase column; no free RNA (260 nm) or protein (280 nm) were observed. TEM micrograph (Fig. 1(F)) confirmed the intactness and monodispersity of the CCMV particles.Encapsulation of anti-miR-181a-oligonucleotides within CCMV VLPs
CCMV CPs were used to encapsulate two different formats of anti-miR-181a-oligonucleotides: naked RNA (denoted as RNA: 5′-ACUCACCGACAGCGUUGAAUGUU-3′) and chemically modified antagomiR (denoted as modiRNA, from Horizon, PerkinElmer with sequence and chemical information not disclosed, see Table 1 for more information). RNAs are labile molecules that rapidly degrade through enzymes and hydrolysis. For these reasons, chemically modified RNA structures have been developed less prone to degradation, therefore prolonging longevity in the target cell.50 Here we used a modified anti-miR181a from Horizon that combines proprietary chemical modifications and completely novel secondary structure motif to enhance stability and longevity of the molecule. An alternative to chemical alteration of the RNA is to stabilize the RNA by delivery through nanoparticles.51 Therefore, we hypothesized that CCMV delivery would stabilize the RNA and thus allow to deliver native vs. chemically stabilized modiRNA with matched efficacy. An advantage for delivery of native RNA is that in vivo assembly during co-expression of the target RNA and coat protein (CP) would be feasible (which is a future goal of this work).| anti-miR | sequence |
|---|---|
| Naked anti-miR-181a (denoted as RNA) | 5′-ACUCACCGACAGCGUUGAAUGUU-3′ |
| Manufacturer: Integrated DNA Technologies (IDT) | |
| Molecular weight: 7307.4 g mol−1 | |
Extinction coefficient: 226![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 L (mol−1 cm−1) 800 L (mol−1 cm−1) |
|
| Chemically modified anti-miR-181a (denoted as modiRNA) | Sequence and chemical modifications are proprietary and not provided by the manufacturer |
| Manufacturer: Horizon | |
Molecular weight: 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 384.1 g mol−1 384.1 g mol−1 |
|
Extinction coefficient: 550![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 L (mol−1 cm−1) 900 L (mol−1 cm−1) |
|
| Link: https://horizondiscovery.com/en/gene-modulation/knockdown/mirna/products/miridian-microrna-hairpin-inhibitor?nodeid=mirnamature-mimat0000256&catalognumber=IH-300552-07-0050 | |
CCMV disassembly and assembly was performed as previously reported with reassembly of CP and RNA cargo at a mass ratio of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
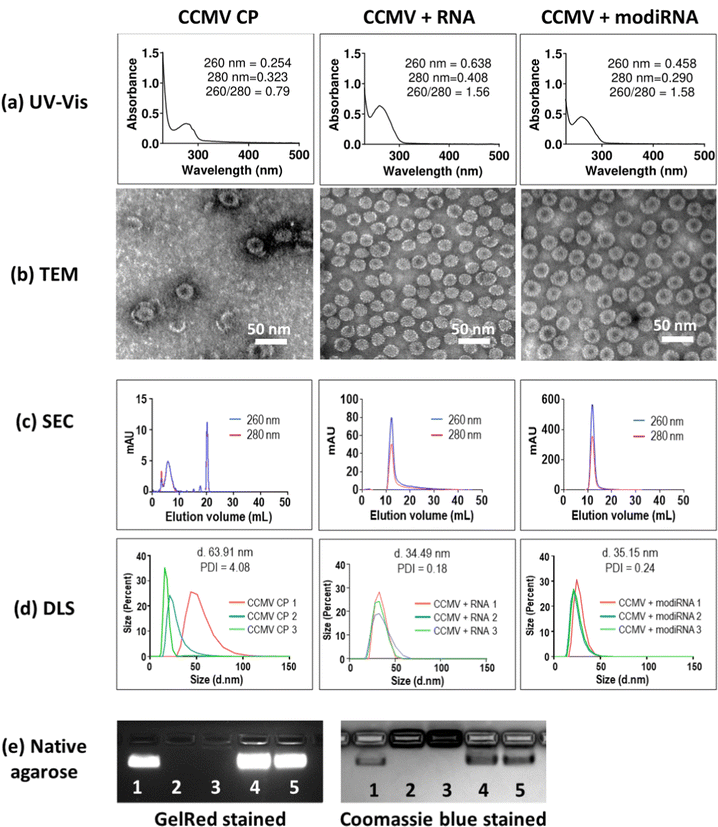
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.52 The encapsulation of target RNA and modiRNA into CCMV VLPs was validated via UV-Vis, TEM, SEC and DLS, and native agarose gel electrophoresis (Fig. 2). CCMV encapsulated with RNA (CCMV + RNA) and modiRNA (CCMV + modiRNA) gave 260/280 ratio around 1.5 similarly to wild type CCMV (∼1.7) indicating successful encapsulation of nucleic acids (Fig. 2(a), CCMV CP panel). In contrary, CCMV CP only assemblies without nucleic acid exhibited a lower 260/280 ratio of 0.79 reflecting the lack of nucleic acids content. CCMV consists of two components: the protein capsid (detected at 280 nm) and the RNA cargo – native or designer RNA (detected at 260 nm). The UV-Vis ratio of 260 nm over 280 nm therefore is characteristic for each VLP assembly and provides information about the purity of the system. Wild type CCMV has a UV-Vis ratio of ∼1.7 at 260/280. The comparison of the UV-Vis 260/280 ratio of wild type and reassembled CCMV VLPs thus provides insights into the intactness of the formulation and whether RNA cargo is present or not.
1.52 The encapsulation of target RNA and modiRNA into CCMV VLPs was validated via UV-Vis, TEM, SEC and DLS, and native agarose gel electrophoresis (Fig. 2). CCMV encapsulated with RNA (CCMV + RNA) and modiRNA (CCMV + modiRNA) gave 260/280 ratio around 1.5 similarly to wild type CCMV (∼1.7) indicating successful encapsulation of nucleic acids (Fig. 2(a), CCMV CP panel). In contrary, CCMV CP only assemblies without nucleic acid exhibited a lower 260/280 ratio of 0.79 reflecting the lack of nucleic acids content. CCMV consists of two components: the protein capsid (detected at 280 nm) and the RNA cargo – native or designer RNA (detected at 260 nm). The UV-Vis ratio of 260 nm over 280 nm therefore is characteristic for each VLP assembly and provides information about the purity of the system. Wild type CCMV has a UV-Vis ratio of ∼1.7 at 260/280. The comparison of the UV-Vis 260/280 ratio of wild type and reassembled CCMV VLPs thus provides insights into the intactness of the formulation and whether RNA cargo is present or not.
TEM imaging confirmed the assembly of VLPs when assembly was initiated in the presence of RNA/modiRNA. In contrast, few VLPs of less regular structure, darker center, and protein stacking was observed when CP only was assembled (Fig. 2(b)). The darker center is explained by uranyl acetate (UAc) stain accumulating in the empty capsid structure; and protein stacking occurs due to the electrostatic interaction between the negatively charge of the CP outer surface and positively charger CP inner surface. This data also aligns with separation by SEC (Fig. 2(c)) and size analysis by DLS (Fig. 2(d)). In SEC, free CP elute from the Superose6 Increase column at 20 mL, while intact VLPs elute at ∼12 mL with co-elution of RNA and protein (measured at 260 nm and 280 nm, respectively); the latter was observed for VLPs containing the target RNA and modiRNA (Fig. 2(c)). DLS confirmed the presence of monodisperse VLPs measuring ∼35 nm in hydrodynamic diameter with narrow size distribution (PDI < 0.25). In stark contrast, the CP only assembly resulted in inconsistent sizing data reflecting a broad range of assemblies and free CPs (Fig. 2(d)). Lastly, native agarose gel electrophoresis was performed. CCMV + RNA (Fig. 2(e) Lane 4) and CCMV + modiRNA (Fig. 2(e) Lane 5) showed a band which migrated similarly to the native CCMV particles (Fig. 2(e) Lane 1). Co-migration of nucleic acid (GelRed Stain) and proteins (stained by Coomassie blue) indicate intact VLPs were formed; free RNA/CP or aggregation was not detected. Reassembly of CCMV CP only (Fig. 2(e) Lane 3) showed no band when stained with GelRed – no RNA was present. The CP only assemblies did not migrate, neither did free CP (Fig. 2(e) Lane 2 + 3). Together, this data confirms that VLPs containing RNA and modiRNA were successfully assembled.
Quantification of miR-181a expression in ovarian cancer cells and knockdown of miR-181a
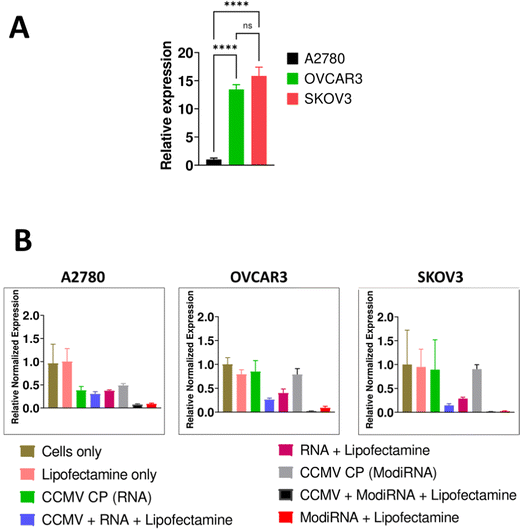
We selected three different ovarian cancer cell lines to assess the levels of miR-181a expression: A2780, OVCAR3, and SKOV3. The more aggressive cell lines OVCAR3 and SKOV3 showed high levels of miR-181a expression, while A2780 had negligible levels of miR-181a (Fig. 3(A)), and this data is consistent with a previous report.3Next, we assessed the efficacy of miR-181a knockdown by delivery of anti-miR-181a-oligonucleotides (RNA vs modiRNA) using CCMV VLPs; in this first experiments we used lipofectamine as transfection agent (Fig. 3(B)). MicroRNA-16 or miR-16 was used as endogenous reference gene to normalize miR-181a expression in the cells, and cells only served as the control to compare the relative expression of miR-181a in the three cancer cells (A2780, OVCAR3, and SKOV3).
Addition of lipofectamine alone did not impact the relative expression of miR-181a across all cancer cells. Similarly, the CCMV CP only controls (= reassembly of CCMV CP devoid of nucleic acids) had no impact on miR-181a expression in SKOV3 and OVCAR3 cells. However, the CCMV CP reduced miR-181a expression in A2780 cells (Fig. 3(B)). It should be noted that miR-181a expression in this cell line were low to start with; it is possible that the CCMV CP assemblies/aggregates, which are of less defined nature (see Fig. 2), have a lower degree of biocompatibility and hence impact cell expression levels. Since we do not propose the application of the CP only assembly controls, and because A2780 cells are not necessarily a target for this approach (because miR-181a levels are now), we did not investigate this further.
When the RNAs (RNA and modiRNA) were encapsulated into and delivered by CCMV (plus lipofectamine), significantly lower relative miR-181a expression levels were achieved when compared to free RNAs delivered via lipofectamine transfection only. miR-181a expression levels were lowered by 36.5%, 16.5%, and 76.5% in SKOV3, A2780, and OVCAR3, respectively, when modiRNAs were encapsulated in CCMV vs. free modiRNA transfected using lipofectamine. Similar observations were made for the non-modified RNA (Fig. 3(B)). This data suggests that CCMV may stabilize the RNA and/or enhance its trafficking, release, and processing within the cell, resulting in enhanced delivery and efficacy. Data also suggest that VLP delivery can also enhance the efficacy of chemically enhanced RNA (modiRNA).
Lipofectamine-free delivery of anti-miR-181a using CCMV and its efficacy on cancer cell invasiveness
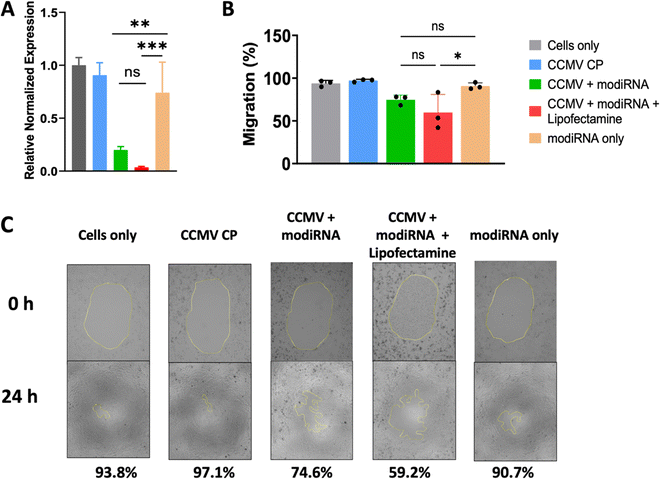
Others have shown that CCMV enables gene delivery to mammalian cells without the use of a transfection agent.53 Therefore, we extended our studies to assess whether CCMV delivery of anti-miR-181a to ovarian cancer cells would be effective in the absence of lipofectamine and whether knockdown led to reduced invasiveness of ovarian cancer cells (determined by cell migration assay). For these studies we chose the SKOV3 cell line, which has the highest miR-181a levels (Fig. 3(A)). We also chose the modiRNA only to exclude rapid RNA degradation as a factor from these experiments.Fig. 4(A) demonstrates that indeed miR-181a levels were significantly lowered when the therapeutic anti-miRs (modiRNA) were delivered using CCMV; free modiRNA afforded negligible knockdown reflecting poor cell uptake of RNA molecules into cells. The addition of lipofectamine to the CCMV delivery agent enhanced efficacy further when compared to CCMV + modiRNA, however this is not relevant for cancer therapy, because the application of lipofectamine is limited to ex vivo applications due to the toxicity of the agent.54
Finally, the efficacy and therapeutic effect of CCMV in delivering anti-miR-181a oligonucleotides to knockdown miR-181a was validated using a cancer cell migration assay (Fig. 4(B) and (C)). Untreated SKOV3 ovarian cancer cells are characterized as highly invasive, which is reflected by the high degree of migration with 93.4% of the area covered with cancer cells after 24 h incubation (Fig. 4(B) and (C)). Similar observations were made for any control samples: treatment with CCMV CP assemblies devoid of anti-miR-181a oligonucleotides or anti-miR-181a oligonucleotides alone (∼97% and ∼91% coverage for CCMV CP only and modiRNA only, respectively). In stark contrast, cell migration was inhibited when the anti-miRs were delivered by CCMV resulting in significantly reduced cell migration; ∼75% for CCMV + modiRNA and ∼59% for CCMV + modiRNA + lipofectamine. Together this data is consistent with results from prior research53 and highlight the potential of CCMV as a gene delivery platform.
Conclusions
Anti-miR-181a oligonucleotides were successfully encapsulated into VLPs from CCMV. Delivery of anti-miR-181a packaged into CCMV afforded efficient knockdown of miR-181a levels in ovarian cancer cells and reduced invasiveness of these cancer cells. Therefore, CCMV is positioned as a platform nanotechnology for gene delivery applications. The plant derived VLPs may offer advantages compared to mammalian viral vectors and synthetic systems in that they can be produced through plant molecular farming or heterologous expression at high yields; being a biologic, the platform has good stability in biological media and plant viruses may not require a cold chain for storge or distribution; lastly, plant viruses are non-infectious toward mammals, therefore adding a layer of safety. While detailed toxicology studies of CCMV as a gene delivery agent must be conducted, preclinical research indicates good biocompatibility and safety of CCMV nanoparticles in mice after intravenous administration.55 In conclusion, plant VLPs such as CCMV are promising platforms for gene delivery and future in vivo work as well as side-by-side comparisons to contemporary viral and non-viral delivery systems are needed to further validate this system.Materials and methods
Production and characterization of CCMV
CCMV was produced under an USDA-approved PPQ526 permit and was purified as previously described.46 Characterizations of CCMV was performed using sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE), UV-Vis spectrophotometry, native agarose gel electrophoresis, dynamic light scattering (DLS), size exclusion chromatography (SEC), and transmission electron microscope (TEM) as previously reported.56Disassembly and assembly of CCMV coat proteins with RNA cargos
CCMV disassembly and re-assembly of VLPs was performed as previously reported.52 CCMV swell when incubated in pH near neutrality and in buffer absence of Mg2+ ion. Disassembled CCMV coat proteins (CPs) were reassembled with RNA cargos (naked RNA, modiRNA, Table 1) using a 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mass ratio as described. The reconstituted CCMV was stored in 0.1 M NaOAc, 1 mM EDTA, pH 4.8 at −80 °C.
1 mass ratio as described. The reconstituted CCMV was stored in 0.1 M NaOAc, 1 mM EDTA, pH 4.8 at −80 °C.
Cell culture
Three different human ovarian cancer cell lines were used in this study including A2780, OVCAR3, and SKOV3. SKOV3 cells were cultured in McCoy's 5A medium, A2780 and OVCAR3 were cultured in DMEM and RPMI-1640, respectively. Cell culture medium was supplemented with 10% (v/v) FBS and 1% (v/v) PenStrep. The cells were incubated at 37 °C in humidified incubator with 5% CO2.In vitro miR-181a knockdown
A total of 2 × 105 cells were seeded in a 24-well treated plate. 20 pmol of anti-miR-181a-oligonucleotides (with or without the CCMV) was added to the cells with or without the lipofectamine. The cells were washed with 1 × PBS and fresh media was introduced one day after the transfection/treatment. Cells were harvested two days after the transfection/treatment. Briefly, the cells were washed twice with 1 × PBS and then 500 μL of trypsin was introduced to the cells for trypsinization. Harvested cells were kept at −80 °C until further processing or used immediately for total RNA extraction.RNA extraction and quantitative real-time PCR (qPCR)
Total RNA extraction was performed using Zymo Quick-RNA Miniprep Plus Kit according to the manufacturer's protocol. The quality and concentration of the total RNA was determined by UV-Vis spectrophotometry using a Nanodrop 2000c/2000 spectrophotometer; the ratio of 260/280 and 260/230 was determined and pure RNA should yield a value of ∼2 for either ratio. Five microliters of the total RNAs were then used for two-step reverse-transcription qPCR to detect and amplify the target miR-181a. The first step reverse transcription was performed using TaqMan™ MicroRNA Reverse Transcription Kit. Then, the transcribed cDNA was amplified using TaqMan Universal Master Mix II (Table 2). All samples were assayed in triplicate using a BioRad CFX96 Touch real-time PCR detection system. Quantification cycle (Cq) values were tabulated by CFX Maestro software. All quantification data were normalized to miR-16 (Applied Biosystem, Table 2).| Primers | Sequence (5-3) | Remarks |
|---|---|---|
| miR-181a RT-PCR primers/probe | Sequence not provided | Assay ID: 000480 |
| https://www.thermofisher.com/order/genome-database/details/microrna/000480?CID=&ICID=&subtype= | ||
| miR-16 RT-PCR primers/probe | Sequence not provided | Assay ID: 000391 |
| https://www.thermofisher.com/order/genome-database/details/microrna/000391?CID=&ICID=&subtype= | ||
Cell migration and invasion assay
For the wound-healing assay, 4 × 105 cells were seeded at full confluency into 6-well plates and serum-starved for 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) h. A sterile 200
h. A sterile 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μL pipette tip was used to scratch the cells to form a wound. The cells were washed with PBS and cultured in medium. Wound closure was visualized with an inverted Nikon Eclipse TS100 phase-contrast microscope and measured using Nikon NIS-Element Basic Research v3.2 software; imaging was performed at 0 and 24 hours. The migration percentage was calculated as follows:
μL pipette tip was used to scratch the cells to form a wound. The cells were washed with PBS and cultured in medium. Wound closure was visualized with an inverted Nikon Eclipse TS100 phase-contrast microscope and measured using Nikon NIS-Element Basic Research v3.2 software; imaging was performed at 0 and 24 hours. The migration percentage was calculated as follows:Author contributions
S. K. C. designed and performed the experimental work. N. F. S. conceived the study and oversaw the design and testing. S. K. C. and N. F. S. wrote the manuscript. All authors read and edited the manuscript.Conflicts of interest
The authors declare the following competing financial interest(s): Dr Steinmetz is a co-founder of and has a financial interest with Mosaic ImmunoEngineering Inc.; Dr Steinmetz serves on the Board and as Acting Chief Scientific Officer; she a paid consultant to Mosaic. Dr Chan declares no competing financial interests.Acknowledgements
This work was funded in part through NIH grants R01-CA253615 and R21-AI161306 as well as support through the Shaughnessy Family Fund for Nano-ImmunoEngineering (nanoIE) at UCSD.References
- R. L. Siegel, K. D. Miller and A. Jemal, Cancer statistics, 2015, Ca-Cancer J. Clin., 2015, 65(1), 5–29 CrossRef PubMed.
- S. J. Chang, M. Hodeib, J. Chang and R. E. Bristow, Survival impact of complete cytoreduction to no gross residual disease for advanced-stage ovarian cancer: a meta-analysis, Gynecol. Oncol., 2013, 130(3), 493–498 CrossRef PubMed.
- A. Parikh, C. Lee, P. Joseph, S. Marchini, A. Baccarini, V. Kolev, C. Romualdi, R. Fruscio, H. Shah, F. Wang, G. Mullokandov, D. Fishman, M. D’Incalci, J. Rahaman, T. Kalir, R. W. Redline, B. D. Brown, G. Narla and A. DiFeo, microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial-mesenchymal transition, Nat. Commun., 2014, 5, 2977 CrossRef PubMed.
- B. L. Theriault and M. W. Nachtigal, Human ovarian cancer cell morphology, motility, and proliferation are differentially influenced by autocrine TGFbeta superfamily signalling, Cancer Lett., 2011, 313(1), 108–121 CrossRef CAS PubMed.
- A. Belur Nagaraj, P. Joseph, E. Ponting, Y. Fedorov, S. Singh, A. Cole, W. Lee, E. Yoon, A. Baccarini, P. Scacheri, R. Buckanovich, D. J. Adams, R. Drapkin, B. D. Brown and A. DiFeo, A miRNA-Mediated Approach to Dissect the Complexity of Tumor-Initiating Cell Function and Identify miRNA-Targeting Drugs, Stem Cell Rep., 2019, 12(1), 122–134 CrossRef CAS PubMed.
- M. Petrillo, G. F. Zannoni, L. Beltrame, E. Martinelli, A. DiFeo, L. Paracchini, I. Craparotta, L. Mannarino, G. Vizzielli, G. Scambia, M. D’Incalci, C. Romualdi and S. Marchini, Identification of high-grade serous ovarian cancer miRNA species associated with survival and drug response in patients receiving neoadjuvant chemotherapy: a retrospective longitudinal analysis using matched tumor biopsies, Ann. Oncol., 2016, 27(4), 625–634 CrossRef CAS PubMed.
- L. Li, Q. H. Xu, Y. H. Dong, G. X. Li, L. Yang, L. W. Wang and H. Y. Li, MiR-181a upregulation is associated with epithelial-to-mesenchymal transition (EMT) and multidrug resistance (MDR) of ovarian cancer cells, Eur. Rev. Med. Pharmacol. Sci., 2016, 20(10), 2004–2010 CAS.
- S. Marchini, R. Fruscio, L. Clivio, L. Beltrame, L. Porcu, I. Fuso Nerini, D. Cavalieri, G. Chiorino, G. Cattoretti, C. Mangioni, R. Milani, V. Torri, C. Romualdi, A. Zambelli, M. Romano, M. Signorelli, S. di Giandomenico and M. D’Incalci, Resistance to platinum-based chemotherapy is associated with epithelial to mesenchymal transition in epithelial ovarian cancer, Eur. J. Cancer, 2013, 49(2), 520–530 CrossRef CAS PubMed.
- Z. X. Yan, Z. Zheng, W. Xue, M. Z. Zhao, X. C. Fei, L. L. Wu, L. M. Huang, C. Leboeuf, A. Janin, L. Wang and W. L. Zhao, MicroRNA181a Is Overexpressed in T-Cell Leukemia/Lymphoma and Related to Chemoresistance, BioMed Res. Int., 2015, 2015, 197241 Search PubMed.
- C. M. Armstrong, C. Liu, W. Lou, A. P. Lombard, C. P. Evans and A. C. Gao, MicroRNA-181a promotes docetaxel resistance in prostate cancer cells, Prostate, 2017, 77(9), 1020–1028 CrossRef CAS PubMed.
- J. Nishimura, R. Handa, H. Yamamoto, F. Tanaka, K. Shibata, K. Mimori, I. Takemasa, T. Mizushima, M. Ikeda, M. Sekimoto, H. Ishii, Y. Doki and M. Mori, microRNA-181a is associated with poor prognosis of colorectal cancer, Oncol. Rep., 2012, 28(6), 2221–2226 CrossRef CAS PubMed.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Non-viral vectors for gene-based therapy, Nat. Rev. Genet., 2014, 15(8), 541–555 CrossRef CAS PubMed.
- N. Nayerossadat, T. Maedeh and P. A. Ali, Viral and nonviral delivery systems for gene delivery, Adv. Biomed. Res., 2012, 1, 27 CrossRef PubMed.
- M. S. Al-Dosari and X. Gao, Nonviral gene delivery: principle, limitations, and recent progress, AAPS J., 2009, 11(4), 671–681 CrossRef CAS PubMed.
- W. F. Anderson, Human gene therapy, Nature, 1998, 392(6679 Suppl), 25–30 CAS.
- J. Couzin and J. Kaiser, Gene therapy. As Gelsinger case ends, gene therapy suffers another blow, Science, 2005, 307(5712), 1028 CrossRef CAS PubMed.
- W. Li, F. Nicol and F. C. Szoka Jr., GALA: a designed synthetic pH-responsive amphipathic peptide with applications in drug and gene delivery, Adv. Drug Delivery Rev., 2004, 56(7), 967–985 CrossRef CAS PubMed.
- Y. W. Cho, J. D. Kim and K. Park, Polycation gene delivery systems: escape from endosomes to cytosol, J. Pharm. Pharmacol., 2003, 55(6), 721–734 CrossRef CAS PubMed.
- O. Zelphati and F. C. Szoka Jr., Mechanism of oligonucleotide release from cationic liposomes, Proc. Natl. Acad. Sci. U. S. A., 1996, 93(21), 11493–11498 CrossRef CAS PubMed.
- Y. Xu and F. C. Szoka Jr., Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection, Biochemistry, 1996, 35(18), 5616–5623 CrossRef CAS PubMed.
- A. Akinc, M. Thomas, A. M. Klibanov and R. Langer, Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis, J. Gene Med., 2005, 7(5), 657–663 CrossRef CAS PubMed.
- A. Kichler, C. Leborgne, E. Coeytaux and O. Danos, Polyethylenimine-mediated gene delivery: a mechanistic study, J. Gene Med., 2001, 3(2), 135–144 CrossRef CAS PubMed.
- N. D. Sonawane, F. C. Szoka Jr. and A. S. Verkman, Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes, J. Biol. Chem., 2003, 278(45), 44826–44831 CrossRef CAS PubMed.
- A. Bonsted, B. O. Engesaeter, A. Hogset, G. M. Maelandsmo, L. Prasmickaite, C. D’Oliveira, W. E. Hennink, J. H. van Steenis and K. Berg, Photochemically enhanced transduction of polymer-complexed adenovirus targeted to the epidermal growth factor receptor, J. Gene Med., 2006, 8(3), 286–297 CrossRef CAS PubMed.
- J. Kloeckner, L. Prasmickaite, A. Hogset, K. Berg and E. Wagner, Photochemically enhanced gene delivery of EGF receptor-targeted DNA polyplexes, J. Drug Targeting, 2004, 12(4), 205–213 CrossRef CAS PubMed.
- M. D. Shin, S. Shukla, Y. H. Chung, V. Beiss, S. K. Chan, O. A. Ortega-Rivera, D. M. Wirth, A. Chen, M. Sack, J. K. Pokorski and N. F. Steinmetz, COVID-19 vaccine development and a potential nanomaterial path forward, Nat. Nanotechnol., 2020, 15(8), 646–655 CrossRef CAS PubMed.
- X. Huang, E. Kon, X. Han, X. Zhang, N. Kong, M. J. Mitchell, D. Peer and W. Tao, Nanotechnology-based strategies against SARS-CoV-2 variants, Nat. Nanotechnol., 2022, 1027–1037 CrossRef CAS PubMed.
- A. A. Aljabali, S. Shukla, G. P. Lomonossoff, N. F. Steinmetz and D. J. Evans, CPMV-DOX Delivers, Mol. Pharmaceutics, 2013, 10(1), 3–10 CrossRef CAS PubMed.
- M. Manchester and P. Singh, Virus-based nanoparticles (VNPs): platform technologies for diagnostic imaging, Adv. Drug Delivery Rev., 2006, 58(14), 1505–1522 CrossRef CAS PubMed.
- A. Schneemann and M. J. Young, Viral assembly using heterologous expression systems and cell extracts, Adv. Protein Chem., 2003, 64, 1–36 CrossRef CAS PubMed.
- P. Iyer, J. M. Ostrove and D. Vacante, Comparison of manufacturing techniques for adenovirus production, Cytotechnology, 1999, 30(1–3), 169–172 CrossRef CAS PubMed.
- M. A. D’Aoust, P. O. Lavoie, J. Belles-Isles, N. Bechtold, M. Martel and L. P. Vezina, Transient expression of antibodies in plants using syringe agroinfiltration, Methods Mol. Biol., 2009, 483, 41–50 CrossRef PubMed.
- M. A. D’Aoust, M. M. Couture, N. Charland, S. Trepanier, N. Landry, F. Ors and L. P. Vezina, The production of hemagglutinin-based virus-like particles in plants: a rapid, efficient and safe response to pandemic influenza, Plant Biotechnol. J., 2010, 8(5), 607–619 CrossRef PubMed.
- L. P. Vezina, L. Faye, P. Lerouge, M. A. D’Aoust, E. Marquet-Blouin, C. Burel, P. O. Lavoie, M. Bardor and V. Gomord, Transient co-expression for fast and high-yield production of antibodies with human-like N-glycans in plants, Plant Biotechnol. J., 2009, 7(5), 442–455 CrossRef CAS PubMed.
- M. A. D’Aoust, P. O. Lavoie, M. M. Couture, S. Trepanier, J. M. Guay, M. Dargis, S. Mongrand, N. Landry, B. J. Ward and L. P. Vezina, Influenza virus-like particles produced by transient expression in Nicotiana benthamiana induce a protective immune response against a lethal viral challenge in mice, Plant Biotechnol. J., 2008, 6(9), 930–940 CrossRef PubMed.
- M. X. Tang and F. C. Szoka, The influence of polymer structure on the interactions of cationic polymers with DNA and morphology of the resulting complexes, Gene Ther., 1997, 4(8), 823–832 CrossRef CAS PubMed.
- M. A. Kay, D. Liu and P. M. Hoogerbrugge, Gene therapy, Proc. Natl. Acad. Sci. U. S. A., 1997, 94(24), 12744–12746 CrossRef CAS PubMed.
- J. Gustafsson, G. Arvidson, G. Karlsson and M. Almgren, Complexes between cationic liposomes and DNA visualized by cryo-TEM, Biochim. Biophys. Acta, 1995, 1235(2), 305–312 CrossRef PubMed.
- T. J. Anchordoquy and G. S. Koe, Physical stability of nonviral plasmid-based therapeutics, J. Pharm. Sci., 2000, 89(3), 289–296 CrossRef CAS PubMed.
- M. E. Davis, Non-viral gene delivery systems, Curr. Opin. Biotechnol, 2002, 13(2), 128–131 CrossRef CAS PubMed.
- J. A. Speir, S. Munshi, G. Wang, T. S. Baker and J. E. Johnson, Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy, Structure, 1995, 3(1), 63–78 CrossRef CAS PubMed.
- L. O. Liepold, J. Revis, M. Allen, L. Oltrogge, M. Young and T. Douglas, Structural transitions in Cowpea chlorotic mottle virus (CCMV), Phys. Biol., 2005, 2(4), S166–S172 CrossRef CAS PubMed.
- J. B. Bancroft, C. E. Bracker and G. W. Wagner, Structures derived from cowpea chlorotic mottle and brome mosaic virus protein, Virology, 1969, 38(2), 324–335 CrossRef CAS PubMed.
- C. Pretto and J. C. M. van Hest, Versatile Reversible Cross-Linking Strategy to Stabilize CCMV Virus Like Particles for Efficient siRNA Delivery, Bioconjugate Chem., 2019, 30(12), 3069–3077 CrossRef CAS PubMed.
- A. Nunez-Rivera, P. G. J. Fournier, D. L. Arellano, A. G. Rodriguez-Hernandez, R. Vazquez-Duhalt and R. D. Cadena-Nava, Brome mosaic virus-like particles as siRNA nanocarriers for biomedical purposes, Beilstein J. Nanotechnol., 2020, 11, 372–382 CrossRef CAS PubMed.
- P. Lam and N. F. Steinmetz, Delivery of siRNA therapeutics using cowpea chlorotic mottle virus-like particles, Biomater. Sci., 2019, 7(8), 3138–3142 RSC.
- A. Biddlecome, H. H. Habte, K. M. McGrath, S. Sambanthamoorthy, M. Wurm, M. M. Sykora, C. M. Knobler, I. C. Lorenz, M. Lasaro, K. Elbers and W. M. Gelbart, Delivery of self-amplifying RNA vaccines in in vitro reconstituted virus-like particles, PLoS One, 2019, 14(6), e0215031 CrossRef CAS PubMed.
- O. Azizgolshani, R. F. Garmann, R. Cadena-Nava, C. M. Knobler and W. M. Gelbart, Reconstituted plant viral capsids can release genes to mammalian cells, Virology, 2013, 441(1), 12–17 CrossRef CAS PubMed.
- H. Cai, S. Shukla and N. F. Steinmetz, The Antitumor Efficacy of CpG Oligonucleotides is Improved by Encapsulation in Plant Virus-Like Particles, Adv. Funct. Mater., 2020, 30, 15 Search PubMed.
- J. Song and C. Yi, Chemical Modifications to RNA: A New Layer of Gene Expression Regulation, ACS Chem. Biol., 2017, 12(2), 316–325 CrossRef CAS PubMed.
- K. Paunovska, D. Loughrey and J. E. Dahlman, Drug delivery systems for RNA therapeutics, Nat. Rev. Genet., 2022, 23(5), 265–280 CrossRef CAS PubMed.
- R. D. Cadena-Nava, M. Comas-Garcia, R. F. Garmann, A. L. Rao, C. M. Knobler and W. M. Gelbart, Self-assembly of viral capsid protein and RNA molecules of different sizes: requirement for a specific high protein/RNA mass ratio, J. Virol., 2012, 86(6), 3318–3326 CrossRef CAS.
- M. V. Villagrana-Escareno, E. Reynaga-Hernandez, O. G. Galicia-Cruz, A. L. Duran-Meza, V. De la Cruz-Gonzalez, C. Y. Hernandez-Carballo and J. Ruiz-Garcia, VLPs Derived from the CCMV Plant Virus Can Directly Transfect and Deliver Heterologous Genes for Translation into Mammalian Cells, BioMed Res. Int., 2019, 2019, 4630891 Search PubMed.
- M. Breunig, U. Lungwitz, R. Liebl and A. Goepferich, Breaking up the correlation between efficacy and toxicity for nonviral gene delivery, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(36), 14454–14459 CrossRef CAS.
- C. R. Kaiser, M. L. Flenniken, E. Gillitzer, A. L. Harmsen, A. G. Harmsen, M. A. Jutila, T. Douglas and M. J. Young, Biodistribution studies of protein cage nanoparticles demonstrate broad tissue distribution and rapid clearance in vivo, Int. J. Nanomed., 2007, 2(4), 715–733 CAS.
- S. K. Chan, P. Du, C. Ignacio, S. Mehta, I. G. Newton and N. F. Steinmetz, Biomimetic Virus-Like Particles as Severe Acute Respiratory Syndrome Coronavirus 2 Diagnostic Tools, ACS Nano, 2021, 15(1), 1259–1272 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |