Open Access Article
Open Access ArticleOrganelle-targeted gene delivery in plants by nanomaterials
Simon Sau Yin
Law
 *a,
Takaaki
Miyamoto
*a,
Takaaki
Miyamoto
 a and
Keiji
Numata
a and
Keiji
Numata
 *ab
*ab
aBiomacromolecules Research Team, RIKEN Center for Sustainable Resource Science, Wako, Saitama 351-0198, Japan. E-mail: simonsauyin.law@riken.jp; keiji.numata@riken.jp
bDepartment of Material Chemistry, Kyoto University, Nishikyo-ku, Kyoto 615-8510, Japan
First published on 15th May 2023
Abstract
Genetic engineering of plants has revolutionized agriculture and has had a significant impact on our everyday life. It has allowed for the production of crops with longer shelf lives, enhanced yields and resistance to pests and disease. The application of nanomaterials in plant genetic engineering has further augmented these programs with higher delivery efficiencies, biocompatibility and the potential for plant regeneration. In particular, subcellular targeting using nanomaterials has recently become possible with the cutting-edge developments within nanomaterials, but remains challenging despite the promise in organellar engineering for the introduction of useful traits and the elucidation of subcellular interactions. This feature article provides an overview of nanomaterial delivery within plants and highlights the application of recent progress in nanomaterials for subcellular organelle-targeted delivery.
Introduction
Plant genetic engineering has emerged as a promising method to tackle climate change and its associated problems with a growing population.1,2 It involves the genetic modification and manipulation of plants to improve their traits. This field is critical in developing crops that are resistant to pests, diseases, and environmental stressors, as well as crops with enhanced yield and nutrients.3 The use of modern biotechnological techniques including gene editing and recombinant DNA technology has revolutionized traditional breeding programs and has paved the way for the development of new plant varieties.4,5 This has not only revolutionized agriculture but has also had a profound impact on food security and tackling sustainability targets in the world.Regardless of the targeted result, one of the critical first steps of plant genetic engineering research involves the delivery of genes into the plant. Since the first transgenic plants were generated in the 1980s, researchers have endeavored to develop and advance new gene delivery systems for plants.6–8 Recently, gene delivery in plants has expanded from simple delivery of DNA to introduce a foreign gene into the genome of a plant to the delivery of genome editing proteins such as CRISPR/Cas9 or RNA silencing.9,10
One of the aims of gene delivery is to provide plants with improved traits and it is accomplished traditionally through a range of techniques, including Agrobacterium-mediated transformation and biolistics, but has now recently expanded to the use of nanomaterial-based (NM) carriers.11,12 Furthermore, the ability to target subcellular organelles has been demonstrated in animal cells, but faces further challenges within plants due to the complex plant cell environment and the presence of the cell wall.13,14 This is further compounded by the high copy number of chloroplasts and mitochondria, which are critical for metabolism in plants. Despite these challenges, progress has been made in adapting NM-mediated delivery for organelle-selective targeting.
In this feature article, we review the major organelle targets within plants and the associated challenges regarding plant organelle delivery with a focus on the physical and chemical barriers preventing efficient delivery. We then examine the major classes of NMs that have demonstrated delivery and uptake of cargo within plant cells highlighting their organelle specificity based on their physicochemical properties. We also specifically outline delivery into the three main targets for plant organelle transformation: nuclei, mitochondria, and chloroplasts. Although several other review articles have covered the topic of NM-mediated delivery for plants extensively,15–18 we aim to provide a comprehensive overview on organelle-targeted delivery methods, which are highly relevant for plant bioengineering.
Plants and their organelles
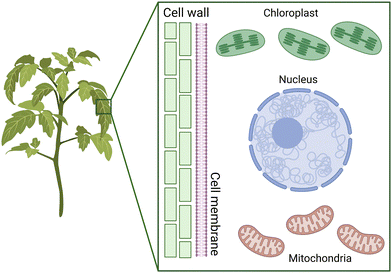
Plants are essential living organisms that play a crucial role in sustaining life on Earth. They are autotrophic, meaning that they can produce their food through photosynthesis, using energy from sunlight to convert carbon dioxide and water into sugars. Plants also play a vital role in regulating the Earth's atmospheric composition, contributing to the production of oxygen and the absorption of carbon dioxide. They form the foundation of food supplies and are critical in feeding the growing population. Furthermore, plants provide a range of vital resources, making them an integral part of human society.Plant cells are characterized by the presence of a cell wall and chloroplasts, and to a lesser extent the existence of a large central vacuole (Fig. 1). They are surrounded by a cell wall, which provides support and protection against mechanical and osmotic stress. Plant cell walls are primarily composed of cellulose, the most abundant macromolecule on Earth.19,20 These cellulose fibers are long, linear polymers made up of hundreds of glucose molecules that aggregate into bundles called microfibrils, which are then embedded in a hydrated network of other polysaccharides.19,20 The cell wall precursor components are synthesized within the cell and then assembled by enzymes that are associated with the cell membrane. Despite the central vacuole shrinking during droughts, the presence of the cell wall helps in maintaining the structural integrity of the plant's stems, leaves, and other structures.20
Along with the nucleus, mitochondria and chloroplasts represent the major targets for genetic transformation and thus gene delivery of exogenous DNA (Fig. 1). Chloroplasts and mitochondria in plants are responsible for photosynthesis and respiration, respectively. These organelles contain their own genomes and are believed to have originated from endosymbiotic events that integrated bacteria into eukaryotes. As a result, these genomes have the potential to improve the efficiency of these processes for bioengineering applications. Additionally, both organelles play a crucial role in producing proteins and small molecules that have therapeutic applications.
Mitochondria are present in almost all eukaryotic organisms, including plants, animals, fungi, protozoa, and protists, and play a crucial role in cellular metabolism.21 Mitochondria are organelles that generate energy for the cell through the process of cellular respiration. They convert the energy stored in sugars and other organic molecules into ATP for the cell.22,23 Mitochondria regulate the production of ATP through oxidative phosphorylation, manage carbon and nitrogen metabolism in the tricarboxylic acid cycle, and influence stress responses through oxidative pathways.22 The capability to genetically engineer plant cell mitochondria has numerous significant applications in both cell biology and agricultural biotechnology.21
Typically, plant mitochondria are organelles that are spherical or rod-shaped and measure around 1–3 μm in length and roughly 0.5 μm in diameter.22 A plant cell typically contains many mitochondria, with estimates suggesting several hundred. These organelles share a similar size and shape with certain types of bacteria, likely since they evolved from endosymbiotic bacteria. The mitochondria contain a double membrane that separates the inner mitochondrial matrix with the intermembrane space. The high pH of the mitochondrial matrix creates the trans-membrane electrochemical gradient that drives ATP synthesis for the plant cell.
In plants, mitochondria have also been associated with important agronomic traits, including cytoplasmic male sterility, which has been utilized in crop breeding and hybrid maintenance.24 Therefore, the development of a robust mitochondrial delivery system could be used to engineer these traits into new plant species and shorten breeding programs. Furthermore, the role of mitochondria in energy generation and respiratory pathways also produces unique metabolites and compounds with applications in therapeutics and natural product synthesis.
Chloroplasts are responsible for the process of photosynthesis, the process in which plants convert light energy into chemical energy in the form of sugars. Chloroplasts contain pigments called chlorophyll that capture light energy and use it to drive the synthesis of sugars from carbon dioxide and water.
Chloroplasts, like mitochondria, are enclosed by a double membrane referred to as the chloroplast envelope. These large organelles are approximately 5 to 10 μm long and have a third membrane system known as the thylakoid membrane, in addition to the inner and outer membranes.25,26 The thylakoid membrane is composed of a network of flattened discs called thylakoids, which are frequently organized in stacks known as grana. This complex three-membrane structure separates chloroplasts into three distinct internal compartments: the intermembrane space between the two membranes of the chloroplast envelope; the stroma, which is situated inside the envelope but outside the thylakoid membrane; and the thylakoid lumen, the site of photophosphorylation during photosynthesis.26
Chloroplasts are particularly effective at producing recombinant proteins, achieving higher product yields than the nucleus. However, introducing new genes to these genomes through plant breeding approaches is not possible since these organelles are maternally inherited. Similar to mitochondria, the outer membrane of the chloroplast envelope is permeable to small molecules. However, the inner membrane is impervious to ions and metabolites, which can only enter the chloroplasts through specific membrane transporters such as the TIC/TOC transporter for chloroplast-targeted proteins.27
Although several techniques such as biolistics and nanotechnologies have been developed for chloroplast and mitochondria transformation, the low efficiency and lack of reliable selection systems have hindered their widespread use. Furthermore, the unique complexities of each of these organelles highlight the challenges associated with targeted delivery into each of these organelles.
Challenges for plant organelle delivery
Although the general uptake mechanisms including the main endocytosis pathways for plants and animals are shared, the receptors and cargo involved vary significantly.28 Our understanding of plant cellular uptake has lagged behind those identified in animal cells, but receptors and transporters related to plant-relevant compounds including ethylene, boron, ammonium and important plant hormones such as auxins have been identified.29–32 Similar to animal cells, clathrin-mediated endocytosis (CME) is the major pathway in the uptake of exogeneous materials including proteins and lipids and is mediated by clathrin-coated pits that sort the intended cargo with clathrin adaptors.31 Also, similar to the uptake pathways found in mammalian cells, clathrin-independent endocytosis mediated by Flottilin-1 has been identified in plants suggesting that membrane microdomains also play an important role in plants.33 Despite these similarities, cargo delivery within plants has had much more limited success than its counterpart within mammalian cell systems.Various techniques are available for genetic material delivery within microbial and animal cells, including chemical treatment, electroporation, sonoporation, microinjection, biolistics, and nanotechnology. However, these methods have not been translated to success within plant systems likely due to differences in biological barriers. Methods like PEG treatment and electroporation covered in the later sections have only worked in protoplasts and not intact plants, likely due to the presence of the cell wall.34,35 Other methods, like sonoporation and injection, have been demonstrated to be applicable for intact plants but have not seen adoption due to tissue damage and low efficiencies.36,37
The challenge of delivering cargo past the plant cell wall can be likened to the physical tissue-based barriers encountered in the traditional mammalian drug delivery such as the vasculature and the blood-tumour barrier.38,39 Both of these obstacles share restrictive size and charge thresholds that prevent the passage of most biomolecules. However, despite advances within NM design including the development of advanced targeting and triggered release systems, localized delivery within these rather inaccessible tissues remain a challenge for both plant and mammalian tissues.40 Other challenges also complicate delivery within both plant and mammalian systems, including protecting the cargo from degradation, targeting specific tissues or cells, bypassing intracellular sequestration and finally releasing cargo at the destination.40
These challenges are further exacerbated in plant organelle delivery in part by fundamental differences in biological barriers in different systems. For example, the cuticle in plants is a physical tissue barrier that must be bypassed for intracellular delivery. Physical barriers, such as the cell wall, cell membrane, and intracellular membranes, must be bypassed and the cargo must be protected from intracellular degradation pathways before delivery. Although plant physiological responses to carriers are different from animal immune responses, strategies for endosomal escape mediated by cationic polymers have also shown to be useful in plant systems.41
The cell wall remains a rather formidable barrier for exogeneous cargo delivery and has a size exclusion limit estimated at 20–50 nm depending on the species, which is two orders of magnitude smaller than cell membrane size exclusion limits (∼500 nm).19,42 Delivery past the cell wall using existing tools requires special attention and development of novel tools to bypass it and simple adoption of animal-cell based delivery systems is likely to fail. However, recent progress has been made in loosening this barrier with chemical treatment to improve delivery efficiencies.43
Furthermore, for organelle-specific delivery, both the chloroplast and mitochondria contain outer and inner membranes, which further complicates the delivery of exogeneous materials. In mitochondria, an outer mitochondrial membrane is separated from an inner mitochondrial membrane by a thin layer, called the intermembrane space and must be bypassed for mitochondrial targeted delivery.23 Similar to mitochondria, chloroplasts are surrounded by two membranes. The outer membrane is permeable to small organic molecules, whereas the inner membrane is less permeable and studded with transport proteins.26 The innermost matrix of chloroplasts, called the stroma, contains metabolic enzymes and multiple copies of the chloroplast genome responsible for most of its metabolic activities. Both of these organelles possess their own protein importation machinery that mediates the entry of unfolded proteins.27 Recently, successful genetic material delivery methods have taken advantage of peptides, which target these receptors as a way to selectively target organelles within plants.44
Nanomaterials for plant delivery
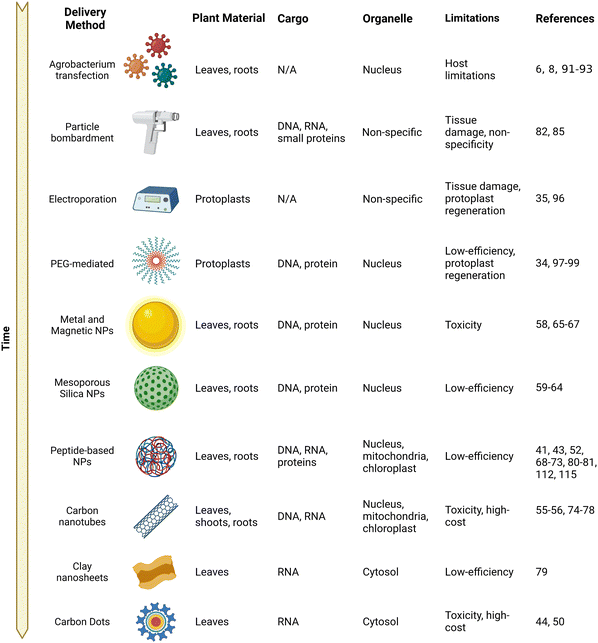
NMs have attracted considerable attention from researchers due to the capability to tune their physical, chemical, and biological properties. NMs have found particularly interesting applications at the interface of materials chemistry and biology. NM-mediated DNA or drug delivery has been demonstrated in both animal cell lines and tissues and intact plants.45–49 The latter has been traditionally challenging due to the extensive physical cell wall barrier that restricts access of other traditional delivery methods. Although plant transformation techniques are relatively mature and have been established approximately three decades ago with the invention of Agrobacterium and biolistic approaches covered in the later sections, transformation remains a bottleneck because many plant species and crop genotypes are recalcitrant to Agrobacterium or respond poorly to regeneration and has otherwise slowed down plant breeding programs. Furthermore, organelle-based transformation has remained rather elusive prior to the advent of NM-based delivery methods.The flexibility and high delivery efficiencies afforded by NMs have brought them to the forefront of delivery into plants. The two main pathways of NM entry into leaves and plant cells are believed to be through their cuticles and stomata.50–52 The overall leaf surface and the particle size of the NMs have been also hypothesized to play an important role in mediating entry into plant cells. Particles with a size smaller than 50 nm have been found to freely penetrate pollen cells, while nanoparticles with a size smaller than 20 nm in at least one dimension are able to enter plant cells.42,53,54 The shape and surface charges of the nanoparticles can also affect their entry into plant cells and increased efficiency has been observed when the net zeta potential of the NM is above 30 mV.55–57
A variety of NMs have been confirmed to deliver a wide range of cargo ranging from chemicals such as pesticides to proteins and nucleic acids for gene expression (Fig. 2). Although an exclusion limit has been proposed for passage across the plant cell wall, the exact mechanisms of entry pathways into plant cells are not yet fully understood for a majority of NMs. A lipid exchange envelope penetration (LEEP) model has been developed to explain the penetration of the high-aspect nanotubes into the double lipid layer of plant cells.54,55 Gold NMs including nanorods and nanospheres have been proposed to embed themselves into the cell wall depending on their shape and do not require physical entry for delivery of their cargo.58 However, the exact subcellular mechanisms are not well described for many NMs. With these physical limitations in mind, a wide variety of NMs have been developed for delivery within plant cells and are highlighted in the following sections.
Mesoporous silica nanoparticles
Mesoporous silica nanoparticles (MSNs) were among the first NMs with potential for specific delivery within plant organelles.59–61 They are NMs made of silica arranged in a honeycomb-like fashion and due to their porous structure, they have a high surface area that is readily modifiable with functional groups due to their exposed silanol groups. Silica particles are also known for their high biocompatibility and inert properties. Furthermore, in MSNs, their chemically and thermally stable mesoporous structures with variable pore sizes (2–10 nm in diameter) have made them ideal for hosting guest molecules of various sizes, shapes and functionalities with potential for gene and chemical delivery.In 2007, Torney and colleagues designed an MSN-based system containing DNA with its chemical inducer that was capped allowing for controlled release and induction of gene expression upon successful delivery within the plants.60 The loading of DNA into the MSNs protected them from unwanted cleavage from nucleases and this system was shown to be efficient in delivery into protoplasts (plant cells without cell walls). Further modification of the MSNs with a gold nanoparticle cap gave them sufficient density to be propelled using particle bombardment, and they were successfully delivered into intact tobacco cotelydons for gene expression. Immature embryos from maize were also bombarded with the capped MSNs and were allowed to proliferate to form a callus. After ten days, GFP expression could be observed in the proliferating regions, suggesting that stable integration of DNA in callus cells could be achieved using MSNs as a delivery vehicle.
Several years later, another study by Chang et al. demonstrated that organically functionalized MSNs could deliver genetic material into intact A. thaliana nuclei by simple incubation.62 Other studies have also demonstrated the potential of MSNs and their ability to deliver both DNA and proteins into intact plants and suggest that MSNs could be an easy-to-use delivery vehicle for delivery into plants.59,63,64
Metal and magnetic nanomaterials
Metal nanoparticles (NPs) such as gold, silver, platinum, and palladium have unique optical properties and have become an important tool for delivering drugs, genes, and other therapeutic agents within mammalian cells.65 They usually have a metal core containing an inorganic metal or metal oxide being made up of organic or inorganic materials or metal oxides that allow for functionalization with cargo.In plants, metal NPs have been used as vehicles in conjugation with other NMs such as MSNs. In 2013, Hao and colleagues synthesized a magnetic gold NP composite functionalized with PEG, which allowed for conjugation of DNA on the surface.66 Upon the application of an external magnetic field, the NPs could be delivered into canola protoplasts. This was further developed as a delivery method into pollen, which has a rather large aperture and a permeable cell wall, and given the term, magnetofection, by Zhao et al. as reported in 2017.67 They were able to deliver magnetic NPs functionalized with polyethyleneimine and exogenous DNA within cotton pollen through an external magnetic field capable of both transient expression and stable integration of the transgene.67
A recent study by Zhang et al. demonstrated that the size and morphology of gold NPs play an important role in determining their entry across the cell wall.58 They prepared gold NPs with either spherical (5–20 nm) or rod (13 × 68 nm) morphology and functionalized with single-stranded DNA or RNA with a thiol modifier. Smaller round gold NPs were unable to cross the cell wall barrier but could associate with the cell wall, whereas higher aspect gold nanorods were able to traverse through the cell wall. Interestingly, both particles were able to successfully deliver siRNA into the plant cells for gene silencing of a GFP reporter construct with the round gold NPs achieving higher delivery efficiencies despite not being able to efficiently enter the plant cell. These results suggest that the morphology of metal NPs affects their role in plant gene delivery and could guide future NP design for efficient uptake.
Peptide-based nanomaterials
Peptide-based NMs are a class of NMs that are derived from or contain peptides. These NMs have unique properties such as biocompatibility, biodegradability, and target specificity that make them attractive for a variety of applications in biomedicine, drug delivery, and biotechnology.68 Additionally, the use of naturally occurring amino acids in the synthesis of peptide-based NMs reduces the relative risk of toxicity compared to other synthetic NMs.69,70Peptide-based NMs can be designed to self-assemble into various structures, such as nanofibers, nanotubes, and nanoparticles, providing tunable properties for specific applications. For example, they can be designed to mimic natural peptides that target specific cell types, making them useful for targeted drug delivery. In recent years, there has been a growing interest in the use of peptide-based NMs for various applications, such as targeted cancer therapy, regenerative medicine, and tissue engineering. The ability to design and synthesize peptide-based NMs with specific properties makes them a promising tool in plant biotechnology.
Sequences can also be engineered to bind specific cargo for delivery or adopt a specific physical orientation. For example, polycation peptides are capable of condensing various sizes and types of DNA and forming peptide/DNA complexes, protected from nuclease-mediated degradation. Cell-penetrating peptides (CPPs) have been shown to traverse through the plasma membrane in a wide range of plant species and tissues.71–73
One of the earliest studies of cargo delivery into plants by peptide-based NMs was performed by Chugh and Eudes.72 They demonstrated delivery of both DNA and intact proteins into permeabilized wheat immature embryos using the TAT protein transduction domain. Tat CPP was able to complex with pDNA containing the reporter construct GUS and protect it from degradation. The pDNA could be successfully delivered into the nucleus for gene expression. Furthermore, they were also able to complex the GUS protein with the same Tat CPP and it could be transported into the wheat embryo, suggesting that peptide-based NMs have tremendous flexibility in the cargo they can carry.
Carbon nanotubes
Carbon NMs are a group of nanoscale materials with unique properties and characteristics that make them ideal for a wide range of applications, including drug delivery. Carbon NMs include nanotubes, graphene, and fullerenes, which are all composed of carbon atoms arranged in various geometric configurations. In particular, carbon nanotubes (CNTs) have emerged as a highly efficient method of gene delivery in plants with tremendous delivery efficiencies and organelle specificity.55,56,74CNTs are long thin cylindrical molecules consisting of rolled up sheets of graphene. They have a high surface area-to-volume ratio, and are generally <2 nm in diameter and generally up to a few micrometers in length. Additionally, their chemical and mechanical stability, biocompatibility, and tunable surface functionalization make them highly flexible for targeted delivery within cells. Furthermore, CNTs have strong intrinsic near-infrared (NIR) fluorescence as well as Raman emission within the cell-silent window and thus allow for tracking of cargo–nanoparticle complexes deep in plant tissues.75–77
One of the first reports of CNTs as potential carriers for intact plant cells was by Liu and colleagues in 2009.78 They showed that FITC-labelled SWNTs were able to enter tobacco Bright Yellow (BY-2) cells, a popular plant cell model, through simple incubation. An intense fluorescence signal could be observed within the cytosol of the incubated cells, suggesting that CNTs could readily translocate into intact plant cells. Furthermore, ssDNA could complex on the surface of the CNTs through aromatic interactions and subsequent results indicated that the CNT/DNA complex was also able to enter the plant cells under similar conditions.78
More recently, the use of polymer conjugated CNTs has been shown to selectively deliver DNA and RNA into specific organelles within the plants. Demirer et al. were among the first to describe the use of DNA grafted on PEI-modified CNTs for delivery within plant nuclei in 2019 (Fig. 3).55 Remarkably, it was demonstrated that this method could be applied to multiple plant species. Recent developments have emerged using CNTs and they have been demonstrated as extremely versatile carriers capable of delivery to all of the major organelle targets within plants as outlined in greater detail within the following organelle-specific sections.55,56,74 These results highlighted the potential of CNTs as carriers for subcellular delivery within plants.
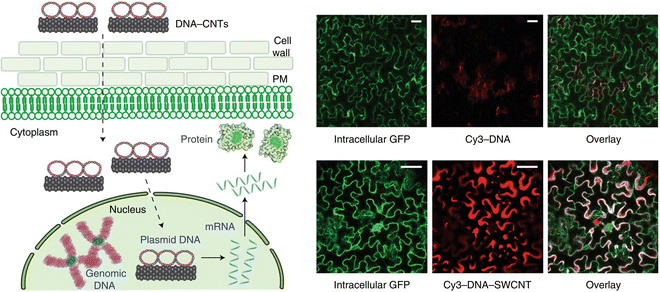 | ||
| Fig. 3 CNT-mediated delivery and expression of exogeneous pDNA in plants. In one of the earliest reports of DNA expression using carbon nanotubes within intact plants, Demirer and colleagues described the use of DNA grafted on PEI-modified carboxylated CNTs for delivery of pDNA into the nuclei of leaves in mature leaves in multiple plant species. DNA is grafted on the surface of the CNTs via π–π stacking or electrostatic interactions with the PEI on the surface of the CNTs. They then demonstrated successful DNA delivery in a variety of plants including N. benthamiana, arugula, wheat and cotton. Cy3-DNA could be observed internalized when delivered in complex with CNTs in N. benthamiana as shown in the accompanying fluorescence micrographs. The DNA is then able to be released for transient expression of a GFP reporter construct. Scale bars represent 50 μm. Reproduced with permission from Springer Nature, rights to the material are owned by a third party.55 | ||
Other nanomaterials
Recently, in the past five years, other novel applications of NMs such as clay nanosheets and carbon dots have been developed as potential carriers for gene delivery within plants.Clay nanosheets have shown promise for the delivery of RNA for gene silencing through siRNA pathways. Clay nanosheets are sheet-like nanoparticles made of clay and inorganic layers that are approximately 30–80 nm in diameter. A study in 2017 by Mitter and colleagues demonstrated the potential use of clay nanosheets for RNAi delivery in plants.79 Double-stranded RNA (dsRNA) could be directly grafted on the surface of the nanosheets and is protected from degradation when the NM is delivered on the leaves of A. thaliana. Upon release, the dsRNA was able to provide RNAi-based systemic protection against targeted viruses even when challenged 20 days after a single spray.
Carbon dots (CD) are small carbon-based NPs with fluorescence that are known for their unique optical properties with applications for photos and imaging. CDs were also demonstrated as potential siRNA carriers by Schwartz et al. in 2020.50 They used PEI-functionalized CDs to bind dsRNA on the surface to protect them from degradation from RNases. The relatively small size (∼20 nm) of the CDs allowed for their entry into intact N. benthamiana and tomato plant leaves upon adaxial spray. GFP silencing of a transgenic line was detected upon spraying with CDs carrying the silencing siRNA targeting the GFP gene. Furthermore, CDs were also efficacious at silencing endogenous genes when used with an siRNA targeting magnesium chelatase, an enzyme in chlorophyll synthesis. In a recent study by Santana and coworkers, CDs have shown potential for chloroplast-targeted vehicles as well.44
Organelle-targeted delivery methods
The development of delivery systems capable of subcellular targeting has tremendous promise in the field of plant bioengineering due to the importance of the chloroplast and mitochondria in plant metabolism and their potential for bioengineering. Recent efforts have been made to target individual organelles including the chloroplast and mitochondria but most research focused on delivery into the nucleus due to its importance for gene expression. Some selectivity has been afforded by the use of targeting peptides or electrostatic interactions for mitochondria and chloroplasts, respectively, but highly specific delivery remains challenging.56,74,80,81 Here, we highlight current methods of gene delivery in the nucleus, mitochondria and chloroplast along with the latest developments in organelle-targeted delivery developed by the Numata group.Nucleus-targeted gene delivery
The nucleus has long been the primary target for genetic modification in plants because most genetic information is contained in chromosomal DNA. To date, much effort has been made to develop DNA delivery methods for nuclear transformation of plants. Existing delivery methods are typically classified into three categories: biological (Agrobacterium-mediated DNA transfer and viral transfection), physical (particle bombardment and electroporation), and chemical approaches (PEG-mediated protoplast transfection).6,8,82–86 In the last few decades, reports on NM-mediated DNA delivery to the plant nucleus have increased rapidly as outlined in the previous section. Since several excellent reviews have summarized the current status of DNA delivery methods for nuclear genome engineering, we describe only an overview of the traditional methods and NMs applicable for DNA delivery to the plant nucleus.18,87–90Agrobacterium-mediated DNA transfer is the dominant method for nuclear transformation of plants.6,8 This method is based on the ability of a soil bacterium, Agrobacterium tumefaciens, to transfer T-DNA, a small region of its tumor-inducing plasmids, into the nuclear genome of host plants. By replacing pathogenic genes in the T-DNA region with transgenes, Agrobacterium can be used as a safe DNA delivery system for transformation of host plants.6Agrobacterium-mediated T-DNA transfer is driven by several virulence genes that support the production and transport of T-DNA and virulence proteins from bacterial cells into plant cells, targeted T-DNA import into the plant nucleus, and T-DNA integration and expression in the plant nuclear genome.91 The expression of these virulence genes is induced by phenolic signaling molecules that are produced in wounded dicotyledonous plants but not in monocots, resulting in the inability of Agrobacterium to infect monocots.92 This limited host range can be overcome by the use of phenolic inducers, such as acetosyringone, which promote virulence gene expression and facilitate Agrobacterium-mediated transformation of various monocotyledonous plants.93 However, many economically important cultivars are still recalcitrant to Agrobacterium-mediated transformation, highlighting the need for the development of new transformation techniques.
Viral vectors are another biological delivery system for plant genetic modification.94 They have long been used for transient heterologous protein expression and gene silencing in plants. Recently, viral vectors have been gaining interest as an efficient tool for DNA-free plant genome editing. However, the use of viral vectors presents many challenges, including their narrow host range, the very limited size of the cargo DNA, and concerns about the integration of toxic viral genes into the host genome or the spread of infection.95
Particle bombardment is a physical approach used to deliver cargo into plants and is as popular as the Agrobacterium-mediated method.82 It involves the formation of metal particles, such as gold or tungsten, functionalized with different cargoes such as DNA. A high-pressure gas then accelerates the particles to high velocity at the epidermal tissue of plant cells.85 The DNA is then able to penetrate both the plant cell wall and the plasma membrane and enter the nucleus. Although particle bombardment can be used as a plant species-independent DNA delivery method, its application in commercial breeding remains limited. This is due to its tendency to integrate multiple copies of the transgene into the plant genome at random locations and chromosomal disruptions caused by highly pressurized particles. Because several parameters, including gas pressure, particle size, and dosing frequency, determine transformation efficiency and toxicity, extensive optimization is often required for each target plant and application.
Finally, electroporation involves the application of an electrical pulse to plant cells to create small pores through which the desired DNA can be introduced, which was originally developed in 1982 for protoplast transformation.35,96 However, electroporation has been standardized for several plant species, including tobacco, rice, wheat, and maize. Although it is a fast and relatively inexpensive method, electroporation is not widely used for plant transformation because it is limited to a few plant species that do not have thick cell walls. In addition, strong electric field pulses damage both the delivered DNA and the target cells, reducing overall efficiency.
Polyethylene glycol (PEG) is a petroleum-derived polyether polymer available in a range of molecular weights. The polymer is hydrophilic and has low biotoxicity. One of its primary uses in gene delivery is as a transfection agent to increase the permeability of the plasma membrane and improve the transmissibility of charged macromolecules such as DNA.34,97–99 PEG-induced protoplast, plant cells with the cell wall enzymatically removed, transfection was the earliest method used in plant transformation.99 The PEG solution reverses the permeability of the cell membrane, allowing cargo such as nucleic acids and proteins to enter the cell and nucleus. This method is easy to perform on many plant species, but regeneration of the transfected protoplasts into mature plants is time-consuming, laborious, and has low efficiency. However, recent studies have expanded the use of PEG-mediated delivery of genome-editing enzymes, reviving interest in this method for plant genetic engineering.98
DNA delivery to the plant nucleus has also been achieved by many types of NMs such as mesoporous silica, magnetic nanoparticles, carbon dots, CNTs, and peptide-based nanocomplexes as outlined in the previous sections. These NMs typically incorporate cationic polymers to form electrostatic complexes with the DNA cargo, protecting it from enzymatic degradation. Although little is known about the mechanisms by which NMs achieve nuclear targeting and import of cargo DNA, NM-based delivery systems have the potential to deliver DNA cargo to intact plant cells in a species-independent manner without the use of specialized equipment or preparation of protoplasts that are difficult to regenerate. Despite these potential advantages over the conventional methods, stable transformants obtained using NMs have been relatively limited. This is likely due to their insufficient DNA delivery efficiency and/or their inability to efficiently integrate the foreign DNA into the nuclear genome.
To overcome these limitations, NM design needs to be optimized for the cell wall translocation, cell entry, cytosolic transfer, nuclear targeting, and cargo DNA release at the target site. The aspect ratios of CNTs and gold nanorods have been proposed to contribute to efficient cell wall permeation, suggesting that nanoparticle morphology plays an important role in determining nuclear delivery efficiencies.55,100 NMs have been shown to enter plant cells via energy-independent direct membrane permeation and/or energy-dependent endocytic pathways. The former mechanism has been proposed for highly charged NMs, while the latter has been frequently observed for peptide-based complexes.52,54,101–104
For gene delivery into intact plants, the Numata group has developed the use of peptide-based NMs to specifically deliver nucleic acid cargo into the nuclei of intact plants.52,70,73 To facilitate access into the plant cells, most peptides contain a CPP or CPP-like domain and are either coincubated or fused with another peptide to facilitate cargo complexation (Fig. 4). To enhance endocytic uptake, a synthetic peptide that could induce a macropinocytosis-like cell entry mechanism in plants was developed.105 The use of the macropinocytosis-inducing peptides enabled significantly enhanced cellular uptake of a peptide/DNA complex, resulting in improved transfection in plant calli.103 One of the major barriers to endocytic uptake is endosomal entrapment and vacuolar degradation of the cargo molecule. This barrier has been mitigated by the use of an endosome-disrupting peptide, which can facilitate the translocation of DNA from endosomes to the cytosol in plants, resulting in enhanced DNA delivery to the nucleus41 (Fig. 5). A nuclear localization signal (NLS, a short peptide that mediates nuclear transport of proteins) has also been used to aid in the nuclear targeting of NMs, but its contribution was mitigated by inadequate cellular uptake and cytosolic translocation of NMs.70,106 The cargo DNA must be released from the NMs to ensure the transgene expression. Although a stimulus-responsive DNA-binding peptide has been reported to release the DNA in response to reducing conditions within plant cells, NM-based delivery systems for efficient DNA release have not been sufficiently explored in plants.107 In most cases, the DNA delivery efficiency of NMs is not sufficient and elucidation of the detailed mechanisms of nanoparticle–plant interactions may provide the effective design principles for next generation delivery platforms.
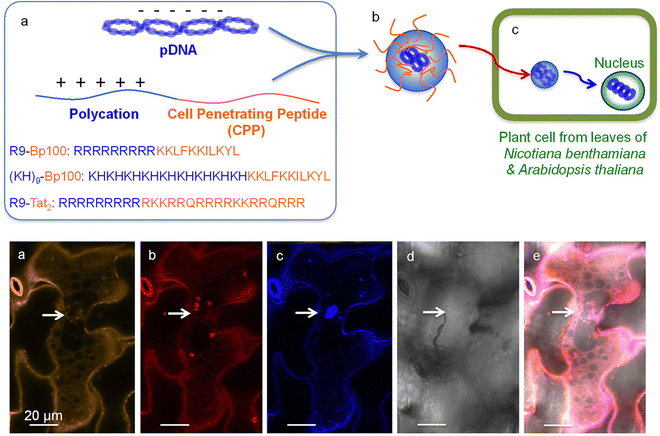 | ||
| Fig. 4 Peptide-based delivery system for gene delivery within N. benthamiana and A. thaliana. Lakshmanan et al. described one of the earliest applications of a fusion peptide system for plants by combining cell penetrating peptides (BP100 or Tat2) and a polycation repeat peptide (R9 or KH9) as a peptide-based delivery system into plant nuclei. The Cy3-labelled pDNA could be observed within the leaf cells in proximity to the nuclei upon the delivery of the complex with observation of the expression of the delivered GFP construct. Scale bars represent 20 μm. Adapted with permission from the American Chemical Society.73 | ||
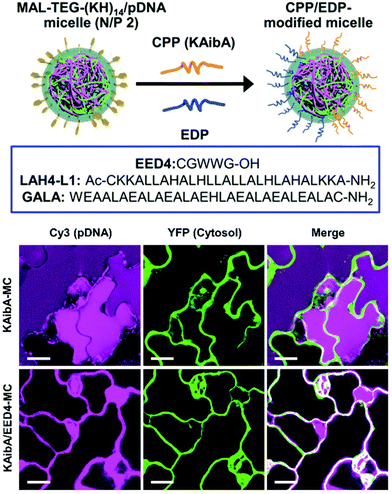 | ||
| Fig. 5 Endosomal disrupting peptide (EDP) for endosomal escape for improved delivery efficiency. A surface modifiable micelle complex composed of a maleimide-containing polycation peptide (MAL-TEG-(KH)14 could be decorated with different CPPs or EDPs on the surface of the micelle to improve their endosomal escape properties. The polycationic is able to condense DNA within the interior of the micelle. Following transfection in the leaves of A. thaliana cotelydons, a significant increase in cytosolic translocation could be observed in the presence of the EDP. Scale bars represent 20 μM. Adapted with permission from the Royal Society of Chemistry.41 | ||
Mitochondria-targeted gene delivery
Research into mitochondria-targeted delivery has been relatively limited due to the challenges in manipulating mitochondrial genomes in whole cells. Most mitochondrial genetic transformations were reported first in isolated mitochondria, likely due to the fact that isolated mitochondria maintain natural competence to import double-stranded linear DNA and allow for incorporation of exogenous DNA. The double membrane and relatively small size of the organelle, in addition to its large cellular population and the subsequent difficulty in obtaining a homoplasmic transformant, are major obstacles in mitochondrial transfection in both plants and animal cells. Furthermore, subgenomes are commonly found in mitochondria with high-frequency intermolecular and intramolecular recombination of large repeated sequences that can be further subdivided, compounding the transfection problem.108For plants, transformations through electroporation of isolated wheat mitochondria were used to study mitochondrial transcription and RNA maturation processes.109 A mitochondria-targeted adeno-associated virus has also recently proved to be effective as a DNA delivery method in a mouse model system and until recently, mitochondria of Chlamydomonas reinhardtii and two yeast species (Saccharomyces cerevisiae and Candida glabrata) were the first exceptions regarding the transformability of isolated mitochondria and were transformed within intact cells using biolistic methods.110
Several critical hurdles have since been identified as being important in plant mitochondrial transfection in addition to their overall complexity and instability. One of the first reports of plant mitochondrial transformation was in 2003 by Koulintchenko and colleagues using isolated plant mitochondria.108,111 dsDNA could be uptaken into plant mitochondria through an active, transmembrane potential-dependent mechanism without sequence specificity. The uptaken DNA is able to be integrated into the mitochondrial genome through homologous recombination. Following recombination into the mitochondrial genome, the exogenous DNA can be transcribed and processed into mature RNA molecules if the gene is controlled by a mitochondrial promoter.
For delivery of DNA into intact plants, the Numata group has developed a method of intracellular delivery of exogenous DNA into the mitochondria of infiltrated A. thaliana leaves using a combination of mitochondria-targeting (Cytcox) and CPPs (Fig. 6). The delivered DNA is capable of both transient expression and genetic integration (Fig. 6). This was initially demonstrated using rationally designed peptide sequences that could target the mitochondria and self-assemble with DNA. A peptide containing a mitochondria-targeting component consisting of the first twelve amino acids of the presequence capable of directing proteins into the mitochondrial matrix was combined with a polycationic peptide containing histidine and lysine to condense pDNA.113,114 This peptide was able to complex pDNA and when combined with a CPP, delivery and expression of a pDNA containing a GFP reporter construct could be observed within the mitochondria of A. thaliana leaves upon infiltration.115 Follow-up studies using this peptide showed that the delivered pDNA was also capable of integration into the mitochondrial genome and a reporter GFP construct could be actively translated upon integration.112
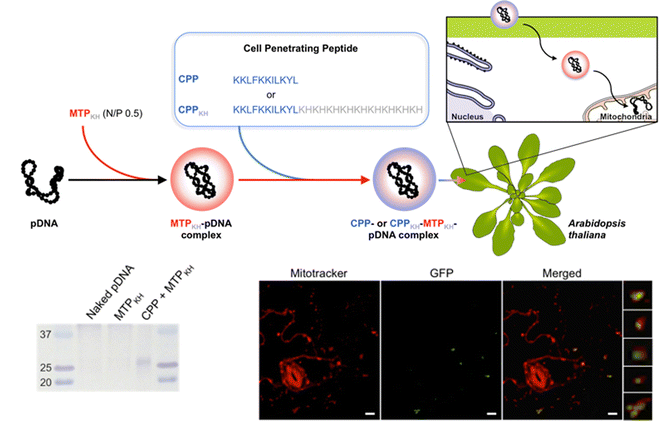 | ||
| Fig. 6 Mitochondrial DNA delivery within A. thaliana using a peptide-based approach. Chuah et al. described the use of a peptide–pDNA complex approach for mitochondrial delivery of exogenous DNA. pDNA complexed with a mitochondria targeting peptide and a CPP were used to deliver pDNA into mitochondria within intact A. thaliana through vacuum infiltration. The gene expression of the reporter construct, GFP, could be detected within Mitotracker-labelled mitochondria using confocal microscopy and by western blot analysis. Scale bars represent 10 μm. Adapted with permission from Springer Nature.80 | ||
Furthermore, the use of carbon nanotubes functionalized with the same mitochondrial targeting and DNA binding sequences showed that delivery efficiencies could be enhanced almost 30-fold compared with the peptide only approaches as demonstrated in a recent study by Law et al.56 (Fig. 7). The CNTs could be loaded with DNA for delivery into intact A. thaliana mitochondria. The higher delivery efficiencies translated into greater stability in the genetic integration and expression of a GFP reporter construct could be detected seven days post-infiltration. Furthermore, the integration of a folate reductase gene also demonstrated increased intracellular folate levels leading to greater root growth, which highlights the potential of this system for further bioengineering applications.
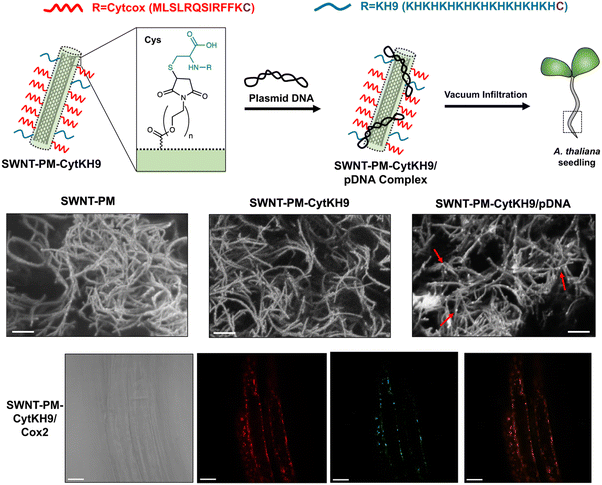 | ||
| Fig. 7 CNT-mediated delivery of DNA into intact plant mitochondria. Law et al. described the use of CNTs with a polymer coating that could be conjugated with a mitochondria-targeting (Cytcox) and a DNA complexing polycationic peptide (KH9) that was used to deliver pDNA into the mitochondria of intact A. thaliana. The CNTs could be observed as long thin strands with pDNA complexed on the surface upon conjugation with the peptides and in complex with pDNA. The CNTs could efficiently complex with pDNA and deliver it within the roots of A. thaliana for gene expression within the mitochondria. Scale bars in the SEM micrographs and confocal microscopy images represent 200 nm and 20 μm, respectively. Adapted with permission from Springer Nature.56 | ||
In addition to genetic integration and expression of foreign DNA, mitochondria genome engineering has expanded into editing of the existing mitochondrial genome through transcription activator-like effector nucleases (TALENs).24,116 Kazama and coworkers recently reported the use of mitochondrial targeted TALENs to stably knock out genes associated with cytoplasmic male sterility in rice and rapeseed.24 Double stranded breaks could be introduced that are repaired via homologous recombination with deletion of the surrounding sequences. A follow up study by the same group also showed that this could be applied to A. thaliana mitochondria, suggesting that this method can likely be used for genetic manipulation of the mitochondrial genome in a variety of other plants.116
Chloroplast-targeted gene delivery
Chloroplasts are essential organelles that are responsible for photosynthesis and play a central role in primary and secondary metabolism in plants. The genetic manipulation of chloroplasts has been used to improve photosynthesis and carbon assimilation, as well as to provide resistance to insects, herbicides, and abiotic stresses.117,118 Chloroplast genetic transformation represents a promising strategy for using plants as sustainable bioreactors to produce pharmaceuticals, antibodies, and vaccines due to its superiority over nuclear transformation.119 Notably, chloroplast transformation allows for high levels of transgene expression, as each plant cell contains numerous chloroplasts with a high copy number of the chloroplast genome.120 Moreover, chloroplast transgene expression is unaffected with gene silencing, which is often found in nuclear transformation.121 Additionally, the compartmentalization of toxic transgene products in chloroplasts can minimize adverse effects. Since most angiosperm species exhibit maternal inheritance of chloroplast genomes, chloroplast transformation avoids transgene transfer via pollen, significantly mitigating the environmental risks of transgenic plants.122Over the last three decades, particle bombardment has been the prevailing technique for introducing foreign DNA in chloroplast transformation.123 With this method, gold or tungsten particles coated with DNA containing two homology arms, whose DNA sequence is identical to the target chloroplast genome, are forcefully propelled into the plant cell. When the particles happen to hit the chloroplasts by chance, the DNA is incorporated into the chloroplast genome via homologous recombination. However, this method cannot target specific organelles, is plagued by low transformation efficiency, and requires specialized equipment. Consequently, only a meagre ten plant species have demonstrated stable and reproducible chloroplast transformation using this method.124 Another alternative for chloroplast transformation is PEG-mediated transfection of protoplasts, plant cells that lack a cell wall.125 However, the use of this approach is hindered by the difficulty of regenerating whole plants from protoplasts.
In contrast to the previously mentioned conventional methods, the utilization of engineered NMs has simplified delivery of foreign DNA selectively to chloroplasts, obviating the need for specialized equipment that employs high-pressure or the laborious and time-consuming handling of protoplasts. Despite these potential advantages, NMs successfully used for the delivery of DNA to chloroplasts remain limited to single-walled carbon nanotubes (SWCNTs) and peptide-based nanocomplexes.43,44,74,81,126 SWCNTs have demonstrated high efficiency in penetrating plant cells with cell walls, although the mechanisms and reasons for their successful translocation across the cell wall and membrane remain poorly understood.55
Kwak and coworkers have reported the utilization of chitosan-functionalized SWCNTs to deliver plasmid DNA (pDNA) to chloroplasts in intact plants.74 In this system, chitosan was employed for electrostatic complexation with pDNA at weakly acidic pH (pH < 6.0), while the pDNA was released in the relatively basic pH conditions of the chloroplast stroma (pH ∼ 8.0) in response to the deprotonation of the amino groups of chitosan. The chitosan-conjugated SWCNTs were shown to efficiently deliver pDNA and facilitate reporter transgene expression in chloroplasts of various plant species, such as Eruca sativa, Nasturtium officinale, Nicotiana tabacum, Spinacia oleracea, and Arabidopsis thaliana, according to confocal laser scanning microscopy (CLSM) observations. SWCNTs may be employed as a potential method of chloroplast transformation in future studies, although evidence of transgene integration into the chloroplast genome is yet to be provided.
The Numata group has also adopted peptide-based nanocomplexes for targeted DNA delivery to chloroplasts in intact plants (Fig. 8a). To achieve this, a gene carrier peptide was designed by merging a polycationic DNA-binding domain ((KH)9: KHKHKHKHKHKHKHKHKH) with a chloroplast-targeting peptide obtained from a 34 kDa polypeptide present on the outer envelope membranes of A. thaliana chloroplasts (OEP34: MFAFQYLLVM).112 Electrostatic interactions facilitate the formation of an ionic complex between the designed peptide and pDNA, enabling selective delivery to chloroplasts in plants. Notably, this peptide-based system enabled transgene integration into the chloroplast genome through homologous recombination, as well as transient transgene expression in chloroplasts. Subsequent research confirmed that the peptide-based system could be extended to other types of plastids, such as tomato fruit chromoplasts and tomato root and potato tuber amyloplasts, in addition to the chloroplasts of Arabidopsis and tobacco leaves.81 More recently, the peptide-based DNA delivery system has been used in combination with a selection system reliant on a spectinomycin/streptomycin-resistance marker gene (aadA) for the stable plastid transformation of tobacco (N. tabacum), rice (Oryza sativa), and kenaf (Hibiscus cannabinus).126 The peptide facilitated the integration of foreign DNA encoding the marker gene into the plastid genome of each plant species through homologous recombination and its transmission to the next generation. However, the resulting transformants exhibited heteroplasmy, emphasizing the requirement for more efficient DNA delivery systems.
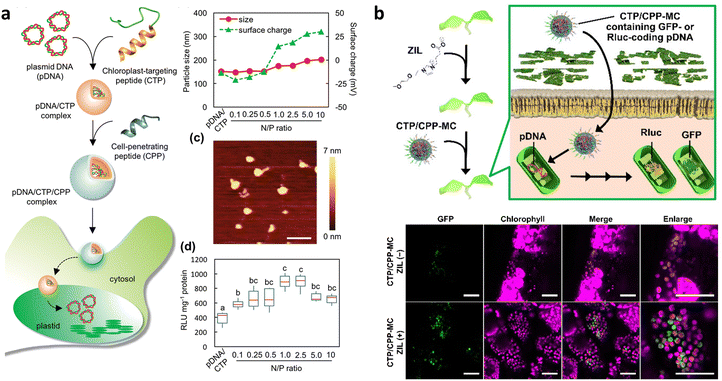 | ||
| Fig. 8 Chloroplast-targeted delivery methods using peptide/pDNA complexes. (a) A chloroplast-targeting peptide and CPP complexed with pDNA was developed by Thagun et al. for delivery of pDNA into the plastids of multiple plant species including tomato, N. benthamiana, and A. thaliana. The complexes were roughly 150–200 nm in diameter as observed by dynamic light scattering and atomic force microscopy depending on the ratio of peptide to pDNA. Expression of a Renilla luciferase reporter construct showed an optimum N/P ratio (# of amine groups in peptide/# of phosphate groups in DNA) of roughly 1. Scale bars in the AFM image represent 500 nm. (b) Miyamoto and colleagues applied the use of a zwitterionic liquid to loosen the cell wall of epidermal cells in A. thaliana cotelydons to enhance the transfection efficiency of pDNA containing a GFP reporter construct within chloroplasts using a micelle complex containing a chloroplast-targeting peptide. An increase in the GFP reporter construct could be observed within the leaves of A. thaliana upon treatment with the zwitterionic liquid. Scale bars represent 40 μm. Adapted with permission from Wiley Publications.43,81 | ||
Multiple barriers in plant cells must be overcome to achieve effective chloroplast-targeted DNA delivery. A live imaging study has shown that a large proportion of the peptide/pDNA complexes are trapped in the cell wall and vacuole lumen.17 The size of the complex, approximately 150 nm in mean diameter, exceeds the size exclusion limit of the cell wall.42,53 Endosomal entrapment of the complex during endocytic cellular uptake also leads to its accumulation and degradation in the vacuole lumen.103 Recently, a novel approach was proposed to improve cell wall permeability and overcome vacuolar entrapment.43 This approach combined the use of a cell wall-loosening reagent, zwitterionic liquid (ZIL),21 and an endosome-escaping micelle, which displayed multiple functional peptides for endosome-disruption and chloroplast-targeting.41 Pretreatment of A. thaliana seedlings with ZIL resulted in increased cell wall permeability through partial dissolution of cellulose without cytotoxicity. The ZIL pretreatment allowed the micelle to cross the cell wall, and the chloroplast-targeting and endosome-disrupting peptides on the micelle surface enhanced the efficiency of pDNA delivery to the chloroplasts (Fig. 8b).43
Santana and colleagues have proposed another approach to achieve effective chloroplast-targeted delivery, using a tertiary complex of a chloroplast-targeting cationic peptide, a polyethyleneimine-coated SWCNT, and pDNA44 (Fig. 9). This tertiary complex exhibited more efficient translocation to the chloroplasts of Arabidopsis plants than the SWCNT/pDNA complex lacking the targeting peptide. However, the tertiary complex showed a similar level of transgene expression as the SWCNT/pDNA complex, suggesting that the cationic targeting peptide may interfere with pDNA expression in chloroplasts, possibly due to the insufficient release of pDNA from the tertiary complex. Overall, continuous efforts to improve DNA delivery efficiency are necessary for ensuring stable chloroplast transformation.
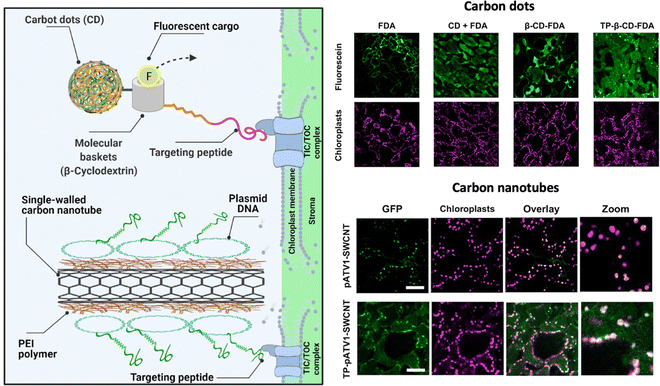 | ||
| Fig. 9 NM-mediated chloroplast delivery by the inclusion of a chloroplast targeting peptide. Santana and colleagues described the application of NMs including carbon dots and CNTs functionalized with a chloroplast targeting peptide that could mediate cargo delivery within chloroplasts in A. thaliana leaves. Uptake of a fluorescent dye in the chloroplasts could be observed when delivered by carbon dots conjugated with a molecular basket and a biorecognition motif for the TIC/TOC complex. A similar approach was used with PEI-conjugated CNTs that were complexed with pDNA and conjugated with a peptide containing a DNA binding domain and the same biorecognition motif. Expression of the reporter GFP construct could be observed after 7 days of exposure. Scale bars represent 50 μm. Adapted with permission from the American Chemical Society.44 | ||
Future outlook
NM-mediated organelle-targeted gene delivery has rapidly advanced in the last decade and has allowed researchers to overcome the once formidable cell wall barrier and reduce the shortcomings associated with the previously developed transgene delivery systems. Significant advancements in a wide variety of NMs have been made to target specific organelles within plants, while maintaining high biocompatibility and a low barrier-to-entry. In this article, we have reviewed the latest developments within organelle-targeted delivery within plants and the potential applications of NMs as targeted delivery vehicles.However, there are still many challenges to overcome before reaching widespread commercial applications of NMs in agriculture and industrial settings. Some of these include their long-term toxicity including their bioaccumulation and feasibility for large-scale application for genetic transformation. Furthermore, the exact mechanisms that mediate NM interactions with plant cells remain poorly understood and remains a bottleneck for rational design of NMs for specific and targeted delivery.
Practically, challenges also expand to the rules for designing structures and functions to enable direct germline editing required for stable transformation, as well as delivering large cargos like DNA plasmids larger than 10 kilobases and functional proteins. While preliminary work has been done on transforming maize pollen, more research is needed to establish the reliability and species amenability of direct germline transformation. In addition to pollen, other germline tissues such as ovules or pluripotent tissues like meristems could also be targeted for NM-mediated delivery. Progress towards delivering large cargos has been slow, but methods such as biolistic delivery and PEG transfection of proteins could provide nanomediated strategies for DNA-free, non-biolistic plant genome editing with site-specific nucleases for efficient molecular breeding programs.
In terms of organelle specific transformation in plants, the greatest challenge remaining is the lack of selectivity and difficulty in delivery. This is further compounded by the lack of selection systems for obtaining transformants and difficulty in obtaining homoplasmic mitochondria or chloroplast transformants. However, these are slowly being resolved as newer generations of NMs have drastically improved delivery efficiencies and selectivity for the target organelle. Cell-penetrating peptides have been successful in delivering genetic material as well as proteins to specific organelles. Combining the internalization capability of cell-penetrating peptides with the ability of NPs to protect cargos from degradation is a promising approach to improving delivery efficiencies. Further development and application of these NMs along with a greater understanding of internalization mechanisms and physiochemistry relationships would likely bring NM-mediated gene delivery into a commercially viable method for producing transgenic plants in the near future.
Author contributions
All authors contributed to the writing of this article.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
We acknowledge funding support by Grants-in-Aid from the Japan Science and Technology Agency Exploratory Research for Advanced Technology (JST-ERATO; Grant No. JPMJER1602 to K. N.), COI-NEXT (to K. N.), MEXT Data Creation and Utilization-type MaTerial R&D project (to K. N.), and the Japan Society for the Promotion of Science for Scientific Research (JSPS-KAKENHI; Grant No. JP19K15411 and JP22K14575 to T. M.). Fig. 1 and 2 were created with BioRender.com.Notes and references
- B. Ouyang, X. Gu and P. Holford, Plant Growth Regul., 2017, 83, 171–173 CrossRef CAS
.
- T. Jenkins, A. Bovi and R. Edwards, Philos. Trans. R. Soc., A, 2011, 369, 1826–1839 CrossRef CAS PubMed
.
- T. M. Chaloner, S. J. Gurr and D. P. Bebber, Nat. Clim. Change, 2021, 11, 710–715 CrossRef
.
- A. Furtado, J. S. Lupoi, N. V. Hoang, A. Healey, S. Singh, B. A. Simmons and R. J. Henry, Plant Biotechnol. J., 2014, 12, 1246–1258 CrossRef CAS
.
- M. F. Serag, N. Kaji, S. Habuchi, A. Bianco and Y. Baba, RSC Adv., 2013, 3, 4856–4862 RSC
.
- M. Bevan, Nucleic Acids Res., 1984, 12, 8711–8721 CrossRef CAS PubMed
.
- M. W. Bevan, R. B. Flavell and M.-D. Chilton, Nature, 1983, 304, 184–187 CrossRef CAS
.
- M.-D. Chilton, D. A. Tepfer, A. Petit, C. David, F. Casse-Delbart and J. Tempé, Nature, 1982, 295, 432–434 CrossRef CAS
.
- N. Wada, R. Ueta, Y. Osakabe and K. Osakabe, BMC Plant Biol., 2020, 20, 234 CrossRef CAS PubMed
.
- K. Belhaj, A. Chaparro-Garcia, S. Kamoun, N. J. Patron and V. Nekrasov, Curr. Opin. Biotechnol, 2015, 32, 76–84 CrossRef CAS PubMed
.
- J. B. Ristaino, P. K. Anderson, D. P. Bebber, K. A. Brauman, N. J. Cunniffe, N. V. Fedoroff, C. Finegold, K. A. Garrett, C. A. Gilligan, C. M. Jones, M. D. Martin, G. K. MacDonald, P. Neenan, A. Records, D. G. Schmale, L. Tateosian and Q. Wei, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(23), e2022239118, DOI:10.1073/pnas.2022239118
.
- T. Li, B. Liu, M. H. Spalding, D. P. Weeks and B. Yang, Nat. Biotechnol., 2012, 30, 390–392 CrossRef CAS PubMed
.
- B. L. Weis, E. Schleiff and W. Zerges, Biochim. Biophys. Acta, Mol. Cell Res., 1833, 2013, 260–273 Search PubMed
.
- A. Saminathan, M. Zajac, P. Anees and Y. Krishnan, Nat. Rev. Mater., 2022, 7, 355–371 CrossRef
.
- S. Ahmar, T. Mahmood, S. Fiaz, F. Mora-Poblete, M. S. Shafique, M. S. Chattha and K.-H. Jung, Front. Plant Sci, 2021, 12, 663849, DOI:10.3389/fpls.2021.663849
.
- S. Vats, S. Kumawat, J. Brar, S. Kaur, K. Yadav, S. G. Magar, P. V. Jadhav, P. Salvi, H. Sonah, S. Sharma and R. Deshmukh, Plant Nano Biol., 2022, 1, 100001 CrossRef
.
-
M. A. Gad, M. Li, F. K. Ahmed and H. Almoammar, in Multifunctional Hybrid Nanomaterials for Sustainable Agri-Food and Ecosystems, ed. K. A. Abd-Elsalam, Elsevier, 2020, pp. 135–153 Search PubMed
.
- S. K. Jat, J. Bhattacharya and M. K. Sharma, J. Mater. Chem. B, 2020, 8, 4165–4175 RSC
.
- K. Houston, M. R. Tucker, J. Chowdhury, N. Shirley and A. Little, Front. Plant Sci, 2016, 7, 984, DOI:10.3389/fpls.2016.00984
.
- B. Zhang, Y. Gao, L. Zhang and Y. Zhou, J. Integr. Plant Biol., 2021, 63, 251–272 CrossRef PubMed
.
- J. B. Spinelli and M. C. Haigis, Nat. Cell Biol., 2018, 20, 745–754 CrossRef CAS PubMed
.
- J. R. Friedman and J. Nunnari, Nature, 2014, 505, 335–343 CrossRef CAS PubMed
.
- M. Giacomello, A. Pyakurel, C. Glytsou and L. Scorrano, Nat. Rev. Mol. Cell Biol., 2020, 21, 204–224 CrossRef CAS PubMed
.
- T. Kazama, M. Okuno, Y. Watari, S. Yanase, C. Koizuka, Y. Tsuruta, H. Sugaya, A. Toyoda, T. Itoh, N. Tsutsumi, K. Toriyama, N. Koizuka and S. Arimura, Nat. Plants, 2019, 5, 722–730 CrossRef CAS PubMed
.
- L. A. Staehelin, Photosynth. Res., 2003, 76, 185–196 CrossRef CAS PubMed
.
- S. Murakami and L. Packer, J. Cell Biol., 1970, 47, 332–351 CrossRef CAS PubMed
.
- C. Andrès, B. Agne and F. Kessler, Biochim. Biophys. Acta, Mol. Cell Res., 1803, 2010, 715–723 Search PubMed
.
- L. Fan, R. Li, J. Pan, Z. Ding and J. Lin, Trends Plant Sci., 2015, 20, 388–397 CrossRef CAS PubMed
.
- Q. Wang, Y. Zhao, W. Luo, R. Li, Q. He, X. Fang, R. D. Michele, C. Ast, N. von Wirén and J. Lin, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 13204–13209 CrossRef CAS PubMed
.
- J. Takano, M. Tanaka, A. Toyoda, K. Miwa, K. Kasai, K. Fuji, H. Onouchi, S. Naito and T. Fujiwara, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 5220–5225 CrossRef CAS PubMed
.
- S. Di Rubbo, N. G. Irani, S. Y. Kim, Z.-Y. Xu, A. Gadeyne, W. Dejonghe, I. Vanhoutte, G. Persiau, D. Eeckhout, S. Simon, K. Song, J. Kleine-Vehn, J. Friml, G. De Jaeger, D. Van Damme, I. Hwang and E. Russinova, Plant Cell, 2013, 25, 2986–2997 CrossRef CAS PubMed
.
- S. Robatzek, D. Chinchilla and T. Boller, Genes Dev., 2006, 20, 537–542 CrossRef CAS PubMed
.
- R. Li, P. Liu, Y. Wan, T. Chen, Q. Wang, U. Mettbach, F. Baluska, J. Samaj, X. Fang, W. J. Lucas and J. Lin, Plant Cell, 2012, 24, 2105–2122 CrossRef CAS PubMed
.
- W. Kofer, C. Eibl, K. Steinmüller and H.-U. Koop, In Vitro Cell. Dev. Biol.: Plant, 1998, 34, 303–309 CrossRef CAS
.
- M. E. Fromm, L. P. Taylor and V. Walbot, Nature, 1986, 319, 791–793 CrossRef CAS PubMed
.
- G. Neuhaus and G. Spangenberg, Physiol. Plant., 1990, 79, 213–217 CrossRef CAS
.
- Y. Liu, H. Yang and A. Sakanishi, Biotechnol. Adv., 2006, 24, 1–16 CrossRef CAS PubMed
.
- J. Li and K. Kataoka, J. Am. Chem. Soc., 2021, 143, 538–559 CrossRef CAS PubMed
.
- M. K. Danquah, X. A. Zhang and R. I. Mahato, Adv. Drug Delivery Rev., 2011, 63, 623–639 CrossRef PubMed
.
- E. Blanco, H. Shen and M. Ferrari, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS PubMed
.
- T. Miyamoto, K. Tsuchiya and K. Numata, Nanoscale, 2021, 13, 5679–5692 RSC
.
- E. Corredor, P. S. Testillano, M.-J. Coronado, P. González-Melendi, R. Fernández-Pacheco, C. Marquina, M. R. Ibarra, J. M. de la Fuente, D. Rubiales, A. Pérez-de-Luque and M.-C. Risueño, BMC Plant Biol., 2009, 9, 45 CrossRef PubMed
.
- T. Miyamoto, K. Tsuchiya, K. Toyooka, Y. Goto, A. Tateishi and K. Numata, Angew. Chem., Int. Ed., 2022, 61, e202204234 CrossRef CAS PubMed
.
- I. Santana, S.-J. Jeon, H.-I. Kim, M. R. Islam, C. Castillo, G. F. H. Garcia, G. M. Newkirk and J. P. Giraldo, ACS Nano, 2022, 16, 12156–12173 CrossRef CAS PubMed
.
- T. c Yih and M. Al-Fandi, J. Cell. Biochem., 2006, 97, 1184–1190 CrossRef CAS PubMed
.
- E. Keles, Y. Song, D. Du, W.-J. Dong and Y. Lin, Biomater. Sci., 2016, 4, 1291–1309 RSC
.
- A. Z. Wilczewska, K. Niemirowicz, K. H. Markiewicz and H. Car, Pharmacol. Rep., 2012, 64, 1020–1037 CrossRef CAS PubMed
.
- Q. Jiang, S. Liu, J. Liu, Z.-G. Wang and B. Ding, Adv. Mater., 2019, 31, 1804785 CrossRef CAS PubMed
.
- M. Malmsten, Curr. Opin. Colloid Interface Sci., 2013, 18, 468–480 CrossRef CAS
.
- S. H. Schwartz, B. Hendrix, P. Hoffer, R. A. Sanders and W. Zheng, Plant Physiol., 2020, 184, 647–657 CrossRef CAS PubMed
.
- A. Pérez-de-Luque, Front. Environ. Sci, 2017, 5, 12, DOI:10.3389/fenvs.2017.00012
.
- K. Midorikawa, Y. Kodama and K. Numata, Sci. Rep., 2019, 9, 271 CrossRef PubMed
.
- T. Eichert, A. Kurtz, U. Steiner and H. E. Goldbach, Physiol. Plant., 2008, 134, 151–160 CrossRef CAS PubMed
.
- M. H. Wong, R. P. Misra, J. P. Giraldo, S. Y. Kwak, Y. Son, M. P. Landry, J. W. Swan, D. Blankschtein and M. S. Strano, Nano Lett., 2016, 16, 1161–1172 CrossRef CAS PubMed
.
- G. S. Demirer, H. Zhang, J. L. Matos, N. S. Goh, F. J. Cunningham, Y. Sung, R. Chang, A. J. Aditham, L. Chio, M.-J. J. Cho, B. Staskawicz and M. P. Landry, Nat. Nanotechnol., 2019, 14, 456–464 CrossRef CAS PubMed
.
- S. S. Y. Law, G. Liou, Y. Nagai, J. Giménez-Dejoz, A. Tateishi, K. Tsuchiya, Y. Kodama, T. Fujigaya and K. Numata, Nat. Commun., 2022, 13, 2417 CrossRef CAS PubMed
.
-
M. Gumustas, C. T. Sengel-Turk, A. Gumustas, S. A. Ozkan and B. Uslu, in Multifunctional Systems for Combined Delivery, Biosensing and Diagnostics, ed. A. M. Grumezescu, Elsevier, 2017, pp. 67–108 Search PubMed
.
- H. Zhang, N. S. Goh, J. W. Wang, R. L. Pinals, E. González-Grandío, G. S. Demirer, S. Butrus, S. C. Fakra, A. Del Rio Flores, R. Zhai, B. Zhao, S.-J. Park and M.
P. Landry, Nat. Nanotechnol., 2022, 17, 197–205 CrossRef CAS PubMed
.
- S. Martin-Ortigosa, D. J. Peterson, J. S. Valenstein, V. S.-Y. Lin, B. G. Trewyn, L. A. Lyznik and K. Wang, Plant Physiol., 2014, 164, 537–547 CrossRef CAS PubMed
.
- F. Torney, B. G. Trewyn, V. S.-Y. Lin and K. Wang, Nat. Nanotechnol., 2007, 2, 295–300 CrossRef CAS PubMed
.
- S. Martin-Ortigosa, J. S. Valenstein, W. Sun, L. Moeller, N. Fang, B. G. Trewyn, V. S.-Y. Lin and K. Wang, Small, 2012, 8, 413–422 CrossRef CAS PubMed
.
- F.-P. Chang, L.-Y. Kuang, C.-A. Huang, W.-N. Jane, Y. Hung, Y. C. Hsing and C.-Y. Mou, J. Mater. Chem. B, 2013, 1, 5279–5287 RSC
.
- L. Wu, H. Pan, W. Huang, M. Wang, Z. Hu and F. Zhang, Environ. Technol. Innovation, 2022, 28, 102890 CrossRef CAS
.
- M. Zolghadrnasab, A. Mousavi, A. Farmany and A. Arpanaei, Ultrason. Sonochem., 2021, 73, 105507 CrossRef CAS PubMed
.
- V. Chandrakala, V. Aruna and G. Angajala, Emergent Mater., 2022, 5, 1593–1615 CrossRef CAS PubMed
.
- Y. Hao, X. Yang, Y. Shi, S. Song, J. Xing, J. Marowitch, J. Chen and J. Chen, Botany, 2013, 91, 457–466 CrossRef CAS
.
- X. Zhao, Z. Meng, Y. Wang, W. Chen, C. Sun, B. Cui, J. Cui, M. Yu, Z. Zeng, S. Guo, D. Luo, J. Q. Cheng, R. Zhang and H. Cui, Nat. Plants, 2017, 3, 956–964 CrossRef CAS PubMed
.
-
Biopolymer Science for Proteins and Peptides, ed. K. Numata, Elsevier, 2021 Search PubMed
.
- J.-A. Chuah, A. Matsugami, F. Hayashi and K. Numata, Biomacromolecules, 2016, 17, 3547–3557 CrossRef CAS PubMed
.
- T. Miyamoto, K. Tsuchiya and K. Numata, Biomacromolecules, 2019, 20, 653–661 CrossRef CAS PubMed
.
- C. P. Cerrato, K. Künnapuu and Ü. Langel, Expert Opin. Drug Delivery, 2017, 14, 245–255 CrossRef CAS PubMed
.
- A. Chugh and F. Eudes, FEBS J., 2008, 275, 2403–2414 CrossRef CAS PubMed
.
- M. Lakshmanan, Y. Kodama, T. Yoshizumi, K. Sudesh and K. Numata, Biomacromolecules, 2013, 14, 10–16 CrossRef CAS PubMed
.
- S. Y. Kwak, T. T. S. Lew, C. J. Sweeney, V. B. Koman, M. H. Wong, K. Bohmert-Tatarev, K. D. Snell, J. S. Seo, N. H. Chua and M. S. Strano, Nat. Nanotechnol., 2019, 14, 447–455 CrossRef CAS PubMed
.
- M. S. Dresselhaus, G. Dresselhaus, R. Saito and A. Jorio, Phys. Rep., 2005, 409, 47–99 CrossRef
.
- A. E. Porter, M. Gass, K. Muller, J. N. Skepper, P. A. Midgley and M. Welland, Nat. Nanotechnol., 2007, 2, 713–717 CrossRef CAS PubMed
.
- H. C. Nerl, C. Cheng, A. E. Goode, S. D. Bergin, B. Lich, M. Gass and A. E. Porter, Nanomedicine, 2011, 6, 849–865 CrossRef CAS PubMed
.
- Q. Liu, B. Chen, Q. Wang, X. Shi, Z. Xiao, J. Lin and X. Fang, Nano Lett., 2009, 9, 1007–1010 CrossRef CAS PubMed
.
- N. Mitter, E. A. Worrall, K. E. Robinson, P. Li, R. G. Jain, C. Taochy, S. J. Fletcher, B. J. Carroll, G. Q. (Max) Lu and Z. P. Xu, Nat. Plants, 2017, 3, 1–10 Search PubMed
.
- J.-A. Chuah, T. Yoshizumi, Y. Kodama and K. Numata, Sci. Rep., 2015, 5, 7751 CrossRef CAS PubMed
.
- C. Thagun, J.-A. Chuah and K. Numata, Adv. Sci., 2019, 6, 1902064 CrossRef CAS PubMed
.
-
J. R. Kikkert, J. R. Vidal and B. I. Reisch, Transgenic Plants, Humana Press, New Jersey, 2005, vol. 286, pp. 061–078 Search PubMed
.
- T. M. Klein, E. C. Harper, Z. Svab, J. C. Sanford, M. E. Fromm and P. Maliga, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 8502–8505 CrossRef CAS PubMed
.
- T. M. Klein, L. Kornstein, J. C. Sanford and M. E. Fromm, Plant Physiol., 1989, 91, 440–444 CrossRef CAS PubMed
.
- J. C. Sanford, T. M. Klein, E. D. Wolf and N. Allen, Part. Sci. Technol., 1987, 5, 27–37 CrossRef CAS
.
- Y.-C. Wang, T. M. Klein, M. Fromm, J. Cao, J. C. Sanford and R. Wu, Plant Mol. Biol., 1988, 11, 433–439 CrossRef CAS PubMed
.
- H. Zhi, S. Zhou, W. Pan, Y. Shang, Z. Zeng and H. Zhang, Int. J. Mol. Sci., 2022, 23(15) DOI:10.3390/ijms23158501
.
- Y. Yan, X. Zhu, Y. Yu, C. Li, Z. Zhang and F. Wang, Adv. Mater., 2022, 34, 2106945 CrossRef CAS PubMed
.
- Z. Lv, R. Jiang, J. Chen and W. Chen, Plant J., 2020, 104, 880–891 CrossRef CAS PubMed
.
- F. J. Cunningham, N. S. Goh, G. S. Demirer, J. L. Matos and M. P. Landry, Trends Biotechnol., 2018, 36, 882–897 CrossRef CAS PubMed
.
- H.-H. Hwang, M. Yu and E.-M. Lai, Arabidopsis Book, 2017, 15, e0186, DOI:10.1199/tab.0186
.
- G. W. Bolton, E. W. Nester and M. P. Gordon, Science, 1986, 232, 983–985 CrossRef CAS PubMed
.
- I. Godwin, G. Todd, B. Ford-Lloyd and H. J. Newbury, Plant Cell Rep., 1991, 9, 671–675 CrossRef CAS PubMed
.
- S. S.-A. Zaidi and S. Mansoor, Front. Plant Sci., 2017, 8, 539, DOI:10.3389/fpls.2017.00539
.
- P. Abrahamian, R. W. Hammond and J. Hammond, Annu. Rev. Virol., 2020, 7, 513–535 CrossRef CAS PubMed
.
-
M. Fromm, J. Callis, L. P. Taylor and V. Walbot, Methods in Enzymology, Academic Press, 1987, vol. 153, pp. 351–366 Search PubMed
.
- A. Hayashimoto, Z. Li and N. Murai, Plant Physiol., 1990, 93, 857–863 CrossRef CAS PubMed
.
- S. Wu, H. Zhu, J. Liu, Q. Yang, X. Shao, F. Bi, C. Hu, H. Huo, K. Chen and G. Yi, BMC Plant Biol., 2020, 20, 425 CrossRef CAS
.
- T. Golds, P. Maliga and H.-U. Koop, Nat. Biotechnol., 1993, 11, 95–97 CrossRef CAS
.
- A. Avellan, J. Yun, Y. Zhang, E. Spielman-Sun, J. M. Unrine, J. Thieme, J. Li, E. Lombi, G. Bland and G. V. Lowry, ACS Nano, 2019, 13, 5291–5305 CrossRef CAS PubMed
.
- T. T. S. Lew, M. H. Wong, S.-Y. Kwak, R. Sinclair, V. B. Koman and M. S. Strano, Small, 2018, 14, 1802086 CrossRef PubMed
.
- K. Oikawa, A. Tateishi, M. Odahara, Y. Kodama and K. Numata, Front. Plant Sci., 2021, 12, 759871, DOI:10.3389/fpls.2021.759871
.
- T. Miyamoto, K. Tsuchiya and K. Numata, Biomacromolecules, 2020, 21, 2735–2744 CrossRef CAS PubMed
.
- B. Guo, J. Itami, K. Oikawa, Y. Motoda, T. Kigawa and K. Numata, PLoS One, 2019, 14, e0214033 CrossRef CAS PubMed
.
- T. Miyamoto, K. Toyooka, J.-A. Chuah, M. Odahara, M. Higchi-Takeuchi, Y. Goto, Y. Motoda, T. Kigawa, Y. Kodama and K. Numata, JACS Au, 2022, 2, 223–233 CrossRef CAS PubMed
.
- R. Ni, R. Feng and Y. Chau, ACS Appl. Nano Mater., 2023, 6(5), 3191–3201, DOI:10.1021/acsanm.2c04279
.
- J.-A. Chuah and K. Numata, Biomacromolecules, 2018, 19, 1154–1163 CrossRef CAS PubMed
.
- D. Mileshina, M. Koulintchenko, Y. Konstantinov and A. Dietrich, Nucleic Acids Res., 2011, 39, e115 CrossRef CAS PubMed
.
- J.-C. Farré and A. Araya, Nucleic Acids Res., 2001, 29, 2484–2491 CrossRef PubMed
.
-
N. Bonnefoy, C. Remacle and T. D. Fox, Methods in Cell Biology, Academic Press, 2007, vol. 80, pp. 525–548 Search PubMed
.
- M. Koulintchenko, Y. Konstantinov and A. Dietrich, EMBO J., 2003, 22, 1245–1254 CrossRef CAS PubMed
.
- T. Yoshizumi, K. Oikawa, J. A. Chuah, Y. Kodama and K. Numata, Biomacromolecules, 2018, 19, 1582–1591 CrossRef CAS PubMed
.
- E. C. Hurt, U. Müller and G. Schatz, EMBO J., 1985, 4, 3509–3518 CrossRef CAS PubMed
.
- E. C. Hurt, B. Pesold-Hurt, K. Suda, W. Oppliger and G. Schatz, EMBO J., 1985, 4, 2061–2068 CrossRef CAS PubMed
.
- J.-A. Chuah, Y. Horii and K. Numata, J. Visualized Exp., 2016, 118, 54972, DOI:10.3791/54972
.
- S. Arimura, H. Ayabe, H. Sugaya, M. Okuno, Y. Tamura, Y. Tsuruta, Y. Watari, S. Yanase, T. Yamauchi, T. Itoh, A. Toyoda, H. Takanashi and N. Tsutsumi, Plant J., 2020, 104, 1459–1471 CrossRef CAS PubMed
.
- D. R. Ort, S. S. Merchant, J. Alric, A. Barkan, R. E. Blankenship, R. Bock, R. Croce, M. R. Hanson, J. M. Hibberd, S. P. Long, T. A. Moore, J. Moroney, K. K. Niyogi, M. A. J. Parry, P. P. Peralta-Yahya, R. C. Prince, K. E. Redding, M. H. Spalding, K. J. van Wijk, W. F. J. Vermaas, S. von Caemmerer, A. P. M. Weber, T. O. Yeates, J. S. Yuan and X. G. Zhu, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 8529–8536 CrossRef CAS PubMed
.
- S. Jin and H. Daniell, Trends Plant Sci., 2015, 20, 622–640 CrossRef CAS PubMed
.
- P. Maliga and R. Bock, Plant Physiol., 2011, 155, 1501–1510 CrossRef CAS PubMed
.
- M. Oey, M. Lohse, B. Kreikemeyer and R. Bock, Plant J., 2009, 57, 436–445 CrossRef CAS PubMed
.
- R. Bock, Curr. Opin. Biotechnol, 2014, 26, 7–13 CrossRef CAS PubMed
.
- S. Ruf, D. Karcher and R. Bock, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 6998–7002 CrossRef CAS PubMed
.
- R. Bock, Annu. Rev. Plant Biol., 2015, 66, 211–241 CrossRef CAS PubMed
.
- G. M. Newkirk, P. de Allende, R. E. Jinkerson and J. P. Giraldo, Front. Plant Sci., 2021, 12, 691295, DOI:10.3389/fpls.2021.691295
.
- A. U. Kumar and A. P. K. Ling, J. Genet. Eng. Biotechnol., 2021, 19, 148 CrossRef PubMed
.
- M. Odahara, Y. Horii, J. Itami, K. Watanabe and K. Numata, Front. Plant Sci., 2022, 13, 989310, DOI:10.3389/fpls.2022.989310
.
| This journal is © The Royal Society of Chemistry 2023 |