Background-filtered telomerase activity assay with cyclic DNA cleavage amplification†
Hyogu
Han
ab,
Chihyun
Park
c,
Chang Yeol
Lee
 *d and
Jun Ki
Ahn
*d and
Jun Ki
Ahn
 *a
*a
aMaterial & Component Convergence R&D Department, Korea Institute of Industrial Technology (KITECH), Ansan 15588, Korea
bDepartment of Chemistry, Gangneung-Wonju National University, Gangneung 25457, Korea
cDaejeon District Office, National Forensic Service, Daejeon 34054, Korea
dBionanotechnology Research Center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Daejeon 34141, Korea
First published on 19th September 2023
Abstract
Overexpression of telomerase incites the abnormal proliferation of cancer cells. Thus, it has been regarded as a cancer biomarker and a potential therapeutic target. Existing assays suggest a promising sensing scheme to detect telomerase activity. However, they are complicated in terms of assay preparation and implementation. We herein report a Quenching-Exempt invader Signal Amplification Test, termed ‘QUEST’. The assay leverages on a high turnover, specific cleaving enzyme, flap endonuclease I (FEN1), and graphene oxide (GO) for background (BG) filtering. In response to the target, FEN1 significantly boosts the signal with invader signal amplification. To distinguish the target signal, GO filters out the BG. It captures residual reporter invader probes (RP) to quench undesired signals. QUEST is straightforward without any pre-preparatory steps and washing/separation. Its probe design is simple and cost-effective. With QUEST, we investigated telomerase activities in various cell lines. Notably, we discriminated cancer cell lines from normal cell lines. In addition, a candidate inhibitor for telomerase was screened, which showed the promising potential of QUEST in real applications.
Introduction
Telomerase elongates the termini of chromosomes with telomere sequences (TTAGGG)n. Its overexpression suppresses timely telomere shortening that induces cellular senescence and hampers the normal cell cycle, with cells being abnormally immortalized.1–3 Most of the cancer cells overexpress telomerase, and it has been regarded as a significant biomarker for cancer diagnosis and a potential therapeutic target.4,5The telomeric repeat amplification protocol (TRAP) based on the polymerase chain reaction (PCR) is the gold standard for telomerase detection.6 Real-time signal detection, time-resolved fluorescence assay, and enzyme-linked immunosorbent assay (ELISA) were introduced to improve the TRAP.6–8 However, the need for precise temperature control and relevant resources limited their wide use. To circumvent this issue, various detection modes adopted isothermal nucleic acid (NA) assay schemes such as isothermal NA amplification, nanomolecular machines, and clustered regularly interspaced short palindromic repeats/CRISPR-associated protein (CRISPR/Cas) systems (Table S1†).9–16
Although promising, they have a few drawbacks: (i) preparing assays is complicated and laborious. Electrodes or particles are supposed to be modified with DNA probes with several washing steps. (ii) Detection procedures are complicated. They proceed with several separate intervening steps for reagent addition or particle washing/separation, which would increase the likelihood of false results. (iii) Assay components whose cost-to-benefit ratios are poor are involved, such as a CRISPR complex (i.e. Cas protein/guide RNA) and a lateral flow assay kit, and (iv) assays based on isothermal NA amplification are vulnerable to non-specific amplification, compromising the reliability.
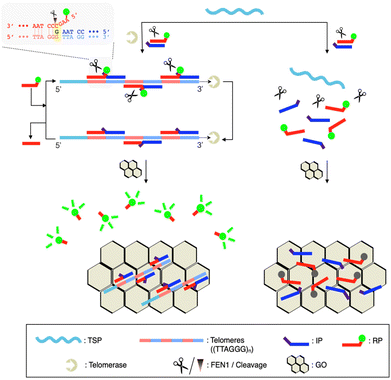
Here, we have developed a Quenching-Exempt invader Signal Amplification Test, termed ‘QUEST’, for telomerase activity detection (Scheme 1). In particular, we designed a reporter invader probe (RP) having a short protruding sequence (flap sequence; 4 mer).17 As the telomerase concatenates a telomerase substrate primer (TSP) with telomeres ((TTAGGG)n), pairs of invader detection probes bind to the extended sequences and form junction structures where a base at the 5′-end crosses the junction site by a single base. Flap endonuclease I (FEN1) recognizes such structures and catalytically cuts out flap sequences having fluorophores (F). Graphene oxide (GO) is then added to capture unreacted DNA and turn off the fluorescence signal.17,18 However, the flap sequences are short enough to exempt themselves from quenching by GO. Thus, high signal is retained. Notably, QUEST is seamless. It concurrently executes the telomere extension reaction and invader signal amplification, minimizing hands-on steps. Its probe design is simple and cost-effective. By using GO as a quencher (Q), the assay eliminates the need for dual-labeling with F/Q. FEN1 has a high turnover rate and is quite specific to the junction structure, which would ensure high sensitivity and specificity.19,20
Experimental
Materials
We purchased oligonucleotides from Integrated DNA Technology Inc. (Coralville, IA, USA); details are given in Table S2.† Flap endonuclease I (FEN1; M0645S) and HotStart Taq DNA polymerase were purchased from New England Biolabs Inc. (Beverly, MA, USA). Graphene oxide (GO) was purchased from Graphene Supermarket (Ronkonkoma, NY, USA) (Fig. S7†). Nuclease-free water (DW) was purchased from Invitrogen Co. (Carlsbad, CA, USA). 1× CHAPS lysis buffer, tris(hydroxymethyl)amino-methane (Tris), MgCl2, KCl, ethylene glycol-bis(2-aminoethyl-ether)-N,N,N′,N′-tetraacetic acid (EGTA), 3′-azido-3′-deoxythymidine (AZT), SYBR Green I, and Tween 20 were purchased from Sigma Aldrich (St Louis, MO, USA).Cell culture and preparation of cell extracts
We cultured HeLa, MRC-5, A549, MCF-7 and HepG2 cell lines (American Type Culture Collection) in DMEM (Welgene, Korea) supplemented with 10% fetal bovine serum (RMBIO, Missoula, MT, USA) and 1% penicillin–streptomycin (Gibco, Scotland) under a humidified atmosphere with 5% CO2 at 37 °C. 106 cells in the exponential growth phase were collected and washed three times with ice-cold 1× PBS (4000g, 3 min). Cell pellets were then resuspended in 200 μL of ice-cold 1× CHAPS lysis buffer (0.5% CHAPS, 1 mM MgCl2, 1 mM EGTA, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 5 mM mercaptoethanol, 10% glycerol, 10 mM Tris-HCl, pH 7.5). After 30 min of incubation on ice, the lysed sample was centrifuged at 4 °C (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 20 min) to remove cell debris and the supernatant was transferred to a clean tube. The cell extract was stored at −80 °C until use.
000g, 20 min) to remove cell debris and the supernatant was transferred to a clean tube. The cell extract was stored at −80 °C until use.
Protocols of QUEST for telomerase assay and inhibitor screening
Telomeric repeat amplification protocol
Using a TRAPeze Telomerase Detection Kit (MilliporeSigma, Burlington, MA, USA), we conducted the TRAP according to the manufacturer's protocol with slight modification. We prepared 50 μL of TRAP solution by mixing 400 nM TSP, 1 μL of TRAP primer mix, 2 U of HotStart Taq DNA polymerase, 1× SYBR Green I, 1× dNTP, and 104 cell extract of the intended cell line in 1× TRAP reaction buffer (20 mM Tris-HCl, 1.5 mM MgCl2, 63 mM KCl, 0.05% Tween 20, 1 mM EGTA, pH 8.3 at 1×). After 30 min of telomerase elongation at 30 °C, we executed the PCR according to the following protocol: 95 °C, 2 min, 35 cycles of 94 °C, 15 s; 59 °C, 30 s; 72 °C, 1 min, and 72 °C, 1 min. Fluorescence signals of SYBR Green I were then measured to reflect the telomerase activity.Gel electrophoresis
We loaded the reaction solutions in 1× loading buffer (Bioneer, Korea) on a 15% polyacrylamide gel and ran the electrophoresis at 110 V for 3 h in 1× TBE running buffer. After GelRed (Biotium, Fremont, CA, USA) staining, we obtained the gel image using a GelDoc™ EZ Image (Bio-Rad, Hercules, CA, USA).Results and discussion
Overall scheme and feasibility of QUEST
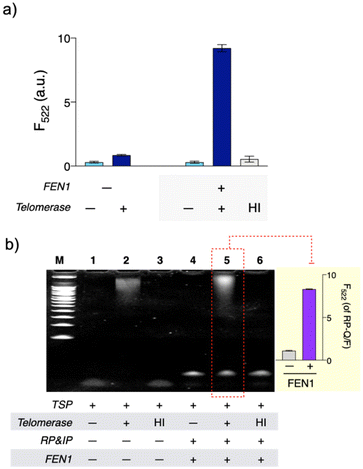
QUEST proceeds with telomerase-triggered invader signal amplification and BG filtering with GO (Scheme 1). In the presence of telomerase, the TSP is concatenated with telomeres that recruit invader probe pairs (IP and RP) to form junction structures. At such structures, a base at the 5′-end of the IP invades beyond the junction site by a single base, enabling FEN1 to initiate cyclic cleavage reactions. The reaction proceeds at 37 °C at which FEN1 retains substantial activity (Fig. S1†). The hybridization sequence of the RP was intentionally designed such that its melting temperature (37.2 °C, IDT Oligo Analyzer) with the extended TSP would be close to the reaction temperature (37 °C).17,21,22 The RP becomes less complementary to the extended TSP by a single base upon the cleavage reaction (light gray box, Scheme 1). In addition, the number of intact RPs is higher than that of cleaved RPs. These conditions facilitate the intact RP to displace the cleaved RP for the cyclic cleavage reactions.21,22 Thus, a large number of flap sequences are released and go through GO filtering for signaling. As the flap length was designed to be too short to adsorb onto GO (4 mer),17 the free flap sequences retain high fluorescence.We verified the feasibility by carrying out QUEST under varying conditions (Fig. 1). In the absence of FEN1, the signal was significantly quenched even in the presence of telomerase. In contrast, a high signal was attained by telomerase when FEN1 took part in the assay. Heat-inactivated telomerase was added to confirm that the signal essentially came from telomere elongation. As we expected, the signal was almost quenched (Fig. 1(a)).
Furthermore, in order to verify the cyclic cleavage reaction, we designed a model experiment (Fig. S2†). A TSP appended with (TTAGGG)4 (pre-concatenated TSPmini, Fig. S2(b)†) was used as a template for the FEN1-catalyzed reaction to restrict the reaction such that only a single molecule of the IP and RP–Q/F can bind to a single pre-concatenated TSPmini. The expected form of cleaved RP–Q/F, RPcut–F (Fig. S2(b)†), was prepared to be concentration-matched with TSPmini samples. Their signals were assumed as the signals for a one-to-one cleavage reaction and subsequently compared with those of the FEN1-catalyzed reaction. At all concentrations, signals of the FEN1-catalyzed reaction were much higher than those of controls in which the same concentrations of RPcut–F as those of the pre-concatenated TSPmini were included (*p < 0.005, Fig. S2(c)†). This result supports that FEN1 executes multiple cleavage reactions for a single template.
We further investigated the assay with gel electrophoresis (Fig. 1(b)). With telomerase, bands for the extended TSP (lane 2) and a complex formed between the extended TSP and invader probe (TSP/IP/RP; lane 5) were observed whereas none of these were present without telomerase or with heat-inactivated telomerase (lanes 1, 3, 4, and 6, Fig. 1(b)). To check the FEN1 reaction, we intended to use fluorescence intensities rather than bands, considering that the flap sequence is too short to (i) be detected and (ii) provide the complex (TSP/IP/RP) with enough size difference after the FEN1 reaction in gel electrophoresis. So, we used another type of RP (RP–Q/F). In RP–F/Q, the F is internally labeled, while the Q is labeled at the 5′-end (Table S2†). As FEN1 cuts out Q-labeled fragments, intense fluorescence was observed (lane 5 and inset, Fig. 1(b)). As we envisioned, FEN1 played a key role in the telomere-fueled cyclic cleavage reaction.
Optimization of QUEST
To maximize the performance of QUEST, we optimized key reaction conditions (Fig. S3 and S4†). Excess amounts of the invader probes would enhance the signal, but also increase the BG as they unnecessarily occupy vacant sites of GO to compromise its filtering efficiency. QUEST requires a sufficient amount of FEN1 to promote the cyclic cleavage reaction. In addition, an appropriate supply of dNTP is required because an excess of dNTP would inhibit target's elongation activity. Accommodating such a consideration, we chose the optimal reagent concentrations showing the highest signal-to-noise ratios (SNRs): [RP]: 100 nM, [IP]: 50 nM, [FEN1]: 0.4 U μL−1, [GO]: 0.04 mg mL−1, and [dNTP]: 250 μM (Fig. S3†). We further tried to shorten the assay time, while maintaining high sensitivity (Fig. S4†). 60 min of QUEST reaction and 10 min of GO incubation enabled the assay to exhibit good detection performance.Sensitivity of QUEST
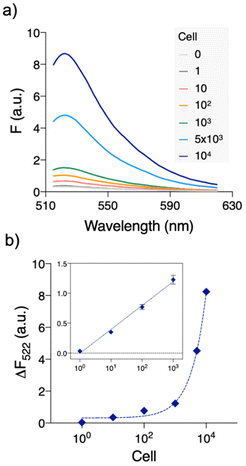
Under the optimized conditions, we evaluated the sensitivity of QUEST (Fig. 2). Serially diluted HeLa cell extracts were prepared and tested for telomerase activity. The fluorescence signals were well commensurate with the number of cells. The signals were linearly correlated with the logarithmic cell numbers over 1–1000 cells (ΔF522 = 0.400 × log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N + 0.315, R2 = 0.9937, where N is the number of cells; inset in Fig. 2(b)). Based on the 3σ/S rule for the limit of detection (LOD), where σ is the standard deviation at the lowest concentration of the linear range and S is the slope, the LOD was 2 cells (36 cells per mL). This value is lower than or comparable to those of the previous telomerase assays (Table S1†). Moreover, QUEST is straightforward. It can be executed without starting material preparation and extra hands-on steps such as washing/separation.
N + 0.315, R2 = 0.9937, where N is the number of cells; inset in Fig. 2(b)). Based on the 3σ/S rule for the limit of detection (LOD), where σ is the standard deviation at the lowest concentration of the linear range and S is the slope, the LOD was 2 cells (36 cells per mL). This value is lower than or comparable to those of the previous telomerase assays (Table S1†). Moreover, QUEST is straightforward. It can be executed without starting material preparation and extra hands-on steps such as washing/separation.
QUEST application to cell line test and inhibitor screening
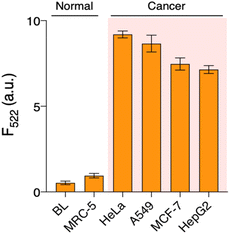
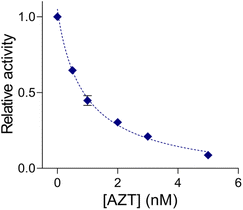
To pursue its practical application, we applied QUEST to other types of cell lines (normal: MRC-5, cancer: A549 (lung), MCF-7 (breast), and HepG2 (liver)). Their cell extracts were prepared and telomerase activities were investigated by QUEST. As shown in Fig. 3, the signal from the normal cell extract was comparable to that of blank whereas cancer cell extracts resulted in higher signals. The results were consistent with those of the standard TRAP assay, ensuring the reliability of QUEST (Fig. S5†). Also, this suggests the great potential of QUEST in cancer diagnosis.Lastly, we implemented QUEST to screen candidate inhibitors of telomerase activity (Fig. 4). We selected AZT as a model inhibitor and confirmed that it did not affect the QUEST reaction (Fig. S6†). Then, we assessed telomerase activities against different AZT concentrations, measuring the signals and converting them into relative activities (see the Experimental section). With increasing AZT concentration, relative telomerase activities decreased. The half maximal inhibitory concentration (IC50), which is the inhibitor concentration that reduces the enzyme activity to 50%, was about 0.91 nM. The result was in accordance with those from previous reports.23 The assay has the capability to screen candidate telomerase inhibitors and would be advanced to be an analytical tool for the discovery of new anticancer drugs.
Conclusions
We developed QUEST for sensitive telomerase activity detection. QUEST seamlessly combined invader signal amplification and GO-based BG filtering to take advantage of beneficial features of the key components. FEN1 has a high turnover rate and delicate junction structure recognition, conferring high sensitivity and specificity on QUEST. GO discriminates short DNA from longer ones. That is, it does not capture short flap sequences from the QUEST reaction, but captures unreacted F-labeled DNA (i.e. RP). With such a setup, QUEST assayed the telomerase activity in the HeLa cell extract down to 2 cells (36 cells per mL) within 70 min. It was able to distinguish the telomerase activities from those of other cell lines and screen candidate inhibitors against telomerase. In addition, QUEST has the following merits: (i) the entire assay procedure is streamlined. It does not need material preparation such as probe conjugation and washing and several intervening steps for reagent loading and washing/separation and (ii) probe design is simple and cost-effective. GO, as a universal quencher, expels the need for dual-labeling of the probe with F/Q.Meanwhile, we could advance the system by the following endeavors. The first is to introduce a secondary invader probe to redouble signal amplification. By designing the flap sequence to bind to the secondary invader probe to initiate further cyclic cleavage reactions, higher sensitivity would be attained. The second is to challenge the cellular imaging of telomerase activity with further optimization. GO is known to be biocompatible and stably carry single-stranded DNA and enzymes with non-covalent interactions.24–26 In particular, GO can directly enter cells via endocytosis.27,28 Thus, the QUEST assay components would be dispatched into cells by GO to initiate the cellular imaging of telomerase activity. Lastly, the fluorescence polarization (FP) mode would replace the current signal mode due to its benefits. As a ratiometric signal mode, it is tolerant to external noise, which would allow for robust signal detection and a higher SNR.29 In addition, the FP mode only requires the F for signal detection so it would enable the assay to be implemented without any Q. Thus, the assay procedure and probe design would be simplified. Being empowered by such improvements, the core detection principle of QUEST would be widely applied to develop various biosensors where target-induced concatenation precedes.
Author contributions
H. H.: data curation, formal analysis, investigation, validation, visualization, writing – original draft, and writing – review and editing; C. P.: data curation, formal analysis, visualization, writing – original draft, and writing – review and editing; C. Y. L.: conceptualization, formal analysis, methodology, project administration, supervision, writing – original draft, and writing – review and editing; J. K. A.: funding acquisition, methodology, project administration, resources, supervision, writing – original draft, and writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study has been conducted with support from the Korea Institute of Industrial Technology through “Development of fiber-based technology for reduction of hazardous substances in the air (kitech EO-23-0005)”. This work was also supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through the Crop Viruses and Pests Response Industry Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (321104-3).References
- J. Feng, W. D. Funk, S. S. Wang, S. L. Weinrich, A. A. Avilion, C. P. Chiu, R. R. Adams, E. Chang, R. C. Allsopp, J. Yu, S. Le, M. D. West, C. B. Harley, W. H. Andrews, C. W. Greider and B. Villeponteau, Science, 1995, 269, 1236–1241 CrossRef CAS PubMed
.
- G. B. Morin, Cell, 1989, 59, 521–529 CrossRef CAS PubMed
.
- E. Savoysky, K. I. Akamatsu, M. Tsuchiya and T. Yamazaki, Nucleic Acids Res., 1996, 24, 1175–1176 CrossRef CAS PubMed
.
- C. B. Harley, Nat. Rev. Cancer, 2008, 8, 167–179 CrossRef CAS PubMed
.
- E. Hiyama and K. Hiyama, Oncogene, 2002, 21, 643–649 CrossRef CAS PubMed
.
- B.-S. Herbert, A. E. Hochreiter, W. E. Wright and J. W. Shay, Nat. Protoc., 2006, 1, 1583–1590 CrossRef CAS PubMed
.
- M. Gabourdes, V. Bourgine, G. Mathis, H. Bazin and B. Alpha-Bazin, Anal. Biochem., 2004, 333, 105–113 CrossRef CAS PubMed
.
- D. Kong, Y. Jin, Y. Yin, H. Mi and H. Shen, Anal. Bioanal. Chem., 2007, 388, 699–709 CrossRef CAS PubMed
.
- X. Chen, Y. Deng, G. Cao, X. Liu, T. Gu, R. Feng, D. Huo, F. Xu and C. Hou, Anal. Chim. Acta, 2021, 1146, 61–69 CrossRef CAS PubMed
.
- Y. Guo, S. Liu, H. Yang, P. Wang and Q. Feng, Anal. Chim. Acta, 2021, 1144, 68–75 CrossRef CAS PubMed
.
- Y. Li, H. Han, Y. Wu, C. Yu, C. Ren and X. Zhang, Biosens. Bioelectron., 2019, 142, 111543 CrossRef CAS PubMed
.
- F. Ma, S. Wei, J. Leng, B. Tang and C. Zhang, Chem. Commun., 2018, 54, 2483–2486 RSC
.
- D. Wang, R. Guo, Y. Wei, Y. Zhang, X. Zhao and Z. Xu, Biosens. Bioelectron., 2018, 122, 247–253 CrossRef CAS PubMed
.
- F. Meng, H. Chai, X. Ma, Y. Tang and P. Miao, J. Mater. Chem. B, 2019, 7, 1926–1932 RSC
.
- X. Liu, F. Meng, R. Sun, K. Wang, Z. Yu and P. Miao, Chem. Commun., 2021, 57, 2629–2632 RSC
.
- C. Jiang, F. Meng, D. Mao, Y. Tang and P. Miao, ChemBioChem, 2021, 22, 1302–1306 CrossRef CAS PubMed
.
- C. Y. Lee, H. Jang, H. Kim, Y. Jung, K. S. Park and H. G. Park, Microchim. Acta, 2019, 186, 330 CrossRef PubMed
.
- J. Song, C. Y. Lee and H. G. Park, Chem. Commun., 2022, 58, 2279–2282 RSC
.
- F. Rashid, P. D. Harris, M. S. Zaher, M. A. Sobhy, L. I. Joudeh, C. Yan, H. Piwonski, S. E. Tsutakawa, I. Ivanov, J. A. Tainer, S. Habuchi and S. M. Hamdan, eLife, 2017, 6, 1–23 CrossRef PubMed
.
- M. A. Sobhy, M. Tehseen, M. Takahashi, A. Bralić, A. De Biasio and S. M. Hamdan, Comput. Struct. Biotechnol. J., 2021, 19, 4456–4471 CrossRef CAS PubMed
.
- B. Zou, Y. Ma, H. Wu and G. Zhou, Angew. Chem., Int. Ed., 2011, 50, 7395–7398 CrossRef CAS PubMed
.
- V. Lyamichev, A. L. Mast, J. G. Hall, J. R. Prudent, M. W. Kaiser, T. Takova, R. W. Kwiatkowski, T. J. Sander, M. de Arruda, D. A. Arco, B. P. Neri and M. A. D. Brow, Nat. Biotechnol., 1999, 17, 292–296 CrossRef CAS PubMed
.
- C. He, Z. Liu, Q. Wu, J. Zhao, R. Liu, B. Liu and T. Zhao, ACS Sens., 2018, 3, 757–762 CrossRef CAS PubMed
.
- K. H. Liao, Y. S. Lin, C. W. MacOsko and C. L. Haynes, ACS Appl. Mater. Interfaces, 2011, 3, 2607–2615 CrossRef CAS PubMed
.
- Y. Zhang, J. Zhang, X. Huang, X. Zhou, H. Wu and S. Guo, Small, 2012, 8, 154–159 CrossRef CAS PubMed
.
- Y. Wang, L. Tang, Z. Li, Y. Lin and J. Li, Nat. Protoc., 2014, 9, 1944–1955 CrossRef CAS PubMed
.
- L. Zhang, J. Peng, M. F. Hong, J. Q. Chen, R. P. Liang and J. D. Qiu, Analyst, 2018, 143, 2334–2341 RSC
.
- Y. Jiang, Z. Guo, M. Wang, J. Cui and P. Miao, Nanoscale, 2022, 14, 612–616 RSC
.
- C. Y. Lee, I. Degani, J. Cheong, J.-H. Lee, H.-J. Choi, J. Cheon and H. Lee, Biosens. Bioelectron., 2021, 178, 113049 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr04132h |
| This journal is © The Royal Society of Chemistry 2023 |