 Open Access Article
Open Access ArticleA light-operated integrated DNA walker–origami system beyond bridge burning†
Xiao Rui
Liu
a,
Iong Ying
Loh
a,
Winna
Siti
a,
Hon Lin
Too
ab,
Tommy
Anderson
a and
Zhisong
Wang
 *ab
*ab
aDepartment of Physics, National University of Singapore, 117542, Singapore. E-mail: phywangz@nus.edu.sg
bIntegrative Sciences and Engineering Programme, NUS Graduate School, National University of Singapore, 117542, Singapore
First published on 11th April 2023
Abstract
Integrating rationally designed DNA molecular walkers and DNA origami platforms is a promising route towards advanced nano-robotics of diverse functions. Unleashing the full potential in this direction requires DNA walker–origami systems beyond the present simplistic bridge-burning designs for automated repeatable operation and scalable nano-robotic functions. Here we report such a DNA walker–origami system integrating an advanced light-powered DNA bipedal walker and a ∼170 nm-long rod-like DNA origami platform. This light-powered walker is fully qualified as a genuine translational molecular motor, and relies entirely on pure mechanical effects that are complicated by the origami surface but must be preserved for the walker's proper operation. This is made possible by tailor-designing the origami for optimal match with the walker to best preserve its core mechanics. A new fluorescence method is combined with site-controlled motility experiments to yield distinct and reliable signals for the walker's self-directed and processive motion despite origami-complicated fluorophore emission. The resultant integrated DNA walker–origami system provides a ‘seed’ system for future development of advanced light-powered DNA nano-robots (e.g., for scalable walker-automated chemical synthesis), and also truly bio-mimicking nano-muscles powered by genuine artificial translational molecular motors.
New conceptsA new conceptual line for developing advanced nano-robotics is the ‘marriage’ between the field of DNA molecular motors and the field of DNA origami, with the former for automated nanometres-resolved motility and the latter providing a submicroscale platform of rich designability and functionalizability. A conspicuous bottleneck for this line of development is a lack of integrated DNA origami–motor systems. Such systems must involve genuine molecular motors in order to attain the capability of automatic repeatable operation or collective motor action for amplified mechanical outputs and scalable robotic functions. The accumulated research in this direction, focusing on DNA walker–origami systems by far, has yet to fully break the bottleneck. This is because these reported systems involve either directionless or ‘bridge-burning’ DNA walkers that still fall short of genuine translational molecular motors by the strictest definition. Developing the bottleneck-breaking DNA origami–motor systems faces the challenge of engineering sophisticated molecular mechanics on DNA origami. This is a new regime to explore, especially for light-powered DNA origami–motor systems as their entire operation rely on pure mechanical effects. This study demonstrates such a light-operated DNA origami-translational motor system, thus opening a door to complex DNA-based nano-robotics and related developments, e.g., active materials. |
Introduction
Integrating artificial DNA molecular walkers1 and DNA origami platforms2 is a promising strategy to develop advanced nano-robotics. The reported DNA walker–origami robotic systems cover autonomous navigation of prescriptive landscapes,3 nanoscale assembly lines,4 cargo sorting,5 muscle-like multilayer sliding nanosystems6 or linear nano-actuators,7 plasmonic nano-optics8 by controlled motion below the diffraction limit, etc. In these nano-robotic systems, both the walkers and origami platforms are rationally designed, and constructed from DNA molecules (deoxyribonucleic acids, or similar ribonucleic acids) by sequence-programed self-assembly. Such integrated DNA walker–origami systems combine the dynamic nanoscale positioning capability of molecular walkers and the rich designability of DNA origami over a larger size of tens to hundreds of nanometers, potentially enabling diverse robotic functions. A DNA walker–origami system also possesses a big capacity for functional expansion as the DNA origami surface can be site-selectively functionalized to host many mechanisms at different locations. However, the DNA origami-based operation is reported to date for DNA molecular walkers that are either directionless5,9 or gain a direction by denying the walker's access to the traversed track (managed by chemical damage,3,10,11 physical coverage,12,13 or manual sequential removal/recovery of walker-track linkages4,6–8,14–18). These directional walkers follow the so-called burning-the-bridge strategy, thus impeding repeatable automatic operation of these walker–origami systems and limiting their functional scalability and nano-robotic applications. Indeed, these bridge-burning walkers are still short of the strictest definition of genuine translational molecular motors that should be capable of automatic self-directed motion on an intact and fully accessible track (thus allowing repeated robotic operation by motor reloading or collective action of multiple motors on an origami-based track for amplified mechanical outputs).Advanced non-bridge-burning DNA walkers that are qualified as genuine translational molecular motors have been reported.19–31 Their integration with DNA origami, though crucial for advancing DNA-based nano-robotics, is not achieved yet. The reported bridge-burning DNA walker–origami systems rely on creating or introducing new DNA species on a track to ‘force’ a directional cascade. Such ‘brutal’ direction-rectifying mechanisms remain largely robust on origami. A non-bridge-burning DNA walker involves no track-modifying new species, and its direction on an invariable track arises from delicate molecular mechanical effects23,26,32–34 that can be complicated by an origami surface. Such an advanced walker often gains a net direction by mechanically amplifying a local asymmetry of a periodic track. This mechanical amplification ensures not only a walker's direction but also intra-walker coordination22,24 necessary for the walker's processive on-track motion, typically through fine-tuned free-energy gaps22,23,35 between walker-track binding states. Such a sophisticated mechanics-centred synergic rectification1,22,34 is sensitive to the track's small asymmetric features and the walker-track size match.23,24 It is non trivial to adapt the size-sensitive synergic rectification to a two-dimensional DNA origami surface from a one-dimensional straight track along a single DNA duplex or a single DNA strand (whence all the reported non-bridge-burning DNA walkers19–31 are developed; straight within a walker's size or a few times longer). This is because the origami surface has stronger electrostatic repulsion for a DNA walker, blocks half of its circumferential freedom on the track, and often does not allow the track's perfect straight longitudinal arrangement despite remarkable design freedom for the origami lattice. Therefore, the synergic mechanics and free-energy gaps critical for a non-bridge-burning walker become far more complicated on a DNA origami surface than on a single-duplex track. Enabling the origami-based synergic motor mechanics represents a new regime for engineering sophisticated molecular mechanics, especially for advanced light-operated DNA walker–origami systems that rely entirely on pure mechanical effects. Experimental exploration of this new molecular mechanics regime involves many technical details of a complex walker–origami system, and the challenging costly walker–origami co-optimization concerning assembly procedures, design parameters, operation conditions, etc.
In this study, we report a light-operated integrated DNA walker–origami system with its light-driven walker fully qualified as a genuine translational molecular motor. This DNA motor–origami system goes beyond bridge-burning designs, and operates automatically by light-activated mechanical effects. To our best knowledge, this is the first successful integration between DNA origami and genuine artificial DNA molecular motors. This is made possible by tailor-designing a new origami (rod-shaped, ∼170 nm-long) specifically for the walker, plus systematic optimization of the integrated walker-track system to best preserve the walker's core mechanics. A new fluorescence method is combined with site-controlled motility experiments to yield distinct and reliable signals for the walker's directional and processive motion despite complications of fluorophore emission from the origami surface. The resultant integrated DNA walker–origami system provides a ‘seed’ system for future development of advanced light-powered DNA nano-robots capable of repeated operation and diverse functions (e.g., for scalable walker-automated chemical synthesis36,37), and also bio-mimicking nano-muscles powered by genuine artificial translational molecular motors.
Results and discussion
The DNA walker and origami
The DNA walker is a bipedal translational molecular motor previously demonstrated21,23,24 for light-powered self-directed motion along a linear duplex track. As shown in Fig. 1(A), the motor has two identical single-stranded legs that each consist of a 20 nt-long D2 segment tethered with 9 light-responsive azo-moieties and a 5 nt-long non-azo D1 segment. A shown in Fig. 1(B), the track is a periodic array of identical binding sites that are each made of two nearby single-stranded overhangs D2* and D1* with their nucleotide sequences complementary to those of the leg's D2 and D1 segment. The D2–D2* duplex, which is the major binding component between the motor's legs and a binding site, is destabilized by ultraviolet (UV) light irradiation and stabilized by visible light irradiation. Under alternating UV and visible light, the motor walks hand-over-hand processively on the periodic track towards its plus end (defined by a horizontal arrow pointing from D1* to the longer D2* within a binding site). Fig. 1(B) shows the motor's walking scheme as established by previous studies.21,23,24 To gain a unique direction along a periodic track without ‘burning’ it, the motor relies on a mechanics-mediated symmetry breaking mechanism22–24 that is inspired by biological molecular motors35,38–40 and sensitive to the intra-site D1*–D2* displacement (i.e., the small local asymmetry critical for this motor) and to the size match between the motor and the track's inter-site distance. When the overhangs are supported by a single-duplex track, previous studies23,24 find a reasonably good size match as 20 bp for the motor's inter-pedal duplex bridge (as compared to 25 bp and 30 bp variations) versus 15 bp for the intra-site D1*–D2* separation and 75 bp for the inter-site D2*–D1* separation (with a total of 90 bp for the track's binding site period).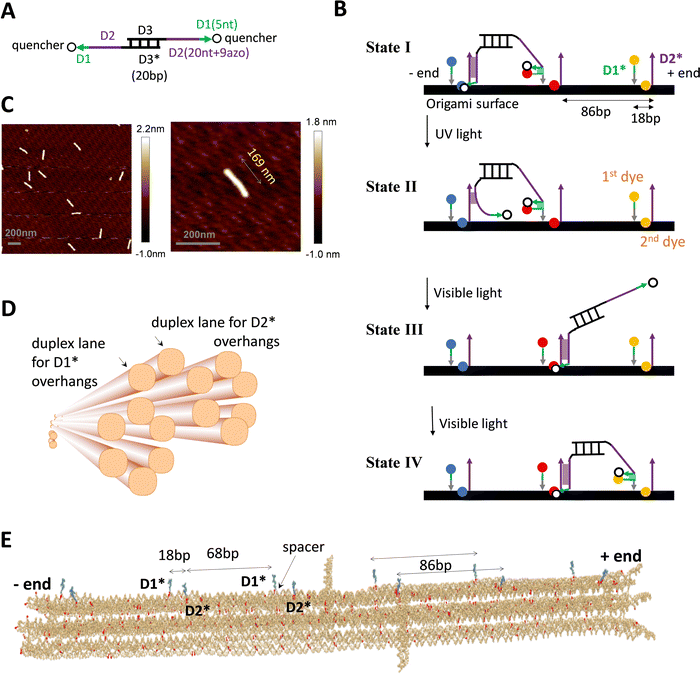 | ||
| Fig. 1 DNA motor and origami. (A) and (B) The light-powered DNA motor (panel A) and its walking scheme (panel B, adapted from ref. 21; ‘bp’ for base pair, ‘nt’ for nucleotide; arrows for each strand pointing from 5′ and 3′; asterisks in D1* or D2* indicating nucleotide sequences complementary to that of D1 or D2). The bipedal motor carries azobenzene moieties in the legs for optical driving (in the 20 nt D2 segment, which is linked to the central duplex bridge via a 4 nt spacer for flexibility). The UV irradiation induces trans-to-cis photoisomerization of the azobenzenes to open the D2–D2* duplex for rear leg dissociation off the track while the front leg remains on the track due to the non-azo D1–D1* duplex (state II in panel B, rear or front leg relative to the motor's direction towards the track's plus end as indicated). The visible light induces the reverse cis-to-trans photoisomerization for the front leg's forward migration within a bi-overhang binding site (consisting of neighbouring D1* and D2* overhangs), and further for the forward binding of the dissociated leg (state III and IV). For the purpose of fluorescence detection, the motor carries two quenchers (BHQ2, see panel A), and the origami-based track is labelled by three types of dyes (TYE, ATTO488, ATTO647, represented by spheres of different colours in panel B, and tethered on D1* overhang or below D2* overhang via a nearby staple strand of the origami, see Fig. S1D, ESI†). (C) AFM images of the rod-shaped origami, yielding an average length of 169 nm for the origami (with a standard deviation of 5 nm). (D) The origami design, as shown by a schematic illustration (generated from Cadnano software) highlighting the origami's cross section and the two duplex lanes hosting D1* and D2* overhangs (i.e., track for the motor). (E) Realistic structure of the origami with six bi-overhang binding sites (generated from a simulation using oxView server). The periodic arrangement of D1* and D2* overhangs is shown; each D1* has a spacer at the bottom as indicated. | ||
The single-stranded D1* and D2* overhangs may be created on a DNA origami surface as protruding extension of staple strands that hybridize with a long scaffold strand to form the origami's main body. To support the motor's translational motion, these overhangs must be arranged in a linear periodic array on the origami lattice, ideally along a single duplex lane. However, the choice of location for these protruding overhangs is constrained by the two specific inter-overhang separations (15 bp and 75 bp) matching the size-sensitive motor, and further constrained by the origami lattice design that is limited by the origami's shape, stability, and fabrication quality. Under these constraints, replicating the motor's previous near perfect linear single-duplex track on a DNA origami is difficult, if not possible.
To match the light-powered DNA motor, a new rod-shaped DNA origami is specially designed and fabricated in this study (Fig. 1(C) and (D)). This origami accommodates a quasi-linear zigzag array of D1* and D2* overhangs along two neighbouring duplex lanes to best preserve the motor-track size match. Besides, the two track-hosting duplex lanes are above the origami's other duplex lanes to minimize their possible interference with the motor's operation. Specifically, the rod-like origami is made of 14 parallel helical lanes that are arranged in three overlapped hexagonal bundles (Fig. 1(D)). The two top duplex lanes in the central hexagonal bundle accommodate D2* and D1* overhangs, respectively. The D2* array and the D1* array both have the same 86 bp period that is close to the original duplex track period (90 bp). The adjacent D1* and D2* that form a binding site are now on different duplex lanes and are displaced by 18 bp along the longitudinal axis of the origami rod. The resultant intra-site D1*–D2* separation is larger than the original value of 15 bp for the previous single-duplex track. As a compensation, each staple strand for the D1* overhang contains an extra flexible spacer between the D1* segment and the scaffold-hybridizing segment, i.e., near the bottom of each D1* overhang (Fig. 1(E)). The spacer allows the adjacent D1* and D2* overhangs to get closer than the 18 bp gap for sake of the downhill D1*-to-D2* migration by the motor's leg, i.e., the transition from state II to state III in Fig. 1(B), which is crucial for the motor's plus-end-directed motion.
The detailed design and all the staple sequences for the origami are presented in Fig. S1 and Table S1 (ESI†). The origami's fabrication is elaborated in Methods. Fig. 1(C) shows the atomic force microscopy (AFM) images of the fabricated origami rod, which has a length of 169 nm ± 5 nm (see a typical length profile in Fig. S2, ESI† and length estimation in Methods). The fabricated origami has a reasonably good quality as reflected from the low length fluctuation (<3%), and also from the rather uniform shape of individual rods in the AFM images (Fig. 1(C)). Six binding sites are created in total on the ∼170 nm long origami rod, with Fig. 1(E) showing the realistic structure of the track-carrying origami generated from a simulation using the oxDNA server.41 The full 6-site track is ∼152 nm long from the first to last overhang along the longitudinal axis of the origami rod. This 6-site track and its truncated 5-site and 3-site variations are fabricated and used in this study for different scenarios of motility detection (see Fig. 2(A) for the different tracks).
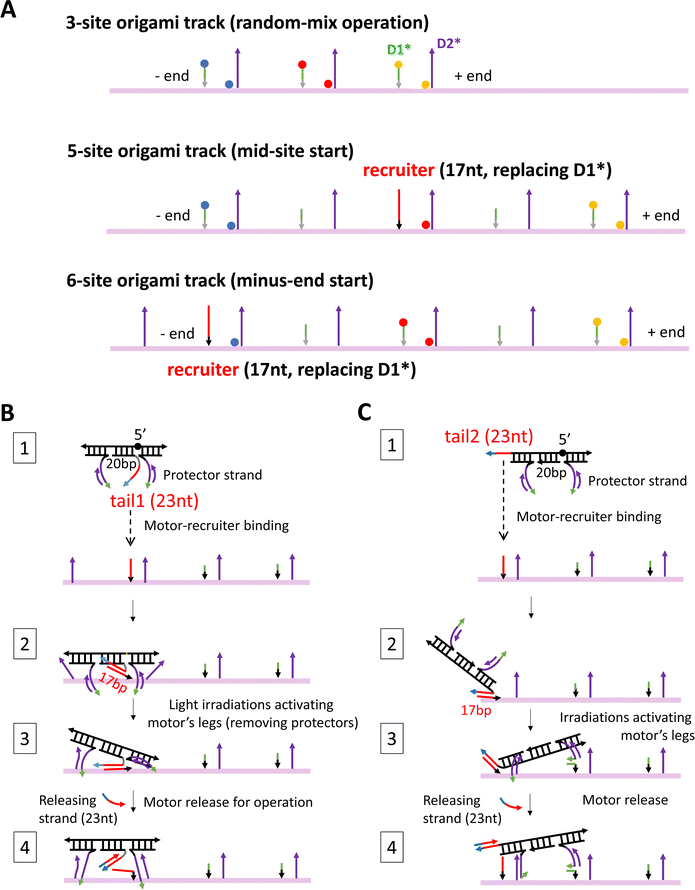 | ||
| Fig. 2 Three origami tracks (A) and two methods for site-specific motor introduction to the origami (B) and (C). The three tracks in panel A are truncated versions of the full 6-site track in Fig. 1(E). The three tracks are labelled with TYE dyes always at the middle site, but either with ATTO488 at the minus-end site and ATTO647 at the plus-end site (labelling scheme 1) or with the same two types of dyes at exchanged positions (labelling scheme 2). The average between the two labelling schemes yields the fluorescence signals in Fig. 3–5. The 5-site and 6-site tracks in panel A have one D1* replaced by a recruiter for site-specific motor introduction. For the same purpose, the motor is modified into a four-strand construction with a tail that can hybridize with a track's recruiter (panels B and C, state 1). For a motor with both legs covered by protector strands, the tail-recruiter hybridization ensures the site-selective motor–origami binding (state 2). The protector strands are removed by alternating UV-visible light irradiations, thus activating the motor's legs for binding with nearby overhang sites (state 3). The 23 nt-long tail has not only a recruiter-binding domain (17 nt long, highlighted by red colour) but also a 6 nt single-stranded toehold domain (highlighted in cyan) for the final motor release via a toehold-mediated strand displacement (state 4, produced by adding a 23 nt-long releasing strand that is complementary to the entire tail). The tail is introduced to the motor at two positions, leading to two methods for site-specific motor placement to the origami tracks. States 2–4 in panels B and C schematically illustrate the likely pathways for the two placement methods. The four-strand motor still has two identical legs that are now connected through an inverted DNA strand with a reverse a 5′–5′ linkage in the middle (highlighted by black sphere in state 1, panels B and C). | ||
For fluorescence detection of the motor's motion on the origami, a track is labelled with three types of dyes (TYE, ATTO488, ATTO647) at different sites, and the motor is labelled with two quenchers (BHQ2). The labelling schemes for the motor and tracks are shown in Fig. 1(A), (B) and 2(A). To fully report the motor's presence at a bi-overhang site, two identical dyes are used per site (represented by spheres of same colour in Fig. 1(B)), with the first dye tethered to the tip of D1* overhang and the second dye placed near the bottom of D2* overhang (tethered to a nearby staple strand without a protruding overhang, see Fig. S1D, ESI†). As shown in Fig. 1(B), this labelling scheme allows highly efficient contact quenching when the motor's quencher-labelled leg hybridizes with either D1* or D2* overhang.
A new fluorescence method for detecting light-powered motor motility on DNA origami
Ensemble fluorescence experiments based on site-specific dye/quencher labelling is a convenient method for detecting a DNA motor's directional motility, especially when compared to costly single-molecule methods.9,10,15,29 The fluorescence method is complicated by a DNA origami surface and particularly for the light-powered motor in this study. This is because the fluorescence of dyes on an origami-based track is affected not only by the motor-carried quenchers, but also by nucleotide-induced quenching42 from the origami substrate and further by the UV and visible light irradiations for the motor's operation (e.g., photobleaching25,27,28,30 as commonly observed for light-powered DNA molecular motors). Compared to the motor's previous single-duplex track, the origami has much higher nucleotide density, likely resulting in serious nucleotide-induced quenching. This is confirmed by a track-only control experiment in which the origami-based 5-site track with dye exchange between its two ends is subject to a series of alternating UV-visible light irradiations designed for the motor's operation. The fluorescence data are collected from the two differently labelled tracks in two separate experiments, but using the same amount of track samples at the same concentration. The fluorescence at the start of the light operation has a similar level between the two experiments for TYE dye that labels the track's middle site, and also for ATTO647 dye that is exchanged between the plus-end and minus-end sites of the track (Fig. S3, ESI†). However, the fluorescence of ATTO488 dye, when its location is changed from the minus-end to plus-end site, rises by ∼47%. This dramatic pre-operation fluorescence difference is caused purely by the dye's different locations on the origami surface as the minus-end and plus-end sites have identical overhangs. This indicates existence of a serious origami-induced quenching that is site-sensitive and dye-dependent. For the same track-only experiments (Fig. S3, ESI†), the fluorescence after the first UV irradiation shows minor photobleaching (i.e., monotonic decrease with more UV-visible light cycles) for ATTO647 and TYE regardless of their location, but a slight monotonic increase for ATTO488 at the plus-end site and even a non-monotonic change for the same dye at the minus-end site. The abnormal ATTO488 behaviors imply a dynamic interplay, again being dye-dependent and site-sensitive, between the light operation and the origami-induced quenching effect.In this study, we introduce a new fluorescence method that allows reliable conclusions on the motor's directional motion despite the origami-related complications. Specifically, we swap dyes between the minus-end and plus-end sites of an origami-based track and fabricate two sets of origami for the two labelling schemes, then conduct separate fluorescence experiments for the motor's operation using the two origami sets by the same amount at the same concentration. Finally, we do site-specific average between the two experiments to remove dye dependence. Namely, we average the fluorescence collected from the minus-end dyes in both experiments, and from the plus-end dyes in both experiments. This dual-experiment average is between two different types of dyes due to the dye swap, and is done not using the dye-dependent and site-sensitive absolute fluorescence intensity from each experiment but using each dye type's fluorescence normalized to its own value at the start of the motor's light operation. This site-specific average of the normalized fluorescence is the final operation-dependent fluorescence signal that is used to conclude whether the motor's direction of motion is towards the track's plus end or minus end. The resultant minus-end signal and plus-end signal both have a unity value at the start of the motor's operation, then diverge with a successively widening gap by cyclic light operation if the motor moves directionally on the track. This gap may be quantified as the minus-end signal minus the plus-end signal. If a motor is plus-end-directed (or minus-end-directed), this –/+ signal gap should be positive and increase (or negative and decrease) with more cycles of light operation despite abnormal behaviours of individual dyes (e.g., ATTO488 in Fig. S3, ESI†).
The conclusion on a motor's direction from the −/+ signal gap is not affected by the dye choice because this gap is a fluorescence difference for the same combination of two dye types (swapped between a track's plus-end and minus-end sites). The plus-end or minus-end signal is still affected by the photobleaching but on the equal footing, resulting in cancellation in the signal gap. Through the fluorescence normalization to its own initial value per site (before the dual-experiment average), the −/+ signal gap largely removes influence of the site-sensitive origami-based quenching. Therefore, the −/+ signal gap is a reliable indicator of on-origami motional direction of the light-powered DNA motor.
In this study, the new fluorescence method is further combined with different site-controlled motility experiments for distinct signals of the motor's on-origami directional motion. These motility experiments have different motor distributions before the start of light operation: the motor is initially distributed randomly on a 3-site track (by simple motor–origami mixing), or placed at the middle site of a 5-site track, or placed at the minus end of a 6-site track (by a site-selective procedure). As shown in Fig. 2(A), the three origami-based tracks are fabricated as truncated versions of the full 6-site track in Fig. 1(E). For sake of consistence, the three tracks have the same two dye types swapped between minus-end and plus-end sites (ATTO647, ATTO488), and the same third dye type for an intermediate site (TYE). The experimental methods for preparing the different motor distributions are schematically illustrated in Fig. 2(B) and (C), and explained in later sections and Methods. For sake of methodological optimization (especially for site-selective motor placement), three motor versions are fabricated and used for the motility experiments: one is the motor's original bi-strand design (see Fig. 1(A), for random-mix experiments), the other two are adapted four-strand designs (Fig. 2(B) and (C), for site-controlled experiments).
Motility data from random mix of the motor and origami track
Fig. 3 shows typical fluorescence signals from the dual-experiment average for the motor's operation on the truncated 3-site track on the origami (i.e., 2nd to 4th sites from the minus end in Fig. 1(E); with pre-average fluorescence given in Fig. S4A and C, ESI†). The data are collected from light operation of motor-track binding complexes prepared by mixing the fabricated motor and origami track, during which the motor randomly binds the track's three sites (as indicated by the decreasing fluorescence for all the three sites during the motor–origami mixing, see Fig. S4, ESI†). Under the cyclic light operation, the motor's averaged signals for the minus and plus end indeed diverge and develop a clear gap. Although either signal is not monotonic with increasing operational time (Fig. 3(A) and (C); reminding of abnormal fluorescence behaviors in the track-only control in Fig. S3, ESI†), the −/+ signal gap shows a smooth monotonic increase indicating the motor's plus-end directed motion (Fig. 3(B) and (D)). This confirms the ability of the new fluorescence method to filter out origami-related complications. The final magnitude of the −/+ signal gap at the end of 9 alternating UV-visible light cycles differs for different values of the origami–motor molar ratio but is all positive, indicating a robust direction of the motor on the origami track (Fig. 3(E)).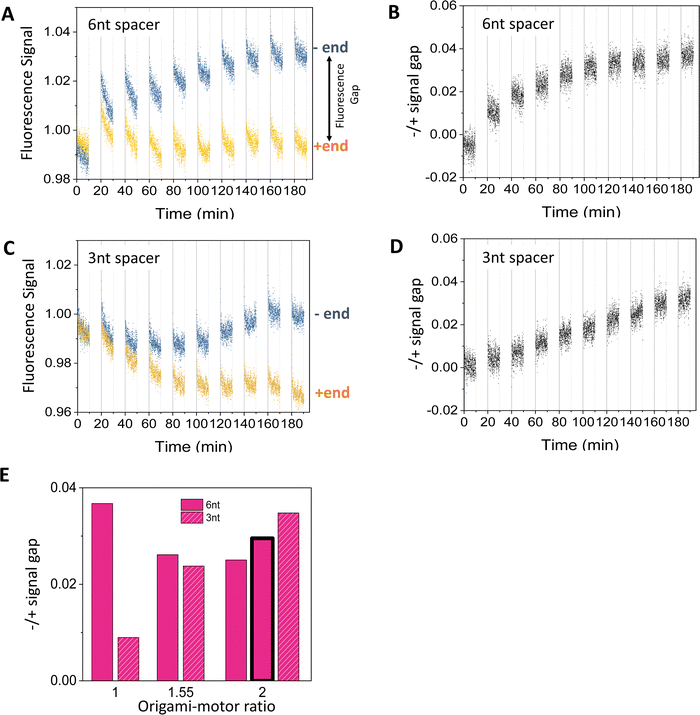 | ||
| Fig. 3 Motility experiments of the motor on a 3-site origami track started from a random motor–origami mix (see track in Fig. 2(A), upper plot). (A)–(D) Fluorescence signal versus time for different spacer lengths below D1* overhangs (6 nt for panels (A and B), and 3 nt for panels (C and D). Along the time axis, the durations with data are the visible light irradiations and the gaps are the UV irradiations during which the fluorescence data are not collected. Each UV or visible light lasts 10 minutes. The signal gap between the minus and plus ends in panels B and D are extracted from the data in panels A and C, respectively (with the gap marked in panel A). The data are obtained at an origami–motor ratio of 1 for panels A and B and 2 for panels C and D, all at 8 nM origami concentration. (E) The signal gap at the end of 9 UV-visible light cycles versus the origami–motor ratio. The data in all the panels of this figure are obtained using the motor of bi-strand construction (as shown in Fig. 1(A)), except for the data represented by the bar with bold outline in panel E that are obtained using the motor of four-strand construction (as shown in Fig. 2(B), state 1) on the 3-site track with the 6 nt spacer below D1*. The similar signal gaps from the two motor versions indicate that they both work well on the origami. | ||
The −/+ signal gap shows different features when the flexible spacer below the D1* is 3 nt or 6 nt long. The gap for the 3 nt spacer keeps rising linearly over the 9 UV-visible light cycles (Fig. 3(D)), whilst the gap for the 6 nt spacer rises first and saturates after the 6th cycle (Fig. 3(C)). The final gap after the 9 cycles is roughly comparable between the 3 nt and 6 nt spacers for origami–motor ratio of 1.55 and 2, but is apparently bigger for the 6 nt spacer for equal origami–motor ratio (Fig. 3(E)). This suggests that both spacer lengths are ok to facilitate the motor's directional motion on the origami rod though the longer 6 nt spacer is slightly better. Nevertheless, entirely deleting the spacer results in near zero gap (Fig. S5, ESI†), indicating necessity of a finite spacer below the short D1* overhang for the motor's on-origami operation.
The fluorescence method in this study differs from a control-based method24–28,30 in previous studies of DNA motors on short single-duplex tracks. For these minimal DNA tracks labelled with fluorescent dyes, the quenching is dominated by the motor-carried quenchers, and the track-only control typically exhibits a monotonic fluorescence decrease (often minor) due to photobleaching even for light-powered DNA motors24,25,27,28,30 (including the present one, see ref. 24). A dye's fluorescence from an operated motor-track mix divided by the fluorescence from the track-only control largely removes the influence of photobleaching and dye dependence, and allows a reasonable estimation of the motor's on-track population. This control-calibrated signal typically shows monotonic increase (or decrease) for a track's minus end (or plus end) for a plus-end-directed motor. The control-based method is not applicable to the present study considering the non-monotonic and even abnormal rising fluorescence from the track-only control (Fig. S3, ESI†). The new method is control-free as the final −/+ signal gap is determined from operation experiments alone. The dye swap further removes the influence of dye choice from the −/+ signal gap. Therefore, the new fluorescence method likely provides a general method for reliable direction detection for complex DNA walker–origami systems, especially light-powered ones. Nevertheless, we should point out that this method likely underestimates on-origami translocation of the present motor as the serious pre-existent origami-induced quenching competes with the motor-carried quenchers to reduce a dye's effectiveness for reporting the motor's motion. We also note that the previous control-based fluorescence method without the dye swap might still work for DNA walker–origami systems powered not by light but by other means (e.g., chemical fuels, hence free of complicated dynamic interplay from alternating UV-visible light operations).
The necessity of dye swap, which is crucial for the new fluorescence method, can be further clarified by examining emission patterns of individual dyes from operation experiments. As an example, Fig. S4A and C (ESI†) presents the normalized fluorescence for each dye that is used to calculate the swap-based average fluorescence signals in Fig. 3(A) and (B). The normalized fluorescence for a minus-end dye or a plus-end dye is still affected by the operation-induced photobleaching and its dynamic interplay with the origami-induced quenching, and may have abnormal temporal patterns. For example, a slight fluorescence decrease for a minus-end dye is possible even for this plus-end-directed motor due to the photobleaching effect, as shown in Fig. S4C (ESI†). A slight abnormal fluorescence increase for a plus-end dye is possible too as shown in Fig. S4C (ESI†). These dye behaviours prevent unambiguous determination of the motor's direction. Furthermore, the opposite fluorescence trends for the minus and plus ends emerge for the same motor by swapping the dyes between the two sites (see Fig. S4A versus Fig. S4C, ESI†), indicating a dye dependence. To remove this dye dependence, we average the normalized fluorescence from the two minus-end dyes and also average the normalized fluorescence from the two plus-end dyes. The two averaged signals, one for the track's minus end and one for the plus end as shown in Fig. 3(A), are from the same two dyes due to the dye swap. Hence the gap between the two averaged signals, i.e., the −/+ signal gap, is independent of the dye choice and reflects the motor's direction reliably. If the −/+ signal gap is above zero and accumulates over the cycles of light irradiations, the motor moves from the minus end to the plus end. Such a neat fluorescence pattern is observed throughout this study, with two examples shown in Fig. 3(B) and (D). Besides, the signals from the swap-based average clearly rises during each UV light and drops during each visible light (Fig. 3(A) and (C)). This periodic rise-drop pattern matches the motor's response to the light operation, namely UV-induced leg dissociation (e.g., state I to II in Fig. 1(B)) and the visible-light-restored leg binding (state III to IV in Fig. 1(B)). Hence these signals originate unambiguously from the motor's light-powered motility on the origami.
Motility data for mid-site start of the motor
The motor's plus-end-directed motion is further confirmed by fluorescence experiments on the 5-site track (see Fig. 2(A); i.e., 2nd to 6th sites from minus end in Fig. 1(E)). As shown in Fig. 2(B) and (C), the motor is initially prepared with its two legs covered by protector strands and introduced to the middle site through a recruiter overhang, then activated for leg-track binding by peeling off the protectors via alternating UV and visible light irradiations, and finally freed from the recruiter by a releasing strand for light-powered motion. The typical fluorescence signals for the mid-site-started motor under the light operation are shown in Fig. 4(A) and (B) (again from the dual-experiment average for plus-end or minus-end sites under swapped dye labelling, plus similar average for the mid-site though for the same dye). Over 9 cycles of light operation, the signals for the mid-site, plus end and minus end change in apparently different patterns. While the mid-site signal rises monotonically, the plus-end signal drops monotonically from unity, and the minus-end signal remains close to unity. The same patterns for the three sites are observed for two different procedures to introduce the motor to the recruiter (Fig. 4(A) and (B)). The patterns for the three sites altogether suggest that the motor leaves the middle site and moves forward to the plus end but not backward to the minus end. These data provide a compelling evidence for unidirectional motion of the motor since this bipedal motor, with identical legs and initially introduced to the mid-site of the 5-site track, has an equal number of binding sites on both sides, thus would have virtually equal chance to move to the plus or minus end along the periodic track if the motor were directionless. It is interesting to note that a bridge-burning DNA walker, if started from the middle of a linear periodic track with identical binding sites, often has equal chance to move either forward or backward. Therefore, the mid-site start experiment highlights the difference between bridge-burning walkers and the present motor: the former have an adventitious direction and the latter an inherent direction when both are placed on periodic tracks.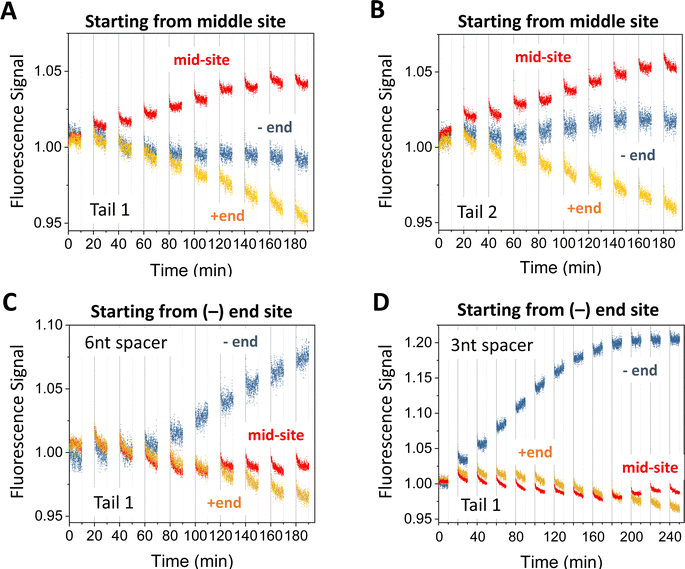 | ||
| Fig. 4 Motility experiments of the motor started from the middle of a 5-site origami track (A) and (B) or from the minus end of a 6-site track (C) and (D). The two tracks are illustrated in Fig. 2(A) (middle and lower plots). The UV-visible light operation is the same as for Fig. 3. The data in panels A and B are obtained under the same condition but using different tails for site-specific motor introduction (as shown in Fig. 2(B) and (C)). The data in panels C and D are obtained for Tail 1 for different spacer lengths below D1* (as indicated; 6 nt spacer for panels A and B). All the data in this figure are obtained using protector 1 at 8 nM origami concentration (for origami–motor ratio 2 for panels A–C and 1 for panel D). | ||
Motility data for minus-end start of the motor
When a similar preparation procedure is used to start the motor from the minus end of the 6-site track (Fig. 2(A)), the fluorescence signals from the dual-experiment average for the minus end, plus end, and a middle site show new patterns that are distinctly different from those of the mid-site start experiments despite the same dye positions on the 5-site or 6-site track for the two types of site-specific experiments (see Fig. 2(A), middle plot versus bottom plot). The plus-end signal still drops but to a lesser extent for the same number of operation cycles (see Fig. 4(C)versusFig. 4(A)). The minus-end signal changes from the flat pattern into a relatively big monotonic increase (Fig. 4(C) and (D)). The new patterns for the plus and minus ends suggest that the motor leaves the minus end and arrives at the plus end (by more consecutive steps than in the mid-site start experiments, hence less drop of the plus-end signal). This again confirms the motor's direction from the minus to plus end.The mid-site signal also changes from the monotonic increase for the mid-site start to an initial drop followed by a recovering rise for the minus-end start. This V-shaped drop-rise pattern becomes clearer when the number of operation cycles is increased from 9 to 12 (from Fig. 4(C)–(D); for the same UV duration and visible light duration per cycle). This V-shaped pattern is rather common for the minus-end start experiments for multiple variations of the preparation procedure (Fig. 5). Notably, the V-shaped pattern is more apparent when the −/+ signal gap is bigger (see Fig. 5(A) (C)–(E)), and disappears for small −/+ signal gaps (Fig. 5(B) (F)). The drop-rise pattern of the mid-site signal plus its positive correlation with the motor's directional signal (i.e., the −/+ signal gap) suggests that the motor starting from the minus end arrives at the mid-site first (hence initial drop of its signal), then passes it towards the plus end (hence later recovery of the mid-site signal). In other words, this is a processive motor that translocates itself along the 6-site track from the minus end to the plus end through the middle site. Indeed, the motor in the minus-end start experiments makes three or four consecutive steps (∼29 nm each) to produce the plus-end signal drop shown in Fig. 4(C) and (D) (three or four steps depending on the motor introduction schemes as shown in Fig. 2(B) and (C)). This finding of motor processivity is consistent with an early experimental finding23,24 that the motor possesses two directional biases, i.e., selective rear leg dissociation under UV (from state I to II in Fig. 1(B)), and forward binding of the dissociated leg under visible light (state III to IV). The two biases combine to ensure43,44 the motor's inter-leg coordination for processive hand-over-hand on-track walking.
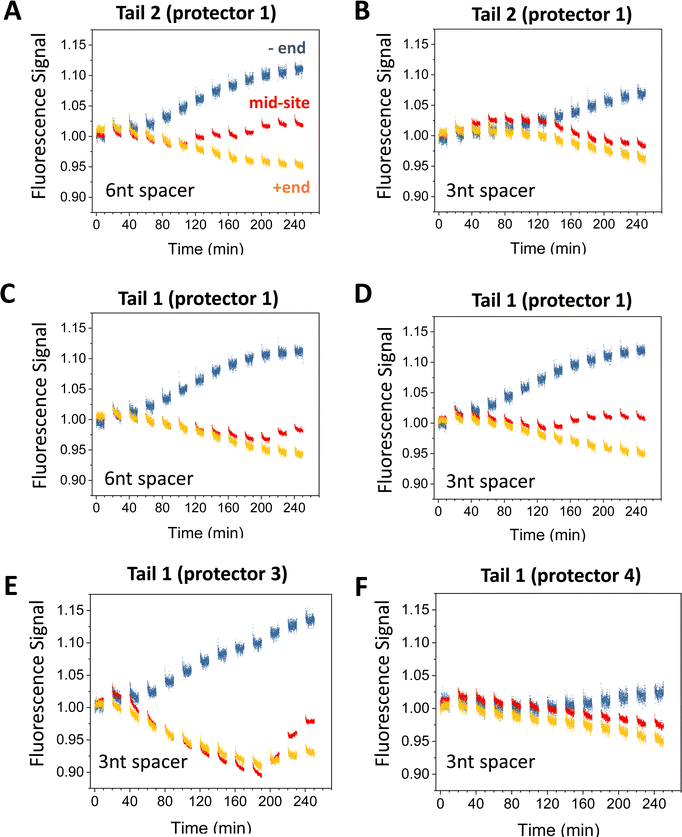 | ||
| Fig. 5 Motility experiments of the motor started from the minus end of a 6-site track under different combinations of tail, protector, and spacer length under D1* (as indicated). The track is illustrated in Fig. 2(A) (lower plot). All the data in this figure are obtained at 8 nM origami concentration for origami–motor ratio 2. | ||
Origami-based genuine translational molecular motor
The random-mix experiments and both sets of site-specific experiments altogether demonstrate an origami-based controlled nanoscale motion that is qualified as the action of genuine translational molecular motors by the strictest standard. Following ref. 21, a molecular motor should be able to generate a ‘directed’ non-equilibrium distribution of itself from the otherwise uniform distribution on a periodic track by the pure motor operation without changing the track – in a way like a macroscopic car running on a road. Indeed, the DNA walker–origami system in this study has a distinct thermodynamic feature that it possesses the same intrinsic equilibrium before and after the motor operation due to the unchanged system identity, but generates a truly nonequilibrium distribution by the self-directed motor operation. The previously reported DNA walker–origami systems gain a directional motion by changing the track chemically or physically (so-called bridge-burning). The system change results in a new thermodynamic equilibrium for the walker's on-track distribution to steer a direction for the walker. This is like digging the road in front of a car for its weight-induced sliding down the slope – a scenario short of the strictest motor definition.This study thus crosses an important scientific threshold for DNA origami-based nano-robotics. This scientific progress delivers new technological capabilities. Due to the thermodynamically less advanced motion control, the bridge-burning DNA walker–origami systems have serious technological limitations for nano-robotic applications, e.g., lack of repeatable automatic operation. Integration of DNA origami and genuine DNA translational molecular motors is necessary to overcome these limitations for advanced DNA nano-robotics, but faces the challenge of engineering sophisticated mechanics-centered control of molecular motion on DNA origami. This bottleneck is now broken by this study.
Specifically, the key for the present motor's control of direction is to adjust the motor's size (e.g., central duplex bridge) to fine-tune the free-energy hierarchy of the motor's inter-site binding states towards an asymmetric ground state (state I in Fig. 1(B)), in which the bipedal motor has one leg bound with the D2* overhang of a site and the other leg bound with the D1* of a front site. Thus, only the rear leg is selectively dissociated by the UV light (state II in Fig. 1(B)), though the two identical legs have equal chance for UV-induced trans-to-cis photoisomerization for their azo-carrying D2 segments. Under the visible light, both legs again have equal chance for cis-to-trans photoisomerization to recover their hybridization ability. When the recovery is earlier for the front leg than the dissociated leg, the front leg undergoes a downhill intra-site migration from the D1* to D2* overhang to bias the dissociated leg for its forward binding to the front site (state III to state IV in Fig. 1(B)). Then this round of UV/visible light irradiations produces a full forward step for the motor. When the recovery of hybridization ability is earlier for the dissociated leg, it binds backward before the front leg's migration, resulting in a futile step. In both scenarios, the asymmetric inter-site state is resumed with the motor's center of mass either displaced forward or recovered, but not displaced backward (hence a ratchet effect). The resumed asymmetric state makes the motor ready for a new forward-stepping attempt when another round of UV-visible light is applied. The same stepping cycle will occur between any three identical bi-overhang sites as shown in Fig. 1(B), resulting in the motor's processive steps on any longer track towards the same direction (i.e., plus end of the track). As found in ref. 24, the motor under the visible light actually has more than 50% chance for forward binding despite the equal chance of visible light-induced photoisomerization for both legs. This is because the front leg's intra-site migration is local (over a small distance of ∼18 bp) and faster than the other leg's inter-site backward binding (over a distance of ∼68 bp). Although quantifying the binding bias is not possible in this study (due to the control-free fluorescence method that loses the motor's on-track population), there are signs of suppressed backward binding, e.g., from the minus-end start experiments (Fig. 4(C) and (D)). The motor's backward binding to the starting site (minus end) is signaled by the fluorescence drop during a visible light immediate after a UV-induced fluorescence rise. However, the minus-end fluorescence in Fig. 4(C) and (D) shows minimal drop, and sometimes a flat or even rising pattern.
In this study, we tailor-design a new DNA origami for the optimal motor-track size match from the previous studies.23,24 This largely preserves the mechanics-centered control of the motor's motion as reflected from the origami-based motility experiments. In particular, the random-mix experiments start from the equilibrated motor distribution on the 3-site origami track (prepared by long incubation of motor–origami mix), but result in a non-equilibrium distribution with surplus motor accumulation at the plus-end site by pure light operation of the motor–origami system. This is a direct experimental evidence for the present DNA walker–origami system meeting the strictest standard of genuine molecular motors.
Site-specific motor introduction to DNA origami
The site-specific introduction is reasonably successful in this study as suggested by the apparently more fluorescence drop at the desired site than at other dye-labelled sites after the motor placement procedure is completed. For an example of minus-end motor placement (i.e., the same experiment as for Fig. 4(D)), Fig. S6 (ESI†) shows the fluorescence data throughout the placement procedure (plus raw data from the ensuing operation before the dual-experiment average). The initial motor-recruiter binding brings the motor selectively to the minus-end recruiting site as reflected by the obvious fluorescence quenching at this site but minimal fluorescence change at the plus-end site and mid-site (Fig. S6A and C, ESI†). This fluorescence difference is well preserved by the later light-induced activation for the motor's final settlement on the 6-site origami track. Hence the leg-exposed motor finally binds the track mostly at the recruiting site, ready for the motor's release from recruiter and ensuing site-controlled operation. The same pattern of fluorescence difference is observed for both labelling schemes of the 6-site track with swapped dyes between its minus-end and plus-end sites (see Fig. S6A versus Fig. S6C, ESI†), indicating successful site-selective motor placement.In this study, the experimental procedure for the motor's site-controlled introduction is designed based on two adapted motor constructions, as elaborated below. To avoid the motor's uncontrolled binding to the track's identical sites, the motor is pre-annealed with a protector strand to cover both legs, and the protected motor is guided to the target site (minus-end or middle site) via a recruiter overhang that is just the D1* overhang of the target site but elongated (to 17 nt) and mutated into a new sequence (see Fig. 2(A) for 5-site and 6-site tracks with the recruiter). The recruiter recognizes and captures the motor by hybridizing with a tail overhang (23 nt long) that is introduced to the motor at one end or between the two legs (called tail 1 and tail 2, as illustrated in state 1, Fig. 2(B) and (C)). To accommodate the tail, the motor is adjusted from its original bi-strand construction (see Fig. 1(A), for random-mix experiments of this study) into two four-strand constructions for the site-specific experiments (see state 1, Fig. 2(B) and (C); working equally well on the origami tracks as the bi-strand version as indicated by the data in Fig. 3(E)). After the tail-guided motor-recruiter binding (state 2, Fig. 2(B) and (C)), alternating UV and visible light irradiations are applied to activate the motor for binding the two nearby sites (state 3). This is because the UV light dissociates the protector into the bulk solution and the exposed legs bind the nearby sites. The activated motor is finally released from the origami-bound recruiter by a 23 nt-long release strand that breaks the 17 bp recruiter-tail duplex by forming a 23 bp duplex with the tail (state 4). Now the motor is ready for on-track motion under a new series of alternating UV and visible light.
The motor introduction procedure is tested and optimized rather extensively for the two motor tails and four types of protector strands. The motor's final settlement on the origami tracks has different pathways for the two tails due to their different location on the motor. As shown in Fig. 2(B), the tail 1 places the motor into a state in which the motor's two legs bind the two D2* overhangs, likely incompletely, on both sides of the recruiter. This state is accessible to the motor as found in previous studies,24 and readily evolves into the walking mode by UV-induced dissociation of either leg. The simultaneous dissociation of both legs is not impossible but the chance is low due to the modest UV intensity21 used in this study. The tail 2 places the motor from a recruiter towards either the plus end or minus end of the track. The motor adopts the symmetric state in the latter case. In the former case, as shown in Fig. 2(C), the motor adopts the most desired asymmetric state for on-track walking (i.e., the state in Fig. 1(B), top plot, ready for selective rear leg dissociation under the UV light). For the four types of protector strands, the protector 1 and 2 are 15 nt long and cover incompletely the leg's 20 nt-long D2 segment with its 5 nt-long exposure near 5′ end and 3′, respectively; the protector 3 is 20 nt long to cover the full D2 segment, and the protector 4 is 25 nt long to further cover the whole leg (D2 plus 5 nt-long D1). For sake of effective dissociation by light irradiations, protector 4 is not entirely complementary to the leg, with a point mutation every 5 nucleotides.
Fig. 5 present more fluorescence data from the minus-end start experiments for different combinations of tail types and protector types. Fig. 6 is a summary of the −/+ signal gap for a broader comparison covering both the minus-end start and mid-site start experiments. If both experiments have a similar level of motor translocation from the initial site to the plus end, the −/+ signal gap for the minus-end start should be two times that for the mid-site start. This expectation matches the data roughly (see Fig. 6(A)versusFig. 6(B)). Tail 1 and tail 2 both work well for the protector 1, 2, and 3, with the −/+ signal gap comparable for the protector 1 and protector 2 (Fig. 5(A)–(D) and 6(A)) and slightly larger for the protector 3 (Fig. 5(E) and 6(B)). For the minus-end start experiments, these tail-protector combinations all lead to signal gaps apparently larger than those from the random-mix experiments despite the shorter track for the latter experiments (see Fig. 5versusFig. 3). This is consistent with the success of site-controlled motor placement. However, the longest protector 4 leads to the −/+ signal gaps for the minus-end start experiments as small as for the random-mix experiments (Fig. 5(F)versusFig. 3(C) and (D)). This implies failure of the protector 4 to protect the motor's legs, probably because the protector-leg duplex is not stable enough due to the 4 mutations in this protector.
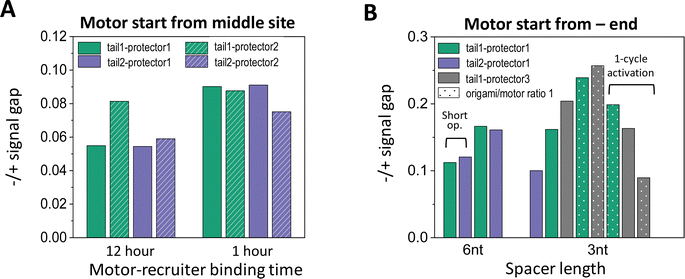 | ||
| Fig. 6 (A) Effectiveness of site-specific motor introduction (as assessed by fluorescence signal gap between minus and plus ends) for key parameters of the motor placement procedure (shown in Fig. 2(C) and (D)). (A) Motor introduction to the middle site of a 5-site track (as shown in Fig. 2(A), middle plot). All data in this histogram plot are obtained for 6 nt spacer below D1*, and with the motor activated for leg-site binding by 6 cycles of alternating UV and visible light (10 minutes per UV and 5 minutes per visible light, except for the leftmost two bars for 30 minutes per UV). (B) Motor introduction to the minus-end site of a 6-site track (as shown in Fig. 2(A), lowest plot). All data in this histogram plot are obtained using the 1 hour motor-recruiter binding time followed by the 6 light cycles for motor activation (10 minutes per UV and 5 minutes per visible light), except for the rightmost 3 bars (by a single 60 minutes UV and a 20 minutes visible light for motor activation). For both histogram plots in this figure, the −/+ signal gap is obtained at the end of 13 cycles of alternating UV-visible light operation (10 minutes per UV, 10 minutes per visible light) after the motor's release for on-track motion (all for origami–motor ratio 2, except ratio 1 for dotted bars in panel B and 10-cycle light operation for the leftmost 2 bars in the same panel). | ||
Like the random-mix experiments, the site-specific motor placement works for both 3 nt and 6 nt spacers under the D1* overhangs (see Fig. 6(B)versusFig. 3(E)). For some tail-protector combinations, the divergence between the minus-end and plus-end signals occurs later for the 3 nt spacer than for the 6 nt spacer (Fig. 5(B) and (F)). This difference between the spacers is also observed in the random-mix experiments (Fig. 3(A) and (C)), and results in different patterns for the −/+ signal gap (Fig. 3(B) and (D)). Changing the origami–motor molar ratio slightly changes the −/+ signal gap for the minus-end start experiments (Fig. 6(B)), again similar to the random-mix experiments (Fig. 3(E)). For origami–motor ratio 2, the 3 nt spacer leads to a rather big −/+ signal gap for a random-mix experiment (Fig. 2(E)), and for a minus-end start experiment (Fig. 5(E) for tail 1 and protector 3). In the latter experiment, the big signal gap coincides with a particularly apparent V-shape pattern of the mid-site signal, and even the plus-end signal starts to show a mild V-shape pattern after 10 cycles of UV-visible light, probably due to saturated motor accumulation at the plus end and ensuing motor derailment by further light operation. When the optical activation of the motor for leg-site binding is changed from multiple UV-visible light cycles to a single cycle of elongated UV followed by visible light, the −/+ signal gap drops slightly, suggesting less effective minus-end placement (Fig. 6(B)). Elongating the motor-recruiter binding time from 1 hour to 12 hour does not improve the −/+ signal gap; it slightly drops instead (Fig. 6(A)).
Conclusion
This study has demonstrated the light-driven on-origami operation of an advanced non-bridge-burning DNA bipedal walker. This rationally designed walker is a genuine translational molecular motor that exploits pure mechanical effects for light-powered automatic translocation on the ∼170 nm-long rod-like origami platform. The resultant integrated DNA motor–origami system provides a ‘seed’ system for advanced light-powered DNA nano-robots or artificial nano-muscles capable of repeated and automatic operation beyond the capacity of previous bridge-burning walker–origami systems. A rather straightforward nano-robotic application is molecular walker-automated chemical synthesis,36,37,45 which exploits a walker's precise-positioning capability to encode the sequence for a synthesis product, and is previously demonstrated using bridge-burning walkers36,37,45 on non-origami tracks (hence the walker-track system not re-usable and synthesis unscalable). The integrated DNA motor–origami system from this study is directly usable as a light-operated nano-robotic platform for repeatable and potentially scalable synthesis (by site-controlled motor reloading; hence the motor placement methods from this study will become useful too). This origami-based platform has a big room to accommodate extra mechanisms to enhance and expand the synthesis function. Another possible application of the system from this study is artificial muscles, which are previously demonstrated using nanomotor bi-state switching components,46,47 light-powered artificial molecular rotors,48 or bridge-burning DNA walker–origami systems6,7 operated by manually administered strands. The motor–origami system from this study is readily usable for developing advanced automatic DNA nano-muscles powered by light. These nano-muscles will be truly biomimicking artificial muscles powered by rationally designed translational molecular motors, and may serve as driving elements in many nano/micro-robots or be further assembled into larger active materials via DNA nanotechnology.Methods and materials
Design and fabrication of DNA origami
The origami is based on M13mp18 scaffold (7249 nucleotides) and designed using Cadnano software.49 The details of the designed origami structure are presented in Fig. S1 (ESI†), where the protruding overhangs for the full 6-site track (Fig. 1(E)) are also shown. Table S1 (ESI†) lists the nucleotide sequences of the staple strands for the origami and its variations for different tracks (Fig. 2(A)). All the origami strands were purchased from Integrated DNA Technologies Inc. For the origami fabrication, the staple strands (including those modified for the track overhangs) were first mixed with the scaffold at a molar ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 in TE buffer with 12.5 mM MgSO4. The sample was then placed in the thermal cycler to anneal under a gradual cooling from 95 °C to 20 °C over 14 hours. Specifically, the origami annealing protocol starts at 25 °C for 30 minutes, followed by sharp rise to 95 °C and held for 20 minutes. Then the temperature drops sharply to 90 °C and continues to decrease by 0.1 °C per 25 s until 80 °C, and further decrease by a slower rate of 0.1 °C per 72 s to 20 °C. The origami was purified by transferring the annealed sample to a 100k Amicon Ultra Ultracel centrifugal filter unit (placed in an Amicon tube, topped up with 5 mM MgSO4 in TE until the 0.5 mL mark for buffer exchange). The sample was then spun at 5000g for 10 minutes before the filter was removed and the flow-through poured away (for size exclusion column purification). The filter was placed back in the tube, followed again by buffer top-up (470 μL of 5 mM MgSO4 in TE) and centrifuging (5000g for 10 minutes). These processes were repeated another two times (i.e., 4 cycles of 5000g centrifuging in total). Finally the column was taken out and inverted in a fresh Amicon tube for one more centrifuging (1000g for 2 minutes) to obtain the purified origami sample.
2 in TE buffer with 12.5 mM MgSO4. The sample was then placed in the thermal cycler to anneal under a gradual cooling from 95 °C to 20 °C over 14 hours. Specifically, the origami annealing protocol starts at 25 °C for 30 minutes, followed by sharp rise to 95 °C and held for 20 minutes. Then the temperature drops sharply to 90 °C and continues to decrease by 0.1 °C per 25 s until 80 °C, and further decrease by a slower rate of 0.1 °C per 72 s to 20 °C. The origami was purified by transferring the annealed sample to a 100k Amicon Ultra Ultracel centrifugal filter unit (placed in an Amicon tube, topped up with 5 mM MgSO4 in TE until the 0.5 mL mark for buffer exchange). The sample was then spun at 5000g for 10 minutes before the filter was removed and the flow-through poured away (for size exclusion column purification). The filter was placed back in the tube, followed again by buffer top-up (470 μL of 5 mM MgSO4 in TE) and centrifuging (5000g for 10 minutes). These processes were repeated another two times (i.e., 4 cycles of 5000g centrifuging in total). Finally the column was taken out and inverted in a fresh Amicon tube for one more centrifuging (1000g for 2 minutes) to obtain the purified origami sample.
Origami characterization by atomic force microscopy (AFM) and oxDNA simulation
AFM imaging of the samples was performed using the Bruker Dimension Icon machine in tapping mode in air with Tespa-V2 tips (Bruker). 20 μL of annealing buffer (12.5 mM MgSO4 in TE) was first deposited onto a freshly cleaved mica surface and left to rest for 5 minutes. Then approximately 150 μL of the purified origami sample (diluted to ∼1 ng μL−1) was pipetted onto the mica surface. This was left to rest for more than 15 minutes, then washed with 1 mL of deionized water five times. The disc is finally dried with nitrogen gas for AFM imaging. The origami length (shown in Fig. 1(C)) was determined from AFM image of individual origami rods, by longitudinal scans for a rod to first identify a plateau of typically stable height over the rod's main body (see Fig. S2, ESI†) and then obtain the rod's length from the plateau profile as the distance between the half-height points at the rod's two ends. The origami was simulated using oxView server41 (at 1 M Na+, 25 °C) to generate the realistic structure shown in Fig. 1(E). The simulation was started with the system relaxation, followed by molecular dynamic simulation (for a total of 1 × 109 simulation steps; time interval as 0.003 simulation unit per step).Light operation and fluorescence detection
The procedures largely follow those in ref. 21 and 24, using a UV-VIS spectrophotometer (Shimadzu Corp.). For motility experiments started from random motor–origami binding, the motor of a bi-strand construction (Fig. 1(A)) was used, and annealed from two azo-tethered strands at equimolar ratio in TE buffer with 200 nM NaCl (azo-strands purchased from Nihon Techno Service Co. Ltd, with sequences given in Table S2, ESI†). The pre-operation motor–origami system was prepared by incubating the motor together with the origami in 12.5 mM MgSO4 in TAE (10×) buffer over 2 h, with the motor–origami binding monitored by applying 15 short visible light irradiations (10 seconds each, with 5 minutes of no irradiation in between) to minimize effects of photobleaching. For motility experiments started from a site-specific motor–origami binding, the pre-operation motor–origami system was prepared by a different procedure (elaborated in the following section). The light operation driving the motor's on-track motion consists of alternating UV-visible light irradiations (10 minutes per UV and 10 minutes per visible light throughout this study). The UV is at 350 nm wavelength for trans-to-cis photoisomerization of azo-moieties in the motor's legs. The visible light is both for motor driving (backward cis-to-trans photoisomerization of azo-moieties) and for fluorescence detection (with excitation and emission wavelengths matching those of the fluorescent dyes in kinetic mode at 5 nm slit width: 502 nm, 549 nm and 649 nm for excitation of ATTO488, TYE and ATTO647, respectively; 522 nm, 563 nm and 662 nm for their emission). Throughout this study, a low concentration of origami (8 nM) was used for motor operation to reduce aggregation. All the motor operation experiments were done at 25 °C. Besides, the UV irradiations used for the typical light operation in this study appear to have a negligible influence to structural integrity of the DNA origami as the AFM images for the origami before and after 12 rounds of UV-visible light irradiations show no observable difference (Fig. S7, ESI†).Site-specific motor introduction
To prepare a site-specific motor–origami binding system, a four-strand motor was first annealed together with protector strands in TE buffer with 200 nM NaCl (using equimolar ratio for the four constituent strands but 5 times for protector to ensure leg protection). The annealed motor with protection was incubated with the recruiter-carrying origami for motor-recruiter binding (for 1 h typically, or >12 h for test purpose, see Fig. 6(A)). The resultant motor–origami complex underwent alternating UV-visible light irradiations to remove the protector and activate the motor's legs for binding at the targeted site of the track (6 UV-visible light cycles, 30 or 10 minutes per UV and 5 minutes per visible light). Finally, the motor was set free from the recruiter by adding the releasing strand in excess (10 releasing strand per motor) for an hour before the start of motor operation by extra UV-visible light cycles. Four types of protector strands and two four-strand motors with different tail position (Fig. 2(B) and (C)) were tested for the site-selective motor placement. The nucleotide sequences for the motor strands and the protector strands are given in Tables S3–S5 (ESI†).Author contributions
X. R. L. and Z. W. designed the experiments, analyzed the data, and wrote the manuscript. X. R. L. conducted the experiments. I. Y. L., W. S. and T. A. participated the experiments and data analysis. H. L. T. performed the oxDNA simulation. Z. W. conceived the idea, planned and oversaw the project.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Ministry of Education of Singapore under grant no. A-0004192-00-00, A-0008378-00-00, A-8000628-00/01-00, A-8000982-00-00 (to Z. S. Wang). We thank Deborah Fygenson for designing the DNA origami for this study.References
- Z. Wang, R. Hou and I. Y. Loh, Nanoscale, 2019, 11, 9240–9263 RSC.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- K. Lund, A. J. Manzo, N. Dabby, N. Michelotti, A. Johnson-Buck, J. Nangreave, S. Taylor, P. Pei, M. N. Stojanovic, N. G. Walter, E. Winfree and H. Yan, Nature, 2010, 465, 206–210 CrossRef CAS PubMed.
- H. Gu, J. Chao, S. J. Xiao and N. C. Seeman, Nature, 2010, 465, 202–206 CrossRef CAS PubMed.
- A. J. Thubagere, W. Li, R. F. Johnson, Z. Chen, S. Doroudi, Y. Lim Lee, G. Izatt, S. Wittman, N. Srinivas, D. Woods, E. Winfree and L. Qian, Science, 2017, 357, 1095–1103 CrossRef PubMed.
- P. Zhan, S. Both, T. Weiss and N. Liu, Nano Lett., 2019, 19, 6385–6390 CrossRef CAS PubMed.
- E. Benson, R. C. Marzo, J. Bath and A. J. Turberfield, Small, 2021, 17, 2007704 CrossRef CAS PubMed.
- C. Zhou, X. Duan and N. Liu, Nat. Commun., 2015, 6, 8102 CrossRef CAS PubMed.
- J. Li, A. Johnson-Buck, Y. Renee Yang, W. M. Shih, H. Yan and N. G. Walter, Nat. Nanotechnol., 2018, 13, 723–729 CrossRef CAS PubMed.
- S. F. J. Wickham, M. Endo, Y. Katsuda, K. Hidaka, J. Bath, H. Sugiyama and A. J. Turberfield, Nat. Nanotechnol., 2011, 6, 166–169 CrossRef CAS PubMed.
- Y. Yang, M. A. Goetzfried, K. Hidaka, M. You, W. Tan, H. Sugiyama and M. Endo, Nano Lett., 2015, 15, 6672–6676 CrossRef CAS PubMed.
- J. Valero, N. Pal, S. Dhakal, N. G. Walter and M. Famulok, Nat. Nanotechnol., 2018, 13, 496–503 CrossRef CAS PubMed.
- J. Valero and M. Famulok, Angew. Chem., Int. Ed., 2020, 59, 16366–16370 CrossRef CAS PubMed.
- T. E. Tomov, R. Tsukanov, M. Liber, R. Masoud, N. Plavner and E. Nir, J. Am. Chem. Soc., 2013, 135, 11935–11941 CrossRef CAS PubMed.
- M. Liber, T. E. Tomov, R. Tsukanov, Y. Berger and E. Nir, Small, 2015, 11, 568–575 CrossRef CAS PubMed.
- T. E. Tomov, R. Tsukanov, Y. Glick, Y. Berger, M. Liber, D. Avrahami, D. Gerber and E. Nir, ACS Nano, 2017, 11, 4002–4008 CrossRef CAS PubMed.
- Y. Yang, R. Tashiro, Y. Suzuki, T. Emura, K. Hidaka, H. Sugiyama and M. Endo, Chem. – Eur. J., 2017, 23, 3979–3985 CrossRef CAS PubMed.
- D. C. Khara, J. S. Schreck, T. E. Tomov, Y. Berger, T. E. Ouldridge, J. P. K. Doye and E. Nir, Nucleic Acids Res., 2018, 16, 1553–1561 CrossRef PubMed.
- S. J. Green, J. Bath and A. J. Turberfield, Phys. Rev. Lett., 2008, 101, 238101 CrossRef CAS PubMed.
- J. Bath, S. J. Green, K. E. Allen and A. J. Turberfield, Small, 2009, 5, 1513–1516 CrossRef CAS PubMed.
- J. Cheng, S. Sreelatha, R. Z. Hou, A. Efremov, R. C. Liu, J. R. van der Maarel and Z. S. Wang, Phys. Rev. Lett., 2012, 109, 238104 CrossRef PubMed.
- Z. S. Wang, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 17921–17926 CrossRef CAS PubMed.
- J. Cheng, S. Sreelatha, I. Y. Loh, M. Liu and Z. S. Wang, Methods, 2014, 67, 227–233 CrossRef CAS PubMed.
- M. H. Liu, R. Z. Hou, J. Cheng, I. Y. Loh, S. Sreelatha, J. N. Tey, J. Wei and Z. S. Wang, ACS Nano, 2014, 8, 1792–1803 CrossRef CAS PubMed.
- I. Y. Loh, J. Cheng, S. R. Tee, A. Efremov and Z. S. Wang, ACS Nano, 2014, 8, 10293–10304 CrossRef CAS PubMed.
- M. H. Liu, J. Cheng, S. R. Tee, S. Sreelatha, I. Y. Loh and Z. S. Wang, ACS Nano, 2016, 10, 5882–5890 CrossRef CAS PubMed.
- Q. Y. Yeo, I. Y. Loh, S. R. Tee, Y. H. Chiang, J. Cheng, M. H. Liu and Z. S. Wang, Nanoscale, 2017, 9, 12142–12149 RSC.
- Y. H. Chiang, S. L. Tsai, S. R. Tee, O. L. Nair, I. Y. Loh, M. H. Liu and Z. S. Wang, Nanoscale, 2018, 10, 9199–9211 RSC.
- X. Hu, X. Zhao, I. Y. Loh, J. Yan and Z. Wang, Nanoscale, 2021, 13, 13195–13207 RSC.
- X. R. Liu, X. Hu, I. Y. Loh and Z. Wang, Nanoscale, 2022, 14, 5899–5914 RSC.
- M. Skugor, J. Valero, K. Murayama, M. Centola, H. Asanuma and M. Famulok, Angew. Chem., Int. Ed., 2019, 58, 6948–6951 CrossRef CAS PubMed.
- R. Z. Hou, I. Y. Loh, H. Li and Z. S. Wang, Phys. Rev. Appl., 2017, 7, 024020 CrossRef.
- S. R. Tee, X. Hu, I. Y. Loh and Z. S. Wang, Phys. Rev. Appl., 2018, 9, 034025 CrossRef CAS.
- H. Xu, R. Hou, H. Li and Z. Wang, J. Appl. Phys., 2020, 128, 164701 CrossRef CAS.
- Z. S. Wang, M. Feng, W. W. Zheng and D. G. Fan, Biophys. J., 2007, 93, 3363–3372 CrossRef CAS PubMed.
- Y. He and D. R. Liu, Nat. Nanotechnol., 2010, 5, 778–782 CrossRef CAS PubMed.
- B. Lewandowski, G. De Bo, J. W. Ward, M. Papmeyer, S. Kuschel, M. J. Aldegunde, P. M. E. Gramlich, D. Heckmann, S. M. Goldup, D. M. D’Souza, A. E. Fernandes and D. A. Leigh, Science, 2013, 339, 189–193 CrossRef CAS PubMed.
- D. G. Fan, W. W. Zheng, R. Hou, F. Li and Z. S. Wang, Biochemistry, 2008, 47, 4733–4742 CrossRef CAS PubMed.
- W. W. Zheng, D. Fan, M. Feng and Z. S. Wang, Phys. Biol., 2009, 6, 036002 CrossRef PubMed.
- Y. Xu and Z. S. Wang, J. Chem. Phys., 2009, 131, 245104 CrossRef PubMed.
- J. Bohlin, M. Matthies, E. Poppleton, J. Procyk, A. Mallya, H. Yan and P. Šulc, Nat. Protoc., 2022, 17, 1762–1788 CrossRef CAS PubMed.
- S. A. E. Marras, F. R. Kramer and S. Tyagi, Nucleic Acids Res., 2002, 30, e122 CrossRef PubMed.
- A. Efremov and Z. S. Wang, Phys. Chem. Chem. Phys., 2011, 13, 5159–5170 RSC.
- A. Efremov and Z. S. Wang, Phys. Chem. Chem. Phys., 2011, 13, 6223–6233 RSC.
- J. Echavarren, M. A. Y. Gall, A. Haertsch, D. A. Leigh, J. T. J. Spence, D. J. Tetlow and C. Tian, J. Am. Chem. Soc., 2021, 143, 5158–5165 CrossRef CAS PubMed.
- Y. Liu, A. H. Flood, P. A. Bonvallet, S. A. Vignon, B. H. Northrop, H. R. Tseng, J. O. Jeppesen, T. J. Huang, B. Brough, M. Baller, S. Magonov, S. D. Solares, W. A. Goddard, C. M. Ho and J. F. Stoddart, J. Am. Chem. Soc., 2005, 127, 9745–9759 CrossRef CAS PubMed.
- T. Mirfakhrai, J. D. W. Madden and R. H. Baughman, Mater. Today, 2007, 10, 30–38 CrossRef CAS.
- Q. Li, G. Fuks, E. Moulin, M. Maaloum, M. Rawiso, I. Kulic, J. T. Foy and N. Giuseppone, Nat. Nanotechnol., 2015, 10, 161–166 CrossRef CAS PubMed.
- S. M. Douglas, A. H. Marblestone, S. Teerapittayanon, A. Vazquez, G. M. Church and W. M. Shih, Nucleic Acids Res., 2009, 37, 5001–5006 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nh00565d |
| This journal is © The Royal Society of Chemistry 2023 |
