 Open Access Article
Open Access ArticleNanoparticle-based applications by atmospheric pressure matrix assisted desorption/ionization mass spectrometry
Yihan
Wang
,
Shunxiang
Li
* and
Kun
Qian
 *
*
State Key Laboratory of Systems Medicine for Cancer, School of Biomedical Engineering, Institute of Medical Robotics and Shanghai Academy of Experimental Medicine, Shanghai Jiao Tong University, Shanghai, 200030, China. E-mail: lishunxiang@sjtu.edu.cn; k.qian@sjtu.edu.cn
First published on 30th October 2023
Abstract
Recently, the development of atmospheric pressure matrix assisted desorption/ionization mass spectrometry (AP MALDI MS) has made contributions not only to biomolecule analysis but also to spatial distribution. This has positioned AP MALDI as a powerful tool in multiple domains, thanks to its comprehensive advantages compared to conventional MALDI MS. These developments have addressed challenges associated with previous AP MALDI analysis systems, such as optimization of apparatus settings, synthesis of novel matrices, preconcentration and isolation strategies before analysis. Herein, applications in different fields using AP MALDI MS were described, including peptide and protein analysis, metabolite analysis, pharmaceutical analysis, and mass spectrometry imaging.
Introduction
Matrix assisted laser desorption/ionization at atmospheric pressure (AP MALDI) is based on the same principle as MALDI implemented in vacuo, where the sample is blended with a matrix and irradiated with a pulsed laser beam.1 Nowadays, AP MALDI has not only become a solid platform for biological analysis, including peptides, proteins, metabolites, and drugs, but also offers a comprehensive approach for spatial distribution. Simultaneously, AP MALDI offers advantages over vacuum MALDI, including time and volatility constraints as well as the capability for analysing materials under more physiologically relevant circumstances, which is crucial in the developing region of compound-specific tissue imaging.2 Nevertheless, AP MALDI still has drawbacks that could decrease its sensitivity. First, its ion transmission efficiency is lower than vacuum MALDI due to ionization of the analyte at atmospheric pressure. Second, the formation of matrix–matrix and analyte–matrix clusters can have a harmful influence. With the aim of enhancing the performance of detection, researchers have made contributions in diverse fields.AP MALDI combined with mass spectrometry was first reported in 1999, the technique was demonstrated for the investigation of the peptide composition of paralytic poisons, conotoxins, from a molluscivorous snail.3 In 2000, Laiko and co-workers formally unravelled atmospheric pressure matrix-assisted laser desorption/ionization mass spectrometry as an extension of conventional vacuum MALDI, and it has become a powerful technique for multiple applications, including the identification of proteins, the structural analysis of oligosaccharides, the characterization of enzymes immobilized on solid supports, and the study of phosphopeptides, owing to its ability to detect intact molecular masses.4–8
In summary, the developments of AP MALDI adhibition in the past two decades can be grouped into four aspects: peptide and protein detection, metabolite identification, pharmaceutical analysis, and mass spectrometry imaging.
AP MALDI
AP interface
The atmospheric pressure interface (API) is a standard feature of modern laboratory mass spectrometers, which was invented to transfer ions from an atmospheric pressure region into a reduced pressure region.9 Such interfaces can be connected to various atmospheric pressure ionization sources, including electrospray ionization (ESI), atmospheric pressure chemical ionization (APCI), and AP MALDI, enabling their integration with a variety of separation devices, such as gas/liquid chromatography.10–12 It is also essential for mass spectrometers to have an atmospheric interface to take advantage of the new category of direct ambient ionization methods, such as desorption electrospray ionization (DESI), direct analysis in real time (DART), atmospheric pressure dielectric barrier discharge ionization (DBDI), electrospray-assisted laser desorption/ionization (ELDI), and atmospheric pressure solids analysis probes (ASAPs).13–17 A typical atmospheric pressure interface has a constantly open orifice and a series of differential pumping stages, with a capillary or a small aperture to allow ions to be delivered into the first differential pumping stage and a skimmer at the entrance to the second stage.18 A rough pump is usually utilized to evacuate the first region to around 1 Torr, while multiple turbomolecular pumps or a single pump with split flow is employed to pump the subsequent regions. The final pressure accomplished in the mass analyser is usually 10–5 Torr and the largest losses in ion delivery appear in the first and second stages, resulting in 2 orders of magnitude and 1 order of magnitude, respectively. This results in an overall efficiency of no more than 0.1% for ion transfer through the atmospheric pressure interface (Scheme 1). | ||
| Scheme 1 An overview of applications based on atmospheric pressure matrix assisted laser desorption/ionization mass spectrometry. | ||
MALDI
Matrix assisted laser desorption ionization mass spectrometry was developed by two independent research groups and became commercially available in the early 1990s.19 With technical improvements, MALDI has become an irreplaceable tool for biomolecule analysis.The ionization process of MALDI occurs in the mixture solution of the analyte, matrix, and co-crystallization, typically resulting in the generation of singly charged ions. However, the mechanism of ion generation is still not fully understood with two theories persisting: the first is the gas phase protonation model and the second is known as the ‘Lucky Survivor’ model.20–23 The ‘Lucky Survivor’ model is based on the premise that analyte ions are pre-formed before ablation, while the gas phase protonation model suggests that neutral analyte molecules are protonated after ablation. Both models make the same assumption that gas phase chemistry and definitely the recombination processes with the oppositely charged species during rapid plume expansion play a crucial role in the ionization process and account for the predominantly singly charged ions observed.24–26
Distinct from traditional mass spectrometry (e.g., gas/liquid chromatography MS, GC/LC MS), spectroscopy (e.g., Raman spectroscopy and near infrared spectroscopy), and nuclear magnetic resonance (NMR), MALDI displays desirable advantages. First, MALDI has simple sample pretreatment, superior to the necessary isotopic treatment of NMR and rigorous pretreatment of 0.5–1.0 h for target enrichment in GC/LC MS.27,28 Besides, Raman spectrum requires each droplet to be deposited onto microscope slides covered with aluminum foil with the aim of decreasing the fluorescence interference.29 The detection speed of MALDI shows more advantages than GC/LC MS, which enables the high throughput for biomolecule analysis. Furthermore, MALDI is considered to be a “soft” technique, which allows the desorption and ionization of intact molecular analyte species and thus their successful mass-spectrometric analysis.
AP MALDI
In 1999, Jean-Luc Wolfender and his group first identified the tyrosine sulfation in Conus pennaceus conotoxins by using AP MALDI MS, paving the way for AP MALDI applications. In addition to the advantages traditionally associated with MALDI, such as reduced sample clean-up needs, simplicity of sample preparation, and the ability to re-analyze samples from a previously investigated spot, AP MALDI has some unique advantages. For example, liquid matrices are more easily implemented and measurements often exhibit higher reproducibility compared to vacuum MALDI.30–32 Furthermore, an important characteristic of the AP MALDI processes is that, following ionization, ions are rapidly thermalized by collisional cooling with atmospheric gases, resulting in lower effective ion temperatures.33,34 The rapid thermalization acquired under atmospheric pressure conditions has proven helpful for studying labile biomolecules with minimal unwanted fragmentation. However, in the ionization process of AP MALDI, there is a tendency to have the undesirable side effect of forming matrix–matrix and analyte–matrix clusters and undergoing ion loss when ions transfer from atmospheric pressure to the vacuum region, resulting in limited sensitivity for AP MALDI. In recent years, with the continuous development of AP MALDI-related technologies by scientists, its sensitivity has been improved to some extent and it has gained increasingly widespread applications (Fig. 1).35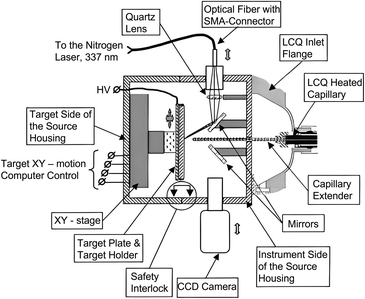 | ||
| Fig. 1 A scheme of the atmospheric pressure MALDI ion source interfaced with an LCQ ion trap mass spectrometer. Reprinted with permission from ref. 35, Copyright 2002 Elsevier B.V. | ||
Applications using AP MALDI
Protein and peptide detection
With the improvement of detection methods, proteomics has been introduced as an innovative perspective for disease identification because protein is the material basis of life and the main participant in life processes, which could reflect changes in the human body. Nowadays, peptides and protein detection are studied by AP MALDI from four perspectives: extraction and separation before analysis, cleavage sites, optimization of mass spectrometers, and novel matrix synthesis.Extraction and separation before analysis
Extraction and separation before analysis was confirmed to be a viable approach for eliminating unwanted biomolecules and therefore enhancing the targeted m/z signals. Recently, noble metal nanoparticles have been widely applied as microprobes in biological analysis, intending to preconcentrate and isolated expected biomolecules. In 2005, Yan Wang and co-workers improved AP MALDI with solid-phase microextraction (SPME) and evaluated its properties by analyzing four peptide mixtures, including angiotensin II, bradykinin, angiotensin I and glucoprotein b.36 In the same year, Putty-Reddy Sudhir's group employed gold nanoparticle-assisted single drop microextraction (SDME-AuNPs) before AP MALDI MS to identify 0.2 μM Met-enkephalin and 0.17 μM Leu-enkephalin in water solution, respectively.37 However, silver nanoparticles were also used as electrostatic probes in single drop microextraction (SDME-AgNPS) by this group in 2008, intending to improve the detection sensitivity for peptide analysis in AP MALDI MS. After optimizing the experimental conditions, including extraction solvent, extraction time, stirring rate, and pH, Met-enkephalin and Leu-enkephalin were successfully extracted from a preconcentrated mixture and AP MALDI was applied for analysis.38 Further, solid-phase extraction–elution on diamond (SPEED) was also combined with AP MALDI-Fourier transform ion cyclotron resonance MS by Sahadevan Sabu's group in 2007, in order to enhance ion transmission efficiency. With the rapid and convenient SPME method, analyte molecules (e.g., angiotensin I, gramicitracin S, bradykinin, and equine cardiomyoglobin Lys C digest) were enriched in highly diluted solution and compared with electrospray ionization, SPEED AP MALDI resulted in better sensitivity (Fig. 2a).39 In 2008, Kamlesh's team developed modified silver nanoparticles as a hydrophobic affinity probe for analyzing peptides. Compared to traditional AP MALDI MS, a 266- to 388-fold improvement in the limit of detection (LOD) of 0.16, 0.13, and 0.16 μM in water, urine, and plasma for gramicidin was achieved, respectively.40 In the next year, 2009, on-chip solid-phase extraction preconcentration/focusing substrates combined with AP MALDI ion trap MS was conducted by Arti Navare et al. for high sensitivity biomolecule analysis including angiotensin I, neuropeptide, trypsin digest, and derived myoglobin digestives. Compared with conventional AP MALDI MS, sensitivity increases by about two orders of magnitude, and the LOD of the standard peptide was enhanced to 5 fmol μL−1. Owing to the improved sensitivity, sequencing of dilute solutions of a derivatized tryptic digest was performed by tandem mass spectrometry (MS/MS).30 Nevertheless, Pavel Ryumin's team ameliorated AP MALDI by coupling nano ultrahigh-performance for protein identification in 2017, and storage of MALDI samples on fully spotted target plates was possible for months without significant sample degradation.41 So far, multiple kinds of nanoparticle have been employed as in the AP MALDI domain to enhance performance. However, a strategy for signal enhancing from one perspective cannot meet the diverse needs.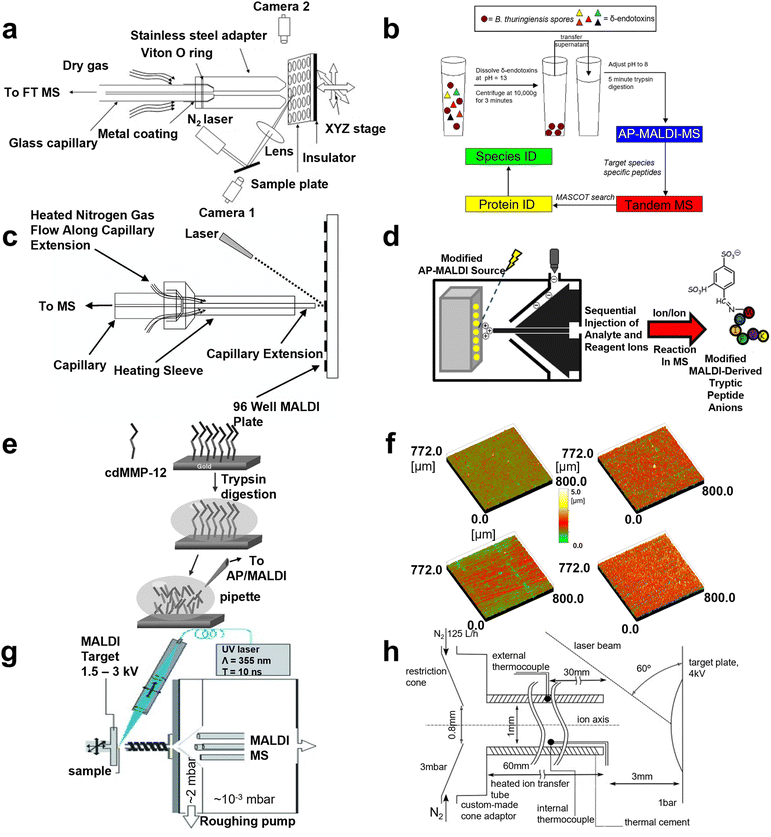 | ||
| Fig. 2 A schematic illustration of AP MALDI MS for peptide and protein detection. (a) A schematic diagram of the home-made AP MALDI source. Reprinted with permission from ref. 39, Copyright 2007 Elsevier Inc. (b) The workflow of the AP-MALDI based method for species-level detection of B. Thuringiensis spores. Reprinted with permission from ref. 44, Copyright 2010 Elsevier B.V. (c) A schematic diagram of the AP-MALDI source. Reprinted with permission from ref. 46, Copyright 2003 John Wiley and Sons. (d) The workflow of modified AP MALDI to detect peptide ions. Reprinted with permission from ref. 49, Copyright 2012 American Chemical Society. (e) A scheme of the procedure applied for in situ tryptic digestion and AP/MALDI-MS characterization. Reprinted with permission from ref. 7, Copyright 2006 John Wiley and Sons. (f) Surface roughness images determined by 3D-confocal-white-light microscopy of different target surfaces. Reprinted with permission from ref. 50, Copyright 2011 Elsevier B.V. (g) The structure of MALDI mass spectrometry using liquid UV-MALDI matrices and a heated ion-transfer tube. Reprinted with permission from ref. 51, Copyright 2013 WILEY-VCH. (h) A schematic of the AP-MALDI ion source and modified AP-to-vacuum interface used for the production of multiply protonated molecules. Reprinted with permission from ref. 26, Copyright 2016 Elsevier Inc. | ||
Cleavage site
Researchers have investigated the cleavage sites of proteins so as to evaluate the sequencing ability of AP MALDI MS, which can help achieve more precise analysis. The complete characterization of the proteolytic fragments produced by the interaction of the insulin degrading enzyme (IDE) with bovine insulin was studied by Guiseppe Grasso and co-workers in 2007, where insulin solution (m/z range of 2020–3500) was identified after interaction with IDE.42 Then, the result was checked by chemical modifications, including reduced alkylation reactions, simplified spectra, and cutting sites. In 2009, this team also studied MS detection of amyloid β-peptides via MALDI, AP MALDI, and ESI techniques to select the best analysis method.43 Moreover, in 2010, the targeted proteomics method for species-level identification of bacillus thuringiensis spores by AP MALDI was employed by Jennifer Nguyen's group and 75.4% specificity for thuringiensis and 60.3% specificity for anthrax were obtained, respectively (Fig. 2b).44 Additionally, Rima Ait-Belcasem et al. optimized in-source decay (ISD) and pseudo-MS3 of a peptide and protein in AP MALDI in 2016, resulting in 100% for bradykinin and 68% for thymin β4 coverage, representing a peptide and a small protein and 40% coverage for cytochrome c representing a large protein.45 MALDI MS has numerous advantages for protein cleavage sites but there still remains room for researchers to improve.Optimization of a mass spectrometer
More recently, researchers have devoted their attention to the comprehensive optimization of mass spectrometers, including hardware improvement, immobilized enzyme application, and target plate evaluation. For the hardware improvement, in 2003, Christine A. et al. optimized the ion sampling hardware and employed a counter current heated drying gas to inhibit the introduction of low molecular weight ions, for the sake of enhancing the low sensitivity of trypsin digest applying AP MALDI compared with other kinds of analyte (Fig. 2c).46 The mixture of trypsin digest (e.g., bovine aipotransferrin, BSA, and equine myoglobin) was identified successfully. In the same year, John T. Melh et al. also compared automated protein identification using AP MALDI ion trap (IT) MS with μLC MS/MS and automated MALDI TOF MS, finding that sample throughput was increased 10-fold in AP MALDI ITMS. Then, Coomassie blue stained gels combined with AP MALDI IT MS were found to be a useful platform for rapid protein identification.5 AP MALDI sources were combined with ion mobility time of flight (IM TOF) MS by Wes E. Steiner and co-workers to identify dipeptide and biogenic amine mixtures in 2004. Enhanced sample ionization efficiency created by this combination provided an overall elevation in signal intensity of ∼1.3 orders in magnitude. Combinations of three dipeptides (including, Gly–Lys, Ala–Lys, and Val–Lys) and nine biogenic amines (including, dopamine, serotonin, B-phenylethylamine, tyramine, octopamine, histamine, tryptamine, spermidine, and spermine) were resolved in less than 18 ms.47 In 2010, the AP MALDI MS ion source can generate laser spray ionization (LSI) using laser fluence and reflective geometry conditions and switch singly charged ions immediately by changing the voltage applied on the target.48 With zero voltage or around zero voltage, the multiply charged ions of the peptide and protein were observed. Nevertheless, Alexander Misharin's team designed a field-deployable ion trap MS with an atmospheric pressure interface in 2012 and a low femtomole peptide, polyproline, and angiotensin II mixture was characterized to measure the sensitivity which was the same as the commercial quadrupole ion trap MS.9 Meanwhile, John R. Stutzman and Scott A. McLuckey employed a dual-source interface attached to a hybrid triple-quadrupole/linear ion trap tandem mass spectrometer to generate gas-phase modification of peptide ions.49 The structural characterization of the modified and unmodified versions of the ions was achieved via ion trap collision-induced dissociation (CID, Fig. 2d). As to immobilized enzyme application, in situ characterization of anchored matrix metalloproteinases (MMP) by AP MALDI was accomplished by Giuseppe Grasso's group in 2006, elucidating that anchoring on the solid phase could not alter the properties of the protein interaction.7 High coverage of cleavage illustrated that MMP is desirable for protein digestion in AP MALDI MS (Fig. 2e). In terms of target plate evaluation, in 2011, the capability for different materials of the target plate was studied, while work was conducted focusing on different target surfaces, (gold-covered stainless steel (SS), titanium nitride-covered SS, hand-polished SS, and microdiamond-covered hard metal).50 A single peptide, peptide mixture, and trypsin digest were analyzed using diverse plates, evaluating the sensitivity and resolution, which suggested that the gold-covered SS target surface was the first choice for AP MALDI MS proteomics (Fig. 2f). Generally, different perspectives according to AP MALDI were considered in order to eliminate the background noise and enhance the desired signals.Novel matrix synthesis
In the MALDI MS system, an appropriate matrix can promote nanoplatforms to display excellent performance so multiple matrices were employed and evaluated to enhance the analyte signals. AP MALDI has been proven to facilitate the formation of multiply charged ions, which increase the degree of fragments, and many researchers demonstrated that a liquid matrix can lead to multiply charged ions rather than singly charged ions. In 2013, Prof. Rainer Cramer applied glycerol and triethylamine based on DHB and CHCA, to enable high and prolonged ion yields of multiply charged peptide and protein ions via AP ultraviolet (UV) MALDI MS (Fig. 2g).51 Further, the optimization of liquid AP MALDI MS equipment coupling with ion mobility separation including the temperature of ion transmission region, geometry of ion source, and residence time was achieved by Pavel Ryumin and his co-workers in 2016, which afforded a 14-fold increase for ionization efficiency. Thereby, peptide and protein (e.g., angiotensin I and bradykinin) solution concentrations as low as 2 fmol μL−1 were detected successfully (Fig. 2h).26 Moreover, this team also developed a novel matrix in 2018 for peptide and protein detection using liquid AP MALDI MS, and the analytical property of the matrix was evaluated with mainly multiply charged ion production.52 Numerous studies have demonstrated that an applicable matrix can facilitate the platform to perform better consequences, efforts should be dedicated to this field in the near future.Proteomics has been widely studied in recent decades as it provides significant evidence for disease diagnosis and disease mechanism investigation. Meanwhile, AP MALDI MS has become an emerging tool in this field. However, there are still many aspects, such as the tedious pretreatment, for researchers to develop in order to improve the efficacy of AP MALDI MS towards clinical applications.
Metabolite identification
Metabolomics is considered to be an irreplaceable platform of biological sciences, which is the closest to the phenotype and the most predictive of phenotype compared with other omics such as genomics, transcriptomics, and proteomics.53 With the emerging of metabolism, AP MALDI has been widely used to detect metabolites in recent years.Direct analysis
Direct analysis of metabolites employed by AP MALDI has been undertaken recently. In 2006, polypeptide antibiotics were detected by Ernst Pittenauer et al. through AP MALDI coupled with an ion trap. Compared with vacuum MALDI IT/RTOF, TOF/reflectron TOF, and ESI IT MS, AP MALDI MS offered the best performance considering the collision induced dissociation spectra of singly charged polypeptide antibiotic precursor ions.54 In 2009, Giuseppe Grasso's team immobilized IDE on a gold surface to interact with insulin delivered by a microfluidic system, then AP MALDI was employed to detect the IDE activity.55 Further, a standard lipid sample, choline phosphate (PC), was detected by Bindesh Shrestha's group using AP infrared MALDI MS to evaluate the analytical capability in 2010, while different lipids (cholesterol, PC, phosphatidylethanolamine, and sphingomyelin) and metabolites in tissue sections of a mouse brain were analyzed, depicting that this method could not only minimize or eliminate artifacts linked to freeze-thawing and post-mortem tissue disintegration but could also provide the substantial capability of studying biological samples with minimal pretreatment.56 In 2012, Emi Ito's group combined AP MALDI with quadrupole ion trap TOF shifting positive and negative ion mode, which was applied to identify the structural characterization of monosialogangliosides (e.g., GM1 and GM2), disialogangliosides, (e.g., GD2, GD1a, and GD1b), and trisialoganglioside (e.g., GT1a). In this system, the negative ion mass spectra of MS, MS2, and MS3 provided sufficient information for the determination of molecular weights, oligosaccharide sequences, and ceramide structures.57 In addition, real samples were also detected recently, which were an extension for AP MALDI applications. In 2020, Cristian Piras et al. classified sheep and goat milk with 100% accuracy via AP MALDI MS. Pure goat milk and sheep milk containing 10% goat milk were also discriminated between with 98% accuracy. Accordingly, β-lactoglobulin was considered to be the biomarker for classification (Fig. 3a).58 However, the feasibility of using AP MALDI with ion trap MS in the analysis of plant oligosaccharides was validated by Sunli Chong and co-workers in 2011. They found that this approach was able to detect xylooligosaccharides (XOS) with a chain length of up to ten xylopyranosyl residues. Specifically, AP MALDI ITMS seemed to be more suitable for the detection of acetylated XOS compared with conventional MALDI MS.59 Giuseppe Grasso et al. noninvasively characterized carbonaceous ink from Renaissance documents by AP MALDI MS in 2017, and the consequences were supported by the validation of Raman spectrometry (Fig. 3b).60 Although direct analysis has afforded better results than NMR, LC/GC MS, and Raman spectroscopy, there are still drawbacks that need to be resolved to improve the MS performance.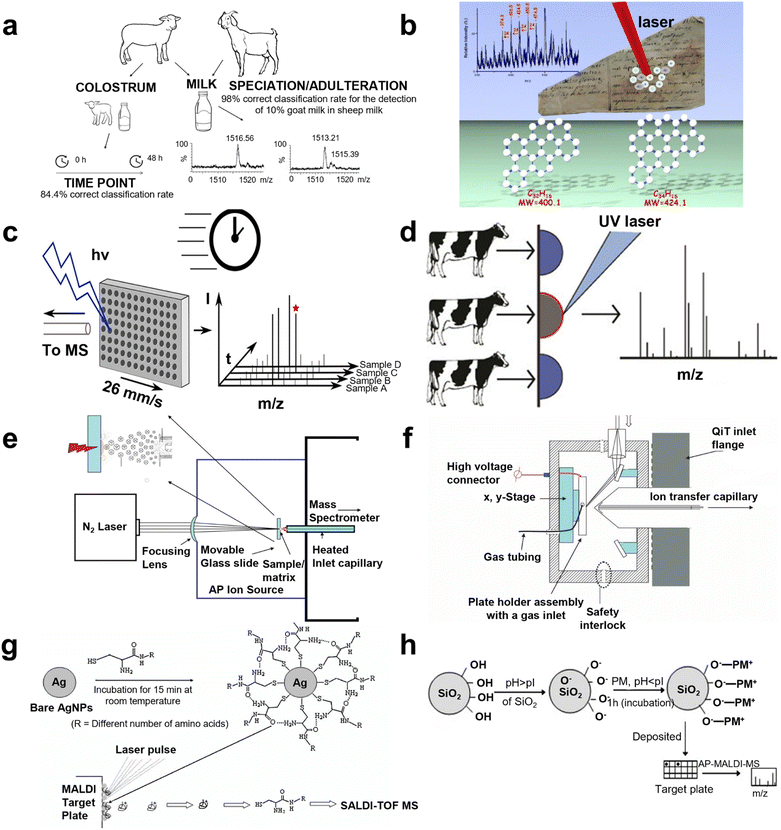 | ||
| Fig. 3 A schematic illustration of AP MALDI MS for metabolite identification. (a) The workflow for goat and sheep milk identification using liquid AP MALDI MS. Reprinted with permission from ref. 58, Copyright 2020 Licensee MDPI. (b) The scheme for AP MALDI MS of the non-invasive characterization of carbonaceous ink from Renaissance documents. Reprinted with permission from ref. 60, Copyright 2017 Springer Nature. (c) The scheme for liquid AP MALDI MS towards ultrahigh-throughput analysis. Reprinted with permission from ref. 66, Copyright 2020 American Chemical Society. (d) The scheme of mastitis diagnosis using milk by liquid AP MALDI MS. Reprinted with permission from ref. 64, Copyright 2019 American Chemical Society. (e) The representation of field-free transmission geometry AP MALDI source design. Reprinted with permission from ref. 2, Copyright 2010 American Chemical Society. (f) The schematic diagram of the inline pneumatically assisted (PA) AP MALDI ion source mounted on a quadrupole ion trap (QiT) mass spectrometer. Reprinted with permission from ref. 68, Copyright 2010 John Wiley and Sons. (g) Representation of AgNPs as preconcentrating probes and the matrix for the SALDI TOF MS analysis of biothiols. Reprinted with permission from ref. 75, Copyright 2008 John Wiley and Sons. (h) A schematic illustration of the attachment of the peptide mixture (PM) on the surface of SiO2 nanoparticles. Reprinted with permission from ref. 76, Copyright 2008 John Wiley and Sons. | ||
With the development of technology and the investigation of researchers, improvements for enhancing AP MALDI performance in metabolic detection have been conducted with different optimizations according to matrices, apparatus settings, and preconcentration methods.
Optimization of the matrix
In terms of matrix exploration, Ryuji Hiraguchi and co-workers tested the features of diverse matrices AP MALDI which employed a 6–7 μm-band mid-infrared tunable laser in 2015.61 Water was considered to be a desirable matrix in this approach, indicating that this method has the potential application for organic solvent and non-volatile buffer solutions. Further, Kasai Hiroko's team synthesized a new matrix, 5-(3-trifluoromethyl benzylidene) thiazolidine-2,4-dione (3-CF3-BTD) to apply in AP MALDI MS in 2016, which could detect dopamine.62 Notably, Oliver J. Hale and her teammates conducted preliminary optimization of ESI-like ions employing the negative mode in liquid AP MALDI MS in 2018, and peptide, protein, DNA, and lipid were identified using the optimal method. In this experiment, analysis of lipid and DNA benefited from the complementary lipid species and the stronger DNA intensity of the signal.63 However, Oliver's group also detected milk samples by liquid AP MALDI MS, intending to diagnose cow mastitis in 2019. Samples were classified according to mastitis status using multivariate analysis, achieving 98.5% accuracy (100% specificity) determined by “leave 20% out” cross-validation and α-s1-casein was found to be the diagnosis biomarker (Fig. 3d).64 In the same year, genetically different bacterial strains were discriminated between by Sophie E. Lellman and Rainer Cramer through liquid AP-MALDI MS, including clinically relevant species such as Escherichia coli, Staphylococcus aureus, and Klebsiella pneumoniae. Each species produced a unique lipid profile in the m/z range of 400–1100, allowing species differentiation and a classification accuracy of 98.63% was achieved by principal component analysis (PCA).65 Furthermore, in 2020, advancing liquid AP MALDI MS towards ultrahigh-throughput analysis for a peptide, lipid, and antibiotic was constructed with <5 s per sample by Henriette Krenkel's group. The ultimate limits of this method in sample throughput can be conservatively estimated to be beyond 10–20 samples per second, which showed highly competitive results (Fig. 3c).66 However, many researchers have determined that an inorganic matrix can reduce the low-mass interferences arising from the organic matrix, so in recent years, metal nanoparticles have emerged as a new choice in AP MALDI systems.Optimization of apparatus settings
As to the optimization of apparatus settings, Victor V. Laiko and co-workers invented a pneumatically assisted (PA) AP MALDI ion source to transfer ions from the atmospheric pressure ionization region to a high vacuum region in 2000, which improved the analysis for a peptide, protein, and oligosaccharide. This ionization approach enables the production of protonated molecular ions for small proteins such as insulin, but tended to form clusters with the matrix material and opened a new chapter for MALDI MS.4 In 2007, an infrared laser was employed in AP MALDI MS by Li Yue's group to stimulate peptides, carbohydrates, and other small biomolecules (e.g., sucrose, reserpine, bradykinin, substance P, and insulin).67 Further, field-free transmission geometry AP MALDI was designed by Sarah and co-workers to identify a lipid, carbohydrate, peptide, and polymer in 2010. Different matrices and preparation experiments were conducted to detect singly and multiply charged ions, while cytochrome c and ubiquitin were obtained in low femtomole amounts and angiotensin I and II were obtained in attomole amounts (Fig. 3e).2 In the same year, an inline pneumatically assisted AP MALDI ion source was developed by Arti T. Navare's team and the capability of the ion carrier, including nitrogen, helium, and sulfur hexafluoride, was tested through experiments of standard peptide and hydroquinone. The best performances were given by nitrogen (Fig. 3f).68 In 2011, U. Hochkirch's group used electron spin resonance (ESR) as a complementary method to examine the distribution profile of the main biotransformation product of nitroxide radicals, CAT-1, while AP MALDI MS was utilized to detect CAT-1 and its diamagnetic metabolites in the different skin layers, discovering information about the concentration of the corresponding hydroxylamine over the sample extension.69 AP visible-wavelength MALDI in transmission geometry MS implement was developed by Raymond E. West III et al. with a LOD of picomole quantities of oligosaccharides and femtomole quantities of peptides in 2013.70 Nevertheless, a novel method utilizing the doping of liquid MALDI samples with divalent metal chloride salts was presented by Oliver J. Hale et al. in 2017, which produced ions with the formula [L + M]2+ (L = lipid and M = divalent metal cation). Pseudo-MS3+ experiments such as in-source decay-CID and ion mobility-enabled time-aligned parallel (TAP) MS provided diagnostic product ion spectra for determining the location of double bonds on the acyl chain and were applied to identify and characterize lipids extracted from soya milk.71 However, in 2018, Bingming Chen et al. combined high-resolution AP MALDI with quadrupole-orbitrap MS, which provided an in situ analysis platform for biomolecules applying multi-mode ionization, including MALDI, LSI, and matrix assisted ionization (MAI) and acquisition, including full MS, targeted MS/MS, data dependent acquisition, and parallel reaction monitoring, aiming to achieve in-depth characterization. This design expanded the detection range and enhanced the fragment efficiency, owing to the multiply charged ion production.72 Anh Tran's team constructed rapid detection of a viral envelop lipid (Influenza A virus, IVA) using a lithium adduct applying AP MALDI MS in 2019. Eight subclasses, including 132 kinds of lipids were identified.73 The construction of the AP MALDI selected reaction monitoring (SRM) method by Vishal Mahale's group in 2022 provided a rapid and quantitative platform for analysis of aflatoxin M1 in milk. The calibration curve displayed excellent linearity (R2 = 0.99) with good recoveries for quality control samples (97–106%) and this method's property was validated by ultrahigh-performance liquid chromatography with fluorescence (UHPLC-FLD).74Optimization of the preconcentration method
For the preconcentration method before analysis, Kamlesh Shrivas and Huifen Wu employed different functional groups capped with silver nanoparticles as affinity probes in AP MALDI ITMS for rapid analysis of sulfur drugs and biothiols in human urine in 2008 (Fig. 3g).75 They also applied AgNPs as the matrix of AP MALDI MS to reduce the side effects arising from a traditional organic matrix. In the same year, Kavita Agrawal and Huifen Wu used bare silica nanoparticles as concentrating and affinity probes for rapid analysis of aminothiols, lysozyme, and peptide mixtures in urine and plasma through AP MALDI ITMS, with enhancing the ionization efficiency of analytes. The limits of detection of the peptide were 4.7 to 360 nM in water, 28 to 620 nM in plasma, and 9.5 to 520 nM in urine, respectively, and homocysteine was successfully discriminated from cysteine (Fig. 3h).76 The nanoparticles displayed excellent capacity for preconcentration, which indicated their potential in AP MALDI systems.So far, metabolites have been emerging as an innovative perspective for detecting disease and depicting the mechanism of disease towards clinical adhibition, owing to the location of metabolites being more distal compared to proteomics and transcriptomics, which could offer more direct information about the human body. Although the detection effect has been improved with the utilization of AP MALDI MS in recent years, there still remain some drawbacks which limit the application of metabolomics by AP MALDI MS in clinical application.
Pharmaceutical detection
Pharmaceutical detection, such as pharmacokinetics, has become much more significant in many domains for determining the clinical medication regimen, predicting the efficacy and toxicity of a drug and rational drug usage. Notably, AP MALDI MS, emerging as an innovative method, has been applied for drug identification from four perspectives: direct analysis, optimization of a mass spectrometer, matrix innovation, and extraction and isolation before analysis.Direct analysis
In terms of direct analysis, in 2008, Bindesh Shrestha and co-workers analyzed different forms of pharmaceuticals, excreted xenobiotics, and endogenous metabolites by AP IR MALDI, suggesting that AP IR MALDI can recover oligomer size distribution changes in excreted samples. Diagnosis of intestinal permeability can be established based on this method.77 However, mass spectra of 39 kinds of seized drugs produced by LC-ESI MS/MS were identified via AP MALDI MS by Pekka Östman et al. in 2012, elucidating that different mass spectrometers could identify an unknown compound with a library established by different ion sources.78Optimization of a mass spectrometer
As to the optimization of a spectrometer, Piia K. Salo's group applied two-dimensional ultra-thin-layer chromatography (2D-UTLC) coupled with AP MALDI MS for bioanalysis in 2007 and benzodiazepines were studied as model substances in human urine.79 First detected by 2D-UTLC, a picomole LOD of benzodiazepine was achieved and then identified by AP MALDI with the aim to enhance the sensitivity. Notably, in 2017, an endogenous substance in a biosample was confirmed to have an influence on the MS results. A chromatography-free AP MALDI high-resolution MS (HRMS) was constructed by Vishal Mahale et al. for simultaneous determination of triazines and triazoles in grapes and 15–20 ng g−1 of LOD and LOQ with <5 ppm of quality error was accomplished.80Matrix innovation
For matrix innovation, Kamlesh Shrivas' group utilized oxidized multiwalled carbon nanotubes (O-MWCNT) as preconcentration probes for quantitative determination of cationic surfactants (CS) in water samples using AP MALDI MS in 2008. The acceptable relative recovery percentage achieved 90.5–97.8% with relative standard deviation (RSD) <15.4%, indicating the high reliability of this separation method.81 Further, Sijian Chen's team also applied four kinds of MWCNT including Fe3O4-droped MWCNT, oxidized MWCNT, β-cyclodextrin-coated MWCNT, and intrinsic MWCNT, to detect nine kinds of pesticides with high performance, finding that the structure of the pesticide could influence the results of analysis in 2023. Experiments confirmed that a matrix can help pesticides to dissociate, while the pesticide would not decompose due to the matrix remaining.82Extraction and isolation before analysis
Besides, extraction and isolation strategies were also employed. In 2017, Yuan Chin et al. applied single drop microextraction as a separation method combined with AP MALDI MS for quantitative analysis of quinidine in fish tissue, showing an absorption rate >72% and a regression ecoefficiency of 0.99, demonstrating the disabled reproducibility and feasibility.83Recently, pharmaceuticals have received more and more attention from the public due to their use not only in medical treatment but also in medical study. However, there are only a few works in drug detection applying AP MALDI MS, which limit the evolution of pharmaceuticals.
Mass spectrometry imaging
Imaging mass spectrometry (IMS) enables spatial visualization of proteins, lipids, and metabolite distribution in tissues, making it a powerful tool for molecule analysis in various fields. We have compiled an overview of IMS applications based on AP MALDI, including lipid visualization, metabolite identification, biomedical analysis, and botanical analysis.Lipid visualization
For lipid visualization, Janne Bredehöft and co-workers visualized lipids and profiled in the vascular organ of the lamina terminalis (OVLT) murine brains during systemic lipopolysaccharide (LPS)-induced inflammation using AP scanning microprobe MALDI MSI in 2019 (Fig. 4b).84 In 2021, another work reported “matrix-free” imaging using the AP transmission mode (TM) MALDI source with low-temperature plasma (LTP) postionization. Direct MSI analysis of murine testis with no sample preparation after tissue sectioning enabled imaging of a range of lipid classes at pixel sizes of 25 μm (Fig. 4a).85 Moreover, in the same year, the distribution of lipids in murine was visualized by Max A. Müller et al. through a high-repetition-rate laser in an AP MALDI MSI system, providing a lateral resolution of 5 μm.86 Besides, lipids in the kidney tissue of a mouse with acute cadmium poisoning were located by Ting Zeng and co-workers with 40 lipid classes visualization in 2022.87 Varun Krishnan and co-workers applied IMS to detect PC and phosphatidylserine (PS) in murine ocular tissue in 2023, which allowed researchers to pinpoint areas of lipid deficiencies or accumulations associated with ocular disorders such as age-related macular degeneration and diabetic retinopathy.88 As to human section visualization, N. Desbenoit and co-workers detected serial cryo-sections of human colon cancer through both time-of-flight secondary ion MS and AP MALDI MS in 2017. Fatty acids, cholesterol, phosphatidylcholine, and sphingomyelin were successfully visualized with complementary data and high spatial correlation from mass spectra, achieving multimodal imaging.89 Kaija Schaepe's team analyzed lipids in native human bone sections to generate high-resolution label-free imaging in 2018. This approach preserved the lipid content (e.g., glycero-, glycerophospho-, and sphingolipids) and the heterogeneous structure of osseous tissue, which enabled consecutive detection of the same sample by TOF-SIMS and AP-SMALDI Orbitrap MSI.90 Recently, many reports focusing on Schistosoma were conducted via AP MALDI MSI. In 2020, lipidomic analysis of Schistosoma mansoni was implemented by Patrik Kadesch et al. via AP scanning microprobe MALDI MSI, demonstrating the tissue- and sex-specificity of lipids.91 In 2022, Katja R. Wiedemann's team characterized changes in the lipid profile of hamster liver after Schistosoma mansoni infection by LC MS/MS, 372 biomarkers were detected. AP MALDI MSI was then employed to analyze these biomarkers, showing the distribution in tissue sections.92 In the same year, 2022, AP scanning microprobe MALDI was established in blood flukes and liver flukes by Saleh M. Khalil's group to identify endogenous lipids that mark characteristic tissues such as the gastrodermis, the tegument, and the parenchyma.93 In addition, in 2015, the phospholipid topography of whole-body sections of the Anopheles stephensi mosquito was characterized by high-resolution AP MALDI MSI with a dedicated sample preparation method.94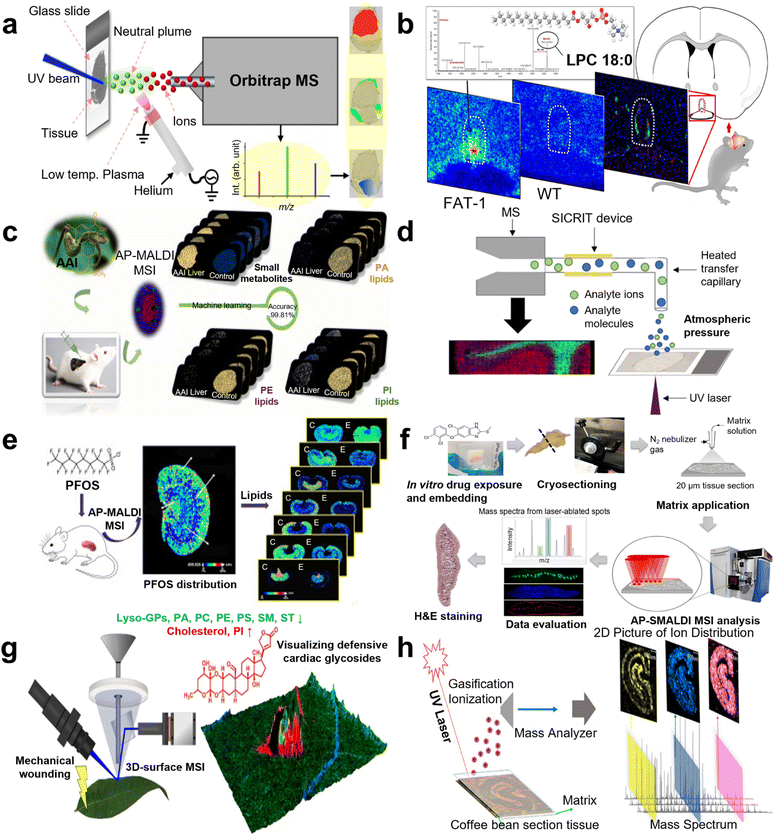 | ||
| Fig. 4 A schematic illustration of AP MALDI MS for mass spectrometry imaging (MSI) applications. (a) The workflow of direct tissue MSI by AP UV MALDI MS with a low-temperature plasma (LTP) for postionization. Reprinted with permission from ref. 85, Copyright 2020 American Chemical Society. (b) A scheme for visualizing and profiling lipids in mouse brains using AP MALDI MSI. Reprinted with permission from ref. 84, Copyright 2019 American Chemical Society. (c) A scheme of metabolic study of an aristolochic acid I-exposed mice liver by AP MALDI MSI and machine learning. Reprinted with permission from ref. 104, Copyright 2022 Elsevier B.V. (d) A scheme of the second-generation transmission mode (TM) AP MALDI imaging platform with in-line plasma postionization using commercially available soft ionization by chemical reaction in transfer (SICRIT) device. Reprinted with permission from ref. 101, Copyright 2020 American Chemical Society. (e) The workflow of spatially revealing perfluorooctane sulfonate-induced nephrotoxicity in mouse kidneys using AP MALDI MSI. Reprinted with permission from ref. 111, Copyright 2022 Elsevier B.V. (f) The workflow of drug imaging and lipid analysis with AP-SMALDI MSI in F. hepatica. Reprinted with permission from ref. 93, Copyright 2022 Springer Nature. (g) The workflow of 3D-surface MALDI MSI for visualizing plant defensive cardiac glycosides in Asclepias curassavica. Reprinted with permission from ref. 120, Copyright 2021 Springer Nature. (h) The workflow of spatial distribution of endogenous molecules in coffee beans by AP MALDI MSI. Reprinted with permission from ref. 118, Copyright 2020 American Chemical Society. | ||
Metabolite distribution
Metabolite distribution was depicted by AP MALDI MSI. AP MALDI imaging of neuropeptides in the mouse pituitary gland was achieved by Sabine Guenther's group with 5 μm spatial resolution and high mass accuracy in 2011.95 Nevertheless, single cell detection was demonstrated by Yvonne Schober and co-workers with the combination of high spatial resolution (7 μm pixel), high mass accuracy (<3 ppm rms), and high mass resolution (R = 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at m/z = 200) in the MS imaging in 2012.96 Dhaka Ram Bhandari and his co-workers employed AP high-resolution scanning microprobe MALDI MSI in 2015 for metabolite localization in whole-body sections and individual organs of the rove beetle Paederus riparius. A spatial resolution of 250 μm was accomplished, and assigned key lipids for specific organs to describe their location in the body were described without any labeling.97 In 2017, another AP MALDI mass spectrometry imaging setup was reported with a lateral resolution of 1.4 μm, a mass resolution greater than 100
000 at m/z = 200) in the MS imaging in 2012.96 Dhaka Ram Bhandari and his co-workers employed AP high-resolution scanning microprobe MALDI MSI in 2015 for metabolite localization in whole-body sections and individual organs of the rove beetle Paederus riparius. A spatial resolution of 250 μm was accomplished, and assigned key lipids for specific organs to describe their location in the body were described without any labeling.97 In 2017, another AP MALDI mass spectrometry imaging setup was reported with a lateral resolution of 1.4 μm, a mass resolution greater than 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, and an accuracy below ±2 ppm. They showed that the setup can be used to detect metabolites, lipids, and small peptides, as well as to perform tandem MS experiments with 1.5 μm2 sampling areas. To showcase these capabilities, they identified subcellular lipid, metabolite, and peptide distributions that differentiate, for example, cilia and oral grooves in Paramecium caudatum.98 In 2018, Shelley N. Jackson and his team conducted AP MALDI MSI to detect gangliosides in the murine brain using 2,6-dihydroxy acetophenone (DHA), which is a novel volatile solid organic matrix. This combination apparently decreased the fragments of ganglioside, owing to the sublimation reduction of DHA.99 In 2019, the build of a new AP TM MALDI MSI ion source was reported by Rory T. Steven's team with plasma ionization enhancement while this novel ion source was used to analyze a selection of increasingly complex systems from molecular standards to murine brain tissue sections. Significant enhancement of detected ion intensity is observed in both positive and negative ion modes in all systems, with up to 2000-fold increases observed for a range of tissue endogenous species.100 However, to improve the detection of certain ions, Efstathios A. Elia et al. developed a second-generation transmission mode AP MALDI imaging platform with in-line plasma postionization in 2020 to detect different metabolites, including amino acids, fatty acids, and raffinose (Fig. 4d).101 In 2021, venom gland mass spectrometry imaging of saw-scaled viper, Echis carinatus sochureki, was realized by Parviz Ghezellous's group with a high lateral resolution of 12 μm via autofocusing AP MALDI MS.102 Novelly, C. Jacques visualized sun filter skin spatial distribution and penetration and detected endogenous metabolites induced by a sun filter in human skin in 2022.103 Metabolic study of aristolochic acid I (AAI)-exposed murine liver section by AP MALDI MSI was conducted by Wenjing Guo's group in 2022, demonstrating the distinct metabolic alternations. Then, AAI-exposed liver was discriminated from a healthy control with a high accuracy of 99.81% by machine learning (LASSO), and metabolomics pathway analysis (MetPA) was conducted to identify influenced pathways (Fig. 4c).104
000, and an accuracy below ±2 ppm. They showed that the setup can be used to detect metabolites, lipids, and small peptides, as well as to perform tandem MS experiments with 1.5 μm2 sampling areas. To showcase these capabilities, they identified subcellular lipid, metabolite, and peptide distributions that differentiate, for example, cilia and oral grooves in Paramecium caudatum.98 In 2018, Shelley N. Jackson and his team conducted AP MALDI MSI to detect gangliosides in the murine brain using 2,6-dihydroxy acetophenone (DHA), which is a novel volatile solid organic matrix. This combination apparently decreased the fragments of ganglioside, owing to the sublimation reduction of DHA.99 In 2019, the build of a new AP TM MALDI MSI ion source was reported by Rory T. Steven's team with plasma ionization enhancement while this novel ion source was used to analyze a selection of increasingly complex systems from molecular standards to murine brain tissue sections. Significant enhancement of detected ion intensity is observed in both positive and negative ion modes in all systems, with up to 2000-fold increases observed for a range of tissue endogenous species.100 However, to improve the detection of certain ions, Efstathios A. Elia et al. developed a second-generation transmission mode AP MALDI imaging platform with in-line plasma postionization in 2020 to detect different metabolites, including amino acids, fatty acids, and raffinose (Fig. 4d).101 In 2021, venom gland mass spectrometry imaging of saw-scaled viper, Echis carinatus sochureki, was realized by Parviz Ghezellous's group with a high lateral resolution of 12 μm via autofocusing AP MALDI MS.102 Novelly, C. Jacques visualized sun filter skin spatial distribution and penetration and detected endogenous metabolites induced by a sun filter in human skin in 2022.103 Metabolic study of aristolochic acid I (AAI)-exposed murine liver section by AP MALDI MSI was conducted by Wenjing Guo's group in 2022, demonstrating the distinct metabolic alternations. Then, AAI-exposed liver was discriminated from a healthy control with a high accuracy of 99.81% by machine learning (LASSO), and metabolomics pathway analysis (MetPA) was conducted to identify influenced pathways (Fig. 4c).104
Biomedicine analysis
MSI is a cutting-edge method in the fields of life sciences and biomedicine, which integrates molecular imaging and advanced image analysis. Thereby, it can visualize the molecular distribution of a large number of endogenous and exogenous compounds simultaneously in tissues without the addition of labels. In drug distribution detection, an in vivo experiment in a rodent model utilizing AP MALDI MSI was conducted by Akos Vegvari's team in 2017, revealing the distribution of tiotropium with 10 μm resolution.105 Meanwhile, AP MALDI imaging mass microscope combined with a relative exposure approach was employed by Tai Rao et al. to study pharmacokinetics via octreotide analysis in mouse target tissues.106 In 2020, three kinds of MSI, desorption electrospray ionization (DESI) MSI, MALDI MSI, and AP MALDI MSI, were employed by Ariful Islam's team for rap mapping of imipramine, chloroquine, and their metabolites in mice kidneys and brains. AP MALDI MSI displayed the best performance and MS/MS was applied for double-check identification.107 Furthermore, in the same year, a study of phospholipid imaging of zebrafish exposed to fipronil was conducted by Wenjie Liu and his group, depicting the alternation of the phospholipid metabolism.108 Ting Zeng's group integrated omics analysis and AP MALDI MSI in 2021 to investigate cadmium toxicity in female mice, uncovering the toxicological mechanism via representative lipid observation.109 Annika S. Mokosch's team investigated the potential uptake and distribution of imatinib in adult schistosomes including its distribution kinetics in 2021.110 Yanyan Chen's team also imaged murine kidneys to reveal the perfluorooctane sulfonate-induced nephrotoxicity, elucidating changes of 42 lipid species in 2022 (Fig. 4e).111 Lavinia Morosi and her team developed an innovative quantitative measurement and validated it for pioglitazone analysis in neoplastic kidney tissues and Carolin M. Morawietz and her team conducted in vitro spatial visualization of triclabendazole (TCBZ) uptake and distribution in Fasciola hepatica with 100 μm of resolution in 2022 (Fig. 4f).93,112Botany detection
Additionally, botany has been studied by AP MALDI MSI in recent years. In 2007, the feasibility of AP infrared MALDI imaging was undertaken by Yue Li et al. according to plant metabolomics, observing carbohydrate, oligosaccharide, amino acid, organic acid, and lipid mass peaks in both positive and negative modes.67 This team also visualized the spatial distribution of metabolites in 2008, including GABA, glutamine, hexose sugars, toluidine blue O, and sucrose in white lily flowers, depicting the accessibility of AP MALDI in botany.113 Besides, in 2014, Bin Li's group imaged natural products in glycyrrhiza glabra rhizome at the cellular level, while AP MALDI MSI at 10 μm pixel size was performed to unravel the spatio-chemical distribution of major secondary metabolites in the root of Paeonia lactiflora in 2016.114,115 In 2017, Ralf W. Kessler's group implemented in situ monitoring of molecular changes during cell differentiation processes in marine macroalgae through AP MALDI MSI, determining representative biomarkers in different developmental stages.116 In 2018, the comparison of vacuum and AP MALDI MSI was implemented by Caitlin Keller and co-workers for detection of metabolites involved in salt stress in Medicago truncatula with the identification of LC tandem MS. This work showed better performance in AP MALDI, providing robust evidence for plant applications.117 Furthermore, in 2020, Li Na and co-workers achieved spatial distribution of endogenous molecules (e.g., sucrose, caffeine, and caffeoylquinic acid) in coffee beans with PCA analysis, elucidating differences in chemical composition and relative content (Fig. 4h).118 Distribution of phytochemicals in the dried root of Isatis tinctoria was carried out by Li Xing Nie et al. and 118 compositions were identified without extraction and isolation in 2021. Using partial least squares regression (PLS), samples collected from four habitats were differentiated unambiguously.119 Moreover, Domenic Dreisbach and co-workers visualized plant defensive cardiac glycosides in Asclepias curassavica through three dimension-surface MALDI MSI in 2021 (Fig. 4g). Variation in oligosaccharide profiles was observed by Jonatan R. Granborg's team in different regions of maize kernels after treatment with xylanases both by AP MALDI MSI in 2023.120,121In summary, atmospheric pressure mass spectrometry imaging has been extensively employed from lipid, metabolite, biomedical, and botany perspectives, which allows the construction of analyte distribution reflecting the perturbations in different tissues, resulting in a powerful tool for biomarker visualization in many domains.
Other applications
In addition to the aforementioned applications, the advantages of AP MALDI MS have led to its increasing use in the fields of food science and environmental analysis.For environmental analysis, in 2022, Lamprini and colleagues utilized AP MALDI to analyze and evaluate engine oil, successfully identifying pure additives. Further, this work revealed that AP MALDI is an efficient technique for directly determining lubricant additives from commercial oil products.122 In 2023, Maurice and co-workers developed and optimized 48 well graphene nanoplatelets (GNP) doped AP MALDI target plates for the fast-screening analysis of additives in electronics and plastic consumer products, while a total of 56 additives including antioxidants, flame retardants, plasticizers, UV-stabilizers, and UV-filters were identified.123 This helps in understanding the level of pollution in the environment and taking necessary measures to protect the environment and human health.
In the field of food science, in 2022, Diming Tan and his group utilized AP MALDI MS to characterize the diversity of esters and partially monitor the fate of propionyl-fructooligosaccharides (FOS). Their aim was to utilize ester bonds to link propionic acid (PA) and a prebiotic FOS to form propionyl-FOS capable of delivering PA to the colon.124 Furthermore, in 2023, Francesco and co-workers subjected fruit seeds belonging to the pomegranate cultivar “Granata” to extraction and oily component analysis through AP MALDI MS to gain information about their composition.125 This contributes to enhancing food safety and preventing outbreaks of foodborne diseases.
It is worth noting that while AP MALDI shows promising potential in these fields, its application in food science and environmental analysis is still relatively new and requires further research and development to refine analytical methods and enhance accuracy and repeatability.
Conclusions and perspectives
AP MALDI has attracted the attention of scientists due to its unique advantages. Over the past 20 years, its analytical performance has been continuously improved, leading to a series of advancements in various analysis domains, including peptide and protein analysis, metabolite detection, pharmaceutical identification, and mass spectrometry imaging.In this review, we have summarized the major applications of AP MALDI in the aforementioned fields. With the increasing demand for these applications, we believe that in the near future, further research in this field will be carried out, including but not limited to (1) more high sensitivity AP MALDI analysis system development; (2) innovative matrix synthesis applied in AP MALDI; (3) AP MALDI MS systems with low costs and smaller sizes; and (4) high spatial resolution performance for AP MALDI MS imaging. Nevertheless, our work did not record applications of environmental analysis and food science. However, considering the evident virtues of AP MALDI, the prevailing employment of AP MALDI is imperative. We believe our review will contribute towards a comprehensive summary of AP MALDI applications.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge financial support from the National Key R&D Program of China (2022YFE0103500, 2021YFF0703500). This work was also sponsored by Medical-Engineering Joint Funds of Shanghai Jiao Tong University (YG2021ZD09, YG2022QN107, YG2023ZD08) and Open Program from the National Institute of Metrology of China (AKYKF2306).Notes and references
- C. Creaser and L. Ratcliffe, Curr. Anal. Chem., 2006, 2, 9–15 CrossRef CAS.
- S. Trimpin, E. D. Inutan, T. N. Herath and C. N. McEwen, Anal. Chem., 2010, 82, 11–15 CrossRef CAS PubMed.
- J. L. Wolfender, F. Chu, H. Ball, F. Wolfender, M. Fainzilber, M. A. Baldwin and A. L. Burlingame, J. Mass Spectrom., 1999, 34, 447–454 CrossRef CAS.
- V. V. Laiko, M. A. Baldwin and A. L. Burlingame, Anal. Chem., 2000, 72, 652–657 CrossRef CAS PubMed.
- J. T. Mehl, J. J. Cummings, E. Rohde and N. N. Yates, Rapid Commun. Mass Spectrom., 2003, 17, 1600–1610 CrossRef CAS PubMed.
- C. S. Creaser, J. C. Reynolds and D. J. Harvey, Rapid Commun. Mass Spectrom., 2002, 16, 176–184 CrossRef CAS PubMed.
- G. Grasso, M. Fragai, E. Rizzarelli, G. Spoto and K. J. Yeo, J. Mass Spectrom., 2006, 41, 1561–1569 CrossRef CAS PubMed.
- S. C. Moyer, R. J. Cotter and A. S. Woods, J. Am. Soc. Mass Spectrom., 2002, 13, 274–283 CrossRef CAS PubMed.
- A. Misharin, K. Novoselov, V. Laiko and V. M. Doroshenko, Anal. Chem., 2012, 84, 10105–10112 CrossRef CAS PubMed.
- S. Reis-Dennis, Monash Bioeth. Rev., 2020, 38, 83–86 CrossRef PubMed.
- D. I. Carroll, I. Dzidic, R. N. Stillwell, K. D. Haegele and E. C. Horning, Anal. Chem., 2002, 47, 2369–2373 CrossRef.
- K. Tanaka, H. Waki, Y. Ido, S. Akita, Y. Yoshida, T. Yoshida and T. Matsuo, Rapid Commun. Mass Spectrom., 1988, 2, 151–153 CrossRef CAS.
- Z. Takats, J. M. Wiseman, B. Gologan and R. G. Cooks, Science, 2004, 306, 471–473 CrossRef CAS PubMed.
- R. B. Cody, J. A. Laramee and H. D. Durst, Anal. Chem., 2005, 77, 2297–2302 CrossRef CAS PubMed.
- N. Na, M. Zhao, S. Zhang, C. Yang and X. Zhang, J. Am. Soc. Mass Spectrom., 2007, 18, 1859–1862 CrossRef CAS PubMed.
- J. Shiea, M. Z. Huang, H. J. Hsu, C. Y. Lee, C. H. Yuan, I. Beech and J. Sunner, Rapid Commun. Mass Spectrom., 2005, 19, 3701–3704 CrossRef CAS PubMed.
- C. N. McEwen, R. G. McKay and B. S. Larsen, Anal. Chem., 2005, 77, 7826–7831 CrossRef CAS PubMed.
- L. Gao, R. G. Cooks and Z. Ouyang, Anal. Chem., 2008, 80, 4026–4032 CrossRef CAS PubMed.
- H. Ehring, M. Karas and F. Hillenkamp, Org. Mass Spectrom., 1992, 27, 472–480 CrossRef CAS.
- B. C. Lund, T. E. Abrams and A. A. Gravely, J. Rehabil. Res. Dev., 2011, 48, vii–ix Search PubMed.
- R. Knochenmuss and R. Zenobi, Chem. Rev., 2003, 103, 441–452 CrossRef CAS PubMed.
- R. Knochenmuss, Analyst, 2006, 131, 966–986 RSC.
- M. Karas and R. Kruger, Chem. Rev., 2003, 103, 427–440 CrossRef CAS PubMed.
- M. Karas, M. Glückmann and J. r. Schäfer, J. Mass Spectrom., 2000, 35, 1–12 CrossRef CAS.
- R. Knochenmuss and L. V. Zhigilei, Anal. Bioanal. Chem., 2012, 402, 2511–2519 CrossRef CAS PubMed.
- P. Ryumin, J. Brown, M. Morris and R. Cramer, Methods, 2016, 104, 11–20 CrossRef CAS PubMed.
- B. Reif, S. E. Ashbrook, L. Emsley and M. Hong, Nat. Rev. Methods Primers, 2021, 1, 2 CrossRef CAS PubMed.
- O. Beckonert, H. C. Keun, T. M. Ebbels, J. Bundy, E. Holmes, J. C. Lindon and J. K. Nicholson, Nat. Protoc., 2007, 2, 2692–2703 CrossRef CAS PubMed.
- L. Cui, H. J. Butler, P. L. Martin-Hirsch and F. L. Martin, Anal. Methods, 2016, 8, 481–487 RSC.
- A. Navare, M. Nouzova, F. G. Noriega, S. Hernandez-Martinez, C. Menzel and F. M. Fernandez, Rapid Commun. Mass Spectrom., 2009, 23, 477–486 CrossRef CAS PubMed.
- K. Turney and W. W. Harrison, Rapid Commun. Mass Spectrom., 2004, 18, 629–635 CrossRef CAS PubMed.
- G. Luo, I. Marginean and A. Vertes, Anal. Chem., 2002, 74, 6185–6190 CrossRef CAS PubMed.
- D. O. Konn, J. Murrell, D. Despeyroux and S. J. Gaskell, J. Am. Soc. Mass Spectrom., 2005, 16, 743–751 CrossRef CAS PubMed.
- V. Gabelica, E. Schulz and M. Karas, J. Mass Spectrom., 2004, 39, 579–593 CrossRef CAS PubMed.
- V. M. Doroshenko, V. V. Laiko, N. I. Taranenko, V. D. Berkout and H. S. Lee, Int. J. Mass Spectrom., 2002, 221, 39–58 CrossRef CAS.
- Y. Wang, B. B. Schneider, T. R. Covey and J. Pawliszyn, Anal. Chem., 2005, 77, 8095–8101 CrossRef CAS PubMed.
- P. R. Sudhir, H. F. Wu and Z. C. Zhou, Anal. Chem., 2005, 77, 7380–7385 CrossRef CAS PubMed.
- P. R. Sudhir, K. Shrivas, Z. C. Zhou and H. F. Wu, Rapid Commun. Mass Spectrom., 2008, 22, 3076–3086 CrossRef CAS PubMed.
- S. Sabu, F. C. Yang, Y. S. Wang, W. H. Chen, M. I. Chou, H. C. Chang and C. C. Han, Anal. Biochem., 2007, 367, 190–200 CrossRef CAS PubMed.
- K. Shrivas and H. F. Wu, Anal. Chem., 2008, 80, 2583–2589 CrossRef CAS PubMed.
- P. Ryumin, J. Brown, M. Morris and R. Cramer, Int. J. Mass Spectrom., 2017, 416, 20–28 CrossRef CAS.
- G. Grasso, E. Rizzarelli and G. Spoto, J. Mass Spectrom., 2007, 42, 1590–1598 CrossRef CAS PubMed.
- G. Grasso, P. Mineo, E. Rizzarelli and G. Spoto, Int. J. Mass Spectrom., 2009, 282, 50–55 CrossRef CAS.
- J. Nguyen and S. C. Russell, J. Am. Soc. Mass Spectrom., 2010, 21, 993–1001 CrossRef CAS PubMed.
- R. Ait-Belkacem, M. Dilillo, D. Pellegrini, A. Yadav, E. L. de Graaf and L. A. McDonnell, J. Am. Soc. Mass Spectrom., 2016, 27, 2075–2079 CrossRef CAS PubMed.
- C. A. Miller, D. Yi and P. D. Perkins, Rapid Commun. Mass Spectrom., 2003, 17, 860–868 CrossRef CAS PubMed.
- W. E. Steiner, B. H. Clowers, W. A. English and H. H. Hill Jr, Rapid Commun. Mass Spectrom., 2004, 18, 882–888 CrossRef CAS PubMed.
- E. Inutan and S. Trimpin, J. Am. Soc. Mass Spectrom., 2010, 21, 1260–1264 CrossRef CAS PubMed.
- J. R. Stutzman and S. A. McLuckey, Anal. Chem., 2012, 84, 10679–10685 CrossRef CAS PubMed.
- E. Pittenauer, A. Kassler, R. Haubner and G. Allmaier, J. Proteomics, 2011, 74, 975–981 CrossRef CAS PubMed.
- R. Cramer, A. Pirkl, F. Hillenkamp and K. Dreisewerd, Angew Chem. Int. Ed. Engl., 2013, 52, 2364–2367 CrossRef CAS PubMed.
- P. Ryumin and R. Cramer, Anal. Chim. Acta, 2018, 1013, 43–53 CrossRef CAS PubMed.
- J. Cao, Y. Wang, Y. Zhang and K. Qian, Adv. Intell. Syst., 2022, 4, 2100191 CrossRef.
- E. Pittenauer, M. Zehl, O. Belgacem, E. Raptakis, R. Mistrik and G. Allmaier, J. Mass Spectrom., 2006, 41, 421–447 CrossRef CAS PubMed.
- G. Grasso, A. I. Bush, R. D'Agata, E. Rizzarelli and G. Spoto, Eur. Biophys. J., 2009, 38, 407–414 CrossRef CAS PubMed.
- B. Shrestha, P. Nemes, J. Nazarian, Y. Hathout, E. P. Hoffman and A. Vertes, Analyst, 2010, 135, 751–758 RSC.
- E. Ito, A. Tominaga, H. Waki, K. Miseki, A. Tomioka, K. Nakajima, K. Kakehi, M. Suzuki, N. Taniguchi and A. Suzuki, Neurochem. Res., 2012, 37, 1315–1324 CrossRef CAS PubMed.
- C. Piras, C. Ceniti, E. Hartmane, N. Costanzo, V. M. Morittu, P. Roncada, D. Britti and R. Cramer, Proteomes, 2020, 8, 20 CrossRef CAS PubMed.
- S. L. Chong, T. Nissila, R. A. Ketola, S. Koutaniemi, M. Derba-Maceluch, E. J. Mellerowicz, M. Tenkanen and P. Tuomainen, Anal. Bioanal. Chem., 2011, 401, 2995–3009 CrossRef CAS PubMed.
- G. Grasso, M. Calcagno, A. Rapisarda, R. D'Agata and G. Spoto, Anal. Bioanal. Chem., 2017, 409, 3943–3950 CrossRef CAS PubMed.
- R. Hiraguchi, H. Hazama, K. Masuda and K. Awazu, J. Mass Spectrom., 2015, 50, 65–70 CrossRef CAS PubMed.
- H. Kasai, M. Nakakoshi, T. Sugita, M. Matsuoka, Y. Yamazaki, Y. Unno, H. Nakajima, H. Fujiwake and M. Tsubuki, Anal. Sci., 2016, 32, 907–910 CrossRef CAS PubMed.
- O. J. Hale, P. Ryumin, J. M. Brown, M. Morris and R. Cramer, Rapid Commun. Mass Spectrom., 2021, 35(1), e8246 CrossRef CAS PubMed.
- O. J. Hale, M. Morris, B. Jones, C. K. Reynolds and R. Cramer, ACS Omega, 2019, 4, 12759–12765 CrossRef CAS PubMed.
- S. E. Lellman and R. Cramer, Clin. Chem. Lab. Med., 2020, 58, 930–938 CrossRef CAS PubMed.
- H. Krenkel, E. Hartmane, C. Piras, J. Brown, M. Morris and R. Cramer, Anal. Chem., 2020, 92, 2931–2936 CrossRef CAS PubMed.
- Y. Li, B. Shrestha and A. Vertes, Anal. Chem., 2007, 79, 523–532 CrossRef CAS PubMed.
- A. T. Navare and F. M. Fernandez, J. Mass Spectrom., 2010, 45, 635–642 CrossRef CAS PubMed.
- U. Hochkirch, W. Herrmann, R. Stosser, M. Linscheid and H. H. Borchert, Anal. Bioanal. Chem., 2011, 401, 901–907 CrossRef CAS PubMed.
- R. E. West III, E. W. Findsen and D. Isailovic, J. Am. Soc. Mass Spectrom., 2013, 24, 1467–1476 CrossRef PubMed.
- O. J. Hale and R. Cramer, Anal. Bioanal. Chem., 2018, 410, 1435–1444 CrossRef CAS PubMed.
- B. Chen, C. OuYang, Z. Tian, M. Xu and L. Li, Anal. Chim. Acta, 2018, 1007, 16–25 CrossRef CAS PubMed.
- A. Tran, I. A. Monreal, E. Moskovets, H. C. Aguilar and J. W. Jones, J. Am. Soc. Mass Spectrom., 2021, 32, 2322–2333 CrossRef CAS PubMed.
- V. Mahale, M. Gupta, M. Dhanshetty, S. Chawan, E. Moskovets, K. Banerjee, N. Bhattacharya and V. Panchagnula, J. AOAC Int., 2022, 105, 1043–1050 CrossRef PubMed.
- K. Shrivas and H. F. Wu, Rapid Commun. Mass Spectrom., 2008, 22, 2863–2872 CrossRef CAS PubMed.
- K. Agrawal and H. F. Wu, Rapid Commun. Mass Spectrom., 2008, 22, 283–290 CrossRef CAS PubMed.
- B. Shrestha, Y. Li and A. Vertes, Metabolomics, 2008, 4, 297–311 CrossRef CAS.
- P. Östman, R. A. Ketola and I. Ojanpera, Drug Test. Anal., 2013, 5, 68–73 CrossRef PubMed.
- P. K. Salo, S. Vilmunen, H. Salomies, R. A. Ketola and R. Kostiainen, Anal. Chem., 2007, 79, 2101–2108 CrossRef CAS PubMed.
- V. Mahale, A. Singh, G. S. Phadke, A. D. Ghanate, D. P. Oulkar, K. Banerjee and V. Panchagnula, J. AOAC Int., 2017, 100, 640–646 CrossRef CAS PubMed.
- K. Shrivas and H. F. Wu, Anal. Chim. Acta, 2008, 628, 198–203 CrossRef CAS PubMed.
- S. Chen, H. Zhao, L. Jiao, Z. Wang, M. Zhao, L. Tian, Y. Xiu and S. Liu, Chin. Chem. Lett., 2023, 34, 107286 CrossRef CAS.
- Y. C. Chen, H. N. Abdelhamid and H. F. Wu, Mass Spectrom. Lett., 2017, 8, 8–13 CrossRef CAS.
- J. Bredehoft, D. R. Bhandari, F. J. Pflieger, S. Schulz, J. X. Kang, S. Laye, J. Roth, R. Gerstberger, K. Mayer, B. Spengler and C. Rummel, ACS Chem. Neurosci., 2019, 10, 4394–4406 CrossRef PubMed.
- B. Yan, T. Murta, E. A. Elia, R. T. Steven and J. Bunch, J. Am. Soc. Mass Spectrom., 2021, 32, 429–435 CrossRef CAS PubMed.
- M. A. Müller, M. Kompauer, K. Strupat, S. Heiles and B. Spengler, J. Am. Soc. Mass Spectrom., 2021, 32, 465–472 CrossRef PubMed.
- T. Zeng, R. Zhang, Y. Chen, W. Guo, J. Wang and Z. Cai, Talanta, 2022, 245, 123466 CrossRef CAS PubMed.
- V. Krishnan, S. D. Meehan, C. Hayter and S. K. Bhattacharya, Methods Mol. Biol., 2023, 2625, 149–161 CrossRef CAS PubMed.
- N. Desbenoit, A. Walch, B. Spengler, A. Brunelle and A. Rompp, Rapid Commun. Mass Spectrom., 2018, 32, 159–166 CrossRef CAS PubMed.
- K. Schaepe, D. R. Bhandari, J. Werner, A. Henss, A. Pirkl, M. Kleine-Boymann, M. Rohnke, S. Wenisch, E. Neumann, J. Janek and B. Spengler, Anal. Chem., 2018, 90, 8856–8864 CrossRef CAS PubMed.
- P. Kadesch, T. Quack, S. Gerbig, C. G. Grevelding and B. Spengler, PLoS Neglected Trop. Dis., 2020, 14, e0008145 CrossRef CAS PubMed.
- K. R. Wiedemann, A. Peter Ventura, S. Gerbig, M. Roderfeld, T. Quack, C. G. Grevelding, E. Roeb and B. Spengler, Anal. Bioanal. Chem., 2022, 414, 3653–3665 CrossRef CAS PubMed.
- C. M. Morawietz, A. M. Peter Ventura, C. G. Grevelding, S. Haeberlein and B. Spengler, Parasitol. Res., 2022, 121, 1145–1153 CrossRef PubMed.
- S. M. Khalil, A. Rompp, J. Pretzel, K. Becker and B. Spengler, Anal. Chem., 2015, 87, 11309–11316 CrossRef CAS PubMed.
- S. Guenther, A. Römpp, W. Kummer and B. Spengler, Int. J. Mass Spectrom., 2011, 305, 228–237 CrossRef CAS.
- Y. Schober, S. Guenther, B. Spengler and A. Rompp, Anal. Chem., 2012, 84, 6293–6297 CrossRef CAS PubMed.
- D. R. Bhandari, M. Schott, A. Rompp, A. Vilcinskas and B. Spengler, Anal. Bioanal. Chem., 2015, 407, 2189–2201 CrossRef CAS PubMed.
- M. Kompauer, S. Heiles and B. Spengler, Nat. Methods, 2017, 14, 90–96 CrossRef CAS PubMed.
- S. N. Jackson, L. Muller, A. Roux, B. Oktem, E. Moskovets, V. M. Doroshenko and A. S. Woods, J. Am. Soc. Mass Spectrom., 2018, 29, 1463–1472 CrossRef CAS PubMed.
- R. T. Steven, M. Shaw, A. Dexter, T. Murta, F. M. Green, K. N. Robinson, I. S. Gilmore, Z. Takats and J. Bunch, Anal. Chim. Acta, 2019, 1051, 110–119 CrossRef CAS PubMed.
- E. A. Elia, M. Niehaus, R. T. Steven, J. C. Wolf and J. Bunch, Anal. Chem., 2020, 92, 15285–15290 CrossRef CAS PubMed.
- P. Ghezellou, S. Heiles, P. Kadesch, A. Ghassempour and B. Spengler, J. Am. Soc. Mass Spectrom., 2021, 32, 1105–1115 CrossRef CAS PubMed.
- C. Jacques, F. Crépel, D. El Assad, T. B. Angerer, J. Bour, C. Jeanjean-Miquel, D. Redoules, D. Bacqueville, F. Pamelard, S. Bessou-Touya, G. Frache and H. Duplan, J. Controlled Release, 2022, 347, 78–88 CrossRef CAS PubMed.
- W. Guo, Z. Shi, T. Zeng, Y. He, Z. Cai and J. Zhang, Talanta, 2022, 241, 123261 CrossRef CAS PubMed.
- A. Vegvari, T. E. Fehniger, M. Dahlback, G. Marko-Varga and K. Strupat, Curr. Anal. Chem., 2017, 13, 182–186 CrossRef CAS.
- T. Rao, Y. Shao, N. Hamada, Y. Li, H. Ye, D. Kang, B. Shen, X. Li, X. Yin, Z. Zhu, H. Li, L. Xie, G. Wang and Y. Liang, Anal. Chim. Acta, 2017, 952, 71–80 CrossRef CAS PubMed.
- A. Islam, T. Sakamoto, Q. Zhai, M. M. Rahman, M. A. Mamun, Y. Takahashi, T. Kahyo and M. Setou, Pharm., 2022, 15, 1314 CAS.
- W. Liu, H. Nie, D. Liang, Y. Bai and H. Liu, Talanta, 2020, 209, 120357 CrossRef CAS PubMed.
- T. Zeng, W. Guo, L. Jiang, Q. Luo, Z. Shi, B. Lei, J. Zhang and Z. Cai, Sci. Total Environ., 2021, 801, 149803 CrossRef CAS PubMed.
- A. S. Mokosch, S. Gerbig, C. G. Grevelding, S. Haeberlein and B. Spengler, Anal. Bioanal. Chem., 2021, 413, 2755–2766 CrossRef CAS PubMed.
- Y. Chen, L. Jiang, R. Zhang, Z. Shi, C. Xie, Y. Hong, J. Wang and Z. Cai, Sci. Total Environ., 2022, 838, 156380 CrossRef CAS PubMed.
- L. Morosi, C. Matteo, M. Meroni, T. Ceruti, I. Fuso Nerini, E. Bello, R. Frapolli, M. D'Incalci, M. Zucchetti and E. Davoli, Talanta, 2022, 237, 122918 CrossRef CAS PubMed.
- Y. Li, B. Shrestha and A. Vertes, Anal. Chem., 2008, 80, 407–420 CrossRef CAS PubMed.
- B. Li, D. R. Bhandari, C. Janfelt, A. Rompp and B. Spengler, Plant J., 2014, 80, 161–171 CrossRef CAS PubMed.
- B. Li, D. R. Bhandari, A. Rompp and B. Spengler, Sci. Rep., 2016, 6, 36074 CrossRef CAS PubMed.
- R. W. Kessler, A. C. Crecelius, U. S. Schubert and T. Wichard, Anal. Bioanal. Chem., 2017, 409, 4893–4903 CrossRef CAS PubMed.
- C. Keller, J. Maeda, D. Jayaraman, S. Chakraborty, M. R. Sussman, J. M. Harris, J. M. Ane and L. Li, Front. Plant Sci., 2018, 9, 1238 CrossRef PubMed.
- N. Li, J. Dong, C. Dong, Y. Han, H. Liu, F. Du and H. Nie, J. Am. Soc. Mass Spectrom., 2020, 31, 2503–2510 CrossRef CAS PubMed.
- L. X. Nie, J. Dong, L. Y. Huang, X. Y. Qian, C. J. Lian, S. Kang, Z. Dai and S. C. Ma, Front. Pharmacol., 2021, 12, 685575 CrossRef CAS PubMed.
- D. Dreisbach, G. Petschenka, B. Spengler and D. R. Bhandari, Anal. Bioanal. Chem., 2021, 413, 2125–2134 CrossRef CAS PubMed.
- J. R. Granborg, S. G. Kaasgaard and C. Janfelt, J. Cereal Sci., 2023, 109, 103586 CrossRef CAS.
- L. Ramopoulou, L. Widder, J. Brenner, A. Ristic and G. Allmaier, Rapid Commun. Mass Spectrom., 2022, 36, e9271 CrossRef CAS PubMed.
- M. de Jonker, P. E. G. Leonards, M. H. Lamoree and S. H. Brandsma, Toxics, 2023, 11, 108 CrossRef CAS PubMed.
- D. Tan, W. Chen, Z. Yang, M. Rong, R. Huang, X. Wu, W. Bai and L. Tian, Food Hydrocolloids, 2023, 142, 108782 CrossRef CAS.
- F. Cairone, C. Salvitti, A. Iazzetti, G. Fabrizi, A. Troiani, F. Pepi and S. Cesa, Foods, 2023, 12, 1592 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |



