Open Access Article
Open Access ArticleIncorporation and modification of fatty acids in cyanobacterial natural products biosynthesis
Pedro N.
Leão
 *a,
Teresa P.
Martins
*a,
Teresa P.
Martins
 ab,
Kathleen
Abt
ab,
João P. A.
Reis
ab,
Sandra
Figueiredo
a,
Raquel
Castelo-Branco
ac and
Sara
Freitas
ac
ab,
Kathleen
Abt
ab,
João P. A.
Reis
ab,
Sandra
Figueiredo
a,
Raquel
Castelo-Branco
ac and
Sara
Freitas
ac
aInterdisciplinary Centre of Marine and Environmental Research (CIIMAR/CIMAR), University of Porto, 4450-208 Matosinhos, Portugal. E-mail: pleao@ciimar.up.pt
bICBAS – Institute of Biomedical Sciences Abel Salazar, University of Porto, 4050-313 Porto, Portugal
cDepartment of Biology, Faculty of Sciences, University of Porto, 4169-007 Porto, Portugal
First published on 24th March 2023
Abstract
Fatty acid-derived alkyl chains are often found in natural products, where they can exert a number of different functions, most notably biological membrane interactions. Such alkyl chains are difficult to modify regio- and stereoselectively, since most positions are distant from any directing functional group. Chemical and biochemical diversification of these moieties is therefore a challenge, and most organisms do not modify alkyl moieties to a great extent. Still, one particular group of microorgansims – cyanobacteria – display not only a large number of fatty acid-incorporating natural products, but also modify these to a great extent. Here, we provide an overview of the unique fatty acid metabolism of cyanobacteria in the context of natural products biosynthesis. We cover the diverse range of fatty acid incorporation mechanisms that these organisms use to recruit and commit fatty acids to natural products biosynthetic pathways. A variety of alkyl chain decorations and modifications that are found in cyanobacterial natural products are highlighted, illustrating the rich enzymatic arsenal that these organisms have evolved to diversify fatty acid-derived alkyl chains.
Introduction
A number of bioactive small molecules, including drugs, contain fatty acyl-derived moieties, which are important for interactions with membranes, signaling, bioactivity or biofilm formation.1–3 For example, a number of lipopeptide antibiotics require an N-acylated tail for activity,4 colibactin uses an acyl chain as part of a pro-drug mechanism5 and acylation is proposed to prevent certain siderophores from diffusing away from the cells before binding iron.6 Despite the well-established importance of such lipid tails, medicinal chemistry efforts do not systematically employ conjugation of lipid moieties such as medium- and long-chain fatty acids to pharmacophores.7,8 Even in biological chemistry, modification of lipid tails is usually restricted to chain length and unsaturation. The reason behind this is likely the same in the fume hood as in the living cell – once generated, alkyl chains are difficult to selectively modify due to a high C–H bond energy and the distance to directing functional groups. Most organisms lack the biochemical tools to modify such saturated fatty acyl groups to a great extent, other than unsaturation and functionalization in alpha or beta positions. These modifications are often introduced during biosynthesis or degradation of fatty acids and take advantage of carbonyl chemistry and the acidity of the alpha-carbon. Still, some widespread desaturases act regioselectively on fully saturated alkyl chains.9 Perhaps to circumvent the challenges with installing functionalities in specific alkyl chain positions, many organisms – in particular bacteria and fungi – have evolved polyketide synthases that modify the alkyl chain as it grows. These pathways enable considerable structural diversification of the growing chain, but are difficult to engineer and to use in biocatalysis/metabolic engineering as they are part of megaenzyme complexes that require multiple proteins/domains to interact spatially to perform their function.10 Nevertheless, notable advances have been made in this field, such as the recent reports on fluorine incorporation into PKS pathway products.11,12Members of the bacterial phylum Cyanobacteria frequently incorporate fatty acids into their natural products.13,14 A large variety of acyl or alkyl moieties, ranging usually from C4 to C18 are commonly found in cyanobacterial natural products.13 In principle, these could be generated by dedicated polyketide synthase (PKS) assembly line enzymes or by acyl-groups recruited from primary metabolism. Increasing studies on secondary metabolite biosynthetic gene clusters (BGCs) associated with cyanobacterial natural products, have made clear that most of these acyl moieties are either fully-derived from fatty acids or from PKS-mediated elongation of slightly shorter fatty acids.15 In fact, BGCs found in cyanobacterial genomes commonly encode fatty acid incorporating enzymes, most notably fatty acyl-AMP ligases (FAALs).14 The genomic contexts of such fatty acid-incorporation genes often encode a rich diversity of biosynthetic enzymes, such as halogenases, desaturases, cytochrome P450s, methyltransferases, oxidoreductases, among others.14,16,17 In line with this, cyanobacterial natural products incorporating fatty acid-derived alkyl chains commonly exhibit rich decorations and structural features in such moieties.
Here, we present an account of unique aspects of fatty acid primary and secondary metabolism in cyanobacteria. We focus on the different ways by which these organisms incorporate fatty acids into their natural product scaffolds and the myriad of modifications that underline the structural diversity found in these lipidic natural products.
The uniqueness of cyanobacterial fatty acid metabolism
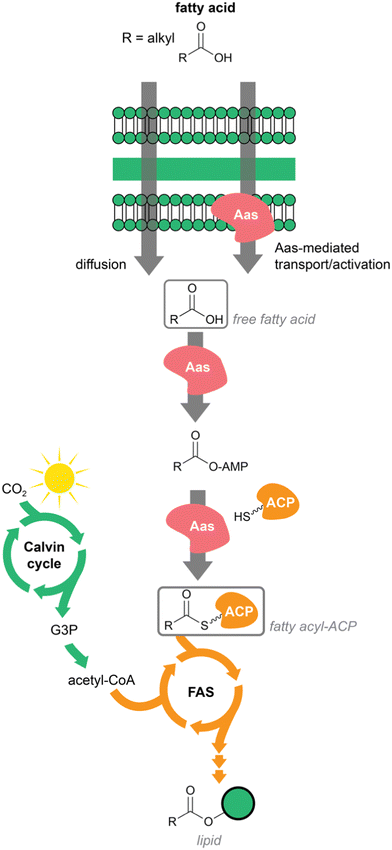
Cyanobacteria are photoautotrophic prokaryotes. They can fix atmospheric carbon and do not require any other external carbon source. Fixed carbon in the form of sugar recirculates through multiple pathways, including fatty acid biosynthesis. In addition, cyanobacteria can also internalize exogenous fatty acids. Smaller fatty acids diffuse freely through the membrane, while longer fatty acids are translocated through the action of acyl–acyl carrier protein (ACP) synthetase (Aas).18 This ATP-dependent enzyme generates acyl-ACPs from fatty acids internalized through its action or those that were internalized by diffusion. Fatty acyl–ACPs of different chain length can then be elongated through the regular fatty acid biosynthesis pathway.18 Aas enzymes carry out a reaction similar to FAALs and both are distinguishable from the related Fatty Acyl CoA Ligases (FACLs), despite the fact that all are members of the adenylate-forming enzyme superfamily.19,20 To our knowledge, Aas-encoding genes are not typically present in BGCs. Across the tree of life, fatty acyl–ACPs are used to generate membrane lipids of different sorts; in cyanobacteria these include several glycolipids that make up the extensive thylakoid membrane system21 (Fig. 1).Many cyanobacteria also generate alkanes and alkenes from fatty acyl–ACPs, through the action of acyl–ACP reductase and aldehyde-deformylating oxygenase (Aar/Ado pathway).22 This route is unique to cyanobacteria and distributed throughout the phylum.22,23 1-Alkenes can also be generated in cyanobacteria from fatty acyl–ACPs through an alternative pathway, the olefin synthase (Ols).24,25 Hydrocarbon production has interest for biotechnological purposes and these unique cyanobacterial pathways have been studied extensively under this scope.26
However, perhaps the most striking feature of cyanobacterial fatty acid metabolism is their apparent lack of a beta-oxidation pathway. Beta-oxidative enzymes have not been reported from cyanobacteria.14,27,28 In a recent study from our group,14 it was not possible to find in cyanobacteria the full set of beta-oxidative genes from E. coli; in most cyanobacterial genomes, the majority of these functions were lacking altogether. The reason behind the absence of this fundamental degradative pathway is unclear, but the widespread absence of pathway orthologs suggests that it is deep-rooted in cyanobacterial history.
Overall, the ability of cyanobacteria to generate a replenishing pool of fatty-acid biosynthesis units from photosynthesis-fixed carbon, to acquire fatty acids exogenously, coupled with the absence of beta-oxidation, has likely shaped their secondary metabolism. In particular, the peculiarities of cyanobacterial fatty acid metabolism, notably the apparent lack of canonical degradation for energy, could explain why fatty acyl moieties are so common in cyanobacterial natural product scaffolds (Scheme 1).
Incorporation of fatty acids into cyanobacterial secondary metabolites
Fatty acids can be recruited to secondary metabolite biosynthesis by a variety of enzymes.14 Genes encoding these functions are often found in BGCs, allowing for the straightforward prediction of fatty acid incorporation. Often, transfer of a fatty acid onto a secondary metabolite biosynthetic pathway-specific ACP initiates biosynthesis. However, in other cases, fatty acid incorporation events can occur at later biosynthetic steps and may not even require a cognate ACP, instead using primary metabolism thioesters (which in cyanobacteria have invariably been found to be of the fatty acyl–ACP type instead of fatty acyl-CoA thioesters, further supporting absence of beta-oxidation in these organisms). In cyanobacteria, FAALs are the most commonly found fatty acid incorporating enzymes encoded in secondary metabolite BGCs.14,15 Lipoyl transferases and GNAT (Gcn5-related-N-acetyltransferase)-family enzymes are also capable of decorating natural product scaffolds or their biosynthetic precursors with fatty acyl moieties, while BrtB catalyzes the incorporation of non-activated fatty acid carboxylates29–31 Dialkylresorcinol/pyrone-forming enzymes and certain type III PKSs are proposed to recruit fatty acyl–ACP thioesters or their derivatives from primary metabolism and commit them to a specific secondary metabolite pathway.17 Starter C-domains can also be found in cyanobacterial BGCs32 and have been shown in other bacteria to N-acylate amino acids with beta-hydroxy fatty acyl groups33 (Fig. 2).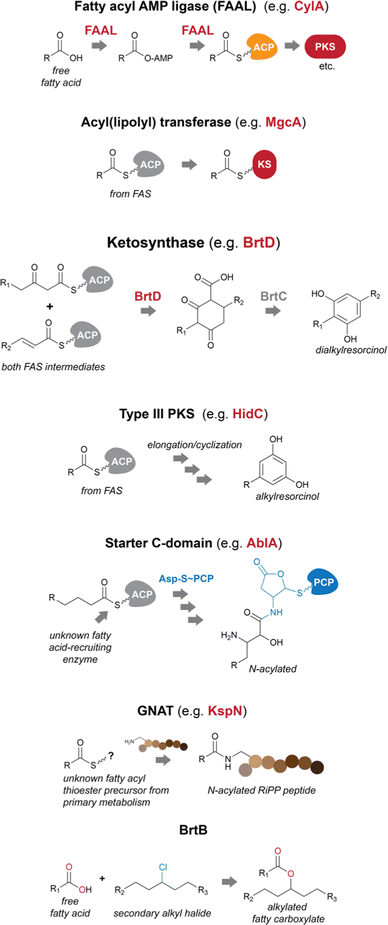 | ||
| Fig. 2 Biosynthetic strategies used by cyanobacteria to recruit and incorporate fatty acid residues into natural products. PKS – polyketide synthase; KS – ketosynthase domain. | ||
Fatty acyl AMP ligases (FAALs)
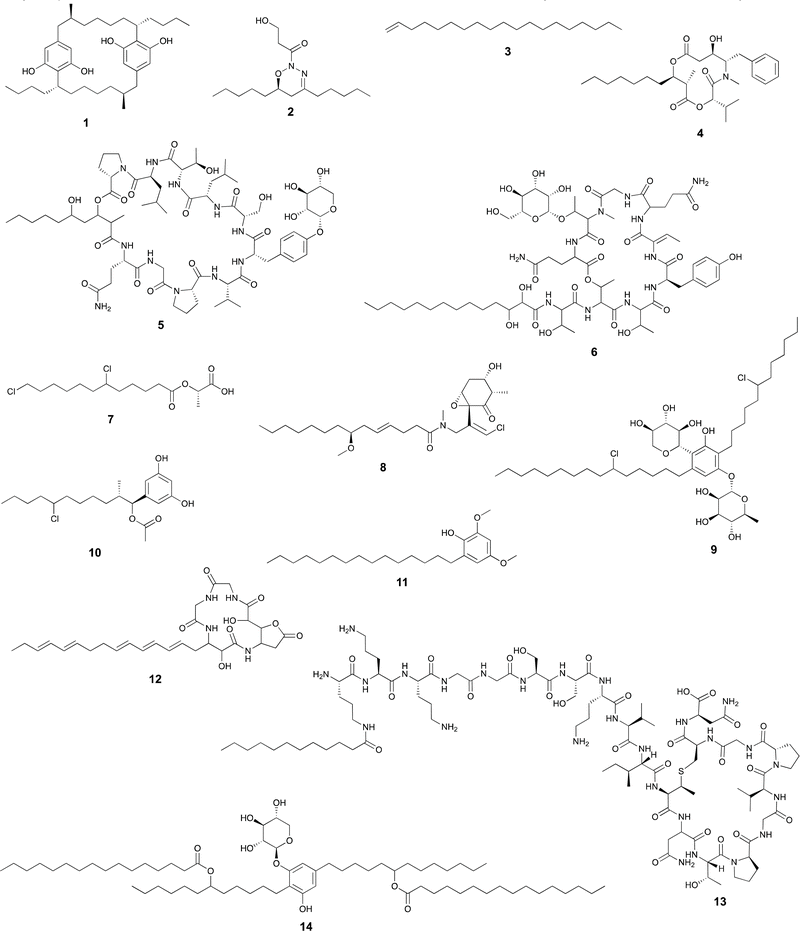
FAALs are widespread in cyanobacterial genomes, with an average of over one homolog per publicly available genome.14 These are members of the acyl-adenylating enzyme superfamily and activate fatty acids by generating an acyladenylate (acyl-AMP) intermediate.34 FAALs then acylate ACPs using the activated acyl-AMPs as substrates.34 At this stage, the fatty acid residue is committed to a secondary metabolite biosynthesis pathway, as the ACP interacts with downstream enzymatic machinery. FAALs bear structural resemblance to FACLs, which acylate CoA instead of an ACP and are members of the same superfamily.20 It has recently been shown that FAALs contain a non-functional CoA-binding pocket and use an alternative pocket to enable discrimination between holo-ACP and CoA and proceed with holo-ACP acylation.20 The ACP-tethered fatty acyl moiety can then be further modified by the action of polyketide synthases (PKSs) or other ketosynthases (KSs), non-ribosomal peptide synthetases (NRPSs), all of which make use of carrier-protein thioester enzymatic logic. The cylindrocyclophanes (e.g. cylindrocyclophane F 1), nocuolin A (2), 1-alkenes (e.g.3) and a wide range of lipopeptides (e.g. hapalosin A, 4) are examples of cyanobacterial natural products whose biosynthesis involves FAAL-mediated activation and incorporation of fatty acids.15,24,35–37 FAALs can be found in cyanobacterial genomes as single domain enzymes, with their partner ACP as FAAL-ACP didomain proteins, or integrated in multidomain megaenzymes.Usually, cyanobacterial FAALs are responsible for the incorporation of a single fatty acid residue into each secondary metabolite end product. The resulting acyl-ACP moiety can then be elongated by PKS modules before further modification, which can include interfacing with NRPS modules (as in the biosynthesis of hapalosins37 – e.g.4 – or desmamides38 – e.g. desmamide A, 5) to generate lipopeptides15 or a type III PKS to generate alkylresorcinols such as 1.35 It has been proposed that a FAAL-generated acyl-ACP interfaces directly with a starter C-domain (NRPS biosynthesis) to generate the hassallidins (e.g. hassallidin A, 6, see section below on starter C-domains).32 In the biosynthesis of the chlorosphaerolactylates (e.g. chlorosphaerolactylate A, 7), a non-elongating PKS is predicted to shuttle the acyl chain to a depsipeptide synthetase (NRPS-like) module.39 In contrast with these single-incorporation examples, we have recently shown that in the biosynthesis of the cyanobacterial 1,2,3-oxadiazine nocuolin A (2), a single FAAL – NocH – activates two different fatty acids (hexanoic and octanoic acids).36 The two acyl-ACPs are then used by the KS NocG to generate a beta-keto acid featuring a linear 13-carbon chain.36
BGCs that encode FAALs usually code for enzymes that modify and decorate acyl chains, including halogenases, cytochrome P450s, desaturases and other.16,40–43 Such modifications can influence the biological activity of the resulting natural product e.g.44
Acyl(lipoyl) transferases
Malyngamides (e.g. malyngamide I, 8)45 are a large group of highly modified marine-derived cyanobacterial lipids. Bioinformatics analysis of their putative BGCs in the genus Okeania indicated the presence, in each BGC, of a gene encoding a lipoyl transferase of the lipoate-protein ligase A (LplA) superfamily.29 The encoded enzyme – MgcA – is proposed to transfer an octanoyl moiety from octanoyl-ACP (from fatty acid biosynthesis) directly onto the KS domain of the ensuing PKS module, which would enable further elongation and processing through canonical PKS/NRPS to generate the final malyngamides.29Homologs of this enzyme were found in cyanobacteria and heterotrophic bacteria, including the Bacillus subtilis LipM, involved in N-acylation of protein lipoyl domains. These were not found in secondary metabolite biosynthesis genomic contexts. Thus, the authors propose that MgcA represents an example of neofunctionalization of LipM to serve a role in the biosynthesis of natural products.29
Dialkylresorcinol/pyrone-forming enzymes
Dialkylresorcinol (DAR) biosynthesis from the head-to-head condensation of two fatty acid-derived precursors was characterized by Bode and co-workers.46 The authors showed that DarB, a member of a phylogenetically congruent clade of widely distributed KSs generated carboxy-cyclohexanediones in Chitinophaga pinensis. These were proposed to be oxidized to resorcinols through the action of the oxidoreductase DarA that was found adjacent to the KSs in DAR-producing bacteria. This initial report indicated the presence of such two-gene cassettes in cyanobacterial BGCs.46The discovery of the bartolosides (e.g., bartoloside B, 9),47 cyanobacterial glycosylated and halogenated DARs, paved the way for the study of BrtD, the DarB homolog encoded in the bartoloside-producing Synechocystis salina LEGE 06155. BrtD was shown to form a carboxy-cyclohexanedione from 2-octadecenoyl- and 3-oxo-hexadecanoyl-ACP thioesters. In the same study, BrtC (DarA homolog) was shown to be an aromatase generating the DAR backbone of the bartolosides.47
DarB/DarA homologs are found in several orphan cyanobacterial BGCs of diverse architecture, suggesting that additional DAR classes are to be found in these organisms.17 Furthermore, in several BGCs, DarB homologs are also found in the absence of a DarA partner.17 In this case, it is unclear whether the final products contain cyclohexanedione moieties.46
DarB and BrtD recruit fatty acid-derived thioesters from primary metabolism. However, DarB recruits both short and long-chain fatty acids to generate DARs,46 while BrtD generates much longer DARs by recruiting, for example, C16 and C18 fatty acid derivatives in the biosynthesis of 9.47 Therefore, while it could be expected that the presence of DarB homologs in a BGC results in fatty acid-derivative recruitment, the structural basis and sequence determinants for selectivity are unclear at this stage. This hinders structural predictions from the bioinformatics analysis of such BGCs.
Type III PKSs
Bacterial type III PKSs perform both extension and cyclization of a variety of substrates.48 The structures generated by these enzymes include quinones, pyrones, phloroglucinols and resorcinols.48 In cyanobacteria, only two families of type III PKS products have been reported so far (cylindrocyclophanes such as 1/cylindrofridins such as 10 and hierridins, e.g.11),17,35,49,50 both involving generation of alkylresorcinol products. This is in contrast with an estimated prevalence of these genes in ∼17% of cyanobacterial genomes.51 There is also considerable phylogenetic diversity associated with cyanobacterial type III PKSs, including homologs that clade with characterized alkylpyrone-generating type III PKSs from heterotrophic bacteria.17,51In cylindrocyclophane/cylindrofridin biosynthesis, a dedicated FAAL activates and loads decanoic acid onto a pathway-specific ACP. This carrier protein-tethered fatty acyl thioester is then elongated by type I PKS machinery before further elongation and cyclization into an alkylresorcinol by the type III PKS enzyme.35,49,52
In contrast, the hierridin BGC (hid) does not contain an ACP and instead we have proposed50 that hexadecanoyl-ACP from fatty acid biosynthesis is recruited by the type III PKS HidC into the hierridin pathway. This substrate would be extended by three acetate units (from malonyl-CoA) before cyclization into an alkylresorcinol by the same enzyme and further modification by other pathway enzymes. A large number of BGCs that are similar to hid and do not contain an ACP in their genomic contexts are found distributed in free-living, planktonic cyanobacteria with smaller genomes and are expected to generate similar alkylresorcinols.50,51
Likewise, type III PKS-encoding BGCs in cyanobacteria that are expected to generate alkylpyrones, are also lacking a dedicated ACP and likely also recruit fatty acyl-ACP substrates from primary metabolism.17,51
Starter C-domains
In NRPS biosynthesis, canonical C-domains perform the N-to-C condensation of an amino acyl or peptidyl-carrier protein (or domain) donor and an amino acyl-carrier protein (or domain) acceptor.10 Other C-domain types can perform additional functions, including cyclization and dehydration, epimerization and ester bond formation.53 Starter C-domains are yet another subclass of C-domains that generate lipopeptides by performing a condensation reaction using a fatty acyl thioester acceptor instead of an amino acyl-carrier protein thioester.33 These domains are usually in the first NRPS module acting in the biosynthesis of such lipopeptides. Both CoA and ACP fatty-acyl thioesters have been shown to N-acylate the amino acyl-carrier protein in the first NRPS module.2In cyanobacteria, the involvement of a starter C-domain in the biosynthesis of the lipoglycopeptides hassallidins (e.g.6) has been proposed to follow the action of the fatty acid-recruiting FAAL HasG, which is part of the hassallidins BGC (has).32 However, in the biosynthesis of the anabaenolysins (e.g. anabaenolysin A, 12), antifungal lipopeptides produced by Anabaena spp., a starter C-domain is expected to perform the N-acylation of an aspartyl-carrier protein thioester from the first NRPS module (AblA).40 The abl BGC does not appear to encode a fatty acid activating enzyme, but the initial NRPS contains an ACP domain which is expected to be loaded with the acylating fatty acid and, therefore, be the acceptor substrate of the reaction of the starter C-domain.40
GNATs
A recent report31 has expanded the arsenal of fatty acid-incorporating enzymes in natural products biosynthesis to include enzymes of the GNAT superfamily. GNAT enzymes commonly participate in different biological processes, through acylation (usually acetylation, but sometimes other groups) of different substrates such as proteins or sugar residues.54 Acyl-CoAs are the reported source of the transferred acyl groups, and while N-acylation is by far the most commonly reported activity for these enzymes, O-acylation has also been observed54 and in the biosynthesis of the cyanobacterial natural product curacin, S-acetylation was attributed to a GNAT domain.55A GNAT enzyme from a streptomycete was also shown to transfer a PKS-derived long acyl chain (from acyl-ACP) to the N-terminus of a peptidic intermediate originating from ribosomally synthesized and post-translationally modified peptide (RiPP) machinery.56 In their report, Hubrich et al.31 show another way by which RiPP biosynthetic pathways can lead to lipopeptides. Specifically, the authors have shown that certain GNAT enzymes, part of (RiPP) BGCs (KspN from a cyanobacterium and PhaN from an alphaprotebacterium), can selectively N-acylate lysine or ornithine side chains in peptide scaffolds. It was proposed that the dodecanoic acid residue found in the cyanobacterial pathway end-product (kamptornamide, 13) is derived from the primary metabolite pool (i.e., an acyl-CoA or -ACP species).31 Because RiPP pathways are amenable to combinational biosynthesis through simple genetic manipulation, a variety of lipopeptide structures can now in principle be engineered in a straightforward manner.31
BrtB
The above-mentioned fatty acid incorporation reactions make use of thioester chemistry, resulting in fatty acylation. Our group has recently uncovered a new type of fatty acid incorporation into secondary metabolites that, instead, uses the fatty carboxylate as a nucleophile and a secondary alkyl chloride as the electrophile. This reaction is carried out by BrtB, an enzyme encoded in the bartoloside BGC (brt).30 BrtB catalyzes the esterification of fatty acids onto the alkyl chains of the bartolosides, generating bartoloside esters such as metabolite 14. It shows remarkable promiscuity towards the fatty acid nucleophile, being capable of esterifying short-, medium- and long-chain fatty acids.30 The only characterized homolog of BrtB, CylK is also capable of transferring a lipidic substructure to alkyl halides, namely an alkylresorcinol, in cylindrocyclophane biosynthesis.52Modification of fatty acyl moieties in cyanobacterial natural products
Quite often, fatty acid-derived moieties are modified during the biosynthesis of cyanobacterial secondary metabolites. These modifications occur with high selectivity and in positions that can be distant from the acyl carbonyl (Scheme 2 and 3) and influence biological activity e.g.44Unsaturation
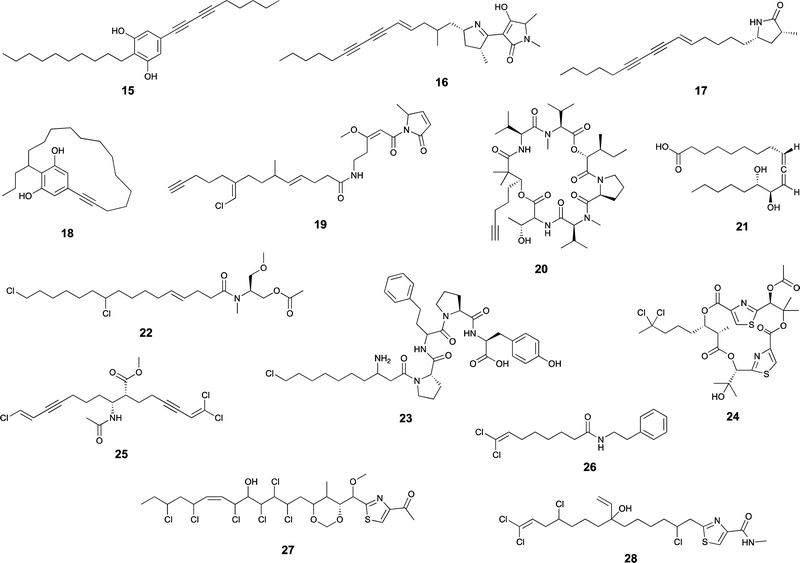
Desaturases are common in a range of organisms and generate unsaturated versions of fatty acyl moieties, generating alkenes or alkynes.9,57,58 For example, acyl-lipid desaturases are well-known regioselective enzymes found in cyanobacteria and other organisms that act on the acyl chains of membrane lipids, and therefore modulate membrane fluidity in response to changing environmental conditions.59 Many desaturases act on cyanobacterial secondary metabolite biosynthesis as well. Microcarbonin A (15) features a conjugated diyne in one of the chains of its dialkylresorcinol core.60 Its putative BGC contains two desaturases.17,46 Metabolites such as 12 contain long fatty acyl moieties (C18-derived) that contain various degrees of unsaturation and at different positions, probably through the action of the desaturase AblC.40 Additional examples of cyanobacterial metabolites with long alkyl chains featuring unsaturations compatible with the action of a desaturase include fischerellins A (16)61 and B (17)62 and nostocycline (18).63Terminal alkynes and alkenes can also be generated in fatty acyl moieties through the action of desaturases.58 This is the case of the well-studied biosynthesis of the terminal alkyne moiety in the jamaicamides (e.g. jamaicamide B, 19),64,65 in which the desaturase JamB acts sequentially on hexanoyl-JamC(ACP) to generate first 1-hexenoyl-JamC and then 1-hexyonyl-ACP (Fig. 3). Terminal olefins can also be generated by the Ols pathway, as mentioned above, which employs an FAAL and type I PKS machinery to elongate/decarboxylate a fatty acyl-ACP substrate.24,25 Marine cyanobacterial cyclic lipopeptides of mixed PKS/NRPS origin, belonging to the so-called kulolide superfamily of natural products (e.g. viequeamide A, 20),66 typically feature a fatty acid-derived residue. This can be saturated, a terminal alkene or a terminal alkyne and the three versions are usually co-produced.66 The biosynthesis of the unsaturated versions in this superfamily of cyclic peptides expected to occur in a similar fashion to the jamaicamides.67 The same has been proposed for the terminal alkyne moieties of the linear lipopeptides vatiamides.68 A cyanobacterial allenic fatty acid – puna’auic acid (21) – has been reported from a marine cyanobacterium. Its biosynthesis is proposed to involve an enyne precursor, which would then be converted to a propargyl epoxide and reduced to the allene.69
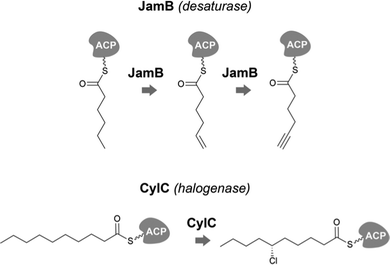 | ||
| Fig. 3 Examples of biochemically-characterized fatty acyl modification reactions in cyanobacterial secondary metabolite biosynthesis. | ||
Halogenation
Cyanobacteria are rich in enzymatic chemistry resulting in halogenated natural products.16,70,71 Different classes of halogenating enzymes are encoded in cyanobacterial BGCs, namely flavin-dependent, nonheme iron/2-oxo-glutarate (2OG)-dependent and dimetal carboxylate halogenases.16,71 From these, dimetal carboxylate and nonheme iron/2OG-dependent halogenases have been shown to act on alkyl moieties.16 Of particular interest for biocatalysis are regio- and stereoselective halogenations on terminal or mid-chain unactivated sp3-hybridized carbons. These are observed commonly in a number of cyanobacterial metabolites containing fatty acid-derived alkyl moieties, like, for example, the columbamides (e.g. columbamide A, 22),72 bartolosides (e.g.9),47 microginins (e.g. microginin 739, 23),43,73 cylindrocyclophanes/cylindrofridins (e.g.1/10),17,49,74 among others. In most such metabolites, chlorine or bromine atom installation can be linked to dimetal carboxylate halogenases, based on BGC annotation.16 That is the case of CylC (Fig. 3), which acts during the biosynthesis of 1/10 and related metabolites and has been characterized biochemically.52 But, for example the generation of the 5,5-dichloro group in hexanoyl-ACP during hectochlorin (24) biosynthesis, is attributed to the nonheme iron/2OG-dependent halogenase HctB.75,76 Mid-chain gem-dichlorination has been observed not only in hectochlorin and related metabolites but also in a bartoloside – suggesting that the dimetal carboxylate halogenase BrtJ can also generate this group.77Gem-dichlorinations have been most often reported in terminal positions (i.e. dichloromethyl group) in marine and freshwater cyanobacterial metabolites (e.g. microginins, columbamides, cylindrofridins) associated with BGCs encoding dimetal carboxylate halogenases.16 (Di)halovinylidenes are additional structural motifs associated with fatty acid halogenation. These rare moieties can be seen in the taveuniamides (e.g. taveuniamide E, 25)78 and caracolamide A (26)79 and in sponge-derived azirines e.g.80 that have been recently linked to a cyanobacterial biogenetic origin.81 Recently reported cyanobacterial metabolites such as the aranazoles (e.g. aranazole C, 27)41 or fischerazoles (e.g. fischerazole A, 28)42 illustrate the rich regioselective decoration of fatty acid-derived alkyl moieties with halogens. This is particularly remarkable in the biosynthesis of the aranazoles, where six halogenated positions are likely generated by the six nonheme iron/2OG-dependent halogenases encoded in the putative aranazole BGC (arz).41Oxygenation
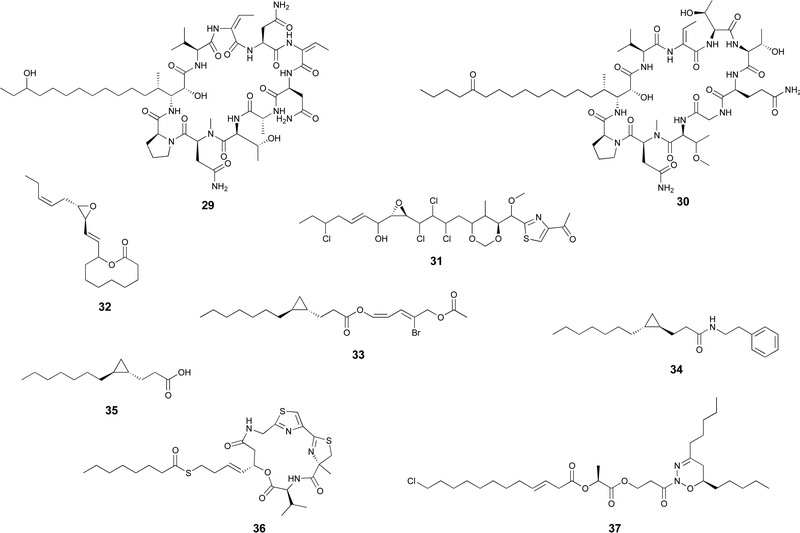
Certain cyanobacterial lipopeptides feature oxygenation in their fatty-acid derived moieties. For example, the related puwainaphycins and minutissamides (e.g. puwainaphycin A, 29 and minutissamide D, 30)82,83 contain mid-chain hydroxy and keto groups, which are likely attributable to an oxygenation event catalyzed by the cytochrome P450 monoxygenase PuwJ.44,84Epoxide groups are common modifications in marine lipids, such as in the algal oxylipins.85 Still, in cyanobacteria, this is a rare moiety in fatty acid-derived alkyl chains, and, to our knowledge, has been reported only in aranazole D (31).41 Gloeolactone (32)86 also features an epoxide but it is unclear if this compound is fully derived from a fatty acid or that it is a fatty acid-polyketide hybrid. While Piel and co-workers, who reported the arz BGC, have not speculated on the origin of the epoxide,41 it encodes two putative P450 oxygenases and one putative Rieske oxygenase, which could be linked to this structural modification.
Additional modifications
Apart from these more commonly observed modifications, a number of additional transformations involving fatty acyl moieties have been reported. Grenadadiene (33) and grenadamide (34)87 feature a dodecanoic acid-derived cyclopropanated acyl moiety, likely of similar biosynthetic nature that the cyanobacterial lyngbyoic acid (35).88 These modifications are well-known from Mycobacterium tuberculosis cell-wall mycolic acids, in which case they are generated through olefin methylation and cyclization by a specific type of SAM-dependent enzyme (cyclopropane synthase).89,90Metabolites with unusual cyclizations that appear associated to fatty acid-derived moieties include 18, 32 and 2. In the case of 18, the cyclization between the resorcinol and the mid-chain position is likely generated by an intramolecular version of the reaction catalzyed by CylK in cylindrocyclophane biosynthesis, which should involve a cryptic halogenation.17 The biosynthesis of 32 has not been proposed but it is plausible that either a C18 fatty acid generates the entire carbon skeleton or that a C10 fatty acyl precursor is elongated by PKS machinery before thioesterase-catalyzed cyclization with the C-1-derived alcohol acting as a nucleophile. During the biosynthesis of 2, the 13-carbon linear chain is further modified to generate the unusual 1,2,3-oxadiazine moiety, whose biosynthesis remains to be elucidated.36
In most natural products, fatty acyl moieties tend to be found as esters or amides. However, this is not the case of the anticancer lead compound largazole (36), a cyanobacterial lipopeptide whose lipid tail features an unusual decanoyl thioester moiety.91
Capturing natural products diversity associated with fatty acids
A faster access to lipidic natural products has the potential to accelerate the discovery of drug-like metabolites8 and reveal enzymes that can selectively modify alkyl moieties and thus be of interest for biocatalytic applications.Precursor supplementation
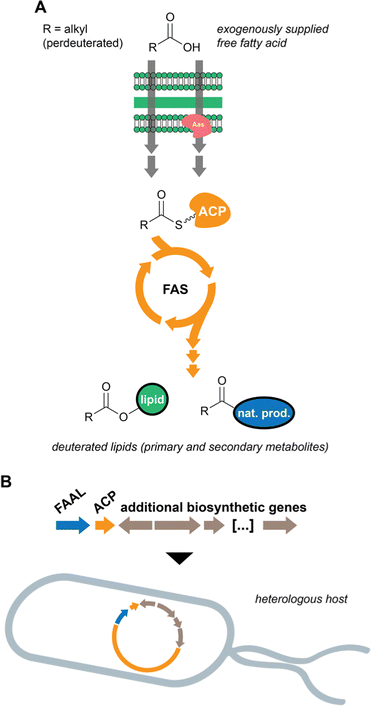
In previous work, we considered that the diversity of functions associated with fatty acid-incorporating enzymes encoded in cyanobacterial BGCs could be regarded as an opportunity to discover unusual natural product structures. Hence, we developed a metabolomics-based method that allowed us to detect such compounds, some of which we have reported.14,42 The method involves supplementation of cyanobacterial cultures with perdeuterated fatty acids, namely d11-hexanoic acid. The deuterons eventually get incorporated into lipids, including natural products, and the label is not scrambled due to the absence of fatty acid degradation (Fig. 4(A)). Mass spectrometry-based comparative metabolomics is then used to detect mass shifts from a non-supplemented control. As anticipated, the discovered compounds contain multiple decorations and hide unusual chemistry. In particular, the nocuolactylates (e.g. nocuolactylate A, 37)14 and the fischerazoles (e.g.28)42 are highly unusual fatty acid-incorporating structures that have been uncovered using this strategy.Heterologous expression of orphan BGCs
Heterologous expression of cyanobacterial BGCs has not progressed as fast as for other groups of chemically rich bacteria (most notably actinobacteria e.g.92). In recent years, however, considerable progress has been made and several BGCs found in the genomes of cyanobacteria have been successfully expressed in heterologous hosts, cyanobacterial or not (Fig. 4(B)).93 Hapalosin A, a product of hybrid PKS/NRPS biosynthesis, was successfully expressed in E. coli using Direct Pathway Cloning Combined with Sequence- and Ligation-Independent Cloning (DiPaC-SLIC).37 Octanoic acid is incorporated into the hapalosins through the action of a FAAL enzyme.14,37 Another FAAL-containing BGC – mic, associated with the microginin family of compounds – was again successfully expressed in E. coli using DiPaC-SLIC to generate natural and non-natural variants of these metabolites.43 The dimetal carboxylate halogenase that generates the dichloromethyl terminus in several microginins (e.g.23) was functional in E. coli and appeared to prefer bromine to generate dibromomethyl microginin analogs.43 Using the cyanobacterium Nostoc sp. PCC 7120 as a heterologous host, a range of halogenated columbamides (e.g.22), including non-natural analogues, were produced in yields comparable to the natural producer (Moorena bouillonii PNG5-198).94Conclusions
The ecophysiological uniqueness of cyanobacteria has shaped their natural products biochemistry. The availability of fatty acids in cyanobacterial cells, resulting from photosynthetic carbon fixation, apparently unabated by a catabolic pathway such as beta-oxidation, has turned these into important substrates for several biosynthetic pathways. In particular, natural products biosynthesis in cyanobacteria makes common use of fatty acid substrates and modifies them to a great extent using a rich combination of enzymes. Over the years, the discovery of cyanobacterial natural products that feature fatty acid residues and the study of their biosynthesis have revealed multiple instances of complex and intricate biochemical events. Still, the phylum Cyanobacteria remains to be extensively explored for their natural products and genome and metagenome data associated with these organisms contains a wealth of BGCs that are not associated with known natural products. Many of such BGCs can be predicted to incorporate fatty acid residues. Therefore, there are ample opportunities for accessing new chemical space within these organisms but also for finding interesting enzymatic chemistries that act selectively on barren alkyl chains. In addition, a better understanding of fatty acid incorporating enzymes, in particular how selectivity as to chain length is achieved, will lead to better predictions of natural products structures from sequence data, something that is currently lacking.95Conflicts of interest
There are no conflicts to declare.Acknowledgements
Funding from Fundação para a Ciência e a Tecnologia through grants PTDC/BIA-BQM/5760/2020, UIDB/04423/2020, and UIDP/04423/2020 is acknowledged. This work has also received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement no 952374 and from the European Research Council under grant agreement no 759840.Notes and references
- S. Vanni, L. Riccardi, G. Palermo and M. De Vivo, Acc. Chem. Res., 2019, 52, 3087–3096 CrossRef CAS PubMed.
- Y.-H. Chooi and Y. Tang, Chem. Biol., 2010, 17, 791–793 CrossRef CAS PubMed.
- A. González, S. Bellenberg, S. Mamani, L. Ruiz, A. Echeverría, L. Soulère, A. Doutheau, C. Demergasso, W. Sand, Y. Queneau, M. Vera and N. Guiliani, Appl. Microbiol. Biotechnol., 2013, 97, 3729–3737 CrossRef PubMed.
- J. Micklefield, Chem. Biol., 2004, 11, 887–888 CrossRef CAS PubMed.
- C. A. Brotherton and E. P. Balskus, J. Am. Chem. Soc., 2013, 135, 3359–3362 CrossRef CAS PubMed.
- G. Xu, J. S. Martinez, J. T. Groves and A. Butler, J. Am. Chem. Soc., 2002, 124, 13408–13415 CrossRef CAS PubMed.
- M. Avadisian and P. T. Gunning, Mol. BioSyst., 2013, 9, 2179 RSC.
- J. Morstein, A. Capecchi, K. Hinnah, B. Park, J. Petit-Jacques, R. C. Van Lehn, J.-L. Reymond and D. Trauner, J. Am. Chem. Soc., 2022, 144, 18532–18544 CrossRef CAS PubMed.
- P. H. Buist, Nat. Prod. Rep., 2004, 21, 249 RSC.
- M. A. Fischbach and C. T. Walsh, Chem. Rev., 2006, 106, 3468–3496 CrossRef CAS PubMed.
- A. Rittner, M. Joppe, J. J. Schmidt, L. M. Mayer, S. Reiners, E. Heid, D. Herzberg, D. H. Sherman and M. Grininger, Nat. Chem., 2022, 14, 1000–1006 CrossRef CAS PubMed.
- S. Sirirungruang, O. Ad, T. M. Privalsky, S. Ramesh, J. L. Sax, H. Dong, E. E. K. Baidoo, B. Amer, C. Khosla and M. C. Y. Chang, Nat. Chem. Biol., 2022, 18, 886–893 CrossRef CAS PubMed.
- K. Tidgewell, B. R. Clark and W. H. Gerwick, in Comprehensive Natural Products II Chemistry and Biology, ed. L. Mander and H.-W. Lui, Elsevier, Oxford, 2010, vol. 2, pp.141–188 Search PubMed.
- S. A. C. Figueiredo, M. Preto, G. Moreira, T. P. Martins, K. Abt, A. Melo, V. M. Vasconcelos and P. N. Leão, Angew. Chem., Int. Ed., 2021, 60, 10064–10072 CrossRef CAS PubMed.
- D. P. Fewer, J. Jokela, L. Heinilä, R. Aesoy, K. Sivonen, T. Galica, P. Hrouzek and L. Herfindal, Physiol. Plant., 2021, 173, 639–650 CrossRef CAS PubMed.
- N. Eusebio, A. Rego, N. R. Glasser, R. Castelo-Branco, E. P. Balskus and P. N. Leão, BMC Genomics, 2021, 22, 633 CrossRef CAS PubMed.
- T. P. Martins, C. Rouger, N. R. Glasser, S. Freitas, N. B. de Fraissinette, E. P. Balskus, D. Tasdemir and P. N. Leão, Nat. Prod. Rep., 2019, 36, 1437–1461 RSC.
- J. Beld, R. Abbriano, K. Finzel, M. Hildebrand and M. D. Burkart, Mol. BioSyst., 2016, 12, 1299–1312 RSC.
- J. Beld, K. Finzel and M. D. Burkart, Chem. Biol., 2014, 21, 1293–1299 CrossRef CAS PubMed.
- G. S. Patil, P. Kinatukara, S. Mondal, S. Shambhavi, K. D. Patel, S. Pramanik, N. Dubey, S. Narasimhan, M. K. Madduri, B. Pal, R. S. Gokhale and R. Sankaranarayanan, eLife, 2021, 10, e70067 CrossRef CAS PubMed.
- D. Petroutsos, S. Amiar, H. Abida, L.-J. Dolch, O. Bastien, F. Rébeillé, J. Jouhet, D. Falconet, M. A. Block, G. I. McFadden, C. Bowler, C. Botté and E. Maréchal, Prog. Lipid Res., 2014, 54, 68–85 CrossRef CAS PubMed.
- A. Schirmer, M. A. Rude, X. Li, E. Popova and S. B. del Cardayre, Science, 2010, 329, 559–562 CrossRef CAS PubMed.
- S. Klahn, D. Baumgartner, U. Pfreundt, K. Voigt, V. Schon, C. Steglich and W. R. Hess, Alkane biosynthesis genes in cyanobacteria and their transcriptional organization, Front. Bioeng. Biotechnol., 2014, 2, 24 Search PubMed.
- R. C. Coates, S. Podell, A. Korobeynikov, A. Lapidus, P. Pevzner, D. H. Sherman, E. E. Allen, L. Gerwick and W. H. Gerwick, PLoS One, 2014, 9, e85140 CrossRef PubMed.
- T. Zhu, T. Scalvenzi, N. Sassoon, X. Lu and M. Gugger, Appl. Environ. Microbiol., 2018, 84, e00425-18 CrossRef PubMed.
- Y. Hayashi and M. Arai, Microb. Cell Fact., 2022, 21, 256 CrossRef CAS PubMed.
- D. Kaczmarzyk and M. Fulda, Plant Physiol., 2010, 152, 1598–1610 CrossRef CAS PubMed.
- S. von Berlepsch, H.-H. Kunz, S. Brodesser, P. Fink, K. Marin, U.-I. Flügge and M. Gierth, Plant Physiol., 2012, 159, 606–617 CrossRef CAS PubMed.
- N. A. Moss, T. Leão, M. R. Rankin, T. M. McCullough, P. Qu, A. Korobeynikov, J. L. Smith, L. Gerwick and W. H. Gerwick, ACS Chem. Biol., 2018, 13, 3385–3395 CrossRef CAS PubMed.
- J. P. A. Reis, S. A. C. Figueiredo, M. L. Sousa and P. N. Leão, Nat. Commun., 2020, 11, 1–9 CrossRef PubMed.
- F. Hubrich, N. M. Bösch, C. Chepkirui, B. I. Morinaka, M. Rust, M. Gugger, S. L. Robinson, A. L. Vagstad and J. Piel, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2113120119 CrossRef CAS PubMed.
- J. Vestola, T. K. Shishido, J. Jokela, D. P. Fewer, O. Aitio, P. Permi, M. Wahlsten, H. Wang, L. Rouhiainen and K. Sivonen, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E1909–E1917 CrossRef CAS PubMed.
- C. Rausch, I. Hoof, T. Weber, W. Wohlleben and D. H. Huson, BMC Evol. Biol., 2007, 7, 78 CrossRef PubMed.
- P. Arora, A. Goyal, V. T. Natarajan, E. Rajakumara, P. Verma, R. Gupta, M. Yousuf, O. A. Trivedi, D. Mohanty, A. Tyagi, R. Sankaranarayanan and R. S. Gokhale, Nat. Chem. Biol., 2009, 5, 166–173 CrossRef CAS PubMed.
- H. Nakamura, H. A. Hamer, G. Sirasani and E. P. Balskus, J. Am. Chem. Soc., 2012, 134, 18518–18521 CrossRef CAS PubMed.
- T. P. Martins, N. R. Glasser, D. J. Kountz, P. Oliveira, E. P. Balskus and P. N. Leão, ACS Chem. Biol., 2022, 17, 2528–2537 CrossRef CAS PubMed.
- P. M. D’Agostino and T. A. M. Gulder, ACS Synth. Biol., 2018, 7, 1702–1708 CrossRef PubMed.
- S. Freitas, R. Castelo-Branco, A. Wenzel-Storjohann, V. M. Vasconcelos, D. Tasdemir and P. N. Leão, J. Nat. Prod., 2022, 85, 1704–1714 CrossRef CAS PubMed.
- K. Abt, R. Castelo-Branco and P. N. Leão, J. Nat. Prod., 2021, 84, 278–286 CrossRef CAS PubMed.
- T. K. Shishido, J. Jokela, C.-T. Kolehmainen, D. P. Fewer, M. Wahlsten, H. Wang, L. Rouhiainen, E. Rizzi, G. D. Bellis, P. Permi and K. Sivonen, Proc. Natl. Acad. Sci. U. S. A., 2015, 201510432 Search PubMed.
- P. Moosmann, R. Ueoka, M. Gugger and J. Piel, Org. Lett., 2018, 20, 5238–5241 CrossRef CAS PubMed.
- S. Figueiredo, K. Abt, T. Martins, I. Lacomba, A. Forero, C. Jiménez, J. Rodríguez and P. Leão, Fischerazoles A-C, cyanobacterial polychlorinated lipids featuring fatty acyl chain rearrangement, ChemRxiv, 2022, preprint DOI:10.26434/chemrxiv-2022-v8zlp.
- N. Eusébio, R. Castelo-Branco, D. Sousa, M. Preto, P. D’Agostino, T. A. M. Gulder and P. N. Leão, ACS Synth. Biol., 2022, 11, 3493–3503 CrossRef PubMed.
- K. Saurav, A. Caso, P. Urajová, P. Hrouzek, G. Esposito, K. Delawská, M. Macho, J. Hájek, J. Cheel, S. Saha, P. Divoká, S. Arsin, K. Sivonen, D. P. Fewer and V. Costantino, ACS Omega, 2022, 7, 11818–11828 CrossRef CAS PubMed.
- J. S. Todd and W. H. Gerwick, Tetrahedron Lett., 1995, 36, 7837–7840 CrossRef CAS.
- S. W. Fuchs, K. A. J. Bozhüyük, D. Kresovic, F. Grundmann, V. Dill, A. O. Brachmann, N. R. Waterfield and H. B. Bode, Angew. Chem., Int. Ed., 2013, 52, 4108–4112 CrossRef CAS PubMed.
- P. N. Leão, H. Nakamura, M. Costa, A. R. Pereira, R. Martins, V. Vasconcelos, W. H. Gerwick and E. P. Balskus, Angew. Chem., Int. Ed., 2015, 54, 11063–11067 CrossRef PubMed.
- D. Yu, F. Xu, J. Zeng and J. Zhan, IUBMB Life, 2012, 64, 285–295 CrossRef CAS PubMed.
- M. Preisitsch, T. H. J. Niedermeyer, S. E. Heiden, I. Neidhardt, J. Kumpfmüller, M. Wurster, K. Harmrolfs, C. Wiesner, H. Enke, R. Müller and S. Mundt, J. Nat. Prod., 2016, 79, 106–115 CrossRef CAS PubMed.
- M. Costa, I. E. Sampaio-Dias, R. Castelo-Branco, H. Scharfenstein, R. Rezende de Castro, A. Silva, M. P. C. Schneider, M. J. Araújo, R. Martins, V. F. Domingues, F. Nogueira, V. Camões, V. M. Vasconcelos and P. N. Leão, J. Nat. Prod., 2019, 82, 393–402 CrossRef CAS PubMed.
- J. S. Larsen, L. A. Pearson and B. A. Neilan, Genome Biol. Evol., 2021, 13, evab056 CrossRef PubMed.
- H. Nakamura, E. E. Schultz and E. P. Balskus, Nat. Chem. Biol., 2017, 13, 916–921 CrossRef CAS PubMed.
- K. Bloudoff and T. M. Schmeing, Biochim. Biophys. Acta, Proteins Proteomics, 2017, 1865, 1587–1604 CrossRef CAS PubMed.
- A. Salah Ud-Din, A. Tikhomirova and A. Roujeinikova, Int. J. Mater. Sci., 2016, 17, 1018 Search PubMed.
- L. Gu, T. W. Geders, B. Wang, W. H. Gerwick, K. Håkansson, J. L. Smith and D. H. Sherman, Science, 2007, 318, 970–974 CrossRef CAS PubMed.
- R. Kozakai, T. Ono, S. Hoshino, H. Takahashi, Y. Katsuyama, Y. Sugai, T. Ozaki, K. Teramoto, K. Teramoto, K. Tanaka, I. Abe, S. Asamizu and H. Onaka, Nat. Chem., 2020, 12, 869–877 CrossRef CAS PubMed.
- J. Shanklin, J. E. Guy, G. Mishra and Y. Lindqvist, J. Biol. Chem., 2009, 284, 18559–18563 CrossRef CAS PubMed.
- X. Li, J.-M. Lv, D. Hu and I. Abe, RSC Chem. Biol., 2021, 2, 166–180 RSC.
- N. Murata and H. Wada, Biochem. J., 1995, 308, 1–8 CrossRef CAS PubMed.
- D. Beresovsky, O. Hadas, A. Livne, A. Sukenik, A. Kaplan and S. Carmeli, Isr. J. Chem., 2006, 46, 79–87 CrossRef CAS.
- L. Hagmann and F. Jüttner, Tetrahedron Lett., 1996, 37, 6539–6542 CrossRef CAS.
- U. Papke, E. M. Gross and W. Francke, Tetrahedron Lett., 1997, 38, 379–382 CrossRef CAS.
- A. Ploutno and S. Carmeli, J. Nat. Prod., 2000, 63, 1524–1526 CrossRef CAS PubMed.
- D. J. Edwards, B. L. Marquez, L. M. Nogle, K. McPhail, D. E. Goeger, M. A. Roberts and W. H. Gerwick, Chem. Biol., 2004, 11, 817–833 CrossRef CAS PubMed.
- X. Zhu, J. Liu and W. Zhang, Nat. Chem. Biol., 2015, 11, 115–120 CrossRef CAS PubMed.
- P. D. Boudreau, T. Byrum, W.-T. Liu, P. C. Dorrestein and W. H. Gerwick, J. Nat. Prod., 2012, 75, 1560–1570 CrossRef CAS PubMed.
- J. Almaliti, K. L. Malloy, E. Glukhov, C. Spadafora, M. Gutiérrez and W. H. Gerwick, J. Nat. Prod., 2017, 80, 1827–1836 CrossRef CAS PubMed.
- N. A. Moss, G. Seiler, T. F. Leão, G. Castro-Falcón, L. Gerwick, C. C. Hughes and W. H. Gerwick, Angew. Chem., Int. Ed., 2019, 58, 9027–9031 CrossRef CAS PubMed.
- E. Roulland, H. Solanki, K. Calabro, M. Zubia, G. Genta-Jouve and O. P. Thomas, Org. Lett., 2018, 20, 2311–2314 CrossRef CAS PubMed.
- K. Kleigrewe, L. Gerwick, D. H. Sherman and W. H. Gerwick, Nat. Prod. Rep., 2016, 33, 348–364 RSC.
- V. Agarwal, Z. D. Miles, J. M. Winter, A. S. Eustáquio, A. A. El Gamal and B. S. Moore, Chem. Rev., 2017, 117, 5619–5674 CrossRef CAS PubMed.
- K. Kleigrewe, J. Almaliti, I. Y. Tian, R. B. Kinnel, A. Korobeynikov, E. A. Monroe, B. M. Duggan, V. Di Marzo, D. H. Sherman, P. C. Dorrestein, L. Gerwick and W. H. Gerwick, J. Nat. Prod., 2015, 78, 1671–1682 CrossRef CAS PubMed.
- S.-K. Zervou, S. Gkelis, T. Kaloudis, A. Hiskia and H. Mazur-Marzec, Chemosphere, 2020, 248, 125961 CrossRef CAS PubMed.
- B. S. Moore, J. L. Chen, G. M. L. Patterson, R. E. Moore, L. S. Brinen, Y. Kato and J. Clardy, J. Am. Chem. Soc., 1990, 112, 4061–4063 CrossRef CAS.
- S. M. Pratter, J. Ivkovic, R. Birner-Gruenberger, R. Breinbauer, K. Zangger and G. D. Straganz, ChemBioChem, 2014, 15, 567–574 CrossRef CAS PubMed.
- A. V. Ramaswamy, C. M. Sorrels and W. H. Gerwick, J. Nat. Prod., 2007, 70, 1977–1986 CrossRef CAS PubMed.
- T. B. Afonso, M. S. Costa, R. Rezende de Castro, S. Freitas, A. Silva, M. P. C. Schneider, R. Martins and P. N. Leão, J. Nat. Prod., 2016, 79, 2504–2513 CrossRef CAS PubMed.
- R. T. Williamson, I. P. Singh and W. H. Gerwick, Tetrahedron, 2004, 60, 7025–7033 CrossRef CAS.
- C. B. Naman, J. Almaliti, L. Armstrong, E. J. Caro-Díaz, M. L. Pierce, E. Glukhov, A. Fenner, C. Spadafora, H. M. Debonsi, P. C. Dorrestein, T. F. Murray and W. H. Gerwick, J. Nat. Prod., 2017, 80, 2328–2334 CrossRef CAS PubMed.
- C. K. Skepper and T. F. Molinski, J. Org. Chem., 2008, 73, 2592–2597 CrossRef CAS PubMed.
- S. P. Gunasekera, S. Kokkaliari, R. Ratnayake, T. Sauvage, L. A. H. dos Santos, H. Luesch and V. J. Paul, Molecules, 2022, 27, 1717 CrossRef CAS PubMed.
- J. M. Gregson, J.-L. Chen, G. M. L. Patterson and R. E. Moore, Tetrahedron, 1992, 48, 3727–3734 CrossRef CAS.
- H.-S. Kang, A. Krunic, Q. Shen, S. M. Swanson and J. Orjala, J. Nat. Prod., 2011, 74, 1597–1605 CrossRef CAS PubMed.
- J. Mareš, J. Hájek, P. Urajová, J. Kopecký and P. Hrouzek, PLoS One, 2014, 9(11), e111904 CrossRef PubMed.
- G. Pohnert and W. Boland, Nat. Prod. Rep., 2002, 19, 108–122 RSC.
- D. B. Stierle, A. A. Stierle, T. Bugni and G. Loewen, J. Nat. Prod., 1998, 61, 251–252 CrossRef CAS PubMed.
- N. Sitachitta and W. H. Gerwick, J. Nat. Prod., 1998, 61, 681–684 CrossRef CAS PubMed.
- J. C. Kwan, T. Meickle, D. Ladwa, M. Teplitski, V. Paul and H. Luesch, Mol. BioSyst., 2011, 7, 1205–1216 RSC.
- R.-Z. Liao, P. Georgieva, J.-G. Yu and F. Himo, Biochemistry, 2011, 50, 1505–1513 CrossRef CAS PubMed.
- K. Takayama, C. Wang and G. S. Besra, Clin. Microbiol. Rev., 2005, 18, 81–101 CrossRef CAS PubMed.
- K. Taori, V. J. Paul and H. Luesch, J. Am. Chem. Soc., 2008, 130, 1806–1807 CrossRef CAS PubMed.
- H.-S. Kang and E.-S. Kim, Curr. Opin. Biotechnol, 2021, 69, 118–127 CrossRef CAS PubMed.
- D. Dhakal, M. Chen, H. Luesch and Y. Ding, J. Ind. Microbiol. Biotechnol., 2021, 48, kuab003 CrossRef CAS PubMed.
- A. Taton, S. Rohrer, B. Diaz, R. Reher, A. M. Caraballo Rodriguez, M. L. Pierce, P. C. Dorrestein, L. Gerwick, W. H. Gerwick and J. W. Golden, ACS Chem. Biol., 2022, 17, 1910–1923 CrossRef CAS PubMed.
- S. L. Robinson, B. R. Terlouw, M. D. Smith, S. J. Pidot, T. P. Stinear, M. H. Medema and L. P. Wackett, J. Biol. Chem., 2020, 295, 14826–14839 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |