Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Switch-on luminescent sensing of unlabelled bacterial lectin by terbium(III) glycoconjugate systems†
Karolina
Wojtczak
 a,
Eva
Zahorska
a,
Eva
Zahorska
 bcd,
Ian J.
Murphy
a,
Finnja
Koppel
e,
Gordon
Cooke
bcd,
Ian J.
Murphy
a,
Finnja
Koppel
e,
Gordon
Cooke
 e,
Alexander
Titz
e,
Alexander
Titz
 bcd and
Joseph P.
Byrne
bcd and
Joseph P.
Byrne
 *af
*af
aSchool of Biological and Chemical Sciences, University of Galway, University Road, Galway, Ireland. E-mail: Joseph.byrne@ucd.ie
bChemical Biology of Carbohydrates, Helmholtz Institute for Pharmaceutical Research Saarland, Helmholtz Centre for Infection Research, Saarbrücken D-66123, Germany
cDepartment of Chemistry, Saarland University, Saarbrücken D-66123, Germany
dDeutsches Zentrum für Infektionsforschung (DZIF), Standort Hannover-Braunschweig, Braunschweig, Germany
eSchool of Chemical & BioPharmaceutical Sciences, Technological University Dublin, Dublin, Ireland
fSchool of Chemistry, University College Dublin, Belfield, Dublin 4, Ireland
First published on 8th June 2023
Abstract
Interactions of lectins with glycoconjugate-terbium(III) self-assembly complexes lead to sensing through enhanced lanthanide luminescence. This glycan-directed sensing paradigm detects an unlabelled lectin (LecA) associated with pathogen P. aeruginosa in solution, without any bactericidal activity. Further development of these probes could have potential as a diagnostic tool.
Pseudomonas aeruginosa (PA) is a Gram-negative bacterium for which the WHO has declared an urgent need for novel antibiotics and diagnostics.1 It expresses two highly conserved characteristic carbohydrate-binding proteins: lectins LecA and LecB, which are crucial in host-adhesion and biofilm development.2 PA is a leading cause of nosocomial infections, with its societal and economic burden increasing with rising antimicrobial resistance.3 The current clinical gold standard for diagnosis of infectious diseases is a bacterial culture, which can be costly and take days to confirm diagnosis.4 More rapid detection paradigms would aid in the effective and targeted use of antibiotics, and characteristic bacterial lectins offer an underexploited target for detection. While fluorescently-tagged lectins have been used to detect and identify glycans due to the high specificity and selectivity for their particular carbohydrate ligand,5–8 the inverse approach of sensing the native unlabelled lectin itself via luminescent glycoconjugate probes and sensors is much less frequently reported in the literature.9–12 Sensing bacterial lectins by employing the specificity and selectivity of these ligand–protein interactions to give a visual output could open the door to a new paradigm for pathogen detection.
Lanthanide-based luminescent systems possess some particular advantages for development of probes for biomolecules.13 Ln(III) emission is long-lived, with narrow bands corresponding to Laporte-forbidden f–f transitions, giving luminescent signals that can be distinguished from background fluorescence in biological media by time-gated measurement.14 Ln(III) complexes, excited via a chromophore, can also be very sensitive to their environment and emission intensity can be modulated by changes in the coordination sphere, solvent properties/pH or presence of ionic or molecular analytes.15,16
Examples of luminescent sensing of pathogenic carbohydrate-binding proteins are known. For example, upon interaction with the bacterium E. coli, mannose-coated gold nanomaterials undergo fluorescence changes due to interaction with mannose-specific adhesin FimH.17 Polymer fibres functionalised with tetraphenylethyelene–mannoside conjugates also turn-on by Aggregation Induced Emission (AIE) in the presence of FimH, and whole bacteria.18 However, to our knowledge, no luminescent glycocluster molecular sensor has been reported for unlabelled bacterial lectins from PA.
Recently, we reviewed ligand design strategies for inhibition of PA's lectins, in hopes of finding non-bactericidal anti-adhesion therapies for PA infections that would not add to the problem of antibiotic resistance.19 Among the vast range of structures reported for targeting galactophilic LecA, with up to nanomolar affinities, are calixarenes, glycopeptide dendrimers, and carbohydrate-centred clusters as well as covalently-binding imaging agents.11,20,21 Several of these also demonstrate biofilm-inhibition activity. Shared characteristics among the most potent examples are: multivalency to exploit the glycocluster effect, galactoside epitopes functionalised at the anomeric position, and long, flexible linkers to a scaffold that allows bridging between the two vicinal binding sites on the lectin tetramer and/or chelation between different tetramers.19
Metal complexes are poorly represented as inhibitors for PA lectins, despite the potential for new modes of action and properties like luminescence and magnetism. These could be advantageous for sensing, combined with high affinity. We reported Ru(II)-centred glycoclusters following these design characteristics, as the first glycoconjugate metal complex to inhibit PA biofilm formation, without being bactericidal or bacteriostatic.22 Following from this, here we design, on the same principles, luminescent terbium(III) complexes that could be used to sense LecA, as a probe for unlabelled bacterial lectin.
Several examples of metal complex sensors for lectins of vegetal origin are known, but their interactions with bacterial lectins have not been explored. A glycoconjugate Tb(III) complex designed to sense plant lectin ConA (α-mannoside specific) through a LRET mechanism has been reported. This required the lectin to be labelled with Rhodamine-B-isothiocyanate, a methodology that would not be applicable in clinical contexts.23 Other examples include dinuclear Re(I) glycodendrons that exhibit turn-on behaviour in the presence of their targets as a result of a hydrophobic micro-environment upon carbohydrate recognition;10 and an Ir(III) mannoside complex.24 Molecules based on highly conjugated aromatic scaffolds like diketopyrrole manno- and galactosides25 have also been developed to sense lectins by AIE. Attempts to adapt an AIE approach for LecA and LecB have been unsuccessful, even though molecules made for this purpose were nM inhibitors of their target lectin. Their emission signal was either too weak to produce meaningful data or overlapped with the intrinsic fluorescence of the cells.26 Furthermore, Ru(II) glycodendrimer complexes with various degrees of multivalency of mannose and galactose were reported by Seeberger and co-workers to interact with three different lectins resulting in relative changes in fluorescence quantum yields that could be used to create a molecular logic gate system.27 This sets a precedent for the use of changes in emission of inorganic glycocomplexes to detect and differentiate between different lectin–sugar interactions.
The new glycoconjugate ligands presented herein are designed with dual roles of multidentate lanthanide chelation, and high-affinity lectin binding. Consequently, the ligands were built upon a 2,6-dipicolinic acid (dpa) scaffold, well known for Ln coordination;28,29 and display galactoside epitopes linked via the anomeric position to a flexible triethylene glycol spacer, following examples of high affinity LecA inhibitors. In brief, dpa-dichloride was reacted with propargyl amine to introduce alkyne ‘arms’ into precursor 1,30 which was reacted by CuACC ‘click’ chemistry with acetyl-protected azido-glycosides to link carbohydrate motifs to the scaffold with simultaneous introduction of two triazole rings. Divalent compounds 2GalOAc and 2ManOAc were quite polar and were conveniently separated from unreacted starting materials by gradient flash chromatography. Ligands 2 underwent Zemplén deacetylation to yield compounds 3 as very hygroscopic white solids, which were oils under ambient conditions, and were easily soluble in water and aqueous buffer. 1H NMR analysis showed a single set of signals, with doublets at 4.20 and 4.75 ppm, for 3Gal and 3Man respectively, demonstrating coupling constants of 7.5 and 1.5 Hz, consistent with the assigned β-galactoside (Gal) and α-mannoside (Man) stereochemistry, Scheme 1.
To assess the ability of these multidentate glycoconjugate ligands to coordinate lanthanide(III) ions, kinetic self-assembly was studied by titration of 3Gal with Tb(III) in aqueous solution, Fig. 1. Specific aliquots of Tb(CF3SO3)3 stock solution were added to a solution of 3Gal (1 × 10−5 M) and changes to UV-Vis absorption and phosphorescence spectra were monitored. Hyperchromic shifts at 225 and 275 nm were observed, while the intensity of the characteristic Tb(III) emission spectrum increased steadily. Global stability constants were estimated by fitting the absorbance data using non-linear regression analysis program ReactLab Equilibria (Jplus Consulting Pty Ltd), for the formation of the Tb:3Galn (n = 1,3) self-assemblies in solution: log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β1
β1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 = 5.5 and log
1 = 5.5 and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β1
β1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 = 16.4 ± 0.1, which are comparable to related dpa-amide systems.29,31 The speciation diagram (ESI,† Fig. S2) calculated from these data supports formation of a monoleptic complex in solution at suitable Tb
3 = 16.4 ± 0.1, which are comparable to related dpa-amide systems.29,31 The speciation diagram (ESI,† Fig. S2) calculated from these data supports formation of a monoleptic complex in solution at suitable Tb![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand ratios.
ligand ratios.
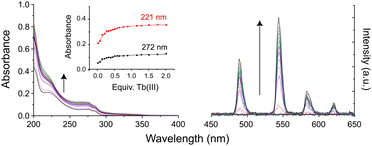 | ||
| Fig. 1 Changes in (left) UV-Vis absorption and (right) phosphorescence spectra of an aqueous solution of 3Gal on addition of Tb(CF3SO3)3. Inset: Binding isotherms. | ||
Ligands 3 were reacted with Tb(CF3SO3)3 in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio to yield solutions of Tb.3, which display green luminescence under UV irradiation, Fig. 2a. Comparison of 1H NMR spectra in D2O to those of the ligands showed significant broadening and slight shifts for all resonances due to the paramagnetic lanthanide ion (Fig. S23 and S24, ESI†). Both complexes Tb.3Gal and Tb.3Man gave characteristic Tb(III) emission bands at 488, 545, 586 and 620 nm, corresponding to each of the 5D4 →7F6–3 transitions.
1 ratio to yield solutions of Tb.3, which display green luminescence under UV irradiation, Fig. 2a. Comparison of 1H NMR spectra in D2O to those of the ligands showed significant broadening and slight shifts for all resonances due to the paramagnetic lanthanide ion (Fig. S23 and S24, ESI†). Both complexes Tb.3Gal and Tb.3Man gave characteristic Tb(III) emission bands at 488, 545, 586 and 620 nm, corresponding to each of the 5D4 →7F6–3 transitions.
For these complexes, in all cases, decay curves of the phosphorescence fit to a single exponential function, indicating a single species in solution. The luminescence lifetimes of complexes Tb.3 in both H2O and D2O solutions were measured to probe the presence of water molecules in the coordination sphere, which can quench Tb(III)-centred emission (Table S1, ESI†). Very high values of apparent hydration state, q,32 were calculated for Tb.3: q ≈ 10 for the galactoside, and 9 for the mannoside (q = 5{(τH2O−1 − τD2O−1) − 0.06}. In addition to the chelating groups in the ligand scaffold, this would indicate unusually large coordination numbers. The presence of many unprotected carbohydrate alcohol groups in the highly flexible ligand structure raises questions about whether this q value is a true reflection exclusively of coordinated H2O, or if sugar epitopes also play a role in quenching luminescence. To evaluate this possibility, complexes of acetyl-protected ligands 2 were also prepared and both shown to have more typical q values (ca. 6) and were consistent for both galactoside and mannoside complexes. The luminescence quantum yield, Φ, of Tb.3Gal was only 7.5%, half that of the mannoside analogue (Φ = 13%). The deviation in both apparent hydration state and quantum yield between Tb.3Gal and Tb.3Man points to greater quenching by the galactose moiety. We hypothesise that binding of the carbohydrate epitope by a receptor will interrupt this quenching and lead to responsive luminescence enhancement.
Before probing luminescent sensing, the affinity of divalent galactoside 3Gal and its Tb(III) complex for PA's galactophilic lectin LecA were determined by a competitive assay based on fluorescence polarisation.33 Both compounds had micromolar affinities in a similar range, and relative potency per galactoside (r.p./n) was enhanced compared to monovalent control due to the ‘glycocluster effect’.34 Ligand 3Gal has IC50 = 163 ± 41 μM (r.p./n = 3.3) and Tb.3Gal 131 ± 26 μM. (r.p./n = 4.1). These data indicate that complexation does not impede lectin binding in solution. Enhancement of LecA's hydrophobic interactions could be made by tailored modification of the aglycon,19,35,36 and indeed we are investigating such structural modifications.
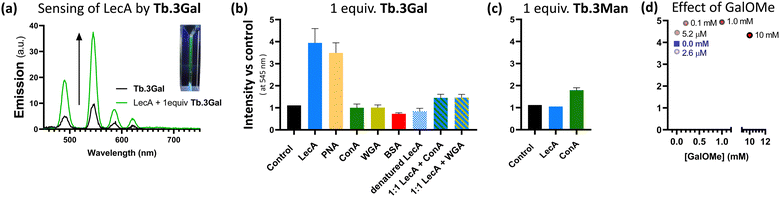
Having designed complexes Tb.3 with the intention of detecting carbohydrate-binding proteins, and established that they can bind to the target bacterial lectin, we chose a panel of isolated lectins to screen for sensing behaviour. In the presence of galactose-binding lectin LecA, Tb.3Gal displayed more than threefold enhancement of the time-gated phosphorescence intensity at 545 nm compared to a control, Fig. 2a and b. Interaction of the complex with carbohydrate-binding domains of the lectin results in dramatic and reproducible ‘switch-on’ sensing by the system. Luminescence lifetimes do not significantly change upon sensing (Table S2, ESI†). Mannose-binding lectin ConA, does not elicit any ‘switch-on’ response with Tb.3Gal, however the mannoside analogue Tb.3Man does demonstrate a doubling of emission intensity with ConA, Fig. 2c. These results highlight that carbohydrate-selectivity of the lectins underpins the sensing paradigm of these lanthanide complexes. Furthermore, peanut agglutinin (PNA, another galactophilic lectin) also led to threefold emission enhancement, while wheat germ agglutinin (WGA, binds sialic-acid and GlcNAc) did not elicit a sensor response. In addition to mismatched lectin–sugar pairs leading to no change in emission, Tb.3Gal also did not sense denatured lectins, indicating that correct protein folding is required for the interactions that drive sensing. Non-specific protein BSA also had no enhancing effect, rather a slight reduction. Addition of competitive ligand GalOMe to ‘switched-on’ Tb.3Gal solutions did not reverse sensing behaviour, even up to 10 mM, Fig. 2d.
An analogous complex, Tb.4Gal, synthesised without a spacer between the triazole and galactose motif (Scheme 1), did not show significant sensing behaviour with either LecA or PNA, despite bearing the appropriate saccharide (ESI,† Fig. S5), indicating that ligand design is important to the sensing ability of lanthanide glycoconjugate complexes. We previously showed how the presence of a flexible linker impacts ability of metal complexes to interact with PA lectins,22 and optimisation of linker arms to facilitate chelate binding of neighbouring carbohydrate-recognition domains has played an important role in developing high-affinity ligands for these lectins.19,37,38
Neither 3Gal nor its Tb(III) complex showed in vitro inhibition of PA growth at concentrations up to 10 mM, nor did crystal violet assays show evidence of biofilm inhibition (see ESI†). Thus, despite their interactions with the target lectin and sensing behaviour, these divalent systems are non-toxic and not antivirulent towards the target bacterium, and have potential for future development towards inert diagnostic probes.
Complex Tb.3Gal shows promise for detecting bacterial lectin LecA in the presence of other proteins. In a solution containing 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixtures of LecA and another lectin with different selectivity (ConA or WGA), a 1.5-fold enhancement in emission was observed, giving clear and reproducible indication of the presence of LecA, Fig. 2b. The more modest intensity enhancement is speculated to be due to the overlap between the UV/Vis absorbance profile of the metal complex and the protein analytes (ESI,† Fig. S6), leading to less efficient sensitisation of Tb(III) emission. The role of competing protein–protein interactions also cannot be discounted. We are developing a second generation of sensor complexes to further improve the system by enhancing binding affinity through careful linker design, and to include conjugated aromatic systems in the scaffold in order to redshift the absorbance maxima. Further optimising this sensing paradigm for detection of characteristic bacterial proteins in complex mixtures and competitive media will be vital to future development of these probes, including the ability to work in the presence of mammalian carbohydrate-binding proteins, such as galectins.
1 mixtures of LecA and another lectin with different selectivity (ConA or WGA), a 1.5-fold enhancement in emission was observed, giving clear and reproducible indication of the presence of LecA, Fig. 2b. The more modest intensity enhancement is speculated to be due to the overlap between the UV/Vis absorbance profile of the metal complex and the protein analytes (ESI,† Fig. S6), leading to less efficient sensitisation of Tb(III) emission. The role of competing protein–protein interactions also cannot be discounted. We are developing a second generation of sensor complexes to further improve the system by enhancing binding affinity through careful linker design, and to include conjugated aromatic systems in the scaffold in order to redshift the absorbance maxima. Further optimising this sensing paradigm for detection of characteristic bacterial proteins in complex mixtures and competitive media will be vital to future development of these probes, including the ability to work in the presence of mammalian carbohydrate-binding proteins, such as galectins.
In summary, we have designed a luminescent glycoconjugate molecular sensor system, which shows reliable enhancement of Tb(III)-centred emission in the presence of unlabelled lectins, including bacterial lectin LecA associated with the critical priority pathogen PA. This ‘switch-on’ response of Ln(III)-luminescence is driven by selective carbohydrate-protein interactions and we have shown that sensing can occur in the presence of competitors.
The authors thank Prof Paul V. Murphy and his group for support. We acknowledge financial support by Science Foundation Ireland (SFI Starting Investigator Research Grant, 18/SIRG/5501, JPB, KW), UoG undergraduate research (IJM), and Dr P. Farras and Dr W. Tong for access to Fluorimeter (SFI, 16/RI/3401).
Conflicts of interest
There are no conflicts to declare.Notes and references
- WHO, Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis, World Health Organisation, Geneva, 2017 Search PubMed.
- A. Imberty, M. Wimmerová, E. P. Mitchell and N. Gilboa-Garber, Microbes Infect., 2004, 6, 221–228 CrossRef CAS PubMed.
- Antimicrobial Resistance Collaborators, Lancet, 2022, 399, 629–655 CrossRef PubMed.
- T. H. Boyles and S. Wasserman, S. Afr. Med. J., 2015, 105, 419 CrossRef.
- A. Hoang, E. Laigre, D. Goyard, E. Defrancq, F. Vinet, P. Dumy and O. Renaudet, Org. Biomol. Chem., 2017, 15, 5135–5139 RSC.
- E. K. C. Tulin, C. Nakazawa, T. Nakamura, S. Saito, N. Ohzono, K. Hiemori, S. Nakakita, H. Tateno, T. Tonozuka and A. Nishikawa, Sci. Rep., 2021, 11, 21973 CrossRef CAS PubMed.
- Y. Choi, U. Park, H.-J. Koo, J. Park, D. H. Lee, K. Kim and J. Choi, Biosens. Bioelectron., 2021, 177, 112980 CrossRef CAS PubMed.
- O. D. Hendrickson, V. D. Nikitushkin, A. V. Zherdev and B. B. Dzantiev, Arch. Microbiol., 2019, 201, 313–324 CrossRef CAS PubMed.
- R. Kikkeri, I. García-Rubio and P. H. Seeberger, Chem. Commun., 2009, 235–237 RSC.
- A. Palmioli, M. Panigati and A. Bernardi, Org. Biomol. Chem., 2018, 16, 8413–8419 RSC.
- S. Wagner, D. Hauck, M. Hoffmann, R. Sommer, I. Joachim, R. Müller, A. Imberty, A. Varrot and A. Titz, Angew. Chem., Int. Ed., 2017, 56, 16559–16564 CrossRef CAS PubMed.
- E. Calatrava-Pérez, J. M. Delente, S. Shanmugaraju, C. S. Hawes, C. D. Williams, T. Gunnlaugsson and E. M. Scanlan, Org. Biomol. Chem., 2019, 17, 2116–2125 RSC.
- D. Parker, J. D. Fradgley and K.-L. Wong, Chem. Soc. Rev., 2021, 50, 8193–8213 RSC.
- J. C. G. Bünzli, J. Lumin., 2016, 170, 866–878 CrossRef.
- H. M. Burke, T. Gunnlaugsson and E. M. Scanlan, Org. Biomol. Chem., 2016, 14, 9133–9145 RSC.
- O. Kotova, S. Comby and T. Gunnlaugsson, Chem. Commun., 2011, 47, 6810–6812 RSC.
- C.-C. Huang, C.-T. Chen, Y.-C. Shiang, Z.-H. Lin and H.-T. Chang, Anal. Chem., 2009, 81, 875–882 CrossRef CAS PubMed.
- L. Zhao, Y. Chen, J. Yuan, M. Chen, H. Zhang and X. Li, ACS Appl. Mater. Interfaces, 2015, 7, 5177–5186 CrossRef CAS PubMed.
- K. Wojtczak and J. P. Byrne, ChemMedChem, 2022, 17, e202200081 CrossRef CAS PubMed.
- M. Bergmann, G. Michaud, R. Visini, X. Jin, E. Gillon, A. Stocker, A. Imberty, T. Darbre and J. L. Reymond, Org. Biomol. Chem., 2015, 14, 138–148 RSC.
- S. Cecioni, R. Lalor, B. Blanchard, J. P. Praly, A. Imberty, S. E. Matthews and S. Vidal, Chem. – Eur. J., 2009, 15, 13232–13240 CrossRef CAS PubMed.
- C. O’Reilly, S. Blasco, B. Parekh, H. Collins, G. Cooke, T. Gunnlaugsson and J. P. Byrne, RSC Adv., 2021, 11, 16318–16325 RSC.
- E. M. Rodríguez, N. Bogdan, J. A. Capobianco, S. Orlandi, M. Cavazzini, C. Scalera and S. Quici, Dalton Trans., 2013, 42, 9453–9461 RSC.
- S. Mandal, R. Das, P. Gupta and B. Mukhopadhyay, Tetrahedron Lett., 2012, 53, 3915–3918 CrossRef CAS.
- J. Wang, Y. Hang and J. Hua, Sens. Actuators, B, 2019, 282, 232–242 CrossRef CAS.
- M. Donnier-Maréchal, S. Abdullayev, M. Bauduin, Y. Pascal, M. Q. Fu, X. P. He, E. Gillon, A. Imberty, E. Kipnis, R. Dessein and S. Vidal, Org. Biomol. Chem., 2018, 16, 8804–8809 RSC.
- R. Kikkeri, D. Grünstein and P. H. Seeberger, J. Am. Chem. Soc., 2010, 132, 10230–10232 CrossRef CAS PubMed.
- D. E. Barry, D. F. Caffrey and T. Gunnlaugsson, Chem. Soc. Rev., 2016, 45, 3244–3274 RSC.
- O. Kotova, J. A. Kitchen, C. Lincheneau, R. D. Peacock and T. Gunnlaugsson, Chem. – Eur. J., 2013, 19, 16181–16186 CrossRef CAS PubMed.
- T. Ziegler and C. Hermann, Tetrahedron Lett., 2008, 49, 2166–2169 CrossRef CAS.
- A. T. O’Neil, N. Zhang, J. A. Harrison, S. M. Goldup and J. A. Kitchen, Supramol. Chem., 2021, 33, 160–173 CrossRef.
- A. Beeby, I. M. Clarkson, R. S. Dickins, S. Faulkner, D. Parker, L. Royle, A. S. de Sousa, J. A. G. Williams and M. Woods, J. Chem. Soc., Perkin Trans. 2, 1999, 493–504 RSC.
- I. Joachim, S. Rikker, D. Hauck, D. Ponader, S. Boden, R. Sommer, L. Hartmann and A. Titz, Org. Biomol. Chem., 2016, 14, 7933–7948 RSC.
- S. Cecioni, J.-P. Praly, S. E. Matthews, M. Wimmerová, A. Imberty and S. Vidal, Chem. – Eur. J., 2012, 18, 6250–6263 CrossRef CAS PubMed.
- F. Casoni, L. Dupin, G. Vergoten, A. Meyer, C. Ligeour, T. Géhin, O. Vidal, E. Souteyrand, J.-J. Vasseur, Y. Chevolot and F. Morvan, Org. Biomol. Chem., 2014, 12, 9166–9179 RSC.
- R. U. Kadam, D. Garg, J. Schwartz, R. Visini, M. Sattler, A. Stocker, T. Darbre and J.-L. Reymond, ACS Chem. Biol., 2013, 8, 1925–1930 CrossRef CAS PubMed.
- C. Ligeour, L. Dupin, A. Angeli, G. Vergoten, S. Vidal, A. Meyer, E. Souteyrand, J. J. Vasseur, Y. Chevolot and F. Morvan, Org. Biomol. Chem., 2015, 13, 11244–11254 RSC.
- S. Cecioni, J.-P. Praly, S. E. Matthews, M. Wimmerová, A. Imberty and S. Vidal, Chem. – Eur. J., 2012, 18, 6250–6263 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, spectroscopic data (emission, NMR), biological data. See DOI: https://doi.org/10.1039/d3cc02300a |
| This journal is © The Royal Society of Chemistry 2023 |