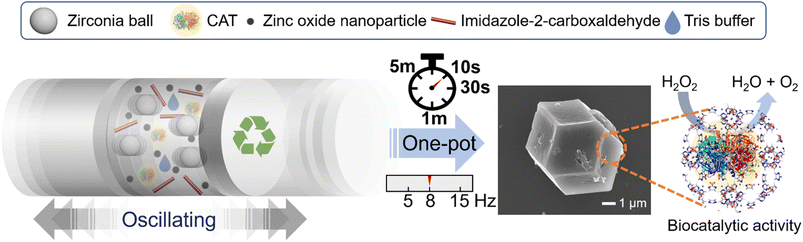
A green and ultrafast one-pot mechanochemical approach for efficient biocatalyst encapsulation in MOFs: insights from experiments and computation†
Phuc Khanh
Lam‡
 a,
Trung Hieu
Vo‡
a,
Trung Hieu
Vo‡
 b,
Jing-Hui
Chen‡
a,
Shang-Wei
Lin‡
b,
Jing-Hui
Chen‡
a,
Shang-Wei
Lin‡
 c,
Chiao-Ling
Kuo
a,
Jian-Jie
Liao
a,
Kuan-Yu
Chen
a,
Sen-Ruo
Huang
c,
Chiao-Ling
Kuo
a,
Jian-Jie
Liao
a,
Kuan-Yu
Chen
a,
Sen-Ruo
Huang
 c,
Dong
Li
a,
Yun-Hsiang
Chang
a,
Hsuan-Yi
Chen
a,
Haw-Ting
Hsieh
d,
Yu-An
Hsu
a,
Heng-Kwong
Tsao
c,
Dong
Li
a,
Yun-Hsiang
Chang
a,
Hsuan-Yi
Chen
a,
Haw-Ting
Hsieh
d,
Yu-An
Hsu
a,
Heng-Kwong
Tsao
 b,
Hsiao-Ching
Yang
*c and
Fa-Kuen
Shieh
b,
Hsiao-Ching
Yang
*c and
Fa-Kuen
Shieh
 *a
*a
aDepartment of Chemistry, National Central University, Taoyuan 32001, Taiwan. E-mail: fshieh@cc.ncu.edu.tw
bDepartment of Chemical and Materials Engineering, National Central University, Taoyuan 32001, Taiwan
cDepartment of Chemistry, Fu Jen Catholic University, New Taipei City, Taiwan. E-mail: hcyang_chem@mail.fju.edu.tw
dDepartment of Electrical Engineering and Computer Sciences, University of California at Berkeley, California 94720, USA
First published on 24th October 2023
Abstract
Mechanochemistry, a sustainable and efficient approach, is now used to encapsulate biocatalysts like enzymes within MOFs (enzyme@MOFs). Yet, achieving ultrafast, eco-friendly, and high-yield synthesis of enzyme@MOFs, with potential for mitigating biocatalyst and industrial MOF production improvement, remains challenging. In this study, we illustrated the enzyme one-pot mechanical encapsulation within zeolitic imidazolate framework-90 (ZIF-90), a MOF subfamily. A substantial yield (ca. 80%) was achieved in only 10 s at 8 Hz or lower frequencies, assisted by a small amount (∼100 μl) of Tris buffer solution. The encapsulated enzyme catalase (CAT), responsible for decomposing hydrogen peroxide, maintains its bioactivity after mechanical treatment. Additionally, the enzyme gains protection from digestion (e.g., proteinase K) due to the size-sheltering effect. Simulations of energy transfer and experimental findings reveal that a buffer solution containing primary amines affects linker deprotonation, thereby facilitating the rapid formation of a well-defined 3D structure of enzyme@ZIF-90 crystals during ultrafast milling reactions. We further demonstrated our mechanical approach by encapsulating bovine serum albumin and Escherichia coli in ZIF-90, highlighting the versatility of this method for crafting MOF biocomposites across diverse industrial applications.
Introduction
Biocatalysts, including enzymes, which are functional proteins known for their outstanding catalytic efficiency, specificity, and selectivity, have been greatly studied for both research purposes and industrial applications.1–3 However, to improve the long-term stability and reusability of biocatalysts, they are often combined with a porous solid through a process called enzyme immobilization.4 Metal–organic frameworks (MOFs) are a class of crystalline porous materials composed of metal ions coordinated to organic ligands, and they are valued for their high porosity, surface area, and tunable properties.5,6 As a result, MOFs have gained considerable attention as potential substrates among various solid supports.7,8 Enzymes can be immobilized on MOFs through two distinct approaches, aimed at reducing leaching: bioconjugation (referred to as enzyme-on-MOFs)9 involving covalently linking the enzyme to the surface of MOFs and encapsulation within MOF frameworks (referred to as enzyme@MOFs) utilizing in situ growth of MOFs surrounding enzymes,10,11 effectively entrapping them within the MOF structure. Inspired by natural biomineralization processes, the encapsulation of enzymes into MOF frameworks, known as enzyme@MOFs, has been successfully demonstrated through de novo methods such as biomimetic mineralization10 and coprecipitation.12 The MOF shell not only serves as an inert host but also enhances the selectivity,11 stability,13 and/or activity12 of the enzymes. However, the MOF material synthesis demands mild water-based conditions to maintain enzymatic activity due to the inherent fragility of enzymes.14 Despite the advancement of various water-based methods for synthesizing MOF materials, the effective encapsulation of enzymes into MOFs using solvent-based approaches while minimizing damage to the enzyme remains a significant challenge. This challenge primarily stems from the demands that lead to enzyme damage, including the use of chemical additives, low pH values, or high temperatures under aqueous conditions necessary for MOF formation.15ZIF materials such as ZIF-8 or ZIF-90 have demonstrated their potential as top candidates for protecting biomacromolecules.16 This superiority stems from their ability to be synthesized under mild conditions in aqueous solutions. Compared to ZIF-8, it becomes apparent that enzymes encapsulated in ZIF-90, enzyme@ZIF-90, exhibit heightened activity due to two key reasons: Firstly, ZIF-90 displays greater hydrophilicity which is favorable for biomolecules.17 Secondly, enzyme deactivation caused by the protonated linker, 2-methylimidazole, (a constituent of ZIF-8), is absent in ZIF-90.18 Enzyme@ZIF-90 materials are obtained through a de novo approach in an aqueous liquid-phase environment, where water plays a vital role in dissolving reagents and achieving a homogeneous mixture.11,16 Nonetheless, this method presents the risk of metal precursors or ligands dissolving, potentially causing a shift in the pH and ionic strength of the aqueous solution. This alteration in conditions, combined with a prolonged synthesis duration, has the potential to induce unfolding of protein structures, resulting in a loss of bioactivity. Therefore, the request for an alternative, ecologically friendly and gentle fabrication approach has gained significance. This approach aims to mitigate solvent-induced effects and reduce synthesis duration when encapsulating enzymes within MOFs.
Mechanochemistry has been demonstrated as a sustainable alternative for crafting MOFs, offering a means to minimize solvent usage and reduce processing time.19,20 Additionally, the mechanical approach applied in MOF synthesis demonstrates scalability21 and boasts considerable yield potential,22 rendering it more pragmatic for industrial applications.23,24 However, the integration of mechanochemistry into the creation of enzyme@MOFs remained invalidated until a significant breakthrough was realized. This breakthrough involved the successful fabrication of a range of enzyme@MOFs utilizing the innovative technique of liquid-assisted grinding (LAG).25,26 However, the complete potential of mechanochemical synthesis for enzyme@MOFs has not yet been fully explored. As an illustration, when metal oxides as precursors are employed instead of salts (such as metal acetate, metal nitrate, and metal carbonate), the reaction process generates only water, while the milling process exclusively yields water byproducts. This significantly diminishes the damage to the enzyme.27,28 Furthermore, although the fabrication process is rapid, requiring only a few minutes, the exploration of ultrafast synthesis procedures (within seconds) that could potentially mitigate the impact of mechanical forces on enzymes and improve enzymatic activity has been conspicuously absent from existing literature.
In this study, we introduce a sustainable and incredibly rapid technique for encapsulating enzymes into ZIF-90, achieving the first mechanical synthesis of both ZIF-90 and enzyme@ZIF-90. The biocatalyst of the CAT enzyme encapsulation process was conducted through a one-step ball-milling procedure at room temperature, augmented by a trace amount of an aqueous Tris buffer solution (Fig. 1). Remarkably, the CAT@ZIF-90 composites can be synthesized using zinc oxide as a precursor in just 10 s, resulting in an impressive yield of approximately 80% (mass of the product/mass of all precursors) with operational frequencies as low as 8 Hz or even lower. Our work stands as the pioneering demonstration that MOF biocomposites can be mechanically synthesized within a mere 10 seconds, using only a trace amount of Tris buffer and a low milling frequency of 8 Hz. The biological activity of encapsulated enzymes was maintained, remaining like, or even surpassing that of the embedded enzymes obtained by the de novo water-based approach. Additionally, the robust MOF shell assumes the role of an intriguing size-sheltering, effectively preserving enzymatic activity and shielding the enzyme from the proteinase K, a broad-range endolytic protease.11 The influence of the Tris buffer solution on the formation of and morphology of enzyme@ZIF-90 crystals was elucidated through simulations of energy transfer mechanisms and experimental studies. These studies encompassed varying concentrations and pH values and comparisons were made against other types of buffer solutions. It suggests that a buffer solution containing primary amines can influence the deprotonation process of linkers, facilitating the swift formation of a well-shaped 3D structure for CAT@ZIF-90 crystals during ultrafast milling reactions. Consequently, this mechanical approach has demonstrated its effectiveness not only in encapsulating another protein, bovine serum albumin (BSA), but also in encapsulating micrometer-sized organisms like living Escherichia coli (E. coli) within MOFs, resulting in E. coli@ZIF-90. The achievement of E. coli@ZIF-90 through mechanical encapsulation represents a significant milestone, marking the first successful demonstration of E. coli's viability using this approach. This highlights the versatility of this method in crafting MOF biocomposites.
Results and discussion
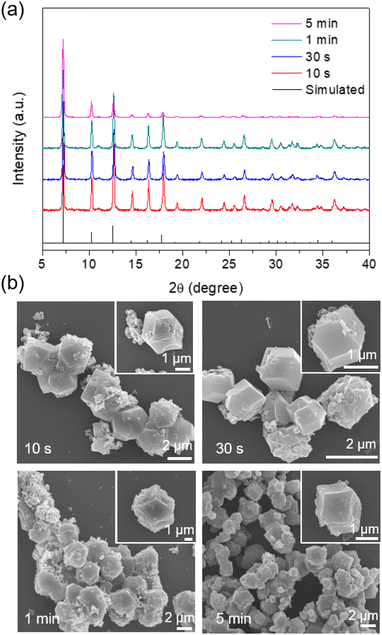
The CAT enzyme, a valuable enzyme utilized for decomposing hydrogen peroxides in wastewater treatment,29 was selected as the model for the encapsulation process. The nano-sized zinc oxide powders, lyophilized CAT, imidazole-2-carboxaldehyde, and trace amounts of Tris buffer solution (or deionized (DI) water) were subjected to ball-milling at 8 Hz for varying durations: 10 s, 30 s, 1 min, and 5 min. Each sample was assigned a label corresponding to its specific synthesis conditions. For instance, the ZIF-90 sample obtained using Tris buffer at 500 mM, pH 7, and a frequency of 8 Hz for 10 s was denoted as CAT@ZIF-90-Tris-pH 7-8 Hz-10 s. The crystallinity and completeness of samples were examined using X-ray diffraction (XRD), while crystal morphology was observed through scanning electron microscope (SEM) images. The XRD patterns of samples synthesized using Tris buffer solution exhibited the typical Bragg diffraction patterns that are characteristic of ZIF-90, as shown Fig. 2a. In contrast, the sample synthesized using DI water displayed diffraction patterns for both 2D and 3D structures (see Fig. S1†). The observed 2D structure might bear resemblance to that found in ZIF-8, characterized by a diamondoid structure known as dia-Zn (MeIm).30 Experiments were also conducted with different Tris concentrations (50, 100, 200 and 250 mM), which resulted in XRD outcomes displaying peaks corresponding to both the 2D and 3D structures of ZIF-90 (Fig. S2†). The XRD pattern exclusively displaying the 3D structure, without the presence of the 2D structure, was achievable only when employing a 500 mM Tris buffer (Fig. 2a). Subsequently, all experiments were carried out using a 500 mM Tris buffer. Upon observing SEM images (see Fig. 2b), it became evident that the morphology of particles prepared with Tris buffer solution became progressively more uniform and well-defined as the ball-milling time increased. To determine whether the enzymes were successfully encapsulated within the frameworks, we conducted sample digestion followed by subsequent analysis using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). As illustrated in Fig. 3a and S3,† the band corresponding to the molecular weight of the monomeric form (60 kDa) is distinctly visible in decomposed CAT@ZIF-90-Tris-pH 7, mirroring the presence of free CAT. This observation strongly suggests that the enzymes were encapsulated during the ball-milling process. To rule out the possibility of CAT molecules being adsorbed onto the external surface of the MOF crystals, we mixed ZIF-90-Tris-pH 7, which was obtained through ball-milling, with CAT. Following the same purification and decomposed procedures, no bands were detected for CAT-on-ZIF-90-Tris-pH 7. This outcome indicates the absence of enzyme attachment to the external surface of the MOF. The loading of CAT, determined by a standard Bradford assay method (Fig. S4†), was approximately 5 wt%.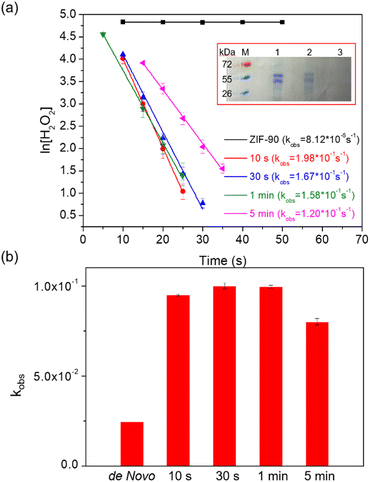 | ||
| Fig. 3 (a) Kinetics of degradation of H2O2 of the CAT@ZIF-90-Tris-pH 7-8 Hz sample at various reaction times without proteinase K; inset: SDS-PAGE gel (M: protein marker, lane 1: free catalase, lane 2: CAT@ZIF-90-Tris-pH 7-8 Hz-10 s, and lane 3: washed CAT-on-ZIF-90-Tris-pH 7-8 Hz-10 s) and (b) comparing the retained activity for CAT@ZIF-90 samples obtained through ball-milling with a frequency of 8 Hz and at various reaction times to that of samples synthesized using a de novo water-based approach,11 after exposure to proteinase K. | ||
To evaluate the encapsulated enzymatic activity, we assessed the peroxide-decomposing performance of the CAT@ZIF-90 composite. This was done by monitoring the change in H2O2 concentration over time using absorbance measurements at 560 nm with xylenol orange (Fig. S5†). The observed rate constants (kobs) for the CAT@ZIF-90-Tris-pH 7-8 Hz sample were determined and are presented in Table S1.† These rate constants (1.98 × 10−1, 1.67 × 10−1, 1.58 × 10−1, and 1.20 × 10−1 for 10 s, 30 s, 1 min, 5 min, respectively) indicate that the enzyme remained undenatured throughout the ball-milling process (see Fig. 3a). The sheltering function was showcased through an incubation experiment involving the CAT@ZIF-90-Tris-pH 7-8 Hz sample and proteinase K, a serine-type protease of size (68.3 × 68.3 × 108.5 Å).31 Notably, this enzyme surpasses the size of the ZIF-90 aperture. Following treatment with proteinase K, as shown in Table S1† and Fig. 3b, the samples retained favorable kobs values of 9.48 × 10−2, 1.03 × 10−1, 9.96 × 10−2, and 8.01 × 10−2 for 10 s, 30 s, 1 min and 5 min, respectively. Although a shift in kobs values was observed due to the presence of partial enzyme encapsulation on the surface of MOF crystals, the effectiveness of the sheltering function remained evident (see Fig. 3b). Comparing the enzymatic activity of CAT@ZIF-90 samples obtained through ball-milling or a de novo water-based approach,11 with or without proteinase K treatment, consistently showed higher biological activity (kobs) for CAT@ZIF-90 prepared via the mechanical method (Fig. S6,†3b and Table S2†). This enhancement in kobs is attributed to the mechanical force, which can induce defects in MOF crystals and potentially increase kobs by enhancing mass transfer. In Fig. S7 and Table S3,† we present the porous characterization of CAT@ZIF-90, prepared via ball-milling, revealing micro- and mesopores.32 Observable hysteresis in the N2 adsorption/desorption isotherms within the P/P0 range of 0.3 to 0.5 confirms the presence of numerous mesoporous defects in the ZIF-90 crystals due to the milling process.33,34 Despite MOF shell defects, CAT@ZIF-90 maintained high enzymatic activity in the presence of proteinase K. Notably, even with just 10 seconds of ball-milling, CAT@ZIF-90 exhibited nearly the same kobs values as longer durations, indicating effective protection without need for extended milling. Milling frequencies of 5 and 15 Hz were employed to explore the influence of various mechanical forces on the synthesis process. The XRD patterns of the samples, showcasing distinct peaks corresponding to ZIF-90, are depicted in Fig. S8 and S9.† Remarkably, despite variations in mechanical energy resulting from changes in frequency (5, 8, and 15 Hz) or milling time (10 s, 30 s, 1 min, and 5 min), there were no observable differences in particle morphology. This resulted in a precision and size uniformity similar to what was achieved by 8 Hz milling (see Fig. S10 and S11†). Significantly, the corresponding kobs values of the samples remained consistent with those synthesized by milling at 8 Hz, as detailed in Table S1.† This indicates that frequencies (5, 8, and 15 Hz) and times (10 s, 30 s, 1 min, and 5 min) explored in this study will not significantly impact crystal formation and the enzyme structure.
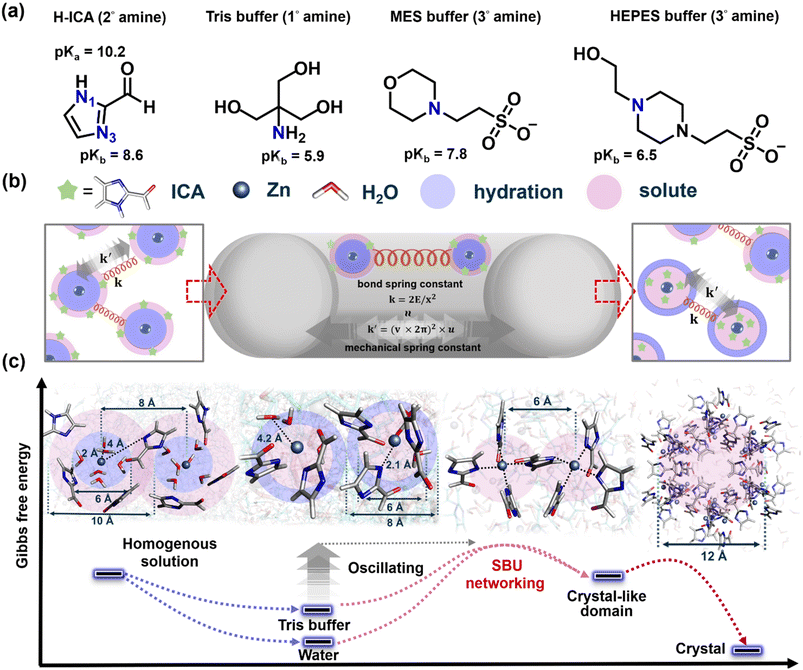
As manifested above, the synthesis of MOFs and enzyme@MOFs is a rapidly growing field with a wide range of potential applications. MOFs are crystalline materials with metal ions or clusters coordinated to organic ligands, in which enzyme@MOFs are MOFs that contain enzymes encapsulated in their structure. The synthesis is often challenging, as it requires the overcoming of significant activation energy barriers. This is because the formation of these materials involves the breaking and reforming of bonds, as well as overcoming repulsive forces between reactant ions. Our experiments have shown that stirring alone is not sufficient to trigger the formation of ZIF-90. However, mechanical force applied through ball-milling can successfully trigger and control the formation of ZIF-90. This emphasizes the role of mechanochemical energy in overcoming the inherent barrier in the desired synthesis process. Fig. 4 clearly illustrates the stepwise synthesis of ZIF-90, highlighting the critical roles of Tris buffer (Fig. 4a) and ball-milling (Fig. 4b). The energy landscape of this reaction process often presents barriers at different precursor stages that cannot be overcome without significant input of energy (Fig. 4c). These barriers are related to the initial breaking and forming of bonds, as well as overcoming repulsive forces between reactant ions.
The ZIF-90 synthesis initial step involves disrupting the water microsolvation shell around the zinc ions. This is done by the Tris buffer because it has primary (1°) amine with a pKb value of 5.9 (Fig. 4a),35 which is a chelating agent that has the strongest capacity to assist the secondary (2°) amine of ICA with a pKb value of 8.6 to displace the water molecules to form the Zn–ICA complex precursor. Once the water microsolvation shell has been disrupted, the zinc ions can then coordinate with the ICA molecules (Fig. 4c). The Tris buffer also helps to trigger deprotonation of N1(H) in the H-ICA (pKa = 10.2).36 This forms the Zn–ICA complex precursor and facilitates the formation of solute-rich regions in the solution more efficiently than in pure water.37 The next step is to replace the zinc hydration clusters with zinc secondary building units (SBUs) and the dehydration of water becomes crucial. This is done by the ball-milling process, which provides the mechanical energy necessary to break zinc–oxygen bonds and facilitate the formation of zinc–nitrogen bonds and generate H2O byproducts. This promotes the assembly of Zn–ICA precursors to overcome the barrier of SBU networking towards the crystal-like domain. Finally, the water molecules are dehydrated, and the ZIF-90 framework crystal is formed.
The mechanochemical transformation of ZIF-90 is a complex process that involves the interplay of mechanical forces and chemical interactions (Fig. 4b). Understanding the underlying transformation mechanism is key to unlocking its potential for precise control of the ball-milling process. The harmonic oscillation model provides a way to understand the interplay between mechanical forces and chemical interactions in the ZIF-90 synthesis. This model treats the chemical reactants and the relevant nucleation precursors as harmonic oscillators. This means that they can store and release energy in the form of vibrations. The frequency of these vibrations is determined by the force constant of the oscillator, which is a measure of the strength of the chemical bonds (or strings).
In the ZIF-90 system (Fig. 4b), the nucleation process is driven by a combination of mechanical forces and chemical interactions. The milling introduces mechanical energy into the bond springs, as shown in the context of a simple harmonic oscillator, eqn (1),
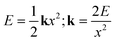 | (1) |
| Chemical energya (kcal mol−1) | 1.0 | 5.0 | 10.0 |
|---|---|---|---|
| a The bond spring constant (k) derived from eqn (1). b The mechanical spring constant (k′) derived from eqn (2). | |||
| k (N m−1) for 2 Å | 3.48 × 10−1 | 1.74 × 100 | 3.48 × 100 |
| k (N m−1) for 4 Å | 8.69 × 10−2 | 4.34 × 10−1 | 8.69 × 10−1 |
| k (N m−1) for 6 Å | 3.86 × 10−2 | 1.93 × 10−1 | 3.86 × 10−1 |
| k (N m−1) for 8 Å | 2.17 × 10−2 | 1.09 × 10−1 | 2.17 × 10−1 |
| k (N m−1) for 10 Å | 1.39 × 10−2 | 6.95 × 10−2 | 1.39 × 10−1 |
| Mechanical frequencyb (Hz) | 5 | 8 | 15 |
|---|---|---|---|
| k′ (N m−1) | 1.60 × 100 | 4.10 × 100 | 1.44 × 101 |
From a mechanical perspective, comprehending the energy introduced through ball-milling can be most effectively achieved by examining the oscillatory characteristics of the process. This understanding takes into account the same eqn (2),
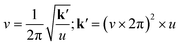 | (2) |
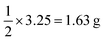 in our experiments), and we derived the force constant for the mechanical energy. This equation shows that the amount of energy introduced by the milling is proportional to the square of the displacement (eqn (1)). This means that even a small displacement can introduce a significant amount of energy into the system. The harmonic oscillation model provides a useful framework for understanding the mechanochemical transformation of ZIF-90. This model can be used to predict the conditions under which the transformation will occur, and it can also be used to optimize the ball-milling process.
in our experiments), and we derived the force constant for the mechanical energy. This equation shows that the amount of energy introduced by the milling is proportional to the square of the displacement (eqn (1)). This means that even a small displacement can introduce a significant amount of energy into the system. The harmonic oscillation model provides a useful framework for understanding the mechanochemical transformation of ZIF-90. This model can be used to predict the conditions under which the transformation will occur, and it can also be used to optimize the ball-milling process.
Intriguingly, for the process to align with the previously mentioned chemical energies, the mechanical frequency “v” must reside within 5–15 Hz. This revelation carries considerable significance, as our ball-milling operates at frequencies of 5, 8, and 15 Hz, falling confidently within this established range. This alignment is not coincidental; the remarkable similarity in magnitudes between the k and k′ values, whether observed chemically or mechanically, strongly suggests an effective energy transfer mechanism in play. Essentially, the mechanical oscillations generated by our ball-milling operating at frequencies of 5, 8, and 15 Hz establish an effective force constant that harmoniously aligns with the necessary chemical potential for ZIF-90 synthesis. Moreover, the alignment of k and k′ values underscores the significance of our chosen ball-milling frequency in the successful synthesis of ZIF-90. The mechanical energy supplied is in perfect harmony with the energy demand for the formation of delicate chemical bonds, thereby facilitating an efficient and rapid synthesis process.
From simulation results, we find that the Tris buffer, specifically its fundamental amine group, is crucial in linker deprotonation. The simulation results highlight the significant role of the Tris buffer in facilitating the deprotonation of linkers, with emphasis on the significance of the basic amine group within the buffer molecule. Moreover, the simulation outcomes are in agreement with the absence of mixed 2D structure impurities in ZIF-90 crystals as the Tris buffer concentration is elevated to 500 mM (Fig. 2a). This emphasizes the notable influence of buffer concentration on the nucleation process. Additionally, we also tested alternative buffers, including HEPES 500 mM at pH 7 (buffer range from pH 6.8 to 8.2, Fig. S12†) and MES 500 mM at pH 7 (buffer range from pH 5.5 to 7.0, Fig. S13†). The samples are subjected to ball-milling at 8 Hz for varying synthesis durations, revealing peaks corresponding to both the 2D and 3D structures of ZIF-90. These buffers contain tertiary (3°) amine groups, which could potentially act as proton acceptors. However, their pKb values exceed that of Tris buffer (HEPES: 6.5, MES: 7.85, Fig. 4a),35 making them less likely to donate protons to the linker molecules. This deficiency leads to incomplete deprotonation, and resulting impurities and prevents the desired 3D structural configuration from forming. These findings validate that the formation of CAT@ZIF-90 via mild mechanical energy exclusively succeeds when utilizing a Tris buffer solution. Taken together, these results confirm that the generation of CAT@ZIF-90 through mild mechanical energy is achievable solely by employing a buffer solution containing the primary amine group (Tris).
Our hypothesis suggests a significant correlation between the morphology of CAT@ZIF-90 crystals and the effectiveness of the deprotonation process. To investigate this relationship, we enhance the basicity of the primary amine by adjusting the pH value. We elevate the pH value of Tris buffer (in its base form) to increase the ratio of its conjugated base form. The heightened concentration of Tris promotes the disruption of the hydration shell, thereby facilitating a more efficient deprotonation mechanism. Given that the amine group is involved in linker deprotonation (Fig. 4a), we explore the impact of pH on the formation of the CAT@ZIF-90 sample. This exploration is conducted by subjecting the sample to ball-milling at various frequencies and durations. A Tris buffer solution with a pH value of 8 was employed for this investigation. The structural analysis of the sample was performed using XRD (Fig. S14–S16†) and SEM (Fig. S17–S19†). The successful encapsulation process of CAT was confirmed using SDS-PAGE, as shown in Fig. S20.† Additionally, the enzymatic activity was evaluated under conditions, with and without proteinase K (Table S4†). Upon adjusting the pH from 7 to 8, the XRD patterns of the sample exhibited improved alignment with simulated ZIF-90 peaks. Concurrently, SEM images revealed a discernible increase in the quantity of well-formed crystals, varying in size from 1 to 3 μm. Furthermore, the enzymatic activity exhibited marginal fluctuation with the increase in pH.
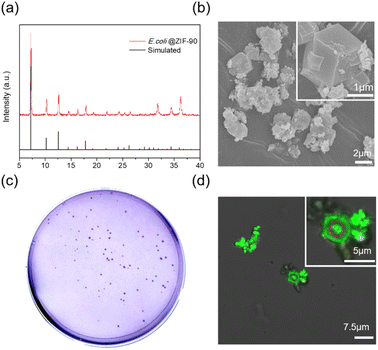
To demonstrate the versatility of this mechanical strategy, successful encapsulation of BSA protein or living E. coli within ZIF-90 crystals was achieved. The crystallinity and morphology of the BSA@ZIF-90 and E. coli@ZIF-90 samples were characterized through XRD and SEM, respectively, as depicted in Fig. 5a and b and S21–S24.† The successful encapsulation process of BSA was confirmed using SDS-PAGE, as shown in Fig. S25.† The loading amount of BSA was approximately 10.2 wt%, determined by a standard Bradford assay method (Fig. S26†). Moreover, following the liberation of the E. coli by eliminating the ZIF shell, their viability was retained intact, as evaluated by plating on LB agar plates (see Fig. 5c). This indicates that the ball-milling process does not inflict harm on the bacteria, highlighting the safety of our ultrafast mechanical approach in encapsulating delicate biomaterials. Fig. 5d, S27 and S28† present a 3D image reconstructed using nondestructive horizontal scanning through confocal laser scanning microscopy. The resultant image displays a viable, rod-shaped E. coli emitting vibrant green light, visibly confined within the single-crystalline ZIF-90 cage, thereby verifying the successful encapsulation. This represents a significant milestone in mechanically obtaining E. coli@ZIF-90, successfully demonstrating E. coli's viability through this mechanical approach for the first time.
Conclusions
This study introduces a novel method for synthesizing MOF biocomposites that is ultrarapid, environmentally friendly and yields high results (about 80%). The one-pot mechanical approach enables the encapsulation of enzymes within ZIF-90 crystals in a mere 10 s when utilizing frequencies of 8 Hz or below. This approach is facilitated with the aid of a trace amount (∼100 μl) of aqueous Tris buffer solution. It is important to highlight that the encapsulated enzymes exhibited significant biological activity. Moreover, the encapsulated enzyme within ZIF-90 maintained its enzymatic activity, even in the presence of proteinase K, attributed to the size-sheltering effect of the MOF shell. Notably, the use of Tris buffer significantly improved the development of a well-defined 3D structure compared to other types of buffer solutions. Furthermore, simulations suggest that the buffer solution containing primary amine facilitates linker protonation through an energy transfer mechanism. The influence of the Tris buffer on linker deprotonation becomes evident in the improved morphology of CAT@ZIF-90 crystals, achieved through meticulous adjustments in concentration and pH values. Our unique mechanical strategy is applicable in the encapsulation of BSA or E. coli bacteria. It indicates that this adaptable strategy underscores the versatility of creating MOF composites for a wide range of applications.Author contributions
P. K. Lam, T. H. Vo, S.-W. Lin, H.-C. Yang and F.-K. Shieh: research design. P. K. Lam, T. H. Vo and S.-W. Lin: manuscript drafting. J.-H. Chen, P. K. Lam, T. H. Vo and H.-Y. Chen: material synthesis of biocatalyst@MOFs. S.-W. Lin and S.-R. Huang: computer simulations. J.-H. Chen, C.-L. Kuo and K.-Y. Chen: enzyme activity experiments. D. Li and Y.-H. Chang: E. coli-related experiments. P. K. Lam, J.-J. Liao, H.-T. Hsieh and Y.-A. Hsu: statistical analyses and manuscript contribution. H.-K. Tsao, F.-K. Shieh and H.-C. Yang: supervision, guidance, manuscript finalization. All authors played a role in manuscript preparation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
F.-K. Shieh, H.-C. Yang and H.-K. Tsao would like to thank the National Science and Technology Council (NSTC), Taiwan for the funding support (111-2113-M-008-005-MY2), (112-2113-M-030-001) and (109-2221-E-008-026-MY3), respectively. We thank Prof. Rong-Jie Chein at Academia Sinica, Taiwan, for conducting the surface area measurement. We also thank Prof. Wen-Tzu Liu and Dr Wong in Chung Yuan Christian University for helping with SEM images. We would like to thank Prof. Chia-Ching Chang for supporting with confocal images in National Yang Ming Chiao Tung University, Taiwan.Notes and references
- J. J. Green and J. H. Elisseeff, Nature, 2016, 540, 386–394 CrossRef CAS PubMed.
- R. A. Sheldon and J. M. Woodley, Chem. Rev., 2018, 118, 801–838 CrossRef CAS PubMed.
- J. Yang and Y.-W. Yang, Small, 2020, 16, 1906846 CrossRef CAS PubMed.
- K. Ariga, Q. Ji, T. Mori, M. Naito, Y. Yamauchi, H. Abe and J. P. Hill, Chem. Soc. Rev., 2013, 42, 6322–6345 RSC.
- H. Furukawa, K. E. Cordova, M. O'Keeffe and O. M. Yaghi, Science, 2013, 341, 1230444 CrossRef PubMed.
- Q. Wang and D. Astruc, Chem. Rev., 2020, 120, 1438–1511 CrossRef CAS PubMed.
- W. Liang, P. Wied, F. Carraro, C. J. Sumby, B. Nidetzky, C.-K. Tsung, P. Falcaro and C. J. Doonan, Chem. Rev., 2021, 121, 1077–1129 CrossRef CAS PubMed.
- X. Lian, Y. Fang, E. Joseph, Q. Wang, J. Li, S. Banerjee, C. Lollar, X. Wang and H.-C. Zhou, Chem. Soc. Rev., 2017, 46, 3386–3401 RSC.
- S.-L. Cao, D.-M. Yue, X.-H. Li, T. J. Smith, N. Li, M.-H. Zong, H. Wu, Y.-Z. Ma and W.-Y. Lou, ACS Sustain. Chem. Eng., 2016, 4, 3586–3595 CrossRef CAS.
- K. Liang, R. Ricco, C. M. Doherty, M. J. Styles, S. Bell, N. Kirby, S. Mudie, D. Haylock, A. J. Hill, C. J. Doonan and P. Falcaro, Nat. Commun., 2015, 6, 7240 CrossRef CAS PubMed.
- F.-K. Shieh, S.-C. Wang, C.-I. Yen, C.-C. Wu, S. Dutta, L.-Y. Chou, J. V. Morabito, P. Hu, M.-H. Hsu, K. C. W. Wu and C.-K. Tsung, J. Am. Chem. Soc., 2015, 137, 4276–4279 CrossRef CAS PubMed.
- F. Lyu, Y. Zhang, R. N. Zare, J. Ge and Z. Liu, Nano Lett., 2014, 14, 5761–5765 CrossRef CAS PubMed.
- F.-S. Liao, W.-S. Lo, Y.-S. Hsu, C.-C. Wu, S.-C. Wang, F.-K. Shieh, J. V. Morabito, L.-Y. Chou, K. C. W. Wu and C.-K. Tsung, J. Am. Chem. Soc., 2017, 139, 6530–6533 CrossRef CAS PubMed.
- V. Gascón, C. Carucci, M. B. Jiménez, R. M. Blanco, M. Sánchez-Sánchez and E. Magner, ChemCatChem, 2017, 9, 1182–1186 CrossRef.
- Z. Chen, X. Wang, R. Cao, K. B. Idrees, X. Liu, M. C. Wasson and O. K. Farha, ACS Mater. Lett., 2020, 2, 1129–1134 CrossRef CAS.
- A. Maleki, M.-A. Shahbazi, V. Alinezhad and H. A. Santos, Adv. Healthcare Mater., 2020, 9, 2000248 CrossRef CAS PubMed.
- D. Ge, M. Li, D. Wei, N. Zhu, Y. Wang, M. Li, Z. Zhang and H. Zhao, Chem. Eng. J., 2023, 469, 144067 CrossRef CAS.
- G. Chen, X. Kou, S. Huang, L. Tong, Y. Shen, W. Zhu, F. Zhu and G. Ouyang, Angew. Chem., Int. Ed. Engl., 2020, 59, 2867–2874 CrossRef CAS PubMed.
- T. Stolar and K. Užarević, CrystEngComm, 2020, 22, 4511–4525 RSC.
- X. Liu, Y. Li, L. Zeng, X. Li, N. Chen, S. Bai, H. He, Q. Wang and C. Zhang, Adv. Mater., 2022, 34, 2108327 CrossRef CAS PubMed.
- H. M. Titi, J.-L. Do, A. J. Howarth, K. Nagapudi and T. Friščić, Chem. Sci., 2020, 11, 7578–7584 RSC.
- S. Darwish, S.-Q. Wang, D. M. Croker, G. M. Walker and M. J. Zaworotko, ACS Sustain. Chem. Eng., 2019, 7, 19505–19512 CrossRef CAS.
- S. Głowniak, B. Szczęśniak, J. Choma and M. Jaroniec, Mater. Today, 2021, 46, 109–124 CrossRef.
- S. Tanaka, in Metal-Organic Frameworks for Biomedical Applications, ed. M. Mozafari, Woodhead Publishing, 2020, pp. 197–222 Search PubMed.
- T.-H. Wei, S.-H. Wu, Y.-D. Huang, W.-S. Lo, B. P. Williams, S.-Y. Chen, H.-C. Yang, Y.-S. Hsu, Z.-Y. Lin, X.-H. Chen, P.-E. Kuo, L.-Y. Chou, C.-K. Tsung and F.-K. Shieh, Nat. Commun., 2019, 10, 5002 CrossRef PubMed.
- R. Gao, N. Zhong, L. Tong, X. Kou, W. Huang, H. Yang, S. Huang, J. Wu, G. Chen and G. Ouyang, Cell Rep. Phys. Sci., 2022, 3, 101153 CrossRef CAS.
- S. Tanaka, K. Kida, T. Nagaoka, T. Ota and Y. Miyake, ChemComm, 2013, 49, 7884–7886 RSC.
- B. Chen, Z. Yang, Y. Zhu and Y. Xia, J. Mater. Chem. A, 2014, 2, 16811–16831 RSC.
- J. Chen, Q. Wang, Z. Hua and G. Du, Enzyme Microb. Technol., 2007, 40, 1651–1655 CrossRef CAS.
- A. D. Katsenis, A. Puškarić, V. Štrukil, C. Mottillo, P. A. Julien, K. Užarević, M.-H. Pham, T.-O. Do, S. A. J. Kimber, P. Lazić, O. Magdysyuk, R. E. Dinnebier, I. Halasz and T. Friščić, Nat. Commun., 2015, 6, 6662 CrossRef CAS PubMed.
- A. Pähler, A. Banerjee, J. K. Dattagupta, T. Fujiwara, K. Lindner, G. P. Pal, D. Suck, G. Weber and W. Saenger, EMBO J., 1984, 3, 1311–1314 CrossRef PubMed.
- M. Gao, J. Wang, Z. Rong, Q. Shi and J. Dong, RSC Adv., 2018, 8, 39627–39634 RSC.
- L. J. Zhiwen Ai, J. Wang and H.-L. Jiang, J. Chin. Chem. Soc., 2022, 4, 3705–3714 Search PubMed.
- H. Yuan, Y. Wu, X. Pan, L. Gao and G. Xiao, Catal. Lett., 2020, 150, 3561–3571 CrossRef CAS.
- N. E. Good, G. D. Winget, W. Winter, T. N. Connolly, S. Izawa and R. M. M. Singh, Biochemistry, 1966, 5, 467–477 CrossRef CAS PubMed.
- M. J. Lee, H. T. Kwon and H.-K. Jeong, Angew. Chem., Int. Ed. Engl., 2018, 57, 156–161 CrossRef CAS PubMed.
- G. C. Sosso, J. Chen, S. J. Cox, M. Fitzner, P. Pedevilla, A. Zen and A. Michaelides, Chem. Rev., 2016, 116, 7078–7116 CrossRef CAS PubMed.
- J. Manz, R. Meyer and H. H. R. Schor, J. Chem. Phys., 1984, 80, 1562–1568 CrossRef CAS.
- J. Emsley, Chem. Soc. Rev., 1980, 9, 91–124 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ta05228a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |