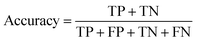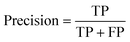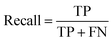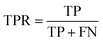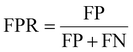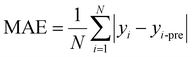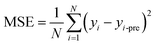Open Access Article
Open Access ArticleUtilizing machine learning to expedite the fabrication and biological application of carbon dots
Yaoyao
Tang
ab,
Quan
Xu
 *ab,
Peide
Zhu
b,
Rongye
Zhu
b and
Juncheng
Wang
*c
*ab,
Peide
Zhu
b,
Rongye
Zhu
b and
Juncheng
Wang
*c
aCollege of Artificial Intelligence, China University of Petroleum-Beijing, Beijing, 102249, China. E-mail: xuquan@cup.edu.cn
bState Key Laboraty of Heavy Oil Processing, China University of Petroleum-Beijing, Beijing, 102249, China
cDepartment of Stomatology, First Medical Center, Chinese PLA General Hospital, Beijing, 100042, China. E-mail: wbhwjc527@126.com
First published on 1st November 2023
Abstract
As a novel type of nanomaterial, carbon dots (CDs) are widely used in biology owing to their optical property, biocompatibility, and intrinsic theranostic properties. Taking advantage of these features, the CDs serve as color agents, fluorescence probes, and anti-cancer drugs. Machine learning (ML) has progressed dramatically, especially for widespread use in the biological field. In this review, we introduce the ML workflow and the leading models in the process and then demonstrate the application of CDs in bioimaging, biosensing, and cancer treatment. Next, we generalize the use in the development of CDs’ combination of ML in complementary aspects. Finally, we briefly summarize the challenges and expectations for the future. This review provides new thoughts and guidance for CDs on the application and integration of machine learning.
1. Introduction
Machine learning (ML), as the domain method of artificial intelligence,1–3 efficiently processes large amounts of data and generates powerful models capable of exploring inherent patterns and accurately predicting outcomes.4,5 Additionally, ML can gradually adapt its models to new data, improving its analytical abilities through self-directed and adaptive learning. These advantages have facilitated the widespread adoption of ML and expanded its potential applications.6 The widespread adoption of machine learning across different fields is exemplified by the successful implementation of voice recognition, machine translation, self-driving cars, and semantic segmentation. With the mainstream technology maturity and success, others have also engaged in the usage of ML in different fields. The combination of ML in the biological field is more extensive. The biological data of images,7,8 tables,9,10 and text11,12 are suitable for ML to train the models, optimize the conditions, predict the results, and enhance the relevant properties.13–15 As computational resources and ML tools are now widely accessible, the expenses and limitations associated with applying ML have been reduced significantly.Carbon dots (CDs) define the zero-dimension carbon materials on account of their less-than-10 nm particle size,16 bringing multiple satisfactory properties,17–22 such as excellent optical properties, chemical stability, solubility, and biocompatibility. More attention has been paid to CDs, and significant progress has been made in the preparation, synthesis,23–28 and applications.29–31 Particularly, CDs act as a powerful tool in the biological field, and have a widespread application in imaging, sensing, and cancer treatment. It furnishes vigorous fluorescence intensity, good chemical stability, and a long fluorescence lifetime, which can serve as fluorescent agents working in the bioimaging field.31 Moreover, due to the large surface area and wealth of chemical groups on the surface, CDs specifically bind to unique substances, enhancing or diminishing the fluorescence intensity depending on the concentration of the target molecule. This optical characteristic is developed so that the fluorescence probe senses the content of pH,32 ions,33–35 and biomolecules.36–38 CDs possess good hydrophilicity owing to their amino, hydroxyl, and carboxyl groups, and can quickly be loaded with drugs. Thus, they have strong potential as superior therapeutic drug delivery agents.39–41
The integration of ML into the biological application of CDs has great capacity, mainly including two major categories: ML guides CDs synthesis; and ML assists in data processing and analysis. The ML algorithm can be widely used in various optimization problems by sampling, and training for limited samples to optimize the synthesis parameters and improve the material properties. For example, Tang et al.42 used ML to guide the synthesis of CDs to enhance the optical property and established a progressive, adaptive model to minimize the number of practical trials. ML, as a data-driven model, quickly and efficiently ascertains the characteristics of the data to improve the accuracy of the analysis results. This paper reviews the CDs’ application in the biological field, and the following section introduces ML in the corresponding combination usage, as displayed in Fig. 1. Finally, the brief conclusion and expectations about ML are shown at the end of every section.
2. Overview of machine learning
ML is an important computational method and a significant part of artificial intelligence (AI). AI is a system that uses computers to simulate human behavior and thought processes, dating from the 1950s.43The birth of AI came from the Dartmouth conference in 1956, and its development phase has gone through three major waves, as shown in Fig. 2. The first breakout stage represents the logical reasoning, whose sign is the making of the simple music Wabot1 robotics. Due to the low computational capacity, the complex and significant problems could not be dealt with, so the development of AI was stalled. The neural network and backpropagation (BP) algorithm significantly facilitated the second breakout stage of AI, which is the expert system based on prior knowledge accumulation. However, the lack of practical application of AI led to a quick decay in it use. Owing the explosive increase in data, powerful computing ability, and constant refining algorithms, AI experienced a burst in growth in the past decade. In particular, Alpha GO defeated the world Go champion Lee Sodel in 2016.44 Nowadays, finance, medical treatment, automotive drive, and other industries45,46 have mature applications and established a relatively complete database.
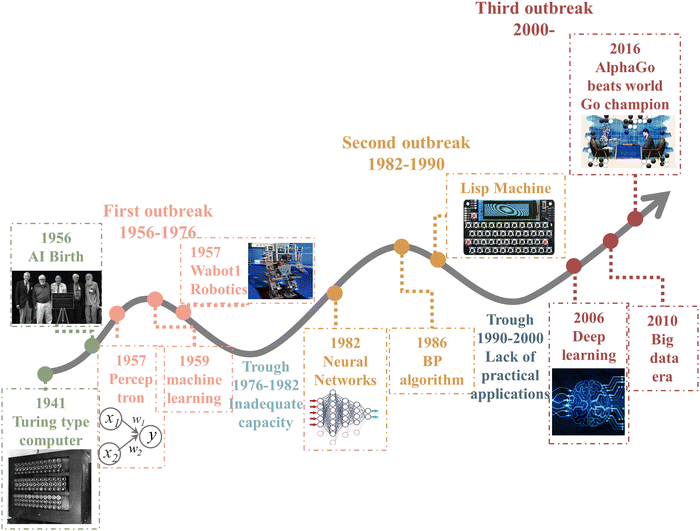 | ||
| Fig. 2 History of artificial intelligence.43–46 | ||
With the recent rapid advances in data availability, arithmetic power, and new algorithms, ML has emerged as one of the key methods for realizing AI, building models by using algorithms to reveal inner connections, which can make better decisions without human intervention. Most industries possess large amounts of data and have recognized the value of ML technology. Banks and other businesses in the financial industry use ML technology to identify important insights in data and prevent fraud, which helps to identify profit opportunities or avoid unknown risks. ML assesses patient health in real-time, and can also help medical professionals analyze data to identify diagnoses and treatments. Websites recommend items you might like based on analyzing previous purchase history by ML. Moreover, we enjoy many conveniences brought by ML, such as text and speech recognition software, web search engines, personalized recommendations for movies, and prediction of delivery times, etc., which is playing an increasingly important role in people's daily lives. Compared with the previous rule, ML can handle massive amounts of data easily to extract patterns and regularities. Furthermore, ML utilizes the information to make predictions and decisions. The process is automated, efficient, and reliable, leading to less time consumption and more rewards.
Applying ML can be divided into four processes: data collecting, feature engineering, model selecting, and model application, as displayed in Fig. 3a. This section reviews the workflows and some common ML models.
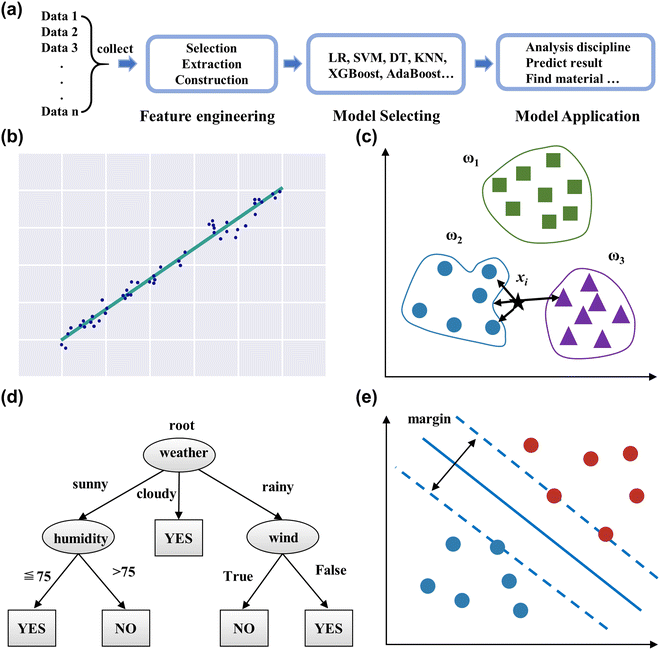 | ||
| Fig. 3 (a) The workflow of ML. Schematic diagram of the linear regression (b), K-nearest neighbor (c), decision tree (d), and support vector machine (e). | ||
2.1 Data collecting
As a data-driven model, ML is restricted by valid datasets for wide application. Still, data in materials science are characterized by high acquisition costs, excessive dispersion, lack of uniform processing standards, etc. The required conditions of data are high quality and integral, which is challenging in applying ML. In the past few decades, databases have been built and gradually replenished to include the physical properties, chemical properties, and crystal structures. Examples include Materials Project, Computational 2D Materials Database, ChemSpider, Inorganic Crystal Structure Database (ICSD), GDB databases, PubChem, etc. The format of data is varied and required according to the demand for ML models. High-throughput computing is a scientific research method to study and predict the properties of materials producing large data with minimal resources and the fastest possible speed. It combines with ML to effectively eliminate the weaknesses of enormous computation resources and accelerate the process of further materials exploration. Moreover, text mining is an important tool in AI, which can handle tens of thousands of text to analyze and discover knowledge.47 Text mining mainly parses unstructured text data to generate high-quality target data through natural language processing methods. Currently, text mining has been applied in the fields of material science,48 political science,49 economics,50 and others.2.2 Feature engineering
Feature engineering is the process of extracting and transforming data into more convenient features, enabling the achievement of optimal performance and improving the prediction accuracy of models. Hence, feature engineering plays a crucial role in applying ML processing and mainly contains feature selection, feature extraction, and feature construction.Feature selection screens useful features, and deletes redundant (and even irrelevant) features from a large number of features.51 If all features are input into the ML model, the problem of dimensional disaster often occurs, and even the model's accuracy can be reduced. Therefore, accomplishing feature selection to exclude invalid features as training data for the model is essential.52 Feature extraction is the process of reducing the dimensionality of the data, retaining significant information, and generating new features.53 The large pristine feature matrix increases computational cost and time, so reducing the dimension is important. Principle component analysis (PCA) and linear discriminant analysis (LDA) show excellent ability and potential to deal with high-dimension data.52 Feature construction processes the original data, combines subsistent features, and generates new features.53 The new introduced features need to be verified to improve the prediction accuracy, rather than adding a useless feature to increase the complexity of the algorithm operation. This required researchers to spend a lot of time on the sample data and think about the nature of the problem, the structure of the data, and how best to use them in the prediction model. For example, the body mass index (BMI) represents the body's fatness and measures whether it is healthy. It is calculated by the mass and height, and constructed as the new feature. The initial data, mass, and height also can denote the body index, but the BMI more directly shows healthy conditions and can help with disease prevention.
2.3 Model selecting
The ML model is divided into supervised and unsupervised learning. The difference between supervised and unsupervised learning is whether it labels the targeted output. In this section, we will review the main model in ML according to its targeted output. The supervised model is commonly used and a comparison is shown in Table 1.| Model | Linear regression | Polynomial regression | Logistic regression | K-Nearest neighbor | Decision tree | Support vector machine | Random forest | Adaptive boosting | Gradient boosting decision tree | XGBoost | LightGBM |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Regression problem | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | |
| Classification problem | √ | √ | √ | √ | √ | √ | √ | √ | √ | ||
| Output | Continuous | Continuous | Discrete | Continuous and discrete | Continuous and discrete | Continuous and discrete | Continuous and discrete | Continuous and discrete | Continuous and discrete | Continuous and discrete | Continuous and discrete |
| Basic principle | Linear function | Polynomial function | Active function | Distance measure | Binary tree | Decision surface | Decision tree | Decision tree | Decision tree | Decision tree | Decision tree |
| Advantage | Easy to understand and avoid overfitting | Easy to implement and understand; fit a nonlinear relationship between variables | Easy to implement and understand; suited for binary classification | Simple technique; suited for multi-model classes | Handle a variety of data; good generalization ability | Not overfitting; appropriate kernel function; handle with high-dimension data | Fast, scalable, robust to noise, does not overfit, easy to interpret and visualize with no parameters to manage | No overfitting and no need to filter features; high accuracy | Handle a variety of data; robust to outliers | Higher precision compared with GBDT; training in parallel; automatic processing of missing value features | Adopt histogram algorithm reduces time complexity; less memory consumption compared to XGBoot |
| Disadvantage | Cannot deal with a non-linear relationship | Easily overfitted data; outlier sensitivity | Difficult to deal with data imbalance; more sensitive to multicollinearity data | outlier sensitivity; intensive computation with huge data | difficult to handle high dimensional data; data fragmentation problem | Cannot solve multiclassification problems | Slow for real-time prediction | Time-consuming for training; data imbalance leads to loss of classification accuracy | Difficult to train data in parallel; computational complexity | Excessive space complexity | More sensitive to noise |
2.3.1.1 Generalize linear regression analysis. The main algorithms for regression analysis contain linear regression, polynomial regression, logistic regressions, ridge regression, lasso regression, etc.
Linear regression (LR) predicts the target variable by fitting a linear function to the data set, which is the basis for the regression problem and the most highly used model in the industry.54 Its function is as follows:
| y = ωT × x |
Polynomial regression develops from linear regression and has more input variables and a higher power exponent,55 which can fit an arbitrary data set without considering the computational effort, overfitting, etc. Logistic regressions belong to the linear classifier. Employing a logistic function,56 the data features are mapped to a probability value in the interval from 0 to 1, compared with 0.5 to the classification.
2.3.1.2 K-Nearest neighbor (KNN). KNN is a basic classification and regression method that can quickly and efficiently tackle predictive classification problems on specific data sets,57 as shown in Fig. 3c. Algorithm steps mainly include calculating the distance of each sample point in the training and test samples, sorting all the distance values above, selecting the first k samples of the minimum distance, and voting based on the classification decision rule to obtain the classification category. The k-value determines the complexity of the model, and requires further validation to select the appropriate value for particular data.
2.3.1.3 Decision tree (DT). Decision trees present a model of decision rules and classification results through a tree structure,58 as shown in Fig. 3d. It consists of nodes and directed edges, which signify the attribute and output result, respectively. The intuitive structure generates visualization results and enhances readability. Moreover, it can handle numerical and categorical features, and is insensitive to missing values and outliers in the data.59
2.3.1.4 Support vector machine (SVM). Support vector machines (SVM) are a binary classification model that seeks the geometrically separated hyperplane with the largest margin to split the training data set,60 as shown in Fig. 3e. SVM requires the sample data to be linearly separable so that a classification hyperplane exists. The kernel function transforms the nonlinear data in the original space and maps it into the high-dimensional space, resulting in linearly separable data possessing an optimal classification hyperplane.61 Since the classifier of the SVM model relies on some support vectors, it leads to strong robustness, which does not increase the computational complexity as the dimensionality of the data increases. SVM also can handle regression problems, as it finds a regression plane to which all the data in a set have the closest distance.
2.3.1.5 Ensemble learning. Ensemble learning refers to building and combining multiple weak learners to form a strong learner with superior performance to fulfill complex tasks. The common ensemble learnings are divided into two categories: bagging and boosting.62
Bagging63 belongs to classical parallel methods whose weak learners can independently train and predict. Bagging uses bootstrap sampling, which should put back the randomly selected data into the data set per round, obtaining the unique dataset for each weak learner. The random forest (RF) is based on the DT utilizing the bagging algorithm to form the strong learner.64 RF has many advantages over DT, such as decreasing the variance, eliminating the overfit characteristic, evaluating the feature importance, etc.
Boosting65 uses weak learners to iterate the learning of the data's intrinsic rules, which is a dynamic process that adjusts the weights for the data and enhances the performance of models. While all the learners are trained, the performance also specifies the weight of every learner. Adaptive Boosting (AdaBoost),66 Gradient Boosting Decision Tree (GBDT), XGBoost, and LightGBM all utilize the boosting method.
The AdaBoost is adaptive to the data, whose weight changes by the weak learners.66 While samples misclassified by the prior learner are strengthened, the whole weighted sample is applied again to train the next learner. GBDT67 calculates the negative gradient to recognize the error and modify the model. XGBoost68 perfects the column subsampling, parallel calculating, and automatic handling of missing values based on GBDT. LightGBM69 handles the huge volume of data in industrial circles, which consumes less memory, possesses faster training speed and accuracy, and supports distributed computing compared with XGBoost.
2.3.2.1 K-Means algorithm. K-Means is a clustering algorithm,70 which is the process of dividing the samples into categories through intrinsic relationships without any prior knowledge of the labels of the samples. The key target of K-means is to divide the given dataset into K clusters, and to give the centroid corresponding to each sample data.
2.3.2.2 Principal component analysis (PCA). Principal component analysis (PCA) is a dimensionality reduction algorithm used to reduce redundancy and compress data sets through feature extraction.71 It maps the original high n-dimensional features to k low-dimensions by the covariance matrix, which are the new orthogonal features also known as principal components. In practical application, reducing the dimension can save us a lot of time and cost within a certain range of information loss and has become a very widely applied method in data preprocessing.
2.3.2.3 Association rule learning. The association rule is used to describe the correlation between two or more things,72 which can be obtained from a large amount of data between valuable data connections. One example is that “if a customer buys beer, s/he is likely to also buy nappies at the same time”. Two parameters, support and trust, are common metrics to measure the effectiveness of the association rule.72
The data division is related to the model evaluation and selection, and is randomly divided into training and test sets according to a certain proportion. Only part of the data is used to train the model, which would affect selected models. Cross-validation is proposed to optimize model validation technology. The most widely used one is K-Fold cross-validation. It divides the data into disjoint and equal numbers of K parts, selects 1 part as the validation set, and the remaining K-1 parts as the training set. After conducting K separate model training and validation, the validation error of this model takes the average of the K validation results. Leave one out cross validation (LOOCV) is a particular instance of K-fold cross validation, which selects one data as the validation set per round. The number of build models is controlled by the size of the data set and the computational cost is tremendous. In practice, cross-validation usually combines with the Grid Search, which tests the performance on the same validation set by permuting different hyperparameters and selects the setting corresponding to the best-performing model.
Suitable evaluation metrics are needed to compare the effectiveness of different models and select the most appropriate model. According to the different types of problem-solving, the evaluation metrics are also different, which are mainly categorized into two types: classification and regression.
The common evaluation metrics for classification problems mainly include accuracy, precision, recall, F1 score, ROC curve, and AUC score. The evaluation indexes of regression problems mainly include MAE and MSE.
In a binary classification problem, samples can be classified according to the combination of their true and predicted categories into four types: true positive (TP), false positive (FP), true negative (TN), and false negative (FN). In general, these are displayed by the confusion matrix.
Accuracy is the percentage of the total sample with correct predictions.
Precision evaluates the accuracy of the positive examples predicted by the model.
Recall represents the probability of being predicted positively in a positive sample.
Precision and recall are contradictory metrics, so the F1 score combines them to find the balance.
The ROC curve is a graphical tool used to represent the performance of a classification model. It shows the relationship between the true positive rate (TPR) and false positive rate (FPR) of the classifier under different thresholds.
The AUC score is the area under the ROC curve, and is used to measure classifier performance. The value of AUC ranges from 0 to 1, which is positively associated with the classifier performance.
For regression problems, the common evaluation metrics are MAE and MSE.
The formula for the mean absolute error (MAE) is
where N represents the number of samples, yi is defined as the ith true value, and yi-pre represents the ith predicted value.
2.4 Model application
ML aims to provide a simple and quick prediction, and decrease the computational and manual trial-and-error costs. This section will introduce two kinds of ML applications in the CDs’ field.ML accelerates the discovery of new recipes, whose material and result can be considered parameters. As we can see in the subsequent section, many models are applied to the synthesis of CDs. The CDs’ recipe and experimental condition are variable, leading to volatile results. So, the CDs' related property is considered the target, such as the quantum yield (QY), emission center, Stokes shift, etc. The effect of fitting the data is excellent for obtaining high-quality CDs. The insight of ML is novel for synthesizing CDs, which provides new ideas for similar materials, and accelerates the fast development in material science.
ML also provides novel analysis methods to enhance interpretability, especially for image data. In general, the 2D image data are set as the input of ML, which can effectively learn the corresponding features. The fluorescence intensity of CDs is related to concentration, which should be measured through a specific instrument. However, ML can directly utilize the fluorescent image to calculate the concentration, which is more convenient than ever. Computer vision is one of the fastest growing and most widely used technologies in the artificial intelligence segment, while ML, as the power tool, will promote development.
3. Bioimaging
If the material has the properties of suitable sensitivity and time/space resolution for fluorescence, then it can have great potential for applications in bioimaging,74 particularly when it comes to cost due to the efficient and cost-effective way to synthesize CDs. This makes it an ideal choice for research in imaging. In this section, we discuss the application of CDs in cell imaging and real-time tracking. Then, the use of ML algorithms to improve the performance of CDs and cell imaging are summarized.3.1 Cell image
The fluorescence spectra of CDs emitting blue light are susceptible to scattering spectral interference because of the absence of a significant Stokes shift.75 Long-wave luminescent (yellow to red) CDs are now beginning to be applied for imaging and analytical detection.76 Jiang et al.77 produced three CDs that successfully created CDs emitting green, yellow, and red luminescence under a single ultraviolet excitation. Confocal micrographs emitted different colors under 405 nm laser excitation, while the MCF-7 cells were incubated in three CDs for 4 h, as shown in Fig. 4a–c. Interestingly, CDs possess up-conversion photoluminescent (UCPL) properties, providing additional application potential for two-photon bioimaging of deep tissues. Liu et al.78 developed a highly efficient and optimal emission wavelength of about 630 nm for red-emitting nitrogen-doped carbonized polymer dots (CPDs). They found that the red fluorescence originated from hydrogen bonds and the π systems in the conjugated aromatic group of CPDs, which had been successfully used as fluorescent probes for bioimaging in vitro and in vivo studies. Effective regulation of the nano-size, surface functional group state, and crystalline structure of CDs will cause changes in their energy band structure, which can effectively adjust the fluorescence emission color of CDs. Wang et al.79 employed polyetherimide (PEI) to modify CDs ((CDs@PEI)) in an aqueous solution to enhance emission. Zeta potential analysis (Fig. 4d) demonstrated that the negative charge of CDs was reversed to the positive charge of CDs@PEI by PEI modification. Fig. 4e and f show that the emission of CDs@PEI aqueous solution was further improved compared to those of the pristine CDs and mixing-with-PEI (CDs + PEI) samples, and the center was redshifted. The technique reverses the surface charge, enhancing the absorption of CDs by cells and allowing for distinct fluorescence imaging.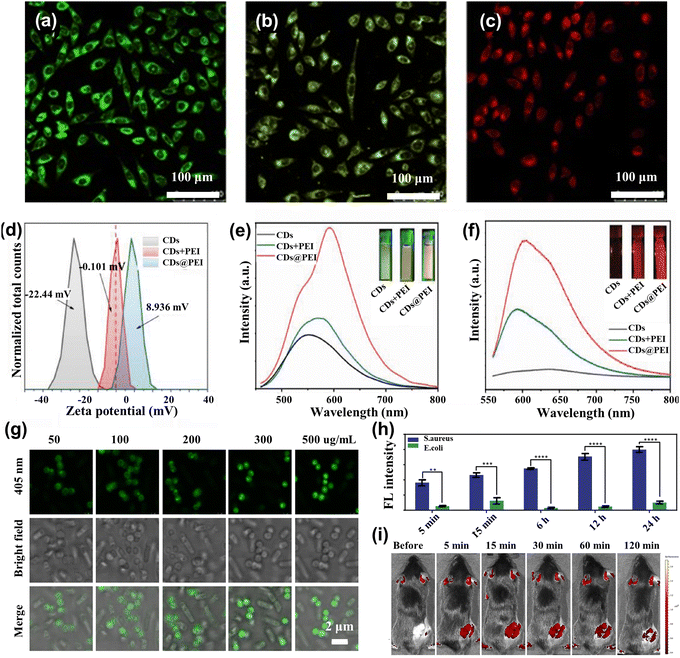 | ||
| Fig. 4 (a)–(c) Confocal fluorescence MCF-7 cells imaging in green, yellow, and red under the 450 nm laser light.77 Reproduced from ref. 47 with permission from John Wiley and Sons, copyright 2015. (d) The zeta potential spectra for CDs, CDs + PEI, and CDs@PEI.79 (e) λ = 450 nm excitation and (f) λ = 550 nm excitation fluorescence spectra of CDs, CDs + PEI, and CDs@PEI aqueous solutions.79 Reproduced from ref. 49 with permission from Elsevier, copyright 2022. (g) Confocal images of the mixture of Escherichia coli and Staphylococcus aureus cells after incubation with 50, 100, 200, 300, and 500 μg mL−1 T-SCDs.82 (h) Average intensity statistics of fluorescence corresponding to confocal images processed by the image.82 Reproduced from ref. 52 with permission from American Chemical Society, copyright 2021. (i) Fluorescence imaging was performed in vivo at 490 nm excitation and 560 nm emission at 5, 15, 30, 60, and 120 minutes before and after injection of N-CQDs.83 Reproduced from ref. 53 with permission from Elsevier, copyright 2023. | ||
Doping heteroatoms also improve the fluorescence properties of CDs. For example, Liu et al.80 proposed the synthesis of nitrogen (N) and sulfur (S)-doped CDs by hydrothermal method. The biocompatibility and excellent fluorescence emission indicated that the NS-CDs can be used for fluorescence imaging. Bouzas-Ramos et al.81 successfully synthesized CDs doped with two elements (nitrogen and lanthanide), Gd and Yb, using a one-pot microwave-assisted hydrothermal method. Especially, the doped-CDs with a QY of 66 ± 7% had intense fluorescence emission, low cytotoxicity, magnetic resonance (MR), and computed tomography (CT) contrast properties for successful application in in vitro fluorescence, MR, and CT cell imaging.
3.2 Real-time track in live cell
A crucial stage in microbiology research and the therapy of bacterial infections is the real-time tracking and detection of live bacteria. Yan et al.82 synthesized three excitation peaks and single-color emission carbon dots (T-SCDs). This material identified Gram-positive bacteria in less than 5 minutes and tracked them in real-time over 24 hours. The fluorescence intensity of S. aureus was amplified with an increase in the concentration of T-SCDs, whereas E. coli had little to no response to the T-SCDs concentration, as shown in Fig. 4g.Similarly, S. aureus exhibited a notable increase in fluorescence intensity with increased incubation time (Fig. 4h), while Gram-negative E. coli did not react to the extended incubation period. T-SCDs were remarkable for the rapid enrichment of Gram-positive bacteria and had good performance in the real-time tracking of Gram-positive bacteria. Bharat Bhushan et al.84 synthesized a simple, inexpensive, and environmentally friendly method to prepare CDs by hydrothermal treatment of commercial casein. The CDs can be specifically labeled for Gram-negative bacteria and were able to enter fibroblasts, remaining effectively labeled in the subsequent 3–4 generations of cells.
Malignant tumors have become a group of common diseases that endanger human health.85 There have been attempts to use CDs to trace tumor cells, label mesenchymal stem cells, pursue intracellular nucleic acids, and target organelles.40 The assembly of CDs bio-probe enables highly sensitive and specific detection, providing a new way for the early diagnosis of malignant tumors. Liu et al.86 injected 100 μL of prepared CDs solution into nude mice for systemic circulation through the tail vein. After 0.5 hours of injection, the strong fluorescence signal observed in the brain suggested that this may have great potential for therapeutic diagnosis of certain brain diseases through real-time monitoring. Zhang et al.83 utilized a molecular fusion strategy to generate a novel range of nitrogen-doped carbon quantum dots (N-CDs). Fig. 4i shows the fluorescence image after the N-CDs were taken into the body, demonstrating the N-CDs were effectively delivered to the tumor site in a mere 5 minutes and able to produce a noteworthy fluorescent signal owing to their ultra-small size (1.5 nm). The fluorescence intensity remained strong after long-term consumption, indicating the superior permeability and retention of N-CQDs, enabling the real-time visualization of the tumor with optimal fluorescence imaging.
3.3 ML assists bioimaging
Red CDs have been extensively used in current research for cellular imaging due to their excellent optical properties. However, their low synthetic efficiency and time-consuming nature are still essential constraints. Luo et al.87 have successfully created an ML model that can accurately forecast the ideal synthesis conditions for the generation of red CDs. One-hot was adopted to transform data features, while principal component analysis (PCA) combined with the XGBoost was applied to extract data features. The logistic regression model achieved good prediction results for determining whether the synthesized CDs were red. This model is used to optimize the production process and ensure that the highest quality red CDs are produced with the most efficient methods, which would help to reduce costs and increase the overall efficiency of the production process. To successfully synthesize a series of CDs with customized optical properties for cellular imaging applications, Hong et al.88 used XGBoost to forecast the maximum fluorescence intensity and emission center of CDs generated from p-benzoquinone (PBQ) and ethylenediamine (EDA) at ambient temperature. The mass of PBQ (MPBQ), the volume of EDA (VEDA), reaction duration, and medium as the typical parameters controlling the fluorescence performance were used as the input feature, while fluorescence intensity and emission center position as the predicted targets were used as output parameters. As shown in Fig. 5a, the authors collected all CDs’ maximum fluorescence intensity values to build the database. CDs-3 are predicted to have excellent optical properties under ML reaction conditions, which are employed for whole-cell imaging (Fig. 5b(i)). The cytoplasmic region mainly showed blue and green fluorescence in Fig. 5b(ii), illustrating that CDs-3 can cross the cell membrane into the cytoplasm quickly, facilitating cell staining and labeling.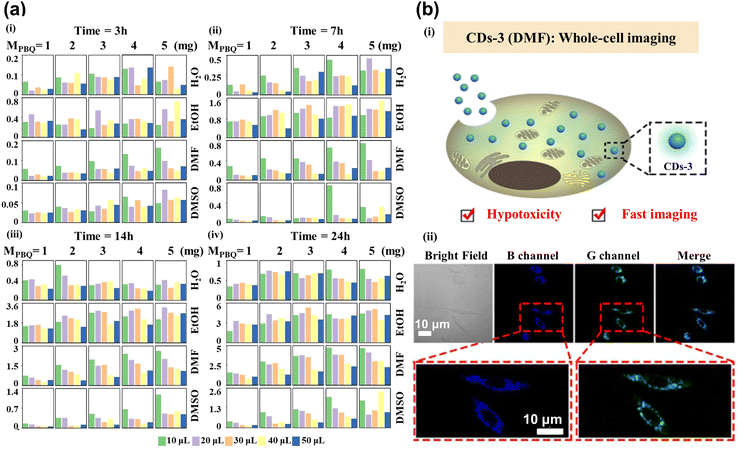 | ||
| Fig. 5 (a) The maximum fluorescence intensity histogram shows the 400 CDs synthesized under different reaction conditions. Each block represents the reaction times of CDs for 3 (i), 7 (ii), 14 (iii), and 24 (iv) h. The column and row were controlled by MPBQ and various solvents, respectively. The colors of different columns in each square lattice show different volumes of VEDA.88 (b) Imaging of whole-cell fluorescence by CDs-3. (i) The diagram shows the internalization of CDs-3 into the cell. (ii) Confocal images from left to right showing the bright field, blue channel, green channel, and merging of cells. Reproduced from ref. 58 with permission from American Chemical Society, copyright 2022. | ||
Bioimaging is currently developing many exciting resources to promote the adoption of supervised machine-learning models. Image noise is inevitable for fluorescence microscopy which can be from the detectors, acquisition speed, imaging resolution, scattering, etc. So, low noise is essential for image analysis. ML-driven analysis can optimize and simplify microscopic image processing, improve signal-to-noise ratios, deconvolve images, and dramatically increase resolution.89 Romain et al.90 explored self-supervised ML methods to denoise images, which is more convenient than supervised ML in the data quality demands. Wang et al.91 combined the transfer learning and U-Net for application in image denoising. The transfer learning possesses prior knowledge about the noise model, which helps to improve the performance of denoising.
Weigert and co-workers developed content-aware image restoration (CARE) networks to restore missing training data.92 Chai et al.93 applied the U-Net to the 3D image to inpaint and denoise, achieving dynamic volumetric imaging and improving the signal-to-noise ratio (SNR).
Image segmentation refers to the separation of objects in a picture from the background, related to the set of pixels. The technology of image segmentation can automatically separate the different tissues in microscopy images, attracting much attention. Schmidt et al.94 utilized the star-convex polygons to localize cell nuclei to enhance the detected performance. Based on the U-Net, the model can segment the overlap nuclei. Researchers have also applied this to 3D images, where the star-convex polyhedral represents the cell nuclei. The method can accurately facilitate the detection and segmentation of cell nuclei in the 3D image.95 Segmenting the membrane is more difficult than nuclei, owing to the various morphologies and different sizes. Khameneh et al.96 combined the superpixel-based tissue classifier (SVM) and modified U-Net to segment cell membranes with 0.94 segmentation and 0.87 classification accuracy. Eschweiler et al.97 modified the 3D U-Net for 3D confocal microscopy images. It showed an advantage in segmentation quality for deep issues without tedious parameter adjustment. The restriction for algorithmic generalization capabilities is the lack of the amount of dataset to train the whole-cell type segmentation model. Stringer et al.98 introduced a generalized model named Cellpose, which is trained on over 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 segmented objects. It can precisely segment cells in various image types, and does not require model retraining or parameter adjustments. The model also supports the 3D image without the labeled 3D data. Cellpose is packed as a software for free usage to support the community contribution online.99,100 Next, Eschweiler and co-operator101 expanded the Cellpose method to improve the classification accuracy in 3D images. The expanded Cellpose could simplify the preparation of training data and instance reconstruction. In addition, Cutler et al.102 proposed another generalized algorithm named Omnipose, which reaches high accuracy of segmentation in cell and expands the applied field to non-bacterial subjects, varied imaging modalities, and three-dimensional objects, demonstrating excellent performance in image segmentation. Organelle segmentation is required in the biological field, while Heinrich prepared work with the matter, which is impeded by the time-consuming manual annotation.103 The open-source web repository, ‘OpenOrganelle’, is created to share data, accelerating the development of segmentation in the biological domain.
000 segmented objects. It can precisely segment cells in various image types, and does not require model retraining or parameter adjustments. The model also supports the 3D image without the labeled 3D data. Cellpose is packed as a software for free usage to support the community contribution online.99,100 Next, Eschweiler and co-operator101 expanded the Cellpose method to improve the classification accuracy in 3D images. The expanded Cellpose could simplify the preparation of training data and instance reconstruction. In addition, Cutler et al.102 proposed another generalized algorithm named Omnipose, which reaches high accuracy of segmentation in cell and expands the applied field to non-bacterial subjects, varied imaging modalities, and three-dimensional objects, demonstrating excellent performance in image segmentation. Organelle segmentation is required in the biological field, while Heinrich prepared work with the matter, which is impeded by the time-consuming manual annotation.103 The open-source web repository, ‘OpenOrganelle’, is created to share data, accelerating the development of segmentation in the biological domain.
3.4 In brief
CDs can be utilized to develop robust and accurate cell imaging and real-time tracking for diagnosis and research due to their excellent optical properties. The need for high-quality CDs is urgent and the ML is capable of guiding the synthesis CDs. By applying ML algorithms to these processes, researchers have been able to enhance the performance of CDs, improving the accuracy and resolution of cell imaging. All mentioned methods are summarized in Table 2. These advances are significant steps forward in nanotechnology and bioimaging, which could lead to more efficient materials and more reliable diagnostic tools.| Model | Result | Ref. |
|---|---|---|
| XGBoost | Produce the highest quality red CDs to increase the cost and have the potential to bioimaging | 87 |
| XGBoost | Maximum fluorescence intensity and emission center's CDs generated and applied in bioimaging | 88 |
| Self-supervised | Denoise images without the data quality demands | 90 |
| U-Net | Denoise images | 91 |
| CARE | Restore the missing training image | 92 |
| U-Net | Inpaint and denoise the 3D image; achieve dynamic volumetric imaging | 93 |
| U-Net | Use star-convex polygons to separate the overlap nuclei | 94 |
| U-Net | Utilize star-convex polyhedral to segment the nuclei in the 3D image | 95 |
| SVM + U-Net | Segment cell membrane with 0.94 segmentation and 0.87 classification accuracy | 96 |
| 3D U-Net | Segment the cell in confocal microscopy image and deep tissue | 97 |
| Cellpose | Segment the cell in various types for 2D and 3D image | 98 |
| Extensive cellpose | Improve the classification accuracy in 3D image | 101 |
| Ominpose | Segment the cell and expand into any 3D objects | 102 |
4. Biosensing
Due to the abundant surface functional groups, CDs are sensitive to tiny disturbances, leading to great potential for sensing environmental changes. The specific binding between CDs and analytes is beneficial for precisely detecting the concentration. In this section, we discuss the use of CDs as probes to detect biological analytes such as pH, metal ions, amino acids, etc. ML is applied and quickly shows the concentration.4.1 pH
pH is involved in various life activities of living organisms, including cell division, wound healing, ion uptake, calcium regulation, etc. Lu et al.104 prepared yellow–green phosphorescent carbon dots (P-CDs) with a good linear relationship in the pH range of 2–12, and retained the strong intensity of fluorescence with a durable pH cycle. As shown in Fig. 6a, the mechanism for detecting the pH was related to the abundant –OH and –COOH, which acted as acceptors for hydrogen ions and hydroxyl radicals, enabling the fast and efficient detection of the acidity and basicity of the solution. Zhao et al.105 assembled an efficient detection platform combining the N-doped blues CDs and the Te-doped CDs, which can effectively and multi-method analyze pH. Huang et al.106 synthesized the CDs, the independent-excitation property at 615 nm. The red CDs (RCDs) can be quenched and recovered by H+ and OH−, respectively. The “off–on” property was due to the combination and break between target ions and the functional group on the surface of RCDs, as shown in Fig. 6b. Wang et al.107 combined two kinds of CDs into the system to detect intracellular pH based on the inner filter effect, as shown in Fig. 6c. The CDs also connected the pH and intracellular polysaccharide contents inside the Pholiota adipose fungus's mycelia to find out the most suitable condition for growth. Samran et al.108 reported on the first synthesis of green CDs (GCDs) and its application with the pH detection for the nucleus in live cells, as shown in Fig. 6d. The fluorescence intensity of GCDs was decreased as the pH varied from 1 to 12, and showed an excellent linear relationship with the pH range from 2–7 and 7–12, respectively. Furthermore, the CDs were successfully applied to A549 cells and zebrafish for pH monitoring and imaging, which had great potential in undertaking the biosensor to understand the nucleus-related physiological process.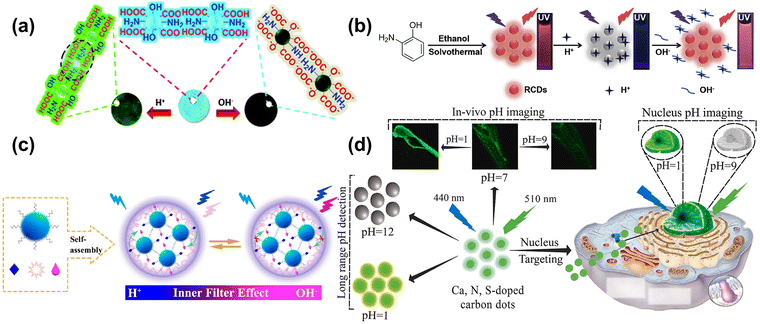 | ||
| Fig. 6 (a) Schematic of P-CDs sensing the pH.104 Reproduced from ref. 59 with permission from the Royal Society of Chemistry, copyright 2019. (b) The schema of synthesizing RCDs and its “off–on” property for the H+/OH−.106 Reproduced from ref. 61 with permission from Elsevier, copyright 2019. (c) Schematic diagram of a mechanism for the pH-based CDs response on the inner filter effect of Aniline Blue on the emission of Rhodamine B.107 Reproduced from ref. 62 with permission from American Chemical Society, copyright 2021. (d) The schematic diagram of Ca, N, and S-CDs with pH-responsive properties applied into pH detection, in vivo pH image, and Nucleus-targeting pH image.108 Reproduced from ref. 63 with permission from Elsevier, copyright 2023. | ||
4.2 Metal ions
The heavy metal ions can interact with the CDs, causing fluorescence quenching. For instance, Nandi et al.109 synthesized nitrogen-doped CDs with dual emission at the peak of 490 nm (green) and 570 nm (yellow) by hydrothermal method. The emission ratios of CDs were sensitive to the Fe3+ content, and the color change under UV lamp irradiation from light green to bright yellow could be observed by naked eye when the Fe3+ content increased, as shown in Fig. 7a. The ratio of yellow and green was selected for monitoring the Fe3+ content with the limit of 7.8 μM, as shown in Fig. 7b. Lesani et al.110 prepared green CDs using phthalocyanine to detect the Fe3+ concentration in MCF-7 live cells based on a stress-induced cell-based model. Li et al.111 obtained gold nanoparticles based on carbon dots to achieve dual ion detection of Cu2+ and Hg2+ dual-mode detection, including fluorescent and colorimetric dual modalities, as shown in Fig. 7c and d. Zhang et al.112 prepared orange-yellow quantum dots through a solvothermal method, which can be used to detect Cu2+. The fluorescence intensity increased with increasing concentration of Cu2+, as displayed in Fig. 7e. The detection of Cu2+ ranges from 0.02 to 30 μM, with a detection limit of 14 nM, as shown in Fig. 7f. Yue et al.113 designed green CDs that detected Ag+ based on the “on–off–on” feature, as shown in Fig. 7g. The surface of the CDs was full of carboxyl, amino, and thiol groups, which can specifically bind to Ag+, causing fluorescence quenching. When the solution containing Cl− was added to the CDs, Ag+ was shed from the surface of the CDs to form AgCl, resulting in the fluorescence recovery.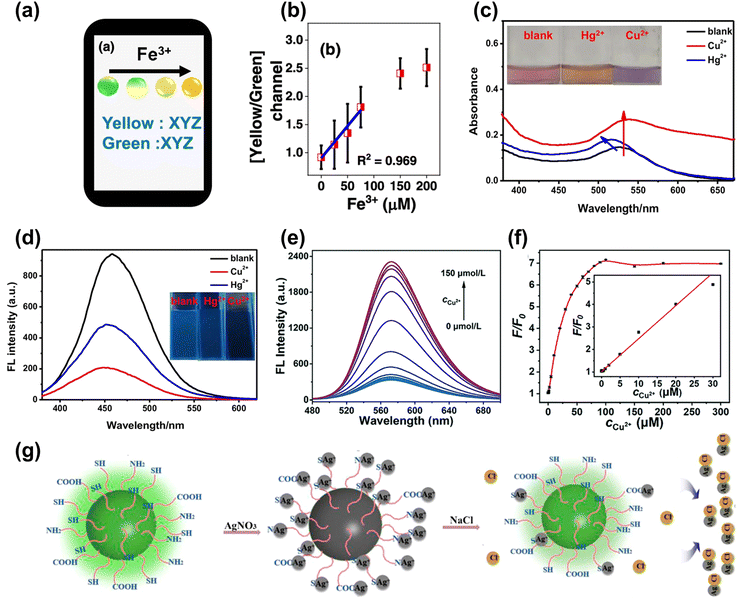 | ||
| Fig. 7 (a) The color change and analysis of CDs with Fe3+ added gradually.109 (b) The curve between the ratio of yellow/green and the concentration of Fe3+. Reproduced from ref. 64 with permission from American Chemical Society, copyright 2022. The absorption (c) and fluorescence spectrum (d) of CDs upon adding the Cu2+ and Hg2+.111 Reproduced from ref. 66 with permission from Springer Nature, copyright 2021. (e) The map of FL spectra for the CDs under different concentrations of Cu2+ (0–150 μmol L−1). (f) The relative changes of the fluorescence intensity (F/F0) of the CDs with different concentrations of Cu2+ (inset: the linear relationship between F/F0 and the concentration of Cu2+).112 Reproduced from ref. 67 with permission from the Royal Society of Chemistry, copyright 2022. (g) The schematic diagram of CDs detecting Cl− based on the “on–off–on” mechanism.113 Reproduced from ref. 68 with permission from Frontiers, copyright 2021. | ||
4.3 Small biological molecules
Amino acids and glucose are essential raw materials for human metabolism and oxidation, as well as for the normal survival of the body. Rossini et al.114 assembled a paper microfluidic device based on the CDs to indirectly detect the glucose and lactate in saliva samples, as shown in Fig. 8a. The responding enzyme can oxidize the glucose and lactate, producing H2O2 and causing quenching.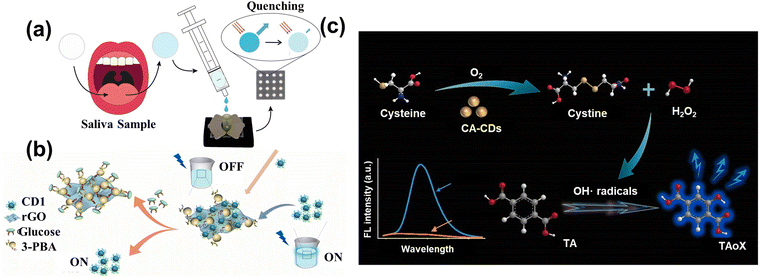 | ||
| Fig. 8 (a) The schematic diagram of CDs sensing glucose and lactate in saliva samples.114 Reproduced from ref. 69 with permission from Elsevier, copyright 2021. (b) The illustration of the preparation of rGO-PBA and use in detecting glucose.116 Reproduced from ref. 71 with permission from Elsevier, copyright 2022. (c) The schematic image of CA-CDs detecting cysteine through oxidation.117 Reproduced from ref. 72 with permission from Elsevier, copyright 2022. | ||
There was a linear relationship between the concentration of the analytes and the intensity of fluorescence. The detection limitations of glucose and lactate were 2.60 × 10−6 and 8.14 × 10−7 mol L−1, respectively. Yang et al.115 fabricated the green hydrophilic CDs to detect glutamic acid and aspartic acid, whose limitations were 1.69 and 1.24 μM, respectively.
To further decrease the cost of the enzyme, some studies utilized the “on–off” mechanism to direct the glucose. Zhou et al.116 used the phenylboronic acid functionalized reduced graphene oxide (rGO-PBA) and the polyhydroxy-modified CDs to detect the glucose. As shown in Fig. 11(b), the modified CDs were attached to the surface of rGO-PBA due to the affinity between the phenylboronic acid and hydroxyl groups. The quenching of fluorescence was related to the photoinduced electron translating from the CDs to graphene oxide. When the glucose was added to the solution, the CDs were shed from the graphene surface by stronger binding, leading to the fluorescence recovery. The additive had a linear relationship with the recovered fluorescence with the limitation of 0.01 M. Furthermore, Lin et al.117 designed the carbon-based enzymes to detect the cysteine (Cys) through the oxidase-mimicking property. CA-CDs were prepared by the one-step solvothermal method using citric acid (CA) as the single carbon source, which had a higher affinity and better catalytic ability with the Cys than the natural enzyme. Fig. 11(c) shows that the Cys was oxidized to form the cysteine and H2O2 by CA-CDs. Subsequently, the Cys can facilitate the decomposition of H2O2 to produce the radical group of OH˙. Then, the solution containing the terephthalic acid (TA) captured the radicals to make the TA-OH, leading the strong fluorescence. Therefore, the combination of CA-CDs and TA accurately detected the Cys with a limitation of 0.036 μM through the intensity of fluorescence. More importantly, this research not only provides a new approach to detecting the concentration of analytes via single simple raw material, but also establishes the foundation for the proposal and synthesis of carbon-based enzymes.
4.4 ML assists biosensing
The preparation of CDs is based on the trial-and-error method, which is subjective, random, and contingent, resulting in challenges to synthesizing the desirable properties. Han et al.118 applied ML to synthesize CDs to enhance the QY. Fig. 9a showcases the main procedure, with five important synthesis features as the input parameters and the QY are output parameters. Based on the ML-assist method, the CDs had a significant improvement in QY of 39.3%. The CDs also can be used as a probe to detect the Fe3+ with the limitation of 0.039 μM due to sensitivity and selectivity. Hong et al.88 utilized ML to guide the synthesis of CDs with high PL intensity and emission center, as displayed in Fig. 9b. The synthesis conditions controlling the optical properties of CDs were considered as the input parameters. According to the trained XGBoost model, the CDs with desired optical properties were obtained. Fe3+ could quench the fluorescence of CDs and was hardly ever influenced by other ions, as shown in Fig. 9c. Xu et al.119 conducted the ML into synthesizing CDs in the microwave to enhance the QY, as described in Fig. 9d. Five experiment-related parameters were viewed as the input feature in the ML model, and the QY of CDs was regarded as the output feature. The ML guiding CDs synthesis enhanced the QY by 200% higher than the pristine average value, showing excellent potential to find out the intrinsic law to increase the performance. The CDs acted as the monitor to detect the H2O2 with the limitation of 0.12 M and displayed the residual H2O2 on the bleaching teeth.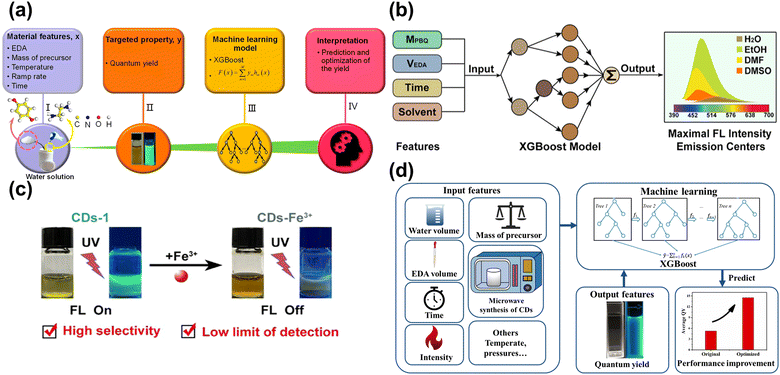 | ||
| Fig. 9 (a) The process of establishing the ML model and application.118 Reproduced from ref. 73 with permission from American Chemical Society, copyright 2020. (b) The input and output features for the XGBoost model.88 Reproduced from ref. 58 with permission from American Chemical Society, copyright 2022. (c) The CDs-1 can be quenched by Fe3+, and has high selectivity and low limit of detection with Fe3+. (d) The schematic diagram of utilizing the XGBoost model to enhance the QY.119 Reproduced from ref. 74 with permission from the Royal Society of Chemistry, copyright 2022. | ||
Compared with traditional sensing methods, the novel approach based on machine learning (ML) can directly and quickly detect to save time. Pandit et al.120 combined the ML and fluorescent array to detect eight proteins based on the different optical patterns. Seven ML algorithms were performed on the data, and the recognition accuracy was greater than that for the traditional linear discriminant analysis (LDA), realizing 100% prediction efficiency. Shauloff et al.121 used interdigitated electrodes (IDEs) coated with CDs to record the capacitance difference to distinguish and sense the bacteria through the CDs matching distinctive gas.
ML was used to study the capacitive response data for sensing different vapors. It exhibited excellent performance in distinguishing the single and mixed groups, which can further detect the kinds of bacteria. The investigation facilitated the real-time detection of bacteria and the application of ML. Xu et al.122 constructed a fluorescence sensor array containing the CDs and lanthanide complex (EDTA-Tb) to sense multiple heavy metal ions through the ML. Different ions combined with the sensor array caused different multi-dimension data (Fig. 10a), while the SX-model can distinguish each other and predict the concentration. The SX model was precise and its accuracy was up to 95.6% in the experimental sample. Liu et al.123 developed a smartphone-based nanoprobe sensing platform using machine learning to detect glutathione (GSH) and azodicarbonamide (ADA). The yolov3 was utilized, belonging to deep learning, to handle the image and establish the model that can relate the concentration of analytes with fluorescence ratio, as shown in Fig. 10b. The novel method was integrated into the WeChat APP, which was convenient for smartphone-based handheld devices. The performance of detection had excellent sensitivity in the concentration range of 0.1–200 and 0.5–160 μM with the limitation of 0.07 and 0.09 μM for GHS and ADA, respectively. Lu et al.124 designed a tri-color fluorescent optical device based on the ML algorithm and smartphone to detect tetracycline antibiotics (TC), as shown in Fig. 10c. The trichromatic sensor consists of blue CDs (BCDs), and the dual-emission red CDs were sensitive to TC. gases, Upon increasing the concentration of TC gradually, the sensor color changed from red to cyan. The YOLO v3 algorithm assisted in the photographing of the solution, which can implement the detection of TCs through the visual method. The technique was integrated into the smartphone, providing real-time and on-time antibiotic detection. Xu and co-workers125 utilized the carbon quantum dots-based fluorescence sensor array to sense the tetracyclines. Tetracyclines mainly contain four kinds: including tetracycline (TC), oxytetracycline (OTC), doxycycline (DOX), and metacycline (MTC). The SVM is applied to handle and analyze the sensor array dataset, which can distinguish the four kinds of tetracyclines. Moreover, the model is extensible, which can sense the binary mixture. In river and milk samples, the model also shows an excellent ability to detect. Wang et al.126 applied the ML to dual-emission fluorescence/colorimetric sensor array to detect the nine antibiotics. The input parameter contains the difference in fluorescent intensity and maximum emission wavelengths, while the antibiotics category and corresponding concentration is the output parameter. All of the processing is optimized by the tree-based pipeline optimization technique (TPOT). The ML can detect nine antibiotics at 0.5–50 μM with 95% accuracy. For the unknown sample, the model also can distinguish the different kinds and quantify the concentration. Jafar et al.127 utilized SVM to sense the concentration of nitrate, while the polynomial kernel showed the highest accuracy. The results showed that the polynomial kernel with the parameter at γ = 0.20 was best, with the MSE of 0.0016 and R2 of 0.93. The ML is integrated into smartphone applications, which is convenient for online detecting. The lifetime of biosensors also increased through applying the SVM, which is usable for up to at least 10 days. Gonzalez-Navarro et al.128 constructed the model between the amperometric response of glucose with dependent variables under different conditions, such as temperature, benzoquinone, and pH. Four kinds of ML regression models are compared, while the radial basis function-based SVM (SVM-R) is an excellent model with an R2 of 0.999. Due to the sensitivity of the sensor response being strongly related to these dependent variables, their interactions should be optimized to maximize the output signal, for which a genetic algorithm and simulated annealing are used, resulting in good generalization error. Rong et al.129 used the SVM to analyze the sense data to detect the small proteins. SVM with four kernels (polynomial, sigmoidal, linear, and radial basis function) were compared to the optimized model, while the radial base function kernel shows the best performance. The model possesses an accuracy of 98% on the bind interactions between the proteins and DNA. For the general test, the relative code shows greater ability than the equivalent circuit analysis. The model can be integrated on the smartphone for quick detection.
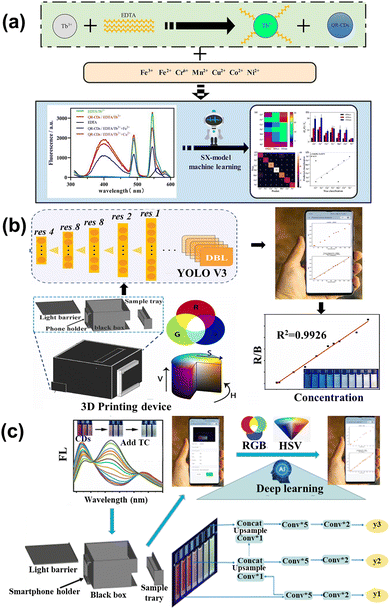 | ||
| Fig. 10 (a) Schematic of the multi-emission array sensor that served to detect heavy metal ions based on the SX model.122 Reproduced from ref. 77 with permission from Elsevier, copyright 2022. (b) The diagram of GSH and ADA determination utilizing Yolo v3 and Wechat.123 Reproduced from ref. 78 with permission from Elsevier, copyright 2022. (c) The illustration of the prepared tri-color CDs and application in TCs detection based on deep learning and smartphone.124 Reproduced from ref. 79 with permission from Elsevier, copyright 2023. | ||
4.5 In brief
The detected method is based on the linear relationship between the fluorescence of CDs and the concentration of analytes, leading to the precise sense. However, the traditional approach is time-consuming and labor-intensive, introducing innovative ideas to facilitate the change. ML, a novel method, brings great potential for promoting development. It can find out the inherent rules to establish a model to simplify the confusing conditions and enhance the promising result, which is a great advantage compared with other ways. According to the ML, the optical properties of CDs can be predicted for the non-experimental CDs. The result is direct and quick to obtain, and saves the cost of trial and error. Furthermore, ML brings a new approach to detecting the concentration through graph processing and the array sensing model. ML not only changes the traditional experiment procedure, but also expands the new method to sense. All related ML models are summarized in Table 3.| Model | Result | Ref. |
|---|---|---|
| XGBoost | Improve the QY to 39.3% and apply CDs to sense Fe3+ | 118 |
| XGBoost | Max PL intensity and emission center and applied CDs to sense Fe3+ | 88 |
| XGBoost | Enhanced the QY 200% higher than the pristine and applied CDs to sense H2O2 | 119 |
| LDA | Detect eight proteins with 100% accuracy | 120 |
| PCA | Detect the kinds of bacteria | 121 |
| SX-model | Sense seven heavy metal ions with 95.6% accuracy in the experimental sample | 122 |
| SX-model | Sense nine antibiotics at 0.5–50 μM with 95% accuracy | 126 |
| SVM | Sense tetracyclines and the binary mixture | 125 |
| YOLO V3 | Detect GSH and ADA in the range of 0.1–200 and 0.5–160 μM with the limitation of 0.07 and 0.09 μM, respectively | 123 |
| YOLO V3 | Detect tetracycline antibiotics | 124 |
| SVM | Sense the concentration of nitrate with R2 of 0.93 | 127 |
| SVM | Sense the concentration of glucose with R2 of 0.9999 | 128 |
| SVM | Detect small proteins with higher accuracy than the equivalent circuit analysis | 129 |
5. Cancer therapy
Cancerous tumors are one of the most complex diseases in the biological system, especially brain tumors related to the human nervous system, which influences linguistic, motor, cognitive, etc. CDs’ large surfaces and excellent hydrophilia have been proven to attach and transport medicine.124 Moreover, the CDs also act as adjuvants in different therapeutic schedules to enhance the treatment performance.5.1 Drug delivery
The huge surface of CDs is fit to load the cancer drug and control the drug release. Marziyeh et al.130 introduced the CDs into the mesoporous silica (MS) to form the drug carriers (MSCDs). The etoposide (ETO) was loaded into the MSCDs, and carboxymethyl β-cyclodextrin (Cβ) as the barrier was attached to the surface of MSCDs (Cβ-MSCDs). Meanwhile, folic acid (FA) was connected to the MSCDs surface (FA-Cβ-MSCDs) to control the place of release for ETO. The study showed that ETO-loaded FA-Cβ-MSCDs can be partially hydrolyzed and consequently release ETO from the nanocarrier for tumor cells (pH = 5.4), as shown in Fig. 11a. It can also effectively inhibit the growth of FA-positive HeLa cells, which can be applied to therapeutic tools. Duan et al.131 assembled DOX into the low-toxicity CDs to form fluorescent therapeutic drug delivery systems, as shown in Fig. 11b. The loading efficiency of DOX could reach 75.3% and the amount of released DOX at pH 5.0 was four times greater than that of pH 7.4. The CDs loading DOX can be taken in by human gastric cancer cells, and the tracking the drug delivery over 48 h is shown in Fig. 11c. Wen et al.132 conducted studies on novel nanohybrid-based CDs with magnetic properties. The nanohybrid showed low toxicity and can be more readily taken up by in vitro magnetic attraction. Due to the enormous surface of nanohybrids, the Pt-based anticancer drug was carried by the CDs, which can be effectively transported into the tumor and absorbed through enhancement by magnetism, resulting in the improved anticancer efficacy. Zhao et al.133 prepared yellow fluorescent nanohybrids (MSNs-CDs) whose raw materials were folic acid (FA) and amino-modified mesoporous silica nanoparticles (MSNs-NH2) by hydrothermal method, as displayed in Fig. 11d. It can be used as the nanocarrier to target the delivery of DOX to the tumor and release the drug in the tumor cell, enhancing the medical effects and decreasing the side effects.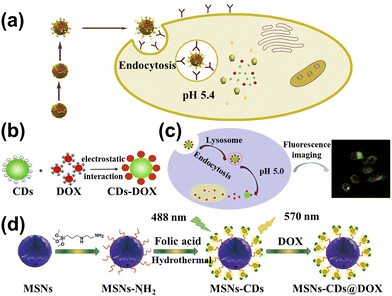 | ||
| Fig. 11 (a) Schematic diagram of the FA-Cβ-MSCDs getting through the cell and releasing ETO.130 Reproduced from ref. 82 with permission from Elsevier, copyright 2018. (b) The principle of CDs combined with DOX to form CDs-DOX.131 (c) The cell uptake of CDs-DOX and application.131 Reproduced from ref. 83 with permission from Elsevier, copyright 2019. (d) The illustration of the MSNs-CDs@DOX preparation and cancer cell targeted-delivery.133 Reproduced from ref. 85 with permission from Springer Nature, copyright 2019. | ||
5.2 Photothermal therapy
Photothermal therapy (PTT) is a treatment method whose materials with high photothermal conversion efficiency convert light energy into heat energy to kill cancer cells under the irradiation of an external light source.134 Generally, the PTT has better operation in the 700–950 nm range belonging in the near-infrared (NIR) region. However, most kinds of CDs have strong absorption in the ultraviolet region due to the π–π* transition.135,136 Doping of heteroatoms, metals, nanoparticles, etc. can effectively improve the optical properties of CDs. In this section, we review the typical heteroatoms, metals, and nanoparticles combined with CDs to act as PTT agents.Permatasari et al.137 synthesized the blue-yellow emission CDs using urea and citric acid as precursors through the microwave-assisted hydrothermal treatment, as displayed in Fig. 12a. The study showed that pyrrolic-N-rich is key for CDs to enhance the near NIR absorption, whose photothermal efficiency was up to 54.2% (Fig. 12b). Kim et al.138 attained sulfur-doped CDs (S-CDs), accessing the Camellia japonica flowers as principle materials acting as the cancer therapeutic agents. The S-CDs had high NIR absorption, whose photothermal conversion efficiency is up to 55.4% under the 808 nm irradiation. The tumor size in mice with S-CD gradually became smaller and formed a black scar under the NIR laser irradiation.
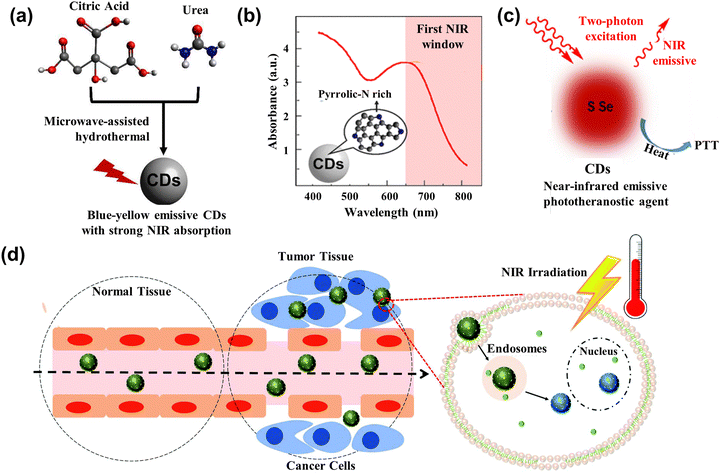 | ||
| Fig. 12 (a) Scheme illustrating the CDs with strong NIR absorption.137 (b) The absorption spectrum of CDs with rich Pyrrolic-N.137 Reproduced from ref. 89 with permission from American Chemical Society, copyright 2018. (c) Illustration of the working mechanism for the two-photon excitation S-Se-CDs.139 Reproduced from ref. 91 with permission from Springer Nature, copyright 2017. (d) Illustration of CD@MSN/ICG targeting the tumor and killing cells based on the PTT.141 Reproduced from ref. 93 with permission from the Royal Society of Chemistry, copyright 2019. | ||
To further enhance the performance of CDs, researchers have also started to dope various metals and their derivatives. Lan et al.139 prepared the S, Se co-doped CDs (S-Se-CDs) through hydrothermal treatment, while polythiophene and diphenyl diselenide acted as precursors in the alkaline solution. The co-doped CDs had two NIR emissions at the peak of 731 nm and 820 nm, and the efficiency of photothermal conversion was 58.2%, as shown in Fig. 12c. Nanoparticles acting as carriers or agents combined with the CDs also can enhance the photothermal property and improve anti-cancer effectiveness. Qian and collaborators140 leveraged the hydrogen bond to assemble the CDs into the framework of mesoporous silica nanoparticles (MSNs), getting the mixture CD@MSNs. The CD@MSNs can degrade in the cell and aggregate the dispersed CDs to enhance the photothermal efficacy, realizing the killing of cancer cells and achieving inhibition of tumor metastasis. Benny Ryplida et al.141 loaded the CDs into MSNs with pH-responsive Indocyanine Green (ICG) through electrostatic interactions. The CD@MSN/ICG released the CDs under the acid pH, which absorbed the excitation and converted it into heat to kill the cancer cells, as displayed in Fig. 12d. Peng and co-works142 utilized the one-step microwave-assisted carbonation method, depositing CDs on Prussian blue nanoparticles (CDs/PBNP). The PBNPs had a photothermal property and the CDs exhibited strong green emission, so the CDs/PBNP can act as an imaging agent and PTT agent.
5.3 Photodynamic therapy
Photodynamic therapy (PDT) is a new area of tumor therapy for malignant tumors and multiple skin diseases.143,144 The principle of PDT is based on photosensitizers (PS) producing the reactive oxygen species (ROS) under light excitation to induce cancer cell apoptosis.145,146 In this section, we review the CDs acting as agents and carriers in the PDT, respectively.CDs are used as the PS and image agents in the PDT field, owing to their non-toxicity and excellent optical properties. Zhao and co-workers doped N and S into CDs, attaining N, S-CDs with red emission.147 These CDs accumulated in the tumor lysosome and mitochondria, and produced singlet oxygen (1O2) to induce cell death under light irradiation. Wang et al.148 synthesized the Cu-doped CDs (Cu-CDs) via the pyrolysis method, which can be used in the imaging-guided PDT for tumor treatment, as shown in Fig. 13a. It showed the high QY of 1O2 (36%) and possessed photoinduced toxicity, which can induce cell apoptosis. To enhance the treatment performance for cancer, the CDs serving as PS targeting the nucleic acid were developed. Xu et al.149 composed the Se and N co-doped CDs (Se/N-CDs), which bonded with RNA and were transported near the nucleus to generate the ROS shown in Fig. 13b. Pang et al.150 designed the novel CDs targeting the nucleolus and producing ROS, which improved the performance under low concentrations of CDs.
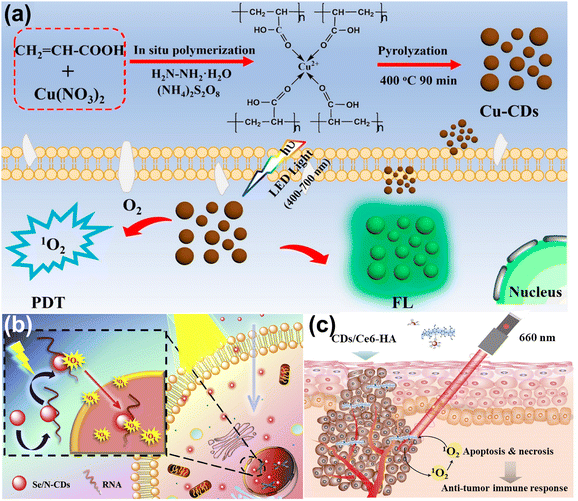 | ||
| Fig. 13 (a) The illustration of Cu-CDs synthesis and imaging-guided PDT application.148 Reproduced from ref. 100 with permission from American Chemical Society, copyright 2019. (b) Schematic of light-induced entry into the nucleus.149 Reproduced from ref. 101 with permission from Elsevier, copyright 2020. (d) The illustration of PDT for CDs/Ce6-HA conjugate.152 Reproduced from ref. 102 with permission from Elsevier, copyright 2015. | ||
CDs act as the carriers, enhancing solubility and biocompatibility, resulting in improving the effect of photodynamics. Yang et al. loaded the insoluble chlorin e6 (Ce6, PS) into CDs to attain the CDs/Ce6, which had good solubility, biocompatibility, and high fluorescence resonance energy transfer (FRET) efficiency.151 The system enhanced the photodynamic process whose 1O2 QY increased by 200% compared with pristine Ce6. Beack et al.152 formed CDs/Ce6-HA conjugate by the dehydration condensation of the amino group of diaminohexane-modified hyaluronate (HA-DHA) and the carboxyl group of CDs/Ce6. The conjugate could target the tumor cell and generate more 1O2 than Ce6 and CDs/Ce6 related to the enhanced solubility (Fig. 13c).
5.4 ML assists in the medical treatment
ML plays an important domain role in the felid of the tumor. In this section, we review the application of ML on tumor discovery, drug design, and drug response prediction.Xie et al.153 utilized the fast correlation-based Filter (FCBF) algorithm to find five important biomarkers to detect early lung cancer in the plasma data. Ail and co-works used an artificial intelligence diagnostic platform to recognize cancer malignancy, invasiveness, and grading on the blue light cystoscopy image.154 The ML models showed a high classification on sensitivity and specificity, up 95.77% and 87.44%, respectively.
This advanced technology exhibited the potential for cancer diagnosis and improved cancer detection rates, and was a beneficial treatment decision for doctors. ML helps discover the relationship between the drug and tumor issue, which is beneficial to the drug design to shorten the drug development cycle. Grisoni et al.155 utilized long short-term memory (LSTM) cells to design the anticancer peptides (ACP), reaching the selective killing of the cancer cell. Half of the ACPs can kill cancer cells without influencing the human erythrocytes whose sensitivity was over three times that of the MCF7 cell. Furthermore, the ensemble model is proposed to design effective ACPs.156 Four counter-propagation artificial neural networks (CPANN) were combined into one ensembled model, where one half belonged to the lung cancer model, and the remaining half belonged to the breast cancer model, as shown in Fig. 14a. The ensembled model de novo designed 14 peptides for testing the MCF7 cell and A549 cell (lung cell). The results showed the six peptides had anticancer ability, five of which were effective against both types of cancer cells.
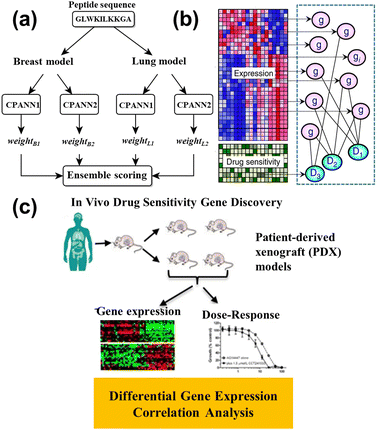 | ||
| Fig. 14 (a) Schematic representation of the ensemble prediction model.156 Reproduced from ref. 108 with permission from Springer Nature, copyright 2019. (b) The illustration of the model for identifying the relationship between gene expression and drug sensitivity.157 Reproduced from ref. 109 with permission from Springer Nature, copyright 2018. (c) Diagram of the correlation analysis and differential expression analysis on patient-derived xenograft (PDX) tumors.158 Reproduced from ref. 110 with permission from Springer Nature, copyright 2020. | ||
Nowadays, some ML models are applied to drug response prediction on patients due to individual variability. Lee et al.157 investigated the relationship between gene expression and the drug's sensitivity to acute myeloid leukemia (AML), as shown in Fig. 14b. They collected genome-wide gene expression profiles and chemotherapy drug sensitivity, and created novel methods for studying the prior anticancer knowledge to identify robust gene expression biomarkers. Kim and partners conducted the correlation analysis between the gene expression and dose–response on the patient-derived xenografts (PDXs) models (Fig. 14c), and built the drug response model via random forest.158 The model predicted the biomarkers for different types of cancers, such as breast cancer, pancreatic cancer, colorectal cancer, or non-small cell lung cancer, for application to drug therapy. Kong et al.159 utilized ridge regression to identify the biomarkers whose data came from organoid culture models. The drug responses were accurately predicted through the identified biomarkers, while the colorectal cancer patient was cured with 5-fluorouracil and the bladder cancer patient was treated with cisplatin, whose numbers were 114 and 77, respectively. The consistency between the experimental and control groups verifies the predicted result. This work combined gene data and ML to efficiently accept drug responses in cancer patients. To explain the drug mechanism in the ML model, Deng et al.160 constructed the deep artificial network (DNN) by introducing a layer of path nodes as a hidden layer that is more interpretable. The performance of DNN was evaluated on the different independent drug sensitivity data sets, and the results showed the DNN model had obvious advantages compared with the extra eight standard regression models. More importantly, they found the activity of disease-related nodes decreased, while the drug input forward propagation, revealing the suppression of the disease path by treating cancer with drugs.
Using a single anticancer drug was uncommon because of how easily and quickly it is for tumor cells to develop drug resistance, so multiple drugs were adopted in the realistic therapeutic schedule. The combination of multiple drugs can produce three influences: additive, antagonistic, and synergistic. The drug synergy is most optimized in these conditions, which can greatly enhance anticancer effect. Predicting drug synergy is important for the application of multiple drugs. Liu and co-workers developed a TranSynergy model based on the knowledge-enable and self-attention transformer.161 In, addition, the Shapley Additive Gene Set Enrichment Analysis (SA-GSEA) method was proposed and applied to deconvolute genes, which improved the interpretability of the model. The combination of TranSynergy and SA-GSEA provided new insight into the discovery of anticancer therapy. Ail et al.162 utilized the silico biological network to recognize the synergistic anticancer drug pair. Based on the drug-perturbed transcriptome profiles and biological network analysis, they proposed six relative network biology features and trained the model on the public drug synergy dataset. The model was capable of discerning whether there was synergy between two drugs, and explaining the situation in terms of the molecular network that passed through them. Regan-Fendt et al.163 designed a new computational approach from gene expression and network, mining to disease analysis and drug combination prediction. The new method considered connectivity mapping and network centrality for transcriptomics. The effect of prediction was tested on the public gene expression data and mutation data, and the drug combination results were confirmed by the high throughput experimental.
5.5 In brief
More and more CDs are being applied to cancer treatment, whose effect is obviously being studied on mice. However, there are some challenges to overcome for clinal use. Therefore, the CDs can further be modified to realize batch synthesis with size control and new properties combining multi-functions to enhance the anticancer effect. ML has offered great convenience for cancer treatment on cancer discovery, drug design, and drug response prediction. The image of cancer tissue is from different departments, and the ML can integrate the information to detect the potential tumor position. With the data increasing on the cancer gene, the ML with strong computational ability is firstly selected to handle this complex data, which shortens the time and cost of new drug discovery. Moreover, multiple drugs simultaneously are used in the clinal treatment whose effect is predictable through the ML, assisting the doctor's decision-making. In the future, ML will continue to play a significant role in the anticancer process and enhance the understanding of cancer. All related ML models are summarized in Table 4.| Model | Result | Ref. |
|---|---|---|
| FCBF | Detect early lung cancer in the plasma data | 153 |
| CNNs | Recognized the tumor with 95.77% sensitivity and 87.44% specificity | 154 |
| LSTM | Design the anticancer peptides and reach the selective killing of the cancer cell. | 155 |
| CPANN | De novo designed 14 peptides for the MCF7 cell and A549 cell | 156 |
| RF | Find the relationship between gene expression and dose–response on the PDXs; predict the biomarkers for different types | 158 |
| Ridge regression | Predict the different drug responses identified biomarkers while the colorectal cancer patient was cured | 159 |
| DNN | Reveal the suppression of the disease path by treating cancer with drugs | 160 |
| TranSynergy | Predict the drug synergy and combine with SA-GSEA to discover anticancer therapy | 161 |
| Silico biological network | Recognize the synergistic anticancer drug pair. | 162 |
| SynGeNet | Analyze disease and predict the drug synergy | 163 |
6 Conclusion and perspective
This review mainly introduces how MLs have facilitated CDs’ application in the biological field. Firstly, owing to their excellent photoluminescence, chemical stability, and low cytotoxicity, the CDs are regarded as a bioimaging agent, biosensor, carrier, and drug to the server in the cell and organism. Then, the ML is introduced into the field to enhance the properties of CDs or change the traditional data analysis method, improving the accuracy and precision of the results. Moreover, the summary in every section shows the excellent potential for incorporating ML.However, two significant problems may impede the development of ML in the biological field. At first, the data from prepared CDs takes a lot of work to gather. Establishing a database is so urgent for the laboratory, which can effectively solve the missing conditions. The database forms unified management and control of data, and dramatically improves data integrity and security. The balance of data is a crucial issue, which directly influences the model performance. Research tends to be biased toward obtaining successful results and previously failed experiments cannot be recorded. So, we should pay more attention to the proportion of data. In addition, current research focuses on changes in the experimental conditions and keeping the recipe the same for synthesizing CDs, which is limited in terms of tuning the material performance. In the future, different precursors and various solvents can be used as experimental conditions to predict the relevant properties by ML. This process requires familiarity with the chemical structure and physical properties of the raw materials, utilizing a set of descriptors or features to represent the material in the dataset.
The automation of acquisition pipelines and the new microscopy technologies have broken the limits of temporal and spatial resolution, dramatically increasing the amount of bioimage data. The major challenge is to interpret these image data sets in a quantitative, automatic, and efficient way. Computer vision and image analysis are applied to microscopic images to extract biological information and generate databases, which expedites the development of bioimaging. For supervised machine learning algorithms, the image itself needs to be labeled, which requires manual operation, and is time-consuming and laborious. In return, the labeled data determine the performance of the model, so special attention should be paid to data selection and labeling.
Biosensors inevitably have some irregular signal noise, which leads to poor stability of biosensors and limits their commercialization. Detecting the corresponding analyte concentration is essential, which is based on analyzing the sensing data. ML can provide a new method of data analysis, perform a series of noise processing on the data, and automatically predict the type or concentration of analytes according to the decision system. In addition, biosensors are easily affected by the sample environment and operating conditions, which greatly disturb the results. ML can detect anomalies and exclude outliers by rule. At present, the combination of biosensors and machine learning for health monitoring is a worthy challenge. Biosensors can continuously detect the corresponding indicators and provide sequential data, while ML analyzes data and evaluates biological health using new algorithms such as RNN, LSTM, etc.
ML has been extensively studied in tumor therapy to diagnose tumors and predict a patient's condition, obtaining excellent success. However, some limitations and obstacles must be overcome before it can be widely used in the clinic. As the amount of CT and MR imaging continues to grow, the management of medical data is a major barrier. Collating the imaging data needs of trained professionals in terms of labeling, annotation, segmentation, quality control, or application, makes the process expensive in both time and cost. It is important to develop automated imaging process software, and the use of data is also restricted due to permission and privacy concerns, so the wide clinical application of ML is hard. ML is a black box model, which cannot be explained to a certain extent, receiving certain limitations for their application. The current development of data visualization can help understand the principle of machine learning to a certain extent, accelerating the integration between ML and cancer therapy.
Conflicts of interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.Acknowledgements
Juncheng Wang acknowledges the support from Beijing Natural Science Foundation (L222109) and Military Health Care Project (22BJZ22). Quan Xu acknowledges the support from the National Natural Science Foundation of China (No. 52211530034) and the Beijing National Science Foundation (No. 3222018).References
- A. Handa, A. Sharma and S. K. Shukla, WIREs Data Mining Knowledge Discovery, 2019, 9, e1306 CrossRef.
- J. Wang, S. Lu, S.-H. Wang and Y.-D. Zhang, Multimedia Tools Appl., 2022, 81, 41611–41660 CrossRef.
- S. K. Selvaraj, A. Raj, R. Rishikesh Mahadevan, U. Chadha and V. Paramasivam, Adv. Mater. Sci. Eng., 2022, 2022, 1949061 Search PubMed.
- C. C. Gray and D. Perkins, Comput. Educ., 2019, 131, 22–32 CrossRef.
- M. P. Than, J. W. Pickering and Y. Sandoval, Circulation, 2019, 140, 899–909 CrossRef PubMed.
- M. I. Jordan and T. M. Mitchell, Science, 2015, 349, 255–260 CrossRef CAS PubMed.
- J. Wäldchen and P. Mäder, Methods Ecol. Evol., 2018, 9, 2216–2225 CrossRef.
- S. Berg, D. Kutra, T. Kroeger, C. N. Straehle, B. X. Kausler, C. Haubold, M. Schiegg, J. Ales, T. Beier, M. Rudy, K. Eren, J. I. Cervantes, B. Xu, F. Beuttenmueller, A. Wolny, C. Zhang, U. Koethe, F. A. Hamprecht and A. Kreshuk, Nat. Methods, 2019, 16, 1226–1232 CrossRef CAS PubMed.
- P. Doupe, J. Faghmous and S. Basu, Value Health, 2019, 22, 808–815 CrossRef PubMed.
- M. J. Willatt, F. Musil and M. Ceriotti, Phys. Chem. Chem. Phys., 2018, 20, 29661–29668 RSC.
- B. P. Yadav, S. Ghate, A. Harshavardhan, G. Jhansi, K. S. Kumar and E. Sudarshan, IOP Conf. Ser.: Mater. Sci. Eng., 2020, 981, 022044 Search PubMed.
- K. Christensen, S. Nørskov, L. Frederiksen and J. Scholderer, Creativity Innovation Manage., 2017, 26, 17–30 CrossRef.
- S. M. Moosavi, A. Chidambaram, L. Talirz, M. Haranczyk, K. C. Stylianou and B. Smit, Nat. Commun., 2019, 10, 539 CrossRef CAS PubMed.
- E. Kim, K. Huang, A. Saunders, A. McCallum, G. Ceder and E. Olivetti, Chem. Mater., 2017, 29, 9436–9444 CrossRef CAS.
- M. Xu, B. Tang, Y. Lu, C. Zhu, Q. Lu, C. Zhu, L. Zheng, J. Zhang, N. Han, W. Fang, Y. Guo, J. Di, P. Song, Y. He, L. Kang, Z. Zhang, W. Zhao, C. Guan, X. Wang and Z. Liu, J. Am. Chem. Soc., 2021, 143, 18103–18113 CrossRef CAS PubMed.
- F. Arcudi, L. Đorđević and M. Prato, Acc. Chem. Res., 2019, 52, 2070–2079 CrossRef CAS PubMed.
- Z. Chen, Y. Liu and Z. Kang, Acc. Chem. Res., 2022, 55, 3110–3124 CrossRef CAS PubMed.
- S. Li, F. Li, Y. Dong, N. Song, L. Pan and D. Yang, Small, 2022, 18, 2106269 CrossRef CAS PubMed.
- Z. Li, L. Wang, Y. Li, Y. Feng and W. Feng, Mater. Chem. Front., 2019, 3, 2571–2601 RSC.
- J. Manioudakis, F. Victoria, C. A. Thompson, L. Brown, M. Movsum, R. Lucifero and R. Naccache, J. Mater. Chem. C, 2019, 7, 853–862 RSC.
- C.-L. Shen, Q. Lou, K.-K. Liu, L. Dong and C.-X. Shan, Nano Today, 2020, 35, 100954 CrossRef CAS.
- X. Song, S. Zhao, Y. Xu, X. Chen, S. Wang, P. Zhao, Y. Pu and A. J. Ragauskas, ChemSusChem, 2022, 15, e202102486 CrossRef CAS PubMed.
- B. Bartolomei, J. Dosso and M. Prato, Trends Chem., 2021, 3, 943–953 CrossRef CAS.
- K. K. Chan, S. H. K. Yap and K.-T. Yong, Nano-Micro Lett., 2018, 10, 72 CrossRef CAS PubMed.
- T. V. de Medeiros, J. Manioudakis, F. Noun, J.-R. Macairan, F. Victoria and R. Naccache, J. Mater. Chem. C, 2019, 7, 7175–7195 RSC.
- J. Guo, H. Li, L. Ling, G. Li, R. Cheng, X. Lu, A.-Q. Xie, Q. Li, C.-F. Wang and S. Chen, ACS Sustainable Chem. Eng., 2020, 8, 1566–1572 CrossRef CAS.
- C. Xia, S. Zhu, T. Feng, M. Yang and B. Yang, Adv. Sci., 2019, 6, 1901316 CrossRef CAS PubMed.
- J. Zhang and S.-H. Yu, Mater. Today, 2016, 19, 382–393 CrossRef CAS.
- Z. Kang and S.-T. Lee, Nanoscale, 2019, 11, 19214–19224 RSC.
- C. Rosso, G. Filippini and M. Prato, ACS Catal., 2020, 10, 8090–8105 CrossRef CAS.
- H. Ali, S. Ghosh and N. R. Jana, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2020, 12, e1617 RSC.
- P. Yang, Z. Zhu, T. Zhang, W. Zhang, W. Chen, Y. Cao, M. Chen and X. Zhou, Small, 2019, 15, 1902823 CrossRef CAS.
- Q. Fan, J. Li, Y. Zhu, Z. Yang, T. Shen, Y. Guo, L. Wang, T. Mei, J. Wang and X. Wang, ACS Appl. Mater. Interfaces, 2020, 12, 4797–4803 CrossRef CAS PubMed.
- N. K. Sahoo, G. C. Jana, M. N. Aktara, S. Das, S. Nayim, A. Patra, P. Bhattacharjee, K. Bhadra and M. Hossain, Mater. Sci. Eng., C, 2020, 108, 110429 CrossRef CAS PubMed.
- L. Bu, J. Peng, H. Peng, S. Liu, H. Xiao, D. Liu, Z. Pan, Y. Chen, F. Chen and Y. He, RSC Adv., 2016, 6, 95469–95475 RSC.
- Y. Liu, X. Gong, W. Dong, R. Zhou, S. Shuang and C. Dong, Talanta, 2018, 183, 61–69 CrossRef CAS PubMed.
- J. Hou, J. Yan, Q. Zhao, Y. Li, H. Ding and L. Ding, Nanoscale, 2013, 5, 9558–9561 RSC.
- P. Zhu, X. Zhao, Y. Zhang, Y. Liu, Z. Zhao, Z. Yang, X. Liu, W. Zhang, Z. Guo, X. Wang, Y. Niu and M. Xu, Front. Bioeng. Biotechnol., 2022, 10, 964814 CrossRef PubMed.
- N. Gao, W. Yang, H. Nie, Y. Gong, J. Jing, L. Gao and X. Zhang, Biosens. Bioelectron., 2017, 96, 300–307 CrossRef CAS PubMed.
- Q. Jia, Z. Zhao, K. Liang, F. Nan, Y. Li, J. Wang, J. Ge and P. Wang, Mater. Chem. Front., 2020, 4, 449–471 RSC.
- L.-j Yue, Y.-y Wei, J.-b Fan, L. Chen, Q. Li, J.-l Du, S.-p Yu and Y.-z Yang, Carbon, 2021, 179, 702 CrossRef.
- B. Tang, Y. Lu, J. Zhou, T. Chouhan, H. Wang, P. Golani, M. Xu, Q. Xu, C. Guan and Z. Liu, Mater. Today, 2020, 41, 72–80 CrossRef CAS.
- J. McCarthy, 2007.
- F. Y. Wang, J. J. Zhang, X. Zheng, X. Wang, Y. Yuan, X. Dai, J. Zhang and L. Yang, IEEE/CAA J. Automatica Sinica, 2016, 3, 113–120 Search PubMed.
- A. Pannu, Artif. Intell., 2015, 4, 79–84 Search PubMed.
- S. Semmler and Z. Rose, Duke L. & Tech. Rev, 2017, 16, 85 Search PubMed.
- Z. Pei, J. Yin, P. K. Liaw and D. Raabe, Nat. Commun., 2023, 14, 54 CrossRef CAS PubMed.
- V. Tshitoyan, J. Dagdelen, L. Weston, A. Dunn, Z. Rong, O. Kononova, K. A. Persson, G. Ceder and A. Jain, Nature, 2019, 571, 95–98 CrossRef CAS PubMed.
- V. Ficcadenti, R. Cerqueti and M. Ausloos, Exp. Syst. Appl., 2019, 123, 127–142 CrossRef.
- J. Currie, H. Kleven and E. Zwiers, AEA Papers Proc., 2020, 110, 42–48 CrossRef.
- I. H. Sarker, Y. B. Abushark, F. Alsolami and A. I. Khan, Journal, 2020, 12, 754 Search PubMed.
- I. H. Sarker, H. Alqahtani, F. Alsolami, A. I. Khan, Y. B. Abushark and M. K. Siddiqui, J. Big Data, 2020, 7, 51 CrossRef.
- I. H. Sarker, Y. B. Abushark and A. I. Khan, Journal, 2020, 12, 754 Search PubMed.
- D. C. Montgomery, E. A. Peck and G. G. Vining, Introduction to linear regression analysis, John Wiley & Sons, 2021 Search PubMed.
- E. Ostertagová, Proc. Eng., 2012, 48, 500–506 CrossRef.
- D. W. Hosmer Jr, S. Lemeshow and R. X. Sturdivant, Applied logistic regression, John Wiley & Sons, 2013 Search PubMed.
- L. E. Peterson, Scholarpedia, 2009, 4, 1883 CrossRef.
- S. B. Kotsiantis, Artif. Intelligence Rev., 2013, 39, 261–283 CrossRef.
- L. Rokach and O. Maimon, Data Mining Knowledge Discovery Handbook, 2005, 165–192 Search PubMed.
- I. Steinwart and A. Christmann, Support vector machines, Springer Science & Business Media, 2008 Search PubMed.
- M. A. Hearst, S. T. Dumais, E. Osuna, J. Platt and B. Scholkopf, IEEE Intell. Syst. Appl., 1998, 13, 18–28 Search PubMed.
- C. D. Sutton, in Handbook of Statistics, ed. C. R. Rao, E. J. Wegman and J. L. Solka, Elsevier, 2005, 24, pp. 303–329 Search PubMed.
- D. Opitz and R. Maclin, J. Artif. Intell. Res., 1999, 11, 169–198 CrossRef.
- L. Breiman, Mach. Learn., 2001, 45, 5–32 CrossRef.
- H. Drucker, C. Cortes, L. D. Jackel, Y. LeCun and V. Vapnik, Neural Comput., 1994, 6, 1289–1301 CrossRef.
- Y. Freund and R. E. Schapire, 1996.
- W. Liang, S. Luo, G. Zhao and H. Wu, Mathematics, 2020, 8, 765 CrossRef.
- K. Song, F. Yan, T. Ding, L. Gao and S. Lu, Comput. Mater. Sci., 2020, 174, 109472 CrossRef CAS.
- Y. Ju, G. Sun, Q. Chen, M. Zhang, H. Zhu and M. U. Rehman, IEEE Access, 2019, 7, 28309–28318 Search PubMed.
- J. MacQueen, 1967.
- H. Hotelling, J. Educ. Psychol., 1933, 24, 417 Search PubMed.
- R. Agrawal, T. Imieliński and A. Swami, presented in part at the Proceedings of the 1993 ACM SIGMOD international conference on Management of data, Washington, D.C., USA, 1993.
- D. H. Wolpert and W. G. Macready, IEEE Trans. Evolutionary Comput., 1997, 1, 67–82 CrossRef.
- H. Li, X. Yan, D. Kong, R. Jin, C. Sun, D. Du, Y. Lin and G. Lu, Nanoscale Horiz., 2020, 5, 218–234 RSC.
- D. Gao, H. Zhao, X. Chen and H. Fan, Mater. Today Chem., 2018, 9, 103–113 CrossRef CAS.
- D. Li, E. V. Ushakova, A. L. Rogach and S. Qu, Small, 2021, 17, 2102325 CrossRef CAS PubMed.
- K. Jiang, S. Sun, L. Zhang, Y. Lu, A. Wu, C. Cai and H. Lin, Angew. Chem., Int. Ed., 2015, 54, 5360–5363 CrossRef CAS PubMed.
- J. Liu, D. Li, K. Zhang, M. Yang, H. Sun and B. Yang, Small, 2018, 14, 1703919 CrossRef PubMed.
- L. Wang, B. Wang, E. Liu, Y. Zhao, B. He, C. Wang, G. Xing, Z. Tang, Y. Zhou and S. Qu, Chin. Chem. Lett., 2022, 33, 4111–4115 CrossRef CAS.
- H. Liu, Y. Zhang and C. Huang, J. Pharm. Anal., 2019, 9, 127–132 CrossRef PubMed.
- D. Bouzas-Ramos, J. Cigales Canga, J. C. Mayo, R. M. Sainz, J. Ruiz Encinar and J. M. Costa-Fernandez, Adv. Funct. Mater., 2019, 29, 1903884 CrossRef.
- C. Yan, C. Wang, T. Hou, P. Guan, Y. Qiao, L. Guo, Y. Teng, X. Hu and H. Wu, ACS Appl. Mater. Interfaces, 2021, 13, 1277–1287 CrossRef CAS PubMed.
- X. Zhang, H. Guo, C. Chen, B. Quan, Z. Zeng, J. Xu, Z. Chen and L. Wang, Appl. Mater. Today, 2023, 30, 101706 CrossRef.
- B. Bhushan, S. U. Kumar and P. Gopinath, J. Mater. Chem. B, 2016, 4, 4862–4871 RSC.
- R. L. Meyers, Surgical Oncology, 2007, 16, 195–203 CrossRef PubMed.
- J. Liu, D. Li, K. Zhang, M. Yang, H. Sun and B. Yang, Small, 2018, 14, 1703919 CrossRef PubMed.
- J. B. Luo, J. Chen, H. Liu, C. Z. Huang and J. Zhou, Chem. Commun., 2022, 58, 9014–9017 RSC.
- Q. Hong, X.-Y. Wang, Y.-T. Gao, J. Lv, B.-B. Chen, D.-W. Li and R.-C. Qian, Chem. Mater., 2022, 34, 998–1009 CrossRef CAS.
- W. Meiniel, J. C. Olivo-Marin and E. D. Angelini, IEEE Trans. Image Process., 2018, 27, 3842–3856 Search PubMed.
- R. F. Laine, G. Jacquemet and A. Krull, Int. J. Biochem. Cell Biol., 2021, 140, 106077 CrossRef CAS PubMed.
- W. Yina, P. Henry, K. Emaad, Z. Shuqin, W. Laura and H. Bo, BioRxiv, 2021, 02, 429188 Search PubMed.
- M. Weigert, U. Schmidt, T. Boothe, A. Müller, A. Dibrov, A. Jain, B. Wilhelm, D. Schmidt, C. Broaddus, S. Culley, M. Rocha-Martins, F. Segovia-Miranda, C. Norden, R. Henriques, M. Zerial, M. Solimena, J. Rink, P. Tomancak, L. Royer, F. Jug and E. W. Myers, Nat. Methods, 2018, 15, 1090–1097 CrossRef CAS PubMed.
- H. Chai-Wei, N. Sumesh, L. Chun-Yu and C. Shean-Jen, Optical Methods for Inspection, Characterization, and Imaging of Biomaterials, 2023, vol. 12622, pp. 80–82.
- U. Schmidt, M. Weigert, C. Broaddus and G. Myers, Cham, 2018, 265–273 Search PubMed.
- M. Weigert, U. Schmidt, R. Haase, K. Sugawara and G. Myers, IEEE/CVF winter conference on applications of computer vision, 2020, pp. 3666–3673.
- F. D. Khameneh, S. Razavi and M. Kamasak, Comput. Biol. Med., 2019, 110, 164–174 CrossRef CAS PubMed.
- D. Eschweiler, T. V. Spina, R. C. Choudhury, E. Meyerowitz, A. Cunha and J. Stegmaier, 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), 2019, pp. 223–227.
- C. Stringer, T. Wang, M. Michaelos and M. Pachitariu, Nat. Methods, 2021, 18, 100–106 CrossRef CAS PubMed.
- M. Pachitariu and C. Stringer, Nat. Methods, 2022, 19, 1634–1641 CrossRef CAS PubMed.
- K. Lee, H. Byun and H. Shim, presented in part at the Proceedings of The Cell Segmentation Challenge in Multi-modality High-Resolution Microscopy Images, Proceedings of Machine Learning Research, 2023, 1-11.
- D. Eschweiler, R. S. Smith and J. Stegmaier, 2022 IEEE International Conference on Image Processing (ICIP), 2022, pp. 191–195.
- K. J. Cutler, C. Stringer, T. W. Lo, L. Rappez, N. Stroustrup, S. Brook Peterson, P. A. Wiggins and J. D. Mougous, Nat. Methods, 2022, 19, 1438–1448 CrossRef CAS PubMed.
- A. Hallou, H. G. Yevick, B. Dumitrascu and V. Uhlmann, Development, 2021, 148, dev199616 CrossRef CAS PubMed.
- C. Lu, Q. Su and X. Yang, Nanoscale, 2019, 11, 16036–16042 RSC.
- H. Zhao, X. Yuan, X. Yang, F. Bai, C. Mao and L. Zhao, Inorg. Chem., 2021, 60, 15485–15496 CrossRef CAS PubMed.
- J. Huang, Y. He, Z. Zhang, B. Lei and W. Wu, J. Lumin., 2019, 215, 116640 CrossRef CAS.
- X. Wang, Y. Wang, W. Pan, J. Wang and X. Sun, ACS Sustainable Chem. Eng., 2021, 9, 3718–3726 CrossRef CAS.
- S. Durrani, Z. Yang, J. Zhang, Z. Wang, H. Wang, F. Durrani, F.-G. Wu and F. Lin, Talanta, 2023, 252, 123855 CrossRef CAS PubMed.
- N. Nandi, K. Choudhury, P. Sarkar, N. Barnwal and K. Sahu, ACS Appl. Nano Mater., 2022, 5, 17315–17324 CrossRef CAS.
- P. Lesani, S. M. Ardekani, A. Dehghani, M. Hassan and V. G. Gomes, Sens. Actuators, B, 2019, 285, 145–155 CrossRef CAS.
- Y. Li, L. Tang, C. Zhu, X. Liu, X. Wang and Y. Liu, Microchim. Acta, 2021, 189, 10 CrossRef PubMed.
- L. C. Zhang, Y. M. Yang, L. Liang, Y. J. Jiang, C. M. Li, Y. F. Li, L. Zhan, H. Y. Zou and C. Z. Huang, Analyst, 2022, 147, 417–422 RSC.
- J. Yue, L. Yu, L. Li, P. Liu, Q. Mei, W.-F. Dong and R. Yang, Front. Chem., 2021, 9, 718856 CrossRef CAS PubMed.
- E. L. Rossini, M. I. Milani, L. S. Lima and H. R. Pezza, Spectrochim. Acta, Part A, 2021, 248, 119285 CrossRef CAS PubMed.
- Y. Z. Yang, N. Xiao, S. G. Liu, L. Han, N. B. Li and H. Q. Luo, Mater. Sci. Eng., C, 2020, 108, 110401 CrossRef CAS PubMed.
- J. Zhou, R. Wang, W. Su, L. Zhang, A. Li and T. Jiao, Colloids Surf., A, 2022, 647, 129122 CrossRef CAS.
- Z. Lin, Q. Zeng, Q. Deng, W. Yao, H. Deng, X. Lin and W. Chen, Sens. Actuators, B, 2022, 359, 131563 CrossRef CAS.
- Y. Han, B. Tang, L. Wang, H. Bao, Y. Lu, C. Guan, L. Zhang, M. Le, Z. Liu and M. Wu, ACS Nano, 2020, 14, 14761–14768 CrossRef CAS PubMed.
- Q. Xu, Y. Tang, P. Zhu, W. Zhang, Y. Zhang, O. S. Solis, T. S. Hu and J. Wang, Nanoscale, 2022, 14, 13771–13778 RSC.
- S. Pandit, T. Banerjee, I. Srivastava, S. Nie and D. Pan, ACS Sens., 2019, 4, 2730–2737 CrossRef CAS PubMed.
- N. Shauloff, A. Morag, K. Yaniv, S. Singh, R. Malishev, O. Paz-Tal, L. Rokach and R. Jelinek, Nano-Micro Lett., 2021, 13, 112 CrossRef CAS PubMed.
- Z. Xu, J. Chen, Y. Liu, X. Wang and Q. Shi, Chem. Eng. J., 2022, 441, 135690 CrossRef CAS.
- T. Liu, S. Chen, K. Ruan, S. Zhang, K. He, J. Li, M. Chen, J. Yin, M. Sun, X. Wang, Y. Wang, Z. Lu and H. Rao, J. Hazard. Mater., 2022, 426, 128091 CrossRef CAS PubMed.
- Z. Lu, S. Chen, M. Chen, H. Ma, T. Wang, T. Liu, J. Yin, M. Sun, C. Wu, G. Su, X. Dai, X. Wang, Y. Wang, H. Yin, X. Zhou, Y. Shen and H. Rao, Chem. Eng. J., 2023, 454, 140492 CrossRef CAS.
- Z. Xu, Z. Wang, M. Liu, B. Yan, X. Ren and Z. Gao, Spectrochim. Acta, Part A, 2020, 232, 118147 CrossRef CAS.
- Z. Xu, K. Wang, M. Zhang, T. Wang, X. Du, Z. Gao, S. Hu, X. Ren and H. Feng, Sens. Actuators, B, 2022, 359, 131590 CrossRef CAS.
- J. Massah and K. Asefpour Vakilian, Biosyst. Eng., 2019, 177, 49–58 CrossRef.
- F. F. Gonzalez-Navarro, M. Stilianova-Stoytcheva, L. Renteria-Gutierrez, L. A. Belanche-Muñoz, B. L. Flores-Rios and J. E. Ibarra-Esquer, Journal, 2016, 16, 1483 Search PubMed.
- Y. Rong, A. V. Padron, K. J. Hagerty, N. Nelson, S. Chi, N. O. Keyhani, J. Katz, S. P. A. Datta, C. Gomes and E. S. McLamore, Analyst, 2018, 143, 2066–2075 RSC.
- M. P. Shirani, B. Rezaei, T. Khayamian, M. Dinari, F. H. Shamili, M. Ramezani and M. Alibolandi, Mater. Sci. Eng., C, 2018, 92, 892–901 CrossRef CAS PubMed.
- Q. Duan, Y. Ma, M. Che, B. Zhang, Y. Zhang, Y. Li, W. Zhang and S. Sang, J. Drug Delivery Sci. Technol., 2019, 49, 527–533 CrossRef CAS.
- Y. Wen, M. Xu, X. Liu, X. Jin, J. Kang, D. Xu, H. Sang, P. Gao, X. Chen and L. Zhao, Colloids Surf., B, 2019, 173, 842–850 CrossRef CAS PubMed.
- S. Zhao, S. Sun, K. Jiang, Y. Wang, Y. Liu, S. Wu, Z. Li, Q. Shu and H. Lin, Nano-Micro Lett., 2019, 11, 32 CrossRef CAS PubMed.
- J. R. Melamed, R. S. Edelstein and E. S. Day, ACS Nano, 2015, 9, 6–11 CrossRef CAS PubMed.
- G. Hu, B. Lei, X. Jiao, S. Wu, X. Zhang, J. Zhuang, X. Liu, C. Hu and Y. Liu, Opt. Express, 2019, 27, 7629–7641 CrossRef CAS PubMed.
- B. Han, W. Wang, H. Wu, F. Fang, N. Wang, X. Zhang and S. Xu, Colloids Surf., B, 2012, 100, 209–214 CrossRef CAS PubMed.
- F. A. Permatasari, H. Fukazawa, T. Ogi, F. Iskandar and K. Okuyama, ACS Appl. Nano Mater., 2018, 1, 2368–2375 CrossRef CAS.
- D. Kim, G. Jo, Y. Chae, S. Subramani, B. Y. Lee, E. J. Kim, M.-K. Ji, U. Sim and H. Hyun, Nanoscale, 2021, 13, 14426–14434 RSC.
- M. Lan, S. Zhao, Z. Zhang, L. Yan, L. Guo, G. Niu, J. Zhang, J. Zhao, H. Zhang, P. Wang, G. Zhu, C.-S. Lee and W. Zhang, Nano Res., 2017, 10, 3113–3123 CrossRef CAS.
- M. Qian, L. Chen, Y. Du, H. Jiang, T. Huo, Y. Yang, W. Guo, Y. Wang and R. Huang, Nano Lett., 2019, 19, 8409–8417 CrossRef CAS PubMed.
- B. Ryplida, G. Lee, I. In and S. Y. Park, Biomater. Sci., 2019, 7, 2600–2610 RSC.
- X. Peng, R. Wang, T. Wang, W. Yang, H. Wang, W. Gu and L. Ye, ACS Appl. Mater. Interfaces, 2018, 10, 1084–1092 CrossRef CAS PubMed.
- S. Monro, K. L. Colón, H. Yin, J. Roque, III, P. Konda, S. Gujar, R. P. Thummel, L. Lilge, C. G. Cameron and S. A. McFarland, Chem. Rev., 2019, 119, 797–828 CrossRef CAS PubMed.
- Y. Liu, X. Meng and W. Bu, Coord. Chem. Rev., 2019, 379, 82–98 CrossRef CAS.
- Q. Li, Y. Li, T. Min, J. Gong, L. Du, D. L. Phillips, J. Liu, J. W. Y. Lam, H. H. Y. Sung, I. D. Williams, R. T. K. Kwok, C. L. Ho, K. Li, J. Wang and B. Z. Tang, Angew. Chem., Int. Ed., 2020, 59, 9470–9477 CrossRef CAS PubMed.
- J. Karges, U. Basu, O. Blacque, H. Chao and G. Gasser, Angew. Chem., Int. Ed., 2019, 58, 14334–14340 CrossRef CAS PubMed.
- S. Zhao, K. Yang, L. Jiang, J. Xiao, B. Wang, L. Zeng, X. Song and M. Lan, ACS Appl. Nano Mater., 2021, 4, 10528–10533 CrossRef CAS.
- J. Wang, M. Xu, D. Wang, Z. Li, F. L. Primo, A. C. Tedesco and H. Bi, Inorg. Chem., 2019, 58, 13394–13402 CrossRef CAS PubMed.
- N. Xu, J. Du, Q. Yao, H. Ge, H. Li, F. Xu, F. Gao, L. Xian, J. Fan and X. Peng, Carbon, 2020, 159, 74–82 CrossRef CAS.
- W. Pang, P. Jiang, S. Ding, Z. Bao, N. Wang, H. Wang, J. Qu, D. Wang, B. Gu and X. Wei, Adv. Healthcare Mater., 2020, 9, 2000607 CrossRef CAS PubMed.
- K. Yang, F. Li, W. Che, X. Hu, C. Liu and F. Tian, RSC Adv., 2016, 6, 101447–101451 RSC.
- S. Beack, W. H. Kong, H. S. Jung, I. H. Do, S. Han, H. Kim, K. S. Kim, S. H. Yun and S. K. Hahn, Acta Biomater., 2015, 26, 295–305 CrossRef CAS PubMed.
- Y. Xie, W.-Y. Meng, R.-Z. Li, Y.-W. Wang, X. Qian, C. Chan, Z.-F. Yu, X.-X. Fan, H.-D. Pan, C. Xie, Q.-B. Wu, P.-Y. Yan, L. Liu, Y.-J. Tang, X.-J. Yao, M.-F. Wang and E. L.-H. Leung, Transl. Oncol., 2021, 14, 100907 CrossRef CAS PubMed.
- N. Ali, C. Bolenz, T. Todenhöfer, A. Stenzel, P. Deetmar, M. Kriegmair, T. Knoll, S. Porubsky, A. Hartmann, J. Popp, M. C. Kriegmair and T. Bocklitz, Sci. Rep., 2021, 11, 11629 CrossRef CAS PubMed.
- F. Grisoni, C. S. Neuhaus, G. Gabernet, A. T. Müller, J. A. Hiss and G. Schneider, ChemMedChem, 2018, 13, 1300–1302 CrossRef CAS PubMed.
- F. Grisoni, C. S. Neuhaus, M. Hishinuma, G. Gabernet, J. A. Hiss, M. Kotera and G. Schneider, J. Mol. Model., 2019, 25, 112 CrossRef PubMed.
- S.-I. Lee, S. Celik, B. A. Logsdon, S. M. Lundberg, T. J. Martins, V. G. Oehler, E. H. Estey, C. P. Miller, S. Chien, J. Dai, A. Saxena, C. A. Blau and P. S. Becker, Nat. Commun., 2018, 9, 42 CrossRef PubMed.
- Y. Kim, D. Kim, B. Cao, R. Carvajal and M. Kim, BMC Bioinf., 2020, 21, 288 CrossRef CAS PubMed.
- J. Kong, H. Lee, D. Kim, S. K. Han, D. Ha, K. Shin and S. Kim, Nat. Commun., 2020, 11, 5485 CrossRef CAS PubMed.
- L. Deng, Y. Cai, W. Zhang, W. Yang, B. Gao and H. Liu, J. Chem. Inf. Model., 2020, 60, 4497–4505 CrossRef CAS PubMed.
- Q. Liu and L. Xie, PLoS Comput. Biol., 2021, 17, e1008653 CrossRef CAS PubMed.
- A. Cuvitoglu, J. X. Zhou, S. Huang and Z. Isik, J. Bioinf. Comput. Biol., 2019, 17, 1950012 CrossRef CAS PubMed.
- K. E. Regan-Fendt, J. Xu, M. DiVincenzo, M. C. Duggan, R. Shakya, R. Na, W. E. Carson, P. R. O. Payne and F. Li, npj Syst. Biol. Appl., 2019, 5, 6 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2023 |