Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A TCF-based fluorescent probe to determine nitroreductase (NTR) activity for a broad-spectrum of bacterial species†
Kai-Cheng
Yan
 ac,
Jordan E.
Gardiner
ac,
Jordan E.
Gardiner
 a,
Adam C.
Sedgwick
a,
Adam C.
Sedgwick
 b,
Naing
Thet
b,
Naing
Thet
 a,
Rachel A.
Heylen
a,
Rachel A.
Heylen
 a,
Tony D.
James
a,
Tony D.
James
 *ad,
A. Toby A.
Jenkins
*ad,
A. Toby A.
Jenkins
 *a and
Xiao-Peng
He
*a and
Xiao-Peng
He
 *ce
*ce
aDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk; a.t.a.jenkins@bath.ac.uk
bChemistry Research Laboratory, University of Oxford, Mansfield Road, OX1 3TA, UK
cKey Laboratory for Advanced Materials and Joint International Research Laboratory of Precision Chemistry and Molecular Engineering, Feringa Nobel Prize Scientist Joint Research Center, School of Chemistry and Molecular Engineering, East China University of Science and Technology, 130 Meilong Rd., Shanghai 200237, China. E-mail: xphe@ecust.edu.cn
dSchool of Chemistry and Chemical Engineering, Henan Normal University, Xinxiang 453007, China
eNational Center for Liver Cancer, The International Cooperation Laboratory on Signal Transduction, Eastern Hepatobiliary Surgery Hospital, the Second Military Medical University, Shanghai 200433, P. R. China
First published on 15th June 2023
Abstract
A nitroreductase (NTR) responsive fluorescent probe with long wavelength fluorescence emission was used to determine the NTR activity of a selection of bacterial species under a range of different bacterial growth conditions ensuring applicability under multiple complex clinical environments, where sensitivity, reaction time, and the detection accuracy were suitable for planktonic cultures and biofilms.
The flavin-containing nitroreductase (NTR) is overexpressed in Enterobacter cloacae and hypoxic human tumours and catalyses the reduction of nitroaromatic compounds to hydroxyl amines or amines, with the assistance of a NADH (Nicotinamide adenine dinucleotide) cofactor.1–3 The NTR enzyme has been used for environmental engineering, in the bioremediation of 2,4,6-trinitrotoluene (TNT) and other nitroaromatic pollutants.4–6 Moreover, important biological functions of NTR in a range of organisms have recently been explored.7,8 NTR detection can thus improve disease diagnosis, and facilitate the development of selective therapeutics activated by NTR.9,10 Numerous NTR sensors have been developed with the majority focused on assessing the hypoxic status of tumours.11–13
The high-expression of NTR in bacteria, especially among Escherichia coli (E. coli) provides us an important marker suitable for pathogen-oriented detection. To date, fluorescence-based systems remain underexplored for sensing NTR, where electrochemical sensors are more commonly used.14–16 While exhibiting acceptable sensitivity, electrochemical methods are restricted in biomedical applications since they require enzyme fixation and are not suitable for imaging and in situ analysis due to complex operational procedures and may cause tissue damage. Fluorescence-based detection has thus gained attention due to the high sensitivity, selectivity, imaging feasibility and biocompatibility, which ensures minimal damage of cells and tissues.17–20
Taking our inspiration from recent fluorescence-based NTR sensors, functionalization of a hydroxyl with p-nitroanisole results in an effective NTR responsive unit.21 1,8-naphthalimide and dicyanomethylene 4H-pyran (DCM) have been used as the cores of NTR-responsive systems by Zhang et al. and Yang et al.,22,23 demonstrating the effective sensing of NTR in solution and in Hep-G2 cells. Although NTR expression among E. coli has been verified, bacterial-based sensing of NTR remains unknown. Herein, we developed a new long-wavelength (560 nm) red-emitting NTR sensor based on 2-dicyanomethylene-3-cyano-4,5,5-trimethyl-2,5-dihydrofuran (TCF) modified using an NTR-responsive nitro locking group (Scheme 1). The selection of red-emitting long-wavelength fluorophore provides significant advantages for sensor performance, including minimal biological damage ensuring biocompatibility, effective penetration through tissues, excellent stability of signals, minimal interference from background fluorescence compared with other fluorescence systems, thus allowing the possibility of biomedical-oriented chemosensors for clinical applications.
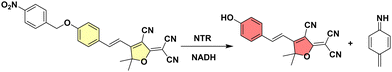 | ||
| Scheme 1 NTR reduces the nitro group of TCF-Nitro (left) to an amine triggering fluorescence “turn-on” due to release of TCF-OH and 4-methylenecyclohexa-2,5-dien-1-imine (right). | ||
Here, we show that TCF-Nitro can be used to sensitively and selectively detect NTR and monitor NTR levels in E. coli. as well as a range of other bacterial species and strains. The TCF-OH fluorophore was modified with a p-nitroanisole NTR-responsive unit (see ESI† for synthesis), as confirmed by both NMR and MS spectroscopy (Fig. S1a, ESI†), which could then be cleaved to produce TCF-OH upon NTR treatment, as confirmed by MS (M + H+ = 304.1083, M + Na+ = 326.0901) (Fig. S1b, ESI†). The as-constructed probe TCF-Nitro was evaluated in aqueous solution, exhibiting a sensitive fluorescence “turn-on” in the presence of the co-factor NADH (response observed for 5 μg mL−1 enzyme) (Fig. S2, ESI†).22,23
Significantly, the response could be quenched using dicoumarin as an NTR inhibitor (Fig. S3, ESI†), which is non fluorescent so does not interfere with the detection (Fig. S4, ESI†). Moreover, TCF-Nitro can generate not only a fluorescence but also a UV-Vis colormetric response upon activation, resulting in a colour change from orange to rose red (Fig. S5, ESI†), a result of the production of TCF-OH (TCF-O−).24 Furthermore, the sensing selectivity and sensitivity were confirmed. Selectivity was demonstrated over a range of interfering agents including γ-glutathione (GSH), vitamin C (VC), H2O2, and a variety of amino acids (Fig. S6, ESI†). A linear response was observed for low NTR concentrations between 0.3–1.5 μg mL−1 (Fig. S7, ESI†). For enzyme-activatable probes, enzymatic kinetics is an important characteristic; significantly, probe TCF-Nitro reacts with NTR completely in 15 min (Fig. S8, ESI†).
Within biological systems (including in vivo systems, mammalian cells, or pathogens including bacteria), the activity of NTR is determined not simply by the amount of NTR present, but by the ability to reduce nitro groups (requiring the cooperation of both endogenous NTR and NADH).22,23 Diagnostic systems that can effectively measure endogenous NTR levels in a range of bacterial species are particularly attractive as they could act as tools to evaluate enzymatic mechanisms amongst different kinds of pathogens, enabling evaluation of their biological and pathological functions, facilitating appropriate clinical treatments resulting in improved prognosis. Therefore, we screened for NTR activity within 15 biologically representative strains which belong to several common and indicative categories of bacteria. To obtain the most useful results, we created a balanced selection, containing common clinically relevant strains including Gram-negative Pseudomonas species, Gram-negative Escherichia species, Gram-positive Staphylococcus species, Gram-positive Streptococcus species, and Gram-positive Enterococcus species. To access the inherent NTR activity presented by microorganisms themselves, probe TCF-Nitro was incubated with the species, to record and compare the signals detected (Fig. S9, ESI†). From the screening results, the NTR activities are not related with the Gram (+/−) types but more with the species themselves. Pseudomonas aeruginosa (Gram −) and Enterococcus faecalis (Gram +) generally do not display NTR activity, while modest levels are found among Streptococcus dysgalactiae NCTC 10238. Especially, the Staphylococcus species Staphylococcus epidermidis (S. epidermidis) RP62A results in a significant NTR response (up to 20-fold), indicating the highest activity. Interestingly, many reports have confirmed the production of NTR by E. coli,25,26 however, amongst the six E. coli strains, we only observed acceptable NTR activity for DH5α (Fig. S9, ESI†). Consequently, we selected the top three strains belonging to different bacterial categories that produced significant signals (DH5α, NCTC 10238, and RP62A) as representatives to conduct subsequent growth-related studies, utilizing TCF-Nitro as an NTR-monitoring tool under a range of conditions. The response of TCF-Nitro was evaluated for fast response (1 h) and standard response (1 d). A fast response enables accurate evaluation within 1 h and is suitable for urgent clinical conditions such as point of care testing (i.e. where a patient's life is in danger).
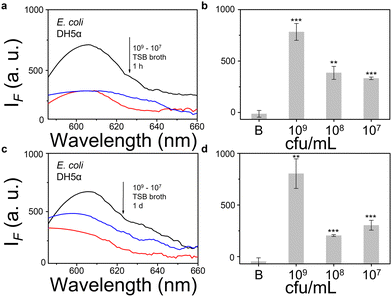
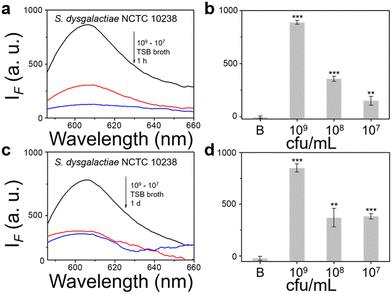
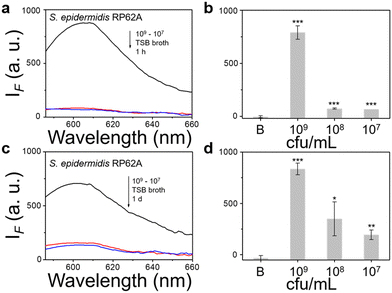
We then evaluated the growth of bacteria under a range of biological environments to elaborate in vivo and clinical applicability, and evaluate the relationship between the growth conditions (e.g. the nutrients absorbed for growth) and the NTR activity level. We used four kinds of commonly used bacterial-culture broths Luria–Bertani (LB), Muller–Hinton (MH), Trypticase Soy Broth (TSB), and Brain–Heart Infusion (BHI) to mimic the different growth conditions, and used a limit of detection method to determine the response level and sensitivity. The DH5α results using TSB broth indicated that TCF-Nitro reacted within 1 h and that further reaction for 1 d did not significantly enhance the response (Fig. 1). In addition, DH5α with MH, LB, and BHI broth cultures indicated that the probe responded to E. coli rapidly, the 1 h results are saturated compared with the results obtained after extended reaction (1 d). Within these results, the detection in LB broth seems to be the most significant (Fig. S10–S15, ESI†). When it comes to the second strain NCTC 10238, the detection performances are rapid (1 h) and significant in the TSB broth (Fig. 2). However, a disappointing result was seen for MH broth where nothing was detected within 1 h. In addition, the probe struggled to achieve completion after 1 d, which is not suitable for practical applications (Fig. S16 and S17, ESI†). The situation improves with LB and TSB broths, where 1 h is enough time for the determination of NTR activity (Fig. S18–S21, ESI†). Finally, we examined the last strain RP62A, which exhibited significant NTR activity within 1 h for TSB, MH, and BHI broths (Fig. 3 and Fig. S22, S23, S26, and S27). The situation in LB broths improves after 1 d, where 1 h reaction confirms the existence of NTR (Fig. S24 and 25, ESI†). In summary, the most obvious and significant NTR activity is observed for RP62A, which is in accordance with the screening results in Fig. S9 (ESI†). To our delight, among the three selected strains TCF-Nitro could provide an acceptable detection performance within 1 h.
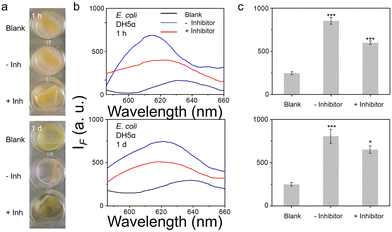
Using dicoumarin (NTR inhibitor) in an inhibition assay,22,23 we confirmed that the signals under all conditions were due to active NTR. We conducted two separate inhibition assays, in one the dicoumarin inhibitor is constantly present in the system during probe incubation and detection, and for the second dicoumarin is removed from the cells before incubation with the probe. What is interesting is that except for two groups (NCTC 10238 in MH broth and in BHI broth), the inhibitor performs well when not removed from the extracellular environment. However, if the inhibitor was washed out the surviving bacteria generate some NTR after 1 d (thus not providing constant inhibition during evaluation), as such the signals rebound (Fig. S28 and S29, ESI†). It appears that S. epidemidis RP62A generally does not produce NTR after inhibition while others produce some NTR.
Since the TCF core is commonly used for microscopy,24,27 we monitored TCF-Nitro using Confocal Laser-Scanning Microscopy (CLSM) to determine the presence of intracellular NTR. Apart from DH5α where fluorescence was clearly seen, both NCTC 10238 and RP62A indicate that NTR tends to be released outside of the cell membrane, which might be an important clue for future mechanism-based pharmacological studies (Fig. S30–S33, ESI†). Biofilms are significant challenges for clinical bacterial treatment and in particular the disinfection/sterilization industry. Biofilms are bacterial communities composed of an extracellular matrix that adheres tightly to the surface of medical devices or other clinically-relevant surfaces and are extremely difficult to eradicate.28
To further mimic clinical biofilm formation, a colony biofilm model was used which provides a more accurate representation of a biofilm in a clinical setting. Colony biofilms were grown on a permeable polycarbonate membrane on broth-saturated agar plate – used to mimic the surface of an infection site (Fig. S34, ESI†). Good results were observed for both E. coli DH5α and S. epidermidis RP62A biofilms after TCF-Nitro treatment, even within a short reaction time (1 h). For NCTC 10238 biofilms, nothing was seen within 1 h but the signal was slightly activated after 1 d, indicating weaker NTR activity (Fig. 4 and Fig. S35, S36, ESI†). Significantly, the activation seems to be enough to cause a colour change with DH5α and RP62A, a slight but detectable rose colour was observed within the positive wells. Hence, a colour change in this assay can be used to indicate activation.
In summary, we have developed an NTR-responsive probe TCF-Nitro based on the red-emitting fluorescent TCF core, which exhibits good sensitivity, selectivity, biocompatibility, and minimal interference from background fluorescence. After screening a range of bacterial species to evaluate their NTR activities using TCF-Nitro, we selected three main strains that exhibit the most significant NTR activity to examine the effect of growth conditions and bacterial concentrations on the levels of NTR, which confirmed that TCF-Nitro could be used to detect NTR under a range of biological conditions (mimicking clinical use). As such our study could offer insight into the development of small-molecule probes for sensing bacterial enzymes based on a variety of other fluorescent dyes.29,30
X. P. H. wishes to thank the Natural National Science Foundation of China (No. 92253306), the Open Funding Project of the State Key Laboratory of Bioreactor Engineering of East China University of Science and Technology, the Fundamental Research Funds for the Central Universities (222201717003) and the Programme of Introducing Talents of Discipline to Universities (B16017) for financial support. A. T. A. J. would like to acknowledge funding from the EPSRC, grant reference: EP/V00462X/1. T. D. J. wishes to thank the Open Research Fund of the School of Chemistry and Chemical Engineering, Henan Normal University for support (2020ZD01). R. A. H. thanks the Annette Trust and EPSRC for funding. A. C. S. would like to thank the Glasstone Research fellowship (University of Oxford) and the major research grant from Jesus College Oxford for financial support.
Conflicts of interest
There are no conflicts to declare.Notes and references
- C. A. Haynes, R. L. Koder, A. F. Miller and D. W. Rodgers, J. Biol. Chem., 2002, 277, 11513 CrossRef CAS PubMed.
- D. W. Bryant, D. R. McCalla, M. Leeksma and P. Laneuville, Can. J. Microbiol., 1981, 27, 81 CrossRef CAS PubMed.
- Z. C. Symons and N. C. Bruce, Nat. Prod. Rep., 2006, 23, 845 RSC.
- W. D. Won, R. J. Heckly, D. J. Glover and J. C. Hoffsomm, Appl. Microbiol., 1974, 27, 513 CrossRef CAS PubMed.
- I. P. Solyanikova, B. P. Baskunov, M. A. Baboshin, A. I. Saralov and L. A. Golovleva, Appl. Biochem. Microbiol., 2012, 48, 21 CrossRef CAS.
- J. C. Spain, Annu. Rev. Microbiol., 1995, 49, 523 CrossRef CAS PubMed.
- A. Carnero and M. Lleonart, BioEssays, 2016, 38, S65 CrossRef PubMed.
- K. Kiyose, K. Hanaoka, D. Oushiki, T. Nakamura, M. Kajimura, M. Suematsu, H. Nishimatsu, T. Yamane, T. Terai, Y. Hirata and T. Nagano, J. Am. Chem. Soc., 2010, 132, 15846 CrossRef CAS PubMed.
- A. F. Bedernjak, P. W. Groundwater and M. Gray, Tetrahedron, 2013, 69, 8456 CrossRef CAS.
- A. L. James, J. D. Perry, C. Jay, D. Monget, J. W. Rasburn and F. K. Gould, Lett. Appl. Microbiol., 2001, 33, 403 CrossRef CAS PubMed.
- J. Sun, Z. Hu, R. Wang, S. Zhang and X. Zhang, Anal. Chem., 2019, 91, 1384 CrossRef CAS PubMed.
- Z. Li, X. H. Li, X. H. Gao, Y. Y. Zhang, W. Shi and H. M. Ma, Anal. Chem., 2013, 85, 3926 CrossRef CAS PubMed.
- K. Ayoub, E. D. van Hullebusch, M. Cassir and A. J. Bermond, Hazard. Mater., 2010, 178, 10 CrossRef CAS PubMed.
- Z. Naal, J. H. Park, S. Bernhard, J. P. Shapleigh, C. A. Batt and H. D. Abruña, Anal. Chem., 2002, 74, 140 CrossRef CAS PubMed.
- J. E. Kwonand and S. Y. Park, Adv. Mater., 2011, 23, 3615 CrossRef PubMed.
- J. Wu, W. Liu, J. Ge, H. Zhang and P. Wang, Chem. Soc. Rev., 2011, 40, 3483 RSC.
- L. Jiang, T. Chen, E. Song, Y. Fan, D. Min, L. Zeng and G.-M. Bao, Chem. Eng. J., 2022, 427, 131563 CrossRef CAS.
- L. Zeng, H. Zeng, L. Jiang, S. Wang, J.-T. Hou and J. Yoon, Anal. Chem., 2019, 91, 12070 CrossRef CAS PubMed.
- Q. Wu, Q.-H. Zhou, W. Li, T.-B. Ren, X.-B. Zhang and L. Yuan, ACS Sens., 2022, 7, 3829 CrossRef CAS PubMed.
- R. Zhong, R. Jiang, J. Zeng, X. Gong, X. Yang, L. He, L. Yuan and D. Cheng, Anal. Chem., 2023, 95, 2428 CrossRef CAS PubMed.
- Q. Yang, Y. Wen, A. Zhong, J. Xu and S. Shao, New J. Chem., 2020, 44, 16265 RSC.
- Z. Zhang, T. Lv, B. Tao, Z. Wen, Y. Xu, H. Li, F. Liu and S. Sun, Bioorg. Med. Chem., 2020, 28, 115280 CrossRef PubMed.
- D. Yang, H. Y. Tian, M. Li, Y. Zhou and J. F. Zhang, Sci. Rep., 2017, 7, 9174 CrossRef PubMed.
- A. C. Sedgwick, H.-H. Han, J. E. Gardiner, S. D. Bull, X.-P. He and T. D. James, Chem. Commun., 2017, 53, 12822 RSC.
- Z. Li, X. H. Gao, W. Shi, X. H. Li and H. M. Ma, Chem. Commun., 2013, 49, 5859 RSC.
- R. H. F. Wong, T. Kwong, K. H. Yau and H. Y. Au-Yeung, Chem. Commun., 2015, 51, 4440 RSC.
- L. Gwynne, G. T. Williams, K.-C. Yan, B. L. Patenall, J. E. Gardiner, X.-P. He, J.-Y. Maillard, T. D. James, A. C. Sedgwick and A. T. A. Jenkins, Biomater. Sci., 2021, 9, 4433 RSC.
- C. Zhang, D.-T. Shi, K.-C. Yan, A. C. Sedgwick, G.-R. Chen, X.-P. He, T. D. James, B. Ye, X.-L. Hu and D. Chen, Nanoscale, 2020, 12, 23234 RSC.
- W.-T. Dou, X. Wang, T. Liu, S. Zhao, J. Liu, Y. Yan, J. Li, C. Zhang, A. C. Sedgwick, H. Tian, J. L. Sessler, D. M. Zhou and X.-P. He, Chemistry, 2022, 8, 1750 CrossRef CAS.
- W.-T. Dou, H.-H. Han, A. C. Sedgwick, G.-B. Zhu, Y. Zang, X.-R. Yang, J. Yoon, T. D. James, J. Li and X.-P. He, Sci. Bull., 2022, 67, 853 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc00462g |
| This journal is © The Royal Society of Chemistry 2023 |