Open Access Article
Open Access ArticleHigh-order framework nucleic acid for targeted-delivery of antisense peptide nucleic acids to overcome drug resistance†
Shu
Xing
ab,
Xiaoqian
Lan
d,
Jiaqian
Zhang
d,
Meng
Li
ab and
Bing
Wang
 *abc
*abc
aNingbo Institute of Materials Technology and Engineering, Chinese Academy of Sciences, Ningbo 315201, China. E-mail: wangbing@nimte.ac.cn
bZhejiang International Scientific and Technological Cooperative Base of Biomedical Materials and Technology, Ningbo Cixi Institute of Biomedical Engineering, Ningbo 315300, China
cUniversity of Chinese Academy of Sciences, Beijing 100049, China
dCollege of Science and Technology, Ningbo University, Ningbo 315300, China
First published on 13th October 2023
Abstract
A sophisticated high-order framework nucleic acid (FNA) was engineered for the targeted delivery and responsive release of environment tolerant antisense peptide nucleic acids (asPNAs). The dendritic FNA-asPNAs system was constructed via simple one-pot modular assembly and demonstrated a good synergistic effect with chemotherapy on drug resistant cancer cells.
Drug resistance is a common but thorny problem in clinical oncology, resulting in over 90% of deaths in patients receiving chemotherapy. Drug resistance can occur through single or multiple mechanisms, including elevated drug efflux, suppressed cell apoptosis, augmented DNA repair and so on.1 Among them, P-glycoprotein (P-gp), a transmembrane efflux pump protein, and B-cell lymphoma 2 (Bcl-2), an anti-apoptotic protein, have been identified as the key resistance mechanisms due to their ability to actively pump drugs out of cells and activate cellular anti-apoptotic defences, respectively. Downregulating the expression levels of these drug resistance-related proteins is a rational way to combat drug resistance. Hence, oligonucleotides such as small interfering RNA (siRNA),2 and antisense oligonucleotides (ASOs)3 have attracted considerable attention and effort in recent decades. Nonetheless, natural oligonucleotides suffer weak binding affinity and susceptibility to nuclease-mediated degradation, resulting in limited bioavailability.
As the third generation of antisense nucleic acids, antisense peptide nucleic acid (asPNA), a chemically modified nucleic acid with a pseudopeptide backbone that replaces the conventional sugar-phosphate backbone of DNA/RNA, was proved with enhanced resistance to natural degradation pathways and exceptionally high affinity and fidelity towards its mRNA target.4 For instance, PNA was reported with only a 20% decrease after 20 hours, compared to siRNA with a half-life of several minutes.5 Meanwhile, the drug resistance reversal efficacy of P-gp asPNA was reported to be 3.1-fold higher than that of ASO with the same sequence in drug-resistant K562 cells.6 These characteristics of asPNA make it an attractive candidate for efficient gene silencing.
Unfortunately, similar to DNA/RNA, cell internalization of asPNA also remains a challenge. Due to its large molecule weight and the lack of positive charges, asPNA exhibits limited permeability across plasma membranes.7 Various types of carriers, including peptides,8 polymers,9 and inorganic nanomaterials,10 have emerged as potential breakthroughs for enhancing cellular uptake of asPNA. However, most carriers suffer from low cargo capacity, limited uniformity, and poor biocompatibility, as well as the incapability to engineer precisely spatially addressable surfaces for the realization of multiple functionalities.
Alternatively, framework nucleic acid (FNA) with a rationally designed geometry, tailorable functionality, and remarkable biocompatibility holds great promise for biomedical applications.11 Different from ssDNA and dsDNA, FNA exhibits high biostability and can enter multiple cell lines. Furthermore, FNA affords an editable size and structure benefitting from its sequence programmability and allows for convenient functionalization of different molecules with defined numbers and positions. However, small-sized FNAs are incapable of complicated formulations and sufficient cargo loading capacity. For instance, tetrahedral FNAs with a size ranging from 5 to 15 nm can only accommodate a restricted number of molecules within a single structure.12 The assembly of basic DNA building units into high-order DNA nanoarchitectures with high yield and relative cost-effectiveness is emerging and offers numerous sites available for multiple modifications.13–15 Therefore, high-order FNAs are expected as superior drug delivery vectors for precise structures, high cargo capacity, and precisely spatially addressable modification.
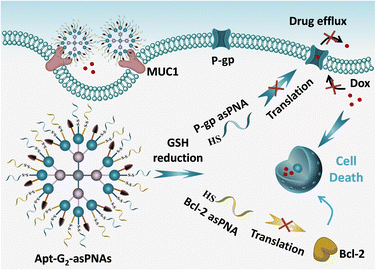
Herein, a multifunctional asPNA delivery system based on high-order FNA is proposed as a strategy to address drug resistance in tumor therapy (Scheme 1). High-order FNA (Apt-G2-asPNAs) with high cargo capacity and multiple modification sites is engineered with MUC1 aptamer and responsive anti-P-gp asPNA and anti-Bcl-2 asPNA. The MUC1 aptamer selectively targets the MUC1 protein overexpressed on Dox-resistant human MCF-7 breast cancer cells (MCF-7ADR), and disulfide linkage modification of asPNAs enable triggered release by the intracellular redox environment in cancer cells. The multifunctional system efficiently delivers asPNA into the target cells, resulting in synergistic inhibition of drug-resistant pathways, restoring the tumor's sensitivity to chemotherapy, and ultimately achieving effective killing of drug-resistant cancers.
 | ||
| Scheme 1 Schematic illustration of RNA silencing and enhanced chemotherapy effect by Apt-G2-asPNAs in drug-resistant cells. | ||
Two asPNAs targeting P-glycoprotein (P-gp) and B-cell lymphoma 2 (Bcl-2) mRNAs were designed and prepared with solid-phase synthesis. Cysteine and fluorophore FAM moieties were modified on either end of the two asPNAs, facilitating subsequent conjugation with mercapto-DNA Linker via the formation of a disulfide bond under DMSO oxidation and enabling optical trace of asPNA. The formation of DNA Linker-asPNA conjugates (DNA-Linker-2-P-gp-asPNA and DNA-Linker-3-Bcl-2-asPNA) was confirmed by reverse-phase HPLC (Fig. S2, ESI†) and denatured polyacrylamide gel electrophoresis (PAGE) (Fig. S3, ESI†). The formation of the disulfide linker and its response to reductants were also verified. Upon addition of reductant TCEP, the bands of DNA Linker-asPNA conjugates disappeared with the recovery of DNA linker bands (Fig. S3, ESI†), which demonstrated the formation of a disulfide bond as the linker and the responsive cleavage for asPNA release.
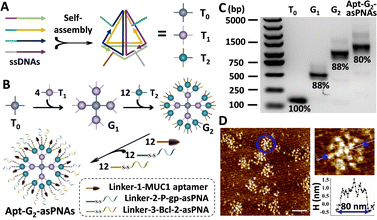
For efficient and targeted delivery of asPNAs, a high-order FNA carrier with targeted functionality was designed. Three 20 bp-edge tetrahedral FNA monomers T0, T1, and T2 were constructed with DNA sequences adapted from a previous report,12,13 and each FNA monomer possessed four single-stranded overhangs along their edges (Fig. 1A and Fig. S4, ESI†). As shown in Fig. 1B, through complementary hybridization of the overhangs, one T0 was used to graft four T1s, resulting in the first-generation dendrimer named G1. Each T1 on the G1 was able to recruit three T2s, forming the second-generation dendrimer structure known as G2. The remaining three different overhangs of each T2 on G2 were designed to hybridize with the corresponding single-stranded DNA Linkers of MUC1 aptamer, P-gp asPNA, and Bcl-2 asPNA, respectively, to prepare the final structure apt-G2-asPNAs.
Following the aforementioned guidelines, tetrahedral FNA monomers T0, T1, and T2 were self-assembled from four oligonucleotides through a one-step annealing process, purified with the established SEC-HPLC method,16 and verified by PAGE (Fig. S5, ESI†). Compared with the DNA tetrahedron without overhangs (T), all three monomers migrated more slowly in electrophoresis and were eluted faster in SEC due to their larger molecule weights. Afterwards, purified T0 and T1 were mixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 molar ratio and incubated at 37 °C for 2 hours to prepare G1. G2 and apt-G2-asPNAs were also formed through a one-step hybridization approach like G1, employing the corresponding stoichiometric proportions. For instance, T0, T1, T2, Linker-1-MUC1 aptamer, DNA-Linker-2-P-gp-PNA, and DNA-Linker-3-Bcl-2-PNA with a molar ratio of 1
4 molar ratio and incubated at 37 °C for 2 hours to prepare G1. G2 and apt-G2-asPNAs were also formed through a one-step hybridization approach like G1, employing the corresponding stoichiometric proportions. For instance, T0, T1, T2, Linker-1-MUC1 aptamer, DNA-Linker-2-P-gp-PNA, and DNA-Linker-3-Bcl-2-PNA with a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 were incubated at 37 °C for 2 hours to yield Apt-G2-PNAs. All as-prepared FNAs were characterized by agarose gel electrophoresis (AGE) (Fig. 1C). Only one major band for each FNA was observed, and the band migration distances decreased with increasing generation and molecule weight, indicating the successful preparation of high-order FNAs. The yield of Apt-G2-asPNAs was estimated as 80% using Image J band intensity analysis, suggesting a hyper-efficient assembly process and pure product even without further purification. The morphology of the FNAs was characterized on an atomic force microscope (AFM). With monomer T0 as a reference, we clearly observed that G1 was composed of five distinct monomers with four arranged around one (Fig. S6, ESI†). Apt-G2-asPNAs (similar to G2 under AFM) were botryoid and compact like a spherical structure (Fig. 1D). Upon morphological examination, it was evident that the Apt-G2-asPNAs/G2 structure boasted a substantial increase in the number of surface binding sites compared to the G1 structure. The diameters of T0, G1, and Apt-G2-asPNAs on the AFM images were measured to be ∼15 nm, ∼59 nm, and ∼80 nm, respectively, which was consistent with electrophoresis results and previous reports.13
12 were incubated at 37 °C for 2 hours to yield Apt-G2-PNAs. All as-prepared FNAs were characterized by agarose gel electrophoresis (AGE) (Fig. 1C). Only one major band for each FNA was observed, and the band migration distances decreased with increasing generation and molecule weight, indicating the successful preparation of high-order FNAs. The yield of Apt-G2-asPNAs was estimated as 80% using Image J band intensity analysis, suggesting a hyper-efficient assembly process and pure product even without further purification. The morphology of the FNAs was characterized on an atomic force microscope (AFM). With monomer T0 as a reference, we clearly observed that G1 was composed of five distinct monomers with four arranged around one (Fig. S6, ESI†). Apt-G2-asPNAs (similar to G2 under AFM) were botryoid and compact like a spherical structure (Fig. 1D). Upon morphological examination, it was evident that the Apt-G2-asPNAs/G2 structure boasted a substantial increase in the number of surface binding sites compared to the G1 structure. The diameters of T0, G1, and Apt-G2-asPNAs on the AFM images were measured to be ∼15 nm, ∼59 nm, and ∼80 nm, respectively, which was consistent with electrophoresis results and previous reports.13
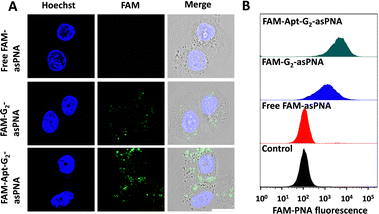
Cell internalization was a crucial prerequisite for asPNA to exert a role in gene regulation. Although the tetrahedral FNA monomers and G1 dendrimers (the overhangs of monomers at the vertex) were reported to enter cells without the assistance of transfection reagents,17,18 the potential of G2 as a novel drug delivery material remained unexplored. Hence, the cell uptake efficiency of asPNA was investigated. FAM labelled P-gp-asPNA (FAM-asPNA) was used to construct FAM-G2-asPNA and FAM-Apt-G2-asPNA. The three FAM labelled materials with a final FAM-P-gp-asPNA concentration of 300 nM were incubated with MCF-7ADR cells for 6 hours for the cell uptake tests on confocal laser scanning microscopy (CLSM) and flow cytometry. As shown in Fig. 2, cells treated with FAM-labelled free asPNA exhibited minimal fluorescence, which was consistent with poor cell uptake of asPNA in the previous report. Conversely, G2-asPNA displayed enhanced asPNA internalization compared to free asPNA and localized within the cytoplasm, suggesting G2-mediated cell internalization. Furthermore, the Apt-G2-asPNA group showed significantly greater asPNA uptake efficiency and cytoplasmic accumulation compared to the G2-asPNA group. This enhancement can be attributed to the targeting capability of MUC1 aptamers, which facilitated selective binding and internalization. Notably, the G2 vector exhibited negligible impact on cell viability even after incubation with MCF-7ADR cells at a concentration of 100 nM for 96 hours (Fig. S7, ESI†), indicating that G2 exhibited excellent biocompatibility. These findings indicated that G2 incorporated with targeting aptamers was an efficient and biocompatible asPNA drug delivery system for potential gene regulation and therapy.
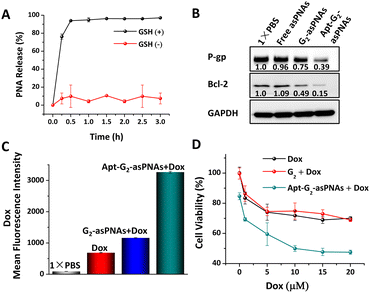
The responsive release of asPNA from Apt-G2-asPNA and following antisense efficiency in drug-resistant cancer cells was further examined. Typically, the concentration of the reducing agent glutathione (GSH) was high in cancer cells (millimolar range).19 5 mM GSH in 1× PBS at pH 7.0 was used to simulate an intracellular reductive environment.2,3 50 nM FAM-labelled Apt-G2-asPNA was incubated with 5 mM GSH at 37 °C to investigate the asPNA release efficiency. After various incubation time intervals, 50 μL of the mixture was subjected to ultrafiltration using 100 kDa centrifugal filters to remove the released FAM labelled asPNA. The resulting residuals were normalized to 100 μL, and the FAM fluorescence intensities and DNA concentrations were measured on a plate reader and an ultra-micro spectrophotometer, respectively. As depicted in Fig. 3A, a rapid asPNA release and nearly complete release at 0.5 h were observed in the presence of GSH, while the DNA concentrations remained relatively unchanged during 3 h incubation (Fig. S8, ESI†). In the absence of GSH, no significant release of asPNA or quantitative changes in DNA were observed. These results demonstrated that the effective responsive release of asPNA from Apt-G2-asPNA can be triggered via GSH reduction. It is worth noting that asPNA release is a crucial prerequisite for asPNA to interact with mRNA targets, thereby leading to the downregulation of downstream protein expression. Consequently, the bioactivity of asPNAs following cell internalization was investigated. After 96 h treatment with free asPNAs, G2-asPNAs, or Apt-G2-asPNAs (300 nM P-gp-asPNA and Bcl-2-asPNA), soluble cell proteins of MCF-7ADR cells were extracted, and western blot analysis was used to evaluate protein expression levels. As shown in Fig. 3B, all treatments involving asPNAs, G2-asPNAs, and Apt-G2-asPNAs exhibited suppression of both P-gp and Bcl-2 protein expression by 4%, 25%, 61% and −9%, 50%, 85%, respectively, with stable GADPH as an internal reference protein. Notably, Apt-G2-asPNAs demonstrated a significantly superior effect in comparison to the other groups, owing to their enhanced ability to target and be taken up by the cells. Furthermore, no obvious non-specific silencing effects were observed using Apt-G2-asPNA containing 1.5 μM (fivefold equivalent excess) P-gp-asPNA alone or Bcl-2-asPNA alone as the negative control for each other (Fig. S9, ESI†). These results highlighted the potential of Apt-G2-asPNAs as a promising antisense material in the treatment of drug-resistant cancers.
To make sure Apt-G2-asPNAs can reverse the drug-resistance of cancer cells, combined therapy of Apt-G2-asPNAs and chemo-drugs was carried out on MCF-7ADR cells. The accumulation of Dox in MCF-7ADR following protein silencing was firstly examined using flow cytometry by detecting Dox fluorescence intensity. After 48-hour incubation with free asPNAs, G2-asPNAs, and Apt-G2-asPNAs, co-incubation with 10 μM Dox was carried out for an additional 6 hours. As displayed in Fig. 3C, the enhanced uptakes of Dox in apt-G2-asPNAs and G2-asPNAs treated cells were determined to be 4.74 and 1.67 times higher compared to the Dox-only treated group, which was in agreement with the P-gp silencing ability in Fig. 3B. The increased drug accumulation was anticipated to overcome tumour drug resistance, which was further demonstrated by proliferation evaluation. Cell viability of MCF-7ADR cells after 96 h treatment with different asPNA-loaded FNAs and Dox from 0–20 μM was measured using the MTT method. As shown in Fig. 3D and Fig. S10 (ESI†), without Dox, Apt-G2-asPNAs showed a slight cytotoxicity compared to non-toxic G2, which should have attributed to the fact that the Bcl-2 silencing promoted cell death because no toxicity was observed on Apt-G2 and Apt-G2-asPNA containing P-gp-asPNA alone.2,20 Along with the increase of Dox concentrations, the G2 + Dox group (cells co-incubated with G2 and free Dox) was similar with that of the Dox group, while the Apt-G2-asPNAs + Dox group showed the highest growth inhibition capability, displaying a measurable IC50 value around 10 μM. These results suggested a synergetic effect achieved by combining Apt-G2-asPNAs with a chemo-drug to recover the drug sensitivity of drug-resistant cancer cells.
In summary, we developed a sophisticated high-order FNA for the targeted delivery and responsive release of environment-tolerant asPNAs. The dendritic hierarchical FNA nanostructure was crafted through a modular assembly of seventeen FNA monomers, boasting a precise structure, impressive cargo-loading capacity, and remarkable biocompatibility. The manifold modification sites of the high-order FNA allow for the functionalization of multiple P-gp and Bcl-2 asPNAs, as well as MUC1 aptamers, thus facilitating cancer cell targeting, heightened internalization, significant downregulation of protein expression levels, and synergistic inhibition of growth when combined with Dox in MCF-7ADR cells. This versatile high-order FNA-asPNA system presents a promising platform to fulfil diverse gene regulation demands, particularly in the context of various forms of drug-resistant cancer.
B. W. gratefully acknowledges support of this work by the start-up funding from the Chinese Academy of Sciences. This research was also supported by the Zhejiang Provincial Natural Science Foundation of China under Grant No. LQ23H180001, Ningbo Natural Science Foundation under Grant No. 2021J035, and Director's Fund of Ningbo Institute of Materials Technology and Engineering, Chinese Academy of Sciences under Grant No. 2022SZKY0304.
Conflicts of interest
There are no conflicts to declare.Notes and references
- C. Holohan, S. Van Schaeybroeck, D. B. Longley and P. G. Johnston, Nat. Rev. Cancer, 2013, 13, 714–726 CrossRef CAS PubMed.
- Z. Wang, L. Song, Q. Liu, R. Tian, Y. Shang, F. Liu, S. Liu, S. Zhao, Z. Han, J. Sun, Q. Jiang and B. Ding, Angew. Chem., Int. Ed., 2021, 60, 2594–2598 CrossRef CAS PubMed.
- Q. Pan, C. Nie, Y. Hu, J. Yi, C. Liu, J. Zhang, M. He, M. He, T. Chen and X. Chu, ACS Appl. Mater. Interfaces, 2020, 12, 400–409 CrossRef CAS PubMed.
- P. E. Nielsen, M. Egholm, R. H. Berg and O. Buchardt, Science, 1991, 254, 1497–1500 CrossRef CAS PubMed.
- V. V. Demidov, V. N. Potaman, M. D. Frankkamenetskii, M. Egholm, O. Buchard, S. H. Sonnichsen and P. E. Nielsen, Biochem. Pharmacol., 1994, 48, 1310–1313 CrossRef CAS PubMed.
- H. L. Wei, Y. J. Wu, T. Jing, D. C. Bai and L. F. Ma, Acta Pharmacol. Sin., 2003, 24, 805–811 CAS.
- T. Shiraishi and P. E. Nielsen, Methods Mol. Biol., 2014, 1050, 193–205 CrossRef CAS PubMed.
- C. J. Cheng, R. Bahal, I. A. Babar, Z. Pincus, F. Barrera, C. Liu, A. Svoronos, D. T. Braddock, P. M. Glazer, D. M. Engelman, W. M. Saltzman and F. J. Slack, Nature, 2015, 518, 107–110 CrossRef CAS PubMed.
- R. Bahal, N. A. McNeer, E. Quijano, Y. Liu, P. Sulkowski, A. Turchick, Y.-C. Lu, D. C. Bhunia, A. Manna, D. L. Greiner, M. A. Brehm, C. J. Cheng, F. Lopez-Giraldez, A. Ricciardi, J. Beloor, D. S. Krause, P. Kumar, P. G. Gallagher, D. T. Braddock, W. M. Saltzman, D. H. Ly and P. M. Glazer, Nat. Commun., 2016, 7, 13304 CrossRef CAS PubMed.
- D. W. Hwang, H. Y. Kim, F. Li, J. Y. Park, D. Kim, J. H. Park, H. S. Han, J. W. Byun, Y.-S. Lee, J. M. Jeong, K. Char and D. S. Lee, Biomaterials, 2017, 121, 144–154 CrossRef CAS PubMed.
- Z. Ge, H. Gu, Q. Li and C. Fan, J. Am. Chem. Soc., 2018, 140, 17808–17819 CrossRef CAS PubMed.
- R. P. Goodman, I. A. T. Schaap, C. F. Tardin, C. M. Erben, R. M. Berry, C. F. Schmidt and A. J. Turberfield, Science, 2005, 310, 1661–1665 CrossRef CAS PubMed.
- J. Li, J. Dai, S. Jiang, M. Xie, T. Zhai, L. Guo, S. Cao, S. Xing, Z. Qu, Y. Zhao, F. Wang, Y. Yang, L. Liu, X. Zuo, L. Wang, H. Yan and C. Fan, Nat. Commun., 2020, 11, 2185 CrossRef CAS PubMed.
- G. Yao, F. Zhang, F. Wang, T. Peng, H. Liu, E. Poppleton, P. Sulc, S. Jiang, L. Liu, C. Gong, X. Jing, X. Liu, L. Wang, Y. Liu, C. Fan and H. Yan, Nat. Chem., 2021, 13, 614 CrossRef CAS PubMed.
- C.-M. Huang, A. Kucinic, J. A. Johnson, H.-J. Su and C. E. Castro, Nat. Mater., 2021, 20, 1264–1271 CrossRef CAS PubMed.
- S. Xing, D. Jiang, F. Li, J. Li, Q. Li, Q. Huang, L. Guo, J. Xia, J. Shi, C. Fan, L. Zhang and L. Wang, ACS Appl. Mater. Interfaces, 2015, 7, 13174–13179 CrossRef CAS PubMed.
- A. S. Walsh, H. Yin, C. M. Erben, M. J. A. Wood and A. J. Turberfield, ACS Nano, 2011, 5, 5427–5432 CrossRef CAS PubMed.
- X. Hu, Y. Huang, H. Zheng, J. Liu, M. Liu, M. Xie, C. Fan and N. Chen, ACS Appl. Mater. Interfaces, 2023, 15, 3839–3850 CrossRef CAS PubMed.
- G. K. Balendiran, R. Dabur and D. Fraser, Cell Biochem. Funct., 2004, 22, 343–352 CrossRef CAS PubMed.
- C. M. Pariante, Eur. J. Pharmacol., 2008, 583, 263–271 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc04298g |
| This journal is © The Royal Society of Chemistry 2023 |