Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
COVID-19 mitigation: nanotechnological intervention, perspective, and future scope
Arpita
Adhikari†
a,
Dibyakanti
Mandal
b,
Dipak
Rana
 *c,
Jyotishka
Nath‡
a,
Aparajita
Bose
d,
Sonika
e,
Jonathan Tersur
Orasugh
*fg,
Sriparna
De
*h and
Dipankar
Chattopadhyay
*c,
Jyotishka
Nath‡
a,
Aparajita
Bose
d,
Sonika
e,
Jonathan Tersur
Orasugh
*fg,
Sriparna
De
*h and
Dipankar
Chattopadhyay
 *a
*a
aDepartment of Polymer Science and Technology, University of Calcutta, 92 A.P.C. Road, Kolkata 700 009, India. E-mail: dcpst@caluniv.ac.in
bDepartment of Microbiology, Faculty of Life Sciences, PDM University, Sector 3A, Bahadurgarh, Haryana 124 507, India
cDepartment of Chemical and Biological Engineering, University of Ottawa, 161 Louis Pasteur St., Ottawa, ON, K1N 6N5, Canada. E-mail: rana@uottawa.ca
dDepartment of Chemistry, University of Calcutta, 92 A.P.C. Road, Kolkata 700 009, India
eDepartment of Physics, Rajiv Gandhi University, Rono Hills, Doimukh, Arunachal Pradesh 791 112, India
fDepartment of Chemical Sciences, University of Johannesburg, Doorfontein, Johannesburg 2028, South Africa
gDST-CSIR National Centre for Nanostructured Materials, Council for Scientific and Industrial Research, Pretoria 0001, South Africa. E-mail: JOrasugh@csir.co.za
hDepartment of Allied Health Sciences, Brainware University, Barasat, Kolkata 700 125, India. E-mail: sd.ah@brainwareuniversity.ac.in
First published on 25th October 2022
Abstract
COVID-19 infections and severe acute respiratory syndrome (SARS) have caused an unprecedented health crisis across the globe with numerous deaths, as well as causing a tremendous economic crash worldwide. To combat this acute pathogenic coronavirus strain, a vital strategy of safe and effective diagnostic and therapeutic measures is highly important and demanding at present. Instead of conventional diagnosis tools, nanotechnology offers inspiring options for therapeutic applications that can ward off the disease from spreading further and remove the threat of this virus causing future pandemics. Physicochemically tuned nanomaterials can be exploited in upgrading detection schemes for viral antigens, nano-vaccines, and inhibitors of the cytokine storm, which are vital in the fight against COVID-19. The one-of-a-kind nanoscale biosynthesis of synthetic nanoparticles can efficiently imitate and interact with the structurally similar spike proteins present on the viral surface. Given this, we envision the precise and concurrent amalgamation of nanoscience and nanotechnology, leading to new avenues that can disrupt the development of viruses and limit the length of the viral lifespan. Current and developing nanotechnology approaches enable the development of therapeutic and precautionary pathways to curb this disease and identify crucial methods in the field of nanoscience for developing upcoming antiviral systems. In this review article, we also present a synopsis of the latest studies on the efficacy of nanoparticles (NPs) as antiviral or diagnostic devices against most viruses. Engineered NPs capable of tempering the patient's immune response can have a pronounced impact in mitigating inflammatory reactions as well as the design of potent nano-vaccines and drugs against viral pandemics such as COVID-19. In summary, state-of-the-art approaches based on nanotechnology can be critically deployed to counter future pandemics including COVID-19 and function at the forefront in tackling various new dangerous viral threats.
1. Introduction
The world is under threat from a relentless minuscule enemy, which was unknown to mankind but is now causing serious illness and infections. As countries across the globe initiate unparalleled techniques to dampen the further spread of COVID-19, a major startling conclusion is that there is still no clear way out of this viral disease. Community scrutiny, rapid testing, contact tracing, social distancing, and swift clinical care are the rudimentary prevention schemes being employed worldwide. However, we do not understand exactly how long these measures need to be imposed or what extent of relaxation will cause the virus to resurge. Numerous contagious diseases spread by viruses have recently come to light, such as SARS-CoV, Ebola virus, and dengue virus, resulting in the demand for advanced antiviral treatments and personal protection equipment (PPE) to combat these contagious agents.1 A cornerstone strategy for maintaining a healthy environment includes appropriate use of disinfectants (cleaning, disinfecting, and hand hygiene) and protective respirator face masks to act as a safeguard for the nose and mouth of the wearer, preventing the spread of droplets carrying viruses or other infectious pathogens. In this context when regular health aid is provided, health professionals prescribe critical care for patients and ensure that the infection is prevented, and infection prevention and control (IPC) measures are carried out and followed in healthcare facilities to curb healthcare-related infections. To prevent infection in healthcare facilities, the WHO suggests the application of contact and droplet precautions by healthcare employees for the treatment of COVID-19 patients.2,3 The WHO also endorses that airborne measures be taken in these settings, emphasizing that the right use of PPE kits is essential, including face protection, goggles, masks and face shields, gloves, gowns, head coverings, and rubber boots. Furthermore, exploration in the fields of nanoscience and nanotechnology can serve to add an extra protective shield to prevent infection colonization in healthcare professionals.4,5 To date, numerous viral diseases have surfaced in the global community, which have turned out to be a serious threat to the population. Besides the common cold, diseases conditions including influenza, herpes virus, viral encephalitis, herpes keratitis, neonatal disseminated infections, prolonged distress, and sometimes persistent infections of hepatitis viruses (namely hepatitis B virus (HBV) and hepatitis C virus (HCV)) or human immunodeficiency virus (HIV) can ultimately lead to severe immunodeficiency diseases, and in some cases even cancer. Thus, considerable struggles have been encountered in attempts to effectively deal with these viral diseases.6,7Nanotechnology and nanoscience as a whole involve the development, progress, and utilization of atomic and molecular structures, which have one of their dimensions in the nanoscale (1–100 nm), involving different constitutions, shapes, structures, size, and surface properties. Nanotechnology-based cutting-edge technology has emerged with the potential to reveal significant alternatives that can constrain the spread of virions. The synergism of nanoparticles in the fields of physics, chemistry, biology, materials science, and medicine encourages the application of this technology in the field of materials design. In the medical sector, nanotechnology-driven biosensing arrays have been extensively adopted for the detection of viruses. Recently, biochemical research has opened a new avenue, wherein novel nanomaterials are fabricated with advanced biocompatibility using virions and virus-like templates. Currently, widespread outbreaks of viral respiratory infections have been reported worldwide. The scientific community is desperately seeking to optimize and design appropriate nano-vaccines and therapeutics, involving the latest nano-based approaches. This versatile nanotechnological paradigm is encouraged by virologists to continuously develop novel delivery devices, which will also be at the vanguard in fighting deadly viruses.
The ongoing pandemic has definitely ravaged the world and there will be many more in the future. Thus, it is necessary for the nano biomaterials research fraternity to come up with appropriate measures to combat COVID-19 and associated pandemic-causing agents in the near future. Nanotechnology and nanoscience-inspired materials such as non-woven disposable materials, antiviral coatings, PPE, multifunctional surfaces, air-conditioning filters, antibacterial textiles, and antiviral fabrics are among the many candidates attracting attention to provide solutions rapidly.8
Nanotechnology-based interventions should be put into full effect to counteract COVID-19 and subsequent outbreaks by various means, as follows: (i) development of contact tracing tools, (ii) fabrication of novel surfaces and surface coatings that aid in virus resistance, virus adhesion, and inactivation, (iii) production of fine mesh face masks and blood filters, (iv) advanced tests for the rapid, sensitive and highly specific detection of infection and immunity, and (v) innovative drugs and vaccines encompassing nanomaterials capable of transferring broad range antiviral cargo or therapies to the lungs. The current crisis has also given us a stern reminder regarding the urgent need to critically analyse unforeseen circumstances stemming from the pandemic. Thus, a roadmap is mandatory to guide the systematic and rapid production of PPE and ventilators. Accordingly, nanotechnology may prove to be useful in aiding the manufacture of equipment by offering strategies for precursor synthesis together with improvement in durability and efficacy. In the following sections, our discussion focuses on the role of nanotechnology in dealing with and mitigating the ill effects of COVID-19 and similar pandemics the future.9,10
1.1. History of outbreaks
The last report of an outbreak was the deadly ‘atypical pneumonia’, which appeared in November 2002, in the region of Guangdong Province, China. Subsequently, a similar occurrence was reported in March 2003, in Hong Kong. Similar types of respiratory disorders were successively reported worldwide including in Taiwan, China, Singapore, Vietnam, the USA, and Canada. This syndrome with new clinical symptoms has been equated with ‘severe acute respiratory syndrome (SARS).11 SARS mainly originated from viruses associated with the Coronaviridae family, known as SARS-coronavirus (SARS-CoV).12 However, sequence data indicates that SARS-CoV is not like previously known coronaviruses.13 On 22nd September 2012, the United Kingdom notified the WHO regarding a case study of a 49 year-old male from Qatar having a travel history to Saudi Arabia and Qatar and suffering from ‘acute respiratory syndrome’ and renal failure.14 The WHO termed the provisional case as “severe respiratory disease associated with novel coronavirus” dated September 25, 2012. On 29th September, this was revised to Severe Acute Respiratory Infections associated with novel coronavirus infection (SARI associated with nCoV). According to laboratory-confirmed reports from May 12, 2013, 34 cases were reported due to Middle East respiratory syndrome coronavirus (MERS-CoV). Also, there were 20 deaths with two cases from previous records of the Jordanian cluster of this class of disease confirmed by the Health Ministry of Jordan in April 2012.Among the various CoV strains, the most infectious strains are alpha-coronaviruses and beta-coronaviruses, which affect mammals, whereas gamma-coronaviruses and delta-coronaviruses affect birds. Before 2019, only 6 CoVs were recognized, among which HCoV-229E, HCoV-OC43, HCoV-NL63, and HKU1 cause mild disease in the upper respiratory area together with fatal infection in infants, youth, and older patients. SARS-CoV and MERS-CoV can contaminate the lower respiratory tract and induce severe respiratory syndrome in people. Most importantly, the new coronavirus 2019-nCoV belongs to beta-coronaviruses, which can significantly affect the lower respiratory tract followed by the development of pneumonia. According to the WHO report on 31st December 2019, the Wuhan Municipal Health Commission of China indicated the existence of a cluster of cases of pneumonia in Wuhan, Hubei Province. Subsequently, a new coronavirus was detected. As of the 3rd of January 2020, about 44 patients having pneumonia of unknown etiology were identified, and the WHO was notified by the national authorities in China. On the 5th of January, the WHO announced that China was dealing with a significant new viral outbreak. At the end of January 2020, 9826 cases were reported globally with 9720 confirmed cases within China. Consulting existing data available on many websites about COVID-19 infections worldwide made it evident that the number of cases was increasing exponentially. As of June 10th, 2020, there were 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 323
323![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 918 confirmed cases including 413
918 confirmed cases including 413![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 733 deaths and 3
733 deaths and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 604
604![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 recovered cases worldwide.15,16
500 recovered cases worldwide.15,16
The mode of transmission of a few CoVs is livestock, birds, bats, mice, whales, and many other wild animals. A report showed that swine acute diarrhea syndrome coronavirus (SADS-CoV) led to a major outbreak (in 2016) of a deadly disease in pigs in Southern China, which caused the death of 24000 piglets.17 This case was the first identified spillover of a bat coronavirus, which triggered deadly disease in livestock. 2019-nCoV and Beluga Whale CoV/SW1 belong to separate genera and diverse host spectra. Hence, the pathogenesis of CoVs needs further research to be identified to understand the specific action of these viruses.18,19
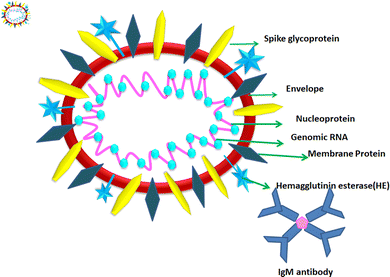
1.2. Structure of COVID-19
According to the genomic and phylogenetic morphology of COVID-19, it is from the Coronaviridae family in the order Nidovirales, which is classified into alpha-coronavirus, beta-coronavirus, gamma-coronavirus, and delta-coronavirus. Among them, the beta-coronavirus genera can affect mammals, causing SARSr-CoV.20,21 Considering its genomic structure, COVID-19 is composed of single-stranded RNA (+ssRNA) (∼30 kb) with a 5′-cap structure and 3′poly-A tail. This is enveloped with a capsid comprising nucleoprotein and matrix protein, containing one of the longest genomes (26.4–31.7 kb) of all known RNA viruses. Also, it possesses various small open reading frames (ORFs), which exist among several genes such as ORF1ab, spike, envelope, membrane, and nucleocapsid. Polypeptides encoded by chymotrypsin-like protease (3CLpro) and one or two papain-like proteases are produced from these ORFs. Additionally, the 4 major structural proteins include spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins and various accessory proteins, such as the HE protein, 3a/b protein, and 4a/b protein, which are predominantly accountable for virus replication and genomic integrity.20,22 Based on the viral entry within the host cell, spike proteins, specifically, glycoprotein (M), play a pivotal role towards therapeutic targets, triggering the engulfment of the virus onto host cell membrane by releasing the NH2 terminal moiety on the outer wall of the virus and long carboxylic cytoplasmic terminus in the virion (Fig. 1).23Meanwhile, the S protein is the prime facilitator of neutralizing antibody activities. Recognition and binding with the host cell and entry between the wall of the virus and the host cell surface are governed by two subunits of the S protein, i.e., S1, and S2. Interestingly, M plays a significant roles in the intracellular formation of virus particles through interaction with S proteins. The spike protein plays a dual role in virus pathogenicity. The S1 subunit consists of a receptor binding domain (RBD), an amino-terminal domain (NTD), and a C-terminal domain. RDB is responsible for binding the receptor and ACE2, which is present on the lung epithelial cells, for entry. Both RBD and NTD are the major contributors to the virus antigenicity, thus and neutralization antibodies are targeted to these regions of the S1 subunit of the spike proteins. Some neutralization antibodies also target CTD of S1. The S2 subunit forms a trimeric core of the spike protein and is mainly involved in virus fusion with the cell membrane.24 COVID-19 disease is associated with hyperinflammation and increased levels of inflammatory cytokines. This is termed the ‘cytokine storm syndrome (CSS)’ and is the cause of severe disease, respiratory failure, and death. Studies showed that the S protein alone can trigger this cytokine storm and different pathways have been proposed. Increased levels of pro-inflammatory cytokines including IL-6, IL-2, IL-1β, IL-8, IL-17, GM-CSF, MCP-1, and TNF-α were reported in COVID-infected individuals. Increased levels of cytokines lead to the recruitment of immune cells, which is the cause of CSS.25,26 Thus, vaccines were designed based on the spike proteins and found to be effective. However, COVID still continues to threaten individuals of all age groups and from all geographical regions even after three years since the pandemic began. India experienced and is still experiencing more than around 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 daily cases and 50–100 deaths. Although effective vaccines have been introduced and the majority of individuals completed two doses of the COVID-19 vaccine, significant virus activity is still being observed. The main reason for this is the emergence of mutant viruses that escape our immune system against the virus. The WHO categorized these COVID-19 variants into three categories, namely, VUI (variant under investigation), VOI (variant of interest), and VOC (variant of concern). All these variants have several mutations within the spike protein (S). These mutations cause changes in the binding affinity of the virus to its receptor angiotensin-converting enzyme 2 (ACE2) and can also alter the levels of an immune response. VOCs are the major cause of serious disease including respiratory distress and deaths among COVID-19-infected individuals. The alpha variant or B1.1.7 was isolated in the UK; the beta variant or B.1.351 first was isolated from South Africa; the gamma variant of P.1 was isolated from Brazil and the delta variant or B.1.617.2 was isolated from India, where the latter is the most deadly strain among them. The D614 mutation in S1 is the signature of this strain. Other common mutations are in the RBD domain of S1, which result in the stronger binding affinity of the virus to ACE2 and also cause hyperinflammation, leading to CSS.27 Recently, the omicron variant B.1.1.529 was identified as the most rapidly spreading variant; however, the severity of the disease caused by this variant is much less compared to other VOCs. In this variant, 30 mutations were reported in the RBD region of S1.28 These mutations caused increased transmissibility and immune evasion, even in vaccinated individuals. Because of the emergence of these variants, vaccine producers and policymakers require changes in existing strategies to control the global COVID-19 pandemic. Within these genera, the exploration and cultivation of various nanoparticles can inhibit the active spread of virions.28 The utilization of nanoparticles can inactivate the infectivity and restrict the adherence property on abiotic surfaces and impact the intracellular environment.
000 daily cases and 50–100 deaths. Although effective vaccines have been introduced and the majority of individuals completed two doses of the COVID-19 vaccine, significant virus activity is still being observed. The main reason for this is the emergence of mutant viruses that escape our immune system against the virus. The WHO categorized these COVID-19 variants into three categories, namely, VUI (variant under investigation), VOI (variant of interest), and VOC (variant of concern). All these variants have several mutations within the spike protein (S). These mutations cause changes in the binding affinity of the virus to its receptor angiotensin-converting enzyme 2 (ACE2) and can also alter the levels of an immune response. VOCs are the major cause of serious disease including respiratory distress and deaths among COVID-19-infected individuals. The alpha variant or B1.1.7 was isolated in the UK; the beta variant or B.1.351 first was isolated from South Africa; the gamma variant of P.1 was isolated from Brazil and the delta variant or B.1.617.2 was isolated from India, where the latter is the most deadly strain among them. The D614 mutation in S1 is the signature of this strain. Other common mutations are in the RBD domain of S1, which result in the stronger binding affinity of the virus to ACE2 and also cause hyperinflammation, leading to CSS.27 Recently, the omicron variant B.1.1.529 was identified as the most rapidly spreading variant; however, the severity of the disease caused by this variant is much less compared to other VOCs. In this variant, 30 mutations were reported in the RBD region of S1.28 These mutations caused increased transmissibility and immune evasion, even in vaccinated individuals. Because of the emergence of these variants, vaccine producers and policymakers require changes in existing strategies to control the global COVID-19 pandemic. Within these genera, the exploration and cultivation of various nanoparticles can inhibit the active spread of virions.28 The utilization of nanoparticles can inactivate the infectivity and restrict the adherence property on abiotic surfaces and impact the intracellular environment.
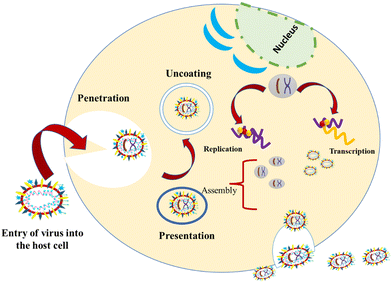
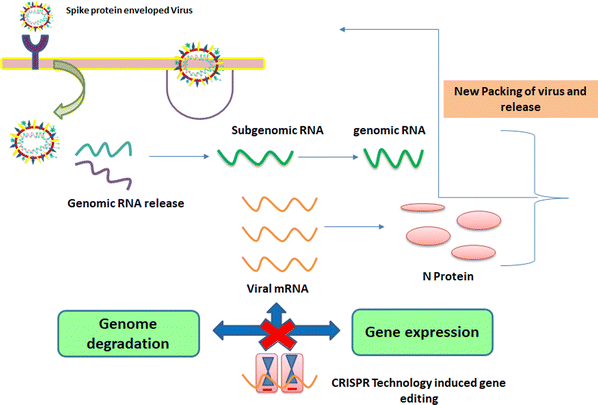
1.3. Mechanism of entry of viruses into host cells
These viruses are sustained via multiplication in host cells. The process of infection includes attachment, penetration, uncoating, replication, assembly, and release. There are several specific receptors identified on the host cell membrane by which viruses enter cells utilizing the attachment proteins within the viral capsid or viral envelop-decorated glycoproteins. Based on the virus specificity, various infectivity schemes were found, wherein RNA viruses such as CoV put their genome to use to synthesize viral genomic RNA and mRNA, eventually resulting in the production of new viruses in the host cells. Bacteriophages gain entry into the host cell via nucleic acids, whereas their capsid remains outside the cell. Few animal and plant viruses gain entry to host cells via endocytosis. Upon entry in the host cell, CoVs use their genomics to synthesize viral genomic RNA and mRNA, and eventually the new virions into the host cells (Fig. 2).29–31A pivotal route of viral entry of the influenza virus, respiratory syncytial virus, and parainfluenza virus is identified as the human respiratory mucosa. The virulence nature and pathogenicity promote infection in the upper respiratory tract, leading to severe illness in the lower respiratory tract and morbidity and mortality. Majorly respiratory tract infections evolved due to SARS-CoV, MERS-CoV, and SARS-CoV-2.32 Due to its fascinating morphology, the entry mechanism in host cells involves the interaction between the host-receptor cells and the angiotensin-converting enzyme 2 (ACE2) protein site, which is associated with host-cellular membrane fusion followed by the release of genetic material into the host cytoplasm.
In the case of SARS-CoV-2, the spike S glycoprotein is also responsible for bonding and entrance into the cell. The unique morphological features of the S protein trigger the attachment, fusion, and entry of the virus in the host cells.33–35 Therefore, blocking the ACE2 receptor binding site, thereby hindering the viral endocytosis pathway, is a compelling route toward drug discovery and therapeutics.
2. Effectiveness of nanomaterials targeting spike proteins (SP) to prevent COVID-19 spread
The antiviral mechanistic approach of nanoparticles should target the viral endocytosis process including attachment, penetration, replication, and budding of viruses. Various morphological architectures possibly influence the blocking of viral replication.29 Importantly, nanoparticles restrict the above-mentioned processes by varying the structural features of the capsid protein, thereby suppressing its virulent property, which may be ascribed to physicochemical methods of reducing the active viral levels (Fig. 3).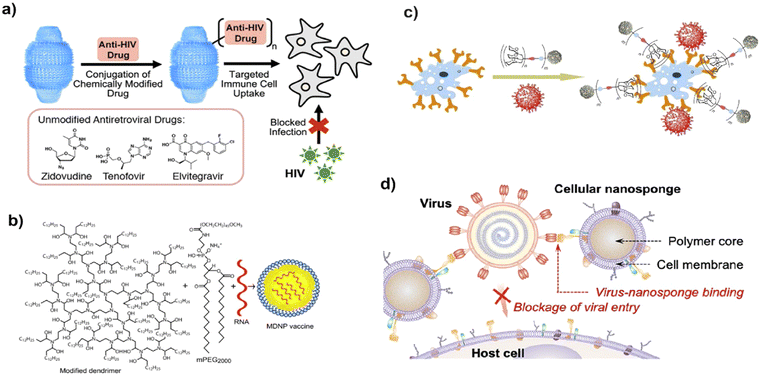 | ||
| Fig. 3 Possible modes of the antivirucidal efficacy of nanotechnology. (a) Effect of antiviral drug delivery against HIV-1 infection, adapted from Fulcher et al.36 Copyright 2019, American Chemical Society. (b) Schematic of vaccine delivery using lipid-derived dendrimer nanoparticles through m-RNA. Reproduced with permission from Chahal et al.37 Copyright 2016, National Academy of Sciences, USA. (c) Blocking approach of virus-like glycodendrinanoparticles to the infection site, adapted from Ribeiro-Viana et al.38 Copyright 2018, Nature Publishing Group. (d) Strategy of mimicking host cell membrane carrying nanodecoy host cell membrane for adsorbing Zika virus, adapted from Zhang et al.39 Copyright 2020, American Chemical Society. | ||
To facilitate their passage in the host cell, viruses use spike proteins triggered by certain enzymes including furin, trypsin, and cathepsin L9.40 Among them, furin has the most accelerating effect on the spike protein located on the novel coronavirus cell membrane.
This enzyme reveals unique and fascinating properties given its ability to exist in human tissues including the lungs, liver, and small intestine, which facilitate the virus invading the host cell in a manner distinct from other SARS family viruses and modes of transmission. A recent investigation pin-pointed the chain of infection of the influenza family virus, wherein the haemagglutinin protein lies on the activation site, lucid from CoV SP, causing alteration in the contagious virus transmission channel. Also, scientific evidence demonstrates that furin and furin-like proteases such as fusogenic envelope glycoprotein of SARS CoV are the most susceptible sites for targeting treatment practice (Fig. 4).
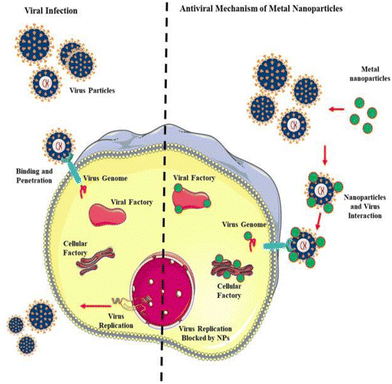 | ||
| Fig. 4 Antiviral mechanism of nanoparticles (NPs). Reproduced with permission from Gurunathan et al.32 Copyright 2020, Multidisciplinary Digital Publishing Institute. | ||
The emergence of COVID-19 variants is a concern. Thus, in this regard, nanotechnology-based approaches are expected to be beneficial, providing better safeguards to tackle this COVID-19 pandemic. Recently, several nanoparticle-based strategies have been developed to prevent the spread of the mutant viruses. One approach is a lipid nanoparticle-based mRNA vaccine called mRNA-LNP, which is based on spike mRNA from new variants. Another effective approach is nanoparticle-based engineered neutralizing antibodies against variants of S proteins, where synthetic antibodies with antigen binding regions specific for new COVID-19 variants are designed.41 Given that all the variant stains of COVID-19 utilize ACE2 as a receptor and do not bind another molecule for entry, nanoparticle-based ACE2 decoys are expected to be more effective in preventing the virus spread. In this technology, the strong interaction of ACE2 receptors of decoy nanoparticles traps the virus of different variants and stops causing serious disease. Another nanotechnology-based method is the nonspecific physical inactivation of virus variants using polymer surfactants and pore-forming peptides. Nanoparticle-based anti-inflammatory drugs also show promise against COVID-19 variants of concern.42
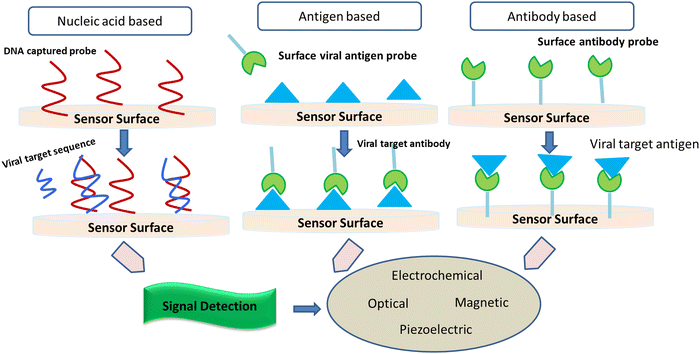
2.1. Implication of biosensors to screen SARS-CoV-2
It is considered that the factors of high transmissibility and the infectious nature of the COVID-19 virus are attributed to its heavy viral load in the upper respiratory area together with the inference that several patients do not show symptoms, thereby spreading the virus. Crucial nanotechnological interventions to fight pandemics include the inhibition of pathogen spreading, activation of suitable diagnostic tools, and proper immunization. For instance, the nanodiagnostic scheme relies on the specific interaction between nanoparticles and the target analyte to yield a quantifiable signal output, thereby catering to the identification of biomarkers or pathogens in their embryonic stages (Fig. 5).Nanobiosensors are a value-added substitute to regular diagnostic devices, which assess single molecules with great sensitivity, specificity, and precision characteristics for clinical and environmental analyses. Au-NP-based nano-carriers and a field-effect transistor (FET) sensor (altered via graphene sheets) that alters a given antibody for the SARS-CoV-2 spike protein were activated to identify SARS-CoV-2 in the culture mode. This sensor exhibited a comparable detection limit (LOD of 1.6 × 101 pfu mL−1) to real pathological samples (LOD: 2.42 × 102 copies per mL).43 Gold NP-studded DNA strands on the surface of electrodes were modified to identify in situ-amplified CTV (recombinase polymerase amplification (RPA) assay for amplification of the P20 gene) via electrochemical impedance spectroscopy (EIS) analysis. Here, the DNA sensor displayed a very low LOD of 1000 fg μL−1.44 Recently, paper/nanomaterial-customized detection techniques, especially, nanoparticle-based lateral-flow tools that may be seen by the bare eyes or administered using an android phone, are the latest known testing devices to filter COVID-19-related biomarkers.45 Specialized research has also portrayed solid-phase isothermal recombinase polymerase amplification (RPA) routes for the detection of the Citrus Tristeza virus (CTV) in plant disease diagnostics.46 This approach uses nanoparticle-engineered biosensors, which if modified appropriately for the detection of other viruses can be adopted, designed, and engineered for the diagnosis of COVID-19.
3. Nanotechnological approach to monitor SARS-CoV2
Different testing methods around the globe have aimed at focusing on parts of the coronavirus genetic profile. Clinical settings have utilized samples triggered by a patient's immune response such as DNA, RNA, antibodies, viral proteins, and intact viral particles to recognize viruses. A host of established methodologies such as serological detection methods, western blot, virus culture, enzyme-linked immunosorbent assay (ELISA), and polymerase chain reaction (PCR) can be used to identify the analytes.47 Generic methodologies are largely ineffective for fast real-time analysis due to complications related to the growth of cultured cells for cytopathological assay, problems associated with the transportation of expensive sophisticated tools at the point of interest, and the requirement of biocontainment and virus isolation. At the beginning of the pandemic, it was reported and globally circulated among health systems that some of the test kits provided results with less reliability and accuracy. Besides, another impending issue was the requirement of large amounts of testing reagents. Thus, this issue created a gap in the whole endeavour. In lieu of well-known diagnostic assays, viral biosensors can be used given that they offer scaled-down, fast, sensitive, cost-effective, and mobile foundations in contrast to standard laboratory methods. Among the various pathogens, viruses are the most important class to cause deadly airborne diseases. Influenza, a very commonplace and highly infectious virus, gets transmitted through small droplets as a result of sneezing, coughing, and talking.48 Another fatally contagious viral infection, the avian influenza, has various subgroups, including H5N1, H5N2, H3N2, and H7N9. This virus mostly infects poultry and cattle, but it is possible for it to be transmitted to people and has incurred severe losses in federal economies globally.49–51SARS (severe acute respiratory syndrome) is a deadly virus-causing syndrome, which results in critical respiratory problems and even death. SARS belongs to the coronavirus family and was first recognized during a major outbreak in China in 2003. SARS was reported to be found in approximately 24 countries in 2003 with a case tally of 8000 and death tally of 774. The confirmatory test for the identification of respiratory virus disease is cell culture.52 In the late 1960s, distinguished research groups carried out the first studies on coronavirus. Researchers independently differentiated a number of strains of a novel virus that expressed significant ether affinity properties. Electrochemical-based biosensors for the identification of influenza virus were one of the most remarkable ones according to the reports in the literature. 6 optical systems were ascertained to be the principal class of sensors for the detection of coronavirus, while a piezoelectric class of sensors was deemed for the identification of SARSCov-1. Besides, an electrochemical system was investigated for the detection of MERS and a different electrochemical detection for the identification of SARS-Cov-2. As an interesting alternative possessing greater sensitivity and rapidity compared to RT-qPCR and serological tests, Huang51 came up with a fibre-optic biosensor that could detect the SARS-Cov antigen, i.e., the nucleocapsid (N) protein. Roh53,54 attained the same detection limit (0.1 pg mL−1) using immobilized quantum dots conjugated with RNA aptamer on a chip to identify the N protein from SARS-Cov. Also, the extent of detection stated above was the lowest with the aid of biosensor devices. Park,53 aiming to immobilize biomolecules, fabricated a surface plasmon-resonance (SPR) platform for the identification of SARS-Cov using the help of gold binding polypeptide (GBP). Park fused GBP with the SARS-Cov membrane envelope (SCVme), which possessed the capability to bond with anti-SCVme antibodies and a modified green fluorescent protein (GBP-E). Thus, this system developed by Park is an interesting system, where the specificity to the gold substrate is high, while keeping the biomolecular activity intact. As a radical effort for an optical sensor, Teengam54 introduced a biosensor for the detection of MERS. The idea for the biosensor was to immobilize pyrrolidinyl peptide nucleic acid (PNA) on a paper-based analytical device (PAD) to recognize synthetic oligonucleotides having a sequence similar to MERS. The mechanism driving the results is the aggregation/de-aggregation of (−) silver nanoparticles with the introduction of (+) DNA followed by the subsequent addition of complementary DNA. The analysis of the results does not need to be done on the typical computing device given that they can be perceived by the naked eye.55 Furthermore, a number of nanoscale devices has been evaluated. Immunocapture approaches amalgamated with antibody-coated chips have been utilized to image viruses56 with the aid of atomic force microscopy (AFM). The limit of detection for coxsackievirus was 106–107 TCID50 per mL (50% tissue culture infective dose). Within a very short time biosensors fabricated from nanowire/carbon nanotubes have transformed from theoretical concepts to practical devices capable of detecting DNA sequences and particular proteins with precision.
This concept of photo-induced nanomaterial possessing CNH for the specific eradication of viruses has been reported. In the mentioned discussion, the T7 bacteriophage was deemed to be the target virus recognizable by a T7 tag antibody. This virus bears a symmetrical structure with an approximate length of 100 nm and width of 55 nm together with long double-strand DNA (dsDNA; about 40 kbp) stationed on the head. This nanoparticle is constituted of a moiety of poly(ethylene glycol) (PEG) and functionalized CNH (CNH-COOH) with T7 tag Ab as the identification site and phospholipid group linked at both ends to enhance its solubility. Briefly, researchers have successfully procured the T7 tag Ab-CNH complex upon NIR laser irradiation T7 phage, which is a model virus successfully eliminated from the complex. Thus, these studies strongly suggest the production of CNH nanomaterials in the near future that exhibit potential for the photo-exothermic eradication of harmful viruses such as SARS, HIV, and avian influenza virus. It was unanimously by researchers that this research made a substantial contribution to antiviral biomedical applications. Ishikawa and co-workers (2009) were pioneers in developing an electrochemical biosensor for the identification of SARS-CoV.57 They developed an FET-based immunosensor, which generated a measurable conductance variation as a result of antigen-antibody binding, producing the analyte amount. The virus antigen nucleocapsid N protein was used as a CoV biomarker.58 Antibody mimic proteins, a viable alternative to conventional antibodies, have been already used as affinity binding agents.57,59 The first amperometric immunosensor for the detection of the MERS-Cov virus was reported by Layqah and Eissa in 2019.60 In their endeavour, spike protein S1 was used as a MERS biomarker. The optimum conditions for this experiment were determined to be 10 g mL−1 for the concentration of antibody and 20 min for binding time, respectively. Square wave voltammetry (SWV) was used to measure the peak current of the ferro-/ferri-cyanide redox probe, and as a result the current signal was obtained.59
In the present scenario, there are two major papers focusing on affinity-based biosensors for the detection of COVID-19. A novel FET-based biosensor for the detection of SARS-Cov-2 was proposed by Seo and co-workers.61 The synthesized device could identify the SARS-CoV-2 antigen spike protein with small concentrations as low as 1 fg mL−1 in phosphate buffer. This device also exhibited no substantial degree of cross-reactivity with the MERS-CoV antigen.61 Qiu and co-workers suggested a dual biosensor by amalgamating LSPR sensing transduction and plasmonic photothermal (PPT) effect on a single chip. The philosophy guiding the sensor chip modification is the utilization of two-dimensional gold nanoislands (AuNIs), which are subsequently activated with complementary DNA (cDNA) receptors with the aid of Au–S bond formation between the thiol groups of cDNA and the AuNIs. This bifunctional biosensor traced a uniform linear range between 0.1 pM and 1 mM with a detection limit of 0.22 pM.62
Recently, PathSensors Inc. reported the development of a “Canary” fast biosensor for the detection of the novel CoV-19. This biosensor uses a cell-based immunosensor, which captures the virus using signal amplification and gives a response in as little as 3–5 min. The primary application and use of the PathSensors device will be for testing environmental samples and keeping track of air in spaces prone to exposure, including hospitals, offices, and food services.63 Graphene and graphene-based materials (GRMs) have been shown to exhibit extraordinary physicochemical, electrical, optical, antiviral, antimicrobial, and many more interesting characteristics, which qualify them as possible contenders for the design and progress of strong-performing components and vehicles that will be needed for the COVID-19 virus and other futuristic disease outbreaks.64 The requirement for rapid home testing kits that are user-friendly is urgent. The cardinal philosophy of POC is for the process to be comfortable and fast. It is desirable for a patient to get immediate medical reports after getting tested, enabling quick treatment from the luxury of their home, which alleviates the pain of traveling to access treatment.
3.1. Nanoparticle inhibition of viral infection
It is well-known that there are 2 classes of antiviral analytes, i.e., virostatic and virucidal, based on their counteracting synergism against a particular virus, impeding viral replication and proliferation.65 Nevertheless, many effective antiviral agents show toxic responses. Thus, to address this issue, research on the antiviral characteristics of nanomaterials has begun. The major question in these systems indeed involves their low toxicity and pure virus curbing routes (Fig. 6). Nanoscale composites are made by merging virostatic and virucidal inactivation processes.66 In nanoscience, several nanomaterials have been explored, wherein non-pharmaceuticals denote a broad spectrum of nanomaterials having therapeutic efficacy, for example, dendrimers, liposomes, micelles, and nanocapsules.67 Metal oxide nanoparticles (MONPs) possess an interesting inhibitory mechanism via the production of reactive oxygen species (ROS) and restricting the S spike protein of the viral reaction with cellular ACE2 receptor.Metal nanoparticles have displayed potency as successful antiviral agents by thwarting viral particle entry in the exterior and restraining viral replication in the interior.68 The efficiency and antiviral action of metal NPs depend on their physical structure and attached metal ions. It should be noted that capped metal NPs have a notably greater reaction with viruses and host cells compared to bare NPs.69 Gold nanoparticles (AuNPs) have also sparked huge interest in the field and nanomedicine as a result of their outstanding electrical, optical, mechanical and biological properties. AuNPs possess outstanding mechanical, biological, electrical, and optical properties, which have attracted significant interest from the industrial sector and nanomedicine.70,71 Gold NPs have attracted broad attention and provide efficient antiviral activity against viruses. Sialic acid-functionalized AuNPs exhibit activity against infections from influenza virus and are superior compared to conventional synthetic drugs such as zanamivir and oseltamivir.72
AuNPs successfully enhance antiviral effects via polyvalent interactions with dendronized AuNPs, thus preventing HIV with more efficacy than solo dendrons. Functionalized AuNPs counteract the action of HIV, HSV, and influenza viruses.73,74 Di Gianvincenzo et al.75 reported that an amphiphilic sulphate-ended ligand coated on gold nanoparticles binds with glycoprotein gp120 on the HIV envelope to restrain the extent of HIV infection in vivo.
Mehranfar and Izadyar reported the properties of AuNPs attached with various moieties, such as 3-mercapto-ethyl sulfonate (MES), undecane sulfonic acid (Mus), octane thiol (OT), and a novel peptide, to hinder the CoV-19 virus using molecular dynamics simulations. Viewing the solid structure of angiotensin-converting enzyme 2 (ACE2), which can attach to the SARS-CoV-2 receptor-binding domain (RBD), about fifteen amino acids of ACE2 have substantial interplay with RBD. Hence, a novel peptide, using the above-mentioned amino acids has been considered and prepared as the functional moiety for AuNP. According to the results, the functionalized AuNPs show highly significant outcomes on the RBD and react quite strongly with this protein in SARS-CoV-2. Considering the above-mentioned nanoparticles, AuNPs were functionalized with a novel peptide coordinated with RBD, which has greater stability compared to ACE2 (human receptor for SARS-CoV-2). Various studies demonstrated strongly that these synthesized AuNPs can be fine contenders for antiviral agents against the COVID-19 virus.76
It has been determined that therapeutically inactive organic molecules when conjugated to gold nanoparticles can become strongly active drugs.74 AuNPs conjugated to peptide triazoles (AuNP-PT) showed a strong level of antiviral efficacy on HIV-1 compared to the parallel peptide triazoles. The stronger virologic activity and parallel irreversible HIV-1 deactivation via AuNP-PT are because of the multi-valent interaction between the nanoparticles and metastable envelope spike (S) proteins on the HIV-1 virus.77 Accordingly, the AuNPs enhanced the immune reaction by lowering the mRNA expression of interleukin (IL)-1 IL-6, the tumour necrosis factor (TNF)-α, IFN-γ, and the inducible nitric oxide synthase, which may be a notable alternative, mainly in antiviral therapeutics.78 Furthermore, gold nanoclusters (AuNCs) restricted and blocked the viral entry site of the respiratory syndrome virus. Precisely, histidine-functionalized AuNCs were active in mitigating the proliferation of the pseudorabies virus (PRV).79 Huang et al. produced a new gold nanorod-based HR1 peptide inhibitor (PIH-AuNRs) for MERS. This is a powerful HR1 inhibitor and functions on a selective basis for MERS-Cov S protein-mediated cell fusion with an IC50 of 1.171 μM. Specifically, PIH-AuNRs show a 10-fold higher inhibitory action than PIH and may present total cell fusion at 1.171 μM. Also, PIH-AuNRs demonstrated noteworthy biostability and biocompatibility in vitro and in vivo, thereby suggesting their wide utilization possibilities for the cure of MERS. Hence, PIH-AuNRs are favourable antiviral devices and may soon have a major effect on the progress of therapeutics for clinical use.80 Au@Ag nanorods (Au@AgNRs), which were made by coating AuNRs with silver, were seen to exhibit efficacy upon the multiplication of porcine epidemic diarrhea virus (PEDV).81 Viral titration analysis showed that Au@AgNRs could prevent PEDV occurrence by a magnitude of 4 orders after 12 h of infection, as confirmed via viral protein expression analysis. This portrayed that Au@AgNRs could suppress the entrance of PEDV and reduce mitochondrial membrane potential as well as caspase-3 activity. In addition, researchers portrayed that a huge rate of virus multiplication may result in the production of reactive oxygen species in cells. The released Ag+ and bare AuNRs by Au@AgNRs after the initiation by ROS have a remarkable antiviral action, which helps in providing large-scale suppression of the PEDV replication process. The results provided evidence that Au@AgNRs may be of use as a promising theranostic route to curb the multiplication of CoVs.
Focus should be placed on the stability of NPs, effective drug release in cells, decreased toxicity of medications after delivery, and targeting to certain tissues to make nanoparticle-based antiviral drug delivery successful. The entrapment of the mononuclear phagocytic system, which is found in tissues such as the liver and spleen, is another obstacle in the successful use of nanoparticles for medication delivery.82 It may be possible to overcome this obstacle by using biodegradable polymers such as PEG (polyethylene glycol) and PVP (polyvinylpyrrolidone). A change in the PEG modification distribution and a decrease in its in vitro toxicity can be achieved with the aid of gold nanorods. Besides, natural polymers such as dextran, chitosan, and pullulan, and surfactants such as sodium oleate and dodecylamine may aid in effective drug delivery.83 Drug distribution to particular targets is a significant difficulty because failing to do so can result in hazardous side effects in other tissues and cells. Accordingly, the use of virosome-based medicine delivery may aid in resolving this problem. Without a nucleocapsid and genetic materials, virosomes resemble enveloped viruses. Extracted glycoproteins have unique receptor binding and fusion capabilities, which enable them to bind to a cell surface receptor and deliver medicines to certain tissues and cells. Virosomes have recently been used to generate effective nanovaccines.84
Current study discoveries are being applied to investigate improved engineering moieties to control virus replications to build broad-spectrum antiviral medicines against different viral mutations. The design will focus mostly on the downsizing of characteristics relevant to various viral presentations.
Although researchers have explored diverse nanoparticles for several viral inhibitions and COVID-19, we believe there is a large scope in this niche for the utilization of noncytotoxic, biocompatible, nanoparticles, which are effective in inhibiting diverse COVID-19 strains.
Given that AgNPs are effective against diverse viral infections including COVID-19 according to the currently available literature, researchers can explore AgNPs in their pristine form, composites, and hybrid form for the inhibition of diverse COVID-19 strains and other viruses.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log) within 15 min.110
log) within 15 min.110
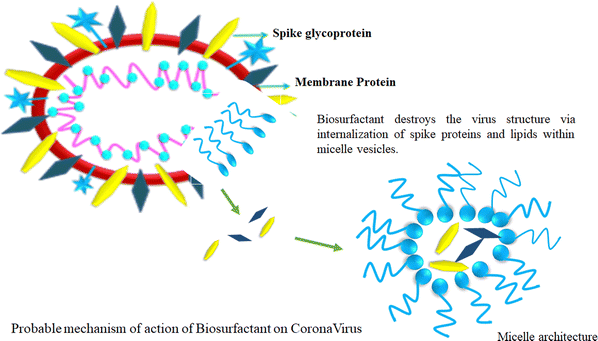
4. Biosurfactants
Biosurfactants have a remarkable trait in which they can form micelles above their critical micelle concentration (CMC). This justifies vigorous study looking at their embryonic applications in the prevention of viral spreading. The amphiphilic structure of biosurfactants suggests that their water-fearing layer can react with the lipid layer of the virus, while concomitantly reacting with hydrophobic substances such as H2O. This unique amphiphilic nature is the cornerstone of the biosurfactant structure, which is very beneficial to disrupt the structure of viruses (Fig. 7).111Biosurfactants possess certain interesting physicochemical properties by which they can maintain their integrity while being delivered as an aerosol. This mode of delivery endows biosurfactants with a drug carrier-like feature considering the lungs as the main area of action. The solubility aspect of biosurfactants in aerosols offers the advantage of enhanced bioavailability post-delivery. Besides, biosurfactants are self-soluble, which augments the proportionality of the drug dosage at the site of action, thus producing better results.112 Biosurfactant-mediated drug delivery offer benefits that are 2-fold in nature. This method of delivery not only provides a safe route for the drug to reach the target but also offers natural antiviral properties at the infection site. Biosurfactants also produce surfactant dysfunction at the alveoli, which is a probable scenario because of the ill effects of COVID-19 infection. Thus, biosurfactants offer healing by killing a host of viruses in the vicinity of a particular site, and consequently provides relief to patients. This aspect has attracted attention from the scientific community who are interested in the further investigation of biosurfactants as a probable treatment mechanism.
4.1. Liposomes
Liposomes are very attractive small spherical vesicles [15–1000 nm size] that originate from the presence of an aqueous core in phospholipids. Liposomes can entrap and produce both water-loving and water-fearing drugs. The liposomal lipid layer can shield the drug from degradation in the gastrointestinal tract and assist in its slow and sustained release.113 Herein, we describe the embryonic use of liposomes as vehicles for the specific delivery of antiviral agents. These may also act as immunomodulators to monocytes and macrophages for prophylaxis and treating viral disease.114 Simico et al. investigated the antiherpetic activity effect in vitro upon the liposomal inclusion of Artemisia arborescens L. essential oil. They prepared multilamellar (MLV) and unilamellar (SUV) (+) liposomes to check the effect of vesicle anatomy and constitution of liposomes on the antiviral action of the vesicle-incorporated oil. They obtained liposomes from hydrogenated (P90H) and dehydrogenated (P90) soy phosphatidylcholine. The antiviral action was investigated for herpes simplex virus type 1 (HSV-1) via a quantitative tetrazolium-based colorimetric method. The observations exhibited that Artemisia essential oil may be included in a significant quantity in the prepared vesicular dispersions. Antiviral screening indicated that the liposomal assimilation of the essential oil increased the in vitro antiherpetic action, chiefly for P90H vesicles (Fig. 8).115 | ||
| Fig. 8 Liposome: a spherical vesicle with an aqueous core formed by a phospholipid and cholesterol bilayer. | ||
Nanocarbon fullerene lipidosomes exhibited activity against the H1N1 influenza virus in vitro.116 Acyclovir-loaded flexible membrane vesicles revealed remarkable antiviral properties in a murine model for HSV-1 infection.117 Stearylamine (SA)-loaded cationic liposomes impeded viral infectivity in the absence of preloaded active pharmaceutical components. The antiviral action of SA liposomes against herpes simplex virus type 1 in A549 cells demonstrated similar activity to the antiviral drug acyclovir.118
5. Carbon-based nanoparticles
Carbon dots (C-dots) obtained from benzoxazine monomer curbed the infectivity of the flavivirus.119 C-dots bond with an amine, curbing HSV-1 from entering the host cells.120 Carbon quantum dots (CQDs) synthesized via the hydrothermal carbonization of citric acid restricted the entrance and multiplication of HCoV-229E.121 Graphene oxide and GQDs curbed the replication of HIV.122 GO-loaded hypericin worked well against the novel duck reovirus (NDRV).120,123 GO with curcumin restricted the respiratory syncytial virus posing as antiviral agents against hand, foot, and mouth disease.124Recent works in the literature have established the efficacy of carbon-based nanoparticles as well as quantum dots such as graphene nanoparticles,125 graphene quantum dots,126 and carbon dots127 for the inhibition of COVID-19. Even though progress has been made in this case, we believe that these findings are still in their infancy and there is more to be explored with these NPs in their pristine, chemically modified, composite, and hybrid forms.
5.1. Carbon nanotubes
Among the numerous nanomaterials, carbon nanotubes (CNTs) have achieved a paramount level of importance in antiviral therapy.128 CNTs are superior to other nanomaterials given that they can traverse cellular walls, transport drugs and molecules, including small interfering RNA, DNA, and proteins within different types of cells, such as T and cancer cells.129 Furthermore, CNTs possess multi-modular conjugation, which permits the inclusion of several functional groups on their exterior. This property gives rise to the supremacy of nanodrugs over the other streamlined agents.130 CNTs have demonstrated their effectiveness as HIV inhibitors through their structure-based design.131It has been reported that reactive oxygen species (ROS) generating compounds such as peroxides are deleterious to various viruses such as coronaviruses. For the fabrication of viral inactivation surfaces, metal-attached single-wall carbon nanotubes (SWCNTs) are very effective for the adsorption of H2O2. Aasi et al.132 investigated the capture of H2O2 molecules on metal-decorated SWCNTs using density functional theory (DFT). They observed that the metal-functionalized SWCNTs exhibited better adsorption capability than pristine SWCNTs. Finally, it was concluded that Ru-SWCNT and Rh-SWCNT composites illustrated excellent H2O2 adsorption. Moreover, they also revealed that Pt- and Cu-attached SWNCT-H2O2 composites have major potency as sieves for the eradication of viruses and their deactivation, and also have a large shelf-life (2.2 × 1012 and 1.9 × 108 years, respectively). This high adsorption of metal-attached SWCNTs and longevity of these nanoparticles imply that they are good contenders for synthesizing PPE kits against viruses.
5.2. Fullerenes
Fullerenes are carbon cage moieties consisting of a fused ring system of pentagons and hexagons. They are superb electron acceptors in the ground state and can reversibly accept up to six electrons.133 Fullerene nanoparticles have attracted substantial consideration by virtue of their distinctive physical and chemical characteristics. The possible reduction of viral activity using fullerenes has been postulated in various studies. Badireddy and team portrayed that under UV light, fullerol yielded singlet oxygen and superoxide. These two ROS deactivated the MS2 bacteriophage.134 Hotze and team shed light on the toxicity route of fullerol upon exposure to viruses. Fullerol conglomerates connect with the viruses and allow the effective passage of 1 oxygen molecule to the capsids, which results in notable oxidative changes to the capsid proteins and small changes to DNA and phospholipids. These changes reduce the ability of the bacteriophage to attach to the receptors on the body of their bacterial hosts.135 The inactivation of the bacteriophage may result from the reduction in the structural integrity of its capsid (deformation or rupture of capsid locally) and reduction in the ejection ability of genomic RNA in the bacterium.136 Fullerenes exhibit very low water solubility, which is the main drawback of their biological application. Hence, efforts should focus on making fullerenes water-soluble.137 Kraevaya et al. reported the preparation of various water-soluble fullerenes having promising anti-HIV activity.138 Du et al. synthesized a fullerene-liposome complex to investigate its antiviral activity against the influenza virus and performed an in vivo mouse model investigation.139 Their conclusions indicated that this complex is a very effective antiviral agent against the influenza virus in vitro and in vivo. They also studied the antiviral activities of fullerene against a few enveloped viruses such as HIV, human cytomegalovirus, SFV, and VZV. Based on their observations, they concluded that the fullerene-liposome may potentially play a crucial role in inhibiting influenza virus infection through a wide array of antiviral activities with low toxicity in the nanomaterial field.139Shoji et al. investigated 12 fullerene derivatives to detect new efficient anti-influenza compounds using an in vitro PA endonuclease inhibition assay. Eight derivatives of fullerene were recognized to inhibit the endonuclease activity of the PA N-terminal domain or full-length PA protein in vitro. Silico docking simulation analysis of C60 fullerene and PA endonuclease was performed, implying that fullerenes can be the active cavity of PA endonuclease.140 Kraevaya et al. reported the design of a water-soluble C60 fullerene derivative attached with 5 residues of phosphonic acid to investigate its antiviral activity.141 Muñoz synthesized a chain of amphiphilic glycodendro142 fullerene monoadduct to investigate the antiviral efficiency of Ebola virus glycoprotein (Ebo GP) pseudo-typed viral particles on Jurkat cells overexpressing DC-SIGN. They discerned an enhancement in the IC50 value compared to various systems furnished with more carbohydrate groups.143
5.3. Graphene
It has been well established in various reports that bacteria coming in contact with a graphene surface [a two-dimensional material] lose their integrity. However, although graphene-based virus-related research has focused on making suitable sensors, the effects of graphene on viruses have not been well-established. Stanford et al. reported a self-cleaning filter by counting laser-induced graphene (LIG), which was capable of capturing bacteria and particulates to reduce the transmission of nosocomial infections. The rapid growth of bacteria was stopped on the filter and by raising the temperature of the filter to above 300 °C, all the pathogens including bacteria, along with molecules were destroyed.144Gedanken et al. synthesized reduced graphene oxide (SMRGO) functionalized with sulfonated magnetic nanoparticles (MNPs) to entrap and demolish type 1 herpes simplex virus (HSV-1) photothermally.145 They observed that SMRGO portrayed great (∼99.99%) photothermal antiviral action by radiating the compound with near-IR light (NIR, 808 nm, 7 min). The distinctive planar structure, capturing efficiency, large surface area, and outstanding photothermal attributes of graphene are responsible for this antiviral activity. In addition, the electrostatic interaction of MNPs with viral agents played a crucial play in significantly reducing infection. GO films enable the easy fabrication of breathable barrier layers in fabrics. Steinberg et al. reported GO membranes for preventive appliances designed to restrict the entry of chemical toxicants or biochemical agents from the environment except for outward perspiration.146 Protective facemasks made of graphene could be recycled by the application of heat or photocatalysis method.147 In addition, graphene-coated textiles and sieves that touch viruses can be sterilized by heat and electronic transduction. Furthermore, the denaturation process of viruses after adsorption on the surface of graphene and low thermal treatment at 56 °C for 30 min was demonstrated. Yang et al. designed a β-cyclodextrin (CD)-decorated graphene oxide (GO) composite, which demonstrated a very good level of antiviral action and could also load curcumin effectively. These researchers employed RSV, a negative-sense single-stranded enveloped RNA virus, as the control to investigate the antiviral properties of multifunctional GO. The results revealed that the composite, which possessed inhibitory and pathological effects on the virus, could avert RSV infection to the host cells by deactivating the virus and preventing viral attachment.124
6. Polymer drug conjugates
A covalently bound therapeutic agent and a polymer constitute a polymer-drug conjugate, where the therapeutic agent can either be a large molecule such as a protein or a small molecule.148 The philosophy behind conjugation is to attain greater efficiency via enlarged plasma stabilization and also safety via targeted delivery.149Many polymers have inbuilt antiviral potential and their complexation to antiviral drugs may work synergistically. Conjugation of interferon α2A and α2A with polyethylene glycol (PEG) worked on HCV.150 Polymers having sialic acid attachment were synthesized and known to be outstanding inhibitors of viral entry via multivalent attachment to hyaluronic acid (HA) and NA on the wall of viruses.151 In 2018, Andersen et al. synthesized albumin–polymer–drug composites to guarantee an increased residential time and lymphatic accumulation, providing a suitable ratio of bare drugs of ART to primary human T cells, which portrayed high shielding from HIV infection compared to individual ART drugs. N-(2-Hydroxypropyl) methacrylamide (PHPMA) was the used polymer.152
When added to the right drug delivery platforms, cell-penetrating peptides (CPPs) such as TAT-peptide have been reported to show promise as immune boosters.153 In comparison to other translocation and delivery methods, CPPs have a lot of benefits, including being simple to create, affordable, and often having little cell toxicity with no immune reaction. To combat COVID-19, it is deemed important to develop drugs against SARS-CoV-2. Available works in literature have shown beyond doubt that polymeric–drug conjugate systems are an effective approach for combating COVID-19.153 However, although these findings are interesting, there is a lot to be investigated within this niche given that it is in its budding phase or development stage. Researchers can wisely adopt diverse CPPs and other polymeric macromolecules in this case towards containing COVID-19.
7. Quantum dots
“Quantum dots (QDs)” or “semiconductor nanomaterials” attached to major fluorescent probes are important for the identification and persistent fluorescence imaging of many cellular activities.154 The major tactic related to suppressing SARS-CoV-2 infections with QDs may interest researchers substantially. The major reason for using QDs is because of their detectability at a specific wavelength of light. Also, QDs may be modified to the required size (1–10 nm) and shape to attack SARS-CoV-2 within a range of 60 and 140 nm in size. Now, the positive charge on the surface of carbon-based QDs is employed to curb the S protein of SARS-CoV-2.155 In addition, the positive surface charges of QDs interplay with the (−) RNA chain of the virus, producing ROS in SARS-CoV-2.156 Carbon dots (CDs) initiate interferon-activated genes, mainly interferon-α production, which are known to curb viral multiplication. Subsequently, the attachment of required moieties with QDs may interplay with entry receptors of CoV-2 and inhibit genomic multiplication.157 The antiviral efficacy of CDs obtained from 4-aminophenyl boronic acid hydrochloride (4-AB/C-dots) portrayed inhibitory action against herpes simplex virus type 1, especially flaring in the initial stage of viral action.158 To inspect the antiviral ability of less toxic QDs, Du et al. explored the viral inhibition effects of CDs minutely.159 The virucidal ability of CDs was seen to be close to their carbon predecessor and formation methods, indicating the requirement of systematic probes of the antiviral action of various carbon dots. This team created a simple but low-cost route to produce stable CDs with antiviral attributes and explored their consequence on viral multiplication via PRV and PRRSV as model structures of DNA and RNA viruses, respectively. The obtained results indicated that CDs can remarkably suppress the replication of both PRV and PRRSV. Therapy using CDs could also drastically initiate the production of endogenous interferon and expression of interferon-stimulating genes (ISGs), which curb virus multiplication.159 Recently, Łoczechin et al. reported that various CDs made via hydrothermal carbonization and complexation with boronic acid (carbon quantum dots-3) show antiviral effects towards the extremely deadly COVID-19 virus in a dosage-dependent way.121 Also, CDs upscaled the expression of several ISGs, which are essential in the CD-based antiviral pathway.160 In addition, Huang et al. developed benzoxazine monomer-obtained carbon dots (BZM-CDs), which exhibited virus-curbing ability towards flaviviruses and other non-walled viruses, such as porcine parvovirus and adenovirus-associated virus.119 Earlier studies showed that the antiviral efficacy of functionalized CDs can be enhanced via suitable surface synthetic changes. For example, Szunerits et al. reported that carbon dots surface-modified with boronic acid or amine groups can fight herpes simplex virus type 1 (HSV-1) by curbing its entry into host cells.120 However, the in vivo toxicity of QDs is a noteworthy drawback of this antiviral agent.Considering the above discussion, the modification of QDs and other experimental pathways using new functional groups against SARS-CoV-2 will give rise to nanotherapeutics for COVID-19 for the obliteration of this pandemic in the near future.161
8. Polymeric nanoformulations
Natural hydrophilic and synthetic hydrophobic polymers are mostly employed for the synthesis of nanoformulations. The use of surface-modified polymers helps to decrease the unspecific reaction with serum proteins and facilitates phagocytosis to manifest commendable changes in the pharmacokinetic profile of drugs. The design of polymer nanocarriers also plays a major part in deciding the manner of drug release with the aid of various sources of stimuli such as chemical, pH variation, magnetic field, outside heat source, and impingement of light. Thus, these methods have improved the bioavailability of drugs by reducing the effects of toxicity at non-specific sites and drug degradation prior to reaching the site of interest.162 Certain essential criteria that need to be considered prior to polymer selection are: (i) nanoparticle size; (ii) drug release profile from the polymer; (iii) physicochemical properties of the drug of interest; (iv) aspects of biodegradation ability, safety, and biocompatibility; (v) drug effectiveness and antigenicity; and (vi) surface morphology and functionalities.163 The various types of polymer-based nanoformulations include nanocapsules, nanospheres, polymeric micelles, and solid polymeric nanoparticles.8.1. Micelles
Micelles have a size in the range of 10 to 100 nm. They are composed of an inner hydrophobic core (holds water-insoluble drugs) that is enveloped in a hydrophilic polymer with an outer layer (such as PEG, which increases their circulation time, and subsequently improves their aggregation). A common example is polymeric micelles, which are well-known drug delivery agents, providing remarkable therapeutic efficacy. The encapsulation of drugs in polymeric micelles is important nanotechnology employed to develop water-soluble and stable drugs. Another reason to use micelles in drug formations is that they dissociate slowly, hence making way for higher drug retention, and also increased aggregation of the drug at the desired location.9. Implication of nanotechnology to mitigate the cytokine storm
Cytokines are a class of signalling molecules that moderate and control the human immunological system. Viruses such as COVID-19 trigger a cytokine storm in the human system called cytokine release syndrome (CRS), which emanates due to unrestricted immune reactions. The inflammatory cytokine storm is the main reason for acute respiratory distress syndrome (ARDS), resulting in a multiple-organ failure, which is one of the primary reasons of death in critical patients. It is noble that extensive current research on drug trials on interleukin (IL)-6, which block its receptor site (Tocilizumab, an anti-IL-6 receptor antibody, and Sarilumab) or IL-6 itself (Siltuximab), are being conducted. On account of the primary nature of macrophages in COVID-19, ACE2-expressing CD68+CD169+ macrophages comprised of the SARS-CoV-2 nucleoprotein antigen portray a higher discharge of IL-6 in the affected lymphatic system.167 The cells and tissues affected by SARS-CoV-2 also exhibit the upregulated expression of Fas, denoting the role of CD169+ macrophages in the significant pathogenesis of viral replication, anomalous inflammation, and lymphocyte apoptosis. Enlarged alveolar exudate caused by the elongated neutrophils and monocytes passing into the lung arterioles containing fibrin deposits was been studied in biopsy slides of affected patients. The state-of-the-art of Nanomedicine may be highly promising in this extensive area by inducing the specificity/efficiency of immunosuppressant delivery-targeting immune cells, followed by minimization of drug overdosage and subsequent possible side effects.168 The distinct architecture of Nanotool design can substantially circumvent the immune system, thereby tuning the surface charge and providing favourable room for drug encapsulation or loading. Carbon-based porous nanostructured materials, mainly graphene, has excellent potential to rummage the CRS from the serum via an extracorporeal perfusion method. In addition, hierarchically porous carbon nanomatrices showed efficacious adsorption of several cytokines, such as IL-6 and TNF-α. Through the impressive application of nanotechnology and nanomedicine, we can envision therapeutic modalities and target specific immune subpopulations by utilizing a fascinating immunomodulatory platform.16910. Reusable and recyclable masks
One-time-use surgical masks or respirators can prevent oral droplets containing viruses from entering the lungs, aiding in the reduction of the chance of being infected, together with the maintenance of proper hygiene practices.170 Nevertheless, conventional masks have a few limitations including surface hydrophobicity, reusability, and recyclability. Thus, to overcome these limitations, the latest nanostructured materials and polymeric fibres, fluorinated polymers, metal nanowires, and graphene have been explored as superhydrophobic coatings (Fig. 9).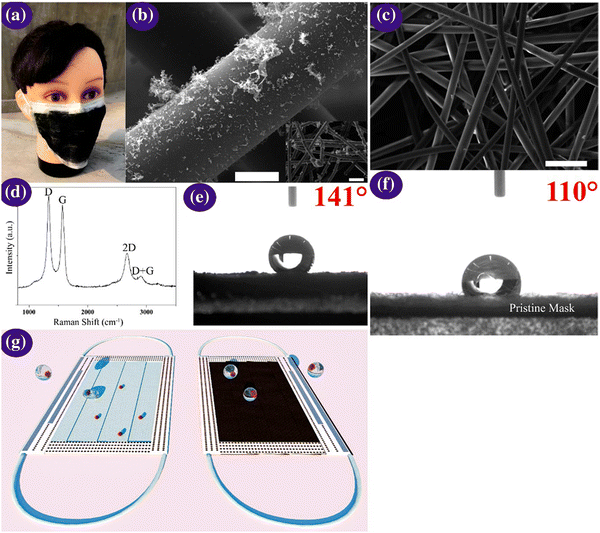 | ||
| Fig. 9 (a) Optical micrographs of the laser-modified graphene mask. (b) Scanning micrographs of the graphene layer nonwoven fibre matrix on a surgical mask; scale bar is 10 μM. The inset is a zoomed-out image, with the scale bar of 100 μM. (c) Scanning micrographs of the nonwoven fibre matrix derived from a surgical mask; the scale bar is 100 μM. (d) Raman spectrum of the graphene layered mask. (e and f) Measurement of contact angle on the graphene layered and bare mask and (g) self-cleaning property analysis of the black graphene-coated mask compared to a pristine surgical mask. Illustration of the self-cleaning properties of the black graphene-coated mask (right) compared to the pristine blue mask (left). Reproduced with permission from Zhong et al.171 Copyright 2020, American Chemical Society. | ||
Super-hydrophobic surface coatings on N95 masks can be made possible via laser-induced graphene materials, which are adaptable, scalable, and cost-effective in comparison to the market predecessors, such as polyimide, SPEEK, and Bakelite. Zhong et al. formulated a double-mode laser-induced forward transfer process for accumulating graphene in a commercial surgical mask.172 The decontamination stratagem is improved via the usage of nanostructured and layered graphene layers, having a static contact angle of more than 140°, wherein the enhanced superhydrophobicity facilitates low adherence of the virus droplets on the mask surface. In addition, solar illumination induces surface sterilization by the prompt increase in the surface temperature to about 80 °C.171 Meanwhile, plasmonic photothermal and superhydrophobic coatings on N95 respirators provide profound protection compared to existing PPE. The coating of silver nanoparticles results in extra shielding through the decontamination effect of the silver ions on microbes. The synergistic landscape of nanostructured composite coatings can encourage the scientific community to miniaturize advanced personal protection equipment (PPE) in the future to fight the COVID-19 pandemic.
11. Exploration of nanovaccines
Nanotechnology has embraced both prophylactic and various therapeutic approaches to combat the outbreaks of fatal diseases and respiratory infectious illnesses. In general, nano-vaccines are a novel troop of vaccines, wherein nanoparticles (NPs) are used as functional vehicles. Conventional vaccines exhibit various constraints regarding weak immunogenicity, inherent in vivo instability, toxicity, and require various multiple administration profiles. Nanotechnology enabled the design of vaccines favouring the cellular immune response, which can assist the advanced phagocytosis process associated with lymphoid tissue. Now, this ultimately leads to enhanced antigen detection to the receptor site and virus penetration.173 The current onset of respiratory contagious diseases is inspiring the development of novel antigen formulations and a new blueprint for viral vaccination based on an emerging nanotechnology-driven approach. Likewise, to upgrade the route of vaccine administration and their delivery to smaller and remote villages in countries around the world, it is necessary to boost the frequency of vaccinations and storage capability of vaccines, which is in the early stages of implementation. Typical vaccines have been fashioned to exploit compromised or live-attenuated organisms (1st generation), but subunit and RNA/DNA vaccines (2nd and 3rd generations, respectively) are pragmatically oriented to advocate preventive immunity from infectious diseases. Surface tailoring with nanocarriers with targeting moieties offers opportunities and facilitates specificity and selectivity in the immune responses by attacking specific surface receptors of various immune cells.174–176The exploration of nano-vaccines minimizes the adversity arising due to cold chain transportation and storage stability given that nanoformulations can be easily lyophilized, leading to an increased shelf-life. Furthermore, the unique size of nanoparticles, which is comparable to that of viruses and bacteria, aids in the prompt recognition of immune systems. Versatile nanostructured carriers, including non-metal NPs, QDs, carbon nanomaterials, polymeric NPs (chitosan, poly(lactide-co-glycolides) (PLGA), poly(glutamic acid) (PGA), metal/metal oxide NPs, silica NPs, carbon black NPs, liposomes, dendrimers, solid lipid nanocarriers, and virus-like particles (VLPs)) have been extensively synthesized and scrutinized to transport various compounds, e.g., drugs, proteins/peptides, DNA/RNA, antibodies, and vaccines, due to their usefulness in antigen therapeutics. Considering the fascinating structural architecture of nanoparticles, they have been widely admired as vaccine adjuvants and utilized in vaccine delivery. In this context, VLPs deliver repetitive immune viral epitopes to initiate a specific, strong immune reaction consisting of humoral or cellular immunities.177,178 Interestingly, the first VLP nano-vaccine licensed for individual convenience was HBsAg (hepatitis B surface antigen). Epaxal®, an FDA-approved liposomal vaccine, is used to prevent HAV and is pragmatic together with immune potentiating reconstituted influenza virus (IRIV).
Liposomes (virosomes) were used to make aluminium-free vaccines made from formalin-deactivated HAV (strain RG-SB). The liposome-based vaccine HepaXen was designed initially with the target to counter hepatitis A, C, and E with a focus on antiviral action. An immune response of 20-fold greater than a popular prophylactic vaccine was attained by a combination of reconnected hepatitis B and plasmid DNA encoding. Another vaccine, Inflexal V, is an approved virosomal adjuvanted vaccine for influenza that possesses good biocompatibility and has the capability to mimic natural infection.
In current trends of vaccination, mainly live-attenuated vaccines exhibit relapse jeopardy to their pathogenic virulence in specific immunocompromised conditions. To combat COVID-19, avant-garde vaccination aims to target the transmembrane spike (S) glycoprotein, which is responsible for host cell penetration. This prompt coupling of human ACE2 with SARS-CoV receptors triggers the chain of transmission in large human populations. Vaccination of mice with CoV S NPs resulted in the production of significant yields of neutralizing antibodies contrary to the homologous virus.179 An adjuvant can play a prime protagonist to instigate the production of neutral antibodies to the COVID-19 S protein. Thus, a nanotechnology-driven innovative approach can open a new avenue of vaccination, which can restrict the CoV pathway and multiplication in in vivo systems. Amongst the wide spectrum of nanoparticles, silver and gold NPs are employed as adjuvants to accelerate the IgA/IgG specific antigen.180,181 Furthermore, the nonvascular architecture of spike proteins (S proteins) can also be explored together with research focused on the development of NP-based vaccination systems against CoV diagnostic theranostic schemes. Considering the published report of the genomic analysis of SARS-CoV-2 on January 11, 2020, extensive investigation has been carried to design vaccines against COVID-19. The first vaccine Phase I human clinical trial was initiated on March 16, 2020,182 and subsequently, 59 novel patients entered the trial quickly as of May 22, 2020.183
Immune-targeted nanotherapeutics can be explored by designing nanomaterials at the nanoscale, which are proficient in magnifying the host's immune reaction, including adjuvants in the milieu of vaccination. Progress of these vaccines will be based on the interactive pattern of viral antigens and immunogenic spike protein (S1), targeted via antibodies of restorative patients who have recovered from the virus.184,185 Successful vaccination requires a suitable type of polymeric conjugate relevant to the target antigen. In this context, lipid and lipid-based NPs can be adopted as a delivery platform for mRNAs or siRNAs to facilitate the design of major viral proteins for vaccines or for disabling viral antigenic target genes.184,185 Several different companies are conducting developmental research on mRNA-based vaccines by entrapping SARS-CoV-2 proteins such as the spike protein encoded in nanoliposomes having particular physical and chemical attributes, which are possibly lower than that known for vaccination from some tumour antigens. On March 16, 2020, Moderna and the Vaccine Research Center at the US National Institutes of Health (NIH), in partnership, began with early volunteers in a Phase I clinical trial, testing and evaluating an mRNA vaccine (mRNA-1273) encapsulated in lipid NPs. This was completed within a given time of only 63 days after sequence selection (NCT04283461). The enrolment of the first group of volunteers (18 to 55 year-old healthy subjects) was completed on April 16, 2020.
Another study inferred the exploration of SARS-CoV-2 nanodrugs by vaccinating camelids (llama or alpaca) with target antigens such as RBD, S1, or S2 of the SARS-CoV-2S protein. Afterward, the vaccinated nanodrug reserves can be entrenched and filtered. The attachment of nanodrugs to the RBD can prevent its receptor from attaching ACE2, thereby restricting viral attachment and entrance. The attachment of nanodrugs to S1 or S2 may prevent the S protein from transforming to its post-fusion conformation, thus restricting membrane fusion and viral entrance.186
Tao et al. aimed at vaccine development against influenza viruses including H1N1, H3N2, and H5N1, utilizing gold nanoparticles coupled with matrix protein 2 (M2e).187 Additionally Novavax, an adjuvant, entered a Phase III clinical trial, showing 89% efficacy to recombinant SARS-CoV-2 glycoprotein-mediated NVX-CoV2373.188 The Novavax COVID-19 vaccine demonstrated 89.3% efficacy in a UK Phase 3 Trial.189 The virus-like protein (VLP) instigates the self-assembly of different genetically engineered viral structural proteins, specifically amplifying the humoral and cell-mediated immunity (AMI and CMI). Currently, extensive research work is underway to develop monodispersed VLP-based vaccine formulations based on commercialized prophylactic against both the hepatitis B virus (HBV) and human papillomavirus (HPV). For instance, Nicotiana benthamiana has been explored for the development of genetic VLP architecture vaccines, modulating the spike proteins of SARS-CoV-2. A trial run has been initiated with vaccine candidates by Medicago and SpyBiotech (Serum Institute of India) for commercialization,190 and SpyBiotech and Serum Institute of India, announced that the first subjects have been dosed in phase I/II trials of a novel virus-like particle vaccine targeting COVID-19.191
Owing to the scalability, sustainability, and prolonged expression of antibodies, nucleic acid-based vaccines and encoded antigenic peptides boost the immune response against viral proteins. However, their major drawbacks include the lack of stability and low cellular internalizations under several physiological conditions. Thus, to address this issue, researchers have focused on the development of DNA vaccines and clinical trial runs were carried out under the active guidance of Inovio Pharmaceuticals/International Vaccine Institute, USA,192 Phase II/III Study of COVID-19 DNA Vaccine (AG0302-COVID19),193 Study of COVID-19 DNA Vaccine (AG0301-COVID-19),193 Cadila Healthcare Limited (India),194 which was predicted to hit the market on June 13 May, 2021,195 Genexine Consortium (South Korea),196 Entos Pharmaceuticals Inc. (Canada),197 GeneOne Life Science, Inc. (South Korea),198 University of Sydney, Bionet Co., Ltd. Technovalia (Australia),199 and Takis/Rottapharm Biotech (Italy).200
Importantly, to bifurcate the insertional mutagenesis, Shim et al. and Zhao et al. prepared polyethyleneimine and polyethyleneimine-stearic acid (PSA) cationic nanomicelles, respectively, for the delivery of mRNA encoding HIV-1 gag to dendritic cells.201 Another vaccine candidate, ARCoV-based on a lipid-nanoparticle, targeted the receptor-binding domain (RBD)-encoded site of SARS-CoV-2, leading to the successive release of antibodies for neutralizations.202
Another prime consequence in the phylogenetic tree map of SARS-CoV-2 is that many variants have emerged, including delta (B.1.617.2), alpha (B.1.1.7), beta (B.1.351), omicron (B.1.1.529) and gamma (P.1). Instead of the variant positions in the S gene, the low immunogenic E sequence may be considered as a prominent targeting site for vaccine development. Current trends imply that the continuous mutation of SARS-CoV2 is quite alarming wherein variants can subsequently infect the vaccinated community, thereby leading to a collapse in herd immunity. Consequently, the NCBI omicron variant with altered genetic mutations showed significant effects and mild breakthrough infections in vaccinated people.203
Alternatively, nasal vaccination is a promising method for stimulating mucosal immunity in a significant manner. Nasopharynx-associated lymphoid tissue (NALT) tactically disseminated in the nasopharynx and oropharyngeal regions and antigens accumulate NALT through dendritic cells and APC (antigen-presenting cells), inhibiting the spike protein and thereby neutralizing SARS-CoV in Dc cells (Table 1).204
| Developer | Commercial name | Vaccine | Efficacy towards variants | Ref. |
|---|---|---|---|---|
| Pfizer-BioNTech COVID-19 Vaccine + Fosun Pharma | Comirnaty | Lipid nanoparticle mRNA vaccines (BNT162b2) | IV NCT04844489; 91% protection against symptomatic; 50% drop after booster for omicron variant | 205 |
| Moderna Therapeutics Inc. and the National Institute of Allergy and Infectious Diseases (NIAID) | Spikevax | Prefusion stabilized S protein mRNA-1273 vaccine | IV NCT04900467 94% against any type of COVID-19, being 98% effective in preventing severe disease and 63% effective | 206 |
| AstraZeneca and University of Oxford | Vaxzevria | ChAdOx1-S Chimpanzee Adenovirus encoding the SARS-CoV-2 spike glycoprotein | IV NCT04760132 76% against symptomatic COVID-19; 40% against delta variant | 207 |
| Janssen Pharmaceuticals, Companies of Johnson Johnson | COVID-19 Vaccine Janssen | Ad26.COV2.S Adenovirus type 26 encoding the SARS-CoV-2 spike glycoprotein | 77% after 28 days from single intramuscular immunization and 100% after two doses | 208 |
| Novavax | Nuvaxvoid | SARS-CoV-2 recombinant spike protein | III NCT04583995; vaccine efficacy of 90%; in severe moderate people 100% | 209 |
| Russian Gamaleya Institute's | Sputnik V | Noviral vector-based vaccine | 80% effective | 210 |
| Bharat Biotech | BBV152 | BBV152 Inactivated vaccine | 70% effective | 190 |
| Sinovac | CoronaVac | CoronaVac Inactivated vaccine | NA | 211 |
| Johnson & Johnson | JNJ-78436735 nonreplicating viral vector | NA | 212 |
Another pivotal aspect regarding the significant effect of a booster dose is illustrated herewith. Various types of COVID-19 vaccines including mRNA, viral vector, inactivated, and protein-based vaccines are effectively practiced and highlighted as boosters in the community. Boosters can facilitate the neutralizing antibody levels (nAb) and their surrogate markers (anti-spike IgG).41 In the case of an immunocompromised individual, a booster dose modulates the cross-reactivity, which further enhances the T cell responses. In summary, the findings signify that the amalgamation between vaccine-induced immunity with the emergence of SARS-CoV-2 variants has ultimately piloted breakthrough COVID-19 infections, especially among elderly and immunocompromised individuals having co-morbidities.41,213
12. Gene editing using CRISPR/Cas technology: cutting-edge technology towards targeting SARS-CoV2
CRISPR/Cas9 technology originated based on a bacterial defence mechanism against invading viruses. This has now cast off to inhibit persistent viral infections by integrating their genome into the human genome of the host cells.214,215 However, genetic remodelling of viruses can evolve to the emergence of new variants highly resistant to CRISPR/Cas9 or other therapeutic agents, thereby boosting the pathogenic index of the virus. Henceforth, variable genomic site-targeting using carrier-free CRISPR/Cas9 schemes can be predominantly in demand for in vivo therapeutic applications (Fig. 10).In 2018, Streptococcus pyogenes Cas9 (SpCas9) mRNA and modified single-guide RNA (sgRNA) were grafted onto lipid-based NPs (LNP-INT01). Afterward, testing was carried out in mouse models, wherein this effective biodegradable nanosystem edited the transthyretin (Ttr) gene in the mouse liver with a reduced 97% efficacy rate in the TTR serum protein levels.216 A similar study revealed the significant contribution of TT3 lipid-like nanomaterial (LLNs)-induced CRISPR/Cas9 to knock down the subtilisin/kexin type 9 (Pcsk9) gene and reduce the developmental expression of HBV DNA in a mouse model. Besides, TT3 lipid-like nanomaterials (LLNs) were also explored and found to instigate the CRISPR/Cas9-mediated knockdown of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene, thereby reducing the expression and genetic deletion of targeted HBV DNA in mice. Upon mutation, Leptotrichia wadei (LwaCas13a)-derived Cas13a and Prevotella sp. (PspCas13b)-derived Cas13b can be employed to target endogenous transcripts in human embryonic kidney HEK293 cells and mammalian cells, respectively. RNA viral genome obliteration is associated with Cas13d, a type VI RNA-guided nuclease, which could cleave the RNA genome of the virus without damaging the human transcriptome.217
Various studies have exploited CRISPR/Cas13d gene-editing technology based on gRNA containing spacer sequences that are complementary to the S gene, ORF1ab, and Cas13d protein, whereas the pCX539 backbone cloned sequences transfected into cultured HEK293T and N2A cells mediated by the Cas13e gene-editing system.218,219 Another avant-garde approach was concocted based on the CRISPR/Cas13d system, wherein the PAC-MAN strategy relies on the 22 CRISPR RNAs (crRNAs) to target all the different RNA-dependent RNA polymerase (RdRP) genes as well as the nucleocapsid gene sequence in the lung epithelial cell line (A549) to treat Covid-19. Further CRISPR/Cas systems for Covid-19 gene editing can be transfected by either AAVs or lipofectamine.220 To tune the immunogenicity and efficiency of AAV in in vivo systems, CRISPR/Cas systems in NPs may lead to the introduction of safer and highly-efficient gene-editing platforms for modulating the spatio-temporal release of therapeutic agents.219,221
13. Nanotechnology approaches for antiretroviral drugs delivery
To date, the present antiviral treatment has still not framed the epitome and adaptability given that ambiguity arises due to the penetrating efficiency in intricate biological barriers. Nanotechnology can be critically scrutinized in this regard. Nanotechnology has promoted remarkable possible opportunities in healthcare settings and in various diagnostics fields. The various fascinating features of NPs include their nanometric size222 and huge surface area to volume ratios to achieve a large drug loading.223 Surface functionalization that enables cellular membrane passage serves to promote NPs as a good candidate as drug delivery vehicles. Also, the versatile properties of NPs, including bioavailability,224 virucidal activity, enhanced biomimetic properties,225 and improved specificity, favour antiviral delivery and sustained drug release to the target site and decrease the emergence of drug resistance, enhance the personalized therapeutic prospect, and reduce adverse drug side effects. To formulate wide-spectrum antiviral agents against various viral mutations, current research findings are being employed to explore advanced engineered moieties to subdue virus replication. The design will be mainly dedicated to the miniaturization of features pertinent to various presentations of the virus.14. Conclusion and future perspective
To mitigate the COVID-19 catastrophe worldwide, which poses an unprecedented threat and has had major negative impacts on the global arenas including healthcare systems, nanotechnological intervention is intrinsically furnishing advanced devices and schematic pathways for the unprecedented search for a full understanding of the pathophysiology of various viruses. The one-of-a-kind architecture of nano-tailored sensing platforms and various nanocarrier systems such as nanotraps, nanorobots, nanobubbles, nanoribbons, and nanodiamonds is making the way in the diagnosis, prevention, and therapeutics of viral infections. Nanotraps are potentially fashioned to rummage the targeted infectious virion and viral proteins via water-insoluble and coulombic interactions. Astonishingly, miniaturized nanorobots consisting of polymeric/biopolymeric materials including biomimetic substances that prompt pathogen detection, genome editing, and act as a therapeutics delivery vehicle can be noteworthy for the early detection and eradication of pathogens. Future endeavours can be triggered to design nanofibers, nanodiamonds, and nanobubbles as nanotraps for the specific delivery of numerous drugs, giving us a wide range of theranostic tools for disease diagnosis and related comorbidities in a heterogeneous population. Further research on an upscale immune reaction and the use of nanoparticles as an adjuvant to antiviral vaccines provide remarkable solutions to prevent and control viral infections.14.1. Future traits
COVID-19 has brought the world to its knees, where viruses such as COVID are known to mutate with time. The progression of this pandemic has not been significantly impacted by various SARS-CoV-2 strains, but it can in the future. David Montefiori questioned whether COVID-19, the fatal virus that caused the epidemic this year, would be evolving as it travelled from person to person. Montefiori is a virologist who has devoted most of his career to understanding how HIV can avoid the immune system due to unintentional mutations. He reasoned that SARS-CoV-2 may have a similar outcome.Montefiori, who oversees the Duke University AIDS vaccine research lab in Durham, North Carolina, contacted Bette Korber, a recognized authority on HIV evolution and a long-time collaborator, in March. As coronavirus spread around the world, Korber, a computational biologist at the Los Alamos National Laboratory (LANL) in New Mexico, had already begun searching thousands of coronavirus genetic sequences for mutations that may have altered the characteristics of this virus.
SARS-CoV-2 is evolving much more slowly than HIV as it spreads. However, for Korber, one mutation jumped out. Korber found that in the gene that codes for the spike protein, which facilitates viral particle cellular entry, a mutation repeatedly showed up in samples from COVID-19 patients. The amino acid aspartate (D, in biochemical shorthand) was routinely being substituted by glycine (G) due to a copying error, which changed a single nucleotide in the 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 903-letter RNA coding of the virus. This substitution occurs at the 614th amino-acid position of the spike protein, which is known as the D614G mutation by virologists. In a preprint uploaded to the bioRxiv server in April, Korber, Montefiori, and others issued a warning that “D614G is increasing in frequency at an alarming rate”. It quickly took over as the main SARS-CoV-2 lineage in Europe before spreading to the US, Canada, and Australia. The research stated that D614G indicated a “more transmissible variant of SARS-CoV-2” that had evolved as a result of natural selection.
903-letter RNA coding of the virus. This substitution occurs at the 614th amino-acid position of the spike protein, which is known as the D614G mutation by virologists. In a preprint uploaded to the bioRxiv server in April, Korber, Montefiori, and others issued a warning that “D614G is increasing in frequency at an alarming rate”. It quickly took over as the main SARS-CoV-2 lineage in Europe before spreading to the US, Canada, and Australia. The research stated that D614G indicated a “more transmissible variant of SARS-CoV-2” that had evolved as a result of natural selection.
The creation of “variants of concern” is the result of novel sets of mutations, where mutations that affect the properties of a virus, such as its transmissibility and antigenicity, may happen as a result of changes in the immunological profiles of the human hosts. Since many of the most recent SARS-CoV-2 variants are linked to increased transmission, concerns have been raised about the types and rates of viral changes.226 The virus has fewer opportunities to mutate the less it distributes.227 SARS-CoV-2 may continue to change in ways that both hasten viral transmission and lessen the effectiveness of vaccines. Infected individuals may also spread the infection even before showing any symptoms. Additionally, many persons who receive a vaccination but subsequently contract an infection may not exhibit any symptoms and will inadvertently continue to spread the virus to others,228 making it difficult to stop its transmission. We will probably be dealing with this pandemic for years to come if we give up on methods to stop the spread and allow the virus to spread unchecked.
It is not strange to see new versions of viruses develop because viruses constantly mutate. Since the start of the pandemic, the mutations-Y145H and A222V-have been identified in numerous coronavirus lineages. The formation of a novel variant is characterized by an accumulation of mutations that dramatically change the characteristics of the virus lineage. Five SARS-CoV-2 variants have been identified as of December 2021, including the alpha variant (previously known as the UK Variant and referred to as B.1.1.7), the beta variant (previously known as the South Africa variant and referred to as B.1.351), the gamma variant (previously known as the Brazil variant and referred to as P.1), the delta variant (previously known as the India variant and referred to as B) (formerly called the South Africa variant and referred to as B.1.1.529). All are said to have transmission rates that are noticeably higher than lineages from prior times.
According to reports, the prevalent delta variant can replicate roughly 1000 times more than its ancestors and exhibits immune-evasive characteristics.229 According to current research, coronavirus antibodies made for the original strain (Wuhan strain) may only be half as potent as those made for the delta variation.230 Even worse is that those who have the delta strain of the virus are shedding a lot more virus. The host's immunity to the virus may be impacted when the viral dosage received is higher.231 Recent research suggests that the effectiveness of the existing vaccinations, which were created to target the spike protein of the original virus, varies depending on the variation.232 The spike protein seems to be reasonably stable even for the delta version, despite the possibility of some slight alterations. Another variation, known as AY.4.2 and dubbed “delta plus” by some, has been discovered in the UK and has the potential to spread more rapidly than the standard delta.233 Although most covid infections in the UK are still caused by the standard delta, “ delta plus” infections are on the rise. Although there is currently no proof that it results in worsening of the sickness, the most recent official data indicate that 6% of covid cases are of this type. Also, the effectiveness of current vaccinations against the delta plus sub-variant is not yet known.
Omicron was found in South Africa,234,235 which exhibits a large number of spike protein mutations and is regarded by the WHO as a “variant of concern.” The spike protein, which the virus uses as a point of entry into our cells and is the target of current vaccinations, has 32 mutations in this version. The receptor-binding domain (RBD) alterations in the omicron variant total 10 compared to just 2 in the widely spread delta variant. Additionally, according to early studies from the University of Hong Kong, the omicron variation infects and replicates in the human bronchus 70 times quicker than its forerunners, yet the infection in the lung is noticeably lower.236 This data confirms the findings that although this variant is more contagious, it results in less severe disease. It is currently undergoing peer review for publication. The spread of this variation around the globe has already been reported by the governments and media of numerous nations. Both the effectiveness of the available vaccinations against this variation and its potential level of hazard is unknown to scientists at this time. In the upcoming weeks, we anticipate some succinct responses.
Uncertainty exists on how vaccination affects virus transmission.237 According to recent research, those who received the delta-type vaccine can contain just as much virus in their noses as those who were not immunized.238 Contrarily, compared to those who are unprotected and infected with the delta form, those who have had vaccinations may have a shorter duration during which they are contagious. For the first week of illness, delta virus loads were comparable between the two groups, but after day 7, in those who had received vaccinations, they rapidly decreased.239
SARS-CoV-2 transmission is not yet seasonal. It has been observed that greater infection not always happens during winter outbreaks in a number of warmer nations, including Iran and India. However, research suggests that the stability and spread of respiratory viruses are enhanced by dry winter air and that breathing dry air during colder weather may weaken the respiratory-tract immune system.240 In addition, people are more prone to stay inside during the winter, which increases the chance of virus transmission by droplets. Accordingly, the containment efforts in the Northern Hemisphere may be hampered by these elements this winter.
Many nations may overlook the discovery of new SARS-CoV-2 variants because they lack the facilities and trained staff necessary to conduct thorough virus genome analyses. Unknown to us, there may be many more varieties circulating worldwide, which will affect the pace of transmission in various regions of the world. Primo or breakthrough infections require careful COVID-19 detection assays, especially considering the constant appearance of novel viral strains. In samples, the PCR test looks for signs of the genetic material of the virus. The PCR test typically looks for a three-part sequence on the spike protein of the virus to confirm the existence of an infection. One of these PCR targets was observed to be increasingly tested negative in samples from England regions with rapidly increasing case numbers, despite the fact that the other two targets in the tests were still functioning. It is rare to have two test sequences function, while the third does not. Investigations revealed that the virus in these samples had changed, where a deletion at the H69 and V70 locations was identified to be the root of the test failures.241
Considering the present challenges posed by COVID-19 with respect to its various variants, it is important to develop vaccines for specific variants and test kits for the diverse variants, given that the many variants may mutate into forms that require specified test approaches to detect and treat them.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors wish to appreciate DST West Bengal, the Centre for Research in Nanoscience and Nanotechnology, and the Department of Polymer Science and Technology University of Calcutta for their financial and technical support.References
- C. Weiss, M. Carriere, L. Fusco, I. Capua, J. A. Regla-Nava, M. Pasquali, J. A. Scott, F. Vitale, M. A. Unal and C. Mattevi, ACS Nano, 2020, 14, 6383–6406 CrossRef CAS PubMed.
- L. Zhang, D. Lin, X. Sun, U. Curth, C. Drosten, L. Sauerhering, S. Becker, K. Rox and R. Hilgenfeld, Science, 2020, 368, 409–412 CrossRef CAS PubMed.
- M. Gandhi, D. S. Yokoe and D. V. Havlir, N. Engl. J. Med., 2020, 382, 2158–2160 CrossRef CAS.
- P. Hassanzadeh, J. Controlled Release, 2020, 328, 112–126 CrossRef CAS.
- WHO Coronavirus (COVID-19) Dashboard, http://covid.who.int (accessed 2021-03-13).
- R. Medhi, P. Srinoi, N. Ngo, H.-V. Tran and T. R. Lee, ACS Appl. Nano Mater., 2020, 3, 8557–8580 CrossRef CAS.
- J. Sharma, K. Shepardson, L. L. Johns, J. Wellham, J. Avera, B. Schwarz, A. Rynda-Apple and T. Douglas, ACS Appl. Mater. Interfaces, 2020, 12, 18211–18224 CrossRef CAS.
- S. Fukushi, A. Fukuma, T. Kurosu, S. Watanabe, M. Shimojima, K. Shirato, N. Iwata-Yoshikawa, N. Nagata, K. Ohnishi and M. Ato, J. Virol. Methods, 2018, 251, 22–29 CrossRef CAS PubMed.
- R. Yan, Y. Zhang, Y. Li, L. Xia, Y. Guo and Q. Zhou, Science, 2020, 367, 1444–1448 CrossRef CAS PubMed.
- J. F.-W. Chan, S. Yuan, K.-H. Kok, K. K.-W. To, H. Chu, J. Yang, F. Xing, J. Liu, C. C.-Y. Yip and R. W.-S. Poon, Lancet, 2020, 395, 514–523 CrossRef CAS.
- H. L. Chan, S. K. Tsui and J. J. Sung, Trends Mol. Med., 2003, 9, 323–325 CrossRef.
- H. F. Rabenau, J. Cinatl, B. Morgenstern, G. Bauer, W. Preiser and H. W. Doerr, Med. Microbiol. Immunol., 2005, 194, 1–6 CrossRef CAS PubMed.
- P. A. Rota, M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Peñaranda, B. Bankamp, K. Maher, M.-H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. T. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Günther, A. D. M. E. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson and W. J. Bellini, Science, 2003, 300, 1394–1399 CrossRef CAS PubMed.
- K. L. Hon, Travel Med. Infect. Dis., 2013, 11, 285–287 CrossRef CAS.
- T. Liu, J. Hu, M. Kang, L. Lin, H. Zhong, J. Xiao, G. He, T. Song, Q. Huang and Z. Rong, Time-varying transmission dynamics of Novel Coronavirus Pneumonia in China, bioRxiv, 2020, preprint, DOI:10.1101/2020.01.25.919787.
- Y. Zhu, L. Chen, H. Ji, M. Xi, Y. Fang and Y. Li, Neurosci. Bull., 2020, 36, 299–302 CrossRef CAS.
- P. Zhou, H. Fan, T. Lan, X.-L. Yang, W.-F. Shi, W. Zhang, Y. Zhu, Y.-W. Zhang, Q.-M. Xie, S. Mani, X.-S. Zheng, B. Li, J.-M. Li, H. Guo, G.-Q. Pei, X.-P. An, J.-W. Chen, L. Zhou, K.-J. Mai, Z.-X. Wu, D. Li, D. E. Anderson, L.-B. Zhang, S.-Y. Li, Z.-Q. Mi, T.-T. He, F. Cong, P.-J. Guo, R. Huang, Y. Luo, X.-L. Liu, J. Chen, Y. Huang, Q. Sun, X.-L.-L. Zhang, Y.-Y. Wang, S.-Z. Xing, Y.-S. Chen, Y. Sun, J. Li, P. Daszak, L.-F. Wang, Z.-L. Shi, Y.-G. Tong and J.-Y. Ma, Nature, 2018, 556, 255–258 CrossRef CAS PubMed.
- P. V. M. Simas, A. C. de Souza Barnabé, R. Durães-Carvalho, D. F. de Lima Neto, L. C. Caserta, L. Artacho, F. A. F. Jacomassa, M. C. Martini, M. M. A. B. Dos Santos and P. A. N. Felippe, Emerging Infect. Dis., 2015, 21, 729 CrossRef CAS PubMed.
- J. Cui, F. Li and Z.-L. Shi, Nat. Rev. Microbiol., 2019, 17, 181–192 CrossRef CAS PubMed.
- S. Van Boheemen, M. De Graaf, C. Lauber, T. M. Bestebroer, V. S. Raj, A. M. Zaki, A. D. Osterhaus, B. L. Haagmans, A. E. Gorbalenya and E. J. Snijder, mBio, 2012, 3, e00473 CrossRef CAS.
- Y. Chen, Q. Liu and D. Guo, J. Med. Virol., 2020, 92, 418–423 CrossRef CAS PubMed.
- M. Czub, H. Weingartl, S. Czub, R. He and J. Cao, Vaccine, 2005, 23, 2273–2279 CrossRef CAS PubMed.
- C. A. De Haan, L. Kuo, P. S. Masters, H. Vennema and P. J. Rottier, J. Virol., 1998, 72, 6838–6850 CrossRef CAS.
- Y. Huang, C. Yang, X. Xu, W. Xu and S. Liu, Acta Pharmacol. Grzech., 2020, 41, 1141–1149 Search PubMed.
- P. Song, W. Li, J. Xie, Y. Hou and C. You, Clin. Chim. Acta, 2020, 509, 280–287 CrossRef CAS.
- T. Gu, S. Zhao, G. Jin, M. Song, Y. Zhi, R. Zhao, F. Ma, Y. Zheng, K. Wang and H. Liu, Front. Immunol., 2021, 11, 621441 CrossRef.
- N. Magazine, T. Zhang, Y. Wu, M. C. McGee, G. Veggiani and W. Huang, Viruses, 2022, 14, 640 CrossRef CAS PubMed.
- Y. Araf, F. Akter, Y. D. Tang, R. Fatemi, M. S. A. Parvez, C. Zheng and M. G. Hossain, J. Med. Virol., 2022, 94, 1825–1832 CrossRef CAS PubMed.
- L. Chen and J. Liang, Mater. Sci. Eng., C, 2020, 112, 110924 CrossRef CAS PubMed.
- M. Yang, K. Sunderland and C. Mao, Chem. Rev., 2017, 117, 10377–10402 CrossRef CAS PubMed.
- M. Oswald, S. Geissler and A. Goepferich, Mol. Pharmaceutics, 2017, 14, 2177–2196 CrossRef CAS.
- S. Gurunathan, M. Qasim, Y. Choi, J. T. Do, C. Park, K. Hong, J.-H. Kim and H. Song, Nanomaterials, 2020, 10, 1645 CrossRef CAS PubMed.
- M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Krüger, T. Herrler, S. Erichsen, T. S. Schiergens, G. Herrler, N.-H. Wu, A. Nitsche, M. A. Müller, C. Drosten and S. Pöhlmann, Cell, 2020, 181, 271–280.e278 CrossRef CAS PubMed.
- X. Ou, Y. Liu, X. Lei, P. Li, D. Mi, L. Ren, L. Guo, R. Guo, T. Chen and J. Hu, Nat. Commun., 2020, 11, 1–12 CrossRef PubMed.
- Q. Wang, Y. Zhang, L. Wu, S. Niu, C. Song, Z. Zhang, G. Lu, C. Qiao, Y. Hu and K.-Y. Yuen, Cell, 2020, 181, 894–904.e899 CrossRef CAS PubMed.
- J. A. Fulcher, K. Tamshen, A. L. Wollenberg, V. A. Kickhoefer, J. Mrazek, J. Elliott, F. J. Ibarrondo, P. A. Anton, L. H. Rome, H. D. Maynard, T. Deming and O. O. Yang, Bioconjugate Chem., 2019, 30, 2216–2227 CrossRef CAS PubMed.
- J. S. Chahal, O. F. Khan, C. L. Cooper, J. S. McPartlan, J. K. Tsosie, L. D. Tilley, S. M. Sidik, S. Lourido, R. Langer and S. Bavari, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E4133–E4142 CrossRef CAS PubMed.
- R. Ribeiro-Viana, M. Sánchez-Navarro, J. Luczkowiak, J. R. Koeppe, R. Delgado, J. Rojo and B. G. Davis, Nat. Commun., 2012, 3, 1–9 Search PubMed.
- Q. Zhang, A. Honko, J. Zhou, H. Gong, S. N. Downs, J. H. Vasquez, R. H. Fang, W. Gao, A. Griffiths and L. Zhang, Nano Lett., 2020, 20, 5570–5574 CrossRef CAS PubMed.
- T. Tang, M. Bidon, J. A. Jaimes, G. R. Whittaker and S. Daniel, Antiviral Res., 2020, 178, 104792 CrossRef CAS.
- U. S. Kumar, R. Afjei, K. Ferrara, T. F. Massoud and R. Paulmurugan, ACS Nano, 2021, 15, 17582–17601 CrossRef CAS PubMed.
- G. Muthiah, A. Sarkar, S. Roy, P. Singh, P. Kumar, K. Bhardwaj and A. Jaiswal, ChemNanoMat, 2022, 8, e202100505 CrossRef CAS.
- G. Seo, G. Lee, M. J. Kim, S.-H. Baek, M. Choi, K. B. Ku, C.-S. Lee, S. Jun, D. Park, H. G. Kim, S.-J. Kim, J.-O. Lee, B. T. Kim, E. C. Park and S. I. Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS.
- M. Khater, A. D. L. Escosura-Muñiz, L. Altet and A. Merkoçi, Anal. Chem., 2019, 91, 4790–4796 CrossRef CAS PubMed.
- F. Biotech, FDA greenlights first smartphone-based home COVID-19 test, https://www.fiercebiotech.com/medtech/fda-greenlights-first-smartphone-based-home-covid-19-test, (accessed 03.09.2022, 2022).
- G. Palestino, I. García-Silva, O. González-Ortega and S. Rosales-Mendoza, Expert Rev. Anti-Infect. Ther., 2020, 18, 849–864 CrossRef CAS.
- B. Udugama, P. Kadhiresan, H. N. Kozlowski, A. Malekjahani, M. Osborne, V. Y. Li, H. Chen, S. Mubareka, J. B. Gubbay and W. C. Chan, ACS Nano, 2020, 14, 3822–3835 CrossRef CAS PubMed.
- J. M. Ferdinands, L. E. W. Olsho, A. A. Agan, N. Bhat, R. M. Sullivan, M. Hall, P. M. Mourani, M. Thompson and A. G. Randolph, J. Infect. Dis., 2014, 210, 674–683 CrossRef.
- W. Chantong and J. B. Kaneene, Southeast Asian J. Trop. Med. Public Health, 2011, 42, 596 Search PubMed.
- R. J. Coker, B. M. Hunter, J. W. Rudge, M. Liverani and P. Hanvoravongchai, Lancet, 2011, 377, 599–609 CrossRef.
- M. Jonduo, S.-S. Wong, N. Kapo, P. Ominipi, M. Abdad, P. Siba, P. McKenzie, R. Webby and P. Horwood, West. Pac. Surveill. Response J., 2013, 4, 11 CrossRef PubMed.
- S. Das, S. Dunbar and Y.-W. Tang, Front. Microbiol., 2018, 9, 2478 CrossRef.
- J. C. Huang, Y.-F. Chang, K.-H. Chen, L.-C. Su, C.-W. Lee, C.-C. Chen, Y.-M. A. Chen and C. Chou, Biosens. Bioelectron., 2009, 25, 320–325 CrossRef CAS.
- C. Roh and S. K. Jo, J. Chem. Technol. Biotechnol., 2011, 86, 1475–1479 CrossRef CAS.
- M. Park, J. Won, B. Y. Choi and C. J. Lee, Exp. Mol. Med., 2020, 52, 963–977 CrossRef CAS PubMed.
- P. Teengam, W. Siangproh, A. Tuantranont, T. Vilaivan, O. Chailapakul and C. S. Henry, Anal. Chim. Acta, 2018, 1044, 102–109 CrossRef CAS PubMed.
- F. N. Ishikawa, H.-K. Chang, M. Curreli, H.-I. Liao, C. A. Olson, P.-C. Chen, R. Zhang, R. W. Roberts, R. Sun and R. J. Cote, ACS Nano, 2009, 3, 1219–1224 CrossRef CAS PubMed.
- Y. Chen, K.-H. Chan, Y. Kang, H. Chen, H. K. Luk, R. W. Poon, J. F. Chan, K.-Y. Yuen, N. Xia and S. K. Lau, Emerging Microbes Infect., 2015, 4, 1–5 Search PubMed.
- R. Funari, K.-Y. Chu and A. Q. Shen, Biosens. Bioelectron., 2020, 169, 112578 CrossRef CAS PubMed.
- L. A. Layqah and S. Eissa, Microchim. Acta, 2019, 186, 224 CrossRef PubMed.
- G. Seo, G. Lee, M. J. Kim, S.-H. Baek, M. Choi, K. B. Ku, C.-S. Lee, S. Jun, D. Park and H. G. Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS PubMed.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2020, 14, 5268–5277 CrossRef CAS.
- I. PathSensors, Announced the Development of a SARS-CoV-2 Biosensor, PathSensors Inc., 2022, https://markets.businessinsider.com/news/stocks/pathsensors-inc-announced-the-development-of-a-sars-cov-2-biosensor-1029025341.
- A. Srivastava, N. Dwivedi, C. Dhand, R. Khan, N. Sathish, M. Gupta, R. Kumar and S. Kumar, Mater. Today Chem., 2020, 18, 100385 CrossRef CAS PubMed.
- Y. Abo-zeid and G. R. Williams, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2020, 12, e1592 Search PubMed.
- R. Parboosing, G. E. Maguire, P. Govender and H. G. Kruger, Viruses, 2012, 4, 488–520 CrossRef CAS.
- R. Bawa, Eur. J. Nanomed., 2010, 3, 34–40 Search PubMed.
- S. Galdiero, A. Falanga, M. Vitiello, M. Cantisani, V. Marra and M. Galdiero, Molecules, 2011, 16, 8894–8918 CrossRef CAS.
- R. A. Schwendener, Ther. Adv. Vaccines, 2014, 2, 159–182 CrossRef CAS PubMed.
- A. Gupta, D. F. Moyano, A. Parnsubsakul, A. Papadopoulos, L.-S. Wang, R. F. Landis, R. Das and V. M. Rotello, ACS Appl. Mater. Interfaces, 2016, 8, 14096–14101 CrossRef CAS PubMed.
- D. Bartczak, O. L. Muskens, T. Sanchez-Elsner, A. G. Kanaras and T. M. Millar, ACS Nano, 2013, 7, 5628–5636 CrossRef CAS PubMed.
- W.-H. Wen, M. Lin, C.-Y. Su, S.-Y. Wang, Y.-S. E. Cheng, J.-M. Fang and C.-H. Wong, J. Med. Chem., 2009, 52, 4903–4910 CrossRef CAS PubMed.
- H. Andresen, M. Mager, M. Grießner, P. Charchar, N. Todorova, N. Bell, G. Theocharidis, S. Bertazzo, I. Yarovsky and M. M. Stevens, Chem. Mater., 2014, 26, 4696–4704 CrossRef CAS.
- M.-C. Bowman, T. E. Ballard, C. J. Ackerson, D. L. Feldheim, D. M. Margolis and C. Melander, J. Am. Chem. Soc., 2008, 130, 6896–6897 CrossRef CAS PubMed.
- P. Di Gianvincenzo, M. Marradi, O. M. Martínez-Ávila, L. M. Bedoya, J. Alcamí and S. Penadés, Bioorg. Med. Chem. Lett., 2010, 20, 2718–2721 CrossRef CAS PubMed.
- A. Mehranfar and M. Izadyar, J. Phys. Chem. Lett., 2020, 11, 10284–10289 CrossRef CAS PubMed.
- A. R. Bastian, A. Nangarlia, L. D. Bailey, A. Holmes, R. V. K. Sundaram, C. Ang, D. R. Moreira, K. Freedman, C. Duffy and M. Contarino, J. Biol. Chem., 2015, 290, 529–543 CrossRef CAS PubMed.
- M. A. Dkhil, A. A. Bauomy, M. S. Diab and S. Al-Quraishy, Int. J. Nanomed., 2015, 10, 7467 CAS.
- Y. Bai, Y. Zhou, H. Liu, L. Fang, J. Liang and S. Xiao, ACS Appl. Nano Mater., 2018, 1, 969–976 CrossRef CAS.
- X. Huang, M. Li, Y. Xu, J. Zhang, X. Meng, X. An, L. Sun, L. Guo, X. Shan and J. Ge, ACS Appl. Mater. Interfaces, 2019, 11, 19799–19807 CrossRef CAS PubMed.
- T. Du, J. Zhang, C. Li, T. Song, P. Li, J. Liu, X. Du and S. Wang, Bioconjugate Chem., 2020, 31, 2553–2563 CrossRef CAS PubMed.
- S. M. Moghimi, A. C. Hunter and J. C. Murray, Pharmacol. Rev., 2001, 53, 283–318 CAS.
- W. H. De Jong, Int. J. Nanomed., 2008, 3, 133–149 CrossRef CAS PubMed.
- K. Asadi and A. Gholami, Int. J. Biol. Macromol., 2021, 182, 648–658 CrossRef CAS PubMed.
- R. M. Rosa, J. C. Silva, I. S. Sanches and C. Henriques, Mater. Lett., 2017, 207, 145–148 CrossRef CAS.
- S. Gaikwad, A. Ingle, A. Gade, M. Rai, A. Falanga, N. Incoronato, L. Russo, S. Galdiero and M. Galdiero, Int. J. Nanomed., 2013, 8, 4303 Search PubMed.
- X. Lv, P. Wang, R. Bai, Y. Cong, S. Suo, X. Ren and C. Chen, Biomaterials, 2014, 35, 4195–4203 CrossRef CAS PubMed.
- D. Xiang, Y. Zheng, W. Duan, X. Li, J. Yin, S. Shigdar, M. L. O’Connor, M. Marappan, X. Zhao and Y. Miao, Int. J. Nanomed., 2013, 8, 4103 CrossRef PubMed.
- C. Balagna, S. Perero, E. Percivalle, E. V. Nepita and M. Ferraris, Open Ceram., 2020, 1, 100006 CrossRef CAS.
- T. Q. Huy, N. T. H. Thanh, N. T. Thuy, P. Van Chung, P. N. Hung, A.-T. Le and N. T. H. Hanh, J. Virol. Methods, 2017, 241, 52–57 CrossRef CAS PubMed.
- Y. Mori, T. Ono, Y. Miyahira, V. Q. Nguyen, T. Matsui and M. Ishihara, Nanoscale Res. Lett., 2013, 8, 1–6 CrossRef PubMed.
- T. Prasad and E. Elumalai, Asian Pac. J. Trop. Biomed., 2011, 1, 439–442 CrossRef CAS PubMed.
- C. Wan, J. Tai, J. Zhang, Y. Guo, Q. Zhu, D. Ling, F. Gu, J. Gan, C. Zhu and Y. Wang, Cell Death Dis., 2019, 10, 1–16 CrossRef PubMed.
- S. D. Kumar, G. Singaravelu, S. Ajithkumar, K. Murugan, M. Nicoletti and G. Benelli, J. Cluster Sci., 2017, 28, 359–367 CrossRef CAS.
- R. Govender, A. Phulukdaree, R. M. Gengan, K. Anand and A. A. Chuturgoon, J. Nanobiotechnol., 2013, 11, 1–9 CrossRef PubMed.
- E. G. Haggag, A. M. Elshamy, M. A. Rabeh, N. M. Gabr, M. Salem, K. A. Youssif, A. Samir, A. B. Muhsinah, A. Alsayari and U. R. Abdelmohsen, Int. J. Nanomed., 2019, 14, 6217 CrossRef CAS PubMed.
- J. Zhong, Y. Xia, L. Hua, X. Liu, M. Xiao, T. Xu, B. Zhu and H. Cao, Artif. Cells, Nanomed., Biotechnol., 2019, 47, 3485–3491 CrossRef CAS PubMed.
- Z. Cheng, X. Zhi, G. Sun, W. Guo, Y. Huang, W. Sun, X. Tian, F. Zhao and K. Hu, J. Med. Virol., 2016, 88, 653–663 CrossRef CAS PubMed.
- Z. Lin, Y. Li, M. Guo, M. Xiao, C. Wang, M. Zhao, T. Xu, Y. Xia and B. Zhu, RSC Adv., 2017, 7, 35290–35296 RSC.
- M. M. Melk, S. S. El-Hawary, F. R. Melek, D. O. Saleh, O. M. Ali, M. A. El Raey and N. M. Selim, Int. J. Nanomed., 2021, 16, 8221 CrossRef CAS PubMed.
- R. Kumar, G. Sahoo, K. Pandey, M. Nayak, R. Topno, V. Rabidas and P. Das, Int. J. Infect. Dis., 2018, 73, 368 CrossRef.
- N. Shionoiri, T. Sato, Y. Fujimori, T. Nakayama, M. Nemoto, T. Matsunaga and T. Tanaka, J. Biosci. Bioeng., 2012, 113, 580–586 CrossRef CAS PubMed.
- X. Hang, H. Peng, H. Song, Z. Qi, X. Miao and W. Xu, J. Virol. Methods, 2015, 222, 150–157 CrossRef CAS PubMed.
- S. Muthamil Selvan, K. Vijai Anand, K. Govindaraju, S. Tamilselvan, V. G. Kumar, K. S. Subramanian, M. Kannan and K. Raja, IET Nanobiotechnol., 2018, 12, 1042–1046 CrossRef PubMed.
- S. L. Warnes, Z. R. Little and C. W. Keevil, mBio, 2015, 6, e01697 CrossRef CAS PubMed.
- S.-r Choi, B. E. Britigan and P. Narayanasamy, Antimicrob. Agents Chemother., 2017, 61, e02505 CAS.
- R. Kumar, M. Nayak, G. C. Sahoo, K. Pandey, M. C. Sarkar, Y. Ansari, V. Das, R. Topno, M. Madhukar and P. Das, J. Infect. Chemother., 2019, 25, 325–329 CrossRef CAS PubMed.
- H. Cui, J. Jiang, W. Gu, C. Sun, D. Wu, T. Yang and G. Yang, Photochem. Photobiol., 2010, 86, 1135–1139 CrossRef CAS PubMed.
- F. Pfaff, B. Glück, T. Hoyer, D. Rohländer, A. Sauerbrei and R. Zell, Lett. Appl. Microbiol., 2019, 69, 302–309 CrossRef CAS PubMed.
- A. H. M. O.-R. Kaaden, General Virology. Measurement of infectivity, Medical microbiology, infection and epidemic theory for veterinarians, biologists, agricultural scientists and interested parties from related fields, 6th edn, 1993 Search PubMed.
- L. Sandeep and S. Rajasree, J. Anal. Pharm. Res., 2017, 4, 10.15406 Search PubMed.
- M. L. Smith, S. Gandolfi, P. M. Coshall and P. K. Rahman, Front. Microbiol., 2020, 11, 1341 CrossRef PubMed.
- T. Li, D. Cipolla, T. Rades and B. J. Boyd, J. Controlled Release, 2018, 288, 96–110 CrossRef CAS PubMed.
- S. C. Mogensen, Microbiol. Rev., 1979, 43, 1–26 CrossRef CAS PubMed.
- C. Sinico, A. De Logu, F. Lai, D. Valenti, M. Manconi, G. Loy, L. Bonsignore and A. M. Fadda, Eur. J. Pharm. Biopharm., 2005, 59, 161–168 CrossRef CAS PubMed.
- H. Ji, Z. Yang, W. Jiang, C. Geng, M. Gong, H. Xiao, Z. Wang and L. Cheng, J. Huazhong Univ. Sci. Technol., Med. Sci., 2008, 28, 243–246 CrossRef CAS PubMed.
- G. Sharma, K. Thakur, A. Setia, B. Amarji, M. P. Singh, K. Raza and O. P. Katare, Drug Delivery Transl. Res., 2017, 7, 683–694 CrossRef CAS PubMed.
- K. Tahara, M. Kobayashi, S. Yoshida, R. Onodera, N. Inoue and H. Takeuchi, Int. J. Pharm., 2018, 543, 311–317 CrossRef CAS PubMed.
- S. Huang, J. Gu, J. Ye, B. Fang, S. Wan, C. Wang, U. Ashraf, Q. Li, X. Wang and L. Shao, J. Colloid Interface Sci., 2019, 542, 198–206 CrossRef CAS PubMed.
- A. Barras, Q. Pagneux, F. Sane, Q. Wang, R. Boukherroub, D. Hober and S. Szunerits, ACS Appl. Mater. Interfaces, 2016, 8, 9004–9013 CrossRef CAS PubMed.
- A. Łoczechin, K. Séron, A. Barras, E. Giovanelli, S. Belouzard, Y.-T. Chen, N. Metzler-Nolte, R. Boukherroub, J. Dubuisson and S. Szunerits, ACS Appl. Mater. Interfaces, 2019, 11, 42964–42974 CrossRef PubMed.
- D. Iannazzo, A. Pistone, M. Salamò, S. Galvagno, R. Romeo, S. V. Giofré, C. Branca, G. Visalli and A. Di Pietro, Int. J. Pharm., 2017, 518, 185–192 CrossRef CAS PubMed.
- X. Du, R. Xiao, H. Fu, Z. Yuan, W. Zhang, L. Yin, C. He, C. Li, J. Zhou and G. Liu, Mater. Sci. Eng., C, 2019, 105, 110052 CrossRef CAS PubMed.
- X. X. Yang, C. M. Li, Y. F. Li, J. Wang and C. Z. Huang, Nanoscale, 2017, 9, 16086–16092 RSC.
- F. De Maio, V. Palmieri, G. Babini, A. Augello, I. Palucci, G. Perini, A. Salustri, P. Spilman, M. De Spirito, M. Sanguinetti, G. Delogu, L. G. Rizzi, G. Cesareo, P. Soon-Shiong, M. Sali and M. Papi, iScience, 2021, 24, 102788 CrossRef CAS PubMed.
- T. Seifi and A. R. Kamali, Med. Drug Discovery, 2021, 11, 100099 CrossRef CAS PubMed.
- A. Kalkal, P. Allawadhi, R. Pradhan, A. Khurana, K. K. Bharani and G. Packirisamy, Sens. Int., 2021, 2, 100102 CrossRef PubMed.
- I. Banerjee, M. P. Douaisi, D. Mondal and R. S. Kane, Nanotechnology, 2012, 23, 105101 CrossRef PubMed.
- Z. Liu, M. Winters, M. Holodniy and H. Dai, Angew. Chem., Int. Ed., 2007, 46, 2023–2027 CrossRef CAS PubMed.
- A. Bianco, Expert Opin. Drug Delivery, 2004, 1, 57–65 CrossRef CAS PubMed.
- D. Iannazzo, A. Pistone, S. Galvagno, S. Ferro, L. De Luca, A. M. Monforte, T. Da Ros, C. Hadad, M. Prato and C. Pannecouque, Carbon, 2015, 82, 548–561 CrossRef CAS.
- A. Aasi, S. M. Aghaei, M. D. Moore and B. Panchapakesan, Int. J. Mol. Sci., 2020, 21, 5211 CrossRef CAS PubMed.
- M. Mohan Gokhale and R. Ravindra Somani, Mini-Rev. Org. Chem., 2015, 12, 355–366 CrossRef.
- A. R. Badireddy, E. M. Hotze, S. Chellam, P. Alvarez and M. R. Wiesner, Environ. Sci. Technol., 2007, 41, 6627–6632 CrossRef CAS PubMed.
- E. M. Hotze, T. Phenrat and G. V. Lowry, J. Environ. Qual., 2010, 39, 1909–1924 CrossRef CAS PubMed.
- M. A. Gacem, H. Gacem and A. Ould-El-Hadj-Khelil, Carbon Nanomaterials for Agri-Food and Environmental Applications, Elsevier, 2020, pp. 505–533 Search PubMed.
- P. Innocenzi and L. Stagi, Chem. Sci., 2020, 11, 6606–6622 RSC.
- O. A. Kraevaya, A. S. Peregudov, I. A. Godovikov, E. V. Shchurik, V. M. Martynenko, A. F. Shestakov, J. Balzarini, D. Schols and P. A. Troshin, Chem. Commun., 2020, 56, 1179–1182 RSC.
- D. Chun-Xian, X. Hai-Rong, J. Hong, L. Qiang, X. Hong and Y. Zhan-Qiu, Sci. Res. Essays, 2012, 7, 705–711 Search PubMed.
- M. Shoji, E. Takahashi, D. Hatakeyama, Y. Iwai, Y. Morita, R. Shirayama, N. Echigo, H. Kido, S. Nakamura and T. Mashino, PLoS One, 2013, 8, e66337 CrossRef CAS PubMed.
- O. A. Kraevaya, A. V. Novikov, A. F. Shestakov, E. S. Ershova, E. A. Savinova, L. V. Kameneva, N. N. Veiko, D. Schols, J. Balzarini and S. V. Kostyuk, Chem. Commun., 2020, 56, 10203–10206 RSC.
- A. Muñoz, B. M. Illescas, J. Luczkowiak, F. Lasala, R. Ribeiro-Viana, J. Rojo, R. Delgado and N. Martín, J. Mater. Chem. B, 2017, 5, 6566–6571 RSC.
- A. Muñoz, D. Sigwalt, B. M. Illescas, J. Luczkowiak, L. Rodriguez-Perez, I. Nierengarten, M. Holler, J.-S. Remy, K. Buffet and S. P. Vincent, Nat. Chem., 2016, 8, 50–57 CrossRef PubMed.
- M. G. Stanford, J. T. Li, Y. Chen, E. A. McHugh, A. Liopo, H. Xiao and J. M. Tour, ACS Nano, 2019, 13, 11912–11920 CrossRef CAS PubMed.
- A. R. Deokar, A. P. Nagvenkar, I. Kalt, L. Shani, Y. Yeshurun, A. Gedanken and R. Sarid, Bioconjugate Chem., 2017, 28, 1115–1122 CrossRef CAS PubMed.
- R. Spitz Steinberg, M. Cruz, N. G. Mahfouz, Y. Qiu and R. H. Hurt, ACS Nano, 2017, 11, 5670–5679 CrossRef CAS PubMed.
- V. Palmieri and M. Papi, Nano Today, 2020, 33, 100883 CrossRef CAS PubMed.
- I. Ekladious, Y. L. Colson and M. W. Grinstaff, Nat. Rev. Drug Discovery, 2019, 18, 273–294 CrossRef CAS PubMed.
- N. Larson and H. Ghandehari, Chem. Mater., 2012, 24, 840–853 CrossRef CAS PubMed.
- X. Chen, X. Chen, W. Chen, X. Ma, J. Huang and R. Chen, J. Med. Virol., 2014, 86, 1705–1713 CrossRef CAS PubMed.
- J. Li, F. Yu, Y. Chen and D. Oupický, J. Controlled Release, 2015, 219, 369–382 CrossRef CAS PubMed.
- A. H. Andersen, C. F. Riber, K. Zuwala, M. Tolstrup, F. Dagnæs-Hansen, P. W. Denton and A. N. Zelikin, ACS Macro Lett., 2018, 7, 587–591 CrossRef CAS PubMed.
- M. A. Ansari, Q. M. S. Jamal, S. Rehman, A. Almatroudi, M. A. Alzohairy, M. N. Alomary, T. Tripathi, A. H. Alharbi, S. F. Adil, M. Khan and M. Shaheer Malik, Arabian J. Chem., 2020, 13, 8069–8079 CrossRef CAS PubMed.
- M. A. Boles, D. Ling, T. Hyeon and D. V. Talapin, Nat. Mater., 2016, 15, 141–153 CrossRef CAS PubMed.
- D. Ting, N. Dong, L. Fang, J. Lu, J. Bi, S. Xiao and H. Han, ACS Appl. Nano Mater., 2018, 1, 5451–5459 CrossRef.
- X. Dong, M. M. Moyer, F. Yang, Y.-P. Sun and L. Yang, Sci. Rep., 2017, 7, 1–10 CrossRef PubMed.
- D. Iannazzo, A. Pistone, S. Ferro, L. De Luca, A. M. Monforte, R. Romeo, M. R. Buemi and C. Pannecouque, Bioconjugate Chem., 2018, 29, 3084–3093 CrossRef CAS PubMed.
- S. Basak and G. Packirisamy, Nano-Struct. Nano-Objects, 2020, 24, 100620 CrossRef CAS.
- T. Du, J. Liang, N. Dong, L. Liu, L. Fang, S. Xiao and H. Han, Carbon, 2016, 110, 278–285 CrossRef CAS.
- H. Liu, Y. Bai, Y. Zhou, C. Feng, L. Liu, L. Fang, J. Liang and S. Xiao, RSC Adv., 2017, 7, 28016–28023 RSC.
- S. Manivannan and K. Ponnuchamy, Appl. Organomet. Chem., 2020, 34, e5887 CrossRef CAS PubMed.
- E. Ratemi, Stimuli Responsive Polymeric Nanocarriers for Drug Delivery Applications, 2018, vol. 1, pp. 121–141 Search PubMed.
- S. Kumar, N. Dilbaghi, R. Saharan and G. Bhanjana, Bionanoscience, 2012, 2, 227–250 CrossRef.
- H. Cabral, K. Miyata, K. Osada and K. Kataoka, Chem. Rev., 2018, 118, 6844–6892 CrossRef CAS PubMed.
- K. Miyata, R. J. Christie and K. Kataoka, React. Funct. Polym., 2011, 71, 227–234 CrossRef CAS.
- Y. S. Ahn, H. J. Baik, B. R. Lee, E. S. Lee, K. T. Oh, D. H. Lee and Y. S. Youn, Macromol. Res., 2010, 18, 747–752 CrossRef CAS.
- H. Al-Lawati, H. M. Aliabadi, B. S. Makhmalzadeh and A. Lavasanifar, Expert Opin. Drug Delivery, 2018, 15, 397–418 CrossRef CAS PubMed.
- Y. Yang, M. Yang, J. Yuan, F. Wang, Z. Wang, J. Li, M. Zhang, L. Xing, J. Wei, L. Peng, G. Wong, H. Zheng, W. Wu, C. Shen, M. Liao, K. Feng, J. Li, Q. Yang, J. Zhao, L. Liu and Y. Liu, Innovation, 2020, 1, 100061 Search PubMed.
- Z. Feng, B. Diao, R. Wang, G. Wang, C. Wang, Y. Tan, L. Liu, C. Wang, Y. Liu and Y. Liu, medRxiv, 2020, preprint, DOI:10.1101/2020.03.27.20045427.
- C. C. Leung, T. H. Lam and K. K. Cheng, Lancet, 2020, 395, 945 CrossRef CAS PubMed.
- H. Zhong, Z. Zhu, J. Lin, C. F. Cheung, V. L. Lu, F. Yan, C.-Y. Chan and G. Li, ACS Nano, 2020, 14, 6213–6221 CrossRef CAS PubMed.
- H. Zhong, Z. Zhu, P. You, J. Lin, C. F. Cheung, V. L. Lu, F. Yan, C.-Y. Chan and G. Li, ACS Nano, 2020, 14, 8846–8854 CrossRef CAS PubMed.
- M.-G. Kim, J. Y. Park, Y. Shon, G. Kim, G. Shim and Y.-K. Oh, Asian J. Pharm. Sci., 2014, 9, 227–235 CrossRef.
- S. Misumi, M. Masuyama, N. Takamune, D. Nakayama, R. Mitsumata, H. Matsumoto, N. Urata, Y. Takahashi, A. Muneoka and T. Sukamoto, J. Immunol., 2009, 182, 6061–6070 CrossRef CAS PubMed.
- D. Raghuwanshi, V. Mishra, M. R. Suresh and K. Kaur, Vaccine, 2012, 30, 7292–7299 CrossRef CAS PubMed.
- B. Lepenies, J. Lee and S. Sonkaria, Adv. Drug Delivery Rev., 2013, 65, 1271–1281 CrossRef CAS PubMed.
- J. M. Ball, D. Y. Graham, A. R. Opekun, M. A. Gilger, R. A. Guerrero and M. K. Estes, Gastroenterology, 1999, 117, 40–48 CrossRef CAS PubMed.
- A. Geldmacher, D. Skrastina, G. Borisova, I. Petrovskis, D. H. Krüger, P. Pumpens and R. Ulrich, Vaccine, 2005, 23, 3973–3983 CrossRef CAS PubMed.
- C. M. Coleman, Y. V. Liu, H. Mu, J. K. Taylor, M. Massare, D. C. Flyer, G. M. Glenn, G. E. Smith and M. B. Frieman, Vaccine, 2014, 32, 3169–3174 CrossRef CAS PubMed.
- D. Sanchez-Guzman, P. Le Guen, B. Villeret, N. Sola, R. Le Borgne, A. Guyard, A. Kemmel, B. Crestani, J.-M. Sallenave and I. Garcia-Verdugo, Biomaterials, 2019, 217, 119308 CrossRef CAS PubMed.
- H. Sekimukai, N. Iwata-Yoshikawa, S. Fukushi, H. Tani, M. Kataoka, T. Suzuki, H. Hasegawa, K. Niikura, K. Arai and N. Nagata, Microbiol. Immunol., 2020, 64, 33–51 CrossRef CAS PubMed.
- W. H. Organisation, NIH Clinical Trial of Investigational Vaccine for COVID-19 Begins., https://www.niaid.nih.gov/news-events/nih-clinical-trialinvestigational-vaccine-covid-19-begins (accessed 2020-05-05).
- W. H. Organisation, Draft Landscape of COVID-19 Candidate Vaccines, https://www.who.int/who-documents-detail/draft-landscape-of-covid-19-candidate-vaccines, (accessed 2020-05-22, 2020).
- L. Du, Y. Yang, X. Zhang and F. Li, Nanoscale, 2022, 14, 1054–1074 RSC.
- F. Amanat and F. Krammer, Immunity, 2020, 52, 583–589 CrossRef CAS PubMed.
- B. Li, X. Zhang and Y. Dong, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2019, 11, e1530 Search PubMed.
- W. Tao, B. L. Hurst, A. K. Shakya, M. J. Uddin, R. S. Ingrole, M. Hernandez-Sanabria, R. P. Arya, L. Bimler, S. Paust and E. B. Tarbet, Antiviral Res., 2017, 141, 62–72 CrossRef CAS PubMed.
- C. Keech, G. Albert, I. Cho, A. Robertson, P. Reed, S. Neal, J. S. Plested, M. Zhu, S. Cloney-Clark and H. Zhou, N. Engl. J. Med., 2020, 383, 2320–2332 CrossRef CAS PubMed.
- Novavax, Novavax COVID-19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 Trial, https://ir.novavax.com/2021-01-28-Novavax-COVID-19-Vaccine-Demonstrates-89-3-Efficacy-in-UK-Phase-3-Trial#:~:text=(Nasdaq%3A%20NVAX)%2C%20a,the%20United%20Kingdom%20(%<?pdb_no 20UK?>20UK<?pdb END?>%20). (accessed 08.09.2022, 2022).
- Y. Li, R. Tenchov, J. Smoot, C. Liu, S. Watkins and Q. Zhou, ACS Cent. Sci., 2021, 7, 512–533 CrossRef CAS PubMed.
- SpyBiotech, SpyBiotech and Serum Institute of India announce that the first subjects have been dosed in a Phase I/II trial of a novel virus-like particle vaccine targeting COVID-19, https://www.globenewswire.com/news-release/2020/09/08/2089755/0/en/SpyBiotech-and-Serum-Institute-of-India-announce-that-the-first-subjects-have-been-dosed-in-a-Phase-I-II-trial-of-a-novel-virus-like-particle-vaccine-targeting-COVID-19.html, (accessed 08.09.2022, 2022).
- INO-4800, INO-4800 COVID-19 Vaccine, https://www.precisionvaccinations.com/vaccines/ino-4800-covid-19-vaccine, (accessed 08.09.2022, 2022).
- I. AnGes, Phase II/III Study of COVID-19 DNA Vaccine (AG0302-COVID19), https://clinicaltrials.gov/ct2/show/NCT04655625).
- ZyCoV-D, ZyCoV-D COVID-19 Vaccine, https://www.precisionvaccinations.com/vaccines/ino-4800-covid-19-vaccine, (accessed 08.09.2022, 2022).
- D. S. Patel, ZyCoV-D, the made-in-India Covid vaccine by Zydus Cadila, likely to hit markets in June, https://theprint.in/health/zycov-d-the-made-in-india-covid-vaccine-by-zydus-cadila-likely-to-hit-markets-in-june/657272/, (accessed 08.09.2022, 2022).
- J. Y. Ahn, J. Lee, Y. S. Suh, Y. G. Song, Y.-J. Choi, K. H. Lee, S. H. Seo, M. Song, J.-W. Oh and M. Kim, medRxiv, 2021, preprint, DOI:10.1101/2021.05.26.21257700.
- M. M. Silveira, G. M. S. G. Moreira and M. Mendonça, Life Sci., 2021, 267, 118919 CrossRef CAS PubMed.
- J. Y. Noh, H. W. Jeong, J. H. Kim and E.-C. Shin, Nat. Rev. Immunol., 2021, 21, 687–688 CrossRef PubMed.
- ClinicalTrials.gov., The Safety and Immunogenicity of a DNA-based Vaccine (COVIGEN) in Healthy Volunteers (COVALIA), https://clinicaltrials.gov/ct2/show/NCT04742842).
- Takis and Rottapharm Biotech announce favourable preliminary results from the phase I clinical trial of COVID-eVax, a DNA-based vaccine candidate against COVID-19, https://www.rottapharmbiotech.com/07-september-2021-press-release/ (accessed 2022-09-09).
- B.-S. Shim, S.-M. Park, J.-S. Quan, D. Jere, H. Chu, M. K. Song, D. W. Kim, Y.-S. Jang, M.-S. Yang and S. H. Han, BMC Immunol., 2010, 11, 1–9 CrossRef PubMed.
- N.-N. Zhang, X.-F. Li, Y.-Q. Deng, H. Zhao, Y.-J. Huang, G. Yang, W.-J. Huang, P. Gao, C. Zhou and R.-R. Zhang, Cell, 2020, 182, 1271–1283.e1216 CrossRef CAS PubMed.
- C. B. Jackson, M. Farzan, B. Chen and H. Choe, Nat. Rev. Mol. Cell Biol., 2022, 23, 3–20 CrossRef CAS PubMed.
- R. W. Chan, S. Liu, J. Y. Cheung, J. G. Tsun, K. C. Chan, K. Y. Chan, G. P. Fung, A. M. Li and H. S. Lam, Front. Immunol., 2021, 4169 Search PubMed.
- W. H. Organization, Interim recommendations for use of the Pfizer–BioNTech COVID-19 vaccine, BNT162b2, under emergency use listing: interim guidance, first issued 8 January 2021, updated 15 June 2021, updated 19 November 2021, World Health Organization, 2021.
- W. H. Organization, Interim recommendations for use of the Moderna mRNA-1273 vaccine against COVID-19: interim guidance, first issued 25 January 2021, updated 15 June 2021, updated 19 November 2021, updated 23 February 2022, World Health Organization, 2022.
- P. SARS-CoV, Variants_of_Concern_VOC_Technical_Briefing_12_England. pdf.
- W. H. Organization, Background document on the Janssen Ad26. COV2. S (COVID-19) vaccine: background document to the WHO Interim recommendations for use of Ad26. COV2. S (COVID-19) vaccine, 17 March 2021, World Health Organization, 2021 Search PubMed.
- E. Yasinski, COVID-19 Vaccine Side Effect Tracker, https://medshadow.org/covid19-vaccine-side-effects-2/#gameleya.
- D. Y. Logunov, I. V. Dolzhikova, D. V. Shcheblyakov, A. I. Tukhvatulin, O. V. Zubkova, A. S. Dzharullaeva, A. V. Kovyrshina, N. L. Lubenets, D. M. Grousova and A. S. Erokhova, Lancet, 2021, 397, 671–681 CrossRef CAS PubMed.
- A. Hasanzadeh, M. Alamdaran, S. Ahmadi, H. Nourizadeh, M. A. Bagherzadeh, M. A. M. Jahromi, P. Simon, M. Karimi and M. R. Hamblin, J. Controlled Release, 2021, 336, 354–374 CrossRef CAS PubMed.
- D. Vasireddy, P. Atluri, S. V. Malayala, R. Vanaparthy and G. Mohan, J. Clin. Med. Res., 2021, 13, 204 CrossRef CAS PubMed.
- R. M. Burckhardt, J. J. Dennehy, L. L. Poon, L. J. Saif and L. W. Enquist, J. Virol., 2022, 96, e01973 CrossRef CAS PubMed.
- M. Doerflinger, W. Forsyth, G. Ebert, M. Pellegrini and M. Herold, Cell. Microbiol., 2017, 19, e12693 CrossRef PubMed.
- C. Lee, Molecules, 2019, 24, 1349 CrossRef PubMed.
- J. D. Finn, A. R. Smith, M. C. Patel, L. Shaw, M. R. Youniss, J. van Heteren, T. Dirstine, C. Ciullo, R. Lescarbeau and J. Seitzer, Cell Rep., 2018, 22, 2227–2235 CrossRef CAS PubMed.
- O. O. Abudayyeh, J. S. Gootenberg, P. Essletzbichler, S. Han, J. Joung, J. J. Belanto, V. Verdine, D. B. Cox, M. J. Kellner and A. Regev, Nature, 2017, 550, 280–284 CrossRef PubMed.
- T. M. Nguyen, Y. Zhang and P. P. Pandolfi, Cell Res., 2020, 30, 189–190 CrossRef CAS PubMed.
- C. Xu, Y. Zhou, Q. Xiao, B. He, G. Geng, Z. Wang, B. Cao, X. Wang, D. Zhou and T. Yuan, Research Square, 2020 DOI:10.21203/rs.3.rs-30924/v1.
- I. Kraja, R. Bing, N. Hiwatashi, B. Rousseau, D. Nalband, K. Kirshenbaum and R. C. Branski, Laryngoscope, 2017, 127, E231–E237 CrossRef CAS PubMed.
- M. Burmistrz, K. Krakowski and A. Krawczyk-Balska, Int. J. Mol. Sci., 2020, 21, 1122 CrossRef CAS PubMed.
- R. Romeo, C. Carnovale, S. V. Giofrè, M. A. Chiacchio, A. Garozzo, E. Amata, G. Romeo and U. Chiacchio, Beilstein J. Org. Chem., 2015, 11, 328–334 CrossRef CAS PubMed.
- D. D. Richman, D. M. Margolis, M. Delaney, W. C. Greene, D. Hazuda and R. J. Pomerantz, Science, 2009, 323, 1304–1307 CrossRef CAS PubMed.
- J. Szebeni, S. M. Wahl, G. V. Betageri, L. M. Wahl, S. Gartner, M. Popovic, R. J. Parker, C. D. Black and J. N. Weinstein, AIDS Res. Hum. Retroviruses, 1990, 6, 691–702 CrossRef CAS PubMed.
- T. Dutta, M. Garg and N. K. Jain, Eur. J. Pharm. Sci., 2008, 34, 181–189 CrossRef CAS PubMed.
- A. Haque and A. B. Pant, J. Autoimmun., 2022, 127, 102792 CrossRef CAS PubMed.
- T. G. Flower, C. Z. Buffalo, R. M. Hooy, M. Allaire, X. Ren and J. H. Hurley, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2021785118 CrossRef CAS PubMed.
- J. Shastri, S. Parikh, V. Aggarwal, S. Agrawal, N. Chatterjee, R. Shah, P. Devi, P. Mehta and R. Pandey, Front. Med., 2021, 1379 Search PubMed.
- K. K. Riemersma, L. A. Haddock III, N. A. Wilson, N. Minor, J. Eickhoff, B. E. Grogan, A. Kita-Yarbro, P. J. Halfmann, H. E. Segaloff, A. Kocharian, K. R. Florek, R. Westergaard, A. Bateman, G. E. Jeppson, Y. Kawaoka, D. H. O’Connor, T. C. Friedrich and K. M. Grande, PLoS Pathog., 2022, 18(9), e1010876 CrossRef CAS PubMed.
- J. P. Moore and P. A. Offit, Jama, 2021, 325, 821–822 CrossRef CAS PubMed.
- H. Kawasuji, Y. Takegoshi, M. Kaneda, A. Ueno, Y. Miyajima, K. Kawago, Y. Fukui, Y. Yoshida, M. Kimura and H. Yamada, PLoS One, 2020, 15, e0243597 CrossRef CAS PubMed.
- K. Sharma, A. Koirala, K. Nicolopoulos, C. Chiu, N. Wood and P. N. Britton, Paediatr. Respir. Rev., 2021, 39, 22–31 Search PubMed.
- S. R. Kannan, A. N. Spratt, A. R. Cohen, S. H. Naqvi, H. S. Chand, T. P. Quinn, C. L. Lorson, S. N. Byrareddy and K. Singh, J. Autoimmun., 2021, 124, 102715 CrossRef CAS PubMed.
- W. H. Organization, World Health Organization, https://www.who.int/news/item/28-11-2021-update-on-omicron, 2021.
- O. J. Wouters, K. C. Shadlen, M. Salcher-Konrad, A. J. Pollard, H. J. Larson, Y. Teerawattananon and M. Jit, Lancet, 2021, 397, 1023–1034 CrossRef CAS PubMed.
- M. Chan, Braz. J. Implantol. Health Sci., 2022, 4, 50–54 CrossRef.
- N. Subbaraman, Nature, 2021, 596, 327–328 CrossRef CAS PubMed.
- P. A. Christensen, R. J. Olsen, S. W. Long, S. Subedi, J. J. Davis, P. Hodjat, D. R. Walley, J. C. Kinskey, M. O. Saavedra and L. Pruitt, Am. J. Pathol., 2022, 192, 320–331 CrossRef CAS PubMed.
- P. Y. Chia, S. W. X. Ong, C. J. Chiew, L. W. Ang, J.-M. Chavatte, T.-M. Mak, L. Cui, S. Kalimuddin, W. N. Chia and C. W. Tan, Clin. Microbiol. Infect., 2022, 28, 612.e611–612.e617 Search PubMed.
- S. Yuan, S.-C. Jiang and Z.-L. Li, Front. Public Health, 2020, 8, 240 CrossRef PubMed.
- A. Bal, G. Destras, A. Gaymard, K. Stefic, J. Marlet, S. Eymieux, H. Regue, Q. Semanas, C. d’Aubarede and G. Billaud, Eurosurveillance, 2021, 26, 2100008 CrossRef CAS PubMed.
Footnotes |
| † Present address: Faculty of Engineering, Indian Institute of Technology, Kharagpur 721 302, India. |
| ‡ Present address: Department of Chemistry, Southern Methodist University, Dallas, TX 75205, USA. |
| This journal is © The Royal Society of Chemistry 2023 |