Open Access Article
Open Access ArticleOne-step simultaneous liquid phase exfoliation-induced chirality in graphene and their chirality-mediated microRNA delivery†
Pranav
ab,
Eswara N. H. K.
Ghali
ab,
Neeraj
Chauhan
ab,
Rahul
Tiwari
ab,
Marco
Cabrera
ab,
Subhash C.
Chauhan
ab and
Murali M.
Yallapu
 *ab
*ab
aDepartment of Immunology and Microbiology, School of Medicine, University of Texas Rio Grande Valley, McAllen, TX, USA. E-mail: murali.yallapu@utrgv.edu; Tel: +1 956-296-1734
bSouth Texas Center of Excellence in Cancer Research, School of Medicine, University of Texas Rio Grande Valley, McAllen, TX, USA
First published on 4th October 2023
Abstract
Graphene (G) has established itself as an exciting prospect for a broad range of applications owing to its remarkable properties. Recent innovations in chiral nanosystems have led to sensors, drug delivery, catalysis, etc. owing to the stereospecific interactions between various nanosystems and enantiomers. As the molecular structure of G itself is achiral introducing chirality in G by simple attachment of a functional group (a chiral ligand) on the G nanosheet may result in more diverse applications. Herein, we demonstrate direct liquid phase exfoliation and chiral induction in G nanosheets abbreviated as L-graphene and D-graphene in the presence of chiral L-tyrosine and D-tyrosine and by applying high-temperature sonication. The obtained exfoliated nanosheets demonstrated stable chirality confirmed by circular dichroism. Fourier transform infrared (FTIR) spectra, Raman spectroscopy, transmission electron microscopy (TEM), X-ray photoelectron spectroscopy (XPS), and differential scanning calorimetry (DSC) showed functional, structural, morphological, surface, and thermal characteristics of L-graphene and D-graphene. The hemo-compatibility of these chiral graphenes was evaluated for the very first time utilizing human red blood cells. Lastly, for the very first time, an attempt was made to explore enantiomeric binding between chiral L-graphene and D-graphene with microRNA (miR-205) and their possibility towards chirality-mediated gene delivery in prostate cancerous cells.
1. Introduction
One of the most intriguing qualities found in nature is chirality, which has the most incredible and diverse technological applications ranging from catalysis to sensing, drug delivery, etc.1 Moreover, chirality has been associated with different biological events such as cell metabolism, cell fate, and, most importantly, the evolution of life on the earth.2 In humans, L-amino acids, L-phospholipids, and D-nucleotides are the basic building blocks that govern most of the cellular and sub-cellular functions3,4 whereas in bacteria the D-amino acids (i.e.D-Ala and D-Glu) are present in the bacterial cell membrane providing resistance suggesting the importance and diversity associated with chirality.5,6Two-dimensional (2D) nanomaterials such as graphene (G) and atomic-layered transition metal dichalcogenides have established themselves at the forefront of different industrial innovations in the areas of electronics to the medical industry.7 Among different 2D nanomaterials, G and its analogues have been the most explored materials since its discovery. Structurally G is an achiral system and by the precise introduction of chirality in it, the formed chiral 2D systems can exhibit superior properties and diverse applications. The structural and chiral post-functionalization are the two major strategies used so far for introducing chirality in G. However, post-functionalization strategies are the most favorable route from an industrial perspective, especially with aqueous forms of G dispersion.1
Chiral post-functionalization can be achieved through covalent or noncovalent functionalization using different chiral systems such as synthetic polymers, small molecules, naturally available macromolecules, etc.8–11 Recently, Geng et al. introduced chirality in the G basal plane using L(D)-tryptophan and thermal fusion.12 In another work, direct exfoliation and introduction chirality in 2D MoS2 were achieved in the presence of chiral L(D)-cysteine and L(D)-penicillamine in an aqueous system.13 Considering the potential applications with chiral 2D nanomaterials, exploring a facile and single-step synthesis method is highly warranted. So far to the best of our knowledge, no report has been found to develop a synthesis method for direct exfoliation as well as induction of chirality in a single step for G.
In the present work, aqueous chiral G abbreviated as L-graphene and D-graphene was produced in a single step by using chiral L-tyrosine and D-tyrosine as a stabilizing and chiral-inducing agent and applying high-temperature sonication. The chirality of the exfoliated L-graphene and D-graphene was assessed with circular dichroism (CD) spectroscopy and their structural, morphological, surface, and thermal studies were performed using Fourier transform infrared (FTIR) spectroscopy, Raman spectroscopy, transmission electron microscopy (TEM), X-ray photoelectron spectroscopy (XPS), and thermogravimetry. Lastly, an attempt was made to explore the chirality-mediated interaction with microRNA (miR-205) and transfection with C4-2B prostate cancer cells.
2. Experimental section
2.1. Materials
Graphite powder (Gr) (CAS number-7782-42-5), L-tyrosine (CAS number-60-18-4) and D-tyrosine (CAS number-556-02-5) were purchased from Sigma Aldrich Co. (St. Louis, MO, USA) and were used as received without further modification/purification. All aqueous solutions were prepared using ultrapure water (resistivity 18.0 MΩ cm, Milli-Q® Water Purification System, EMD Millipore Corporation, Darmstadt, Germany). FAM-miR-205 (catalogue number- AM17121) and mir-205 mimic (has-miR-205-5p, lot number- ASO2IXKX) were purchased from Ambion of Life Technologies. The prostate cancer cell line (C4-2B) was procured from American Type Culture Collection (ATCC, Manassas, VA, USA). These cells were cultured in Roswell Park Memorial Institute-1640 (RPMI-1640) (#2430070, Gibco®, Thermo Fisher Scientific) containing 10% (v/v) fetal bovine serum (FBS) (# catalogue number-FBS-50-HI, Gibco®), and penicillin–streptomycin (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1) (#lot number-1910865, Gibco®) at 37 °C in a humidified 5% CO2–95% air chamber (Sanyo Scientific Ltd, Tokyo, Japan).
000 U mL−1) (#lot number-1910865, Gibco®) at 37 °C in a humidified 5% CO2–95% air chamber (Sanyo Scientific Ltd, Tokyo, Japan).
2.2. Preparation of L-graphene and D-graphene
In a typical synthesis method, 50 mg of graphite powder was mixed with 25 mg of L-tyrosine and D-tyrosine separately and uniformly dispersed in 10 mL Milli-Q water. These solution mixtures were sonicated for 6 h at a temperature of ∼50 to 55 °C. To maintain a temperature of 50–55 °C, a digitally controlled water sonication bath (Branson Ultrasonic water bath, Trainger, Albany, NY) was employed. After sonication, both samples were centrifuged for 10 min at 4000 rpm, and the precipitate was discarded. The supernatant solutions containing L-graphene and D-graphene were stored at room temperature for analysis.2.3. Characterization of L-graphene and D-graphene
To confirm the formation of L-graphene and D-graphene, UV-vis, CD, FTIR, and Raman spectral analysis and electron microscopic techniques (transmission and scanning electron microscopy) were employed. The UV-vis spectra of L-graphene and D-graphene solutions were recorded on Genesys 150 (Thermo Fisher Scientific). The CD spectra were acquired in the range of 180–350 nm utilizing a JASCO J-815 CD spectrophotometer (Jasco International Co., Ltd, Tokyo, Japan). The FTIR spectra of L-graphene and D-graphene were obtained using Spectrum Two FTIR spectrometer (PerkinElmer, USA). The IR spectra were recorded in the range of 4000–400 cm−1 (n = 3). The Raman spectra of the samples were recorded with an iHR 320 Horiba Jobin Yvon spectrometer (HORIBA, Ltd, Kyoto, Japan) using laser excitation with a wavelength of 532![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm and an output power of 25
nm and an output power of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mW. The morphological variation between the exfoliated sheets was evaluated by JEOL JEM-1230 transmission electron microscopy (JEOL Ltd, Tokyo, Japan) and a JSM-7100F scanning electron microscope (JEOL Ltd, Tokyo, Japan). A nano-ZS Malvern Zetasizer (Malvern, Westborough, MA, USA) was used to measure the hydrodynamic diameter and zeta potential of L-graphene and D-graphene solutions. The thermal analysis of L-graphene and D-graphene was done using a PerkinElmer STA 6000 thermal analyzer and differential scanning calorimeter (DSC). Thermograms were obtained by heating the sample at a constant heating rate of 20 °C min−1. A purge of nitrogen gas was used for all runs.
mW. The morphological variation between the exfoliated sheets was evaluated by JEOL JEM-1230 transmission electron microscopy (JEOL Ltd, Tokyo, Japan) and a JSM-7100F scanning electron microscope (JEOL Ltd, Tokyo, Japan). A nano-ZS Malvern Zetasizer (Malvern, Westborough, MA, USA) was used to measure the hydrodynamic diameter and zeta potential of L-graphene and D-graphene solutions. The thermal analysis of L-graphene and D-graphene was done using a PerkinElmer STA 6000 thermal analyzer and differential scanning calorimeter (DSC). Thermograms were obtained by heating the sample at a constant heating rate of 20 °C min−1. A purge of nitrogen gas was used for all runs.
2.4. miR-205 binding with L-graphene and D-graphene
The binding affinity of the miR-205 mimic (mature miRNA sequence: UCCUUCAUUCCACCGGAGUCUG, referred to as miR-205 throughout this work) with L-graphene and D-graphene was investigated using fluorescence spectra (fluorescence quenching) and the gel retardation assay. For this fluorescence binding study, L-graphene and D-graphene solutions (20, 40, 60, 80, and 100 μg mL−1) were incubated with FAM-miR-205 (10 μg mL−1). The fluorescence emission spectra were recorded using the FAM dye in the wavelength range (Ex: 450 nm and Em: 480–600 nm). For gel retardation assay, 5 μg of miR-205 and L-graphene and D-graphene with two different concentrations (10 μg and 15 μg) were mixed and vortexed and incubated at 30 min at room temperature. The complex was then electrophoresed using a 2% w/v agarose gel containing EtBr, 0.5 μg mL−1 in the presence of a routine agarose gel ladder in TAE buffer at 100 V for 1.5 h. The gel was then analyzed using a UV illuminator (Bio-RAD Chem Doc apparatus).2.5. Hemo and cell compatibility assays
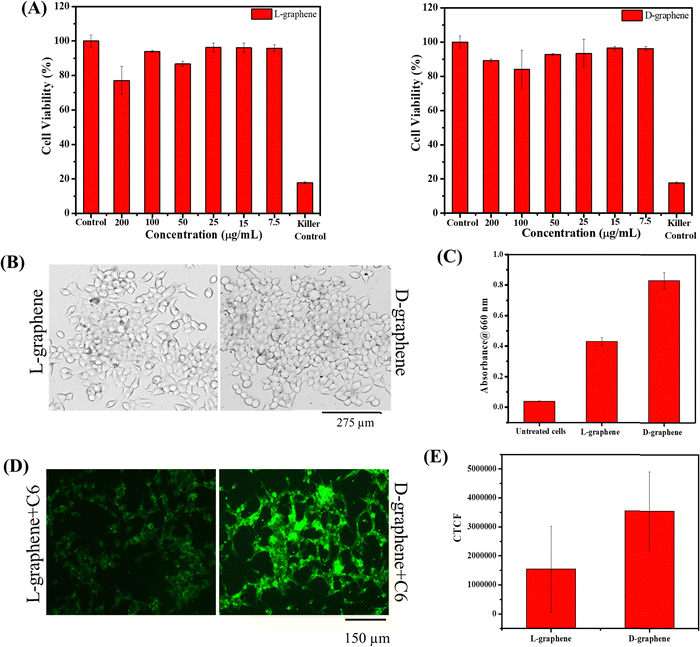
The hemo and cell compatibility assays of L-graphene and D-graphene were investigated using hemolysis and MTT assays. The hemolysis assay utilized human red blood cells whereas the MTT assay involved the use of C4-2B cells. The hemolysis assay was conducted according to our previous reports. In brief, 3 mL of human whole blood from a single donor (Innovative Research, Inc. IWB1K2E10ML, Novi, MI, USA) was centrifuged at 2000 rpm and the obtained RBC cell pellet was resuspended in 10 mL of RPMI-1640 cell culture medium (phenol red free). RBCs (100 μL) were incubated with treatment groups L-graphene and D-graphene (10, 25, 50, and 100 μg). Lipofectamine 2000 was used as a comparative group. Sodium dodecyl sulfate (SDS, 100 μg) and 1X PBS (100 μg) served as positive and negative control groups in this study. These samples were kept in an incubator for 2 h at 37 °C. The extent of hemolysis was measured using the optical density (λmax 570 nm, Varioskan LUX) of supernatant solutions of RBCs (5 minutes at 1000 rpm). For qualitative analysis, RBC smears were made on glass slides to investigate the morphological assessment. The brightfield images of RBCs were acquired using an EVOS M7000 microscope at 40X (Thermo Fisher Scientific, MA, USA).C4-2B cells were used as a model cell line. In brief, C4-2B cells were seeded in a 96-well plate with a cell density of around 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well. After incubating cells for 24 h, RPMI cultured media were replaced with fresh media containing varying concentrations of L-graphene and D-graphene and were incubated for an additional 24 h. Apart from adding L-graphene and D-graphene, a killer control, docetaxel, an anti-cancer drug, was also taken. After that, 20 μL of MTT solution (purchased from Sigma Aldrich with lot number – MKCL9866) in RPMI cultured media (stock solution 5 mg mL−1) was added in each well and incubated for 4 h. Afterwards, the cell supernatant was removed and the formed formazan crystals were dissolved in DMSO (100 μL per well) and absorbance was recorded at 540 nm using a microplate reader (Varioskan LUX from Thermo Scientific). Percentage cell viability was compared with untreated cells.
000 cells per well. After incubating cells for 24 h, RPMI cultured media were replaced with fresh media containing varying concentrations of L-graphene and D-graphene and were incubated for an additional 24 h. Apart from adding L-graphene and D-graphene, a killer control, docetaxel, an anti-cancer drug, was also taken. After that, 20 μL of MTT solution (purchased from Sigma Aldrich with lot number – MKCL9866) in RPMI cultured media (stock solution 5 mg mL−1) was added in each well and incubated for 4 h. Afterwards, the cell supernatant was removed and the formed formazan crystals were dissolved in DMSO (100 μL per well) and absorbance was recorded at 540 nm using a microplate reader (Varioskan LUX from Thermo Scientific). Percentage cell viability was compared with untreated cells.
2.6. Cellular uptake of L-graphene and D-graphene
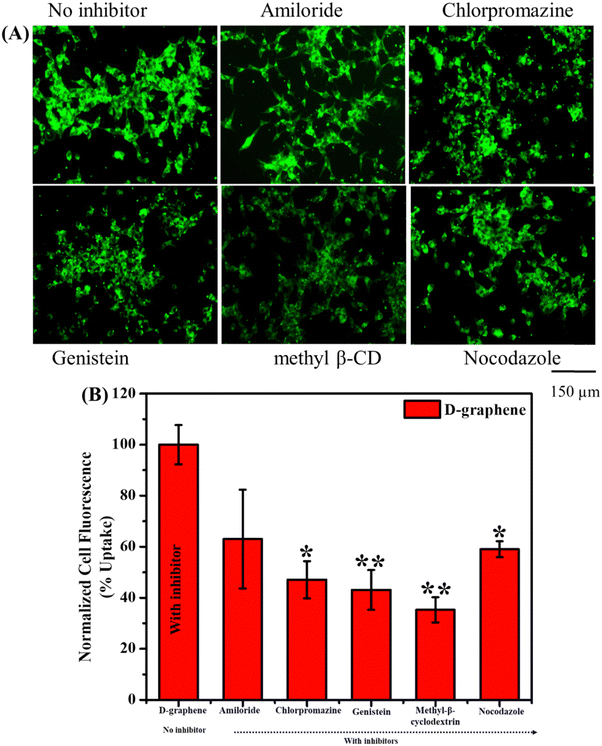
The uptake of L-graphene and D-graphene was studied to understand the possible internalization route and extent of internalization. To start with L-graphene and D-graphene having a concentration of 100 μg mL−1 were treated with C4-2B cells (cell density of 2.5 × 105 cells per well) and incubated for 3 h. After 3 h, the cells were washed thoroughly with 1X PBS to remove any free-floating D-graphene and L-graphene. After washing, the cells were trypsinized and the obtained cell pellet was lysed with a cell lysis buffer (Millipore Sigma, Laemmli 2× concentrated sample buffer). For L-graphene and D-graphene quantification the absorbance of lysed cells was measured at 660 nm. The reason for measuring absorbance at 660 nm is to minimize the interference of protein/amino acid absorbance. It must be mentioned here that, prior to lysis the optical images of C4-2B cells after treatment were also taken to gain an insight into L-graphene and D-graphene inside the cells. Furthermore, for better insight of L-graphene and D-graphene cell uptake, coumarin 6 (C6) dye (10 μg) was mixed with L-graphene and D-graphene having concentrations of 100 μg mL−1 separately and sonicated for 2 h. After 2 h using high centrifugation, the unbound dye was taken out and the remaining samples were used for further investigation. The fluorescent dye enables us to track L-graphene and D-graphene inside the cells. Prior to uptake, successful binding of C6 with L-graphene and D-graphene was confirmed by recording the UV-vis spectra (using Genesys 150) (Thermo Fisher Scientific) in the wavelength range of 260–600 nm and fluorescence spectra (using a microplate reader) (Varioskan LUX from Thermo Scientific) (Ex: 400 nm and Em: 430–790 nm) of C6, L-graphene, D-graphene, L-graphene + C6 and D-graphene + C6. A fluorescence image analysis using an EVOS M 7000 fluorescence microscope was employed to study the uptake pattern of L-graphene and D-graphene. C4-2B cells (cell density of 2.5 × 105 cells per well) were incubated with 10 μg of coumarin 6 dye bound with L-graphene and D-graphene for 3 h. After incubation, cells were washed twice with 1X PBS, and the extent of internalization/uptake of L-graphene and D-graphene was visualized using an EVOS M 7000 fluorescence microscope upon excitation/emission of 488/518 nm (coumarin 6, green). For a better understanding of the internalization mechanism, in another set of experiments, C4-2B cells were pretreated with various endocytosis inhibitors [nocodazole (10 μg mL−1), Mβ-CD (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM), genistein (200
mM), genistein (200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μM), amiloride (10 μg mL−1), and chlorpromazine (10 μg mL−1)] for 60 min at 37 °C and then incubated with 10 μg of coumarin 6 dye bound with L-graphene and D-graphene for 3 h. To further quantify the uptake process flow cytometry was performed using an Attune NxT Acoustic Focusing Cytometer from Thermo Fisher Scientific. Similar incubation and treatment conditions that were used for imaging were used for flow cytometry and after incubation, the cells were washed with PBS and trypsinized. The cell pellet was collected in a phenol-free medium for measurement in the FL1 channel (488 excitation, blue laser, 530 ± 15 nm, FITC/GFP).
μM), amiloride (10 μg mL−1), and chlorpromazine (10 μg mL−1)] for 60 min at 37 °C and then incubated with 10 μg of coumarin 6 dye bound with L-graphene and D-graphene for 3 h. To further quantify the uptake process flow cytometry was performed using an Attune NxT Acoustic Focusing Cytometer from Thermo Fisher Scientific. Similar incubation and treatment conditions that were used for imaging were used for flow cytometry and after incubation, the cells were washed with PBS and trypsinized. The cell pellet was collected in a phenol-free medium for measurement in the FL1 channel (488 excitation, blue laser, 530 ± 15 nm, FITC/GFP).
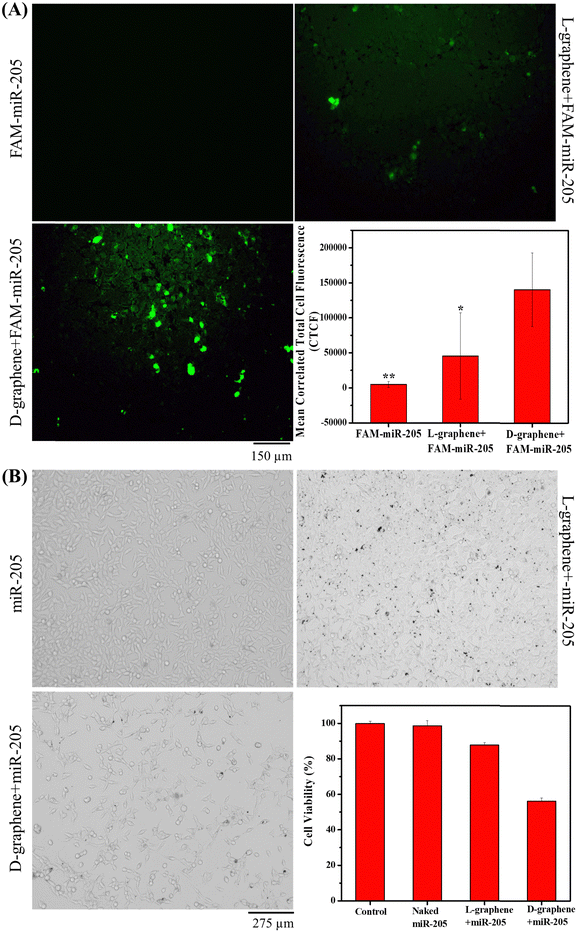
2.7. Transfection efficiency of L-graphene and D-graphene
For the in vitro transfection efficiency study, C4-2B cells (1.5 × 105 per well) were seeded on a 6-well plate for 24 h before transfection. Afterwards, the complete medium was replaced with a serum-free medium for 4 h and then 100 μg of L-graphene and D-graphene loaded with FAM-miR-205 were added separately to each well and incubated for 24 h. To visualize the transfection of FAM-miR-205, after 24 h, the cells were washed with 1X PBS and visualized using a fluorescence microscope at an excitation/emission of 488/518 nm to measure the enhancement in green fluorescence in cells.2.8. Anticancer efficiency of miR-205
The anticancer efficiency of miR-205 was determined using the MTT assay. In brief, the 24 h transfected and untransfected C4-2B cells (5000 cells per well in a 96 well plate) were seeded and after 24 h, the cell viability was determined as mentioned in the cell compatibility assay. The C4-2B cells that are not exposed to any sample served as a control and % cell viability was calculated with respect to the control.2.9. Statistical analysis
All the experimental data were expressed as mean ±standard error (SE) and were analyzed using ANOVA and the t test for the calculation of the significance level of the experimental data. The differences were considered statistically significant at *P < 0.05, **P < 0.01 and ***P < 0.001.3. Results and discussion
3.1. Preparation and characterization of L-graphene and D-graphene
Commercially available bulk graphite (Gr) powder was mixed with L-tyrosine and D-tyrosine separately and sonicated at a temperature (∼50–55 °C) for 6 h as shown in Scheme 1. After sonication the exfoliated colloidal dispersion was separated from large particles by centrifugation and stored at room temperature for further analysis. Here G exfoliated in the presence of L-tyrosine has been mentioned as L-graphene and G exfoliated in the presence of D-tyrosine has been mentioned as D-graphene.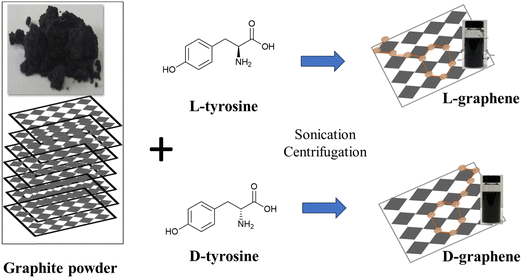 | ||
| Scheme 1 Schematic illustration of exfoliation and induction of chirality in L(D)-graphene by applying high temperature (∼50–55 °C) sonication in the presence of L(D)-tyrosine. | ||
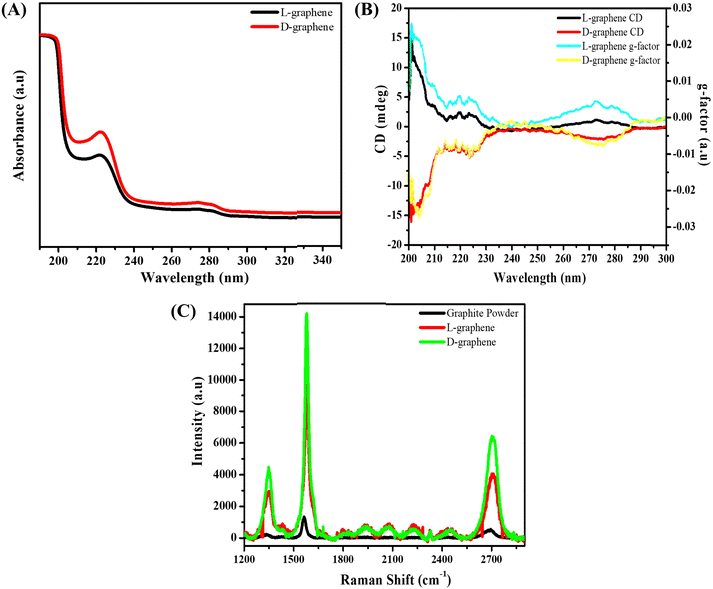
Before studying the generated optically active mode of both L-graphene and D-graphene, CD and UV spectra of L-tyrosine and D-tyrosine were examined as shown in Fig. S1 (ESI†). Both L-tyrosine and D-tyrosine showed three separate exact mirror image CD bands around 210, 240, and 290 nm, respectively. Similarly, three UV bands were observed at the same wavelength i.e., 210, 240, and 290 nm. The presence of all active CD bands at similar UV absorbance peaks arises because of the cotton effect phenomenon causing the UV peak to split into positive and negative CD values.14 Just like L-tyrosine and D-tyrosine, the UV spectrum of L-graphene and D-graphene was recorded between 180 and 350 nm as shown in Fig. 1(A). The three UV peaks in L-graphene and D-graphene get shifted after exfoliation as compared to control L-tyrosine and D-tyrosine to 200, 220, and 280 nm. The generation of all the UV peaks occurred because of π–π* transition occurred between the aromatic moieties. Moreover, the shift in the UV peak position after exfoliation suggests the molecular interaction occurring between the exfoliated sheets and amino acid enantiomers.15 To further assess the interaction between the exfoliated nanosheets and L-tyrosine and D-tyrosine, the fluorescence spectrum of L-tyrosine and D-tyrosine was recorded and compared with L-graphene and D-graphene in the wavelength range of 260 to 400 nm as shown in Fig. S2 (ESI†). The relative fluorescence quenching of L-graphene was around 80% with respect to L-tyrosine and a similar trend was observed with D-graphene and D-tyrosine. Both the enantiomers of tyrosine showed a similar trend suggesting that irrespective of different 3-D arrangements both the enantiomers of tyrosine interacted in a similar manner with the exfoliated sheets.16–18
Since both the enantiomers were able to bind separately with the exfoliated graphite sheet, the CD spectrum of L-graphene and D-graphene was recorded as shown in Fig. 1(B). For both, the enantiomers’ CD spectrum showed the exact mirror image spectrum at optically active wavelengths i.e., 200, 220, and 280 nm. Furthermore, to assess the quality of the obtained CD spectrum of L-graphene and D-graphene, its anisotropy factor (g-factor) was also calculated as shown in Fig. 1(B).19,20
The obtained mirror-imaged CD spectrum and the good g-factor value confirmed the successful chiroptical nature of L-graphene and D-graphene.21,22 Moreover, the chiroptical stability of L-graphene and D-graphene was also checked after 30 days shown in Fig. S3 (ESI†). A slight shift in CD spectrum (Fig. S3B, ESI†) and UV spectrum (Fig. S3A, ESI†) along with the change in g-factor value (Fig. S3C, ESI†) was observed; however, the chiroptical properties of L-graphene and D-graphene were preserved.
After confirming the chirality, the Raman spectrum of L-graphene and D-graphene as shown in Fig. 1(C) was recorded and compared with the Gr powder. Both Gr powder and L-graphene, D-graphene showed three major Raman peaks corresponding to the D-band (1350 cm−1), the G-band (1580 cm−1), and the 2D band (2700 cm−1); however, the ID/IG and I2D/IG values varied after exfoliation i.e. for unexfoliated Gr powder the ID/IG and I2D/IG values were 0.16 and 0.35, respectively, which increased to 0.31 and 0.44 for L-graphene and 0.32 and 0.49 for D-graphene. The increased ID/IG value for both L-graphene and D-graphene occurred because of the edge functionalization from high-temperature sonication leading to the generation of very high shear forces.23 Moreover, the presence of the 2D band confirmed the preservation of aromatic structure after the exfoliation. Also, the I2D/IG value was in line with the earlier report suggesting the formation of multilayer exfoliated sheets.24
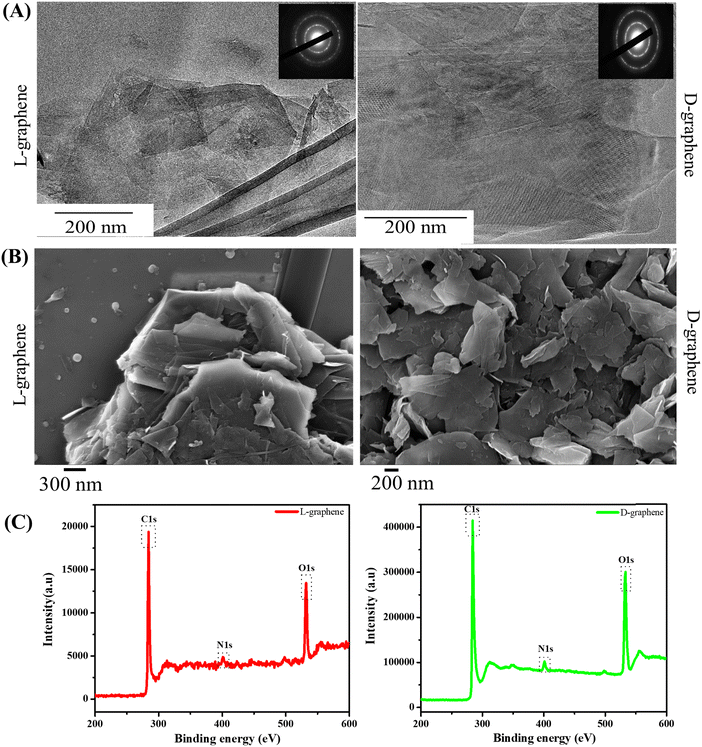
After confirming the chiroptical nature and presence of the signature Raman band, TEM, SEM, and XPS were performed to study the morphological and surface functionalities of L-graphene and D-graphene as shown in Fig. 2.
Fig. 2(A) shows the representative TEM images of L-graphene and D-graphene along with their selected area diffraction (SAED) patterns. For both L-graphene and D-graphene the exfoliated sheets showed disordered and asymmetric flakes and the sheets were entangled with each other however no distinct structural damages were observed. The SAED pattern, as shown in the inset of Fig. 2(A), has very strong diffraction spots illustrating the crystalline nature of the exfoliated nanosheets.25,26 Apart from TEM, SEM was performed to gain better insight into the exfoliated nanosheet morphology. For both L-graphene and D-graphene, exfoliated nanosheets had folded flakes with a wavy morphology. The SEM images obtained confirmed the presence of ultrathin nanosheets as shown in Fig. 2(B) and were consistent with an earlier report.27 The energy-dispersive X-ray (EDX) spectra of L-graphene and D-graphene were recorded to analyze the elemental composition and their relative atomic weight ratios are listed in Table S1 (ESI†). For both L-graphene and D-graphene, nitrogen (N1s) and oxygen (O1s) were found to confirm the successful adsorption of tyrosine enantiomers. Additionally, the weight percentage of N1s and O1s were almost similar in both L-graphene and D-graphene suggesting that both L-tyrosine and D-tyrosine were present in almost equal volume on the exfoliated sheet.
The XPS survey spectrum of L-graphene and D-graphene showed the presence of three core peaks labelled for C1s, N1s and O1s at 284.7 eV, 397.91 eV and 530.29 eV, respectively (Fig. 2(C)).28 In L-graphene C1s, N1s and O1s were present at concentrations of 77.01%, 19.34%, and 3.65% whereas, in D-graphene C1s, N1s and O1s were around 78.18%, 19.31% and 2.24%. Ideally, the pristine G or Gr doesn’t show any N1s peak thus the presence of N1s peak further supported the presence of a tyrosine enantiomer on the nanosheets.28 From the % composition of C1s and O1s, the C/O ratios of L-graphene and D-graphene were found to be around 3.98 and 4.04, respectively. Also, the C1s peak of L-graphene and D-graphene were deconvoluted and plotted (Fig. 2(C)). For L-graphene the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond was around 284.7 eV corresponding to sp2 bonded graphitic carbon atoms. Apart from that another peak around 286.4 eV was also observed which corresponds to C–N, C–O. On the other hand, for D-graphene apart from the C
C bond was around 284.7 eV corresponding to sp2 bonded graphitic carbon atoms. Apart from that another peak around 286.4 eV was also observed which corresponds to C–N, C–O. On the other hand, for D-graphene apart from the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond and the C–N, C–O bond one more peak around 288.1 eV corresponds to amidic C
C bond and the C–N, C–O bond one more peak around 288.1 eV corresponds to amidic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O was also observed. Taken together, the XPS results enabled the successful binding of tyrosine enantiomers on the surface of the exfoliated sheet.13,29,30 For successful direct exfoliation, the solvent/surfactant must match its surface tension with the Gr sheet.31 In the present work high temperature sonication was provided which can disintegrate the π–π interaction of the Gr sheet causing the initiation of the exfoliation process32 and the presence of L-tyrosine and D-tyrosine further gets attached to the exfoliated sheets through the edges of the sheets which are highly unstable and tend to stabilize with the stabilizing agents.33
O was also observed. Taken together, the XPS results enabled the successful binding of tyrosine enantiomers on the surface of the exfoliated sheet.13,29,30 For successful direct exfoliation, the solvent/surfactant must match its surface tension with the Gr sheet.31 In the present work high temperature sonication was provided which can disintegrate the π–π interaction of the Gr sheet causing the initiation of the exfoliation process32 and the presence of L-tyrosine and D-tyrosine further gets attached to the exfoliated sheets through the edges of the sheets which are highly unstable and tend to stabilize with the stabilizing agents.33
Fig. 3(A) and (B) present the FTIR spectra of L-graphene and D-graphene w.r.t. L-tyrosine, D-tyrosine and graphite powder. Both L-graphene and D-graphene exhibited IR peaks at around 3263cm−1, 2040.10 cm−1 and 1685 cm−1 respectively corresponding to N–H/O–H, C–H and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O band. These bands were absent in control graphite powder and were coming after the binding/stabilization of L-tyrosine and D-tyrosine onto the exfoliated sheets. Similarly Fig. 3(C) and (D) show the DSC spectra of L-graphene and D-graphene w.r.t. L-tyrosine, D-tyrosine and graphite. The exothermic peaks of L-graphene and D-graphene were shifted as compared to L-tyrosine, D-tyrosine and graphite powder. However, no major variation in heating pattern was observed based on chirality and no changes occurred because of L-tyrosine and D-tyrosine functionality creating a more oxygenated functional group. Dynamic light scattering (DLS) of L-graphene and D-graphene was performed for measuring the hydrodynamic diameter (DH), polydispersity index (PdI), and zeta potential and the results are summarized in Table S2 (ESI†). The DH values of L-graphene and D-graphene were found to be around 309.43 ± 14.8 and 357.76 ± 4.03 nm, respectively. Also, the PdI values of L-graphene and D-graphene were 0.35 ± 0.072 and 0.386 ± 0.0005, respectively. The PdI values were slightly higher which supported the heterogenous flakes observed from TEM and SEM images. The zeta potentials of L-graphene and D-graphene were around −38.57 ± 0.08 mV and −37.24 ± 0.33 mV. The higher zeta potential value leads to higher stability of the colloidal dispersion. The DH, PdI, and zeta potential of L-graphene and D-graphene were also measured after 30 days to further confirm the colloidal stability of the exfoliated nanosheets. As can be seen from Table S2 (ESI†), a slight variation in all three parameters was observed after 30 days and still L-graphene and D-graphene were stable. This slight variation was well supported by the variation in the CD spectrum of L-graphene and D-graphene reported after 30 days.
O band. These bands were absent in control graphite powder and were coming after the binding/stabilization of L-tyrosine and D-tyrosine onto the exfoliated sheets. Similarly Fig. 3(C) and (D) show the DSC spectra of L-graphene and D-graphene w.r.t. L-tyrosine, D-tyrosine and graphite. The exothermic peaks of L-graphene and D-graphene were shifted as compared to L-tyrosine, D-tyrosine and graphite powder. However, no major variation in heating pattern was observed based on chirality and no changes occurred because of L-tyrosine and D-tyrosine functionality creating a more oxygenated functional group. Dynamic light scattering (DLS) of L-graphene and D-graphene was performed for measuring the hydrodynamic diameter (DH), polydispersity index (PdI), and zeta potential and the results are summarized in Table S2 (ESI†). The DH values of L-graphene and D-graphene were found to be around 309.43 ± 14.8 and 357.76 ± 4.03 nm, respectively. Also, the PdI values of L-graphene and D-graphene were 0.35 ± 0.072 and 0.386 ± 0.0005, respectively. The PdI values were slightly higher which supported the heterogenous flakes observed from TEM and SEM images. The zeta potentials of L-graphene and D-graphene were around −38.57 ± 0.08 mV and −37.24 ± 0.33 mV. The higher zeta potential value leads to higher stability of the colloidal dispersion. The DH, PdI, and zeta potential of L-graphene and D-graphene were also measured after 30 days to further confirm the colloidal stability of the exfoliated nanosheets. As can be seen from Table S2 (ESI†), a slight variation in all three parameters was observed after 30 days and still L-graphene and D-graphene were stable. This slight variation was well supported by the variation in the CD spectrum of L-graphene and D-graphene reported after 30 days.
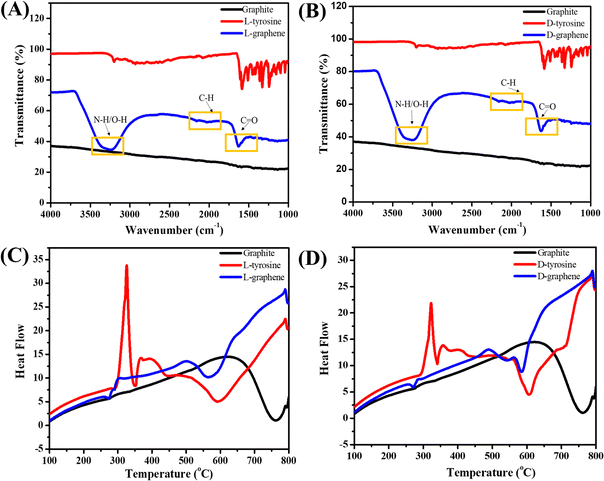 | ||
| Fig. 3 (A) FTIR spectra of L-graphene with L-tyrosine and graphite. (B) FTIR spectra of D-graphene with D-tyrosine and graphite. (C) DSC spectra of L-graphene and (D) DSC spectra of D-graphene. | ||
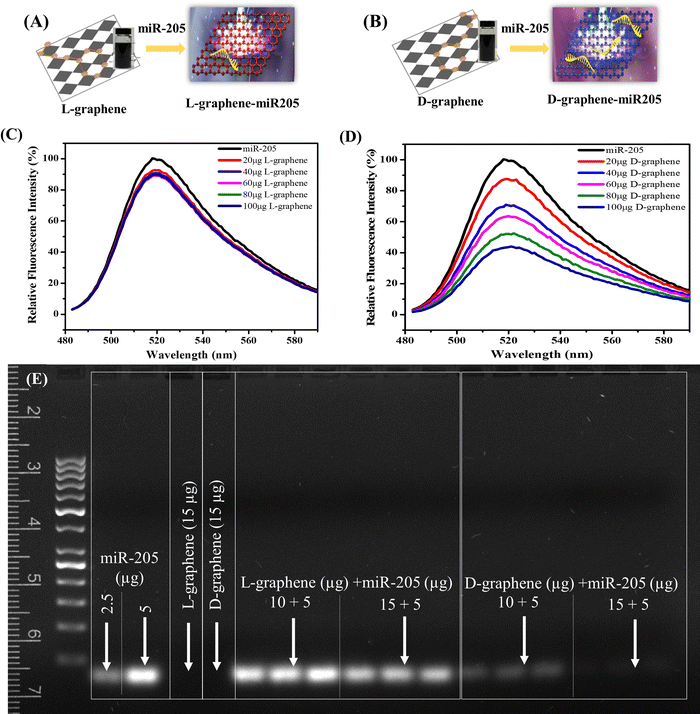
3.2. D-graphene exhibits chirality specific binding with miR-205
The XPS spectrum and zeta potential confirmed that L-tyrosine and D-tyrosine were present in L-graphene and D-graphene. Moreover, no detectable variation in the Raman band and similar morphology supported that exfoliated sheets were similar structurally, and the only variation was with their optically active mode which was seen from their CD spectrum making L-graphene and D-graphene an excellent chiral 2D system. Thus, to explore the chiral application of L-graphene and D-graphene their interaction with nucleic acid was explored.Nano-complexation of L-graphene and D-graphene with miR-205 is presented in Fig. 4(A) and (B). The miR-205 complexation efficiency was investigated employing the fluorescence quenching method (Fig. 4(C) and (D)) and gel retardation assay (Fig. 4(E)). From Fig. 4(C), in the presence of L-graphene with a concentration of 100 μg, around 15% fluorescence quenching of FAM-miR-205 was observed; however, with D-graphene having a concentration of 100 μg, more than 50% fluorescence quenching of FAM-miR-205 was observed as shown in Fig. 4(D).
Moreover, the fluorescence quenching with L-graphene was concentration independent and mainly the quenching occurred because of the FAM-miR-205 physical adsorption on the surface whereas, with D-graphene, concentration-dependent fluorescence quenching was observed and a linear relation between fluorescence quenching and D-graphene concentration was plotted as shown in Fig. S4 (ESI†).34 It is important to note that usually, nucleic acid tends to bind with any nanosystem through electrostatic interactions occurring between the negatively charged nucleic acid and the positively charged nanosystem, however, here despite the negative charge with D-graphene, the FAM-miR-205 tends to bind through chiral–chiral interactions.35
To further authenticate the chiral preferential complexation, miR-205 (5 μg) was mixed with L-graphene and D-graphene with two different concentrations (10 μg and 15 μg) and were subjected to gel electrophoresis. It can be observed that irrespective of concentration L-graphene did not bind much with miR-205 as the miR-205 band clearly observed like the free miR-205; however, with D-graphene complete retardation was found (Fig. 4(E)).
The obtained data confirmed the chirality-based binding between L-graphene and D-graphene with microRNA.35 The plausible explanation of chiral preference binding occurring between D-graphene and miR-205 is because of the geometric matching. MicroRNA has D chirality and thus prefers to bind with D-graphene possibly through hydrogen bonds and hydrophobic interactions between nucleobases of RNA and aliphatic chains and aromatic rings of nanosheets. In the present work, D-tyrosine was present on the nanosheets and governs its chirality and thus may cause less steric hindrance with “D-configured” miR-205 leading to greater affinity.35–37
3.3. L-graphene and D-graphene as transfection reagents
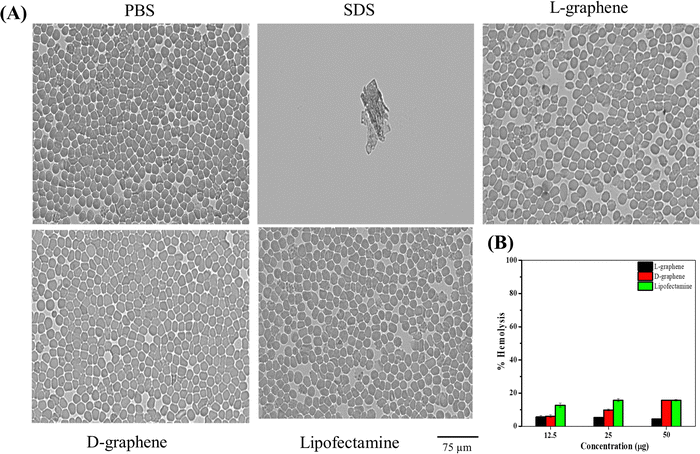
Cellular transfection reagents often cause toxicities. RBCs were chosen to test the toxicity of L-graphene and D-graphene using a hemolysis assay. The reason to choose RBCs is due to their 45% abundance in whole blood. Hemolysis assay refers to the rupture of the membrane of RBCs leading to the release of intracellular contents. Therefore, it is critical to evaluate L-graphene and D-graphene as transfecting agents to deliver miR-205. Lipofectamine®, a commercially used transfection reagent was used as a comparison in this study.Hemolysis was proved by checking the morphological change of RBCs after incubation with L-graphene, D-graphene and Lipofectamine shown in Fig. 5(A). From Fig. 5(A) a negative control group (1X PBS), L-graphene and D-graphene did not alter the membrane or whole cell morphology of RBCs whereas the SDS group shows complete membrane disruption and lysis of RBCs (no intact RBCs were observed). % Hemolysis graph was plotted considering SDS lysis as 100%. Furthermore, % hemolysis of RBCs was evaluated at concentrations 12.5 μg, 25 μg and 50 μg for L-graphene, D-graphene and Lipofectamine shown in Fig. 5(B). It should be mentioned here that the sodium dodecyl sulfate was considered a positive control with 100% hemolysis. Both L-graphene and D-graphene did not show any signs of toxicity at tested concentrations similar to Lipofectamine. This evidence advises us that L-graphene and D-graphene are hemocompatible and can be utilized for miR-205 delivery.
3.4. D-graphene chirality mediated cellular binding on C4-2B cells
Since chiral preferential binding occurring between miR-205 and D-graphene makes them an exciting prospect for gene delivery. Thus, in the present work, miR-205 loaded L-graphene and D-graphene were also checked for intracellular responses with C4-2B prostate cancer cells. Before that, the cell viability of L-graphene and D-graphene was assessed through a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. As shown in Fig. 6(A) both L-graphene and D-graphene didn’t show significant cell death after 24 h with the maximum concentration of 200 μg mL−1.After confirming the cellular viability, the uptake study of L-graphene and D-graphene was performed. 100 μg mL−1 of L-graphene and D-graphene were treated with C4-2B cells for 3 h. Fig. 6(B) presents the optical microscopy images of C4-2B cells with L-graphene and D-graphene. As can be seen from the optical images L-graphene and D-graphene were able to internalize inside cells. D-Graphene tends to get internalized more as compared to L-graphene, but the actual scenario can be obtained by cell lysates that contain graphene, fluorescence, and transfection experiments (Fig. 6(C)–(F) and 7). For L-graphene and D-graphene quantification, the lysed cell's absorbance was recorded at 660 nm. A significant increment in absorbance was observed with D-graphene as compared with L-graphene and untreated cells (Fig. 6(C)). This increment in absorbance further supports the successful and efficient cellular uptake of D-graphene. Furthermore, cell uptake of coumarin 6 dye (C6) encapsulated L-graphene and D-graphene was also verified by the fluorescent microscope. Fig. 6(D) and (E) present superior D-graphene uptake by C4-2B cells.
Usually, the nanomaterial gets internalized into the cells through the endocytosis pathway thus to understand the internalization pathways, inhibitors were used as shown in Fig. 7. The uptake capacity of D-graphene is decreased which can be visualized by a decrease in fluorescence intensity in the presence of inhibitors (Fig. 7(A)). The extent of such a decrease depends on the type of inhibitor that influences the pathway internalization. To investigate the global uptake pattern of this phenomenon, flow cytometry was performed, and results were presented in Fig. 7(B). The % cell uptake value decreased significantly with all the inhibitors. These inhibitors suppress the uptake pathway through various means such as caveolae-/clathrin-mediated endocytosis, microtubule-related internalization, and caveolae-mediated pathways, etc.38–40 In the present work, D-graphene uptake possibly occurred because of the combination of different endocytosis pathways.38,41 Also, cells have the presence of different chiral systems on their surface such as chiral phospholipids membrane, proteins and sugar thus chirality of any external system will affect the uptake mechanism as these systems tend to bind with the cell surface prior to getting internalized.42 A similar pattern of uptake was noticed in the case of L-graphene (Fig. S5, ESI†).
3.5. D-graphene facilitates miR-205 transfection and inhibits the proliferation of cancer cells
After confirming the cell compatibility and cellular uptake of L-graphene and D-graphene with C4-2B cells, the miR-205 transfection in C4-2B cells was examined utilizing FAM-miR-205. Fig. 8(A) confirms that when FAM-miR-205 loaded with D-graphene the green fluorescence coming from C4-2B cells was higher as compared with naked FAM-miR-205 and FAM-miR-205 loaded with L-graphene. The CTCF values were also calculated shown in Fig. 8(A), graph. FAM-miR-205 loaded with D-graphene showed a significant fluorescence difference as compared to naked FAM-miR-205 and FAM-miR-205 loaded with L-graphene. D-Graphene not only facilitates miR-205 transfection into C4-2B cells but also significantly inhibits the growth of the cells compared to L-graphene as is apparent from bright field images (Fig. 8(B)).43,44 This behavior was further evident from the MTT assay (Fig. 8(B), graph).The chiral selective binding of miR-205, chiral selective cell uptake and transfection with L-graphene and D-graphene acknowledge the role of chirality and their decisive role in gene delivery. As we know cellular systems are composed of different levels of chirality, L-graphene and D-graphene can be explored for specific bindings with different cancerous biomarkers such as transferrin, heat shock protein (HSP90), etc. Further studies about proteolytic and immunogenic stabilities of L-graphene, and D-graphene loaded with miR-205 along with their biodegradability investigations may lead to further in vivo and human studies. Thus, these chiral nanosystems have the potential to impact the areas of precision medicine and gene delivery in future.
4. Conclusion
In conclusion, we reported for the first time direct liquid phase exfoliation and induction of chirality in G nanosheets in a single step using L-tyrosine and D-tyrosine as exfoliants and applying high-temperature sonication. Both L-graphene and D-graphene showed an exact mirror image chiral spectrum of each other and an excellent asymmetry value. Also, the exfoliated L-graphene and D-graphene had similar asymmetric morphology arising because of very high sonic forces. Furthermore, the lateral size and zeta potential values of L-graphene and D-graphene were almost the same suggesting that the exfoliation and stabilization mechanisms were similar to both the types of enantiomers of tyrosine. Taking advantage of induced chirality in exfoliated nanosheets we checked its interactions separately with miR-205. Interestingly, we observed that interaction was chirality dependent i.e., D-graphene showed higher interaction, transfection, and delivery of miR-205 as compared with L-graphene in C4-2B cells. To the best of our knowledge this is the first report on chiral interaction between nucleic acid and G nanosheets as well as chirality-based gene delivery with two-dimensional nanosheets and has the capability for chirality-mediated gene transfection in future work through systematic research. This type of direct exfoliation and induction of chirality offer a great industrial prospect ahead and can be extended to other two-dimensional nanomaterials.Author contributions
Conception and design: P, SCC, and MMY; data curation and formal analysis: P, ENHKG, NC, RT, and MC; investigation, validation, and visualization: P, ENHKG, NC, RT, MC, and MMY; resources, funding acquisition, and software: MMY and SCC; writing – original draft: P, ENHKG, RT, and MMY; writing – review & editing: P, SCC, and MMY; supervision and project administration: MMY.Conflicts of interest
The authors report no conflicts of interest in this work.Acknowledgements
The authors acknowledge the start-up research support from the Department of Immunology and Microbiology, School of Medicine, University of Texas Rio Grande Valley given to MJ, MMY, and SCC. This work was partially supported by the National Institute of Health SC1GM139727, the Integrated Cancer Research Core funded by CPRIT Grant #RP210180, and the UT-System Star Award. All authors also want to acknowledge Dr Swati Dhasmana for helping in the transfection experiment, the Kleberg Advanced Microscopy Center (KAMC) for TEM and RAMAN measurement, the Biophysics Instrumentation Core facility for CD spectroscopy, and Dr Karen Lozano and Ms Victoria Padilla for SEM and XPS measurements.References
- B. Zhao, S. Yang, J. Deng and K. Pan, Chiral Graphene Hybrid Materials: Structures, Properties, and Chiral Applications, Adv. Sci., 2021, 8(7), 2003681 CrossRef CAS PubMed.
- Z. Peng, L. Yuan, J. XuHong, H. Tian, Y. Zhang, J. Deng and X. Qi, Chiral nanomaterials for tumor therapy: autophagy, apoptosis, and photothermal ablation, J. Nanobiotechnol., 2021, 19(1), 220 CrossRef PubMed.
- R. A. Rosenberg, D. Mishra and R. Naaman, Chiral Selective Chemistry Induced by Natural Selection of Spin-Polarized Electrons, Angew. Chem., Int. Ed., 2015, 54(25), 7295–7298 CrossRef CAS PubMed.
- G. H. Bird, E. Mazzola, K. Opoku-Nsiah, M. A. Lammert, M. Godes, D. S. Neuberg and L. D. Walensky, Biophysical determinants for cellular uptake of hydrocarbon-stapled peptide helices, Nat. Chem. Biol., 2016, 12(10), 845–852 CrossRef CAS PubMed.
- X. Wang, M. Wang, R. Lei, S. F. Zhu, Y. Zhao and C. Chen, Chiral Surface of Nanoparticles Determines the Orientation of Adsorbed Transferrin and Its Interaction with Receptors, ACS Nano, 2017, 11(5), 4606–4616 CrossRef CAS PubMed.
- T. Ishigami, K. Suga and H. Umakoshi, Chiral Recognition of L-Amino Acids on Liposomes Prepared with L-Phospholipid, ACS Appl. Mater. Interfaces, 2015, 7(38), 21065–21072 CrossRef CAS PubMed.
- K. S. Novoselov, A. K. Geim, S. V. Morozov, D. Jiang, Y. Zhang, S. V. Dubonos, I. V. Grigorieva and A. A. Firsov, Electric Field Effect in Atomically Thin Carbon Films, Science, 2004, 306(5696), 666–669 CrossRef CAS PubMed.
- P. J. Evans, J. Ouyang, L. Favereau, J. Crassous, I. Fernández, J. Perles and N. Martín, Synthesis of a Helical Bilayer Nanographene, Angew. Chem., Int. Ed., 2018, 57(23), 6774–6779 CrossRef CAS PubMed.
- J. D. Roy-Mayhew and I. A. Aksay, Graphene Materials and Their Use in Dye-Sensitized Solar Cells, Chem. Rev., 2014, 114(12), 6323–6348 CrossRef CAS PubMed.
- Y. Shao, M. F. El-Kady, L. J. Wang, Q. Zhang, Y. Li, H. Wang, M. F. Mousavi and R. B. Kaner, Graphene-based materials for flexible supercapacitors, Chem. Soc. Rev., 2015, 44(11), 3639–3665 RSC.
- G. Bottari, M. Á. Herranz, L. Wibmer, M. Volland, L. Rodríguez-Pérez, D. M. Guldi, A. Hirsch, N. Martín, F. D'Souza and T. Torres, Chemical functionalization and characterization of graphene-based materials, Chem. Soc. Rev., 2017, 46(15), 4464–4500 RSC.
- Y. Meng, J. Fan, M. Wang, W. Gong, J. Zhang, J. Ma, H. Mi, Y. Huang, S. Yang, R. S. Ruoff and J. Geng, Encoding Enantiomeric Molecular Chiralities on Graphene Basal Planes, Angew. Chem., Int. Ed., 2022, 61(15), e202117815 CrossRef CAS PubMed.
- F. Purcell-Milton, R. McKenna, L. J. Brennan, C. P. Cullen, L. Guillemeney, N. V. Tepliakov, A. S. Baimuratov, I. D. Rukhlenko, T. S. Perova, G. S. Duesberg, A. V. Baranov, A. V. Fedorov and Y. K. Gun’ko, Induction of Chirality in Two-Dimensional Nanomaterials: Chiral 2D MoS2 Nanostructures, ACS Nano, 2018, 12(2), 954–964 CrossRef CAS PubMed.
- D. W. Miles, R. K. Robins and H. Eyring, Optical rotatory dispersion, circular dichroism, and absorption studies on some naturally occurring ribonucleosides and related derivatives, Proc. Natl. Acad. Sci. U. S. A., 1967, 57(5), 1138–1145 CrossRef CAS PubMed.
- M. Tewari, P. Pareek and S. Kumar, Correlating Amino Acid Interaction with Graphene-Based Materials Regulating Cell Function, J. Indian Inst. Sci., 2022, 102(1), 639–651 CrossRef.
- J. G. Woo, L.-H. Tran, S.-H. Jang, C. Lee and T. J. Kang, Molecular Interactions of Graphene Oxide with Aromatic Amino Acids Tyrosine and Tryptophan, Bull. Korean Chem. Soc., 2015, 36(12), 2959–2961 CrossRef CAS.
- S. Pandit and M. De, Interaction of Amino Acids and Graphene Oxide: Trends in Thermodynamic Properties, J. Phys. Chem. C, 2017, 121(1), 600–608 CrossRef CAS.
- S. Li, A. N. Aphale, I. G. Macwan, P. K. Patra, W. G. Gonzalez, J. Miksovska and R. M. Leblanc, Graphene Oxide as a Quencher for Fluorescent Assay of Amino Acids, Peptides, and Proteins, ACS Appl. Mater. Interfaces, 2012, 4(12), 7069–7075 CrossRef CAS PubMed.
- A. Visheratina, L. Hesami, A. K. Wilson, N. Baalbaki, N. Noginova, M. A. Noginov and N. A. Kotov, Hydrothermal synthesis of chiral carbon dots, Chirality, 2022, 34(12), 1503–1514 CrossRef CAS PubMed.
- A. Döring, E. Ushakova and A. L. Rogach, Chiral carbon dots: synthesis, optical properties, and emerging applications, Light: Sci. Appl., 2022, 11(1), 75 CrossRef PubMed.
- S. Ma, Y.-K. Jung, J. Ahn, J. Kyhm, J. Tan, H. Lee, G. Jang, C. U. Lee, A. Walsh and J. Moon, Elucidating the origin of chiroptical activity in chiral 2D perovskites through nano-confined growth, Nat. Commun., 2022, 13(1), 3259 CrossRef CAS PubMed.
- M. Wakabayashi, S. Yokojima, T. Fukaminato, K.-I. Shiino, M. Irie and S. Nakamura, Anisotropic Dissymmetry Factor, g: Theoretical Investigation on Single Molecule Chiroptical Spectroscopy, J. Phys. Chem. A, 2014, 118(27), 5046–5057 CrossRef CAS PubMed.
- P. Tiwari, N. Kaur, V. Sharma and S. M. Mobin, High-yield graphene produced from the synergistic effect of inflated temperature and gelatin offers high stability and cellular compatibility, Phys. Chem. Chem. Phys., 2018, 20(30), 20096–20107 RSC.
- H. Zhao, B. Xu, J. Ding, Z. Wang and H. Yu, Natural Amino Acids: High-Efficiency Intercalants for Graphene Exfoliation, ACS Sustainable Chem. Eng., 2019, 7(23), 18819–18825 CrossRef CAS.
- G. Wang, J. Yang, J. Park, X. Gou, B. Wang, H. Liu and J. Yao, Facile Synthesis and Characterization of Graphene Nanosheets, J. Phys. Chem. C, 2008, 112(22), 8192–8195 CrossRef CAS.
- D. N. H. Tran, S. Kabiri and D. Losic, A green approach for the reduction of graphene oxide nanosheets using non-aromatic amino acids, Carbon, 2014, 76, 193–202 CrossRef CAS.
- H. Kalil, S. Maher, T. Bose and M. Bayachou, Manganese Oxide/Hemin-Functionalized Graphene as a Platform for Peroxynitrite Sensing, J. Electrochem. Soc., 2018, 165(12), G3133 CrossRef CAS.
- S. Bhattacharya, S. Mishra, P. Gupta, Pranav, M. Ghosh, A. K. Pramanick, D. P. Mishra and S. Nayar, Liquid phase collagen modified graphene that induces apoptosis, RSC Adv., 2015, 5(55), 44447–44457 RSC.
- W. Wang, Y. Zhang and Y.-B. Wang, Noncovalent π⋯π interaction between graphene and aromatic molecule: Structure, energy, and nature, J. Chem. Phys., 2014, 140(9), 094302 CrossRef PubMed.
- S. J. Rodríguez, L. Makinistian and E. Albanesi, Computational study of transport properties of graphene upon adsorption of an amino acid: importance of including –NH2 and –COOH groups, J. Comput. Electron., 2017, 16(1), 127–132 CrossRef.
- E. P. Nguyen, B. J. Carey, T. Daeneke, J. Z. Ou, K. Latham, S. Zhuiykov and K. Kalantar-zadeh, Investigation of Two-Solvent Grinding-Assisted Liquid Phase Exfoliation of Layered MoS2, Chem. Mater., 2015, 27(1), 53–59 CrossRef CAS.
- J. Kim, S. Kwon, D.-H. Cho, B. Kang, H. Kwon, Y. Kim, S. O. Park, G. Y. Jung, E. Shin, W.-G. Kim, H. Lee, G. H. Ryu, M. Choi, T. H. Kim, J. Oh, S. Park, S. K. Kwak, S. W. Yoon, D. Byun, Z. Lee and C. Lee, Direct exfoliation and dispersion of two-dimensional materials in pure water via temperature control, Nat. Commun., 2015, 6(1), 8294 CrossRef CAS PubMed.
- M. Cao, N. Wang, L. Wang, Y. Zhang, Y. Chen, Z. Xie, Z. Li, E. Pambou, R. Li, C. Chen, F. Pan, H. Xu, J. Penny, J. R. P. Webster and J. R. Lu, Direct exfoliation of graphite into graphene in aqueous solutions of amphiphilic peptides, J. Mater. Chem. B, 2016, 4(1), 152–161 RSC.
- P. K. B. Nagesh, P. Chowdhury, E. Hatami, V. K. N. Boya, V. K. Kashyap, S. Khan, B. B. Hafeez, S. C. Chauhan, M. Jaggi and M. M. Yallapu, miRNA-205 Nanoformulation Sensitizes Prostate Cancer Cells to Chemotherapy, Cancers, 2018, 10(9), 289 CrossRef PubMed.
- A. Zoabi and K. Margulis, Differential Interactions of Chiral Nanocapsules with DNA, Int. J. Mol. Sci., 2021, 22(2), 584 CrossRef CAS PubMed.
- T. Sugawara, Y. Suwa, K. Ohkawa and H. Yamamoto, Chiral Biomineralization: Mirror-Imaged Helical Growth of Calcite with Chiral Phosphoserine Copolypeptides, Macromol. Rapid Commun., 2003, 24(14), 847–851 CrossRef CAS.
- Y. Oaki and H. Imai, Amplification of Chirality from Molecules into Morphology of Crystals through Molecular Recognition, J. Am. Chem. Soc., 2004, 126(30), 9271–9275 CrossRef CAS PubMed.
- Y. Gong, P. Chowdhury, P. K. B. Nagesh, M. A. Rahman, K. Zhi, M. M. Yallapu and S. Kumar, Novel elvitegravir nanoformulation for drug delivery across the blood-brain barrier to achieve HIV-1 suppression in the CNS macrophages, Sci. Rep., 2020, 10(1), 3835 CrossRef CAS PubMed.
- M. M. Yallapu, S. Khan, D. M. Maher, M. C. Ebeling, V. Sundram, N. Chauhan, A. Ganju, S. Balakrishna, B. K. Gupta, N. Zafar, M. Jaggi and S. C. Chauhan, Anti-cancer activity of curcumin loaded nanoparticles in prostate cancer, Biomaterials, 2014, 35(30), 8635–8648 CrossRef CAS PubMed.
- L. Zhang, X. Yang, Y. Lv, X. Xin, C. Qin, X. Han, L. Yang, W. He and L. Yin, Cytosolic co-delivery of miRNA-34a and docetaxel with core-shell nanocarriers via caveolae-mediated pathway for the treatment of metastatic breast cancer, Sci. Rep., 2017, 7(1), 46186 CrossRef CAS PubMed.
- H. Yan, M. Cacioppo, S. Megahed, F. Arcudi, L. Dordević, D. Zhu, F. Schulz, M. Prato, W. J. Parak and N. Feliu, Influence of the chirality of carbon nanodots on their interaction with proteins and cells, Nat. Commun., 2021, 12(1), 7208 CrossRef CAS PubMed.
- J. Hu, W. G. Cochrane, A. X. Jones, D. G. Blackmond and B. M. Paegel, Chiral lipid bilayers are enantioselectively permeable, Nat. Chem., 2021, 13(8), 786–791 CrossRef CAS PubMed.
- B.-w Zhao, Z. Zhou and Y. Shen, Effects of chirality on gene delivery efficiency of polylysine, Chin. J. Polym. Sci., 2016, 34(1), 94–103 CrossRef CAS.
- Q. Chen, Q. Yu, Y. Liu, D. Bhavsar, L. Yang, X. Ren, D. Sun, W. Zheng, J. Liu and L. M. Chen, Multifunctional selenium nanoparticles: Chiral selectivity of delivering MDR-siRNA for reversal of multidrug resistance and real-time biofluorescence imaging, Nanomedicine, 2015, 11(7), 1773–1784 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ma00611e |
| This journal is © The Royal Society of Chemistry 2023 |